Abstract
Cells rely on a complex network of spatiotemporally regulated signaling activities in order to effectively transduce information from extracellular cues to intracellular structures. In order to probe this activity architecture, researchers have developed an extensive molecular toolkit of fluorescent biosensors and optogenetic actuators capable of monitoring and manipulating various signaling activities with high spatiotemporal precision. The goal of this review is to provide readers with an overview of basic concepts and recent advances in the development and application of genetically encodable biosensors and optogenetic tools for understanding signaling activity.
Introduction
Cells utilize intracellular signaling networks to sense information from the outside environment and determine their behavior. Despite the complexity of the intracellular milieu, cells faithfully and efficiently decode multiple external signals coming in simultaneously to drive specific responses to environmental changes. In recent decades, detailed studies of these signaling pathways have revealed that the precise spatiotemporal regulation of signaling network components is key to achieving this remarkable functional specificity. These studies have been made possible by the development of powerful molecular tools for probing signaling activities. With the discovery of fluorescent proteins (FPs) and bioluminescent proteins (BPs) leading to the development of various biosensors, researchers have gained the ability to directly visualize dynamic signaling events with high spatiotemporal resolution in living cells. However, observing signaling events is only the first step; understanding their functional importance requires tools to specifically elicit signaling events in living cells, ideally with high spatiotemporal control. Thus, various methods for perturbing signaling activities have been developed in parallel with fluorescent sensors to reveal causal relationships between signaling components. Signaling perturbations can be achieved through protein translocation and enzyme activity control using chemical, optical, and magnetic inputs. Among these, optically inducible tools are widely used in live single-cell studies due to advantages including fewer side effects, more precise spatiotemporal control, and minimal invasiveness. Below, we discuss several genetically encodable tools that have been devised to monitor and elicit cell signaling events. Due to space constraints, we cannot cover the full range of tools and will instead focus on the major classes of technologies and the latest trends in tool development.
Optical sensors to study signaling events
Biosensor basics
Genetically encoded biosensors generally consist of a sensing unit that detects changes in the signal of interest and a reporting unit that converts these changes into a quantitative optical readout ranging from the visible to infrared range [1]. While FPs are most commonly used, self-labeling proteins (SLPs) and BPs can also serve as the genetically encodable “building blocks” for the reporting unit, offering unique advantages [1]. Though only semi-genetically encoded, SLPs open the door to using synthetic fluorophores that often perform favorably versus FPs [2]. Alternatively, BPs such as RLuc, FLuc, and the more recent NLuc do not require external illumination and therefore offer potentially higher signal-to-noise ratios due to reduced background [3]. Researchers also continue to seek novel prototypical FPs and to develop brighter FP variants through mutagenesis and directed evolution [4], in combination with machine learning [5].
Sensing unit designs vary depending on the characteristics of the targeted signaling event. Translocation-based biosensors are constructed by simply tagging the reporting unit to a protein or protein domain that changes localization in response to a specific signaling event. For example, 3’ phosphoinositide (PI) dynamics can be monitored via sensor translocation to and from the plasma membrane [6]. Meanwhile, kinase translocation reporters combine kinase-specific substrates with nuclear localization and/or export sequences to modulate nucleocytoplasmic shuttling of the biosensor upon phosphorylation by a particular kinase. Various kinase activities, including Extracellular signal-Regulated Kinase (ERK), c-Jun N-terminal Kinase (JNK), and Protein Kinase A (PKA), can be monitored this way (Figure 1A) [7]. Another recent innovation is the use of phase separation as the readout, with Zhang et al. developing SPARK (Separation of Phases-based Activity Reporter of Kinase) to monitor PKA and ERK activities [8].
Figure 1.
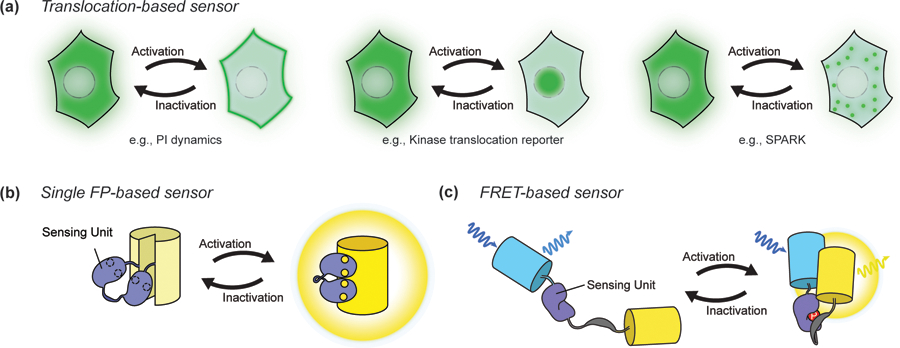
Basic biosensor designs. A. Translocation-based biosensors. Phosphoinositide (PI) sensors utilize lipid-selective binding domains that translocate to or from the membrane to report the production and degradation of specific PIs, such as PIP3 (a). Kinase translocation reporters similarly undergo translocation into or out of the nucleus depending on the phosphorylation of kinase-specific substrate sequences within the reporter (b). SPARK, a phase separation-based kinase reporter, visualizes dynamics of kinase activity with phase separation (c). B. Single FP-based sensor. The insertion of a conformational switch into a single FP can be used to alter chromophore behavior in response to a signaling event. C. FRET-based sensor. Changes in the proximity or orientation between a pair of FPs caused by the sensing unit upon detection of a signaling event results in a change in FRET.
More universal methods for monitoring signaling activities involve designing sensing units that serve as molecular switches by changing conformation in response to a biochemical signal (Figure 1B). A highly versatile approach utilizes the sensing unit to control changes in fluorescence resonance energy transfer (FRET) between two FPs (Figure 1C). Specifically, because FRET depends strongly on the distance and orientation of the donor and acceptor fluorophores, conformational changes induced by a molecular switch can be used to modulate the relative proximity of a FRET-compatible FP pair. This approach has yielded the most versatile family of genetically encoded biosensors, including sensors for monitoring Ca2+ [9] and other ions [10,11], numerous small molecules [12], intracellular messengers [13–17] and the activities of protein kinases [18–24] and other signaling enzymes [25]. Furthermore, the fact that luciferases can also transfer excited-state energy to an acceptor FP has also yielded sensors based on bioluminescence resonance energy transfer (BRET), in which the donor FP of a FRET-based sensor is replaced with a luciferase [26].
The sensing unit can also be directly integrated with a single FP, based on the principle that distorting the FP β-can using a conformational switch will alter chromophore behavior. A popular approach uses circular permutation of the FP to reposition the N- and C-termini within the β-can, followed by attachment of the sensing unit to these new termini. The resulting biosensors can exhibit changes in fluorescence intensity at a fixed wavelength (intensiometric) [27–29] or shift between two maximal excitation or emission wavelengths (ratiometric) [30–32]. Though originally applied to develop genetically encoded Ca2+ indicators (GECIs) [27,28,33], this approach has recently expanded to monitor a wider range of targets such as metabolites [34–37], membrane potential [38], second messengers [29], neurotransmitters [39,40] and kinase activities [30].
These examples only scratch the surface, and more information on biosensor designs not covered here is reviewed by Greenwald et al. [1]. Importantly, the growing need to study signaling activities both at the single-cell level with enhanced spatial and temporal resolution and at the tissue or whole-organism level continues to drive the development of new types of biosensors.
Recent advances in biosensors
Super-resolution compatible biosensors
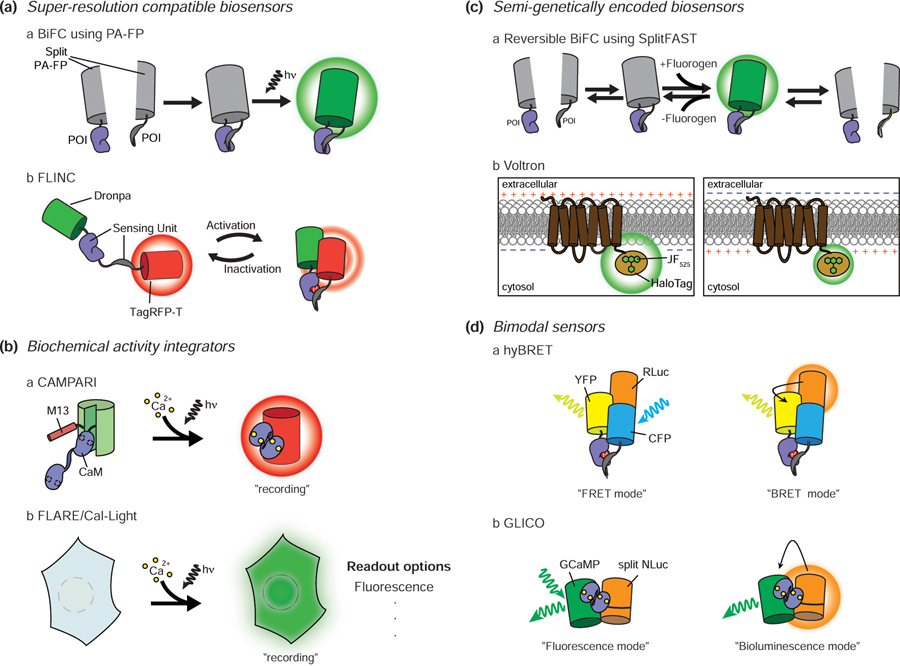
The ability of signaling enzymes to reliably find their specific targets within a crowded cell cannot be explained by diffusion alone. One way that cells achieve this specificity is by utilizing protein-protein interactions (PPIs) to assemble nanometer-scale signaling complexes. However, observing compartmentalized signaling activities within these nanodomains is challenging given the resolution limit of conventional microscopy (about 250nm depending on the wavelength used). Thus, the recent emergence of methods that combine super-resolution imaging techniques such as STORM and PALM with biosensor designs based on photoswitchable FPs has transformed the study of signaling nanodomains. For example, PPIs are frequently observed using biomolecular fluorescence complementation (BiFC), in which complementary, non-fluorescent FP fragments are conjugated to a pair of target proteins of interest (POIs) and will only reconstitute a functional FP if these POIs interact in the cell. Recent studies have utilized photoswitchable FPs such as Dronpa [41], PA-mCherry [42], and mEOS3.2 [43] with BiFC to visualize PPIs in super-resolution (Figure 2Aa), for instance, to demonstrate nanoscale clustering of a Ras/Raf protein complex in mitogen-activated protein kinase (MAPK) signaling [42]. However, the irreversibility of FP fragment complementation can obscure PPI dynamics. Another recent example of a super-resolution-compatible biosensor takes advantage of a novel phenomenon in which the photostable FP TagRFP-T exhibits spontaneous fluorescence fluctuations when placed in very close proximity to the photoswitchable FP Dronpa (Figure 2Ab). By combining this phenomenon, which they termed Fluorescence fLuctuation INcrease by Contact (FLINC), with the sensing units of FRET-based sensor designs, Mo et al. developed FLINC-based biosensors capable of monitoring PKA and ERK activities in super-resolution, which allowed them to observe the nanometer-scale compartmentalization of enzyme activity at a single-pixel level [44]. Although FLINC-based biosensors require more extensive image analysis post-imaging, they offer super-resolution information on dynamic biochemical activities in living cells.
Figure 2.
Recent advances in biosensors. A. Super-resolution compatible biosensors. Bimolecular fluorescence complementation (BiFC) using photoactivatable FPs combined with STORM/PALM imaging enables super-resolution PPI observation (a). TagRFP-T fluorescence shows increasing fluctuations as the proximity between Dronpa and TagRFP-T is closed by the sensing unit. This allows the use of pcSOFI, a super-resolution technique, to observe signaling activity at subdiffraction spatial resolution (b). B. Biochemical activity integrators. CaMPARI is a GCaMP-based Ca2+ sensor that features a green-to-red photoconvertible FP. The sensor undergoes irreversible green-to-red photoconversion only when exposed to both violet light and Ca2+. Thus, the red color persists even when Ca2+ levels drop, allowing post hoc analysis of signaling (a). When FLARE/Cal-Light is exposed to Ca2+ and blue light, a transcription factor that turns on the reporter translocates into the nucleus. Unlike CaMPARI, this system can be used with a variety of readouts (b). C. Semi-genetically encoded biosensors. SplitFAST is suitable for showing dynamic PPIs because of its reversible interaction. Conventional BiFC cannot report PPI dynamics due to the irreversible nature of FP complementation, whereas splitFAST makes this possible (a). New synthetic dyes with improved brightness and stability help to overcome the limitations of the low delivery efficiency of synthetic dyes. Voltron is based on the voltage sensor Ace2N and new bright synthetic dyes such as JF525. This drastically improved in vivo voltage imaging in animals (b). D. Bimodal sensors. hyBRET sensors combine a FRET sensor with a bioluminescent protein (RLuc). In the absence of the luciferase substrate, the signaling activity can be measured in FRET mode, while addition of substrate allows monitoring in BRET mode (a). GLICO combines GCaMP with splitNLuc. In the presence of Ca2+, the GLICO sensing unit undergoes a conformational change that modulates cpGFP fluorescence and leads to splitNLuc complementation. Therefore, Ca2+ levels can be monitored via GFP emission caused by either external illumination (fluorescence mode) or luciferin addition to induce BRET between NLuc and cpGFP (bioluminescence mode) (b). PA: photoactivatable, hn: light, POI: protein of interest.
Biochemical activity integrators
Biosensors typically report biochemical activity changes in real-time, yet continuous recording of activity dynamics is ill-suited for imaging large volumes of tissues such as brain activity mapping, which seeks to identify the specific neuronal subsets that become activated by a given external stimulus or during certain behaviors. This has inspired a new class of biosensor, the biochemical activity integrator, which records a “snapshot” of the signaling activity within a tissue by integrating dynamic changes in a specific biochemical activity over a set time period. For example, Fosque et al. recently developed a Ca2+ integrator to label active neural circuits by engineering a Ca2+ biosensor based on the green-to-red photoconvertible FP mEos2 [45]. The resulting snapshot reporter, named CaMPARI (Ca2+-modulated photoactivatable ratiometric integrator), undergoes green-to-red photoconversion only in the presence of both Ca2+ and violet light (Figure 2Ba). Thus, CaMPARI was able to highlight fly, zebrafish, and mouse neurons with elevated activity (e.g., Ca2+ levels) during the illumination period. Since this system combines the characteristics of an intensity-based Ca2+ sensor with a photoconvertible FP, the change in readout can be observed by the red-to-green fluorescence intensity ratio within 3 min.
Meanwhile, both Wang et al. and Lee et al. also recently reported Ca2+ integrators, called FLARE (Fast Light- and Activity-Regulated Expression) and Cal-Light, respectively, that utilize an alternative design based on a light-and-Ca2+-gated transcription factor (TF) system [46,47] (Figure 2Bb). The optically controlled light-oxygen-voltage (LOV) domain is used to expose a protease recognition site conjugated to a TF upon blue light illumination, while the Ca2+-dependent binding of calmodulin to a calmodulin-binding peptide recruits a protease to cleave and release the TF. Therefore, illuminating cells with blue light allows intracellular Ca2+ elevations to drive the TF into the nucleus and induce expression of a reporter gene expression such as an FP. Unlike CaMPARI, these transcriptional systems are highly adaptable and can be used to not only label but also manipulate specific cell populations. For example, Lee et al. used Cal-Light to drive expression of a yellow-light-gated Cl- channel in neurons that were activated during a learning task, which then allowed them to selectively silence the labeled neurons and dynamically inhibit the learned behavior [47]. While these systems currently require at least 5 min of stimulation to produce meaningful signal changes; thus, further improvements in sensitivity will be needed to allow for better brain mapping in the future. Furthermore, continuous improvement can reduce the basal signals resulting from resting activity and further increase the contrast of these integrator systems.
Semi-genetically encoded biosensors
FP-based biosensors are powerful tools for studying signaling activity but have limitations such as relatively low brightness and poor photostability, as well as irreversible fragment complementation. Semi-genetically encodable SLPs such as HaloTag, SNAP-tag, and CLIP-tag are thus gaining popularity in cell signaling research because they offer a convenient means to covalently label POIs with various synthetic fluorophores directly in situ [2]. The development of bright and photostable synthetic dyes that are compatible with SLP has helped overcome some of the limitations of conventional FP-based biosensors. For example, while BiFC cannot be used to monitor dynamic PPIs due to its irreversibility, Plamont et al. recently reported a new semi-genetically encodable system, called Fluorescence-Activating and Absorption-Shifting Tag (FAST) [48], wherein both the complementation of the splitFAST system and fluorophore binding to the reconstituted FAST are reversible [49] (Figure 2Ca). By taking advantage of this system, the authors were successfully able to monitor the dynamics of Ras/Raf, Mitogen-activated protein Kinase (MEK)/ERK, and ERK/Mitogen-activated protein Kinase Phosphatase (MKP) interactions. Importantly, splitFAST shows some, albeit modest, spontaneous self-association, and will thus require further optimization to allow for more dynamic PPI imaging.
SLPs have also aided in the development of improved genetically encoded fluorescent voltage indicators (GEVIs) to study electrical activity in neurons. In particular, microbial rhodopsin-derived GEVIs critically depend on bright and stable FPs to enhance the sensitivity of electrochromic FRET, wherein voltage-induced changes in the rhodopsin absorption spectrum modulate FRET with an appended FP, for in vivo imaging of large numbers of neurons. For example, Ace2N was engineered using mNeonGreen, the brightest known FP at the time, to yield increased sensitivity [50]. However, Abdelfattah et al. surpassed the performance of even mNeonGreen through the introduction of synthetic dyes. Specifically, they designed a voltage indicator called Voltron by fusing rhodopsin with a HaloTag and using improved rhodamine dyes such as JF549 and JF525 as fluorophores to observe voltage-dependent FRET changes (Figure 2Cb). Because these dyes are three times brighter and eight times more photostable than mNeonGreen, Voltron enabled animal imaging studies that were not possible using conventional FP-based sensors, including long-term (> 15 min) recording of neuronal activity in vivo [51].
Ongoing improvements in synthetic dyes will undoubtedly further expand the applications of SLPs [52]. Notably, Lukinavičius et al. have pushed the limits of SLPs by developing silicon-rhodamine (SiR)-based fluorophores that show excellent cell permeability. Using SiR-SNAP-tag, they observed histones using live-cell STORM and also demonstrated the feasibility of cortical neuron labeling in rat-brain sections [53]. More recently, they have also developed far-red SiR-based fluorescent probes [54], providing researchers with the opportunity to expand multi-color imaging and the SLP-based hybrid sensor toolkit.
Bimodal biosensors
Fluorescence is widely used as a biosensor readout. However, most commonly used fluorophores excite at wavelengths that don’t penetrate deeply into tissues and can also trigger autofluorescence and photobleaching, which diminish the signal. On the other hand, bioluminescence utilizes a chemical substrate rather than external illumination, which increases the signal-to-noise ratio and also enhances tissue penetration, yet most luciferases are dim, and the substrate is short-lived. These complementary properties have inspired the development of various bimodal biosensors capable of both fluorescent and bioluminescent readouts. For example, Komatsu et al. developed a sensor platform called hyBRET, in which RLuc-fused CFP serves as a hybrid donor that can transfer energy to YFP following either CFP excitation or luciferin addition, thereby allowing both FRET and BRET readouts (Figure 2Da) [55]. Similarly, Farhana et al. recently developed Green Luminescent Indicator for Ca2+ Observation (GLICO) by fusing split NLuc to the N- and C-termini of the single FP-based Ca2+ sensor GCaMP [56]. The Ca2+ -dependent conformational change in GLICO can be monitored through either increased GFP intensity under 488-nm excitation (fluorescence mode) or increased NLuc-GFP BRET in the presence of luciferin (bioluminescence mode) (Figure 2Db). Nevertheless, although bimodal biosensors offer more options for measuring signaling activity, even in animal experiments, precise comparisons between the different readout modes are difficult given the differing sensitivities of FRET and BRET.
Optogenetic tools to elicit signaling events
Components and designs of optogenetic tools
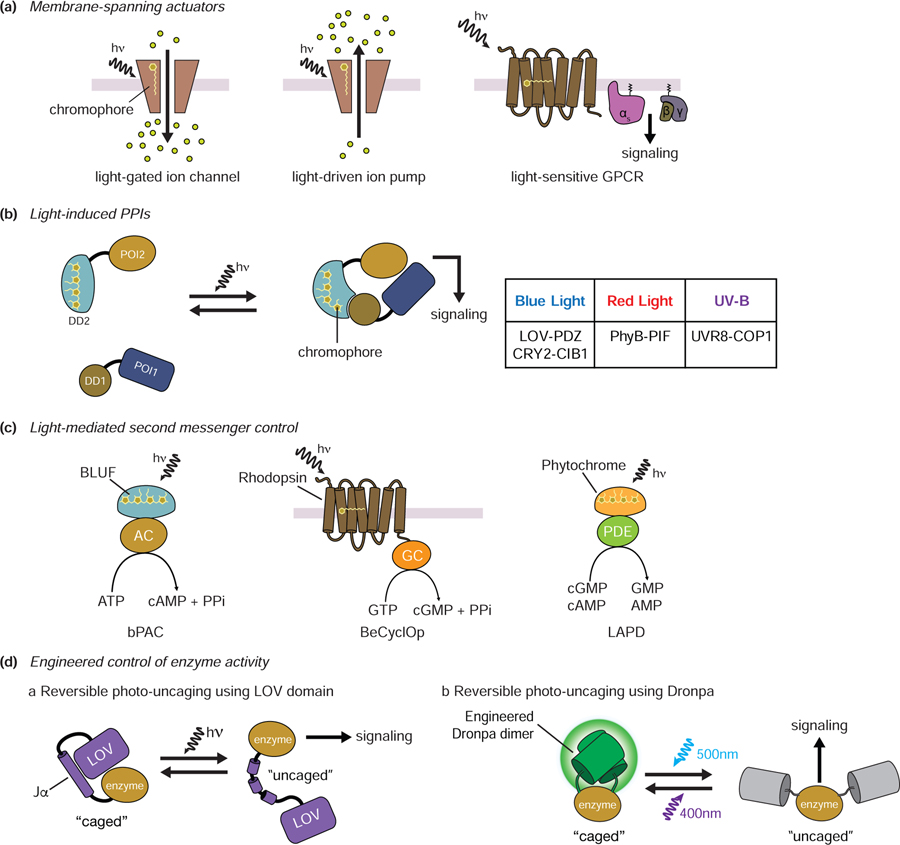
Optically inducible tools to perturb signaling make use of naturally occurring proteins that undergo conformational changes caused by light-induced photochemical reactions [57]. These tools can be roughly divided according to their mode of action: 1) induction of membrane depolarization or cellular signaling cascades; 2) induction of PPIs to modulate protein translocation and/or signaling perturbation, 3) production of second messengers through direct regulation of enzyme activity, and 4) reversible control of protein caging.
Rhodopsin can act as a membrane-spanning actuator to induce membrane depolarization or cellular signaling by playing the role of a light-driven ion pump [58], light-gated ion channel [59], or light-sensitive GPCR depending on the specific configuration [60–64] (Figure 3A). Meanwhile, light-dependent modulation of protein translocation and/or signaling perturbation is primarily achieved using light-induced PPIs between photoreceptors and their effectors. Common examples include the blue-light-activated LOV-PDZ [65] and CRY2-CIB1 systems [66], the red-light-activated PhyB-PIF system [67], and the UV-B-inducible UVR8-COP1 system [68] (Figure 3B). A number of photoactivated adenylyl cyclases, guanylyl cyclases, and phosphodiesterases have been described that allow light-mediated second messenger production. For example, photoactivated adenylyl cyclases (PACs) such as bPACs produce cyclic AMP upon blue-light activation [69]. BeCyclOp, a kind of rhodopsin, is a light-activated guanylyl cyclase and produces cGMP in response to green light [70], whereas the red-light-activated phosphodiesterase (LAPD) degrades cAMP/cGMP [71] (Figure 3C). Interestingly, the Jα helical extension of the Avena sativa LOV domain, which unfolds upon illumination, can also be used to control access to the active site or binding surface of an enzyme, thereby enabling light-induced “caging and uncaging” (Figure 3Da) [72]. Although it is not a photoreceptor, similar optical caging/uncaging of protein activity is also possible using engineered Dronpa (Figure 3Db) [73].
Figure 3.
Components and designs of optogenetic tools. A. Membrane-spanning actuators. Rhodopsin can act as a membrane-spanning actuator in the form of a light-driven ion channel, an ion pump, or a GPCR. The covalently bound chromophore of rhodopsin can trigger a conformational change in rhodopsin upon light illumination. B. Light-induced PPIs. Light-dependent modulation of protein translocation and/or signaling perturbation utilizes light-induced PPIs between photoreceptors and their effectors. Representative light-induced PPIs are shown in the table. C. Light-mediated second messenger control. In bPAC, adenylyl cyclase bound to the BLUF domain is activated by light to produce cAMP. A guanylyl cyclase-containing rhodopsin from Blastocladiella emersonii (BeCyclOp) similarly produces cGMP when activated by light. The red-light-activated phosphodiesterase (LAPD) degrades cGMP and cAMP. D. Engineered control of enzyme activity. The Jα helix of the AsLOV domain is unfolded by light, allowing enzyme “caging” and “uncaging” (a). Photodissociable dimeric Dronpa (pdDronpa1) can form dimers upon 500 nm light and dissociate upon 400nm light. This feature also allows reversible control of protein cagin/uncaging by light (b). hn: light, POI: protein of interest; DD: dimerization domain, AC: adenylyl cyclase, GC: guanylyl cyclase, PDE: phosphodiesterase
Recent advances in optogenetic tools
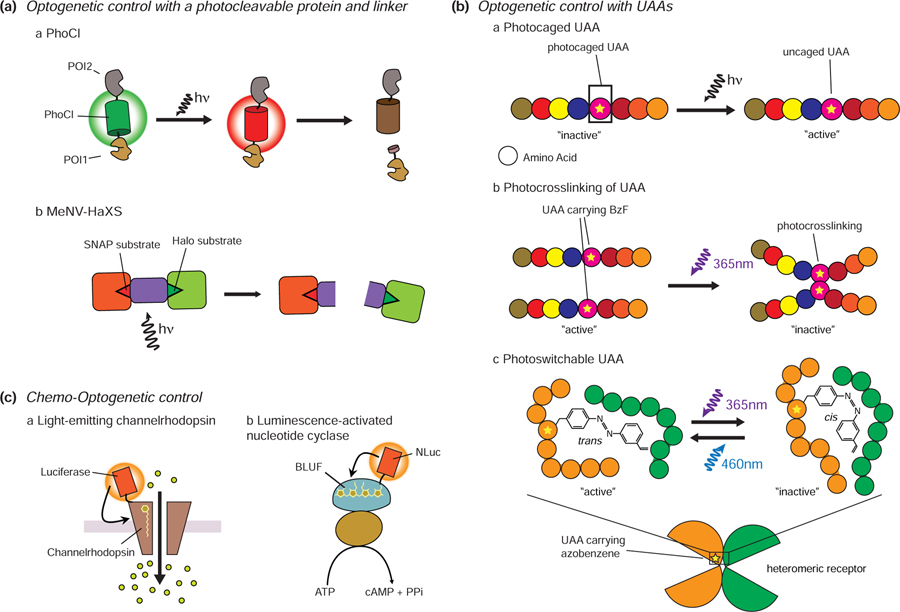
Optogenetic control using photocleavable proteins
The recent development of photocleavable proteins has further extended the optogenetic toolkit. Notably, the green-to-red photoconvertible FP mMaple undergoes a covalent rearrangement of the chromophore in response to violet light that leads to cleavage of the peptide backbone [74], and Zhang et al. sought to utilize this phenomenon as the basis for a novel means of optogenetic control. To do so, they screened a library of mMaple variants and were able to successfully engineer a photocleavable protein (PhoCl) in which exposure to 400 nm light causes the central, chromophore-bearing α-helix to undergo photo-induced cleavage, followed by spontaneous dissociation from the FP β-can (Figure 4Aa). Using this system, Zhang et al. were able to demonstrate light-controlled protein translocation into or out of the nucleus, as well as caging and uncaging of enzyme activity [75]. While the current requirement for prolonged light exposure (more than 6 min for efficient photoconversion/cleavage), as well as slow dissociation rates (t1/2 of 500 s), raise potential concerns for phototoxicity, future PhoCl variants will undoubtedly show improved properties. Similar photocleavable optogenetic control has also been demonstrated through the covalent coupling of photocleavable linkers to SLPs. To this end, Zimmerman and colleagues developed a photocleavable chemical dimerization system by using the photocleavable methyl-6-nitroveratryl (MeNV) group to covalently link HaloTag and SNAP-tag ligands (Figure 4Ab). The resulting hybrid ligand, named MeNV-HaXS, enabled on- and off-switching of specific PPIs via the addition of MeNV-HaXS and subsequent cleavage with 360 nm light [76].
Figure 4.
Recent advances in optogenetic tools. A. Photocleavable protein and linker. The excitation of PhoCl by 400 nm light leads to its photoconversion and backbone cleavage. Proteins conjugated to PhoCl will thus translocate upon PhoCl cleavage (a). MeNV-HaXS is a photocleavable chemical dimerization system. HaloTagged and SNAP-tagged proteins that are tethered by this linker can be dissociated using 360 nm light irradiation (b). B. Optogenetic control with UAAs. Substitution of functionally critical amino acids with photocaged UAAs can be used to inhibit a target protein until light irradiation restores protein function (a). UAAs carrying BzF can induce photocrosslinking upon UV stimulation. The use of photocrosslinking UAAs can enable light-mediated inhibition of protein function (b). Azobenzene moieties can undergo reversible cis/trans photoisomerization in response to blue and UV light. Incorporating UAAs containing azobenzene moieties allows reversible cis/trans photoisomerization of an amino acid side chain, enabling reversible control of protein activity (c). C. Chemo-optogenetic control. Luminopsin combines channelrhodopsin with a bioluminescent protein. Bioluminescence from the luciferase can induce conformational changes in channelrhodopsin without the need for external illumination (a). Similarly, a luminescence-activated adenylyl cyclase also combines a bioluminescent protein (NLuc) with bPAC (b).
Unnatural amino acids (UAAs)
While most current optogenetic approaches work by repurposing naturally occurring photosensitive proteins, the emergence of site-specific protein labeling using UAAs promises to offer a genetically targetable means of optically regulating almost any protein activity. UAAs are site-specifically incorporated into POIs using UAA-charged tRNAs that recognize a specific stop codon, usually UAG [77]. The substitution of functionally critical amino acids with photocaged UAAs such as photocaged cysteines, lysines, and tyrosines can thus be used to inhibit a target protein until light irradiation restores protein function by destroying the photocage [78] (Figure 4Ba). In contrast, the use of photocrosslinking UAAs can enable light-mediated inhibition of protein function [79] (Figure 4Bb). Klippenstein et al. demonstrated this approach using glutamate receptors that incorporate UAAs carrying a p-benzoyl-L-phenylalanine (BzF) photocrosslinker to enable glutamate receptor inactivation upon UV irradiation [80]. In a subsequent study, Klippenstein et al. were able to achieve reversible control of glutamate receptor activity by incorporating photoswitchable UAAs containing azobenzene moieties, which undergo reversible cis/trans photoisomerization in response to blue and UV light [81] (Figure 4Bc). Expanding the use of azobenzene-based photoswitchable UAAs to other receptors may, therefore, offer a powerful new method for reversible control of receptor signaling.
Chemo-optogenetic control of signaling
Optogenetic tools are dependent on external illumination to trigger their biochemical effects and therefore suffer from similar drawbacks as fluorescence. In particular, poor tissue penetration by the activating light, which determines the number and location of cells that are illuminated, can limit the functional range of the photostimulus in optogenetic studies. Thus, researchers have sought alternative means to activate optogenetic tools. In this regard, bioluminescence could be considered an “attractive” alternative light source, as the substrate can be applied to deeper tissues to generate light directly in situ. For example, Berglund et al. fused Gaussia princeps luciferase to the N-terminus of channelrhodopsin and demonstrated effective channel opening in response to luciferase substrate alone [82] (Figure 4Ca). More recently, a tool that combines bPAC and NLuc to control cAMP synthesis has also been reported [83]. NLuc emits light in a dose-dependent manner through the oxidation of luciferin, which subsequently activates bPAC to produce cAMP, thus enabling optically and chemically tuned control of cAMP concentrations (Figure 4Cb). However, there is much room for improvement because these kinds of tools are not easily switched on and off, and the short-lived substrates limit the duration of activation.
Conclusions and future directions
Over the last decade, a large number of genetically encodable biosensors and optogenetic tools for studying cell signaling activity have been developed. These tools have helped researchers to more accurately observe dynamic cell signaling events in live cells and animals. However, there is still plenty of room for tool development, as researchers continue seeking tools that are brighter, more sensitive, faster, and more permeable, depending on the research purpose. More advanced directed evolution systems [84], machine learning [5] and high-throughput screening platforms [85] are expected to yield enhanced toolkits that will power even more sophisticated studies in the near future. Additional multi-component optogenetic systems similar to the design of FLARE [46] are also expected to emerge in the future. Concerns regarding the sizes of these tools may also be alleviated through practical improvement of mini-fluorescence-activating proteins, de novo-designed β-barrels that are only half the size of typical FPs [86], as well as SLPs and fluorescent dyes, in the design of optogenetic tools. Furthermore, though current efforts rely on overexpression, recent advances in the efficiency of homology-directed repair in CRISPR technology [87] or prime editing [88] have made it much easier to insert genetically encodable tools into genomic DNA loci. These efforts are expected to broaden our view of cell signaling to the endogenous level.
Acknowledgments
Funding
This work was supported by the National Institutes of Health (NIH T32 CA009523 to H.N.L. and R01 MH111516, R35 CA197622, R01 DK073368, and R01 GM111665 to J.Z.) and the Air Force Office of Scientific Research (FA9500-18-1-0051 to J.Z.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no competing interests.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References and recommended reading (*)
- **1.Greenwald EC, Mehta S, Zhang J: Genetically Encoded Fluorescent Biosensors Illuminate the Spatiotemporal Regulation of Signaling Networks. Chem Rev 2018, 118:11707–11794.Well organized review of genetically encoded biosensors. This paper provides a detailed description of the history and principles of biosensors, as well as their applications and future development directions.
- 2.Hinner MJ, Johnsson K: How to obtain labeled proteins and what to do with them. Curr Opin Biotechnol 2010, 21:766–776. [DOI] [PubMed] [Google Scholar]
- 3.Mezzanotte L, van ‘t Root M, Karatas H, Goun EA, Löwik CWGM: In Vivo Molecular Bioluminescence Imaging: New Tools and Applications. Trends Biotechnol 2017, 35:640–652. [DOI] [PubMed] [Google Scholar]
- **4.Rodriguez EA, Campbell RE, Lin JY, Lin MZ, Miyawaki A, Palmer AE, Shu X, Zhang J, Tsien RY: The Growing and Glowing Toolbox of Fluorescent and Photoactive Proteins. Trends Biochem Sci 2017, 42:111–129.Here the authors provide a detailed description of the history, latest trends, and future directions in the field of fluorescent proteins.
- 5.Saito Y, Oikawa M, Nakazawa H, Niide T, Kameda T, Tsuda K, Umetsu M: Machine-Learning-Guided Mutagenesis for Directed Evolution of Fluorescent Proteins. ACS Synth Biol 2018, 7:2014–2022. [DOI] [PubMed] [Google Scholar]
- 6.Burd CG, Emr SD: Phosphatidylinositol(3)-phosphate signaling mediated by specific binding to RING FYVE domains. Mol Cell 1998, 2:157–162. [DOI] [PubMed] [Google Scholar]
- 7.Regot S, Hughey JJ, Bajar BT, Carrasco S, Covert MW: High-sensitivity measurements of multiple kinase activities in live single cells. Cell 2014, 157:1724–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Q, Huang H, Zhang L, Wu R, Chung CI, Zhang SQ, Torra J, Schepis A, Coughlin SR, Kornberg TB, et al. : Visualizing Dynamics of Cell Signaling In Vivo with a Phase Separation-Based Kinase Reporter. Mol Cell 2018, 69:347. [DOI] [PubMed] [Google Scholar]
- 9.Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, Tsien RY: Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature 1997, 388:882–887. [DOI] [PubMed] [Google Scholar]
- 10.Wegner SV, Sun F, Hernandez N, He C: The tightly regulated copper window in yeast. Chem Commun (Camb) 2011, 47:2571–2573. [DOI] [PubMed] [Google Scholar]
- 11.Lindenburg LH, Hessels AM, Ebberink EH, Arts R, Merkx M: Robust red FRET sensors using self-associating fluorescent domains. ACS Chem Biol 2013, 8:2133–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan L, Lin W, Zheng K, Zhu S: FRET-based small-molecule fluorescent probes: rational design and bioimaging applications. Acc Chem Res 2013, 46:1462–1473. [DOI] [PubMed] [Google Scholar]
- 13.Nikolaev VO, Bünemann M, Hein L, Hannawacker A, Lohse MJ: Novel single chain cAMP sensors for receptor-induced signal propagation. J Biol Chem 2004, 279:37215–37218. [DOI] [PubMed] [Google Scholar]
- 14.DiPilato LM, Cheng X, Zhang J: Fluorescent indicators of cAMP and Epac activation reveal differential dynamics of cAMP signaling within discrete subcellular compartments. Proc Natl Acad Sci U S A 2004, 101:16513–16518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ponsioen B, Zhao J, Riedl J, Zwartkruis F, van der Krogt G, Zaccolo M, Moolenaar WH, Bos JL, Jalink K: Detecting cAMP-induced Epac activation by fluorescence resonance energy transfer: Epac as a novel cAMP indicator. EMBO Rep 2004, 5:1176–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nikolaev VO, Gambaryan S, Lohse MJ: Fluorescent sensors for rapid monitoring of intracellular cGMP. Nat Methods 2006, 3:23–25. [DOI] [PubMed] [Google Scholar]
- 17.Remus TP, Zima AV, Bossuyt J, Bare DJ, Martin JL, Blatter LA, Bers DM, Mignery GA: Biosensors to measure inositol 1,4,5-trisphosphate concentration in living cells with spatiotemporal resolution. J Biol Chem 2006, 281:608–616. [DOI] [PubMed] [Google Scholar]
- 18.Zhang J, Ma Y, Taylor SS, Tsien RY: Genetically encoded reporters of protein kinase A activity reveal impact of substrate tethering. Proc Natl Acad Sci U S A 2001, 98:14997–15002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bertolin G, Sizaire F, Herbomel G, Reboutier D, Prigent C, Tramier M: A FRET biosensor reveals spatiotemporal activation and functions of aurora kinase A in living cells. Nat Commun 2016, 7:12674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harvey CD, Ehrhardt AG, Cellurale C, Zhong H, Yasuda R, Davis RJ, Svoboda K: A genetically encoded fluorescent sensor of ERK activity. Proc Natl Acad Sci U S A 2008, 105:19264–19269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bakal C, Linding R, Llense F, Heffern E, Martin-Blanco E, Pawson T, Perrimon N: Phosphorylation networks regulating JNK activity in diverse genetic backgrounds. Science 2008, 322:453–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou X, Clister TL, Lowry PR, Seldin MM, Wong GW, Zhang J: Dynamic Visualization of mTORC1 Activity in Living Cells. Cell Rep 2015, 10:1767–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mochizuki N, Yamashita S, Kurokawa K, Ohba Y, Nagai T, Miyawaki A, Matsuda M: Spatio-temporal images of growth-factor-induced activation of Ras and Rap1. Nature 2001, 411:1065–1068. [DOI] [PubMed] [Google Scholar]
- 24.Oliinyk OS, Shemetov AA, Pletnev S, Shcherbakova DM, Verkhusha VV: Smallest near-infrared fluorescent protein evolved from cyanobacteriochrome as versatile tag for spectral multiplexing. Nat Commun 2019, 10:279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shcherbakova DM, Baloban M, Emelyanov AV, Brenowitz M, Guo P, Verkhusha VV: Bright monomeric near-infrared fluorescent proteins as tags and biosensors for multiscale imaging. Nat Commun 2016, 7:12405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dacres H, Michie M, Anderson A, Trowell SC: Advantages of substituting bioluminescence for fluorescence in a resonance energy transfer-based periplasmic binding protein biosensor. Biosens Bioelectron 2013, 41:459–464. [DOI] [PubMed] [Google Scholar]
- 27.Nakai J, Ohkura M, Imoto K: A high signal-to-noise Ca(2+) probe composed of a single green fluorescent protein. Nat Biotechnol 2001, 19:137–141. [DOI] [PubMed] [Google Scholar]
- 28.Zhao Y, Araki S, Wu J, Teramoto T, Chang YF, Nakano M, Abdelfattah AS, Fujiwara M, Ishihara T, Nagai T, et al. : An expanded palette of genetically encoded Ca2+ indicators. Science 2011, 333:1888–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kitaguchi T, Oya M, Wada Y, Tsuboi T, Miyawaki A: Extracellular calcium influx activates adenylate cyclase 1 and potentiates insulin secretion in MIN6 cells. Biochem J 2013, 450:365–373. [DOI] [PubMed] [Google Scholar]
- *30.Mehta S, Zhang Y, Roth RH, Zhang JF, Mo A, Tenner B, Huganir RL, Zhang J: Single-fluorophore biosensors for sensitive and multiplexed detection of signalling activities. Nat Cell Biol 2018, 20:1215–1225.In this paper, the authors developed a suite of single fluorophore-based kinase activity biosensors, including high-performance sensors that shift between two maximum excitation wavelengths upon phosphorylation by a kinase of interest.
- 31.Hanson GT, Aggeler R, Oglesbee D, Cannon M, Capaldi RA, Tsien RY, Remington SJ: Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J Biol Chem 2004, 279:13044–13053. [DOI] [PubMed] [Google Scholar]
- 32.Belousov VV, Fradkov AF, Lukyanov KA, Staroverov DB, Shakhbazov KS, Terskikh AV, Lukyanov S: Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat Methods 2006, 3:281–286. [DOI] [PubMed] [Google Scholar]
- 33.Baird GS, Zacharias DA, Tsien RY: Circular permutation and receptor insertion within green fluorescent proteins. Proc Natl Acad Sci U S A 1999, 96:11241–11246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao Y, Jin J, Hu Q, Zhou HM, Yi J, Yu Z, Xu L, Wang X, Yang Y, Loscalzo J: Genetically encoded fluorescent sensors for intracellular NADH detection. Cell Metab 2011, 14:555–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hung YP, Albeck JG, Tantama M, Yellen G: Imaging cytosolic NADH-NAD(+) redox state with a genetically encoded fluorescent biosensor. Cell Metab 2011, 14:545–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marvin JS, Schreiter ER, Echevarría IM, Looger LL: A genetically encoded, high-signal-to-noise maltose sensor. Proteins 2011, 79:3025–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alicea I, Marvin JS, Miklos AE, Ellington AD, Looger LL, Schreiter ER: Structure of the Escherichia coli phosphonate binding protein PhnD and rationally optimized phosphonate biosensors. J Mol Biol 2011, 414:356–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.St-Pierre F, Marshall JD, Yang Y, Gong Y, Schnitzer MJ, Lin MZ: High-fidelity optical reporting of neuronal electrical activity with an ultrafast fluorescent voltage sensor. Nat Neurosci 2014, 17:884–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feng J, Zhang C, Lischinsky JE, Jing M, Zhou J, Wang H, Zhang Y, Dong A, Wu Z, Wu H, et al. : A Genetically Encoded Fluorescent Sensor for Rapid and Specific In Vivo Detection of Norepinephrine. Neuron 2019, 102:745–761.e748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patriarchi T, Cho JR, Merten K, Howe MW, Marley A, Xiong WH, Folk RW, Broussard GJ, Liang R, Jang MJ, et al. : Ultrafast neuronal imaging of dopamine dynamics with designed genetically encoded sensors. Science 2018, 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hertel F, Mo GC, Duwé S, Dedecker P, Zhang J: RefSOFI for Mapping Nanoscale Organization of Protein-Protein Interactions in Living Cells. Cell Rep 2016, 14:390–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nickerson A, Huang T, Lin LJ, Nan X: Photoactivated localization microscopy with bimolecular fluorescence complementation (BiFC-PALM) for nanoscale imaging of protein-protein interactions in cells. PLoS One 2014, 9:e100589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Z, Xing D, Su QP, Zhu Y, Zhang J, Kong X, Xue B, Wang S, Sun H, Tao Y, et al. : Super-resolution imaging and tracking of protein-protein interactions in sub-diffraction cellular space. Nat Commun 2014, 5:4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *44.Mo GC, Ross B, Hertel F, Manna P, Yang X, Greenwald E, Booth C, Plummer AM, Tenner B, Chen Z, et al. : Genetically encoded biosensors for visualizing live-cell biochemical activity at super-resolution. Nat Methods 2017, 14:427–434.In this work, the authors developed novel super-resolution compatible biosensors to observe the nanometer-scale compartmentalization of enzyme activity dynamics.
- 45.Fosque BF, Sun Y, Dana H, Yang CT, Ohyama T, Tadross MR, Patel R, Zlatic M, Kim DS, Ahrens MB, et al. : Labeling of active neural circuits in vivo with designed calcium integrators. Science 2015, 347:755–760. [DOI] [PubMed] [Google Scholar]
- *46.Wang W, Wildes CP, Pattarabanjird T, Sanchez MI, Glober GF, Matthews GA, Tye KM, Ting AY: A light- and calcium-gated transcription factor for imaging and manipulating activated neurons. Nat Biotechnol 2017, 35:864–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *47.Lee D, Hyun JH, Jung K, Hannan P, Kwon HB: A calcium- and light-gated switch to induce gene expression in activated neurons. Nat Biotechnol 2017, 35:858–863.These two papers report a pair of calcium integrators, called FLARE and Cal-Light, based on a dual, light- and calcium-gated switch design. These reporters were used to label active neuronal populations in mouse brains during complex behavior.
- 48.Plamont MA, Billon-Denis E, Maurin S, Gauron C, Pimenta FM, Specht CG, Shi J, Quérard J, Pan B, Rossignol J, et al. : Small fluorescence-activating and absorption-shifting tag for tunable protein imaging in vivo. Proc Natl Acad Sci U S A 2016, 113:497–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *49.Tebo AG, Gautier A: A split fluorescent reporter with rapid and reversible complementation. Nat Commun 2019, 10:2822.In this paper, the authors reported splitFAST, a rapid and reversible fluorescence complementation system that enables monitoring of PPI dynamics.
- 50.Gong Y, Huang C, Li JZ, Grewe BF, Zhang Y, Eismann S, Schnitzer MJ: High-speed recording of neural spikes in awake mice and flies with a fluorescent voltage sensor. Science 2015, 350:1361–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *51.Abdelfattah AS, Kawashima T, Singh A, Novak O, Liu H, Shuai Y, Huang YC, Campagnola L, Seeman SC, Yu J, et al. : Bright and photostable chemigenetic indicators for extended in vivo voltage imaging. Science 2019, 365:699–704.The authors used improved rhodamine dyes (JF549 and JF525) to develop a novel, semi-genetically encoded voltage indicator called Voltron. The use of bright and stable chemical dyes enabled long-term animal imaging.
- **52.Lavis LD: Teaching Old Dyes New Tricks: Biological Probes Built from Fluoresceins and Rhodamines. Annu Rev Biochem 2017, 86:825–843.A comprehensive review describing the theoretical background, history, applications, and latest trends in the use of small-molecule fluorophores as biological probes.
- 53.Lukinavičius G, Umezawa K, Olivier N, Honigmann A, Yang G, Plass T, Mueller V, Reymond L, Corrêa IR, Luo ZG, et al. : A near-infrared fluorophore for live-cell super-resolution microscopy of cellular proteins. Nat Chem 2013, 5:132–139. [DOI] [PubMed] [Google Scholar]
- 54.Lukinavičius G, Reymond L, Umezawa K, Sallin O, D’Este E, Göttfert F, Ta H, Hell SW, Urano Y, Johnsson K: Fluorogenic Probes for Multicolor Imaging in Living Cells. J Am Chem Soc 2016, 138:9365–9368. [DOI] [PubMed] [Google Scholar]
- *55.Komatsu N, Terai K, Imanishi A, Kamioka Y, Sumiyama K, Jin T, Okada Y, Nagai T, Matsuda M: A platform of BRET-FRET hybrid biosensors for optogenetics, chemical screening, and in vivo imaging. Sci Rep 2018, 8:8984.This paper describes a hybrid sensor, hyBRET, that can be used to monitor signaling activity via both FRET and BRET readouts. Using this sensor, the authors were able to monitor in vivo ERK activity in mice.
- *56.Farhana I, Hossain MN, Suzuki K, Matsuda T, Nagai T: Genetically Encoded Fluorescence/Bioluminescence Bimodal Indicators for Ca. ACS Sens 2019, 4:1825–1834.Here, the authors describe GLICO, a bimodal sensor that combines GCaMP with splitNLuc to enable the monitoring of calcium dynamics via fluorescence or bioluminescence.
- *57.Rost BR, Schneider-Warme F, Schmitz D, Hegemann P: Optogenetic Tools for Subcellular Applications in Neuroscience. Neuron 2017, 96:572–603.This review paper provides an insightful overview of the principles, history, and current trends in the design and application of the optogenetic tool, with a particular focus on applications in neuroscience.
- 58.Inoue K, Ono H, Abe-Yoshizumi R, Yoshizawa S, Ito H, Kogure K, Kandori H: A light-driven sodium ion pump in marine bacteria. Nat Commun 2013, 4:1678. [DOI] [PubMed] [Google Scholar]
- 59.Schneider F, Grimm C, Hegemann P: Biophysics of Channelrhodopsin. Annu Rev Biophys 2015, 44:167–186. [DOI] [PubMed] [Google Scholar]
- 60.Masseck OA, Spoida K, Dalkara D, Maejima T, Rubelowski JM, Wallhorn L, Deneris ES, Herlitze S: Vertebrate cone opsins enable sustained and highly sensitive rapid control of Gi/o signaling in anxiety circuitry. Neuron 2014, 81:1263–1273. [DOI] [PubMed] [Google Scholar]
- 61.Bailes HJ, Zhuang LY, Lucas RJ: Reproducible and sustained regulation of Gαs signalling using a metazoan opsin as an optogenetic tool. PLoS One 2012, 7:e30774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Spoida K, Eickelbeck D, Karapinar R, Eckhardt T, Mark MD, Jancke D, Ehinger BV, König P, Dalkara D, Herlitze S, et al. : Melanopsin Variants as Intrinsic Optogenetic On and Off Switches for Transient versus Sustained Activation of G Protein Pathways. Curr Biol 2016, 26:1206–1212. [DOI] [PubMed] [Google Scholar]
- 63.Airan RD, Thompson KR, Fenno LE, Bernstein H, Deisseroth K: Temporally precise in vivo control of intracellular signalling. Nature 2009, 458:1025–1029. [DOI] [PubMed] [Google Scholar]
- 64.Siuda ER, Copits BA, Schmidt MJ, Baird MA, Al-Hasani R, Planer WJ, Funderburk SC, McCall JG, Gereau RW, Bruchas MR: Spatiotemporal control of opioid signaling and behavior. Neuron 2015, 86:923–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pudasaini A, El-Arab KK, Zoltowski BD: LOV-based optogenetic devices: light-driven modules to impart photoregulated control of cellular signaling. Front Mol Biosci 2015, 2:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD, Tucker CL: Rapid blue-light-mediated induction of protein interactions in living cells. Nat Methods 2010, 7:973–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Levskaya A, Weiner OD, Lim WA, Voigt CA: Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature 2009, 461:997–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jenkins GI: The UV-B photoreceptor UVR8: from structure to physiology. Plant Cell 2014, 26:21–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Blain-Hartung M, Rockwell NC, Moreno MV, Martin SS, Gan F, Bryant DA, Lagarias JC: Cyanobacteriochrome-based photoswitchable adenylyl cyclases (cPACs) for broad spectrum light regulation of cAMP levels in cells. J Biol Chem 2018, 293:8473–8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gao S, Nagpal J, Schneider MW, Kozjak-Pavlovic V, Nagel G, Gottschalk A: Optogenetic manipulation of cGMP in cells and animals by the tightly light-regulated guanylyl-cyclase opsin CyclOp. Nat Commun 2015, 6:8046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gasser C, Taiber S, Yeh CM, Wittig CH, Hegemann P, Ryu S, Wunder F, Möglich A: Engineering of a red-light-activated human cAMP/cGMP-specific phosphodiesterase. Proc Natl Acad Sci U S A 2014, 111:8803–8808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Seifert S, Brakmann S: LOV Domains in the Design of Photoresponsive Enzymes. ACS Chem Biol 2018, 13:1914–1920. [DOI] [PubMed] [Google Scholar]
- *73.Zhou XX, Fan LZ, Li P, Shen K, Lin MZ: Optical control of cell signaling by single-chain photoswitchable kinases. Science 2017, 355:836–842.Here, the authors report photodissociable dimeric Dronpa, an engineered variant of Dronpa which the authors used to design photoswitchable kinases whose activity can be switched on and off by light.
- 74.Mizuno H, Mal TK, Tong KI, Ando R, Furuta T, Ikura M, Miyawaki A: Photo-induced peptide cleavage in the green-to-red conversion of a fluorescent protein. Mol Cell 2003, 12:1051–1058. [DOI] [PubMed] [Google Scholar]
- *75.Zhang W, Lohman AW, Zhuravlova Y, Lu X, Wiens MD, Hoi H, Yaganoglu S, Mohr MA, Kitova EN, Klassen JS, et al. : Optogenetic control with a photocleavable protein, PhoCl. Nat Methods 2017, 14:391–394.This paper reports the development of a novel optogenetic tool in the form of the photocleavable protein, PhoCl. As a proof of concept, the authors were able to demonstrate the application PhoCl light to achieve light-dependent regulation of protein translocation and enzyme activity.
- 76.Zimmermann M, Cal R, Janett E, Hoffmann V, Bochet CG, Constable E, Beaufils F, Wymann MP: Cell-permeant and photocleavable chemical inducer of dimerization. Angew Chem Int Ed Engl 2014, 53:4717–4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Young TS, Schultz PG: Beyond the canonical 20 amino acids: expanding the genetic lexicon. J Biol Chem 2010, 285:11039–11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Karginov AV, Zou Y, Shirvanyants D, Kota P, Dokholyan NV, Young DD, Hahn KM, Deiters A: Light regulation of protein dimerization and kinase activity in living cells using photocaged rapamycin and engineered FKBP. J Am Chem Soc 2011, 133:420–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **79.Nödling AR, Spear LA, Williams TL, Luk LYP, Tsai YH: Using genetically incorporated unnatural amino acids to control protein functions in mammalian cells. Essays Biochem 2019, 63:237–266.A comprehensive review paper describing principles, applications, and recent research trends in the field of unnatural amino acids.
- 80.Klippenstein V, Ghisi V, Wietstruk M, Plested AJ: Photoinactivation of glutamate receptors by genetically encoded unnatural amino acids. J Neurosci 2014, 34:980–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *81.Klippenstein V, Hoppmann C, Ye S, Wang L, Paoletti P: Optocontrol of glutamate receptor activity by single side-chain photoisomerization. Elife 2017, 6.Here, the authors demonstrate optical control of glutamate receptor activity based on the incorporation of photoswitchable UAAs containing azobenzene moieties directly into the primary sequence of a glutamate receptor.
- 82.Berglund K, Birkner E, Augustine GJ, Hochgeschwender U: Light-emitting channelrhodopsins for combined optogenetic and chemical-genetic control of neurons. PLoS One 2013, 8:e59759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *83.Naim N, White AD, Reece JM, Wankhede M, Zhang X, Vilardaga JP, Altschuler DL: Luminescence-activated nucleotide cyclase regulates spatial and temporal cAMP synthesis. J Biol Chem 2019, 294:1095–1103.This very recent paper describes a new chemical and optogenetic tool that combines bPAC and nLuc to generate cAMP via in situ bioluminescence. Interestingly, cAMP production can be tuned as a function of the luciferin concentration.
- 84.Wang T, Badran AH, Huang TP, Liu DR: Continuous directed evolution of proteins with improved soluble expression. Nat Chem Biol 2018, 14:972–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Amano M, Hamaguchi T, Shohag MH, Kozawa K, Kato K, Zhang X, Yura Y, Matsuura Y, Kataoka C, Nishioka T, et al. : Kinase-interacting substrate screening is a novel method to identify kinase substrates. J Cell Biol 2015, 209:895–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dou J, Vorobieva AA, Sheffler W, Doyle LA, Park H, Bick MJ, Mao B, Foight GW, Lee MY, Gagnon LA, et al. : De novo design of a fluorescence-activating β-barrel. Nature 2018, 561:485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu M, Rehman S, Tang X, Gu K, Fan Q, Chen D, Ma W: Methodologies for Improving HDR Efficiency. Front Genet 2018, 9:691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, Chen PJ, Wilson C, Newby GA, Raguram A, et al. : Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019. [DOI] [PMC free article] [PubMed]