Abstract
Abiotic stresses are major constraints limiting crop growth and production. Heat shock factors (Hsfs) play significant roles in mediating plant resistance to various environmental stresses, including heat, drought and salinity. In this study, we explored the biological functions and underlying mechanisms of wheat TaHsfA6f in plant tolerance to various abiotic stresses. Gene expression profiles showed that TaHsfA6f has relatively high expression levels in wheat leaves at the reproductive stage. Transcript levels of TaHsfA6f were substantially up-regulated by heat, dehydration, salinity, low temperature, and multiple phytohormones, but was not induced by brassinosteroids (BR). Subcellular localization analyses revealed that TaHsfA6f is localized to the nucleus. Overexpression of the TaHsfA6f gene in Arabidopsis results in improved tolerance to heat, drought and salt stresses, enhanced sensitivity to exogenous abscisic acid (ABA), and increased accumulation of ABA. Furthermore, RNA-sequencing data demonstrated that TaHsfA6f functions through up-regulation of a number of genes involved in ABA metabolism and signaling, and other stress-associated genes. Collectively, these results provide evidence that TaHsfA6f participates in the regulation of multiple abiotic stresses, and that TaHsfA6f could serve as a valuable gene for genetic modification of crop abiotic stress tolerance.
Keywords: wheat, Hsf, gene expression, transgenic, Arabidopsis, RNA-seq
1. Introduction
Abiotic stresses, such as heat, drought, and salinity, have a tremendous impact on crop growth and production. Plant adaptation to abiotic stresses involves a series of sophisticated regulatory mechanisms. Plant heat shock factors (Hsfs) represent important players in the intricate regulatory network through modulating the expression of genes responsive to a variety of environmental stresses [1]. It was reported that Hsfs regulate the transcription of heat shock protein (Hsp) genes and other stress-inducible genes by recognizing palindromic heat shock elements (HSEs: 5′-AGAAnnTTCT-3′) within the promoter regions of these genes [2,3]. The protein products of Hsp genes function as molecular chaperones and assist in protein folding, assembly, and translocation, thus protecting plants from impairment under stress conditions [4]. To date, Hsf gene families have been identified and analyzed at the genome level in many plant species. For example, 21 Hsf genes were identified in Arabidopsis [5], 25 in rice [6], 25 in maize [7], and 38 in soybean [8].
Similar to other transcription factor families, the Hsf family has common structural features, including a highly conserved DNA-binding domain (DBD) and an oligomerization domain (HR-A/B regions) [5]. The N-terminal DBD allows Hsf proteins to recognize HSEs in target promoter regions and thus to regulate the expression of downstream genes. The HR-A/B domain enables Hsf proteins to form homologous trimers to facilitate binding to Hsp promoters. Moreover, some Hsf proteins also contain nuclear localization signals (NLSs) and a C-terminal activation domain characterized by short peptide motifs (AHA motifs). Based on the characteristics of the HR-A/B domains, plant Hsfs are generally grouped into three classes, HsfA, HsfB, and HsfC [5,9]. The Hsf family has been described in detail in model organism Arabidopsis [5,6]. Arabidopsis HsfA proteins function as positive regulators of heat stress-responsive genes, which results in the accumulation of HSPs and other proteins that increase thermotolerance [1]. HsfB may serve as coregulators or repressors of the HsfA genes, while the functions of HsfC remain unclear [10].
Thus far, studies about plant Hsfs have mainly focused on their roles in heat stress responses. For example, in Arabidopsis, AtHsfA1a and AtHsfA1b were shown to play important roles in the early phase of the heat stress response [5]; AtHsfA1a/b double-knockout mutants are considerably impaired in the early transient mRNA accumulation of HSPs [11]. AtHsfA2 was found to be an essential regulator of plant acquired thermotolerance in the recovery period of heat stress and is directly regulated by HsfA1 [12]. AtHsfA3 transcript levels were induced by heat stress, and the overexpression of AtHsfA3 showed remarkably elevated thermotolerance, while a hsfA3 T-DNA insertion mutant had decreased thermotolerance [13]. More recently, AtHsfA3 has been revealed to play important roles in the regulation of heat stress-induced oxidative stress signaling [14]. Additionally, AtHsfA6b was reported to be involved in ABA signaling and required for ABA-mediated heat stress resistance [15]. Several Hsfs in other plant species, including tomato SlHsfA1 [16], tall fescue FaHsfA2c [17], wheat TaHsfA6e [18], and maize ZmHsf05 [19], have also been proved to participate in thermotolerance.
In addition to heat stress, Hsfs have been demonstrated to play a role in other abiotic stress responses. Arabidopsis HsfA1s are involved in salt, osmotic, and oxidative stresses [20]. Both HsfA1b and HsfA3 participate in drought stress responses [21], while HsfA2 has been shown to confer improved tolerance to several abiotic stresses including salt, osmotic stress, and anoxia stress [12,22]. AtHsfA6b increased tolerance to salt and drought stresses when overexpressed in Arabidopsis [15]. Recently, it was found that the expression of AtHsfA7b is up-regulated by salt stress, and that AtHsfA7b protein directly binds to E-box-like motifs and/or HSEs to regulate a series of genes involved in salt tolerance [23].
The studies mentioned above have clearly indicated that Hsfs mediate cross-talk between heat stress and other abiotic stress responses. Previous studies have demonstrated that TaHsfA6f plays a role in increasing thermotolerance via regulation of heat stress protection genes in wheat [24]. In this study, we explored the biological functions and underlying mechanisms of wheat TaHsfA6f in plant tolerance to multiple abiotic stresses. To this end, we cloned the TaHsfA6f gene from the heat and drought tolerant wheat cv. TAM107. We analyzed the expression patterns of TaHsfA6f during multiple abiotic stresses and phytohormones, and investigated the functions of TaHsfA6f via heterologous overexpression and RNA-seq analyses. It was shown that TaHsfA6f is a positive regulator of multiple abiotic stresses in plants and it may be associated with the ABA signaling pathway. Our results revealed the potential value of TaHsfA6f in the genetic improvement of crop abiotic stress tolerance.
2. Results
2.1. Gene Expression of TaHsfA6f is Up-Regulated by Various Abiotic Stresses and Phytohormones
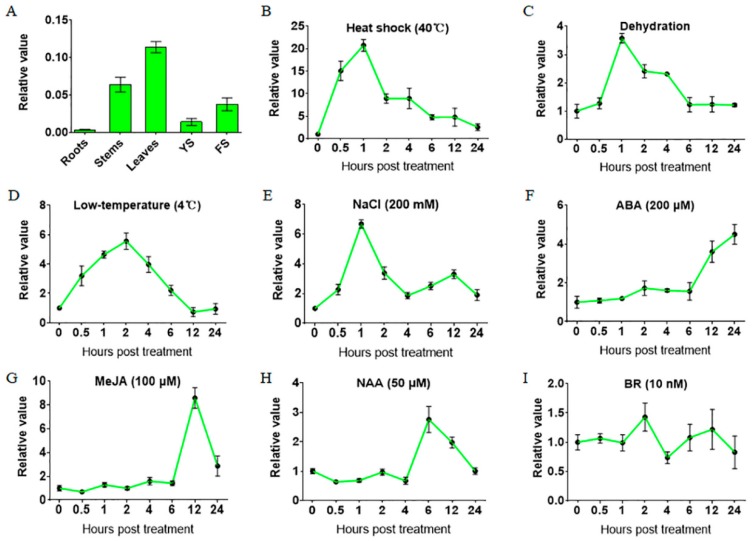
The coding and genomic sequences of the TaHsfA6f gene were obtained by PCR amplification using the cDNA and genomic DNA of the leaves of wheat variety TAM107 as a template, respectively. A sequence alignment between the obtained coding sequence and genomic sequence revealed that the TaHsfA6f gene contains one intron and two exons (Figure S1A). To disclose the phylogenetic relationship of TaHsfA6f, the amino acid sequence of TaHsfA6f was blasted against the UniProt database, and the homologous sequences of TaHsfA6f in other species were obtained. Phylogenetic analyses of TaHsfA6f and the sequences from other species showed that TaHsfA6f and its homologous proteins diverged between monocotyledon and dicotyledon species (Figure S1B). Among the monocotyledon branches, TaHsfA6f had the highest similarity with the barley protein A0A287PMR3. It was also found that the homologous protein of TaHsfA6f in Arabidopsis thaliana is AtHsfA6b (Q9LUH8) (Figure S1B). In order to examine the expression patterns of the TaHsfA6f gene in wheat tissues, we selected five tissues (root, stem, leaf, young spikes and flowering spikes) at the reproductive stage of Chinese spring, and analyzed the gene expression by qPCR. It was found that TaHsfA6f gene was expressed in all detected tissues, with the highest expression level in leaves and the lowest in roots under normal growth conditions (Figure 1A).
Figure 1.
Gene expression patterns of TaHsfA6f. (A) Relative expression levels of TaHsfA6f in various wheat organs. Total RNA was isolated from roots, stems, leaves, young spikes (YS), and flowering spikes (FS). The data represent means of three biological replicates ± SD. (B-I) Gene expression patterns of TaHsfA6f during abiotic stresses and phytohormones. Total RNA was extracted from plants treated with 40 °C heat shock (B), dehydration (C), 4 °C low temperature (D), 200 mM NaCl (E), 200 µM ABA (F), 100 mM MeJA (G), 50 mM NAA (H), and 10 nM BR (I), for the indicated time points. The relative expression level was measured by qPCR and calculated by the comparative CT method. Error bars indicate standard deviations (SD).
Next, we investigated the expression patterns of the TaHsfA6f gene in response to different abiotic stresses and phytohormones. As shown in Figure 1, the TaHsfA6f gene was greatly and quickly (within an hour) up-regulated by heat shock, drought, low temperature, and NaCl treatments. Remarkably, the gene expression of TaHsfA6f was up-regulated by 40 °C heat stress, the highest level being 20-fold greater than the untreated control (0 h), and peak values appearing 1 h after heat stress treatment (Figure 1B). By contrast, the responses of TaHsfA6f to phytohormones were slow. Specifically, the expression of the TaHsfA6f gene was significantly increased at 12 h by both ABA and MeJA, and at 6 h by NAA (Figure 1F–H). No obvious induction of TaHsfA6f gene expression was observed for BR (Figure 1I).
2.2. Subcellular Location of TaHsfA6f Protein
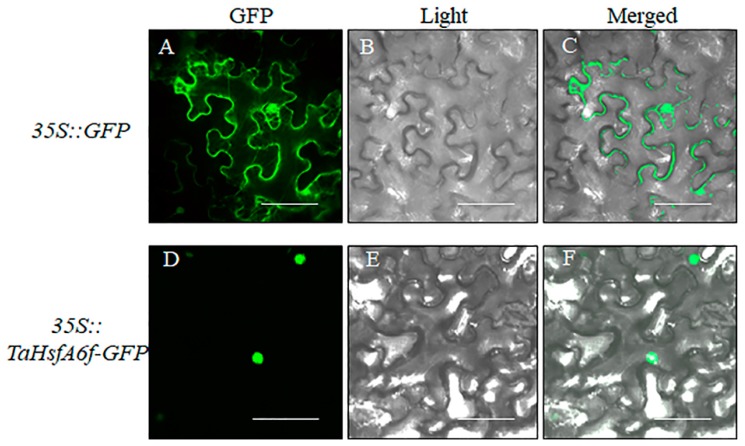
Subcellular localization of TaHsfA6f was performed in Nicotiana benthamiana leaf epidermal cells transformed with constructs containing 35S::TaHsfA6f-GFP and 35S::GFP, respectively. As shown in Figure 2, the green fluorescence of cells with GFP was seen in membrane, nucleus and cytosol, while it was only present in the nucleus of cells with TaHsfA6f-GFP, indicating that TaHsfA6f is a nucleus protein.
Figure 2.
Subcellular localization of TaHsfA6f protein by Nicotiana benthamiana leaf epidermal cells transient expression. 35S::GFP and 35S::TaHsfA6f-GFP plasmids were transformed into Nicotiana benthamiana leaf epidermal cells and signals were visualized with laser confocal-scanning fluorescence microscopy. From left to right, the photographs were taken in the dark-field for green fluorescence (GFP) (A,D), under bright-field for the morphology of the cell (Light) (B,E), and overlays of the GFP signals and bright-field (Merged) (C,F). The scale bars represent 40 μm.
2.3. Overexpression of TaHsfA6f Confers Improved Tolerance to Heat, Drought and Salt Stresses in Arabidopsis
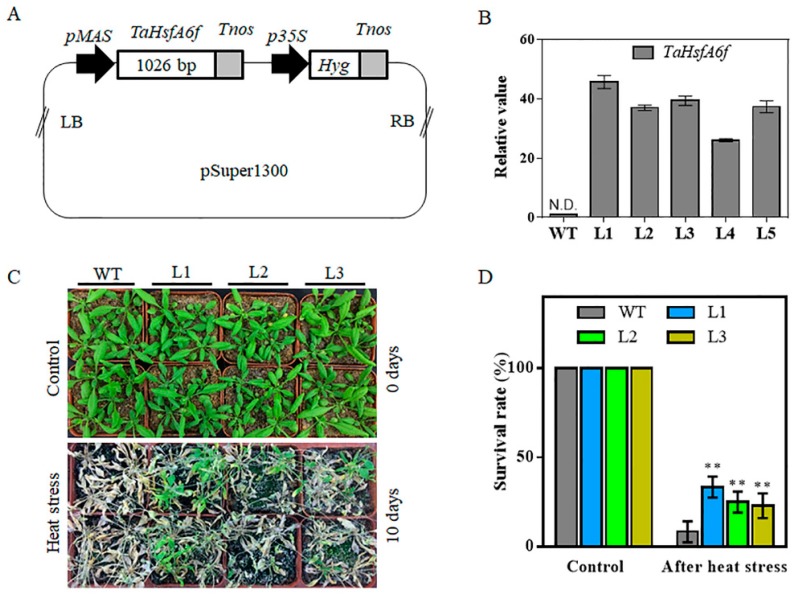
In order to reveal the biological functions of wheat TaHsfA6f gene, we constructed an overexpression vector (pMAS::TaHsfA6f) of TaHsfA6f (Figure 3A), which was transferred into Agrobacterium GV3101, and the positive Agrobacterium strain with TaHsfA6f was then transformed into Arabidopsis. Five single copy transgenic Arabidopsis lines were obtained after hygromycin B screening. The expression of TaHsfA6f in transgenic Arabidopsis was detected in all pMAS::TaHsfA6f transgenic lines but not in the WT plants (Figure 3B). Three independent T3 transgenic lines (L1, L2 and L3) exhibiting the highest TaHsfA6f expression levels were selected for further analyses.
Figure 3.
Thermotolerance of WT and transgenic Arabidopsis plants. (A) Schematic representation of the construct used for Arabidopsis transformation. LB left border, pMAS mannopine synthase promoter, Tnos nopaline synthase gene (NOS) terminator, Hyg Hygromycin-resistance gene, RB right border. (B) Relative expression levels of TaHsfA6f in the leaves of T3 transgenic Arabidopsis lines via qPCR analysis. Each value is the mean of triplicate experiments. Bars indicate standard deviations. WT wild-type, N.D = Not Detected. (C) Comparison of thermotolerance among WT and three transgenic lines (L1, L2, and L3). (D) Survival rates of WT and three transgenic lines after heat stress at 40/32 °C (day/night) for 10 days. For each experiment, 20 plants per line were used; values are means ± SD from three independent measurements (** p < 0.01 by Student’s t-test).
Firstly, we examined whether TaHsfA6f overexpression could improve tolerance to heat stress in transgenic Arabidopsis plants. As shown in Figure 3C,D, under normal conditions (control), no differences were found between transgenic and WT plants and all of them grew well. Under a heat stress of 40/32 °C (day/night) for 10 days, however, the survival rates of transgenic Arabidopsis L1 (33%), L2 (25%) and L3 (23%) were clearly higher than those of WT (8%), indicating that overexpression of the TaHsfA6f gene could improve the heat tolerance of transgenic Arabidopsis.
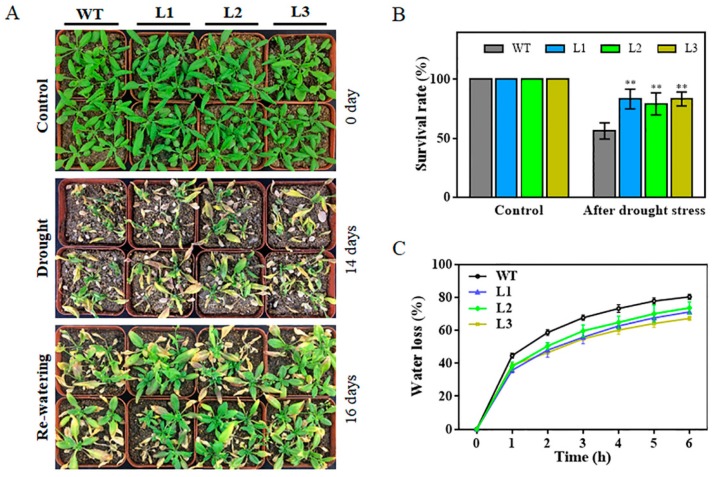
To investigate whether TaHsfA6f overexpression could enhance drought stress tolerance in plants, three-week-old transgenic and WT plants were subjected to drought stress by withholding watering for 14 days, and then a two-day growth recovery was conducted. The three transgenic lines (L1, L2, and L3) exhibited better growth status than WT (Figure 4A) and the survival rates of the transgenic lines were significantly higher than that of the WT (Figure 4B). Water loss rates serve as an important indicator of drought tolerance in plants. Water loss rates of detached leaves were recorded every hour at room temperature for both the WT and the three transgenic lines. As shown in Figure 4C, the transgenic lines exhibited significantly lower water loss rates compared to the WT.
Figure 4.
Comparison of drought tolerance among WT and three TaHsfA6f overexpressing lines. (A) Drought-tolerant phenotypes and (B) survival rates of WT and transgenic lines. Water was withheld for 14 days; afterwards, re-watering was applied and survival rates were calculated two days after re-watering. (C) Water loss rates of WT and transgenic lines. The data presented are means ± SD of three replicates (** p < 0.01 by Student’s t-test).
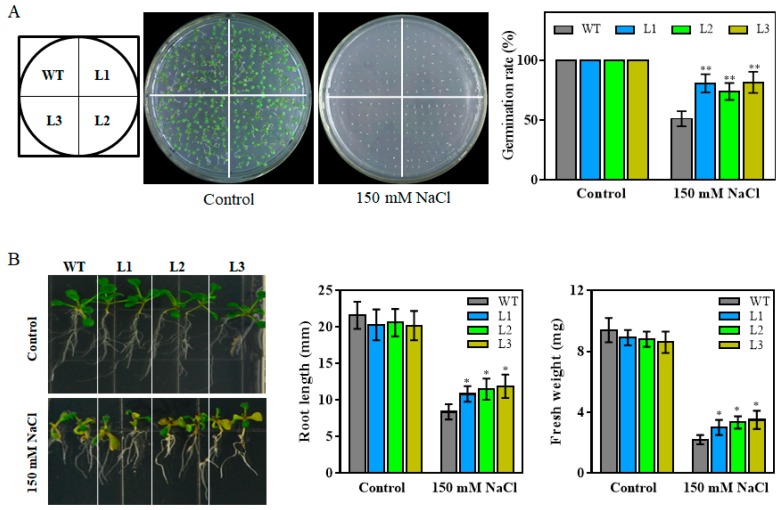
To reveal a potential role of TaHsfA6f in salt stress responses, 150 mM NaCl was applied to media with seeds or seven-day-old seedlings of transgenic and WT plants. As shown in Figure 5A,B, there was no difference between WT and transgenic plants in seed germination rates, root length, and fresh weight under control conditions. However, when grown on 150 mM NaCl-containing 1/2MS media, all transgenic plants displayed increased seed germination rates, root length, and fresh weight compared with WT. These results indicated that TaHsfA6f overexpression in transgenic Arabidopsis plants enhances their tolerance to heat, drought and salt stresses.
Figure 5.
Effect of salt stress on WT and transgenic Arabidopsis plants. (A) Comparison of germination rates among WT and three transgenic lines (L1, L2, and L3) under salt stress (NaCl). Seeds were germinated on 1/2MS plates supplemented with 0 (control) or 150 mM NaCl. The data was recorded after four days of germination. (B) Growth and biomass accumulation of seven-day-old seedlings of WT and three transgenic lines treated with 150 mM NaCl for five days. Values are means ± SD from three independent measurements (* p < 0.05, ** p < 0.01 by Student’s t-test).
2.4. Transgenic Arabidopsis Plants are Hypersensitive to ABA
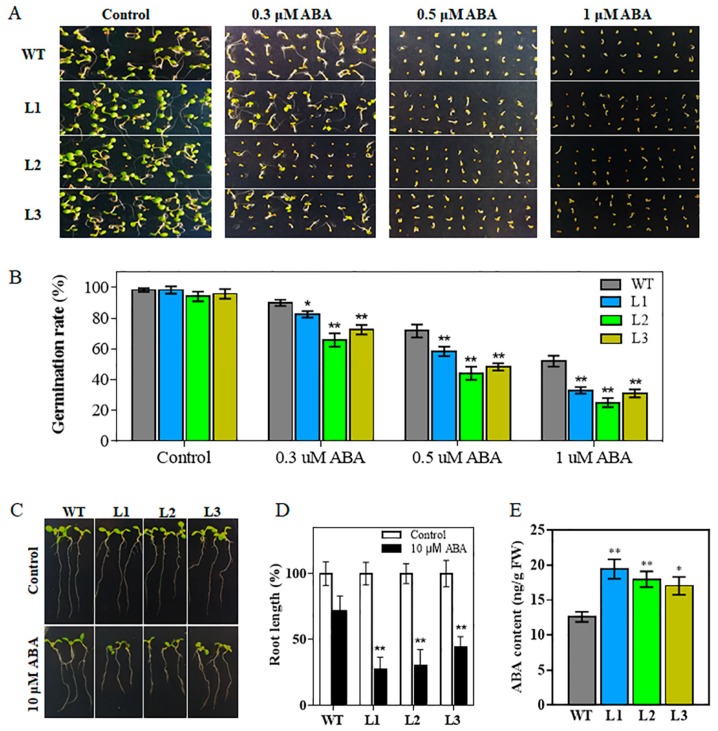
Since the expression of TaHsfA6f was induced by ABA treatment (Figure 1F), we investigated whether TaHsfA6f plays a role in ABA signaling pathway. As shown in Figure 6A,B, there was no noticeable difference in seed germination between transgenic lines (L1, L2, and L3) and WT on ABA-free media (Control). However, on media containing 0.3-, 0.5- or 1-μM ABA, the germination capability of seeds from the transgenic lines was much more reduced than that of the WT seeds (Figure 6A,B). In terms of root elongation, no significant growth difference was observed between transgenic and WT seedlings under control conditions (Figure 6C,D). However, on 10-μM ABA, the root elongation of the transgenic seedlings was reduced compared with that of the WT seedlings (Figure 6C,D). These results showed that transgenic plants are hypersensitive to ABA during seed germination and seedling growth.
Figure 6.
Effect of ABA on TaHsfA6f transgenic Arabidopsis plants. (A–B) Germination assays of TaHsfA6f transgenic plants under ABA treatment. Seed germination ratios of WT and three TaHsfA6f transgenic lines on 1/2MS agar media with different concentrations of ABA. (C) Morphology of WT and three TaHsfA6f transgenic seedlings grown vertically on 1/2MS agar media with or without 10-μM ABA. The photographs were taken three days after seedlings were transferred to the indicated agar media. (D) Root length recorded before photographing. (E) Levels of endogenous ABA in the leaves of WT and the three transgenic lines under normal conditions. Data shown are means ± SD of three independent experiments (* p < 0.05, ** p < 0.01 by Student’s t-test).
2.5. Overexpression of TaHsfA6f Increases Cellular ABA Content
As shown above, overexpression of the TaHsfA6f gene in Arabidopsis increased heat, drought and salt tolerance. ABA plays a pivotal role in plants’ resistance to adverse environments. To study whether TaHsfA6f affects the biosynthesis of ABA to improve abiotic stress tolerance, we measured the content of endogenous ABA using a UPLC–MS/MS system. The content of ABA was significantly higher in transgenic plants (L1, L2, and L3) than that of the WT plants under normal growth conditions (Figure 6E). These results clearly revealed that overexpression of the TaHsfA6f gene enhances cellular ABA levels.
2.6. Transcriptome Analysis of TaHsfA6f Transgenic Arabidopsis
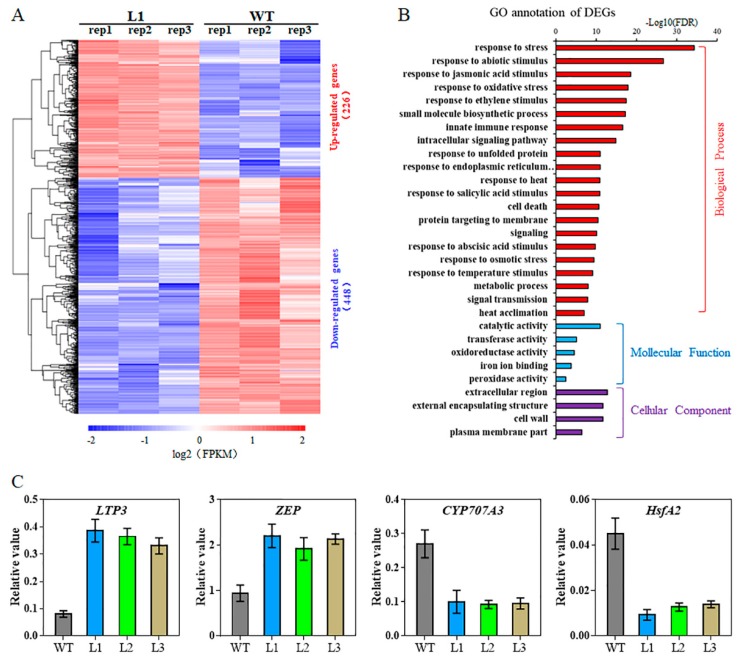
To decipher the possible molecular mechanisms of TaHsfA6f functioning in plant stress responses, genes with altered expression levels were identified in the transgenic plants using the RNA-seq approach. A total of 226 and 448 genes were found to be up- and down-regulated (fold change > 2, FDR < 0.001), respectively, in the transgenic plants compared with WT (Figure 7A and Table S2). Gene Ontology (GO) analyses revealed that the DEGs were highly enriched in the entries of ‘response to stress’ and ‘response to abiotic stimulus’ in the biological process category (Figure 7B). In the molecular function category, DEGs were highly accumulated in ‘catalytic activity’, oxidoreductase activity, and antioxidant activity (Figure 7B). Based on gene annotation, we found that many up-regulated DEGs in the transgenic plants are Hsp genes, and most of them were small Hsp genes (Table 1). In addition, other stress-responsive genes, such as three galactinol synthase genes (GolS1, GolS2, GolS4), two lipid-transfer protein-encoding genes (LTP3 and LTP4), dehydration-responsive element-binding protein 1 genes (DREB1A and DREB1C), ascorbate peroxidase 2 (APX2), and late embryogenesis abundant group3 (LEA3) were up-regulated in the transgenic plants. Interestingly, ZEP, which encodes an enzyme important in ABA biosynthesis, was found to be slightly up-regulated in transgenic plants. CYP707A3 (encoding a protein involved in ABA catabolism) and HsfA2 were found to be down-regulated in transgenic plants. Four genes from Table 1 were randomly selected and further validated using qPCR. For all four genes, the fold changes in gene expression determined by qPCR agreed well with the RNA-seq data (Figure 7C).
Figure 7.
Transcriptomic analyses of TaHsfA6f transgenic Arabidopsis. (A) Heat map of DEGs in leaves of transgenic vs. WT plants. The heat map represents the gene expression levels of 674 significant DEGs (log2Foldchange > 1 and FDR < 0.001) between transgenic (L1: rep1, rep2 and rep3) and WT (WT: rep1, rep2 and rep3) plants. The indicated scale is the log2 value of the normalized level of gene expression. (B) Gene ontology classification for DEGs in TaHsfA6f transgenic and WT plants. (C) qPCR validation of RNA-seq results for DEGs. Error bars indicate standard deviations.
Table 1.
The list of DEGs in TaHsfA6f transgenic Arabidopsis that are highlighted in this paper.
| Gene ID | Gene Symbol | Fold Change | Profile | Description |
|---|---|---|---|---|
| AT4G10250 | HSP22.0 | 673.8 | up | 22.0 kDa heat shock protein |
| AT1G60470 | GOLS4 | 571.1 | up | Galactinol synthase 4 |
| AT4G27670 | HSP25.3 | 475.1 | up | 25.3 kDa heat shock protein |
| AT3G09640 | APX2 | 268.1 | up | L-ascorbate peroxidase 2 |
| AT1G52560 | HSP26.5 | 149.4 | up | 26.5 kDa heat shock protein |
| AT5G59720 | HSP18.1 | 84.1 | up | 18.1 kDa class I heat shock protein |
| AT1G53540 | HSP17.6C | 46.1 | up | 17.6 kDa class I heat shock protein 3 |
| AT4G25200 | HSP23.6 | 31.5 | up | 23.6 kDa heat shock protein |
| AT5G59310 | LTP4 | 19.0 | up | Non-specific lipid-transfer protein 4 |
| AT5G12020 | HSP17.6 | 11.9 | up | 17.6 kDa class II heat shock protein |
| AT4G21320 | HSA32 | 9.7 | up | HEAT-STRESS-ASSOCIATED 32 |
| AT5G12030 | HSP17.7 | 9.0 | up | 17.7 kDa class II heat shock protein |
| AT2G47180 | GOLS1 | 8.8 | up | Galactinol synthase 1 |
| AT2G29500 | HSP17.6B | 6.5 | up | 17.6 kDa class I heat shock protein 2 |
| AT1G02820 | LEA3 | 4.5 | up | late embryogenesis abundant 3 |
| AT5G59320 | LTP3 | 4.4 | up | Non-specific lipid-transfer protein 3 |
| AT5G37670 | HSP15.7 | 4.3 | up | 15.7 kDa heat shock protein |
| AT1G16030 | HSP70-5 | 3.0 | up | Heat shock 70 kDa protein 5 |
| AT2G32120 | HSP70-8 | 3.0 | up | Heat shock 70 kDa protein 8 |
| AT1G56600 | GOLS2 | 2.6 | up | Galactinol synthase 2 |
| AT4G25480 | DREB1A | 2.2 | up | Dehydration-responsive element-binding protein 1A |
| AT4G25470 | DREB1C | 2.2 | up | Dehydration-responsive element-binding protein 1C |
| AT5G66400 | RAB18 | 2.1 | up | Dehydrin Rab18 |
| AT5G67030 | ZEP | 1.8 | up | Zeaxanthin epoxidase, chloroplastic |
| AT5G03760 | CSLA9 | 0.3 | down | Glucomannan 4-beta-mannosyltransferase 9 |
| AT5G45340 | CYP707A3 | 0.3 | down | Abscisic acid 8′-hydroxylase 3 |
| AT2G26150 | HSFA2 | 0.2 | down | Heat stress transcription factor A-2 |
3. Discussion
Wheat (Triticum aestivum L.), belonging to the Poaceae family, is one of the world’s most important staple foods. Wheat production is challenged by various stresses, such as high temperature, drought and salinity. Therefore, it is of great significance to characterize wheat stress response genes for their modification to enhance crop adaptability. A previous study has shown that the wheat TaHsfA6f gene was induced in response to heat stress; TaHsfA6f overexpression transgenic plants showed improved thermotolerance [24]. In this study, we found that TaHsfA6f was quickly (within an hour) and dramatically up-regulated by heat stress (Figure 1B) and overexpression of the TaHsfA6f gene improved the heat tolerance of transgenic Arabidopsis (Figure 3C,D). These results imply that the improvement of heat resistance by TaHsfA6f is conserved between monocot and dicot plant species. More importantly, we found that TaHsfA6f was up-regulated by various abiotic stresses (Figure 1), suggesting a potential role of TaHsfA6f in various stress responses. Indeed, TaHsfA6f transgenic Arabidopsis plants exhibited enhanced tolerance to drought and salt stresses (Figure 4 and Figure 5). Numerous studies have shown that plant hormones play important roles in plants adaptation to a variety of environmental stresses [25]. In the present study, we found that TaHsfA6f responds to ABA, MeJA, and NAA treatments (Figure 1). These results indicated that TaHsfA6f probably participates in a complex regulatory network in plant stress responses.
To explore the molecular mechanism underlying the involvement of TaHsfA6f in plant stress responses, we analyzed differential gene expression between transgenic Arabidopsis and WT plants through RNA-seq. A total of 674 DEGs were found in transgenic Arabidopsis compared to WT plants (Figure 7). As expected, the expression of several Hsp genes, including Hsp22.0, Hsp25.3, Hsp26.5, Hsp18.1, Hsp17.6C, Hsp23.6, Hsp17.6, Hsp17.6B, and Hsp70-5/8 were increased markedly in transgenic plants (Table 1). It was reported that the Hsps function as molecular chaperones to prevent non-native aggregation and facilitate appropriate refolding of heat-damaged proteins [4]. Arabidopsis GolS1, GolS2, GolS4 genes encoding for galactinol synthase, which are key enzymes of raffinose oligosaccharide synthesis, were also up-regulated in transgenic plants (Table 1). Those genes were previously reported to mainly function in drought, high-salinity, and osmotic stress tolerance [26,27,28,29]. Other stress-responsive genes, such as APX2, LTP3, LTP4, and LEA3, were also up-regulated in TaHsfA6f overexpressing plants. Arabidopsis APX2, encoding cytosolic ascorbate peroxidase, plays an important role in the acquisition of thermotolerance [30]. The expression of APX2 contributes to maintaining the activity of the antioxidant system and protects plants against oxidative damage caused by adverse stresses [27,30]. Arabidopsis LTP3 and LTP4 were demonstrated to participate in the transportation of cuticular waxes, which protect plants against biotic and abiotic stresses; overexpression of LTP3 increased drought and freezing tolerance in transgenic Arabidopsis [31]. In addition, overexpression of AtLEA3 enhanced drought tolerance in Arabidopsis [32]. Our results agreed well with previous reports on other HsfAs. It was shown that overexpression of the AtHsfA1, AtHsfA2, and AtHsfA3 activated the expression of a group of Hsps and stress-associated genes, which consequently led to improved abiotic stress tolerance in Arabidopsis [14,20,33].
Intriguingly, the expression of Arabidopsis HsfA2, which is a key regulator in abiotic stress responses, was significantly repressed in TaHsfA6f transgenic plants (Table 1). Previous studies have indicated that Hsfs could form homo- or heterotrimers, which then act on their own promoters and/or promoters of other HSF genes, increasing or decreasing the expression of these genes [34]. In this study, phylogenetic analyses showed that Arabidopsis AtHsfA6b is the closest member to TaHsfA6f (Figure S1B). AtHsfA6b cannot interact with itself, but interacts with the AtHsfA2 in the nucleus [15]. Therefore, we speculated that the repressed expression of AtHsfA2 in TaHsfA6f transgenic plants might be due to the phenomenon that TaHsfA6f interacts with AtHsfA2 to form hetero-oligomers, suppressing AtHsfA2 transcription. Future molecular biological evidence is required.
It is well known that ABA plays a crucial role in abiotic stress tolerance [25]. Several studies have indicated that Hsfs could connect ABA signaling. For instance, HsfA9 functions downstream of ABI3, a transcription factor in the ABA signaling pathway, and contributes to thermotolerance in Arabidopsis [35]. AtHsfA6b was induced by ABA and involved in the ABA signaling pathway and ABA-mediated thermotolerance and drought tolerance [15]. In this study, the expression of TaHsfA6f was induced by ABA treatment (Figure 1F). Overexpression of TaHsfA6f in Arabidopsis enhanced sensitivity to ABA during seed germination and seedling development (Figure 6). Additionally, we observed that the ABA levels in transgenic lines were increased (Figure 6E). RNA-seq analyses showed that the expression of CYP707A3 was significantly down-regulated in the TaHsfA6f transgenic plants (Table 1). It was reported that CYP707A3 is involved in ABA catabolism and functions as a determinant of endogenous ABA levels during dehydration–rehydration responses [36]. Moreover, the expression of AtZEP and APX2 were up-regulated in the TaHsfA6f transgenic plants (Table 1). Overexpression of AtZEP, an enzyme important in ABA biosynthesis, improved drought and salt tolerance and caused an increase in endogenous ABA content [37]. Arabidopsis with constitutively high expression of APX2 showed high ABA levels [38]. In addition, ABA can induce the expression of GolS genes, which contribute to oxidative stress tolerance by acting downstream of ABA signaling [39]. All of these findings suggest the possible involvement of TaHsfA6f in the ABA signaling pathway, and that the improved tolerance to heat, drought, and salt is at least partly attributed to ABA.
In conclusion, our results showed that overexpression of TaHsfA6f altered the expression levels of genes involved in the ABA metabolism and signaling, and other stress-related genes, thereby conferring heat, drought, and salt tolerance in Arabidopsis. It will be interesting to study the potential regulatory network of TaHsfA6f in ABA metabolism and/or ABA signaling pathways.
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
Heat- and drought-tolerant wheat, cv. TAM107, was used for gene cloning and expression analyses. The seeds were grown hydroponically in a growth chamber at 22 °C with a 16-h photoperiod. For salt, and exogenous hormone treatments, 10-day-old wheat seedlings were transferred to growth media supplemented with 200-mM NaCl, 200-µM abscisic acid (ABA), 100-mM methyl jasmonate (MeJA), 50-mM a-naphthaleneacetic acid (NAA), or 10-nM brassinosteroids (BR), respectively. For the dehydration treatment, 10-day-old wheat seedlings were placed on filter papers at room temperature. Cold treatments were conducted by transferring 10-day-old wheat seedlings to a pre-cooled medium and placed in a growth chamber at 4 °C. Heat stress was carried out by placing plants in a 40 °C chamber. Shoots were collected separately at 0, 0.5, 1, 2, 4, 6, 12 and 24 h after each treatment. To study the tissue-specific expression of the TaHsfA6f, roots, stems, leaves, young spikes, and flowering spikes at the reproductive stage were collected from Chinese Spring wheat. All of the samples were immediately frozen in liquid nitrogen and stored at −80 °C for RNA extraction.
The Arabidopsis ecotype Columbia-0 was used as the wild-type. Seeds were surface sterilized with a 5% NaClO solution, washed five times with sterile water, and stratified at 4 °C for two days in the dark. The sterilized seeds were plated on Murashige and Skoog (MS) media containing 0.8% (w/v) agar and then transferred to a growth chamber. Growth conditions were 22 °C under a 16-h photoperiod, with a light intensity of 100 μmol m−2 s−1. After seven days, uniform seedlings were transplanted into soil and grown in the growth chamber.
4.2. Gene Cloning and Sequence Analysis
The TaHsfA6f sequence was retrieved from the IWGSC database (http://www.wheatgenome.org/) and used to design primers. The PCR for amplifying TaHsfA6f was conducted with the following program using high-fidelity Primestar polymerase (TaKaRa, Dalian, China): initial denaturation at 98 °C for 3 min; followed by 98 °C for 15 s, 60 °C for 20 s, 72 °C for 1 min with 34 cycles; and then 3 min at 72 °C. The PCR products were purified and cloned into the pEASY-Blunt cloning vector (TransGen, Beijing, China) and subjected to sequencing. Primers were designed using DNAMAN software. BLAST (https://www.uniprot.org/blast/) was used to search for the homologous sequences of TaHsfA6f in the UniProt databases. The phylogenetic tree was constructed with MEGA 7 software using the neighbor-joining (NJ) method [40]. All of the primers used are listed in Supplementary Table S1.
4.3. RNA Extraction and qPCR Analysis
The total RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. First-strand cDNA was generated using a Reverse Transcription System (Vazyme, Nanjing, China) following the manufacturer’s instructions. A real-time quantitative PCR (qPCR) experiment was performed using the SYBR Green PCR Master Mix kit (Tiangen Biotech, Beijing, China) and the IQ5 light cycler system (Bio-Rad, Hercules, CA, USA). The parameters for qPCR have previously been described [41]. The wheat β-actin and AtActin2 genes were used as internal controls in wheat and Arabidopsis, respectively. The samples were assessed using three biological replicates. The relative gene expression was calculated using the comparative CT method [42].
4.4. Subcellular Localization Analysis
The coding sequence of the TaHsfA6f gene without the terminal codon was introduced in frame to the 5′ end of the green fluorescent protein (GFP) gene sequence in pCAM35s vector. The resulting construct was transformed into the Agrobacterium GV3101, which was used for transient expression in leaf epidermal cells of Nicotiana benthamiana. After Agrobacterium infiltration, plants were allowed to grow for two days at 22 °C with a 16-h-light/8-h-dark photoperiod before visualization under an Olympus FV10-ASW confocal laser scanning microscope.
4.5. Arabidopsis Transformation and Stress Treatments
The coding sequence of TaHsfA6f amplified by PCR from pEASY-Blunt::TaHsfA6f plasmid was cloned into the pSuper1300 vector harboring the mannopine synthase (MAS) promoter. The recombinant plasmid was transformed into the Agrobacterium GV3101, which was then used for transformation of wild type (Col-0) Arabidopsis using the floral-dip method. Transgenic plant seeds were screened on MS media containing 50 mg L−1 hygromycin. Putative positive seedlings were confirmed by qPCR.
For stress treatment experiments, three T3 transgenic lines and WT plants were germinated on MS agar plates for seven days. The healthy uniform seedlings were then transferred to 1/2MS media containing either 0- or 150-mM NaCl and planted vertically for five days, after which the fresh weight and root length of the plants were measured. Additionally, seven-day-old seedlings were transplanted into plastic pots containing the mixed soil of sand, peat, and perlite (1:1:1, v/v/v) and grown in a growth chamber for 14 days under normal growth conditions. The three-week-old plants were used for both heat and drought treatments. For heat stress, transgenic lines and WT plants were grown at 40/32 °C (day/night) with irrigation every day. The treatment lasted for 10 days and then survival rates were measured. For drought stress, water was withheld for 14 days. Afterwards, re-watering was applied, and survival rates were calculated two days after re-watering. Water loss rates were measured according to the method described by Zhao et al. [43]. The above experiments were repeated three times.
For the seed germination assay under stress treatments, the seeds of WT and three transgenic lines were sterilized and germinated on 1/2MS agar plates containing either 150-mM NaCl or ABA at different concentrations (0.3, 0.5 or 1 μM) following the method described previously [41]. Root elongation rate assay was carried out on 1/2MS agar media supplemented with 0- or 10-μM ABA according to Ling’s method [44].
4.6. Quantification of ABA Levels in Arabidopsis
The ABA content was measured using a method described previously [45]. ABA was extracted from flesh rosette leaves of three-week-old WT and transgenic Arabidopsis under normal growth conditions. Three biological replicates were used.
4.7. RNA-seq Analysis
Total RNA was extracted using Trizol reagent (TransGen, Beijing, China) from the leaves of three-week-old Arabidopsis seedlings of a transgenic line (L1) and WT plants under normal growth conditions. cDNA synthesis, libraries construction, sequencing, and raw reads analysis were conducted according to Zhao et al. [43]. The resulting libraries were sequenced on an Illumina HiSeq X Ten platform (Illumina, San Diego, CA, USA), resulting in 150 bp single-end reads. Three biological replicates were used. Approximately 4 GB of clean data were produced for each sample. The clean reads generated were aligned against the reference Arabidopsis genome using HISAT2 [46]. The number of sequencing reads generated from each sample was converted into FPKM (Fragments per Kilobase per Million Mapped Fragments) [47]. Differentially expressed genes (DEGs) were identified using the R package DEGseq [48]. The threshold value was set to |log2ratio| > 1 and false discovery rate (FDR) < 0.001. Gene Ontology (GO) analyses were performed using agriGO (a Web-based tool and database for gene ontology analysis; http://bioinfo.cau.edu.cn/agriGO/) [49]. RNA-Seq data reported here can be accessed with accession number PRJNA622833 in the Sequence Read Archive (SRA) at the National Center for Biotechnology Information (NCBI).
4.8. Statistical Analyses
Statistical analyses were performed in Microsoft Excel 2016. Data are represented as means ± standard deviation (SD). Differences between WT and transgenic lines were evaluated using a Student’s t-test. A single asterisk (*) represents a significant difference at p < 0.05, and a double asterisk (**) represents a significant difference at p < 0.01.
Abbreviations
| ABA | abscisic acid |
| BR | brassinosteroids |
| DEGs | differentially expressed genes |
| FDR | false discovery rate |
| FPKM | Fragments per Kilobase per Million Mapped Fragments |
| GFP | green fluorescent protein |
| Hsf | heat shock factor |
| Hsp | heat shock protein |
| MAS | mannopine synthase |
| MeJA | methyl jasmonate |
| NAA | a-naphthaleneacetic acid |
| qPCR | real-time quantitative PCR |
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/21/9/3121/s1.
Author Contributions
Conceptualization, Y.Z. and W.L.; investigation, H.B., Y.Z., and H.L.; data curation, H.B., Y.Z., and H.L.; writing—original draft preparation, Y.Z.; writing—review and editing, H.B. and W.L.; supervision, H.B.; project administration, Y.Z.; funding acquisition, Y.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (No. 31801363) and the Scientific and Technological Research Project of Henan Province of China (No. 192102110117 and No. 202102110162).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- 1.Von Koskull-Doring P., Scharf K.D., Nover L. The diversity of plant heat stress transcription factors. Trends Plant Sci. 2007;12:452–457. doi: 10.1016/j.tplants.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 2.Rabindran S.K., Haroun R.I., Clos J., Wisniewski J., Wu C. Regulation of heat shock factor trimer formation: Role of a conserved leucine zipper. Science. 1993;259:230–234. doi: 10.1126/science.8421783. [DOI] [PubMed] [Google Scholar]
- 3.Schoffl F., Prandl R., Reindl A. Regulation of the heat-shock response. Plant Physiol. 1998;117:1135–1141. doi: 10.1104/pp.117.4.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parsell D.A., Lindquist S. The function of heat-shock proteins in stress tolerance: Degradation and reactivation of damaged proteins. Annu. Rev. Genet. 1993;27:437–496. doi: 10.1146/annurev.ge.27.120193.002253. [DOI] [PubMed] [Google Scholar]
- 5.Nover L., Bharti K., Doring P., Mishra S.K., Ganguli A., Scharf K.D. Arabidopsis and the heat stress transcription factor world: How many heat stress transcription factors do we need? Cell Stress Chaperones. 2001;6:177–189. doi: 10.1379/1466-1268(2001)006<0177:AATHST>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo J., Wu J., Ji Q., Wang C., Luo L., Yuan Y., Wang Y., Wang J. Genome-wide analysis of heat shock transcription factor families in rice and Arabidopsis. J. Genet. Genom. 2008;35:105–118. doi: 10.1016/S1673-8527(08)60016-8. [DOI] [PubMed] [Google Scholar]
- 7.Lin Y.X., Jiang H.Y., Chu Z.X., Tang X.L., Zhu S.W., Cheng B.J. Genome-wide identification, classification and analysis of heat shock transcription factor family in maize. BMC Genom. 2011;12:76. doi: 10.1186/1471-2164-12-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li P.S., Yu T.F., He G.H., Chen M., Zhou Y.B., Chai S.C., Xu Z.S., Ma Y.Z. Genome-wide analysis of the Hsf family in soybean and functional identification of GmHsf-34 involvement in drought and heat stresses. BMC Genom. 2014;15:1009. doi: 10.1186/1471-2164-15-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kotak S., Port M., Ganguli A., Bicker F., Von Koskull-Doring P. Characterization of C-terminal domains of Arabidopsis heat stress transcription factors (Hsfs) and identification of a new signature combination of plant class A Hsfs with AHA and NES motifs essential for activator function and intracellular localization. Plant J. 2004;39:98–112. doi: 10.1111/j.1365-313X.2004.02111.x. [DOI] [PubMed] [Google Scholar]
- 10.Ikeda M., Mitsuda N., Ohme-Takagi M. Arabidopsis HsfB1 and HsfB2b act as repressors of the expression of heat-inducible Hsfs but positively regulate the acquired thermotolerance. Plant Physiol. 2011;157:1243–1254. doi: 10.1104/pp.111.179036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lohmann C., Eggers-Schumacher G., Wunderlich M., Schoffl F. Two different heat shock transcription factors regulate immediate early expression of stress genes in Arabidopsis. Mol. Genet. Genom. 2004;271:11–21. doi: 10.1007/s00438-003-0954-8. [DOI] [PubMed] [Google Scholar]
- 12.Charng Y.Y., Liu H.C., Liu N.Y., Chi W.T., Wang C.N., Chang S.H., Wang T.T. A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol. 2007;143:251–262. doi: 10.1104/pp.106.091322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoshida T., Sakuma Y., Todaka D., Maruyama K., Qin F., Mizoi J., Kidokoro S., Fujita Y., Shinozaki K., Yamaguchi-Shinozaki K. Functional analysis of an Arabidopsis heat-shock transcription factor HsfA3 in the transcriptional cascade downstream of the DREB2A stress-regulatory system. Biochem. Biophys. Res. Commun. 2008;368:515–521. doi: 10.1016/j.bbrc.2008.01.134. [DOI] [PubMed] [Google Scholar]
- 14.Song C., Chung W.S., Lim C.O. Overexpression of heat shock factor gene HsfA3 increases galactinol levels and oxidative stress tolerance in Arabidopsis. Mol. Cells. 2016;39:477–483. doi: 10.14348/molcells.2016.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang Y.C., Niu C.Y., Yang C.R., Jinn T.L. The heat stress factor HSFA6b connects ABA signaling and ABA-mediated heat responses. Plant Physiol. 2016;172:1182–1199. doi: 10.1104/pp.16.00860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mishra S.K., Tripp J., Winkelhaus S., Tschiersch B., Theres K., Nover L., Scharf K.D. In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Genes Dev. 2002;16:1555–1567. doi: 10.1101/gad.228802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X., Huang W., Liu J., Yang Z., Huang B. Molecular regulation and physiological functions of a novel FaHsfA2c cloned from tall fescue conferring plant tolerance to heat stress. Plant Biotechnol. J. 2017;15:237–248. doi: 10.1111/pbi.12609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar R.R., Goswami S., Singh K., Dubey K., Rai G.K., Singh B., Singh S., Grover M., Mishra D., Kumar S., et al. Characterization of novel heat-responsive transcription factor (TaHSFA6e) gene involved in regulation of heat shock proteins (HSPs)-A key member of heat stress-tolerance network of wheat. J. Biotechnol. 2018;279:1–12. doi: 10.1016/j.jbiotec.2018.05.008. [DOI] [PubMed] [Google Scholar]
- 19.Li G.L., Zhang H.N., Shao H., Wang G.Y., Zhang Y.Y., Zhang Y.J., Zhao L.N., Guo X.L., Sheteiwy M.S. ZmHsf05, a new heat shock transcription factor from Zea mays L. improves thermotolerance in Arabidopsis thaliana and rescues thermotolerance defects of the athsfa2 mutant. Plant Sci. 2019;283:375–384. doi: 10.1016/j.plantsci.2019.03.002. [DOI] [PubMed] [Google Scholar]
- 20.Liu H.C., Liao H.T., Charng Y.Y. The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ. 2011;34:738–751. doi: 10.1111/j.1365-3040.2011.02278.x. [DOI] [PubMed] [Google Scholar]
- 21.Bechtold U., Albihlal W.S., Lawson T., Fryer M.J., Sparrow P.A., Richard F., Persad R., Bowden L., Hickman R., Martin C., et al. Arabidopsis HEAT SHOCK TRANSCRIPTION FACTORA1b overexpression enhances water productivity, resistance to drought, and infection. J. Exp. Bot. 2013;64:3467–3481. doi: 10.1093/jxb/ert185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Banti V., Mafessoni F., Loreti E., Alpi A., Perata P. The heat-inducible transcription factor HsfA2 enhances anoxia tolerance in Arabidopsis. Plant Physiol. 2010;152:1471–1483. doi: 10.1104/pp.109.149815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zang D., Wang J., Zhang X., Liu Z., Wang Y. Arabidopsis heat shock transcription factor HSFA7b positively mediates salt stress tolerance by binding to an E-box-like motif to regulate gene expression. J. Exp. Bot. 2019;70:5355–5374. doi: 10.1093/jxb/erz261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xue G.P., Drenth J., McIntyre C.L. TaHsfA6f is a transcriptional activator that regulates a suite of heat stress protection genes in wheat (Triticum aestivum L.) including previously unknown Hsf targets. J. Exp. Bot. 2015;66:1025–1039. doi: 10.1093/jxb/eru462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verma V., Ravindran P., Kumar P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016;16:86. doi: 10.1186/s12870-016-0771-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taji T., Ohsumi C., Iuchi S., Seki M., Kasuga M., Kobayashi M., Yamaguchi-Shinozaki K., Shinozaki K. Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J. 2002;29:417–426. doi: 10.1046/j.0960-7412.2001.01227.x. [DOI] [PubMed] [Google Scholar]
- 27.Nishizawa A., Yabuta Y., Shigeoka S. Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiol. 2008;147:1251–1263. doi: 10.1104/pp.108.122465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Selvaraj M.G., Ishizaki T., Valencia M., Ogawa S., Dedicova B., Ogata T., Yoshiwara K., Maruyama K., Kusano M., Saito K., et al. Overexpression of an Arabidopsis thaliana galactinol synthase gene improves drought tolerance in transgenic rice and increased grain yield in the field. Plant Biotechnol. J. 2017;15:1465–1477. doi: 10.1111/pbi.12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panikulangara T.J., Eggers-Schumacher G., Wunderlich M., Stransky H., Schoffl F. Galactinol synthase1. A novel heat shock factor target gene responsible for heat-induced synthesis of raffinose family oligosaccharides in Arabidopsis. Plant Physiol. 2004;136:3148–3158. doi: 10.1104/pp.104.042606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Panchuk I.I., Volkov R.A., Schoffl F. Heat stress- and heat shock transcription factor-dependent expression and activity of ascorbate peroxidase in Arabidopsis. Plant Physiol. 2002;129:838–853. doi: 10.1104/pp.001362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo L., Yang H., Zhang X., Yang S. Lipid transfer protein 3 as a target of MYB96 mediates freezing and drought stress in Arabidopsis. J. Exp. Bot. 2013;64:1755–1767. doi: 10.1093/jxb/ert040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang Y., Kang K., Gan L., Ning S., Xiong J., Song S., Xi L., Lai S., Yin Y., Gu J., et al. Drought-responsive genes, late embryogenesis abundant group3 (LEA3) and vicinal oxygen chelate, function in lipid accumulation in Brassica napus and Arabidopsis mainly via enhancing photosynthetic efficiency and reducing ROS. Plant Biotechnol. J. 2019;17:2123–2142. doi: 10.1111/pbi.13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schramm F., Ganguli A., Kiehlmann E., Englich G., Walch D., Von Koskull-Doring P. The heat stress transcription factor HsfA2 serves as a regulatory amplifier of a subset of genes in the heat stress response in Arabidopsis. Plant Mol. Biol. 2006;60:759–772. doi: 10.1007/s11103-005-5750-x. [DOI] [PubMed] [Google Scholar]
- 34.Miller G., Mittler R. Could heat shock transcription factors function as hydrogen peroxide sensors in plants? Ann. Bot. 2006;98:279–288. doi: 10.1093/aob/mcl107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kotak S., Vierling E., Baumlein H., Von Koskull-Doring P. A novel transcriptional cascade regulating expression of heat stress proteins during seed development of Arabidopsis. Plant Cell. 2007;19:182–195. doi: 10.1105/tpc.106.048165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Umezawa T., Okamoto M., Kushiro T., Nambara E., Oono Y., Seki M., Kobayashi M., Koshiba T., Kamiya Y., Shinozaki K. CYP707A3, a major ABA 8′-hydroxylase involved in dehydration and rehydration response in Arabidopsis thaliana. Plant J. 2006;46:171–182. doi: 10.1111/j.1365-313X.2006.02683.x. [DOI] [PubMed] [Google Scholar]
- 37.Park H.Y., Seok H.Y., Park B.K., Kim S.H., Goh C.H., Lee B.H., Lee C.H., Moon Y.H. Overexpression of Arabidopsis ZEP enhances tolerance to osmotic stress. Biochem. Biophys. Res. Commun. 2008;375:80–85. doi: 10.1016/j.bbrc.2008.07.128. [DOI] [PubMed] [Google Scholar]
- 38.Rossel J.B., Walter P.B., Hendrickson L., Chow W.S., Poole A., Mullineaux P.M., Pogson B.J. A mutation affecting ASCORBATE PEROXIDASE 2 gene expression reveals a link between responses to high light and drought tolerance. Plant Cell Environ. 2006;29:269–281. doi: 10.1111/j.1365-3040.2005.01419.x. [DOI] [PubMed] [Google Scholar]
- 39.Van den Ende W., Peshev D., De Gara L. Disease prevention by natural antioxidants and prebiotics acting as ROS scavengers in the gastrointestinal tract. Trends Food Sci. Tech. 2011;22:689–697. doi: 10.1016/j.tifs.2011.07.005. [DOI] [Google Scholar]
- 40.Kumar S., Stecher G., Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao Y., Cheng X., Liu X., Wu H., Bi H., Xu H. The wheat MYB transcription factor TaMYB31 is involved in drought stress responses in Arabidopsis. Front. Plant Sci. 2018;9:1426. doi: 10.3389/fpls.2018.01426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 43.Zhao Y., Tian X., Li Y., Zhang L., Guan P., Kou X., Wang X., Xin M., Hu Z., Yao Y., et al. Molecular and functional characterization of wheat ARGOS genes influencing plant growth and stress tolerance. Front. Plant Sci. 2017;8:170. doi: 10.3389/fpls.2017.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ling Y., Alshareef S., Butt H., Lozano-Juste J., Li L., Galal A.A., Moustafa A., Momin A.A., Tashkandi M., Richardson D.N., et al. Pre-mRNA splicing repression triggers abiotic stress signaling in plants. Plant J. 2017;89:291–309. doi: 10.1111/tpj.13383. [DOI] [PubMed] [Google Scholar]
- 45.Li Y., Wang C., Liu X., Song J., Li H., Sui Z., Zhang M., Fang S., Chu J., Xin M., et al. Up-regulating the abscisic acid inactivation gene ZmABA8ox1b contributes to seed germination heterosis by promoting cell expansion. J. Exp. Bot. 2016;67:2889–2900. doi: 10.1093/jxb/erw131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Siren J., Valimaki N., Makinen V. Indexing graphs for path queries with applications in genome research. IEEE/ACM Trans. Comput. Biol. Bioinform. 2014;11:375–388. doi: 10.1109/TCBB.2013.2297101. [DOI] [PubMed] [Google Scholar]
- 47.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., Van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang L., Feng Z., Wang X., Wang X., Zhang X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- 49.Tian T., Liu Y., Yan H., You Q., Yi X., Du Z., Xu W., Su Z. agriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017;45:W122–W129. doi: 10.1093/nar/gkx382. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.