Abstract
Bacterial pathogens must resist host innate immunity to cause disease. While Gram-negative bacteria have a protective outer membrane, this membrane is subject to host-induced damage that makes these pathogens vulnerable. We developed a high content screening platform that identifies compounds that cause the killing of the bacterial pathogen Salmonella enterica in macrophages. This platform enables the rapid discovery of compounds that work in concert with the macrophage to prevent pathogen survival, as most hit compounds are not active in standard microbiological media and are not pro-drugs. We describe within the platform and the compounds it has found, and consider how they may help us discover new ways to fight infection.
Keywords: Chemical genetics, Gram-negative, high content, innate immunity, macrophage, outer membrane, pathogen, Salmonella
Graphical Abstract
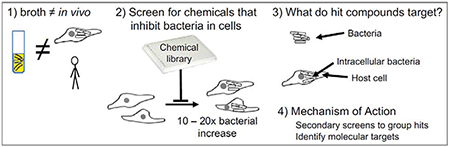
Introduction
“Have you done anything useful yet?” Stan Falkow greeted new trainees this way. And it was perfect, both warm and thought provoking. In Stan’s lab, I learned the power of applying genetics to probe host-pathogen relationships, and this has perhaps led to something useful. My own lab is exploiting chemical genetics to disrupt bacterial survival in cultured, infected cells. Other researchers carried out pioneering in-cell chemical genetics screens for compounds that prevent the cellular invasion, replication and/or survival of Listeria monocytogenes or Mycobacterium tuberculosis using libraries of drugs with known activities [1,2]. We built upon these ideas and screened compounds of unknown activity for their ability to disrupt an established infection by a Gram-negative bacterial pathogen [3]. How Gram-negative bacteria survive within host cells is of particular interest because these pathogens have an outer membrane that is a significant barrier to environmental insults, host defenses and antibiotics. To find new ways to interfere with infections caused by Gram-negative bacterial pathogens, we developed a quantitative, high-content screening platform for chemicals that prevent the replication and/or survival of Salmonella enterica in macrophages.
The crucial factor of soluble innate immunity
To colonize a host, microbes must contend with prophylactic and induced innate immune molecules that make host tissues inhospitable. For instance, serum complement inserts pores into Gram-negative bacterial outer membranes, increasing their susceptibility to antibiotics [4]. Pathogens that survive attack and colonize tissues generally cause damage that the host detects. As the pathogen multiplies, the host senses and responds to molecular signatures unique to microbes. Host recognition of a combination of tissue damage and microbial molecular signatures increases the volume, diversity, and potency of antimicrobials. Soluble innate immune factors are present in all body fluids and also within cells; soluble and cellular innate immunity are normally so effective that most pathogens are eliminated before they multiply and/or cause damage in healthy mammals.
Standard microbiological media, such as Luria-Bertani (LB) and Mueller-Hinton Broth (MHB), were not developed with the intention of modeling conditions bacteria experience during infection [5–7]. While efforts have been made to generate microbiological media that mimics infection conditions, the complexity and dynamics of mammalian systems can only be approximated [8]. Therefore, researchers have attempted to identify compounds that kill microbes in synergy with innate immunity in media, blood components, or insect models. For instance, media conditioned with AMPs or blood synergize with certain antibiotics to reduce the survival of Gram-negative bacteria, including clinical isolates [9,10]. These observations suggest that rapid, broth-based screens of compound libraries in plates could identify small molecules that synergize with innate immunity. An insect model system, the larva of the wax moth Galleria mellonella, may also be useful for identifying antimicrobials that are effective in the context of innate immunity. Soluble insect innate immunity has significant evolutionary conservation with mammals [11], and hurdles to wax moth larvae as a model system are being overcome, particularly with regard to fungal infections [12]. Pseudogymnoascus destructans, the fungus that causes bat white-nose syndrome, was injected into the wax moth larval body cavity followed by a drug library. Two fairly potent inhibitors of fungal colonization were identified [13]. The wax moth larval model has potential as a high-throughput screening platform to identify new chemicals that prevent microbial colonization [14,15] and may synergize with innate immunity. However, while these approaches have demonstrated value in identifying antimicrobials, their common shortcoming is that they cannot recapitulate a normal infection process.
Professional phagocytes, including macrophages, are responsible for engulfing and destroying microbes. They therefore contain vesicles packed with endogenous and inducible innate immune molecules that are quickly delivered to microbe-containing vesicles, called phagosomes. Within phagosomes, a key activity of soluble innate immunity is to damage the microbial envelope, which increases pathogen susceptibility to both host antimicrobial defenses and to clinical antibiotics. For example, antimicrobial peptides (AMPs) damage bacterial outer membranes based on charge distortions, disrupting both barrier function and the activity of efflux pumps, which export of toxic metabolites, anti-microbial agents of host origin, and antibiotics. The AMP-mediated weakening of the outer membrane is demonstrated by the observation that the antibiotic azithromycin is not effective against Gram-negative bacteria in broth unless an AMP is present [16]. In addition to outer membranes, host defenses damage bacterial cell walls, proteins, and lipids with lysozyme, proteases, and redox-reactive agents [17]. These observations suggest that the phagosomes of macrophages provide an environment that could be exploited to identify small molecules that synergize with innate immunity to blunt infection.
A model Gram-negative pathogen
We used macrophages to develop a quantitative, high-content screening platform that identifies compounds that synergize with innate immunity to disrupt an established infection by a Gram-negative bacterium [3]. We use Salmonella enterica serovar Typhimurium (Salmonella), an excellent model pathogen for multiple reasons. First, a number of Salmonella serovars cause natural infections of humans and other animals. Salmonella animal and cell culture infection models are well developed and described, including those for studying gut-limited infection and systemic infection [18]. A strong international Salmonella research community provides a wealth of genetic, biochemical, and on-line tools useful for establishing gene or compound mechanism of action [19]. Decades of research have revealed that Salmonella, like many pathogens, resides extra- or intracellularly, depending on the host organism and the stage of infection [20]. During systemic infection, Salmonella resides largely within cells of the monocyte lineage, including macrophages. In these cell types, the bacterium replicates within phagosomes that are packed with diverse soluble host defense molecules that damage but often do not kill the pathogen [21–23]. Thus, Salmonella infection of cultured macrophages incorporates key innate immune challenges faced by microbes.
From standard assay to high content screen
A classical experimental model for infection biology is the gentamicin protection assay, in which cultured cells are exposed to a bacterium that they engulf or by which they are invaded. Subsequent incubation allows for bacterial replication. To prevent the replication of the remaining extracellular bacteria, or of bacteria released from dying host cells, cells are treated with an antibiotic (e.g., gentamicin) that does not readily cross membranes and thus does not compromise intracellular replication. After a sufficient period of time, the host cells are monitored for bacterial load, traditionally by lysing the host cells and plating for bacterial colony forming units (CFU). The variant of the protocol we use relies upon RAW264.7 cells [24], a well established macrophage-like cell line that is easy to work with, and enables a virulent Salmonella strain [25,26] to accumulate 10-20 fold, supporting four or more rounds of replication.
To make the gentamicin protection assay amenable to automated microscopy, we miniaturized it in 384-well glass bottomed plates (Fig 1). We infect the RAW264.7 cells with an equivalent of 30 bacteria per host cell using a virulent Salmonella strain harboring a chromosomal sifB::gfp reporter [3]. The sifB promoter is induced when Salmonella become established in the macrophage phagosome [27], likely in response to a combination of changes in pH, osmolarity, and nutrient availability [28]. After 45 minutes of infection, the cultures are treated with a dose of gentamicin that is sufficient to diminish the replication of extracellular Salmonella without blunting the replication of intracellular Salmonella (as micropinocytosis may deliver the antibiotic to Salmonella in vesicles). Test or control compounds are added two hours after infection, and at 18 hours plates are processed for automated imaging. To each well, we add the vital dye MitoTracker, which fluoresces based on voltage across the mitochondrial inner membrane. We fix the cells, incubate them with the DNA probe DAPI, and image the plates on an automated fluorescent microscope for GFP, MitoTracker and DAPI. A MATLAB-based algorithm identifies macrophages based on DAPI and MitoTracker red and calculates bacterial load within macrophages based on GFP signal. The Z’-factor, a measure of robustness of the screening platform, at 0.48, is consistent with other cell-based screens [29–31], and we verified hits in a 96-well format, which had a Z’ factor of 0.59. We refer to this entire screening process as SAFIRE, Screen for Anti-infectives using Fluorescence microscopy of IntracellulaR Enterobacteriaceae).
Figure 1. The SAFIRE-CFU screening platform for discovery of chemicals that interfere with bacterial replication or survival in host cells.
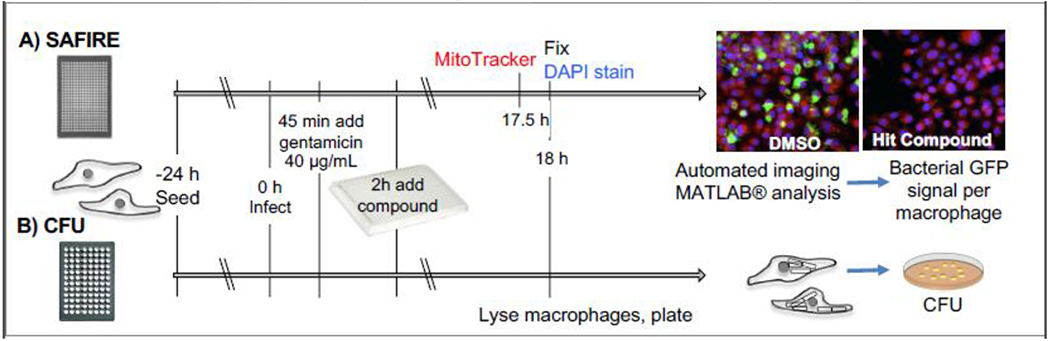
A) Macrophage-like cells (RAW264.7) in a 384-well dish are infected with virulent Salmonella containing chromosomal sifB::gfp (0 h). Gentamicin is added (45 minutes) followed by compounds (2 h). At 17.5 h post-infection, cells are incubated with MitoTracker Red CMXRos, and then fixed in the presence of DAPI (18 h). Automated imaging and MATLAB analyses calculate bacterial signal (green) within macrophage area (red). Validation is performed with the same assay in a 96-well format (not shown). B) To identify SAFIRE hits that prevent bacterial survival, macrophages are lysed and plated for enumeration of colony forming units (CFU).
Key features of the SAFIRE assay
Based on optimization of the pilot screen and on experimentation since carrying out the screen of the Maybridge HitFinder library, the following features of the protocol appear to be critical.
Host cells enable robust bacterial replication. The RAW264.7 cells permit Salmonella to accumulate 10-20-fold within the cell, generating a sufficiently large signal-to-noise ratio for a GFP reporter.
Bacteria are detected with a GFP reporter that is specifically induced inside the phagosome. A chromosomal sifB::gfp reporter [27] minimized background from extracellular bacteria, compared to a chromosomal, constitutive rpsM::gfp reporter [32].
Compounds are added to cells after infection has been established. This approach avoids chemicals that prevent macrophages from engulfing the bacteria [1].
A mitochondrial marker identifies macrophage area. Red fluorescence from MitoTracker more precisely identifies host cells than does the use of just a DNA marker and the assumption that the nucleus is in the center of the cell [33].
Bacterial signal (GFP) is quantified within macrophages. An alternative is to quantify signal across the well, an approach prone to identifying compounds that enhance lysis of host cells and bacterial release [34].
Compounds that are toxic to the host cells are easy to identify based on reduced numbers of macrophages.
Biological Validation of Hits – the Crucial Secondary Screen
While detecting bacteria by GFP is amenable to high content screening, it also has the potential to identify compounds that reduce GFP signal without reducing bacterial survival. Chemicals may interfere, for instance, with gfp transcription, translation, protein folding or fluorescence. We therefore test SAFIRE hits in traditional colony forming unit (CFU) assays, in which lysed macrophages are plated, and recovered bacterial colonies are enumerated. A comparison of CFU and SAFIRE data reveals correlation between the two assays, with a slope of 0.7 (Fig 2). The trendline crosses the Y axis at 34, confirming that CFU is a more stringent criterion than GFP signal. Clinical antibiotics, as expected, reduce both GFP and CFU and thus fall in the upper right corner. Most of the top ~50 compounds we have been studying also derive from this space.
Figure 2. Comparison of compound activity (25 μM) in SAFIRE versus CFU assays.
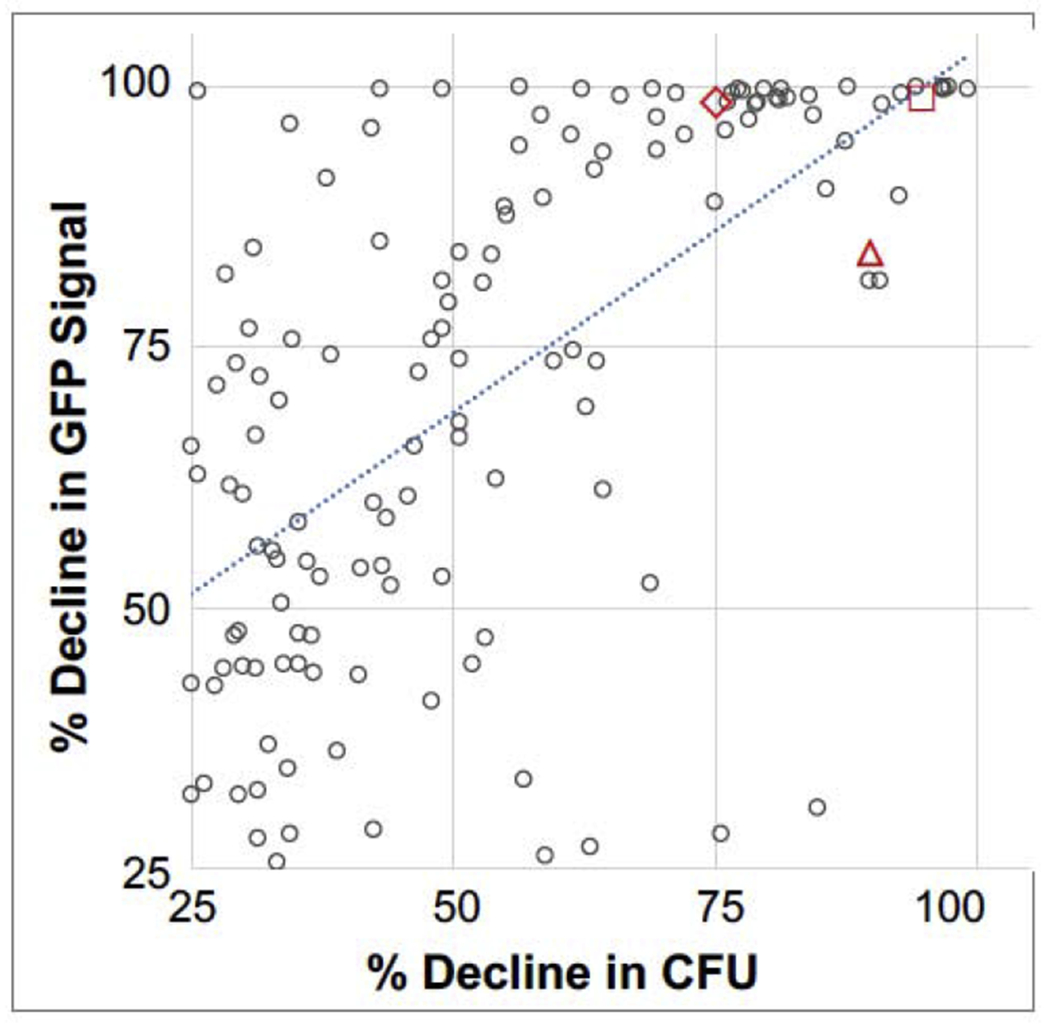
The SAFIRE values of the 133 compounds with a > 25%decline in CFU are plotted.
Antibiotic controls (red): 2 μg/ML rifampicin (triangle), 2 μg/mL ampicillin (square), and 0.2 μg/mL ciprofloxacin (diamond).Trendline, y = 0.697x + 33.876.
Bioinformatic Analyses of Hit Compounds
The structures of validated hits are checked against several databases for known activities and/or similarity to chemicals with known activities. We typically use ChemSpider, PubChem, and ChEMBL [35]. Hit compounds are also examined for similarity to each other, as chemical libraries typically feature analogs that can inform structure activity relationships.
What does SAFIRE/CFU identify?
Compounds identified by a SAFIRE/CFU approach fall into two classes, those that have antibacterial activity in standard microbiological media, and those that are only antibacterial within the host (Fig 3). The first class of compounds include clinical antibiotics. The 14,400 chemicals screened from the Maybridge HitFinder library were ranked based on a combination of SAFIRE and CFU results. The only clinical antibiotic in the Maybridge HitFinder library, chloramphenicol, was the 5th-highest ranked compound. The top 60 compounds were repurchased and 58 were validated with SAFIRE. Only chloramphenicol prevented Salmonella growth in in Mueller Hinton Broth; all other hit compounds prevented Salmonella growth and/or survival only upon treatment of infected host cells [3]. We used a variety of secondary screens to group compounds based on what they may do, much like establishing phenotypic complementation groups in classical genetics [36]. Outlined below are several different classes of compounds we have found or anticipate finding. Since compounds may have multiple targets, these classes are not mutually exclusive.
Figure 3. Hit compounds identified by SAFIRE-CFU.
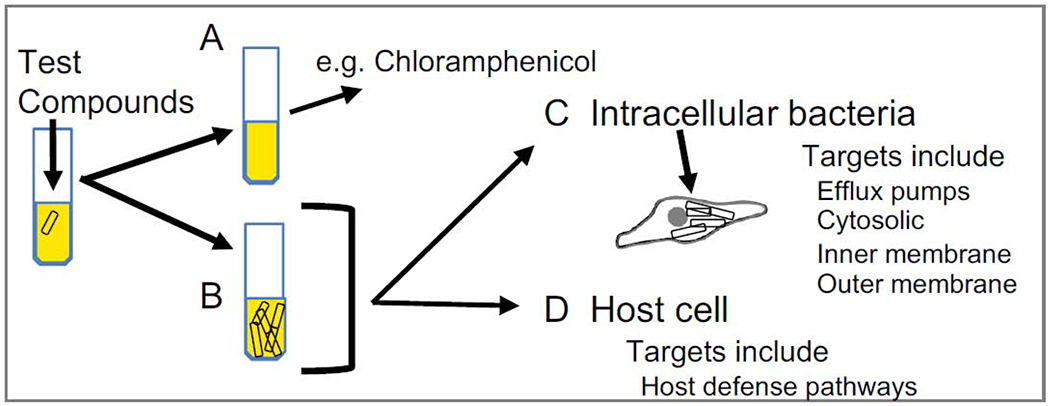
A) Depending on the library screened, hits may include antibiotics or chemicals with antibiotic activity. B) Most hit compounds, like vehicle, do not prevent bacterial growth in standard microbiological media. C) Some of these hits act on bacteria in the host setting. D) Other hits stimulate the host cell to kill bacteria. Pro-drugs activated by the tissue culture medium, host cell, and/or bacteria could also be identified.
Bacterial efflux pump modulators.
We hypothesized that a subset of the hit compounds identified by SAFIRE/CFU may inhibit bacterial efflux pumps. Salmonella encodes nine efflux pumps and at least two of them are required for bacterial survival and replication within macrophages: the AcrAB pump of the resistance-nodulation-development (RND) family [37], and MacAB, an ABC-type pump [38]. In a secondary screen, we used a 96-well assay to distinguish compounds that prevent the accumulation of Hoechst 33342, an efflux pump substrate, as these compounds may inhibit efflux [39]. Three compounds that prevented Hoechst accumulation were subsequently shown to prevent the glucose-dependent export of substrates of the AcrAB efflux pumps. These data indicate that the compounds are efflux pump modulators (EPMs), as the pumps require energy for activity [40]. Subsequent experiments demonstrated that the compounds bind AcrB with affinities comparable to antibiotics exported by AcrAB. Moreover, in macrophages, the EPMs potentiated antibiotics that are established pump substrates. Attempts to increase the potency of these compounds with medicinal chemistry are underway.
Compounds that synergize with innate immune defenses to kill bacteria.
We predict that antibacterial activity of hit compounds in macrophages may be enabled by soluble innate immune defenses. Indeed, the efflux pump modulators, which we screened for based on a different rationale, appear to reduce bacterial load in macrophages in a manner synergistic with host innate immunity: EPM treatment increases bacterial sensitivity to AMPs [3]. This makes sense in part because AMPs are established substrates of efflux pumps, including AcrAB in Enterobacteriaceae [41], and may be exported by bacteria residing in the phagosome. AMPs also weaken the outer membrane and may thereby give the EPMs or other chemicals access to their bacterial targets. To identify such compounds, we use secondary screens for functional synergy with AMPs, which also reveals whether a compound may act in the bacterial cytosol, at the bacterial inner cell membrane, or at the outer membrane.
Compounds with activity in the bacterial cytosol -
The AMP polymyxin B nonapeptide (PMBN) damages bacterial outer membranes sufficiently to enable molecules of modest size to diffuse into the periplasm and find their targets. For example, PMBN allows hydrophobic antibiotics such as novobiocin, to cross the inner membrane and reach an intracellular target, DNA gyrase [42]. In testing whether some compounds reach the cytosol with help from an AMP, it may be of value to combine an AMP with an enzyme that degrades the cell wall, possibly enabling more bulky compounds to access a cell membrane or cytosolic target. Alternatively, other AMPs, or different concentrations of PMBN, could be tested to see whether they allow chemicals access to bacterial targets.
One method to establish whether AMPs allow compounds they potentiate to enter bacterial cells is to incubate bacteria with the AMP and the compound, followed by liquid chromatography with tandem mass spectroscopy [43,44]. Most of the compounds we found in the HitFinder library have significant hydrophobicity and may cross bacterial cell membranes, suggesting the utility of this approach.
Bacterial cell membrane disruptors -
Hydrophobic hit compounds that potentiate AMPs may also or alternatively intercalate into the bacterial cell membrane based on protein or phospholipid binding. Whether their potency in SAFIRE reflect that they reduce bacterial membrane integrity can be established through a variety of membrane disruption assays, including those that monitor ion and/or proton flux, ATP production, and the access of small molecules (e.g., propidium iodide) to the cytosol. Membrane disrupting chemicals may also target host cell membranes, as suggested by host cell toxicity and release of cytoplasmic enzymes. While SAFIRE assays avoid particularly host-toxic compounds, they can identify chemicals that disrupt host cell membranes at higher concentrations than at which we screen, typically 10 – 25 μM. Whether a compound targets a moiety(s) conserved between bacteria and mitochondria can be established by monitoring inner membrane function in macrophages treated with escalating doses of compound. Chemicals that preferentially damage bacterial inner membranes may be of future interest as a tool or for therapeutic development.
Inhibitors of Gram-positive bacterial growth in broth -
Some of the compounds identified by SAFIRE/CFU that appear to act in the bacterial cytosol or at the cell membrane may well have potency in standard microbiological media against Gram-positive bacteria, which lack the outer membrane barrier. This observation would indicate that the compound targets a process that is evolutionarily shared across bacteria, much like most clinical antibiotics. The mechanism of action of compounds with broad-spectrum activity are perhaps most easily studied in Gram-positive model bacteria, avoiding the need to use membrane damaging agents or mutations that weaken the outer membrane to allow the compound to access target(s).
Inhibitors specific to Gram-negative bacteria
If hit compounds lack Gram-positive antibacterial activity, they may act on proteins or processes that are unique to Gram-negative bacteria. For example, the set of hit compounds potentiated by AMPs may include those that damage the outer membrane and thereby allow AMPs to access and kill bacteria at lower concentrations. Secondary screens for outer-membrane damage include assays that monitor access to the periplasm of nitrocefin, a beta-lactam that is cleaved by a periplasmic beta-lactamase to produce a yellow product [45]. In addition, high-resolution microscopy methods, such as SIM and TEM may also reveal that these compounds cause outer membrane shedding or blebbing. To establish whether compound treatment alter the structure of lipopolysaccharide, SDS-PAGE gels are useful [46]. Compounds with activity unique to Gram-negative bacteria may also include those that prevent the deployment of Type Three Secretion systems (T3SS) needed by many pathogens for virulence [47]. For instance, Salmonella survival in macrophages requires a functional T3SS to manipulate the host cell. Inhibitors of T3SS can be detected by monitoring T3SS-dependent protein secretion under specialized broth conditions [48]. Such compounds could become useful experimental tools and/or may have potential for development into narrow-spectrum antibiotics.
Chemicals that target the host
Structural observations of hit compounds from the Maybridge screening library suggest that perhaps half of the compounds found by SAFIRE/CFU target the host. Some of the chemicals are, or resemble, compounds previously demonstrated to contribute to bacterial clearance, such as those that are neuro-active [49,50] or reduce estrogen receptor activity [51]. We have also found that SAFIRE/CFU identifies compounds that stress host cells and trigger host antimicrobial defense responses, specifically, autophagy [52]. Along these lines, an in-cell screen of a drug library for inhibitors of Mycobacterium tuberculosis identified Toll-like receptor agonists and protein kinase inhibitors previously known to restrict intra-vesicular bacteria [2]. Since there are many more molecular surfaces within the much larger and more complex host cell, it may well be that, depending on the library, many of the chemicals that reduce Salmonella load in macrophages target the host [53,54]. Target identification needs to be followed up by experiments establishing whether a particular target molecule contributes to decreased bacterial survival in host cells.
Pro-drugs
Finally, hit compounds may include pro-drugs that become modified and activated in the tissue culture media and/or the host cell. One way to identify pro-drugs is to screen for antibacterial activity that is facilitated by incubation in host cell cytosolic lysates. However, if pro-drugs target the bacteria and/or host pathways that help clear bacteria, more sophisticated approaches will be needed to identify them.
Perspectives
Target identification may be the most difficult part of screens for unknown chemicals that disrupt the host pathogen interaction. Multiple secondary screens are needed to group compounds, followed by more complicated analyses of a few compounds to establish targets and demonstrate their relevance. However, as with most complex biological screens, following up on all hit compounds is not likely worth the effort for most research laboratories. Instead, focusing on the low hanging fruit identified by key secondary screens may be the most efficient way to move science forward. In addition, varying the primary screen is likely to allow for the identification of different kinds of hit compounds. Possible SAFIRE modifications include incorporating multidrug-resistant Salmonella clinical isolates, identifying hit compounds that increase instead of decrease bacterial load, and screening in other cell types or with other intracellular pathogens. It will also be important to explore compound libraries that cover new chemical territory, including synthetic and natural products. Since variations on complex screens will not yield what is wanted, but rather what is asked for, in-cell screening is best viewed as a discovery process, not simply a means to an end.
One question is whether the time and expense of in-cell screens are worth the effort when simpler, whole-cell plate-based assays could be used to identify a specific class of compounds. For instance, the compounds we found that inhibit bacterial efflux pumps could have been identified by screening the same library for Hoechst exclusion [55]. However, SAFIRE can show us unanticipated roles for pathogen and host pathways in maintaining the host-pathogen relationship, roles that will be missed if we only look for what we already know. Thus, SAFIRE as a primary screen may reveal new druggable bacterial and host targets. The macrophage phagosome is likely more complicated than recognized, and continued probing for chemicals that lead to bacterial death in macrophages will contribute to a broader understanding of how host-pathogen relationships are sustained and can be disrupted.
In conclusion, the conditions under which a screen is conducted determine what will be found. The more realistic the model system, the more likely its use will facilitate discovery of new hit compounds even in libraries that have been well-interrogated with target-based biochemical or whole-cell bacterial screens. In addition, realistic model systems have the potential to suggest new microbial targets. Salmonella enterica naturally infect and replicate within macrophages, which represent the contribution of innate immunity to pathogen clearance. While our molecular understanding of infection biology remains limited, and our need for new antimicrobials grows, infection screens are likely to reveal new biology and new avenues for antibiotic discovery.
Acknowledgements
The author thanks Toni Nagy, Tin Tin Su, Jamie Dombach and Paul Muhlrad for critical comments. This work was supported by NIH grants AI121474 (C.S.D. and Toni A. Nagy), AI126453 (C.S.D.), and AI121365 (C.S.D). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest statement
The author declares no competing interests.
REFERENCES
- 1.Lieberman LA, Higgins DE: Inhibition of Listeria monocytogenes infection by neurological drugs. International Journal of Antimicrobial Agents 2010, 35:292–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stanley SA, Barczak AK, Silvis MR, Luo SS, Sogi K, Vokes M, Bray M-A, Carpenter AE, Moore CB, Siddiqi N, et al. : Identification of Host-Targeted Small Molecules That Restrict Intracellular Mycobacterium tuberculosis Growth. PLoS Pathogens 2014, 10:e1003946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reens AL, Crooks AL, Su C-C, Nagy TA, Reens DL, Podoll JD, Edwards ME, Yu EW, Detweiler CS: A cell-based infection assay identifies efflux pump modulators that reduce bacterial intracellular load. PLoS Pathog 2018, 14:e1007115. [DOI] [PMC free article] [PubMed] [Google Scholar]; Describes the development of the SAFIRE screening platform and the identification of small molecules that modulate efflux pumps.
- 4.Heesterbeek D a. C, Martin NI, Velthuizen A, Duijst M, Ruyken M, Wubbolts R, Rooijakkers SHM, Bardoel BW: Complement-dependent outer membrane perturbation sensitizes Gram-negative bacteria to Gram-positive specific antibiotics. Sci Rep 2019, 9:3074. [DOI] [PMC free article] [PubMed] [Google Scholar]; Demonstrates that the complement system membrane attack complex sensitizes Gram-negative bacteria to antibiotics that are typically considered specific for Gram-positive bacteria.
- 5.Bertani G: Lysogeny at Mid-Twentieth Century: P1, P2, and Other Experimental Systems. Journal of Bacteriology 2004, 186:595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nizet V: The Accidental Orthodoxy of Drs. Mueller and Hinton. EBioMedicine 2017, 22:26–27. [DOI] [PMC free article] [PubMed] [Google Scholar]; Meditates on the historical development and limitations of the use of Mueller-Hinton Broth as the worldwide standard for antibiotic susceptibility testing.
- 7.Mueller JH, Hinton J: A Protein-Free Medium for Primary Isolation of the Gonococcus and Meningococcus. Proceedings of the Society for Experimental Biology and Medicine 1941, 48:330–333. [Google Scholar]
- 8.Ersoy SC, Heithoff DM, Barnes L, Tripp GK, House JK, Marth JD, Smith JW, Mahan MJ: Correcting a Fundamental Flaw in the Paradigm for Antimicrobial Susceptibility Testing. EBioMedicine 2017, 20:173–181. [DOI] [PMC free article] [PubMed] [Google Scholar]; Demonstrates that mean growth inhibitory concentrations differ in media that is more representative of the host, compared with in Mueller-Hinton Broth.
- 9.Sakoulas G, Kumaraswamy M, Kousha A, Nizet V: Interaction of Antibiotics with Innate Host Defense Factors against Salmonella enterica Serotype Newport. mSphere 2017, 2:e00410–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ulloa ER, Dillon N, Tsunemoto H, Pogliano J, Sakoulas G, Nizet V: Avibactam Sensitizes Carbapenem-Resistant NDM-1-Producing Klebsiella pneumoniae to Innate Immune Clearance. J Infect Dis 2019, doi: 10.1093/infdis/jiz128. [DOI] [PMC free article] [PubMed] [Google Scholar]; Identifies a Food and Drug Administration–approved non–β-lactam β-lactamase inhibitor that is effective against Klebsiella pneumoniae in the context of soluble and cellular effectors of innate immunity but not in standard media.
- 11.Kounatidis I, Ligoxygakis P: Drosophila as a model system to unravel the layers of innate immunity to infection. Open Biol 2012, 2:120075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Champion OL, Titball RW, Bates S: Standardization of G. mellonella Larvae to Provide Reliable and Reproducible Results in the Study of Fungal Pathogens. J Fungi (Basel) 2018, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]; Takes key steps towards standardizing dosing and scoring in the wax moth model for infection with fungal pathogens.
- 13.Beekman CN, Meckler L, Kim E, Bennett RJ: Galleria mellonella as an insect model for P. destructans, the cause of White-nose Syndrome in bats. PLOS ONE 2018, 13:e0201915. [DOI] [PMC free article] [PubMed] [Google Scholar]; Developed a wax moth model for the fungal pathogen that causes the deadly bat White-nose Syndrome and identified amphotericin B from a small library of known compounds as an inhibitor of infection.
- 14.Adamson DH, Krikstopaityte V, Coote PJ: Enhanced efficacy of putative efflux pump inhibitor/antibiotic combination treatments versus MDR strains of Pseudomonas aeruginosa in a Galleria mellonella in vivo infection model. J Antimicrob Chemother 2015, 70:2271–2278. [DOI] [PubMed] [Google Scholar]
- 15.Entwistle FM, Coote PJ: Evaluation of greater wax moth larvae, Galleria mellonella, as a novel in vivo model for non-tuberculosis Mycobacteria infections and antibiotic treatments. J Med Microbiol 2018, 67:585–597. [DOI] [PubMed] [Google Scholar]
- 16.Lin L, Nonejuie P, Munguia J, Hollands A, Olson J, Dam Q, Kumaraswamy M, Rivera H Jr, Corriden R, Rohde M, et al. : Azithromycin Synergizes with Cationic Antimicrobial Peptides to Exert Bactericidal and Therapeutic Activity Against Highly Multidrug-Resistant Gram-Negative Bacterial Pathogens. EBioMedicine 2015, 2:690–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murphy K, Weaver C: Janeway’s Immunobiology. Garland Science; 2016. [Google Scholar]
- 18.Tsolis RM, Xavier MN, Santos RL, Bäumler AJ: How to become a top model: impact of animal experimentation on human Salmonella disease research. Infect Immun 2011, 79:1806–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perez-Sepulveda BM, Hinton JCD: Functional Transcriptomics for Bacterial Gene Detectives. Microbiol Spectr 2018, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]; Describes useful online resources for the visualization and analysis of expression profiles of coding genes and small RNAs.
- 20.Acheson D, Hohmann EL: Nontyphoidal Salmonellosis. Clinical Infectious Diseases 2001, 32:263–269. [DOI] [PubMed] [Google Scholar]
- 21.Haraga A, Ohlson MB, Miller SI: Salmonellae interplay with host cells. Nature Reviews Microbiology 2008, 6:53–66. [DOI] [PubMed] [Google Scholar]
- 22.Monack DM: Salmonella persistence and transmission strategies. Current Opinion in Microbiology 2012, 15:100–107. [DOI] [PubMed] [Google Scholar]
- 23.Ohl ME, Miller SI: Salmonella: a model for bacterial pathogenesis. Annual review of medicine 2001, 52:259–274. [DOI] [PubMed] [Google Scholar]
- 24.Raschke WC, Baird S, Ralph P, Nakoinz I: Functional macrophage cell lines transformed by Abelson leukemia virus. Cell 1978, 15:261–267. [DOI] [PubMed] [Google Scholar]
- 25.Hoiseth SK, Stocker B a. D: Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 1981, 291:238–239. [DOI] [PubMed] [Google Scholar]
- 26.Smith BP, Reina-Guerra M, Hoiseth SK, Stocker BA, Habasha F, Johnson E, Merritt F: Aromatic-dependent Salmonella typhimurium as modified live vaccines for calves. Am J Vet Res 1984, 45:59–66. [PubMed] [Google Scholar]
- 27.Rollenhagen C, Sörensen M, Rizos K, Hurvitz R, Bumann D: Antigen selection based on expression levels during infection facilitates vaccine development for an intracellular pathogen. Proc Natl Acad Sci USA 2004, 101:8739–8744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fang FC, Frawley ER, Tapscott T, Vázquez-Torres A: Bacterial Stress Responses during Host Infection. Cell Host Microbe 2016, 20:133–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dragiev P, Nadon R, Makarenkov V: Systematic error detection in experimental high-throughput screening. BMC Bioinformatics 2011, 12:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brodin P, Poquet Y, Levillain F, Peguillet I, Larrouy-Maumus G, Gilleron M, Ewann F, Christophe T, Fenistein D, Jang J, et al. : High content phenotypic cell-based visual screen identifies Mycobacterium tuberculosis acyltrehalose-containing glycolipids involved in phagosome remodeling. PLoS Pathog 2010, 6:e1001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Samantaray S, Correia JN, Garelnabi M, Voelz K, May RC, Hall RA: Novel cell-based in vitro screen to identify small-molecule inhibitors against intracellular replication of Cryptococcus neoformans in macrophages. Int J Antimicrob Agents 2016, 48:69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vazquez-Torres A, Jones-Carson J, Bäumler AJ, Falkow S, Valdivia R, Brown W, Le M, Berggren R, Parks WT, Fang FC: Extraintestinal dissemination of Salmonella by CD18-expressing phagocytes. Nature 1999, 401:804–8. [DOI] [PubMed] [Google Scholar]
- 33.Zanella F, Lorens JB, Link W: High content screening: seeing is believing. Trends in Biotechnology 2010, 28:237–245. [DOI] [PubMed] [Google Scholar]
- 34.Ellis MJ, Tsai CN, Johnson JW, French S, Elhenawy W, Porwollik S, Andrews-Polymenis H, McClelland M, Magolan J, Coombes BK, et al. : A macrophage-based screen identifies antibacterial compounds selective for intracellular Salmonella Typhimurium. Nat Commun 2019, 10:197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gaulton A, Hersey A, Nowotka M, Bento AP, Chambers J, Mendez D, Mutowo P, Atkinson F, Bellis LJ, Cibrián-Uhalte E, et al. : The ChEMBL database in 2017. Nucleic Acids Res 2017, 45:D945–D954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hartwell LH: Saccharomyces cerevisiae cell cycle. Microbiology and Molecular Biology Reviews 1974, 38:164–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buckley AM, Webber MA, Cooles S, Randall LP, La Ragione RM, Woodward MJ, Piddock LJV: The AcrAB–TolC efflux system of Salmonella enterica serovar Typhimurium plays a role in pathogenesis. Cellular Microbiology 2006, 8:847–856. [DOI] [PubMed] [Google Scholar]
- 38.Bogomolnaya LM, Andrews KD, Talamantes M, Maple A, Ragoza Y, Vazquez-Torres A, Andrews-Polymenis H: The ABC-Type Efflux Pump MacAB Protects Salmonella enterica serovar Typhimurium from Oxidative Stress. mBio 2013, 4:e00630–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coldham NG, Webber M, Woodward MJ, Piddock LJV: A 96-well plate fluorescence assay for assessment of cellular permeability and active efflux in Salmonella enterica serovar Typhimurium and Escherichia coli. J Antimicrob Chemother 2010, 65:1655–1663. [DOI] [PubMed] [Google Scholar]
- 40.Bohnert JA, Karamian B, Nikaido H: Optimized Nile Red Efflux Assay of AcrAB-TolC Multidrug Efflux System Shows Competition between Substrates. Antimicrob Agents Chemother 2010, 54:3770–3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Joo H-S, Fu C-I, Otto M: Bacterial strategies of resistance to antimicrobial peptides. Philos Trans R Soc Lond, B, Biol Sci 2016, 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ofek I, Cohen S, Rahmani R, Kabha K, Tamarkin D, Herzig Y, Rubinstein E: Antibacterial synergism of polymyxin B nonapeptide and hydrophobic antibiotics in experimental gram-negative infections in mice. Antimicrob Agents Chemother 1994, 38:374–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davis TD, Gerry CJ, Tan DS: General platform for systematic quantitative evaluation of small-molecule permeability in bacteria. ACS Chem Biol 2014, 9:2535–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richter MF, Drown BS, Riley AP, Garcia A, Shirai T, Svec RL, Hergenrother PJ: Predictive compound accumulation rules yield a broad-spectrum antibiotic. Nature 2017, doi: 10.1038/nature22308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hancock RE, Wong PG: Compounds which increase the permeability of the Pseudomonas aeruginosa outer membrane. Antimicrob Agents Chemother 1984, 26:48–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pilonieta MC, Erickson KD, Ernst RK, Detweiler CS: A protein important for antimicrobial peptide resistance, YdeI/OmdA, is in the periplasm and interacts with OmpD/NmpC. J Bacteriol 2009, 191:7243–7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abby SS, Rocha EPC: The Non-Flagellar Type III Secretion System Evolved from the Bacterial Flagellum and Diversified into Host-Cell Adapted Systems. PLOS Genetics 2012, 8:e1002983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu X-J, Grabe GJ, Liu M, Mota LJ, Holden DW: SsaV Interacts with SsaL to Control the Translocon-to-Effector Switch in the Salmonella SPI-2 Type Three Secretion System. mBio 2018, 9:e01149–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang C-Y, Hsu C-Y, Fang C-S, Shiau C-W, Chen CS, Chiu H-C: Loxapine, An Antipsychotic Drug, Suppresses Intracellular Multiple-Antibiotic-Resistant Salmonella enterica Serovar Typhimurium in Macrophages. Journal of Microbiology, Immunology and Infection 2019, doi: 10.1016/j.jmii.2019.05.006. [DOI] [PubMed] [Google Scholar]
- 50.Andersson JA, Fitts EC, Kirtley ML, Ponnusamy D, Peniche AG, Dann SM, Motin VL, Chauhan S, Rosenzweig JA, Sha J, et al. : New role for FDA-approved drugs in combating antibioticresistant bacteria. Antimicrob Agents Chemother 2016, doi: 10.1128/AAC.00326-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim D-K, Jeong J-H, Lee J-M, Kim KS, Park S-H, Kim YD, Koh M, Shin M, Jung YS, Kim H-S, et al. : Inverse agonist of estrogen-related receptor γ controls Salmonella typhimurium infection by modulating host iron homeostasis. Nat Med 2014, 20:419–424. [DOI] [PubMed] [Google Scholar]
- 52.Nagy Toni A., Quintana JL, Reens AL, Crooks AL, Detweiler CS: A Small Molecule Inhibits Salmonella Survival in Macrophages and Mice by Inducing Autophagic Flux. Submitted, [DOI] [PMC free article] [PubMed] [Google Scholar]; Identifies a small molecule inhibitor of Salmonella survival in macrophages that stimulates autophagy.
- 53.Franken H, Mathieson T, Childs D, Sweetman GMA, Werner T, Tögel I, Doce C, Gade S, Bantscheff M, Drewes G, et al. : Thermal proteome profiling for unbiased identification of direct and indirect drug targets using multiplexed quantitative mass spectrometry. Nat Protoc 2015, 10:1567–1593. [DOI] [PubMed] [Google Scholar]
- 54.Webb KJ, Ball KA, Coleman SJ, Jacobsen J, Stowell MHB, Old WM: Rapid discovery of drug target engagement by isothermal shift assay. Pharmacology and Toxicology; 2019. [Google Scholar]; Developed the isothermal shift assay (iTSA), for rapid identification of drug targets in lysates and living cells.
- 55.Blair JMA, Piddock LJV: How to Measure Export via Bacterial Multidrug Resistance Efflux Pumps. mBio 2016, 7:e00840–16. [DOI] [PMC free article] [PubMed] [Google Scholar]


