Abstract
Background:
Pityriasis versicolor (PV) is the most common chronic superficial infection of the stratum corneum, reported in 40–60% of the tropical population. After the description of the new Malassezia species, only a few studies have been conducted from India.
Aims:
Molecular identification, quantification of Malassezia species implicated with PV and correlation to its clinical presentation.
Materials and Methods:
The subjects include 50 PV patients, who attended the dermatology outpatient department of our hospital and 50 healthy individuals. Same size area of the skin was sampled from lesional and non-lesional sites in the patient group and from forehead, cheek, and chest of healthy individuals. Malassezia spp. isolated were identified by conventional method and confirmed by ITS2 PCR-RFLP and sequencing of D1/D2 region of 26S rDNA.
Results:
Eighty percent of patients presented with hypopigmented lesions and 20% with hyperpigmented lesions. From PV lesions, the most frequently isolated species was M. furfur (50%), followed by M. globosa (27.3%), mixture of M. furfur and M. globosa (15.9%), M. sympodialis (4.5%), and M. slooffiae (2.3%). Higher Malassezia density was found in lesional area as compared to non-lesional area of PV patients and in healthy individuals (P 0.0001).
Conclusion:
Although M. furfur was the most prevalent species isolated from both patients and controls, significantly higher isolation of M. globosa from the lesional area compared to non-lesional area indicates its possible role along with M. furfur in causing PV.
Keywords: DNA sequencing, Malassezia, PCR-RFLP, pityriasis versicolor
Introduction
Pityriasis versicolor (PV) is one of the most common chronic superficial fungal infections of the stratum corneum. PV is characterized by hypopigmented or hyperpigmented round or oval macules with fine scales.[1] This cosmetic problem has primary predilection for lipid-rich areas of the human body. Endogenous factors such as genetic predisposition and/or environmental factors such as hot and humid climate influence the prevalence of the disease in a particular geographic location. PV has been reported in 30–40% of the tropical population.[2]
The PV causing Malassezia yeasts are members of human cutaneous flora. Till now, 15 species of Malassezia have been described.[3] The prevalence and distribution of Malassezia species varies according to the geographical location of host, climatic changes, body site, sample collection, and identification method. After the description of the new Malassezia species, only few studies have been conducted from India. The burden of yeast loads in a PV-lesional area reflects its pathogenic role. The non-culture method of quantification clearly indicates that Malassezia load is higher in lesional area than non-lesional area.[4] However, culture-based quantification methods are sparse and results are perplexing.
Therefore, the purpose of our study was to (i) determine the prevalence of Malassezia spp. in lesional and non-lesional area of PV patients and healthy controls by molecular methods; (ii) evaluate the burden of Malassezia load in such subjects.
Materials and Methods
Subjects
Fifty consecutive patients with PV attending the dermatology outpatient department were included in the study. The study protocol was cleared by the Institute's Ethics Committee. The samples were collected after informed consent of the patients. Fifty healthy individuals were included as historical control (data from the previous study).[5] Disease characteristics of PV patients were recorded.
Collection and culture of samples
The samples from PV patients were collected from severely affected lesional sites as well as from non-lesional sites (dorsum of the hand). From healthy individuals, samples were collected from forehead, cheek, and chest. Samples were collected from the defined area (~2 cm diameter) using a sterile cotton swab dipped in PBS (pH 7.9) containing 0.1% (v/v) triton X-100 and inoculated over the surface of modified Dixon's agar plate.[6] The plates were incubated at 30°C in a humid chamber up to 4 weeks.
Calculation of population densities of Malassezia spp.
Quantification of population densities was carried out by counting and grading the colonies on a 90 mm diameter petri plate as 1 to 10 cfu/inch2 = +; 11 to 40 cfu/inch2 = ++; 41 to 100 cfu/inch2 = +++; and >100 cfu/inch2 = ++++.
Identification of Malassezia spp.
Colonies of Malassezia spp. were counted and identified on the basis of phenotypic methods (macroscopic and microscopic features; physiological and biochemical tests).[3,5] Standard strains of M. furfur (Microbial Type Culture Collection, Institute of Microbial Technology, Chandigarh, India, MTCC1374), M. restricta (CBS-KNAW Fungal Biodiversity Centre, Utrecht, The Netherlands, CBS7877), M. obtusa (CBS7876), M. globosa (CBS7966), M. pachydermatis (CBS1879), M. slooffiae (CBS7956), M. sympodialis (CBS7222), M. japonica (CBS9348), and M. yamatoensis (CBS9725) were used as control. Genomic DNA extraction was performed by phenol-chloroform method.[7] The identity of the isolates was further confirmed by PCR-RFLP of the ITS2 region of the rDNA gene; DNA sequencing of 26S region rDNA gene and compared with the sequences of the GenBank DNA database and CBS-KNAW database as per our previous protocol.[3,5,8]
Statistical analysis
Comparison of species distribution in different lesion location was done by two-way ANOVA and analyzed by software GraphPad Prism version 6.01. Comparison of Malassezia isolation rate, Malassezia colony count from lesional and non-lesional site of PV patients, season of onset of PV lesion, size of PV lesions was done by Chi-square test (Software- Epi Info™ version 7.1.0.6 and OpenEpi version 2.3).
Results
Table 1 represents the baseline demographic data. Isolation rate of Malassezia was significantly higher from lesional area of PV patients (n = 44, 88%) than non-lesional area (n = 20, 40%) (P 0.0001). From healthy individuals, Malassezia could be recovered by culture in 60% (n = 30) cases. The spectrum of Malassezia spp. isolated from lesional and non-lesional site of PV patients and healthy individuals are given in Figure 1. Isolation of M. globosa was significantly higher in the lesional area (n = 12, 27%) of PV patients compared to non-lesional area (n = 1, 5%) (P 0.00002). On the basis of colony count grading, Malassezia load from lesional area of PV patients was significantly higher than the non-lesional area (P 0.0001) [Figure 2 and Table 2]. M. furfur was predominantly isolated in both the genders and in all the age groups studied. M. furfur was involved in the lesions in all the sites except from shoulder. M. globosa is also involved in all the lesional sites except chest [Figure 3]. Hypopigmentation was more associated with M. furfur (n = 18, 53%), whereas hyperpigmentation with M. globosa (n = 5, 50%).
Table 1.
Demographic and salient clinical features of pityriasis versicolor patients
| Clinical profile | No. of patients (%) |
|---|---|
| Gender | |
| Male | 37 (74%) |
| Female | 13 (26%) |
| Age range | |
| <16 years | 3 (6%) |
| 16 to 30 years | 35 (70%) |
| 31 to 45 years | 10 (20%) |
| >45 years | 2 (4%) |
| Summer as season of onset | 47 (94%) |
| Seasonal improvement in winter | 41 (82%) |
| Itching | 6 (12%) |
| Hypopigmentation | 40 (80%) |
| Hyperpigmentation | 10 (20%) |
| Lesional site | |
| Neck | 19 (38%) |
| Back | 17 (34%) |
| Arms | 6 (12%) |
| Chest | 5 (10%) |
| Axilla | 2 (4%) |
| Shoulder | 1 (2%) |
| Well marginated lesions of <5 mm in diameter | 37 (74%) |
| Isolation rate of Malassezia | |
| Lesional area | 44 (88%) |
| Non-lesional area | 20 (40%) |
Figure 1.
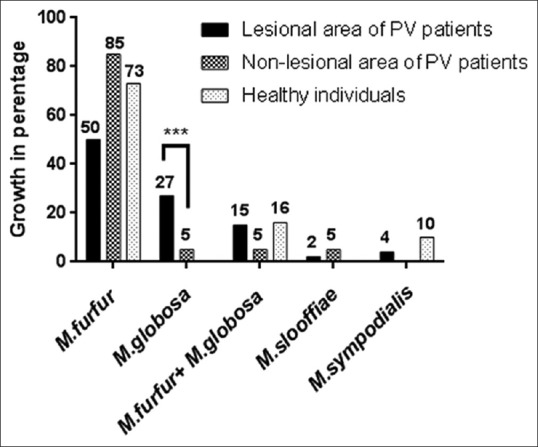
The spectrum of Malassezia spp. isolated from lesional, non-lesional sites of pityriasis versicolor patients and healthy individuals
Figure 2.
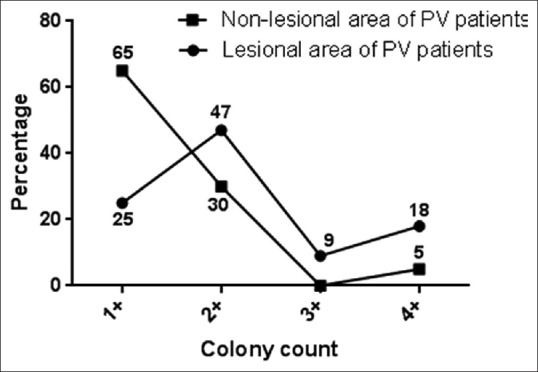
Mean colony count from pityriasis versicolor patient's skin
Table 2.
Malassezia load from lesional area and non-lesional area
| cfu/inch2 | Colony count | Lesional area (n=44) | Non-lesional area (n=20) |
|---|---|---|---|
| 1-10 | + | 9 | 13 |
| 11-40 | ++ | 23 | 6 |
| 41-100 | +++ | 4 | 0 |
| >100 | ++++ | 8 | 1 |
(1–10 cfu/inch2 = +; 11–40 cfu/inch2 = ++; 41–100 cfu/inch2 = +++; and 100 cfu/inch2= ++++). Malassezia count was significantly higher in lesional area
Figure 3.
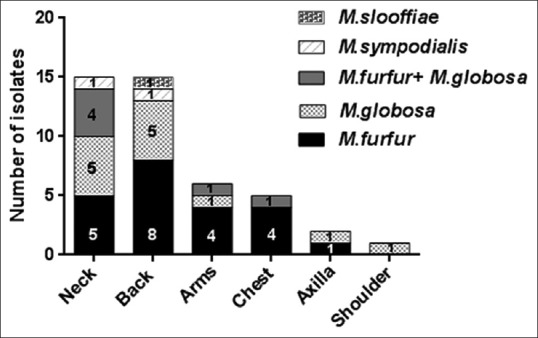
Distribution of Malassezia spp. on different sites of pityriasis versicolor lesions
Representative sequences have been deposited in the GenBank database with the accession numbers JQ271599 to JQ271623.
Discussion
We observed PV was most prevalent in males and between the age range of 16–30 years as reported by other studies in India.[9,10,11,12] The relationship between gender and PV is still ambiguous.[4] Less attention to beauty and skin hygiene and more exposure of male skin to adverse climatic condition in Indian scenario might have resulted in a high prevalence of PV in males. As observed by other investigators, PV was uncommon in children and higher in adolescence.[4] This indicates a higher prevalence of PV when the sebum (the nutrition source for Malassezia on the skin) activity is more.
During the summer months, from April till June, the average temperature and relative humidity at the sample collection region is generally 40°C and 65%, respectively. Summer was the season of onset in 94% cases indicating that hot and humid climate exacerbates the disease. Similar to the previous studies, hypopigmented lesions were more common in our study.[9,10,11] In concordance to worldwide data, maximum lesions were noticed on neck and back (64%), where the sebaceous gland activity is more.[4]
Among the few studies carried out in Indian subcontinent, M. globosa and M. sympodialis were predominately isolated from northern and southern India, respectively.[9,11] However, all these studies employed phenotypic methods for speciation of Malassezia. Owing to the limited number and ambiguity of the phenotypic tests used for differentiating Malassezia species, it is difficult to accurately identify those using phenotypic methods alone. Molecular methods such as employed in this study are essential to confirm the species. Although statistically non-significant, hypopigmented lesions were more associated with M. furfur (53%), whereas hyperpigmented lesions with M. globosa (50%). This result is in contrast to the recent study in which M globosa was isolated from only one patient with hypopigmented lesions.[12]
Similar to the findings of non-culture-based methods, in our culture-based study also Malassezia density was higher in the lesional area than the non-lesional area of PV patients or from healthy individual's skin.[12] In addition, the isolation rate was higher from the lesional area of PV patients. Few studies have stressed that the quantity of the organisms play an important role, among which most of the researchers focused on non-culture-based methods.[13] However, these non-culture-based methods will amplify DNA from both viable and nonviable cells (metabolically inactive). In the present study, quantification of metabolically active Malassezia cells reconfirmed its role in the pathogenesis of PV. Our culture based method indicated the coexistence of Malassezia spp. on the skin. It is important to incubate the culture for a longer period to isolate the Malassezia species separately. These isolates can be further used for the pathogenesis study and antifungal susceptibility testing.
Conclusion
Although M. furfur was the most prevalent species isolated from both patients and controls, significantly higher isolation of M. globosa from the lesional area compared to non-lesional area indicates its possible role along with M. furfur in causing PV.
Limitations of our study: (i) using the healthy individual's data from our previous study; (ii) small number of subjects and small geographic coverage.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Acknowledgments
We duly acknowledge the Indian Council of Medical Research (ICMR), New Delhi, India for the financial support.
References
- 1.Rasi A, Naderi R, Behzadi AH, Falahati M, Farehyar S, Honarbakhsh Y, et al. Malassezia yeast species isolated from Iranian patients with pityriasis versicolor in a prospective study. Mycoses. 2010;53:350–5. doi: 10.1111/j.1439-0507.2009.01727.x. [DOI] [PubMed] [Google Scholar]
- 2.Krisanty RI, Bramono K, Made Wisnu I. Identification of Malassezia species from pityriasis versicolor in Indonesia and its relationship with clinical characteristics. Mycoses. 2009;52:257–62. doi: 10.1111/j.1439-0507.2008.01593.x. [DOI] [PubMed] [Google Scholar]
- 3.Honnavar P, Prasad GS, Ghosh A, Dogra S, Handa S, Rudramurthy SM. Malassezia arunalokei sp.nov, a novel yeast species isolated from seborrheic dermatitis patients and healthy individuals from India. J Clin Microbiol. 2016;54:1826–34. doi: 10.1128/JCM.00683-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sugita T, Velegraki A. Epidemiology of Malassezia-related skin diseases. Berlin, Germany: Springer-Verlag; 2010. [Google Scholar]
- 5.Rudramurthy SM, Honnavar P, Chakrabarti A, Dogra S, Singh P, Handa S. Association of Malassezia species with psoriatic lesions. Mycoses. 2014;57:483–8. doi: 10.1111/myc.12186. [DOI] [PubMed] [Google Scholar]
- 6.Gupta P, Chakrabarti A, Singhi S, Kumar P, Honnavar P, Rudramurthy SM. Skin colonization by Malassezia spp. in hospitalized neonates and infants in a tertiary care centre in North India. Mycopathologia. 2014;178:267–72. doi: 10.1007/s11046-014-9788-7. [DOI] [PubMed] [Google Scholar]
- 7.Rudramurthy SM, Honnavar P, Dogra S, Yegneswaran PP, Handa S, Chakrabarti A. Association of Malassezia species with dandruff. Indian J Med Res. 2014;139:431–7. [PMC free article] [PubMed] [Google Scholar]
- 8.Honnavar P, Chakrabarti A, Dogra S, Handa S, Rudramurthy SM. Phenotypic and molecular characterization of Malassezia japonica isolated from psoriasis vulgaris patients. J Med Microbiol. 2015;64:232–6. doi: 10.1099/jmm.0.000011. [DOI] [PubMed] [Google Scholar]
- 9.Kindo AJ, Sophia SK, Kalyani J, Anandan S. Identification of Malassezia species. Indian J Med Microbiol. 2004;22:179–81. [PubMed] [Google Scholar]
- 10.Dutta S, Bajaj AK, Basu S, Dikshit A. Pityriasis versicolor: Socioeconomic and clinico-mycologic study in India. Int J Dermatol. 2002;41:823–4. doi: 10.1046/j.1365-4362.2002.01645.x. [DOI] [PubMed] [Google Scholar]
- 11.Chaudhary R, Singh S, Banerjee T, Tilak R. Prevalence of different Malassezia species in pityriasis versicolor in central India. Indian J Dermatol Venereol Leprol. 2010;76:159–64. doi: 10.4103/0378-6323.60566. [DOI] [PubMed] [Google Scholar]
- 12.Sharma A, Rabha D, Choraria S, Hazarika D, Ahmed G, Hazarika NK. Clinicomycological profile of pityriasis versicolor in Assam. Indian J Path Microbiol. 2016;62:159–65. doi: 10.4103/0377-4929.182027. [DOI] [PubMed] [Google Scholar]
- 13.Sugita T, Zhang E, Tanaka T, Nishikawa A, Tajima M, Tsuboi R. Recent advances in research on Malassezia microbiota in humans. Med Mycol J. 2013;54:39–44. doi: 10.3314/mmj.54.39. [DOI] [PubMed] [Google Scholar]


