Abstract
Atopic dermatitis (AD) is a highly prevalent, itchy inflammatory skin disorder that is thought to arise from a combination of defective skin barrier and immune dysregulation. Kallikreins (KLK), a family of serine proteases with a diverse array of homeostatic functions including skin desquamation and innate immunity, are speculated to contribute to AD pathogenesis. Their precise role in AD, however, has not been clearly defined. In this study, unbiased RNA-seq analyses identified KLK7 as the most abundant and differentially expressed KLK in both human AD and murine AD-like skin. Further, in mice, Klk7 expression was localized to the epidermis in both steady state and inflammation. However, KLK7 was dispensable for the development of AD-associated skin inflammation. Instead, KLK7 was selectively required for AD-associated chronic itch. Even without alleviation of skin inflammation, KLK7-deficient mice exhibited significantly attenuated scratching, compared to controls, after AD-like disease induction. Collectively, our findings indicate that KLK7 promotes AD-associated itch independently from skin inflammation, and reveal a previously unrecognized epidermal-neural mechanism of AD itch.
INTRODUCTION
Atopic dermatitis (AD) is a chronic and relapsing inflammatory skin disorder that affects 10–20% of the population (DaVeiga 2012; Silverberg 2017). In moderate or severe forms, the crusted, oozing, and itchy skin lesions can dramatically lower quality of life for AD patients (Drucker et al. 2017). Although chronic itch (pruritus) is considered the central and most debilitating symptom of AD, treatments have almost exclusively targeted proinflammatory mediators, rather than specific pruritogenic pathways.
AD is currently thought to arise from a combination of skin barrier dysfunction and immune dysregulation (Brunner et al. 2018; Czarnowicki et al. 2019; Elias et al. 2008). Many recent studies have focused on how epidermal barrier defects (e.g., filaggrin mutations) lead to the initiation of allergic inflammation, characterized by the recruitment and activation of T helper type 2 (Th2) cells, group 2 innate lymphoid cells (ILC2s), and basophils in the affected skin (Esaki et al. 2015; Howell et al. 2007; Kim 2014; Mashiko et al. 2017; Palmer et al. 2006). These cells, in turn, produce the hallmark type 2 effector cytokines, interleukin (IL)-4, IL-5, and IL-13, central to AD immune dysregulation (Brandt and Sivaprasad 2011; Guttman-Yassky et al. 2019). However, despite our increasing understanding of the inflammatory pathogenesis of AD, the molecular mechanisms of AD-associated itch remain poorly defined.
The kallikrein (KLK) family of serine proteases is speculated to be a significant contributor to AD pathology. A number of KLKs are reported to be enriched in lesional AD skin of humans (Komatsu et al. 2007; Komatsu et al. 2005; Morizane et al. 2012; Vasilopoulos et al. 2011) and may contribute to AD associated pruritus in patients (Nattkemper et al. 2018). Moreover, overexpression of KLKs in murine skin can spontaneously lead to an AD-like disease (Briot et al. 2009; Hansson et al. 2002).
In humans, the KLK family is comprised of 15 structurally conserved members (KLK1–15) that are involved in an array of homoeostatic and disease processes across a wide variety of tissues (Shaw and Diamandis 2007; Sotiropoulou et al. 2009). In healthy human skin, serine peptidase activity in the stratum corneum is mainly attributed to the trypsin-like KLK5 (Deraison et al. 2007; Ekholm et al. 2000) and the chymotrypsin-like KLK7 (Borgono et al. 2007; Caubet et al. 2004; Yousef et al. 2000), which help maintain barrier homeostasis by regulating the cleavage of corneodesmosomes. Notwithstanding this, a number of other KLKs may also be present in lower abundance and have related functions (Borgono et al. 2007; Komatsu et al. 2007; Komatsu et al. 2005; Komatsu et al. 2003).
All KLKs are translated as pre-proenzymes and secreted into the extracellular space as inactive zymogens. There, they are activated by a variety of mechanisms including autocatalytic activity, endopeptidases, or by other KLKs (Sotiropoulou et al. 2009). In the skin, KLK activity is further controlled by a number of homeostatic processes, including the endogenous activity of serine protease inhibitors (serpins), such as lymphoepithelial Kazal-type inhibitor (LEKTI) encoded by SPINK5 (Deraison et al. 2007). In the setting of AD, it is hypothesized that over activity of KLKs sets off a cascade of proteolytic activity, which in turn contributes to its pathogenesis.
In lesional AD skin, a number of KLKs, including KLK5 and KLK7, have been shown to be increased (Brunner et al. 2017; Komatsu et al. 2007; Morizane et al. 2012; Vasilopoulos et al. 2011). Furthermore, transgenic overexpression of Klk5 or Klk7 in mice resulted in the spontaneous development of AD-like disease features (Briot et al. 2009; Furio et al. 2014; Hansson et al. 2002). Likewise, loss-of-function mutations in SPINK5 resulted in unregulated epidermal KLK activity and Netherton Syndrome, a severe AD-like syndrome, in both mice and humans (Chavanas et al. 2000; Descargues et al. 2005). In mice, the symptoms of Netherton Syndrome can be prevented by genetic ablation of Klk5 and Klk7 (Kasparek et al. 2017). Although it is becoming increasingly clear that KLK dysregulation contributes to AD pathogenesis, the mechanisms underlying this process remain poorly defined.
In this study, we demonstrate that KLK7, but not KLK5, is upregulated in human and murine lesional AD skin. Further, we show that basal and AD-associated KLK7 expression is restricted to the epidermis, provoking the hypothesis that epithelial cell-derived KLK7 is critically required for the development AD. Surprisingly, we found that KLK7-deficient mice had no improvement in AD-like skin disease severity, but showed markedly attenuated AD-associated itch. These findings demonstrate a previously unrecognized role for KLK7 in mediating itch in the context of AD and provide additional insight into KLK function in disease states.
RESULTS
KLK7 is upregulated in human AD lesions
Both human AD and murine AD-like disease are characterized by several shared histological and immunological features. At the molecular level, these include a variety of KLKs upregulated in lesional AD skin (Komatsu et al. 2007). However, which KLKs are selectively and critically upregulated remains to be defined.
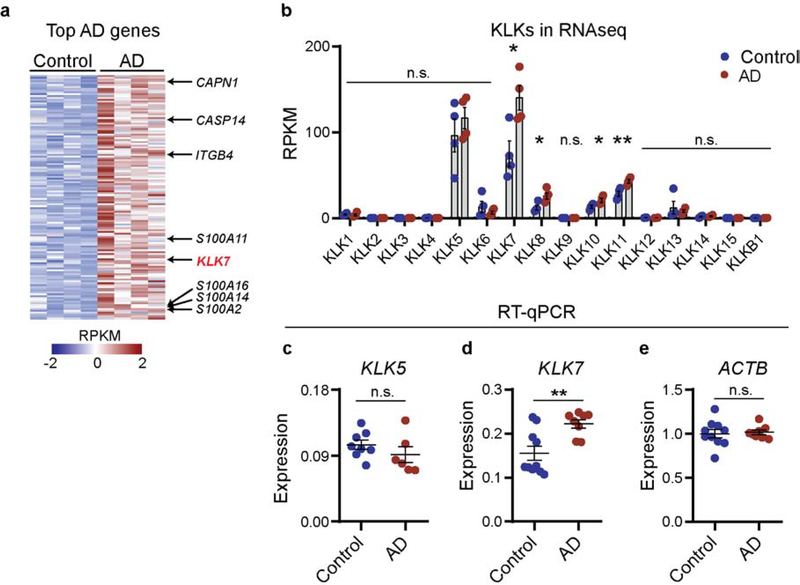
In this study, we reanalyzed a previously published RNA-seq dataset of human control and lesional AD skin for the top differentially expressed genes by fold change and expression level (Oetjen et al. 2017). Unbiased analysis of the most abundant differentially expressed genes (DEGs) identified KLK7 as the only KLK that was differentially upregulated in human AD skin (Figure 1a). Other keratinocyte-associated genes such as S100A family members, calpain 1 (CAPN1), and caspase 14 (CASP14) were also enriched in AD skin (Figure 1a). Comprehensive analysis of all KLKs in this dataset demonstrated that, consistent with prior studies, KLK5 and KLK7 are the most highly expressed KLKs in control human skin, accounting two-third of total KLK transcripts in control biopsies (Figure 1b). In addition, four KLKs - KLK7, KLK8, KLK10, and KLK11 – showed statistically significant expression increases in lesional AD skin, with KLK7 exhibiting the most prominent overexpression. Changes in KLK8, KLK10, and KLK11 expressions were comparatively less pronounced. KLK5 expression, surprisingly, was unchanged.
Figure 1. KLK7 is overexpressed in human lesional atopic dermatitis (AD) skin.
a. Heatmap of z-scored RPKMs of differentially expressed genes with >2-fold change, adjusted p<0.05, and base mean expression level >1000. b. RPKMs of all human KLKs present in skin samples of control and AD individuals. N = 4 per group. c-e. RT-qPCR of c. KLK5 and d. KLK7 in human control and AD skin, normalized to e. ACTB. n = 8–10 per group. Error bars represent SEM. n.s. no significance. * p<0.05. ** p<0.01.
Our finding that, compared to KLK5, KLK7 is selectively overexpressed in lesional AD skin was further confirmed using additional sets of human control and AD skin samples. Using RT-qPCR, we confirmed that KLK7, but not KLK5, was significantly enriched in lesional AD skin (Figure 1c–e). Additionally, we looked specifically at matched lesional (LS) and non-lesional (NL) sites in another cohort of 35 AD patients by RNA-seq (Figure S1a–b). Consistent with the observations from our other cohorts, we found no increases in KLK5 expression in lesional (LS) AD skin in this dataset (Figure S1a). Moreover, we confirmed KLK7 mRNA is significantly elevated in lesional (LS) AD skin, compared to matched non-lesional sites (Figure S1b). However, the variations in KLK7 levels within lesional tissues did not significantly correlate with patient reported itch severity (Figure S1c). Based on these results, we hypothesized that KLK7 may be a key driver in AD disease pathogenesis.
Klk7 is upregulated in murine AD-like lesions
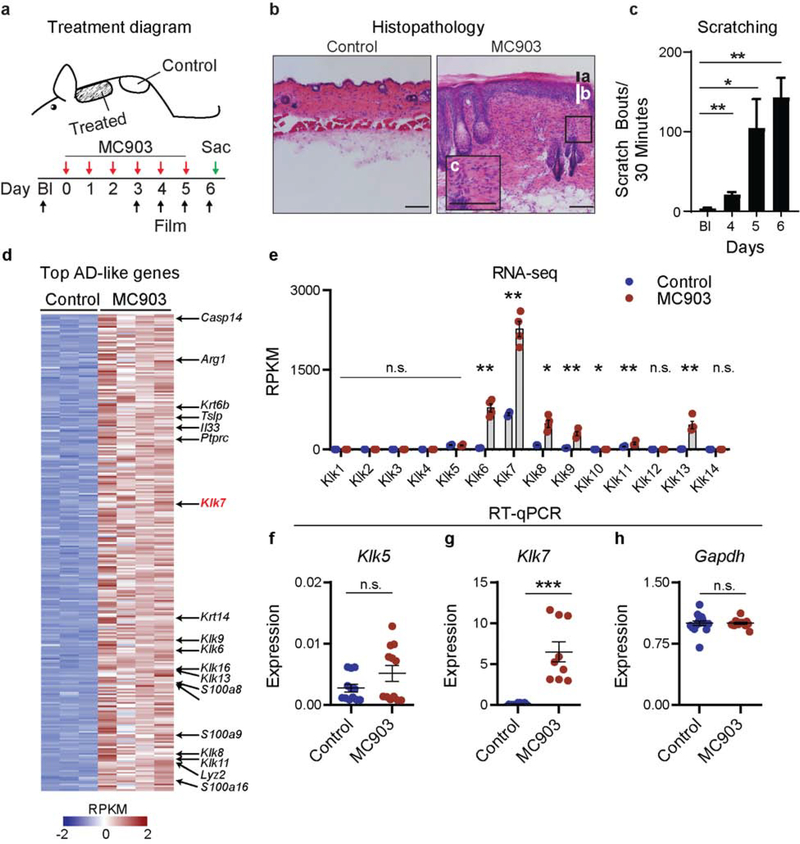
Since KLKs are highly conserved across mammalian species (Pavlopoulou et al. 2010), we sought to explore whether Klk7 expression is also dysregulated in murine AD-like skin. To test this, we employed a well-established model of AD-like disease in which mice are treated with the topical irritant MC903 (calcipotriol) (Li et al. 2006) (Figure 2a). Following MC903 treatment, mice develop histopathologic features of AD (Figure 2b) as well as chronic itch behavior (Figure 2c).
Figure 2. Klk7 is overexpressed in lesional murine AD-like skin.
a. Schematic of murine AD model. Control skin tissues were taken from non-lesional skin caudal to the treated area. b. Representative H&E section of control and lesional mouse skin after six MC903 treatments. Note presence of hyperkeratosis (a), acanthosis (b), and mixed lymphocyte infiltration (c) that define human AD lesions. n = 3 biological pairs. c. Mice develop severe spontaneous scratching behavior directed at the treated site after MC903 treatment. Statistical comparisons are made against baseline scratching. n=6 mice. d-e. RNA-seq of mouse control (ethanol vehicle) and MC903 treated skin, n = 3–4 mice per group. d. Heat map showing z-scored RPKMs of genes differentially expressed by ≥2-fold, adjusted p<0.05, and base mean expression values ≥1000. e. RPKM values of all Klks expressed in skin samples of control- and MC903-treated mice. f-h. RT-qPCR of f. Klk5 and g. Klk7 expression in mouse control- and MC903-treated skin normalized to h. Gapdh. n = 3 biological pairs. Error bars represent SEM. n.s. no significance. * p<0.05. ** p<0.01. *** p<0.001. The scale bars represent 200 μm.
To define the expression status of Klk7 in mouse AD-like skin, we reanalyzed a previously published RNA-seq dataset of MC903-treated murine AD-like skin (Oetjen et al. 2017). Using the same unbiased analysis techniques, we identified Klk7 among the top most abundant differentially expressed genes in murine AD-like skin (Figure 2d). Comprehensive analysis of all KLKs in this dataset demonstrated that, like human skin (Figure 1), Klk7 is the most highly expressed KLK in control skin, accounting for 66.9% ± 1.43% of total Klk transcripts. Moreover, seven KLKs – Klk6, Klk7, Klk8, Klk9, Klk10, Klk11, and Klk13 – showed statistically significant expression increases in murine AD-like skin (Figure 2e). Similar to our findings in human AD skin, Klk7 was the most abundant transcript and demonstrated a 4-fold expression increase in AD-like skin (Figure 2e). Klk5 expression, again, was unchanged in this context (Figure 2e). These findings were further validated with additional samples by RT-qPCR. Klk7, but not Klk5, is significantly enriched in AD-like skin (Figure 2f–h).
Based on the consistent KLK7 upregulation in AD skin of both humans and mice, we sought to investigate the mechanisms by which KLK7 promotes AD-like disease pathogenesis in vivo.
Epidermal KLK7 expression is enhanced in AD-like disease and promotes itch but not inflammation
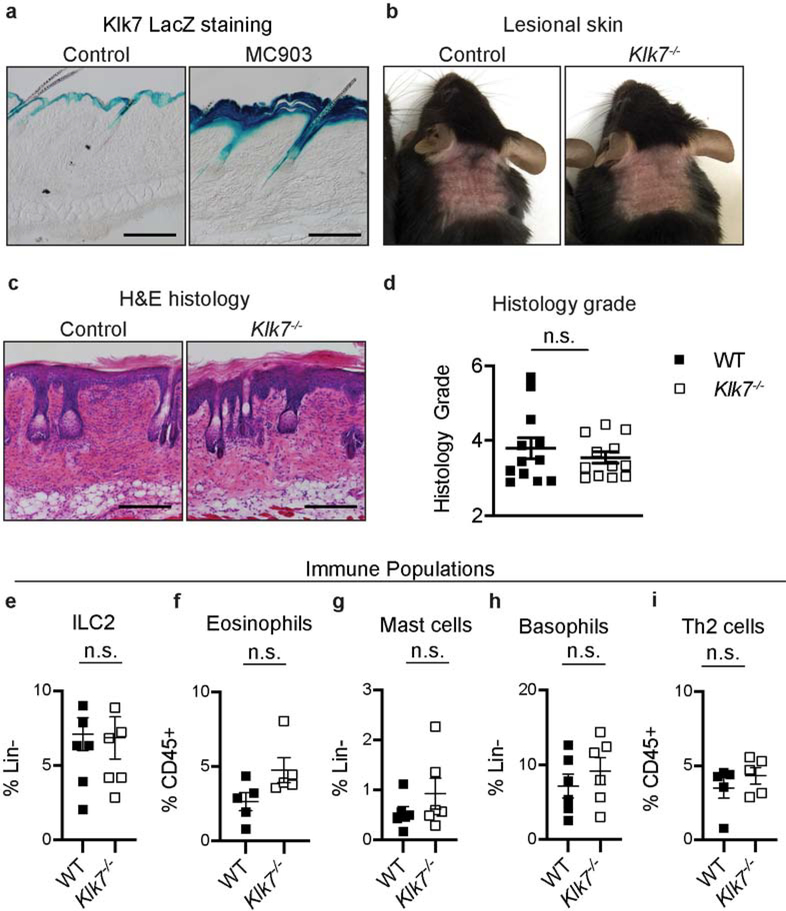
Klk7 expression is highly conserved between mice and humans and is mainly restricted to the skin in both species (Fagerberg et al. 2014; Pavlopoulou et al. 2010). In steady state, murine Klk7 expression is largely restricted to epidermal skin and is undetectable in dermal skin, neural and immune tissues (Figure 3a, Figure S2). After the induction of AD-like disease, Klk7, detected using a LacZ-β-galactosidase reporter, was selectively enhanced in the epidermis, while remaining undetec in the dermis where infiltrating immune cells and peripheral fibers of sensory neurons are present (Figure 3a). Taken together, these findings indicate that the epidermis is the primary source of Klk7 in the skin, in both homeostatic and inflammatory conditions.
Figure 3. KLK7 deficiency does not cause defects in AD-associated immune response.
a. X-gal staining of control untreated nape skin from Klk7LacZ mice and after treatment with MC903. b. Representative images of AD-like disease induction in control wild-type (WT) and Klk7−/− mice after 6 days of MC903 treatment; n > 8 mice per group. c. Representative H&E-stained sections of MC903-treated skin of control and Klk7−/− mice; n = 4 mice per group. d. Histological grading of H&E-stained sections of control and Klk7−/− mouse skin; n = 4 mice per group. e-i. Flow cytometric analysis of AD-associated e. ILC2, f. eosinophil, g. mast cell, h. basophil, and i. Th2 cell frequency in MC903-treated nape skin from control and Klk7−/− mice on day 6 of treatment; n = 5 mice per group. Error bars represent SEM. n.s. no significance. The scale bars represent 200 μm.
To directly investigate the contribution of KLK7 to AD-like skin disease, we generated the MC903-induced AD-like disease model using Klk−/− mice. Surprisingly, compared to wild-type controls, Klk7−/− mice did not exhibit significant reduction in disease severity as measured by redness, edema/papulation, scaling, and lichenification (enhanced skin lines) (Figure 3b). Additionally, histologic analyses did not demonstrate any difference between groups by clinical grading (Figure 3c–d). Flow cytometric analysis of skin from control and Klk7−/− mice also did not reveal any difference in the frequency of key inflammatory cells (Figure 3, e–i).
Additional quantification of Th2 and inflammatory markers did not reveal any detectible differences between MC903 treated control and Klk7−/− mice. Serum IgE and TARC/CCL17 levels, as detected by ELISA, were similar between the genotypes (Figure S3a–b). Il4rα, Il4, and Il13 expression levels in MC903 treated lesional skin, as detected by RT-qPCR, also were not different between control and Klk7−/− mice (Figure S3c–e). Lastly, Tslp expression in the skin was strongly induced by MC903 treatment as expected, but was expressed at similar levels between the genotypes at both basal and diseased states (Figure S3f).
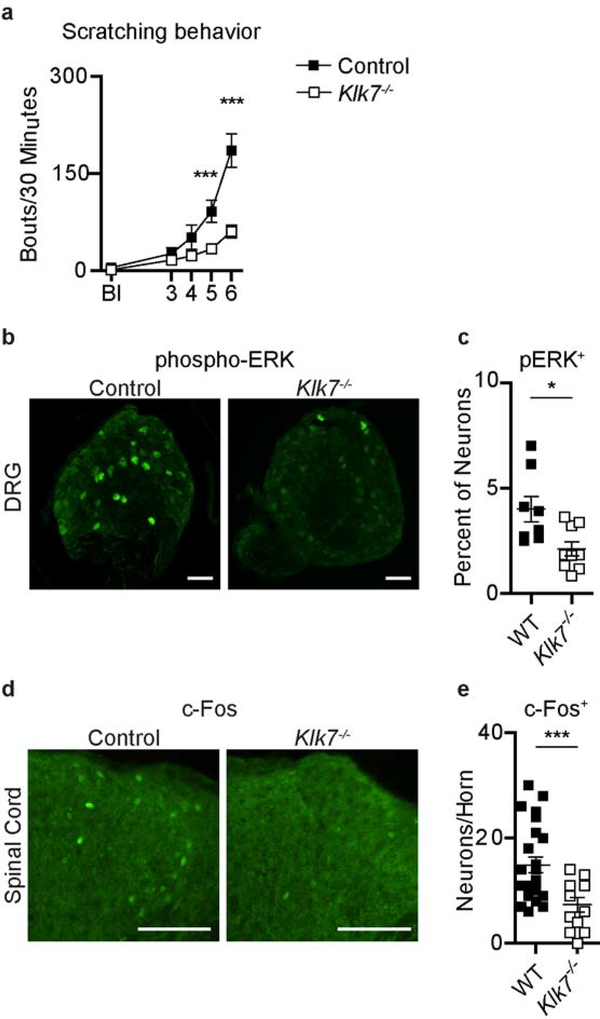
Despite the lack of observable differences in skin inflammation and key inflammatory markers, spontaneous scratching was markedly reduced in Klk7−/− mice compared to controls (Figure 4a). On average, Klk7−/− mice scratched two to three times less than controls and demonstrated a mean of 61.33 ±10.24 bouts/30 minutes compared to 186.13 ± 25.89 bouts/30 minutes for control mice on day 6 of treatment (Figure 4a). These findings were recapitulated in a 12-day ear treatment model, in which Klk7−/− mice exhibited a similar reduction in itch (Figure S4).
Figure 4. KLK7 is required for the development of AD-associated itch.
a. Spontaneous scratching after MC903 nape treatment in control and Klk7−/− mice; n > 8 per group. b-c. b. Representative immunofluorescence images and c. quantification of phosphorylated ERK (pERK) staining in DRGs innervating MC903-treated nape skin of control and Klk7−/− mice; n = 3 mice per group. d-e. d. Representative immunofluorescence images and e. quantification of c-Fos staining in thoracic spinal cords of MC903-treated control and Klk7−/− mice on day 6 of treatment; n = 3 mice per group. Error bars represent SEM. * p<0.05. *** p<0.001. The scale bars represent 200 μm.
Given the striking itch phenotype, we examined whether sensory neuron activation was correspondingly attenuated in Klk7−/− mice under AD-like conditions. Although rarely detectible in primary afferents at baseline, ERK phosphorylation (pERK) can be transiently induced in dorsal root ganglia (DRG) neurons by intense stimulation or under chronic pathological conditions (Gao and Ji 2009). Upper thoracic DRGs that innervate the inflamed skin showed a substantially diminished proportion of pERK immunoreactive neurons in Klk7−/− mice, compared to control mice (2.11 ± 0.34% vs 4.00 ± 0.60%, respectively) (Figure 4b–c), indicating that activation and sensitization of peripheral sensory neurons was significantly attenuated in Klk7−/− mice. Likewise, c-Fos expression, which is induced by strong and sustained activation of spinal dorsal horn neurons (Gao and Ji 2009), was significantly decreased in the thoracic spinal cords of Klk7−/− mice, most notably in lamina I and II where itch-sensing c-fiber DRG neurons project (Han et al. 2013) (Figure 4d–e). These findings indicate that, along with a reduction in behavioral itch (Figure 3i) sensory neuron activation is significantly attenuated in Klk7−/− mice in the context of AD. Collectively, our findings demonstrate that the epidermis is the major source of KLK7 and reveal a previously unrecognized epithelial-neural circuit by which KLK7 specifically mediates AD-associated itch.
DISCUSSION
KLKs are highly conserved across species, widely expressed throughout many organ systems, and exhibit heterogeneity in their composition across tissues (Pavlopoulou et al. 2010; Sotiropoulou et al. 2009). As serine proteases, they are involved in a diversity of both homeostatic and pathologic processes. In the skin, their primary role is to mediate desquamation (Ekholm et al. 2000). Both KLK5 and KLK7 have been shown to be key mediators of corneodesmosomal cleavage and epithelial turnover (Caubet et al. 2004). While a number of KLKs, including KLK5 and KLK7, have been shown to be upregulated in lesional AD skin from patients (Brunner et al. 2018; Komatsu et al. 2007; Morizane et al. 2012; Vasilopoulos et al. 2011), their precise role in AD pathogenesis, however, is unclear. In this study, we provide three conceptual advances that clarify the contributions of KLKs to AD. First, employing unbiased RNA-seq analysis, we discovered that KLK7 is selectively upregulated in lesional AD skin of both mice and humans. Second, we show that, despite its abundant expression in the skin, KLK7 is dispensable for the development of AD-associated inflammation. Third, epidermal expression of Klk7 is selectively required for AD-associated itch. Importantly, these finding demonstrates an emerging paradigm in itch biology, that itch mediators may be separate from the mechanism driving AD-like skin inflammation.
The induction of the type 2 cytokines IL-4 and IL-13 critically promote AD-associated skin inflammation and itch (Oetjen and Kim 2018; Trier and Kim 2018). Additionally, clinical trials with dupilumab, an anti-IL-4Rα monoclonal antibody, have demonstrated rapid and marked improvement of itch symptoms in AD patients (Beck et al. 2014; Guttman-Yassky et al. 2019; Simpson et al. 2016; Thaçi et al. 2016). Recent studies have shown that both IL-4 and IL-13 induce selective expression of KLK7, but not KLK5, in NHEK cells (Morizane et al. 2012). Thus, whether there is a type 2 cytokine-epithelial-KLK circuit promoting itch remains a promising area of future inquiry.
Prior studies have reported broad upregulation of a number of KLKs in AD skin (Komatsu et al. 2007; Vasilopoulos et al. 2011), our gene expression analysis only identified KLK7 as the predominate KLK in both mouse and human AD skin. Although KLK5 has been reported to be upregulated in human AD skin (Komatsu et al. 2007) and implicated in the promotion of AD-like disease in mice (Briot et al. 2009; Furio et al. 2014), we consistently could not detect KLK5 upregulation in AD skin. This may be in part due to the complexity of AD disease, with varying stages (acute vs. chronic), levels of severity, and genetic and age-dependent heterogeneity. Indeed, mutations in filaggrin vary considerably across ethnicities and there is emerging evidence that immune profiles in AD are also sensitive to the genetic background of individuals (Czarnowicki et al. 2019; Kaufman et al. 2018; Leung 2015; Margolis et al. 2014; Osawa et al. 2011). Thus, future studies focused on understanding the differential contributions of KLK5 and KLK7 to skin inflammation and itch are warranted. Additionally, it is widely appreciated that KLKs can activate one another (Sotiropoulou et al. 2009). Thus, understanding how KLK7 may interact with other KLKs to regulate the development of AD-associated itch is an intriguing area of investigation.
Prior studies have shown that overexpression of Klk7 was particularly notable for the development of severe itch associated with cutaneous inflammation (Hansson et al. 2002). Despite these advances, whether KLKs mediated itch occurs indirectly through the induction of skin inflammation or directly by acting on sensory neurons was unknown. Our finding that KLK7 deficiency attenuates AD-associated itch with no effect on inflammation provokes the hypothesis that this phenomenon occurs through direct epidermal-neural mechanisms. It has previously been shown that KLK5 can proteolytically activate PAR2 receptors, which has been implicated in itch (Liu et al. 2011; Shimada et al. 2006; Stefansson et al. 2008). However, KLK7 does not exhibit this function (Figure S5) (Stefansson et al. 2008) or the ability to induce significant behavioral response or neuronal activation in naive mice (Figure S6a–c). Thus, whether KLK7 generates endogenous pruritogens or if KLK7 can proteolytically sensitize neuronal itch receptors in AD skin remains open to further exploration.
In summary, therapeutic agents targeting KLK7 may be able to provide substantial itch relief for AD patients. Due to the specificity of KLK7 expression in epidermal skin, KLK7 antagonists could be used topically on affected AD skin to avoid potential side effects. Moreover, given the unique chymotrypsin-like functionality of KLK7, in contrast to the trypsin-like properties of other KLKs, redundancy and compensation by other KLKs is also less likely following pharmacologic KLK7 inhibition. These findings open exciting avenues for the exploration and development of novel therapeutics that target KLK in the setting of AD and its associated itch.
METHODS
Ethics statement
All animal experiments were performed under protocols approved by the Institutional Animal Care and Use Committee (IACUC) at Washington University School of Medicine (WUSM) and in strict accordance with recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (NIH). Human studies were performed under protocols reviewed and approved by local Institutional Review Boards (IRBs), and biopsies were taken only after obtaining written, informed patient consent.
Animals
C57BL/6NJ wild-type were purchased from The Jackson Laboratory (Bar Harbor, ME). Klk7tm1(KOMP)Vlcg sperm was purchased from the UC Davis KOMP repository (Davis, CA). IVF was performed by the WUSM Molecular Genetics Service Core. PirtGCaMF3/+ mice were gifted by Dr. Xinzhong Dong at Johns Hopkins University. All researchers were blinded to mouse genotypes throughout testing and data quantification.
Human sample collection
AD diagnosis was made by a board-certified dermatologist according to the American Academy of Dermatology recommended criteria (Eichenfield et al. 2014). Biopsy samples were collected from consenting patients in clinic. Control tissues were either obtained from individuals without history of inflammatory skin conditions, or from healthy skin margins from patients undergoing Mohs surgery. All skin samples were de-identified and stored in RNAlater (ThermoFisher, Waltham, MA) at −80°C prior to processing.
Mouse Tissue Collection
For RT-PCR and RT-qPCR, mice were euthanized via CO2 overdose and tissues were dissected and stored in ice cold RNeasy Kit Lysis Buffer (Qiagen, Germany). Samples were processed for RNA extraction immediately after dissection.
For histopathology and LacZ staining, mice were transcardially perfused with ice cold phosphate buffered saline (PBS) and 4% paraformaldehyde (PFA) for fixation. Dissected tissues were fixed on ice as follows: skin in 1% PFA 1 hour, DRG and TG in 2% PFA 30 minutes, spinal cord (SC) in 2% PFA 1 hour, and brain in 2% PFA 2 hours. After fixation, tissues were immersed in 30% sucrose for 24 hours at 4°C and embedded in OCT (Sakura, Torrance, CA) for frozen sectioning. Mouse DRGs and SCs used for immunostaining were fixed in 4% PFA on ice for 90 minutes before sucrose incubation and frozen sectioning. For flow cytometry, skin was harvested without fixation and processed immediately.
For ELISA, mouse serum was isolated from blood collected by orbital bleed by centrifugation at 10,000g for 10 minutes at 4°C on experimental day 6. ELISAs for IgE (Biolegend 432404, San Diego, CA) and TARC/CCL17 (R&D DY529–05, Minneapolis, MN) were performed according to the manufacturers’ instructions.
H&E, X-Gal, and immunofluorescence staining
OCT-embedded mouse samples were sectioned on a Leica (Buffalo Grove, IL) cryostat. Brain sections were processed as floating sections while other tissues were slide-mounted. H&E staining was performed by the WUSM Pulmonary Morphology Core. Images were acquired using a BX63 microscope (Olympus, Waltham, MA). Clinical grade was determined using a previously published protocol (Kim et al. 2014).
Tissues used for LacZ and immunofluorescence staining were allowed to air dry for 2 hours before processing. For LacZ staining, slides were fixed on ice in 1% PFA for 1 minute. Chromogenesis was performed using the X-Gal Staining Assay Kit (Genlantis, San Diego, CA) according to the manufacture’s recommended protocol. Color development time was approximately 6 minutes. Sections were dehydrated and mounted using ThermoFisher Permount.
For pERK and c-FOS immunofluorescence staining, slides were blocked using 10% normal goat serum (NGS) (MilliporeSigma, Burlington, MA) and incubated in primary antibodies overnight at 4°C. Afterwards, slides were incubated in fluorescent secondary antibody for 2 hours at room temperature and mounted using Fluoromount-G (SoutherBiotech, Birmingham, AL). Images were acquired using a Ti-E microscope (Nikon, Melville, NY). Antibodies used were rabbit anti-phospho-p44/42 MAPK (Thr202/Tyr204) mAb (CST, Danvers, MA), rabbit anti-c-Fos pAb (Calbiochem, San Diego, CA), and goat anti-Rabbit IgG (H+L), Alexa Fluor 488 conjugated secondary (ThermoFisher, Waltham, MA). All antibodies were diluted 1:1,000 in 1x PBS with 1% Tween-20 and 1% NGS.
RNA isolation, RT-PCR, and RT-qPCR
RNA-seq performed and analyzed as previously described (Oetjen et al. 2017). Briefly, for RNA-seq, 1 mg of total RNA was enriched with RiboZERO (Illumina, San Diego, CA) and sequenced on an Illumina HiSeq3000. Sequences were aligned with STAR (Dobin et al. 2013), counted with Subread:featureCount (Liao et al. 2014), and differential gene expression determined by DESeq2 package (Love et al. 2014) in R v3.5.
Samples for RT-PCR and RT-qPCR were homogenized in lysis buffer supplied with the Qiagen RNeasy Kit using a bead homogenizer. RNA extraction was performed using the same kit, according to the manufacturer’s recommended protocol. Samples were then treated with ThermoFisher Turbo DNase, and cDNA was generated using approximately 1,000 ng of total RNA and ABI High-Capacity cDNA Reverse Transcription Kit. Reactions without reverse transcriptase were included as negative controls for downstream PCR.
RT-PCR was performed using Qiagen HotStarTaq Polymerase and 10 ng of template. Images presented are representative of results of 30 PCR cycles. RT-qPCR was performed using ABI Fast Sybr Green Master Mix and a Step-One Plus Real-Time PCR System. All reactions were run as technical triplicates using 10 ng of cDNA template and the presented data were normalized to ACTB (for human samples) or Gapdh (for mouse samples) expression. Primers used are listed in Table S1.
MC903 treatment and mouse behavior
The murine AD model was adapted from previous publications (Kim et al. 2014; Kim et al. 2013; Li et al. 2006; Oetjen et al. 2017). In the nape model, the nape and lower back areas (approximately 17 mm x 17 mm each) were shaved under isoflurane anesthesia three days before baseline behavioral recording. Test animals were habituated for two consecutive days immediately preceding baseline recording. To induce AD-like disease, 40 μl of 0.1 mM MC903 (Tocris, Minneapolis, MN) in ethanol was applied topically to the shaved nape skin under anesthesia once every 24 hours, starting 24 hours after baseline recording, for six consecutive days. Spontaneous scratching was scored for 4 consecutive days, starting 4 days after baseline recording. Scratching bouts, defined as a continuous scratch movement directed at the treated skin by the hind paw, were scored from video recordings after the completion of the experiment. For the ear model, mice were treated with 40 μl of 0.05 mM MC903 on each ear under anesthesia for 8 days. Ear thickness was determined daily with a dial caliper.
Flow cytometry
Flow cytometry was performed as previous described (Oetjen et al. 2017). Briefly, harvested tissues were digested in 0.25 mg/mL Liberase TL (Roche, Switzerland) in DMEM media for 90 minutes at 37°C. Afterwards, tissues were mechanically dissociated and passed through 70 μm cell strainers to obtain a single cell suspension. Cells were then stained with ZombieUV (Biolegend, San Diego, CA) at room temperature for 20 minutes, followed by primary antibodies (Table S2) on ice for 30 minutes. Biolegend Streptavidin-FITC and streptavidin-PE secondary stains were performed on ice for 30 minutes. Samples were acquired on an LSRFortessa X-20 (BD, Franklin Lakes, NJ).
Calcium imaging
Calcium imaging was performed as previously described (Huang et al. 2018; Oetjen et al. 2017). In brief, KNRK cells stably expressing hPAR2 receptors (KNRK-PAR2) were seeded onto cover slips pre-coated with 0.1 mg/ml poly-D-lysing (Corning, Corning, NY) and cultured overnight in DMEM complete medium. Primary cultures of DRG neurons from PirtGCaMP3/+ mice were acutely extracted and dissociated in dispase II and collagenase I enzyme mixture from ThermoFisher. Dissociated DRG neurons were then seeded onto coverslips pre-coated with poly-D-lysing and 0.01 mg/ml laminin (Corning) and cultured overnight in DH10 media supplemented with 25 pg/ml NGF (Corning) and 50 pg/ml GDNF (R&D Systems, Minneapolis, MN).
Approximately 24 hours after seeding, KNRK cells were loaded with Fura2-AM (Fisher) and imaged at 340 and 380 nm excitation. DRG neurons were imaged at 488 nm excitation. Images were acquired using a Nikon (Melville, NY) Ti-E microscope with a Photometries (Tucson, AZ) HQ2 camera. rhKLK7 (R&D Systems) and trypsin (ThermoFisher) were bath applied. 340/380 nm ratios and quantified GFP fluorescence were determined using Nikon NIS AR software.
Data analysis
All data are presented as the mean ± SEM. Statistical significance for two groups were determined using two-tailed, unpaired Student’s t test. Differences between three or more groups were tested using one-way ANOVA, followed by two tailed Student’s t tests. Differences were considered significant if p<0.05. Flow cytometry data was analyzed with Treestar (Ashland, OR) Flowjo v10. Graphs were generated using Graphpad (San Diego, CA) Prism 8 and R v3.5 (Vienna, Austria).
DATA AVAILBILITY STATEMENT
Datasets related to this article can be found at the GEO DataSets using accession numbers GSE90883 and TBD, hosted at www.ncbi.nlm.nih.gov.
Supplementary Material
ACKNOWLEDGEMENTS
Research in the Kim Lab is supported by the Doris Duke Charitable Foundation, Celgene Corporation, LEO Pharma, and the National Institute of Arthritis Musculoskeletal and Skin Diseases (NIAMS) of the National Institutes of Health [NIH (K08AR065577 and R01AR070116)]. MRM was supported by the NIH National Institute of Allergy and Infectious Diseases T325T32AI00716340. The Kim Lab has also been generously supported by donations from Anabeth and John Weil and Carole Kroeger. Research in the Liu Lab is supported by the National Institutes of Health (R01AI125743), Brain Research Foundation Fay/Frank Seed Grant, and Pew Scholar Award to QL.
The Klk7tml(KOMP)Vlcg mouse strain used for this research project was generated by the trans-NIH Knock-Out Mouse Project (KOMP) and obtained from the KOMP Repository (www.komp.org). NIH grants to Velocigene at Regeneron Inc (U01HG004085) and the CSD Consortium (U01HG004080) funded the generation of gene-targeted ES cells for 8500 genes, including Klk7, in the KOMP Program and archived and distributed by the KOMP Repository at UC Davis and CHORI (U42RR024244).
Footnotes
CONFLICT OF INTEREST
BSK has consulted for AbbVie, Inc., Concert Pharmaceuticals, Incyte Corporation, Menlo Therapeutics, and Pfizer, Inc. and has been an advisory board participant for Celgene Corporation, Kiniksa Pharmaceuticals, Menlo Therapeutics, Regeneron Pharmaceuticals, Inc., Sanofi, and Theravance Biopharma. BSK is a stockholder of Gilead Sciences, Inc. and Mallinckrodt Pharmaceuticals and founder and Chief Scientific Officer of Nuogen Pharma, Inc. EG has received grants (paid to Mount Sinai Health System), consulting fees, and/or honoraria from AbbVie, Allergan, Anacor, Asana Bioscience, Bristol-Myers Squibb, Celgene Corporation, Celsus Therapeutics, Curadim Pharma, Dermira, Drais, Eli Lilly, Escalier, Galderma, Genentech, Glenmark, Janssen Biotech, Kymab Limited, Kyowa Kirin, Lead Pharma, LEO Pharma, Merck Pharmaceuticals, Novartis, Pfizer, Regeneron, Sanofi, Sienna Biopharmaceuticals, Stiefel/GlaxoSmithKline, Theravance, and Vitae. MLC has consulted for MDOutlook. MRM, LKO, CJG, ABP and QL have no financial disclosures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Beck LA, Thaçi D, Hamilton JD, Graham NM, Bieber T, Rocklin R, et al. Dupilumab Treatment in Adults with Moderate-to-Severe Atopic Dermatitis. N. Engl. J. Med 2014;371(2):130–9 [DOI] [PubMed] [Google Scholar]
- Borgoño CA, Michael IP, Komatsu N, Jayakumar A, Kapadia R, Clayman GL, et al. A potential role for multiple tissue kallikrein serine proteases in epidermal desquamation. J. Biol. Chem 2007; [DOI] [PubMed] [Google Scholar]
- Brandt EB, Sivaprasad U. Th2 Cytokines and Atopic Dermatitis. J. Clin. Cell. Immunol 2011;2(3):1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briot A, Deraison C, Lacroix M, Bonnart C, Robin A, Besson C, et al. Kallikrein 5 induces atopic dermatitis–like lesions through PAR2-mediated thymic stromal lymphopoietin expression in Netherton syndrome. J. Exp. Med 2009; [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner PM, Guttman-Yassky E, Leung DYM. The immunology of atopic dermatitis and its reversibility with broad-spectrum and targeted therapies. J. Allergy Clin. Immunol Elsevier; 2017;139(4):S65–76 Available from: 10.1016/jjaci.2017.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner PM, Israel A, Zhang N, Leonard A, Wen H-C, Huynh T, et al. Early-onset pediatric atopic dermatitis is characterized by TH2/TH17/TH22-centered inflammation and lipid alterations. J. Allergy Clin. Immunol Elsevier; 2018;141(6):2094–106 Available from: 10.1016/jjaci.2018.02.040 [DOI] [PubMed] [Google Scholar]
- Caubet C, Jonca N, Brattsand M, Guerrin M, Bernard D, Schmidt R, et al. Degradation of corneodesmosome proteins by two serine proteases of the kallikrein family, SCTE/KLK5/hK5 and SCCE/KLK7/hK7. J. Invest. Dermatol 2004; [DOI] [PubMed] [Google Scholar]
- Chavanas S, Bodemer C, Rochat A, Hamel-Teillac D, Ali M, Irvine AD, et al. Mutations in SPINK5, encoding a serine protease inhibitor, cause Netherton syndrome. Nat. Genet 2000; [DOI] [PubMed] [Google Scholar]
- Czarnowicki T, He H, Krueger JG, Guttman-Yassky E. Atopic dermatitis endotypes and implications for targeted therapeutics. J. Allergy Clin. Immunol Elsevier; 2019;143(1):1–11 Available from: 10.1016/jjaci.2018.10.032 [DOI] [PubMed] [Google Scholar]
- DaVeiga SP. Epidemiology of atopic dermatitis: A review. Allergy Asthma Proc 2012. p. 227–34 [DOI] [PubMed] [Google Scholar]
- Deraison C, Bonnart C, Lopez F, Celine B, Robinson R, Jayakumar A, et al. LEKTI Fragments Specifically Inhibit KLK5, KLK7, and KLK14 and Control Desquamation through a Ph-dependent Interaction. Mol. Biol. Cell 2007; [DOI] [PMC free article] [PubMed] [Google Scholar]
- Descargues P, Draison C, Bonnart C, Kreft M, Kishibe M, Ishida-Yamamoto A, et al. Spink5-deficient mice mimic Netherton syndrome through degradation of desmoglein 1 by epidermal protease hyperactivity. Nat. Genet 2005; [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drucker AM, Wang AR, Li WQ, Sevetson E, Block JK, Qureshi AA. The Burden of Atopic Dermatitis: Summary of a Report for the National Eczema Association. J. Invest. Dermatol 2017. [DOI] [PubMed] [Google Scholar]
- Eichenfield LF, Tom WL, Chamlin SL, Feldman SR, Hanifin JM, Simpson EL, et al. Guidelines of care for the management of atopic dermatitis: section 1. Diagnosis and assessment of atopic dermatitis. J. Am. Acad. Dermatol 2014;70(2):338–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekholm IE, Brattsand M, Egelrud T. Stratum corneum tryptic enzyme in normal epidermis: a missing link in the desquamation process? J. Invest. Dermatol 2000;114(1):56–63 [DOI] [PubMed] [Google Scholar]
- Elias PM, Hatano Y, Williams ML. Basis for the barrier abnormality in atopic dermatitis: Outside-inside-outside pathogenic mechanisms. J. Allergy Clin. Immunol 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esaki H, Ewald DA, Ungar B, Rozenblit M, Zheng X, Xu H, et al. Identification of novel immune and barrier genes in atopic dermatitis by means of laser capture microdissection. J. Allergy Clin. Immunol 2015;135(1):153–63 Available from: http://www.sciencedirect.com/science/article/pii/S0091674914015796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerberg L, Hallstrom BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, et al. Analysis of the Human Tissue-specific Expression by Genome-wide Integration of Transcriptomics and Antibody-based Proteomics. Mol. & Cell. Proteomics 2014;13(2):397 LP–406 Available from: http://www.mcponline.org/content/13/2/397.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furio L, de Veer S, Jaillet M, Briot A, Robin A, Deraison C, et al. Transgenic kallikrein 5 mice reproduce major cutaneous and systemic hallmarks of Netherton syndrome. J. Exp. Med 2014; [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y-J, Ji R-R. c-Fos or pERK, Which is a Better Marker for Neuronal Activation and Central Sensitization After Noxious Stimulation and Tissue Injury? Open Pain J. 2009; [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman-Yassky E, Bissonnette R, Ungar B, Suarez-Farinas M, Ardeleanu M, Esaki H, et al. Dupilumab progressively improves systemic and cutaneous abnormalities in patients with atopic dermatitis. J. Allergy Clin. Immunol Elsevier; 2019;143(1):155–72 Available from: 10.1016/jjaci.2018.08.022 [DOI] [PubMed] [Google Scholar]
- Han L, Ma C, Liu Q, Weng HJ, Cui Y, Tang Z, et al. A subpopulation of nociceptors specifically linked to itch. Nat. Neurosci 2013; [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson L, Bâckman A, Ny A, Edlund M, Ekholm E, Hammarstrom BE, et al. Epidermal overexpression of stratum corneum chymotryptic enzyme in mice: A model for chronic itchy dermatitis. J. Invest. Dermatol 2002; [DOI] [PubMed] [Google Scholar]
- Howell MD, Kim BE, Gao P, Grant A V., Boguniewicz M, DeBenedetto A, et al. Cytokine modulation of atopic dermatitis filaggrin skin expression. J. Allergy Clin. Immunol 2007;120(1):150–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CC, Yang W, Guo C, Jiang H, Li F, Xiao M, et al. Anatomical and functional dichotomy of ocular itch and pain. Nat. Med 2018; [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasparek P, Ileninova Z, Zbodakova O, Kanchev I, Benada O, Chalupsky K, et al. KLK5 and KLK7 Ablation Fully Rescues Lethality of Netherton Syndrome-Like Phenotype. PLoS Genet. 2017; [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman BP, Guttman-Yassky E, Alexis AF. Atopic dermatitis in diverse racial and ethnic groups—Variations in epidemiology, genetics, clinical presentation and treatment. Exp. Dermatol 2018. [DOI] [PubMed] [Google Scholar]
- Kim BS. Innate Lymphoid Cells in the Skin. J. Invest. Dermatol 2014; [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BS, Siracusa MC, Saenz SA, Noti M, Monticelli LA, Sonnenberg GF, et al. TSLP elicits IL-33-independent innate lymphoid cell responses to promote skin inflammation. Sci. Transl. Med 2013;5(170):170ra16–170ra16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BS, Wang K, Siracusa MC, Saenz SA, Brestoff JR, Monticelli LA, et al. Basophils Promote Innate Lymphoid Cell Responses in Inflamed Skin. J. Immunol. (Baltimore, Md 1950) 2014;193(7):3717–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu N, Saijoh K, Kuk C, Liu AC, Khan S, Shirasaki F, et al. Human tissue kallikrein expression in the stratum corneum and serum of atopic dermatitis patients. Exp. Dermatol 2007; [DOI] [PubMed] [Google Scholar]
- Komatsu N, Saijoh K, Toyama T, Ohka R, Otsuki N, Hussack G, et al. Multiple tissue kallikrein mRNA and protein expression in normal skin and skin diseases. Br. J. Dermatol 2005; [DOI] [PubMed] [Google Scholar]
- Komatsu N, Takata M, Otsuki N, Toyama T, Ohka R, Takehara K, et al. Expression and localization of tissue kallikrein mRNAs in human epidermis and appendages. J. Invest. Dermatol 2003; [DOI] [PubMed] [Google Scholar]
- Leung DYM. Atopic dermatitis: Age and race do matter! J. Allergy Clin. Immunol 2015; [DOI] [PubMed] [Google Scholar]
- Li M, Hener P, Zhang Z, Kato S, Metzger D, Chambon P. Topical vitamin D3 and low-calcemic analogs induce thymic stromal lymphopoietin in mouse keratinocytes and trigger an atopic dermatitis. Proc. Natl. Acad. Sci. U. S. A 2006;103(31):11736–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–30 [DOI] [PubMed] [Google Scholar]
- Liu Q, Weng HJ, Patel KN, Tang Z, Bai H, Steinhoff M, et al. The distinct roles of two GPCRs, MrgprC11 and PAR2, in itch and hyperalgesia. Sci. Signal 2011; [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis DJ, Gupta J, Apter AJ, Ganguly T, Hoffstad O, Papadopoulos M, et al. Filaggrin-2 variation is associated with more persistent atopic dermatitis in African American subjects. J. Allergy Clin. Immunol 2014;133(3):784–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashiko S, Mehta H, Bissonnette R, Sarfati M. Increased frequencies of basophils, type 2 innate lymphoid cells and Th2 cells in skin of patients with atopic dermatitis but not psoriasis. J. Dermatol. Sci 2017; [DOI] [PubMed] [Google Scholar]
- Morizane S, Yamasaki K, Kajita A, Ikeda K, Zhan M, Aoyama Y, et al. TH2 cytokines increase kallikrein 7 expression and function in patients with atopic dermatitis. J. Allergy Clin. Immunol 2012; [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nattkemper LA, Tey HL, Valdes-Rodriguez R, Lee H, Mollanazar NK, Albornoz C, et al. The Genetics of Chronic Itch: Gene Expression in the Skin of Patients with Atopic Dermatitis and Psoriasis with Severe Itch. J. Invest. Dermatol 2018; [DOI] [PubMed] [Google Scholar]
- Oetjen LK, Kim BS. Interactions of the immune and sensory nervous systems in atopy. FEBS J. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oetjen LK, Mack MR, Feng J, Davidson S, Liu Q, Kim Correspondence BS, et al. Sensory Neurons Co-opt Classical Immune Signaling Pathways to Mediate Chronic Itch. Cell. Elsevier Inc; 2017;171:217–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa R, Akiyama M, Shimizu H. Filaggrin Gene Defects and the Risk of Developing Allergic Disorders. Allergol. Int 2011; [DOI] [PubMed] [Google Scholar]
- Palmer CNA, Irvine AD, Terron-Kwiatkowski A, Zhao Y, Liao H, Lee SP, et al. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat. Genet 2006;38(4):441–6 [DOI] [PubMed] [Google Scholar]
- Pavlopoulou A, Pampalakis G, Michalopoulos I, Sotiropoulou G. Evolutionary History of Tissue Kallikreins. PLoS One. 2010; [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw JL V, Diamandis EP. Distribution of 15 human kallikreins in tissues and biological fluids. Clin. Chem 2007; [DOI] [PubMed] [Google Scholar]
- Shimada SG, Shimada KA, Collins JG. Scratching behavior in mice induced by the proteinase- activated receptor-2 agonist, SLIGRL-NH2. Eur. J. Pharmacol 2006; [DOI] [PubMed] [Google Scholar]
- Silverberg JI. Public Health Burden and Epidemiology of Atopic Dermatitis. Dermatol. Clin 2017. p. 283–9 [DOI] [PubMed] [Google Scholar]
- Simpson EL, Bieber T, Guttman-Yassky E, Beck LA, Blauvelt A, Cork MJ, et al. Two Phase 3 Trials of Dupilumab versus Placebo in Atopic Dermatitis. N. Engl. J. Med 2016;375(24):2335–48 [DOI] [PubMed] [Google Scholar]
- Sotiropoulou G, Pampalakis G, Diamandis EP. Functional roles of human Kallikrein-related peptidases. J. Biol. Chem 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson K, Brattsand M, Roosterman D, Kempkes C, Bocheva G, Steinhoff M, et al. Activation of proteinase-activated receptor-2 by human kallikrein-related peptidases. J. Invest. Dermatol 2008; [DOI] [PubMed] [Google Scholar]
- Thaçi D, Simpson EL, Beck LA, Bieber T, Blauvelt A, Papp K, et al. Efficacy and safety of dupilumab in adults with moderate-to-severe atopic dermatitis inadequately controlled by topical treatments: A randomised, placebo-controlled, dose-ranging phase 2b trial. Lancet. 2016;387(10013):40–52 [DOI] [PubMed] [Google Scholar]
- Trier AM, Kim BS. Cytokine modulation of atopic itch. Curr. Opin. Immunol 2018;54:7–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilopoulos Y, Sharaf N, di Giovine F, Simon M, Cork MJ, Duff GW, et al. The 3’-UTR AACCins5874 in the stratum corneum chymotryptic enzyme gene (SCCE/KLK7), associated with atopic dermatitis; causes an increased mRNA expression without altering its stability. J. Dermatol. Sci Elsevier; 2011;61(2):131–3 [DOI] [PubMed] [Google Scholar]
- Yousef GM, Scorilas A, Magklara A, Soosaipillai A, Diamandis EP. The KLK7 (PRSS6) gene, encoding for the stratum corneum chymotryptic enzyme is a new member of the human kallikrein gene family - Genomic characterization, mapping, tissue expression and hormonal regulation. Gene. 2000; [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.