Abstract
During industrialization, humans have been exposed to increasing numbers of foreign chemicals. Failure of the immune system to tolerate drugs, cosmetics and other skin products causes allergic contact dermatitis, a T cell-mediated disease with rising prevalence. Models of αβ T cell response emphasize T cell receptor (TCR) contact with peptide-MHC complexes, but this model cannot readily explain activation by most contact dermatitis allergens, which are non-peptidic molecules. We tested whether CD1a, an abundant MHC I-like protein in human skin, mediates contact allergen recognition. Using CD1a autoreactive human αβ T cell clones to screen clinically important allergens present in skin patch testing kits, we identified responses to balsam of Peru, a tree oil widely used in cosmetics and toothpaste. Additional purification identified benzyl benzoate and benzyl cinnamate as antigenic compounds within balsam of Peru. Screening of structurally related compounds revealed additional stimulants of CD1a-restricted T cells, including farnesol and coenzyme Q2. Certain general chemical features controlled response: small size, extreme hydrophobicity and chemical constraint from rings and unsaturations. Unlike lipid antigens that protrude to form epitopes and contact TCRs, the small size of farnesol allows sequestration deeply within CD1a, where it displaces self-lipids and unmasks the CD1a surface. These studies identify molecular connections between CD1a and hypersensitivity to consumer products, defining a mechanism that could plausibly explain the many known T cell responses to oily substances.
One sentence summary
CD1a, an abundant lipid-antigen presenting molecule in human skin, mediates T cell responses to small contact allergens.
Introduction
The human immune system evolved to respond to foreign microbial antigens, but must also tolerate foreign compounds present in the environment, such as plants and foods. Over the past two centuries, industrialization has introduced the widespread use of chemical extraction techniques and synthetic chemistry methods. Industrial development has greatly increased the range of synthetic or purified botanical compounds to which humans are commonly exposed through pollution, or the intentional use of drugs, fragrances, cosmetics and other consumer products, especially those applied at high concentrations directly on the skin. Accordingly, the incidence of contact dermatitis has risen, especially in industrialized countries (1). Lifetime incidence currently exceeds 50%, making contact dermatitis the most common occupational skin disease (2). The essential pathophysiological feature of contact dermatitis is the allergen-specific nature of immune hypersensitivity reactions. Diagnosis relies on identifying the specific allergens to which a patient was exposed. Physicians measure local skin inflammation to a grid network of allergen patches applied to the skin as a diagnostic test. The mainstay of treatment is avoidance of exposure to named allergens.
Considerable evidence documents a role for αβ T cells in contact dermatitis, which is caused by delayed type hypersensitivity reactions. Gell and Coombs defined Type IV reactions as ‘delayed type’ hypersensitivity because they appear after 72 hours. Type IV reactions are T cell-mediated and are worsened after repeated exposure to allergens (3). During the sensitization phase, naive T cells are activated in a process that involves Langerhans cells and dermal dendritic cells (2). In the elicitation phase, T cells cause inflammatory manifestations in the skin. Biologists’ views of T cell response are strongly influenced by the known mechanisms by which T cell antigen receptors (TCRs) recognize peptide antigens bound to major histocompatibility complex (MHC) I and MHC II proteins (4–6). Yet, most known contact allergens are non-peptidic small molecules, cations or metals that are typically delivered to skin as drugs, oils, cosmetics, skin creams or fragrances (1, 2). Thus, the chemical nature of contact allergens does not match the chemical structures of most antigens commonly recognized within the TCR-peptide-MHC axis.
This apparent disconnect, which represents a core question regarding the origin of delayed type hypersensitivity, might be explained if MHC proteins use atypical binding interactions to display non-peptidic antigens to TCRs. For example, the anti-retroviral drug abacavir binds within the HLA-B*57:01 groove to alter the seating of self-peptides, creating neo-self epitopes (7). Similarly, the MHC class II protein encoded by HLA-DP2, can bind beryllium, thereby plausibly altering the MHC-peptide complex shape to enable binding of an autoreactive TCR (8). Here, autoimmune response to non-peptidic compounds still involves peptides in some way, and is linked to a specific HLA-allomorph that uses a defined structural mechanism. A second general model is that non-peptidic allergens form covalent bonds with peptides in vivo. Such ‘haptenation’ reactions might create hybrid molecules with peptide-based MHC binding moieties and TCR epitopes formed from the haptenizing drug or chemical. This concept derived from Landsteiner’s landmark studies with 2,4-dinitrophenols (9) and evolved into broader predictions that drugs could haptenate peptides or innate receptors (10). Some evidence indicates that drugs can generate immune hypersensitivity reactions via haptenation. For example, sulfamethoxazole, lidocaine, penicillins, lamotrigine, carbamazepine, p-phenylendiamine or gadolinium can bind peptides, MHC proteins or TCRs (11–16). Although the haptenation hypothesis is broadly taught to physicians, the extent to which it accounts for the larger spectrum of contact allergens remains unknown (17).
Both of these models derive from the premise that αβ T cell responses are mediated by MHC-encoded proteins and emphasize atypical modes of peptide presentation. Putting aside this premise, we tested a straightforward model whereby drugs and other non-peptidic contact allergens are presented by a system that evolved to present non-peptidic antigens to T cells (18). CD1 proteins are MHC I-like molecules that fold to form an antigen binding cleft comprised of two pockets, A’ and F’, which are larger and more hydrophobic than the clefts present in MHC I and MHC II proteins (19, 20). Most published studies of human CD1 proteins (CD1a, CD1b, CD1c and CD1d) emphasize display of amphipathic membrane phospholipids and sphingolipids. The alkyl chains bind within and fill up the cleft of CD1, and the polar head groups, comprised of carbohydrates or phosphate esters, protrude through a small portal (F’ portal) to lie on the outer surface of CD1, where they are presented to TCRs (21).
Whereas most known CD1-presented antigens are amphipathic lipids, some evidence suggests that CD1 proteins mediate recognition of non-lipidic, drug-like molecules. For example, CD1d mediates T cell response to phenyl pentamethyldihydrobenzofuran sulfonates (PPBFs) (22), and chemically reactive small molecules can influence CD1-restricted T cell response by an unknown mechanism that might involve induced lipid autoantigen synthesis (23). PPBFs lack aliphatic hydrocarbon chains that define lipids, and they are instead ringed, sulfated small molecules that chemically resemble allergenic drugs, such sulfonamide antibiotics and furosemide. However, PPBF antigens are much smaller than the known volume of CD1d cleft. Unlike amphipathic lipids, they lack a defined lipid anchor and hydrophilic head group (22), raising questions about how PPBFs could bind within CD1d and yet protrude in some way for TCR contact.
Among human CD1 isoforms, we focused on CD1a because it is abundantly expressed on epidermal Langerhans cells and dermal dendritic cells, which are implicated in contact dermatitis (24). Also, CD1a-autoreactive T cells home to the skin, and polyclonal autoreactive T cells derived from blood and skin show higher responses to CD1a as compared to other CD1 proteins (25, 26). In addition, surface CD1a proteins can rapidly capture extracellular antigens using mechanisms that do not require complex mechanisms of antigen processing within the endosomal network (27, 28). Recently, transfer of the human CD1a gene into mice (29) was found to augment intradermal T cell responses to the natural, plant-derived compound, urushiol (30). Actual CD1a-mediated T cell responses to commonly used drugs or contact allergens in consumer goods are, to our knowledge, unknown.
As a screen for the most common and clinically important contact dermatitis antigens, we tested for human T cell response to compounds embedded in the thin-layer rapid use epicutaneous (T.R.U.E.) test (or Truetest), which is broadly used in dermatology and allergy clinics to screen patients for contact dermatitis allergens that are most commonly encountered in medical practice. This approach identified a human T cell response to a tree oil-derived contact allergen known as balsam of Peru. Larger scale screens defined the general chemical requirements for a T cell response to oily substances and discovered additional contact allergens presented by CD1a, including farnesol. The crystal structure of the CD1a-farnesol complex and study of the self-lipids bound to CD1a provided evidence for a molecular mechanism for recognition of a contact allergen, explaining how small antigens sequestered fully within CD1a can lead to T cell responses through absence of interference with CD1a-TCR contact.
Results
Balsam of Peru binds CD1a and activates T cells
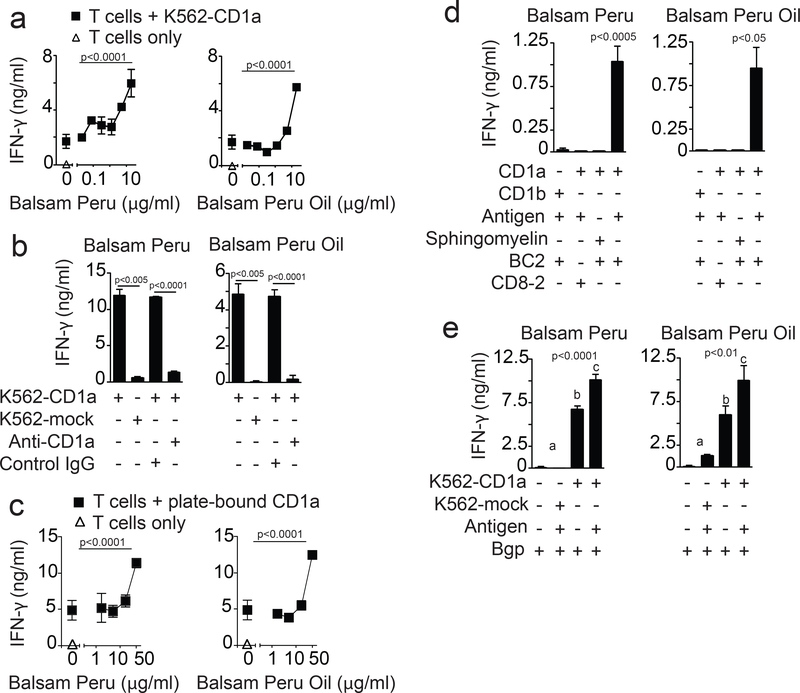
To determine if CD1a can present contact allergens to T cells, we initially used the CD1a-restricted αβ-T cell line known as BC2 for testing response to the T.R.U.E. test panel 1 (Truetest 1) (Fig. S1). BC2 is a T cell line derived from peripheral blood T cells of a blood bank donor, and has previously been shown to be activated by CD1a loaded with small hydrophobic self-lipids (31). Normally, the Truetest panel consists of compounds arrayed on sterile matrix, which is placed on patient skin. Localized erythema occurring in vivo on skin 2–5 days after exposure is considered a positive test, allowing allergen identification based on position in the grid. For testing in vitro, individual allergen patches and untreated patch matrix (control patch) were cut apart with sterile technique. Patches were soaked in media and removed (soaking method) or inserted into wells to contact (contact method) CD1a-transfected K562 (K562-CD1a) antigen presenting cells (APCs). We saw a modest response to K562-CD1a in the absence of added patch material using interferon (IFN)-γ ELISA, as expected based on the known CD1a autoreactivity of the BC2 T cell line (Fig. S1a).
Compared to the control patch, most of the antigen-containing patches, including nickel, potassium dichromate, colophony, lanolin and paraben, showed no effect. A combination of molecules known as ‘fragrance mix 1’ showed slight suppression of cytokine release, consistent with toxicity to cells (Fig. S1a). Cobalt, neomycin and ethylenediamine dihydrochloride showed small increases in IFN-γ at some doses tested, but not reproducibly in subsequent assays. In contrast, balsam of Peru showed a significant response above background (Fig. S1a), which also repeated in subsequent assays (Figs. S1b, Fig. 1a). Response to balsam of Peru was not seen with patch soaking (Fig. S1b), indicating that the stimulatory factor(s) was not physically released from the patch. Overall, the screen suggested a T cell response to balsam of Peru embedded in Truetest patches, leading to focused studies of this natural botanical extract.
Figure 1. Balsam of Peru activates T cells via a CD1a-dependent, APC-independent mechanism.
T cell lines with CD1a autoreactivity (BC2, Bgp) or foreign antigen reactivity (CD8–2) were tested for activation to lipids using IFN-γ ELISA in cellular assays with CD1a-transfected K562 cells (K562-CD1a) or mock transfected K562 cells (K562-mock) (a, b, e) or on streptavidin plates coated with biotinylated CD1 proteins (c, d). Data are representative of three or more experiments each with the mean of triplicate measurements shown with standard deviation. The significance of lipid concentration on IFN-gamma release was tested by one-way ANOVA (panel a, c). Relevant pairwise comparisons were tested using Welch’s t-test (panel b). Post-hoc comparison of marginal means after adjustment by the Sidak method was used to group treatments at the specified significance level following a significant result by two-way ANOVA (panel d). Post-hoc comparison by least-squares means after adjustment by the Sidak method was used to group treatments with non-overlapping marginal means and 95% confidence levels into a, b or c at the specified significance level following a significant result by two-way ANOVA (panel e).
Balsam of Peru is a resin from the South American tree, Myroxylon balsamum, which has a vanilla scent and is used as a fragrance and flavor in many personal care products such as skin creams and toothpaste. Balsam of Peru is a common contact allergen seen in medical practice, where it causes severe skin rash in allergic individuals (32, 33). We tested balsam of Peru extract and oily substances derived therefrom, which is known as balsam of Peru oil. Both preparations are commonly used in consumer products. BC2 T cells were activated by both preparations, establishing a T cell dose response to a common botanical extract used in consumer goods (Fig. 1a).
Given the unusual chemical nature of oily substances found in Balsam of Peru oil, we considered candidate mechanisms of T cell activation other than antigen display by CD1a. In theory, compounds might undergo peptide haptenation reactions for presentation by MHC proteins, but this possibility was less favored since K562 cells express very low or undetectable MHC I and MHC II (25). Oily mixtures might influence cellular lipid production (23) or contain mitogens that cross-link CD3 complexes or broadly activate lymphocytes via TCR-independent mechanisms (34). To determine the cellular and molecular mechanisms of T cell stimulation, we measured T cell activation by K562 APCs and by biotinylated CD1a proteins bound to avidin-coated plates. As assessed with anti-CD1a blocking antibodies and K562 cells lacking CD1a, CD1a was required for the BC2 response to crude balsam of Peru and oils derived therefrom (Fig. 1b–c). Treating plate bound CD1a protein with balsam of Peru was sufficient to activate the BC2 response, albeit at higher doses than with antigen in the presence CD1a-expressing cells (Fig. 1c). Thus, APCs facilitate some aspect of T cell response, but clear activation in APC-free systems ruled out that antigen processing is required. As a specificity control, BC2 did not respond structurally unrelated lipid, sphingomyelin, which is a known ligand for CD1a (Fig. 1d) (35). These results were most consistent with CD1a forming complexes with some molecule in these antigen preparations. Further specificity controls showed that balsam of Peru preparations did not activate a CD1a-restricted T cell clone, CD8–2, that recognizes CD1a presenting a mycobacterial antigen (18, 36) (Fig. 1d). This finding, along with the absolute requirement for CD1a in all recognition events, strongly indicated these substances are not mitogens. However, both balsam of Peru and balsam of Peru oil did activate another CD1a-autoreactive T cell line, Bgp (31). This indicates that balsam of Peru response was not limited to the BC2 T cell line (Fig. 1e).
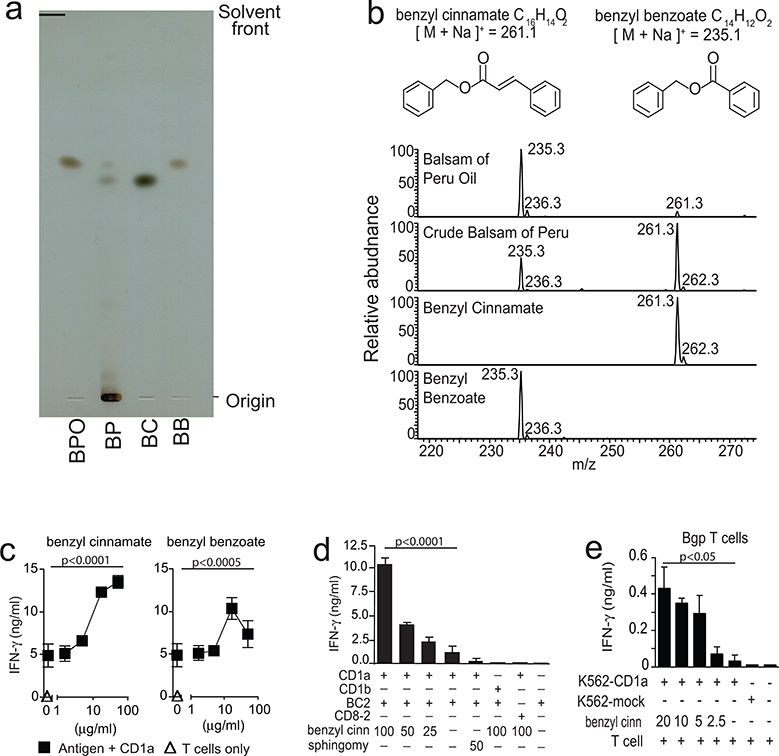
Chemical composition of balsam of Peru
Next, we sought to pinpoint chemical structures of the antigenic substances. Balsam of Peru is a complex botanical extract, with the most abundant components previously reported to be benzyl cinnamate and benzyl benzoate (37). Silica thin-layer chromatography (TLC) showed that crude balsam of Peru contained hydrophilic compounds that remained near the origin, as well as two dark spots that co-migrate with synthetic benzyl benzoate and benzyl cinnamate standards (Fig. 2a). As expected, oils extracted from balsam of Peru lacked the hydrophilic compounds that adhered at the origin. Balsam of Peru oil generated one dark spot that co-migrated with benzyl benzoate. More sensitive methods of positive mode nano-electrospray ionization mass spectrometry (nano-ESI-MS) (Fig. 2b) detected sodium adducts [M+Na]+ of benzyl cinnamate (m/z 261.3) and benzyl benzoate (m/z 235.3) in both preparations. The signal for benzyl benzoate was ~10-fold stronger than for benzyl cinnamate in balsam of Peru oil. Thus, benzyl cinnamate was present in both preparations, but its concentration was below the threshold of detection by TLC.
Figure 2. Chemical analysis of antigenic substances in balsam of Peru.
a. Normal phase silica TLC plate resolves balsam of Peru oil (BPO), crude balsam of Peru (BP), synthetic benzyl cinnamate (BC) and synthetic benzyl benzoate (BB). b. Structures of benzyl cinnamate and benzyl benzoate are shown with the expected mass of sodium adducts [M+Na]+, which were detected in positive-mode nanoelectrospray ionization mass spectrometry. c. T cell clones that are autoreactive to CD1a (BC2) or foreign antigen (CD8–2) were tested for response to antigens (μg/ml) or sphingomyelin (sphingomy) by IFN-γ ELISA in cellular (e) or CD1a-coated plate (c, d) assays. Data are representative of three or more experiments, each shown as the mean of triplicate samples +/− standard deviation. The significance of lipid concentration on IFN-γ release was tested by one-way ANOVA (panel c). The significance of benzyl cinnamate and benzyl benzoate concentration on IFN-γ release and of the effects of CD1b or CD8–2 T-cells were tested by two-way ANOVA (panels d,e).
Benzyl cinnamate and benzyl benzoate are CD1a-presented antigens
False positive results from trace contaminants in natural preparations occur, so we tested whether benzyl benzoate and benzyl cinnamate, provided as purified synthetic molecules, activated CD1a-restricted T cells. We observed T cell activation in response to both synthetic molecules, and the response was dependent on pre-coating the plate with CD1a. We observed a stronger and more potent response to benzyl cinnamate (Fig. 2c), which was then used for further mechanistic studies. Detailed testing of BC2 and CD8–2 activation by benzyl cinnamate confirmed the dose-dependence, CD1a-dependence and TCR-specificity of the T cell response to benzyl cinnamate (Fig. 2d). Sphingomyelin, a known CD1a ligand (31), which has a bulky polar head group, did not activate T cells. Responses to benzyl cinnamate were seen in two T cell lines, BC2 and Bgp (31). Benzyl cinnamate and benzyl benzoate were efficiently presented by plate-bound CD1a proteins after a short co-incubation, demonstrating the lack of a cellular processing requirement (Fig. 2c–d). These findings are most consistent with the formation of CD1a-benzyl cinnamate complexes as the target of T cell response. Thus, tree oils that are known to act as potent contact hypersensitivity agents also function as T cell stimulants that act via CD1a.
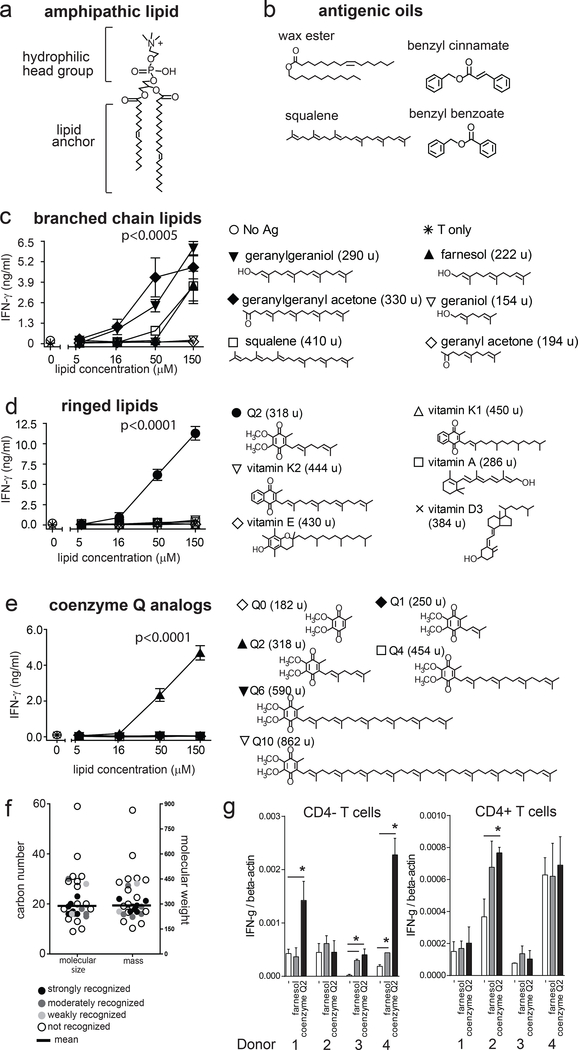
Shared structures and size among oily antigens
The dual benzyl rings present in benzyl cinnamate and benzyl benzoate (Fig. 2b) are chemically different from the alkyl chains present in most CD1-presented antigens. However, they are strikingly similar to the dually ringed structure present in the unusual non-lipidic antigen presented by CD1d known as PPBF (22). All three non-lipidic T cell stimulants are smaller (212–345 Da) than most previously known CD1-presented lipid antigens (~700–1500 Da) (21). Prior CD1-lipid structures (21) established a widely accepted mechanism whereby the acyl chains rest inside the hydrophobic clefts of CD1 proteins, so that hydrophilic head groups protrude outside CD1 and form epitopes that specifically contact TCRs (38) (Fig. 3a). In contrast, the antigenic tree oils identified here lack any identifiable polar group that could function as a TCR epitope (Fig. 3b). Further, the size of the carbon skeletons of benzyl benzoate and benzyl cinnamate (C14–16) are substantially smaller than other CD1 antigens (C20–40) and the estimated capacity of the CD1a cleft (~C36) (19, 39, 40). Because tree oils are apparently too small to fill the CD1a cleft and protrude to the outer surface, we hypothesized that they might not form TCR epitopes and so function outside the main CD1 antigen display paradigm. For example, interactions within the CD1a cleft might alter the shape of CD1-lipid complexes from the inside (41). Alternatively, similar to recent studies of CD1a (31, 35) and CD1c (42), tree oils might displace endogenous lipids, like sphingomyelin, whose large head groups interfere with TCR contact with CD1a. This emerging model is known as “absence of interference” because carried lipids do not contact TCRs directly, but instead bind CD1 in a manner that allows direct contact between CD1 and the TCR (31, 35).
Figure 3. T cell responses to chemically diverse oily substances.
a. Using phosphatidylcholine as an example, CD1 ligands are often composed of head groups and lipid anchors, but (b) recently identified CD1a presented antigens are oils. c. BC2 T cells were tested for cytokine release in response to small hydrophobic molecules pulsed on plate-bound CD1a pre-treated with acidic citrate buffer to strip ligands (31). Tested compounds are classified into groups based on the presence of branched chain unsaturated lipids structurally related to squalene (c), ringed lipids structurally related to benzyl cinnamate (d) or molecules that show branched, polyunsaturated and ringed structure, such as coenzyme Q2 (e). Results of triplicate analyses are shown as mean +/− standard deviation with each compound tested 2 or more times. Post-hoc comparison by marginal means of the interaction term between lipid and concentration after adjustment by the Sidak method was used to group treatments by non-overlapping 95% confidence levels at the specified significance level following a significant result by two-way ANOVA. f. The size of all tested antigens is shown based on the number of carbon atoms (C) or mass (atomic mass units, abbreviated as u), as compared to the volume of the CD1a cleft, which has been measured at 1650 Å3, and can accommodate ~ 36 methylene units (C36) (19, 40). g. Purified T cells (CD4− and CD4+) were incubated overnight with plate-bound CD1a, either mock treated or pre-treated with the indicated antigens (50μg/ml). Real-time PCR of IFN-γ mRNA relative to β-actin.* P<0.05 Student’s t-test 2-sided, antigen-treated compared to mock-treated CD1a.
Testing rings, unsaturations and molecular size
The approach to testing chemical features was guided by the observation that squalene, benzyl benzoate and benzyl cinnamate have ringed or unsaturated structures that chemically constrain molecules, rendering them bulky and rigid. Using tree oils and skin oils as lead compounds (Fig. 3b) to generate a larger test panel (Figs. 3c–e, S2), we surveyed 29 structurally related molecules that differed in size, saturation, branching patterns or ringed structures. Fifteen compounds, including examples among branched (Fig. 3c), ringed (Fig. 3d–e) and saturated or unsaturated fatty acyl compounds (Fig. S2) were recognized. This moderately promiscuous pattern was markedly different from T cell responses to glycolipids such as α-galactosyl ceramide or glucose monomycolate, where altering a single stereocenter on the carbohydrate epitope abolished recognition (43, 44). However, not every oily substance was sufficient to activate T cells.
Considering the particular chemical structures that control response, squalene is a C30 polyunsaturated branched chain lipid antigen (Fig. 3b) (31). We found cross-reactivity to structurally related C20 geranylgeraniol and C23 geranylgeranylactone, as well as C15 farnesol, but not smaller geraniol-based compounds (Fig. 3c). The farnesol response is notable because it is also a contact allergen in Truetest panel 2 (45) (Fig. 1) and so represents another link between contact allergens and CD1a antigens. Further, considering molecules with branched and ringed structures related to benzyl cinnamate, we identified a new antigen, coenzyme Q2 (Fig. 3d). Although coenzyme Q2 has not been described as a contact allergen, idebenone, which has an identical headgroup (2,3-dimethoxy, 5-methyl, 1,4-benzoquinone), but a less hydrophobic lipid tail, comprised of a 10-carbon alkyl chain with a hydroxyl group, is a well-known skin allergen (46–48). Also, in our CD1a plate assays, idebenone stimulated a dose-dependent T cell response, supporting a link between coenzyme Q2-related structures and contact allergens (Fig. S3). Notably, vitamin E, a known skin allergen, did not induce a response in this BC2-based screening. However, this does not exclude the existence of CD1a-restricted T cells to this hydrophobic compound within a polyclonal T cell repertoire.
The identification of a strong stimulatory response to coenzyme Q2 prompted screening of coenzyme Q length analogs, finding optimal response to coenzyme Q2 but not larger or smaller chain length analogs (Fig. 3e). (Fig. 3d). Last, comparison of 12 fatty acyl analogs consistently showed stronger response when the normally charged carboxylate group was capped by a methyl, alkyl or other structure to generate a non-polar molecule (Fig. S2). A weak effect was seen in some cases, where potency was increased by cis-unsaturation.
In summary, compared to highly flexible lipids with saturated alkyl chains, an unsaturation, ringed or branched structure correlated with higher response. However, very highly constrained or bulky structures, such as vitamin A, vitamin D and vitamin E, were not recognized. Considering molecular size, response was optimal with compounds (222–410 Da, C15-C30), which were near the middle of the size range tested (154–862 Da, C9-C59) (Fig. 3f). These optima were considerably smaller than known CD1 antigens (~700–1500 Da). Even the largest stimulatory compound, squalene (C30, 410 Da), was substantially smaller than the predicted number of methylene units (~C36) that would fill the CD1a cleft (1650 Å3) (19, 40). Unlike molecules that form antigenic epitopes for TCRs, no single molecular variant could be assigned as essential for T cell activation.
Last, to determine if the identified link between CD1a and contact allergens is generalizable to polyclonal T cells and among genetically unrelated human donors, we screened purified polyclonal T cells (CD4+ and CD4−) from blood bank donors, and determined their response to plate-bound CD1a pre-loaded with either farnesol or coenzyme Q2. As also seen in clinical evaluation of contact dermatitis patients, not all patients responded to every antigen, but we observed polyclonal responses to both antigens in two or more subjects using sensitive real-time qPCR testing of IFN-γ response (Fig. 3g). Responses were seen in the CD4+ T cell fractions, but were stronger in the CD4− T cell fraction (Fig. 3g). This suggests that the normal T cell repertoire contains T cells that respond to CD1a-contact allergen complexes. Similarly, in a different set of donors, T cell responses were detected to benzyl cinnamate-loaded CD1a (Fig.S4). Together, these results support the broader relevance of these CD1a allergens beyond the specificity of two T cell lines.
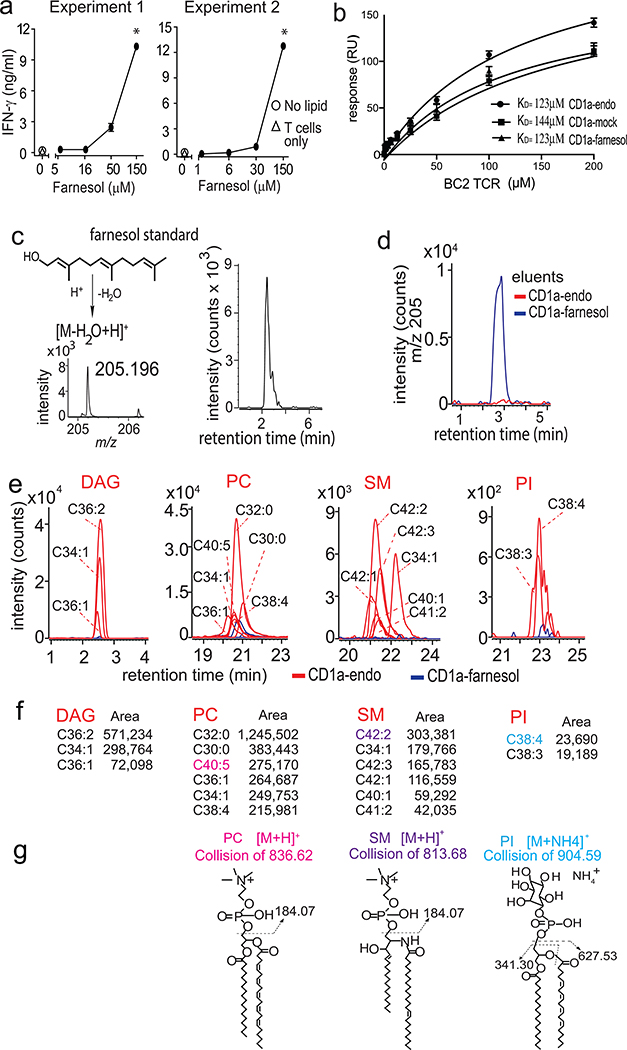
CD1a-lipid binding to the TCR
Farnesol is a common additive to cosmetics and skin creams, where its use requires precaution labeling, based on its recognized role as a contact allergen (45). Farnesol testing is routine in clinical practice, where it is present in the ‘fragrance mix 2’ in Truetest patches. Farnesol can also be tested as a pure compound, generating responses in ~1 % of people with suspected contact dermatitis (45). After the screen identified a farnesol response (Fig. 3c), we observed reproducible and dose-dependent response for BC2 in the CD1a-coated plate assay (Fig. 4a). Thus, farnesol was unlikely to be modified prior to recognition and was likely recognized by the BC2 TCR as a CD1a-farnesol complex.
Figure 4. CD1a-farnesol complexes.
a. IFN-γ release by BC2 T cells in response to CD1a-coated plates treated with farnesol was measured. * The significance of lipid concentration on IFN-γ release was assessed by marginal means with adjustment by the Sidak method after a significant result by ANOVA, treating experiments 1 and 2 as blocks. At the highest concentration of farnesol in both experiments, non-overlapping 95% confidence intervals were observed at p < 0.001 b. Affinity measurements (KD) by surface plasmon resonance in response to the recombinant BC2 TCR binding biotinylated CD1a directly isolated from cells (CD1a-endo), CD1a pre-treated with farnesol (CD1a-farnesol) or CD1a treated with buffer (CD1a-mock). Positive mode HPLC-MS analysis of a farnesol standard (c) and eluents from farnesol-treated CD1a (d) demonstrated ions that matched the expected mass (m/z 205.195) of an indicated dehydration product with a retention time of 2.9 min. e-f. Lipid eluents from CD1a-endo and CD1a-farnesol were analyzed by positive normal phase HPLC-MS QToF mass spectrometry. Ion chromatograms were generated at the nominal mass values of diacylglycerol (DAG), phosphatidylcholine (PC), sphingomyelin (SM) and phosphatidylinositol (PI), which are shown as CX:Y, where X is the number of methylene units in the combined lipid chains and Y is the total number of unsaturations. g. Compound identifications were based on the unknown matching the retention time and mass of standards. Further, one compound in the PC, SM and PI families (shown in color) underwent collision-induced dissociation mass spectrometry analysis to generate the indicated diagnostic fragments.
To test this hypothesis, we loaded farnesol onto biotinylated CD1a monomers, generated fluorescent tetramers and stained the BC2 T cell line and a control line. In several attempts with differing protocols, we failed to detect staining with CD1a-farnesol tetramers above background levels seen with farnesol-treated CD1b tetramers (Fig. S5). Turning to surface plasmon resonance (SPR), we produced the BC2 TCR heterodimer in vitro and measured its binding to untreated CD1a carrying mixed endogenous lipids (CD1a-endo), CD1a that was treated with media (CD1a-mock) and CD1a treated with farnesol (CD1a-farnesol) after coupling to SPR chips. The BC2 TCR bound to all three complexes with low but measurable binding affinities for CD1a-endo (KD = 123 μM), CD1a-mock (KD = 144 μM) and CD1a-farnesol (KD = 123 μM) (Fig. 4b). SPR is known to be more sensitive that tetramer staining (49), so the relatively low affinity interactions likely explained the absent tetramer staining. Yet, interactions are still in the physiological range, demonstrating direct binding between the BC2 TCR and CD1a. However, the cross reactivity of the BC2 TCR to three forms of CD1a left unclear the role of farnesol or other carried lipids in mediating CD1a-TCR interactions.
Lipid analysis of CD1a-lipid complexes
A recently proposed but unproven hypothesis is small hydrophobic lipids could fully sequester within CD1a (31, 50), displacing larger endogenous self-lipids that cover TCR epitopes on the outer surface of CD1a. Therefore, we undertook direct biochemical analysis of CD1a-lipid complexes formed in vitro with detergents and stimulatory substances, analyzing elutable lipids using high performance liquid chromatography-mass spectrometry (HPLC-MS). First, we addressed the trivial possibility that the lack of effect of farnesol treatment on TCR binding to CD1a resulted from the lack of farnesol loading onto CD1a. Analysis of eluents from farnesol-treated CD1a monomers was initially inconclusive because farnesol is a non-polar alcohol and does not readily adduct the cations or anions needed for MS detection. However, building on the fortuitous detection of a positively charged dehydration fragment [M-H20+H]+ generated in the MS source (31), we could reliably detect the equivalent product (m/z 205.196, C15H25+) from a farnesol standard. Subsequently we detected strong signal for this product from farnesol-treated CD1a proteins but not CD1a-endo, directly documenting farnesol in CD1a complexes (Fig. 4d).
Further, the HPLC-MS-based platform allowed broader analysis of the lipid ligands carried in CD1a-endo and CD1a-farnesol complexes. Similar to prior reports (31, 35), we could detect many ions in CD1a-endo eluents, which were self-lipids captured during protein expression in cells. Focusing on specific classes of lipids, including neutral lipids, phospholipids and sphingolipids, we could identify many self-ligands. CD1a-endo complexes carried at least three molecular species of diacylglycerol (DAG), six phosphatidylcholines (PC), six sphingomyelins (SM) and two phosphatidylinositol species. Initially these identifications were based on the expected early (DAG) or later (PI, PC, SM) retention time, as well as match of the detected m/z value with the expected mass of these ligands (Fig. 4e–f). For one lead compound in each class, we confirmed the identification using collision-induced dissociation mass spectrometry (CID-MS), which demonstrated the characteristic phosphocholine, phosphoinositol, sphingolipid or diacylglycerol fragments (Fig. 4g).
Elution analysis of farnesol-treated CD1a directly demonstrated farnesol loading (Fig. 4d). The comparison of CD1a-endo and CD1a-farnesol eluents, showed complete or nearly complete suppression of ion chromatogram signals corresponding to all the 17 tested self-lipids (Fig. 4e, blue). Although the conditions used to load farnesol in vitro are not the same as those in immunological assays, these findings suggest high occupancy of CD1a proteins by farnesol and that farnesol and self-lipids are not simultaneously bound. Together these data support a simple model for the cross-reactivity, where the TCR binds CD1a carrying either farnesol or certain self-lipids that permit recognition. Treatment of CD1a with farnesol displaces lipids with hydrophilic head groups to generated more homogenously liganded CD1a proteins (Fig. 4d–e).
CD1a-farnesol crystal structure overview
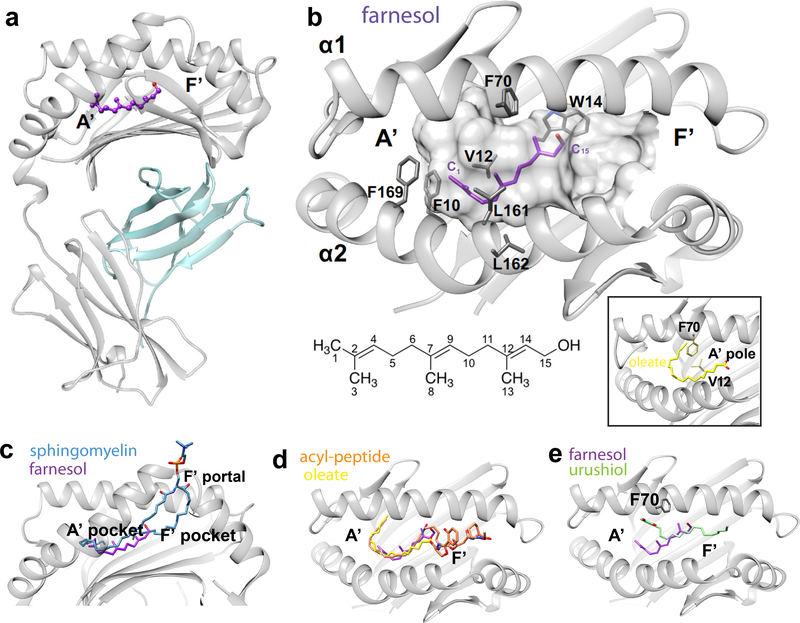
To determine the structural basis of farnesol response, we solved the CD1a-farnesol crystal structure at 2.2 Å resolution (Table S1). The electron density for the bound farnesol and surrounding CD1a residues were unambiguous (Fig. S6), allowing determination of the position and orientation of farnesol within the cleft (Fig. 5a–b). Unlike covalent binding of vitamin B metabolites to MR1 (51) and the predictions of haptenation models, we find no evidence for haptenation of CD1a residues by farnesol.
Figure 5. Crystal structure of CD1a-farnesol complexes.
a. Overview of the binary crystal structure of CD1a (grey)-farnesol (purple)/β2m(cyan). b. Molecular interactions of farnesol (purple) with the hydrophobic residues within CD1a binding cleft (grey surface). The side chains of the residues within 4 Å distance from the lipid are shown. A diagram of trans, trans farnesol with carbon numbering is shown. The A’ pole formed by V12-F70 interaction in the context of oleic acid-bound CD1a pocket (PDB ID: 4X6D) is highlighted in the inset. c-e. Superimposition of CD1a bound to farnesol and sphingomyelin (PDB ID: 4X6F, (35)) (c) lipopeptide (PDB ID: 1XZ0, (40)) (d) and urushiol (PDB ID: 5J1A, (30)) (e).
Instead the striking finding is that farnesol is deeply sequestered within the CD1a cleft, where it is fully inaccessible to TCRs. Most known amphipathic membrane lipids, such as sulfatide or sphingomyelin (19), occupy nearly all of the CD1a cleft and then extend their head groups through a portal (F’-portal) onto the external surface of CD1a (Fig. 5c). In contrast, farnesol occupies only 36% of the cleft. Accordingly, this relatively small ligand could have been seated in many ways within the larger cavity or potentially bound with lipid:CD1 stoichiometry of 2:1 or 3:1 (52). Instead, a preferred seating and orientation of a single molecule is observed at the junction of the A’ and F’ pockets. Unlike CD1b structures in which two lipids bind simultaneously within the cleft (53, 54), electron density corresponding to a second lipid or spacer in the cleft was not observed (Fig. 5a–b). This finding agreed with elution experiments showing substantial exclusion of the measured self-lipids from CD1a complexes (Fig. 4e). Together, the biochemical and structural data indicated that farnesol itself was sufficient to stabilize a partially occupied CD1a cleft.
Farnesol is buried fully within CD1a
In previously solved CD1a structures in complex with oleic acid (35) or an acyl-peptide (40), the flexible fatty acyl chains take a C-shaped conformation around the margin of the curved A’ pocket (Fig. 5d) (19, 35, 40). These lipids encircle a vertical structure known as the A’ pole, which is formed by an interaction of Phe70 and Val12, located in the ceiling and floor of the A ‘pocket, respectively (Fig. 5b, inset) (19, 35, 40). The semi-rigid and branched structure of farnesol does not allow the C-shaped peripheral conformation seen with other lipids and instead lies in the center of the A’ pocket, disrupting the A’ pole. The orientation of farnesol is discernible: the terminal methyl and hydroxyl groups point towards the A’ and F’ pocket, respectively (Fig. 5b). The polar hydroxyl group is situated nearer the solvent-exposed F’ portal of CD1a with ~ 15 percent of its surface water exposed. Farnesol made van der Waals contacts with Phe10, Trp14, Phe70, Val98, Leu161, Leu162 and Phe169 from CD1a. (Fig. 5b, Table S2). Here, Trp14 stacked against the unsaturated hydrocarbons C12 and C14 of farnesol, stabilizing further the bound lipid within the cleft. Interestingly, the same Trp14 residue maintains hydrophobic contacts with sphingosine and acyl chain moieties in the CD1a-sphingomyelin and CD1a-sulfatide structures (19), respectively. Collectively, this positioning mechanism appears to be driven by unsaturations in farnesol, which limit its ability to bend and provide and van der Waals interactions with the inner surface of CD1a.
Parallels in the positioning of CD1a-urushiol and CD1a-farnesol (Fig. 5e) highlight how the positioning of bulky and constrained lipids differs from the seating of acyl chain-containing ligands (Fig. 5d). Although farnesol and urushiol are not located in the same position, they are both situated near the junction of the A’ and F’ pockets (Fig. 5e) and do not take the deep and curved positioning at the rim of the A’ toroid (Fig. 5d). Whereas oleate and acyl peptide wrap around the intact A’ pole (Fig 5d, 5b inset), farnesol and urushiol complexes show a marked repositioning of Phe 70, which disrupts the A’ pole (Fig. 5b, e). Urushiol extends substantially into the F’-pocket so that it approaches the F’ portal of CD1a. It is unknown whether TCRs contact urushiol, but the molecule is adjacent to the surface portal (30) and TCRs can contact lipids located just within the portal (55). In contrast, farnesol is ~ 8Å more deeply positioned, so that it is unequivocally separated from the F’ portal and the TCR contact surface (Fig. 5e).
Overall, the structure-activity relationships (Fig. 3) indicated that many small, hydrophobic, bulky lipids from consumer goods are recognized by T cells. The biochemical (Fig. 4) and structural (Fig. 5) analysis of CD1a-lipid complexes demonstrate that farnesol’s small size and unsaturated structure allow it to interact specifically, but not covalently, within CD1a. This binding interaction stabilizes the CD1a cleft and positions farnesol out of the reach of the TCR, largely or fully displacing lipids that normally emerge to the outer surface of CD1a (19, 35, 40).
Discussion
In 1963 Gell and Coombs classified human disease-related immune manifestations into 4 types of hypersensitivity reactions (3). Despite the early development and descriptive nature of this scheme, the classification system is still widely taught in clinical immunology and medicine. Type I, II and III reactions are rapid and mediated by B cells, whereas the delayed Type IV response is mediated by T cells. Our study sought molecular mechanisms underpinning Type IV hypersensitivity to the most common contact dermatitis allergens in consumer products. Our data provide specific molecular connections between CD1a-reactive T cells and four structurally related contact dermatitis allergens: benzyl benzoate, benzyl cinnamate, farnesol and coenzyme Q-related compounds. Whereas haptens (9), drugs (7), or cations (8) can influence MHC-peptide display, here we detail a straightforward mechanism for T cell activation by small molecules that non-covalently bind CD1a.
In the MHC and CD1 systems, the most common recognition mechanism involves TCR co-contact with an epitope on the carried peptide or lipid and the antigen presenting molecule (21, 56–58). Here we show evidence that the key active components of balsam of Peru and farnesol activate T cells by binding to CD1a without cellular processing. However, both the structural and biochemical data strongly point to a new model of recognition that does not involve TCR contact with epitopes present on the stimulatory small molecules. Antigenic tree oils, PPBF, farnesol, coenzyme Q2 and the other 14 oily stimulants identified here all lack carbohydrate, phosphate or peptidic groups that normally serve as TCR epitopes. We show that the BC2 TCR can cross-react among at least 16 stimulatory compounds, which do not share any single chemical structure that would be a candidate cross-reactive epitope. More conclusively, farnesol resides deeply within the CD1a cleft, essentially ruling out direct contact with the TCR. Sequestration of molecules of a small size is could be a general mechanism of their recognition, since all of the stimulatory molecules are smaller than the CD1a cleft (21, 40, 57).
Prior studies of CD1-lipid complexes have emphasized head group positioning, where the seating of amphipathic lipids in the cleft is guided by carbohydrates or charged moieties that interact near the F’ portal. Alkyl chains have a ‘bland’ repetitive structure, and have been described as sliding within CD1 allowing diversely positioning in the groove (54, 59). Based on this concept, we expected that the small hydrophobic ligands studied here might slide freely or adopt multiple positions in the CD1a cleft. Also, since many of the lipids have a molecular size that is less than half the volume of the CD1a cleft, they might have bound in pairs or together with spacer lipids (52, 53, 60, 61). However, farnesol shows one defined position in the CD1a groove. Both mass spectrometry and crystallographic analysis failed to detect co-binding spacer lipids indicating that partial occupancy by one small lipid is sufficient to stabilize the CD1a cleft.
Comparison of CD1a-farnesol with previously solved CD1a-lipid structures provides insight into the roles of steric hindrance and interior pocket remodeling. CD1a-oleate (35), CD1a-mycobactin-like lipopeptide (40), CD1a-sulfatide (19) and CD1a-sphingomyelin (35) complexes involve lipids with flexible alkyl chains. These alkyl chains insert deeply into CD1a by curling along the outer wall of the A’ pocket and wrapping around the A’ pole to insert fully within the cleft (40). In contrast farnesol is chemically hindered and bulky, based on polyunsaturation and methyl branching. The rigid and bulky moiety in urushiol derives from a substituted catechol ring. These two molecules cannot curl to trace the outer wall of the A’ pocket so do not penetrate deeply, and both sit in a central position within the A’ pocket that prevents the A’ pole form forming. Farnesol is anchored in a specific position by a series of van der Waals interactions with named pocket residues formed by its polyunsaturated and branched structure. While the roles of benzyl rings in benzyl benzoate and benzyl cinnamate are not studied structurally, they also constrain the chemical structure in ways that are also expected to prevent the side-wall curvature (19, 35, 40). More generally, many of the stimulatory lipids identified here and in a recent study (31), including farnesol, squalene, geranylgeraniol, geranylgeranylacetone and coenzyme Q2, are polyunsaturated or branched isoprenoid lipids that could plausibly anchor in CD1a by similar mechanisms.
Lipid antigen binding wholly within CD1a could trigger T cell responses by remodeling the 3-dimensional structure of CD1a, as previously reported for CD1d (62, 63), CD1b (54) and CD1c (41, 64). However, comparing CD1a-farnesol with all CD1a-lipid structures solved to date (19, 35, 40) does not demonstrate a broad or obvious change in CD1a conformation. Also, binding of the BC2 TCR to both CD1a-farnesol and CD1a-endo point away from this explanation. Instead, biochemical analysis of CD1a-endo complexes and the CD1a-farnesol structure both indicate that farnesol displaces endogenous ligands from the cleft. Whereas farnesol can be considered a headless ligand, some amphipathic self-lipid ligands in CD1a-endo structures have head groups comprised of phosphates or sugars that normally cover the exposed surface of CD1a (35). In the case of the sphingomyelin, it blocks autoreactive T cells by interfering with TCR contact with CD1a (31, 35). Our experimental observations rule in key aspects of the “absence of interference” model, where activating substances are sequestered within the CD1a cleft, so that recognition occurs by ejecting self-lipids and freeing up epitopes on the surface of CD1a itself.
As contrasted with MHC I and MHC II, where peptides are broadly exposed over the lateral dimension of the platform, human CD1 proteins have a large roof-like structure above their clefts and a small antigen exit portal at the margin of the platform (65). This creates a potentially large, ligand-free TCR contact surface on CD1 proteins. Evidence for the predominant contact of αβ TCRs with the surface of CD1 proteins in preference to carried lipids, including the extreme case in which TCRs contact CD1 only, is becoming a central theme in CD1 research (65). Recent studies have shown direct TCR contact with the unliganded surface of CD1a and CD1c by autoreactive clones and polyclonal T cells (31, 35, 42). Thus, the stimulatory compounds identified here, which are small and internally sequestered, provide a molecular link to polyclonal autoreactive T cell responses, which are specific for the surface of CD1 rather than the carried lipid.
The presence of CD1a in all individuals prompts the question of why allergic contact dermatitis does not universally develop in everyone. However, inter-individual differences that may play a role include permeability of the skin barrier (66), dose and number of chemical exposures to allergens, regulatory T cell activity (67–69), and inter-individual differences in T cell repertoires. Prior studies show that there is inter-individual variability in the frequency of CD1a-restricted T cells in the blood and skin of healthy individuals, and differences in CD1a autoreactive response rates in skin (25, 66, 70, 71). Increased CD1a-restricted T cells responses were observed in allergic individuals and those with inflammatory skin disease (66, 70, 72), which may be a factor in susceptibility to development of CD1a-mediated allergic contact dermatitis in certain individuals. Consistent with these known patterns of antigen response, our small study of 11 humans demonstrates differing patterns of polyclonal response in each individual rather than a universal response to one antigen, which might be expected from an innate receptor.
Overall, the molecular analysis of tree oils and isoprenoid lipids presented in this manuscript invites focused consideration of the role of CD1a in T cell-mediated skin diseases. In this new view, the pattern of high density CD1a on the Langerhans cell network present throughout the skin could mediate responses to oils naturally produced within the skin or oils that contact the skin through application of commercial skin products containing botanical extracts, synthetic lipids or oils. Other immunogenic oils used in human patients or for experimental biology include the adjuvant MF-59 (squalene) and incomplete Freund’s adjuvant (mineral oil). These immunogens, as well as drug-like small molecules resembling PPBF or sulfonamide antibiotics, could plausibly act through the CD1 system.
Materials and Methods
Study Design
The goal of this study was to determine if known contact allergens can bind to CD1a and stimulate a CD1a-dependent T cell response. This study involved in vitro T cell assays using both CD1a-restricted T cell lines and polyclonal purified T cells from healthy blood bank donors. For T cell recognition, either cell-based assays using CD1a-expressing antigen presenting cells or CD1a plate assays using recombinant plate-bound CD1a were performed. Cytokine release was measured by ELISA, and/or cytokine transcription was measured by real-time qPCR. Complex lipid mixtures, such as balsam of Peru, were purified by thin layer chromatography, and analyzed by nanoelectrospray ionization mass spectrometry. Lipid eluents from CD1a, after displacement by contact allergens, were analyzed by positive normal phase HPLC-MS QToF mass spectrometry. Structural insights into CD1a complexed with the contact allergen farnesol were obtained by X-ray crystallography.
Contact dermatitis antigen screen
The T.R.U.E. (Thin-layer Rapid Use Epicutaneous Patch) test 1 (Truetest 1) is a test routinely used in clinic to diagnosis contact dermatitis in response to the most common allergens (SmartPractice, Phoenix, AZ). The system consists of surgical tape (5.2 × 13.0 cm) that is embedded with antigen patches of 0.81 cm2 with each coated with a polyester film that contains uniformly dispersed specific allergen. Using sterile technique, individual allergen patches were cut and placed directly in the assay wells containing ~ 106 antigen presenting cells and 1 ml T cell media in 24 well plates (contact method) or first extracted by soaking patch in 1 ml media (2 hrs, 37°C), followed by removing the patch and transferring 100 μL of media to T cell assays. Antigen dose was normalized to mm2 of patch exposure. Antigens or extracts were co-cultured with 50,000 CD1a-transfected or mock-transfected K562 cells (25) and a CD1a-dependent T cell line in a 96-well plate. Activation was measured by IFN-γ ELISA (ThermoFisher Scientific).
CD1a assays for T cell antigens
Balsam of Peru, balsam of Peru oil, benzyl cinnamate, and benzyl benzoate or other isolated antigens were dried in clean glass, subjected to water bath sonication in T cell media for 120 sec and cultured with 50,000 CD1a-transfected K562 cells or mock-transfected K562 cells for 3 h at 37°C and then co-cultured with 50,000–200,000 cells per well of an autoreactive T cell line (BC2 or Bgp) (31) or foreign antigen reactive T cells (CD8–2) (18) for 24 h at 37°C in 96-well plates as previously described (31). Activation was measured using IFN-γ ELISA (ThermoFisher Scientific). For blocking experiments, CD1a-transfected K562 cells were pre-incubated for 1 h at 37°C with CD1a-blocking antibody (OKT-6) or isotype-matched control IgG (P3) (10 μg/ml) before the addition of T cells. For plate assays, 96-well streptavidin plates (ThermoFisher Scientific) were incubated for 24 h at room temperature with biotinylated CD1a or CD1b protein (10 μg/ml, NIH Tetramer Core Facility) and anti-CD11a (2.5 μg/ml) in PBS, pH 7.4 as previously described (31). For the acid-stripping protocol (Fig. 4, Fig. 5a, and Fig. S2), after 24 h of coating with protein, plates were washed three times with PBS, followed by washing twice with citrate buffer at a pH of 3.4 for 10 min, followed three washes in PBS before the addition of lipid antigens (30). Peripheral blood mononuclear cells (PBMC) were isolated from buffycoats obtained from the New York Blood Center, as approved by the Institutional Review Board of Columbia University Irving Medical Center. Polyclonal T cell assays were performed using FACS-sorted T cells from PBMC (CD4− and CD4+), and CD1a-coated 96-well plates as described above. Plate-coated CD1a was either treated with buffer only (0.05% CHAPS in PBS) or lipid antigens sonicated in buffer and incubated overnight at 37°C. Plates were washed three times and then purified T cells were added to the wells and incubated overnight at 37°C. RNA was extracted using RNeasy (Qiagen), and first-strand cDNA synthesis was performed using iScript (BioRad).
Lipid sources
Balsam of Peru (W211613), balsam of Peru oil (W211710), benzyl cinnamate (234214), benzyl benzoate (B9550), geranylgeraniol (G3278), farnesol (277541), geranylgeranyl acetone (G5048), geraniol (163333), squalene (S3626), geranyl acetone (250716), vitamin K1 (V3501), vitamin K2 (V9378), vitamin A (R7632), vitamin E (T3251), vitamin D3 (C9756), coenzyme Q2 (C8081), coenzyme Q0 (D9150), coenzyme Q4 (C2470), coenzyme Q6 (C9504), coenzyme Q10 (C9538), palmitoleic acid (P9417), methyl palmitoleate (P9667), cis-11-hexadecenal (249084), palmityl acetate (P0260), palmitoleyl alcohol (P1547), lauryl palmitoleate (P1642), oleamide (O2136), palmitoyl ethanolamide (P0359), tetradecanoic acid ethylamide (R425567), N-oleoyl glycin (O9762), N,N-dimethyl tetradecanamide (S347388), and 1-dodecyl-2-pyrrolidinone (335673) were obtained from Sigma-Aldrich (St. Louis, MO). Coenzyme Q1 (270–294-M002) was obtained from Alexis Biochemicals.
Lipid analysis by TLC
Silica-coated glass thin layer chromatography (TLC) plates (10 cm x 20 cm; Scientific Adsorbents Incorporated) were pre-cleared in chloroform-methanol-water (60:30:6; vol/vol/vol). Samples (10 – 20 μg) were developed with a solvent system-hexane/diethyl ether/acetic acid (70/30/1 (vol/vol/vol)). For visualization, plates were sprayed with a solution of 3% (wt/vol) of cupric acetate in 8% (vol/vol) phosphoric acid, followed by heating for 20–30 min at 150 °C.
Nanoelectrospray-ionization Mass Spectrometry
2 μg/ml of methanol solution was prepared for each reagent, and then 10 μl was loaded onto a glass nanospray tip for positive-mode electrospray-ionization mass spectrometry performed on a LXQ (Thermo), two-dimensional ion-trap mass spectrometer. The spray voltage and capillary temperature were set to 0.8 kV and 200 °C.
High performance liquid chromatography (HPLC)-QTof-Mass spectrometry
CD1a-endo (200 μg) and CD1a-farnesol (200 μg) were transferred to 15-ml glass tubes and treated with chloroform, methanol, and water for lipid extraction according to the method of Bligh and Dyer (73). The lipid-containing organic solvent layer was separated from the top aqueous layer by centrifugation at 850 g for 10 min. For HPLC-MS analysis, the samples were normalized based on the input proteins (20 μM), and 20 μl of eluent was injected to an Agilent 6530 Accurate-Mass Q-TOF spectrometer equipped with a 1260 series HPLC system using a normal phase Inertsil diol column (150 mm × 2.1 mm, 3 micron, GL Sciences) with a guard column (10 mm x 3 mm, 3 micron, GL Sciences), running at 0.15 ml/min according to a published method (74).
Recombinant CD1a expression and purification
The glycoprotein CD1a was expressed in HEK293S GnTI− cells and purified as previously described (35). Following an endoglycosidase H (New England BioLabs) and thrombin treatment, the purified CD1a was first loaded with the ganglioside GD3 (GD3) (Avanti) that was dissolved in a solution containing 2.5% dimethylsulfoxide (DMSO) and 0.5% tyloxapol (Sigma). CD1a was first incubated with GD3 overnight at room temperature at a molar ratio of 1:8. The CD1a sample loaded with GD3 was further purified using ion exchange chromatography (MonoQ 10/100 GL-GE Healthcare). Trans, trans-farnesol (Sigma) was dissolved in a solution containing 2.5% DMSO and 0.5% tyloxapol (Sigma). The CD1a-GD3 sample was then incubated overnight with farnesol at a 1:100 molar ratio and at room temperature. A subsequent ion exchange chromatography (MonoQ 10/100 GL) was performed to remove the excess of farnesol, CD1a-GD3 and tyloxapol.
Expression, refolding and purification of recombinant TCRs
The BC2 TCR was produced using a previously described method (31). Briefly, individual α and β chains of the TCR, with an engineered disulfide bond between the TRAC and TRBC constant domains, were expressed in BL21 E. coli cells as inclusion bodies and solubilised in 8 M urea buffer containing 10 mM Tris-HCl pH 8, 0.5 mM Na-EDTA, and 1 mM dithiothreitol. The TCR was then refolded in buffer that was comprised of 5M urea, 100 mM Tris-HCl pH 8, 2 mM Na-EDTA, 400 mM L-Arg-HCl, 0.5 mM oxidized glutathione and 5 mM reduced glutathione. The refolded solution was dialyzed twice against 10 mM Tris-HCl pH 8.0 overnight. The dialyzed samples were then purified through DEAE cellulose, size-exclusion and anion exchange HiTrap Q chromatography approaches. The quality and purity of the samples were analyzed via SDS-PAGE.
Crystallization, structure determination and refinement
Seeds obtained from previous binary CD1a-antigen crystals (30) were used to grow crystals of the CD1a-farnesol binary complex in 20–25% PEG 1500 / 10% MMT buffer pH 5–6. The crystals were flash-frozen and data were collected at the MX2 beamline (Australian Synchrotron) to a 2.2 Å resolution. All the data were processed with the program XDS (75) and were scaled with SCALA from the CCP4 programs suite (76). Upon successful phasing by molecular replacement using the program PHASER (77) and the CD1a-urushiol structure as search model (30), the farnesol electron density was clearly evident in the unbiased electron density maps in addition to some very weak residual density. An initial run of rigid body refinement was performed using phenix.refine (78). Iterative model improvement was performed using with the program COOT (79) and phenix.refine. The final refinement led to an R/R-free (%) of 20/25. The quality of the structure was confirmed at the Research Collaboratory for Structural Bioinformatics Protein Data Bank Data Validation and Deposition Services website. All presentations of molecular graphics were created with UCSF-Chimera (80).
Surface plasmon resonance
Biotinylated CD1a-endogenous lipids derived from HEK293 cells was incubated over-night with 30-fold molar excess of farnesol solubilized in 2.5% dimethylsulfoxide/0.5% tyloxapol (CD1a-farnesol) or with solvent only (CD1a-mock). The sample was coupled onto research-grade streptavidin-coated chips (SA) to a mass concentration of ~3000 resonance units (RU). Increasing concentrations of the BC2 TCR (0–200 μM) were injected over all flow cells for 30s at a rate of 5 μl/min on a Biacore 3000 in 10 mM Tris-HCl pH 8, 150 mM NaCl buffer. The final response was calculated by subtraction of the response for CD1a-endogenous minus a flow cell containing an unrelated protein. The data were fitted to a 1:1 Langmuir binding model using BIAevaluation version 3.1 software (Biacore AB) and the equilibrium data analyzed using Prism program for biostatistics, curve fitting and scientific graphing (GraphPad).
Statistical analyses
All statistical analyses were performed in R (https://www.R-project.org/). Pairwise t-tests, ANOVA post hoc testing and adjustments of p values for multiple hypothesis testing used base R and the package emmeans (https://CRAN.R-project.org/package=emmeans). Dose response analyses used the package drc to fit log normal or logistic curves to the data and to test fitted models against simplified, pooled models (81). R code is available on request.
Supplementary Material
Acknowledgements
We thank A.G. Kasmar, M.C. Castells, and P. Brennan for advice or critical comments on the manuscript. We thank the staff at the Australian Synchrotron for assistance with data collection, and the NIH Tetramer Core Facility for recombinant biotinylated CD1 protein.
Funding: SN was supported by a NIH training grant (T32 AI007306), and is currently employed by HealthPartners, St. Paul, Minnesota. AdJ is supported by a K01 award from the NIH (K01 AR068475) and an Irving Scholarship from the Irving Institute for Clinical and Translational Research at Columbia University. DBM is supported by the NIH (R01 AR048632) and the Wellcome Trust Collaborative Award. This work was supported by the National Health and Medical Research Council of Australia (NHMRC) and the Australian Research Council (ARC) (CE140100011). JLN is supported by an ARC Future Fellowship (FT160100074); JR is supported by an Australian ARC Laureate Fellowship and the Wellcome Trust Collaborative Award. Research reported in this publication was performed in the CCTI Flow Cytometry Core, supported in part by the Office of the Director, National Institutes of Health under awards S10OD020056.
Footnotes
Competing Interests Statement.
The authors declare that they have no competing interests.
Data and reagent availability statement. Reagents are available to qualified scientists subject to the limitation that cells from primary T cell lines can be limited in number. The data and refined coordinates for the CD1a-farnesol structure were deposited in the Protein Data Bank under 6NUX accession code. All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
References and Notes
- 1.Peiser M, Tralau T, Heidler J, Api AM, Arts JH, Basketter DA, English J, Diepgen TL, Fuhlbrigge RC, Gaspari AA, Johansen JD, Karlberg AT, Kimber I, Lepoittevin JP, Liebsch M, Maibach HI, Martin SF, Merk HF, Platzek T, Rustemeyer T, Schnuch A, Vandebriel RJ, White IR, Luch A, Allergic contact dermatitis: epidemiology, molecular mechanisms, in vitro methods and regulatory aspects. Current knowledge assembled at an international workshop at BfR, Germany. Cellular and molecular life sciences : CMLS 69, 763–781 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaplan DH, Igyarto BZ, Gaspari AA, Early immune events in the induction of allergic contact dermatitis. Nature reviews. Immunology 12, 114–124 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gell PGH, Coombs RRA, Clinical Aspects of Immunology. (Blackwell, Oxford (UK), 1963), vol. 1. [Google Scholar]
- 4.Garboczi DN, Ghosh P, Utz U, Fan QR, Biddison WE, Wiley DC, Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature 384, 134–141 (1996). [DOI] [PubMed] [Google Scholar]
- 5.Garcia KC, Degano M, Stanfield RL, Brunmark A, Jackson MR, Peterson PA, Teyton L, Wilson IA, An alphabeta T cell receptor structure at 2.5 A and its orientation in the TCR-MHC complex. Science 274, 209–219 (1996). [DOI] [PubMed] [Google Scholar]
- 6.Rossjohn J, Gras S, Miles JJ, Turner SJ, Godfrey DI, McCluskey J, T cell antigen receptor recognition of antigen-presenting molecules. Annual review of immunology 33, 169–200 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Illing PT, Vivian JP, Dudek NL, Kostenko L, Chen Z, Bharadwaj M, Miles JJ, Kjer-Nielsen L, Gras S, Williamson NA, Burrows SR, Purcell AW, Rossjohn J, McCluskey J, Immune self-reactivity triggered by drug-modified HLA-peptide repertoire. Nature 486, 554–558 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Clayton GM, Wang Y, Crawford F, Novikov A, Wimberly BT, Kieft JS, Falta MT, Bowerman NA, Marrack P, Fontenot AP, Dai S, Kappler JW, Structural basis of chronic beryllium disease: linking allergic hypersensitivity and autoimmunity. Cell 158, 132–142 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Landsteiner K, Jacobs J, Studies on the Sensitization of Animals with Simple Chemical Compounds. Ii. The Journal of experimental medicine 64, 625–639 (1936). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt M, Raghavan B, Muller V, Vogl T, Fejer G, Tchaptchet S, Keck S, Kalis C, Nielsen PJ, Galanos C, Roth J, Skerra A, Martin SF, Freudenberg MA, Goebeler M, Crucial role for human Toll-like receptor 4 in the development of contact allergy to nickel. Nature immunology 11, 814–819 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Burkhart C, Britschgi M, Strasser I, Depta JP, von Greyerz S, Barnaba V, Pichler WJ, Non-covalent presentation of sulfamethoxazole to human CD4+ T cells is independent of distinct human leucocyte antigen-bound peptides. Clinical and experimental allergy : journal of the British Society for Allergy and Clinical Immunology 32, 1635–1643 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Farrell J, Lichtenfels M, Sullivan A, Elliott EC, Alfirevic A, Stachulski AV, Pirmohamed M, Naisbitt DJ, Park BK, Activation of carbamazepine-responsive T-cell clones with metabolically inert halogenated derivatives. The Journal of allergy and clinical immunology 132, 493–495 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Sieben S, Kawakubo Y, Al Masaoudi T, Merk HF, Blomeke B, Delayed-type hypersensitivity reaction to paraphenylenediamine is mediated by 2 different pathways of antigen recognition by specific alphabeta human T-cell clones. The Journal of allergy and clinical immunology 109, 1005–1011 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Keller M, Lerch M, Britschgi M, Tache V, Gerber BO, Luthi M, Lochmatter P, Kanny G, Bircher AJ, Christiansen C, Pichler WJ, Processing-dependent and -independent pathways for recognition of iodinated contrast media by specific human T cells. Clinical and experimental allergy : journal of the British Society for Allergy and Clinical Immunology 40, 257–268 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Adam J, Pichler WJ, Yerly D, Delayed drug hypersensitivity: models of T-cell stimulation. British journal of clinical pharmacology 71, 701–707 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levine BB, Ovary Z, Studies on the mechanism of the formation of the penicillin antigen. III. The N-(D-alpha-benzylpenicilloyl) group as an antigenic determinant responsible for hypersensitivity to penicillin G. The Journal of experimental medicine 114, 875–904 (1961). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bharadwaj M, Illing P, Theodossis A, Purcell AW, Rossjohn J, McCluskey J, Drug hypersensitivity and human leukocyte antigens of the major histocompatibility complex. Annual review of pharmacology and toxicology 52, 401–431 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Moody DB, Young DC, Cheng TY, Rosat JP, Roura-Mir C, O’Connor PB, Zajonc DM, Walz A, Miller MJ, Levery SB, Wilson IA, Costello CE, Brenner MB, T cell activation by lipopeptide antigens. Science 303, 527–531 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Zajonc DM, Elsliger MA, Teyton L, Wilson IA, Crystal structure of CD1a in complex with a sulfatide self antigen at a resolution of 2.15 A Nature immunology 4, 808–815 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Zeng Z, Castano AR, Segelke BW, Stura EA, Peterson PA, Wilson IA, Crystal structure of mouse CD1: An MHC-like fold with a large hydrophobic binding groove. Science 277, 339–345 (1997). [DOI] [PubMed] [Google Scholar]
- 21.Salio M, Silk JD, Jones EY, Cerundolo V, Biology of CD1- and MR1-restricted T cells. Annual review of immunology 32, 323–366 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Van Rhijn I, Young DC, Im JS, Levery SB, Illarionov PA, Besra GS, Porcelli SA, Gumperz J, Cheng TY, Moody DB, CD1d-restricted T cell activation by nonlipidic small molecules. Proceedings of the National Academy of Sciences of the United States of America 101, 13578–13583 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Betts RJ, Perkovic A, Mahapatra S, Del Bufalo A, Camara K, Howell AR, Martinozzi Teissier S, De Libero G, Mori L, Contact sensitizers trigger human CD1-autoreactive T-cell responses. European journal of immunology 47, 1171–1180 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dougan SK, Kaser A, Blumberg RS, CD1 expression on antigen-presenting cells. Current topics in microbiology and immunology 314, 113–141 (2007). [DOI] [PubMed] [Google Scholar]
- 25.de Jong A, Pena-Cruz V, Cheng TY, Clark RA, Van Rhijn I, Moody DB, CD1a-autoreactive T cells are a normal component of the human alphabeta T cell repertoire. Nature immunology 11, 1102–1109 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Lalla C, Lepore M, Piccolo FM, Rinaldi A, Scelfo A, Garavaglia C, Mori L, De Libero G, Dellabona P, Casorati G, High-frequency and adaptive-like dynamics of human CD1 self-reactive T cells. European journal of immunology 41, 602–610 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Sugita M, Grant EP, van Donselaar E, Hsu VW, Rogers RA, Peters PJ, Brenner MB, Separate pathways for antigen presentation by CD1 molecules. Immunity 11, 743–752 (1999). [DOI] [PubMed] [Google Scholar]
- 28.Manolova V, Kistowska M, Paoletti S, Baltariu GM, Bausinger H, Hanau D, Mori L, De Libero G, Functional CD1a is stabilized by exogenous lipids. European journal of immunology 36, 1083–1092 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Kobayashi C, Shiina T, Tokioka A, Hattori Y, Komori T, Kobayashi-Miura M, Takizawa T, Takahara K, Inaba K, Inoko H, Takeya M, Dranoff G, Sugita M, GM-CSF-independent CD1a expression in epidermal Langerhans cells: evidence from human CD1A genome-transgenic mice. The Journal of investigative dermatology 132, 241–244 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim JH, Hu Y, Yongqing T, Kim J, Hughes VA, Le Nours J, Marquez EA, Purcell AW, Wan Q, Sugita M, Rossjohn J, Winau F, CD1a on Langerhans cells controls inflammatory skin disease. Nature immunology 17, 1159–1166 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Jong A, Cheng TY, Huang S, Gras S, Birkinshaw RW, Kasmar AG, Van Rhijn I, Pena-Cruz V, Ruan DT, Altman JD, Rossjohn J, Moody DB, CD1a-autoreactive T cells recognize natural skin oils that function as headless antigens. Nature immunology 15, 177–185 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fransway AF, Zug KA, Belsito DV, Deleo VA, Fowler JF Jr., Maibach HI, Marks JG, Mathias CG, Pratt MD, Rietschel RL, Sasseville D, Storrs FJ, Taylor JS, Warshaw EM, Dekoven J, Zirwas M, North American Contact Dermatitis Group patch test results for 2007–2008. Dermatitis : contact, atopic, occupational, drug : official journal of the American Contact Dermatitis Society, North American Contact Dermatitis Group 24, 10–21 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Wolf R, Orion E, Ruocco E, Baroni A, Ruocco V, Contact dermatitis: facts and controversies. Clinics in dermatology 31, 467–478 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Swamy M, Beck-Garcia K, Beck-Garcia E, Hartl FA, Morath A, Yousefi OS, Dopfer EP, Molnar E, Schulze AK, Blanco R, Borroto A, Martin-Blanco N, Alarcon B, Hofer T, Minguet S, Schamel WW, A Cholesterol-Based Allostery Model of T Cell Receptor Phosphorylation. Immunity 44, 1091–1101 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Birkinshaw RW, Pellicci DG, Cheng TY, Keller AN, Sandoval-Romero M, Gras S, de Jong A, Uldrich AP, Moody DB, Godfrey DI, Rossjohn J, alphabeta T cell antigen receptor recognition of CD1a presenting self lipid ligands. Nature immunology 16, 258–266 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosat JP, Grant EP, Beckman EM, Dascher CC, Sieling PA, Frederique D, Modlin RL, Porcelli SA, Furlong ST, Brenner MB, CD1-restricted microbial lipid antigen-specific recognition found in the CD8+ alpha beta T cell pool. Journal of immunology 162, 366–371 (1999). [PubMed] [Google Scholar]
- 37.Hamilton T, de Gannes GC, Allergic contact dermatitis to preservatives and fragrances in cosmetics. Skin therapy letter 16, 1–4 (2011). [PubMed] [Google Scholar]
- 38.Young DC, Moody DB, T-cell recognition of glycolipids presented by CD1 proteins. Glycobiology 16, 103R–112R (2006). [DOI] [PubMed] [Google Scholar]
- 39.Moody DB, Zajonc DM, Wilson IA, Anatomy of CD1-lipid antigen complexes. Nature reviews. Immunology 5, 387–399 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Zajonc DM, Crispin MD, Bowden TA, Young DC, Cheng TY, Hu J, Costello CE, Rudd PM, Dwek RA, Miller MJ, Brenner MB, Moody DB, Wilson IA, Molecular mechanism of lipopeptide presentation by CD1a. Immunity 22, 209–219 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Mansour S, Tocheva AS, Cave-Ayland C, Machelett MM, Sander B, Lissin NM, Molloy PE, Baird MS, Stubs G, Schroder NW, Schumann RR, Rademann J, Postle AD, Jakobsen BK, Marshall BG, Gosain R, Elkington PT, Elliott T, Skylaris CK, Essex JW, Tews I, Gadola SD, Cholesteryl esters stabilize human CD1c conformations for recognition by self-reactive T cells. Proceedings of the National Academy of Sciences of the United States of America 113, E1266–1275 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wun KS, Reijneveld JF, Cheng TY, Ladell K, Uldrich AP, Le Nours J, Miners KL, McLaren JE, Grant EJ, Haigh OL, Watkins TS, Suliman S, Iwany S, Jimenez J, Calderon R, Tamara KL, Leon SR, Murray MB, Mayfield JA, Altman JD, Purcell AW, Miles JJ, Godfrey DI, Gras S, Price DA, Van Rhijn I, Moody DB, Rossjohn J, T cell autoreactivity directed toward CD1c itself rather than toward carried self lipids. Nature immunology 19, 397–406 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kawano T, Cui J, Koezuka Y, Toura I, Kaneko Y, Motoki K, Ueno H, Nakagawa R, Sato H, Kondo E, Koseki H, Taniguchi M, CD1d-restricted and TCR-mediated activation of valpha14 NKT cells by glycosylceramides. Science 278, 1626–1629 (1997). [DOI] [PubMed] [Google Scholar]
- 44.Moody DB, Reinhold BB, Guy MR, Beckman EM, Frederique DE, Furlong ST, Ye S, Reinhold VN, Sieling PA, Modlin RL, Besra GS, Porcelli SA, Structural requirements for glycolipid antigen recognition by CD1b-restricted T cells. Science 278, 283–286 (1997). [DOI] [PubMed] [Google Scholar]
- 45.Schnuch A, Uter W, Geier J, Lessmann H, Frosch PJ, Contact allergy to farnesol in 2021 consecutively patch tested patients. Results of the IVDK. Contact dermatitis 50, 117–121 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Mc Aleer MA, Collins P, Allergic contact dermatitis to hydroxydecyl ubiquinone (idebenone) following application of anti-ageing cosmetic cream. Contact dermatitis 59, 178–179 (2008). [DOI] [PubMed] [Google Scholar]
- 47.Natkunarajah J, Ostlere L, Allergic contact dermatitis to idebenone in an over-the-counter anti-ageing cream. Contact dermatitis 58, 239 (2008). [DOI] [PubMed] [Google Scholar]
- 48.Sasseville D, Moreau L, Al-Sowaidi M, Allergic contact dermatitis to idebenone used as an antioxidant in an anti-wrinkle cream. Contact dermatitis 56, 117–118 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Zhou D, Mattner J, Cantu C 3rd, Schrantz N, Yin N, Gao Y, Sagiv Y, Hudspeth K, Wu YP, Yamashita T, Teneberg S, Wang D, Proia RL, Levery SB, Savage PB, Teyton L, Bendelac A, Lysosomal glycosphingolipid recognition by NKT cells. Science 306, 1786–1789 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Kronenberg M, Havran WL, Immunology: oiling the wheels of autoimmunity. Nature 506, 42–43 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Corbett AJ, Eckle SB, Birkinshaw RW, Liu L, Patel O, Mahony J, Chen Z, Reantragoon R, Meehan B, Cao H, Williamson NA, Strugnell RA, Van Sinderen D, Mak JY, Fairlie DP, Kjer-Nielsen L, Rossjohn J, McCluskey J, T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature 509, 361–365 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Huang S, Cheng TY, Young DC, Layre E, Madigan CA, Shires J, Cerundolo V, Altman JD, Moody DB, Discovery of deoxyceramides and diacylglycerols as CD1b scaffold lipids among diverse groove-blocking lipids of the human CD1 system. Proceedings of the National Academy of Sciences of the United States of America 108, 19335–19340 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gadola SD, Zaccai NR, Harlos K, Shepherd D, Castro-Palomino JC, Ritter G, Schmidt RR, Jones EY, Cerundolo V, Structure of human CD1b with bound ligands at 2.3 A, a maze for alkyl chains. Nature immunology 3, 721–726 (2002). [DOI] [PubMed] [Google Scholar]
- 54.Garcia-Alles LF, Collmann A, Versluis C, Lindner B, Guiard J, Maveyraud L, Huc E, Im JS, Sansano S, Brando T, Julien S, Prandi J, Gilleron M, Porcelli SA, de la Salle H, Heck AJ, Mori L, Puzo G, Mourey L, De Libero G, Structural reorganization of the antigen-binding groove of human CD1b for presentation of mycobacterial sulfoglycolipids. Proceedings of the National Academy of Sciences of the United States of America 108, 17755–17760 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gras S, Van Rhijn I, Shahine A, Cheng TY, Bhati M, Tan LL, Halim H, Tuttle KD, Gapin L, Le Nours J, Moody DB, Rossjohn J, T cell receptor recognition of CD1b presenting a mycobacterial glycolipid. Nature communications 7, 13257 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zajonc DM, The CD1 family: serving lipid antigens to T cells since the Mesozoic era. Immunogenetics 68, 561–576 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Borg NA, Wun KS, Kjer-Nielsen L, Wilce MC, Pellicci DG, Koh R, Besra GS, Bharadwaj M, Godfrey DI, McCluskey J, Rossjohn J, CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature 448, 44–49 (2007). [DOI] [PubMed] [Google Scholar]
- 58.Garcia KC, Degano M, Pease LR, Huang M, Peterson PA, Teyton L, Wilson IA, Structural basis of plasticity in T cell receptor recognition of a self peptide-MHC antigen. Science 279, 1166–1172 (1998). [DOI] [PubMed] [Google Scholar]
- 59.Moody DB, How T cells grasp mycobacterial lipid antigens. Proceedings of the National Academy of Sciences of the United States of America 114, 13312–13314 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Garcia-Alles LF, Versluis K, Maveyraud L, Vallina AT, Sansano S, Bello NF, Gober HJ, Guillet V, de la Salle H, Puzo G, Mori L, Heck AJ, De Libero G, Mourey L, Endogenous phosphatidylcholine and a long spacer ligand stabilize the lipid-binding groove of CD1b. The EMBO journal 25, 3684–3692 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cotton RN, Shahine A, Rossjohn J, Moody DB, Lipids hide or step aside for CD1-autoreactive T cell receptors. Current opinion in immunology 52, 93–99 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McCarthy C, Shepherd D, Fleire S, Stronge VS, Koch M, Illarionov PA, Bossi G, Salio M, Denkberg G, Reddington F, Tarlton A, Reddy BG, Schmidt RR, Reiter Y, Griffiths GM, van der Merwe PA, Besra GS, Jones EY, Batista FD, Cerundolo V, The length of lipids bound to human CD1d molecules modulates the affinity of NKT cell TCR and the threshold of NKT cell activation. The Journal of experimental medicine 204, 1131–1144 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wun KS, Cameron G, Patel O, Pang SS, Pellicci DG, Sullivan LC, Keshipeddy S, Young MH, Uldrich AP, Thakur MS, Richardson SK, Howell AR, Illarionov PA, Brooks AG, Besra GS, McCluskey J, Gapin L, Porcelli SA, Godfrey DI, Rossjohn J, A molecular basis for the exquisite CD1d-restricted antigen specificity and functional responses of natural killer T cells. Immunity 34, 327–339 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Scharf L, Li NS, Hawk AJ, Garzon D, Zhang T, Fox LM, Kazen AR, Shah S, Haddadian EJ, Gumperz JE, Saghatelian A, Faraldo-Gomez JD, Meredith SC, Piccirilli JA, Adams EJ, The 2.5 A structure of CD1c in complex with a mycobacterial lipid reveals an open groove ideally suited for diverse antigen presentation. Immunity 33, 853–862 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Van Rhijn I, Godfrey DI, Rossjohn J, Moody DB, Lipid and small-molecule display by CD1 and MR1. Nature reviews. Immunology 15, 643–654 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jarrett R, Salio M, Lloyd-Lavery A, Subramaniam S, Bourgeois E, Archer C, Cheung KL, Hardman C, Chandler D, Salimi M, Gutowska-Owsiak D, de la Serna JB, Fallon PG, Jolin H, McKenzie A, Dziembowski A, Podobas EI, Bal W, Johnson D, Moody DB, Cerundolo V, Ogg G, Filaggrin inhibits generation of CD1a neolipid antigens by house dust mite-derived phospholipase. Science translational medicine 8, 325ra318 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Luckey U, Schmidt T, Pfender N, Romer M, Lorenz N, Martin SF, Bopp T, Schmitt E, Nikolaev A, Yogev N, Waisman A, Jakob T, Steinbrink K, Crosstalk of regulatory T cells and tolerogenic dendritic cells prevents contact allergy in subjects with low zone tolerance. The Journal of allergy and clinical immunology 130, 781–797 e711 (2012). [DOI] [PubMed] [Google Scholar]
- 68.Braun A, Dewert N, Brunnert F, Schnabel V, Hardenberg JH, Richter B, Zachmann K, Cording S, Classen A, Brans R, Hamann A, Huehn J, Schon MP, Integrin alphaE(CD103) Is Involved in Regulatory T-Cell Function in Allergic Contact Hypersensitivity. The Journal of investigative dermatology 135, 2982–2991 (2015). [DOI] [PubMed] [Google Scholar]
- 69.El Beidaq A, Link CW, Hofmann K, Frehse B, Hartmann K, Bieber K, Martin SF, Ludwig RJ, Manz RA, In Vivo Expansion of Endogenous Regulatory T Cell Populations Induces Long-Term Suppression of Contact Hypersensitivity. Journal of immunology 197, 1567–1576 (2016). [DOI] [PubMed] [Google Scholar]
- 70.Subramaniam S, Aslam A, Misbah SA, Salio M, Cerundolo V, Moody DB, Ogg G, Elevated and cross-responsive CD1a-reactive T cells in bee and wasp venom allergic individuals. European journal of immunology 46, 242–252 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hardman CS, Chen YL, Salimi M, Jarrett R, Johnson D, Jarvinen VJ, Owens RJ, Repapi E, Cousins DJ, Barlow JL, McKenzie ANJ, Ogg G, CD1a presentation of endogenous antigens by group 2 innate lymphoid cells. Science immunology 2, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cheung KL, Jarrett R, Subramaniam S, Salimi M, Gutowska-Owsiak D, Chen YL, Hardman C, Xue L, Cerundolo V, Ogg G, Psoriatic T cells recognize neolipid antigens generated by mast cell phospholipase delivered by exosomes and presented by CD1a. The Journal of experimental medicine 213, 2399–2412 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bligh EG, Dyer WJ, A rapid method of total lipid extraction and purification. Canadian journal of biochemistry and physiology 37, 911–917 (1959). [DOI] [PubMed] [Google Scholar]
- 74.Layre E, Sweet L, Hong S, Madigan CA, Desjardins D, Young DC, Cheng TY, Annand JW, Kim K, Shamputa IC, McConnell MJ, Debono CA, Behar SM, Minnaard AJ, Murray M, Barry CE 3rd, Matsunaga I, Moody DB, A comparative lipidomics platform for chemotaxonomic analysis of Mycobacterium tuberculosis. Chemistry & biology 18, 1537–1549 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kabsch W, Xds. Acta crystallographica. Section D, Biological crystallography 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS, Overview of the CCP4 suite and current developments. Acta crystallographica. Section D, Biological crystallography 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ, Phaser crystallographic software. Journal of applied crystallography 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD, Towards automated crystallographic structure refinement with phenix.refine. Acta crystallographica. Section D, Biological crystallography 68, 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Emsley P, Lohkamp B, Scott WG, Cowtan K, Features and development of Coot. Acta crystallographica. Section D, Biological crystallography 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE, UCSF Chimera--a visualization system for exploratory research and analysis. Journal of computational chemistry 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 81.Ritz C, Baty F, Streibig JC, Gerhard D, Dose-Response Analysis Using R. PloS one 10, e0146021 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.