Abstract
The increasing number of deaths due to the COVID-19 pandemic has raised serious global concerns. Increased testing capacity and ample intensive care availability could explain lower mortality in some countries compared to others. Nevertheless, it is also plausible that the SARS-CoV-2 mutations giving rise to different phylogenetic clades are responsible for the apparent death rate disparities around the world. Current research literature linking the genetic make-up of SARS-CoV-2 with fatalities is lacking. Here, we suggest that this disparity in fatality rates may be attributed to SARS-CoV-2 evolving mutations and urge the international community to begin addressing the phylogenetic clade classification of SARS-CoV-2 in relation to clinical outcomes.
Keywords: COVID-19, SARS-CoV-2, Coronavirus, S-Protein, D614G
The rapid spread of novel SARS-CoV-2 across the globe has become a significant threat to humankind (Wang et al., 2020). Since the emergence of SARS-CoV-2, the emphasis on combatting the COVID-19 pandemic focused on social distancing and lockdown of cities to avoid medical care bottlenecks and to spread-out critical care over time. However, the escalating death toll in some countries when compared with others is alarming (World Health Organization 2020). Several factors, including but not limited to, age and comorbid health conditions, have been significantly linked to higher fatalities, especially in Italy (Onder et al., 2020). Such differences in fatality rates could be linked to genetic variations in SARS-CoV-2, but the literature about this is lacking.
Currently, complete SARS-CoV-2 sequences using NGS technology are being deposited at GISAID (Shu and McCauley, 2017). These sequences are being extensively scrutinized to understand their transmission, evolutionary and genetic relationships through phylogeny (Hadfield et al., 2018). However, it is unclear whether the SARS-CoV-2 genetic mutations influence its transmissibility and/or its virulence, which could account for higher fatalities in some countries. Here, we utilized the phylogenetic tree of 4,246 SARS-CoV-2 genomes obtained from GISAID (Shu and McCauley, 2017) and predict a possible link between higher fatalities and SARS-CoV-2 codon 614 in Spike protein (S), which replaces Aspartic acid (D) with Glycine (G) at this site. The SARS-CoV-2 genomes with the S-D614G variant have been classified under a larger phylogenetic clade G constituting most strains from Europe (Figure 1 A) (Shu and McCauley, 2017, Hadfield et al., 2018). Notably, mutations in the S protein likely induce conformational modifications that alter antigenicity (Phan, 2020), plausibly by mimicking the open status and facilitate the exposure of the cleavage domain to proteases, FURIN, or TMPRSS2, and could be sufficient to speed up the cleavage (Figure 2 ).
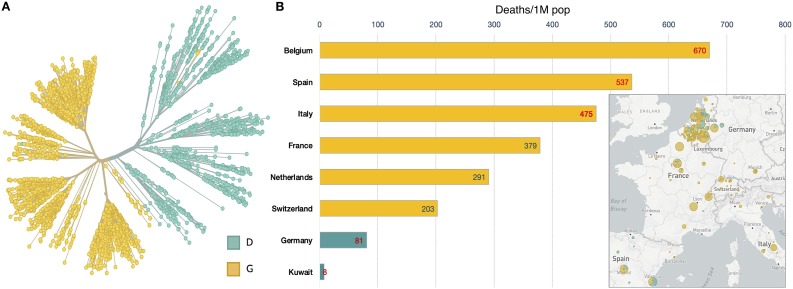
Figure 1.
(A) Unrooted phylogenetic tree with clear distinction of the 614G and 614D clades at the S protein site (https://nextstrain.org/ncov; last accessed May 2, 2020). (B) Bar plot showing the total number of deaths per million populaces (https://www.worldometers.info/coronavirus/; last accessed May 2, 2020). The map inside the box plot presents the distribution of S-614G and the wild-type S-614D in European populations (https://nextstrain.org/ncov; last accessed May 2, 2020) labeled in the y-axis of the bar plot.
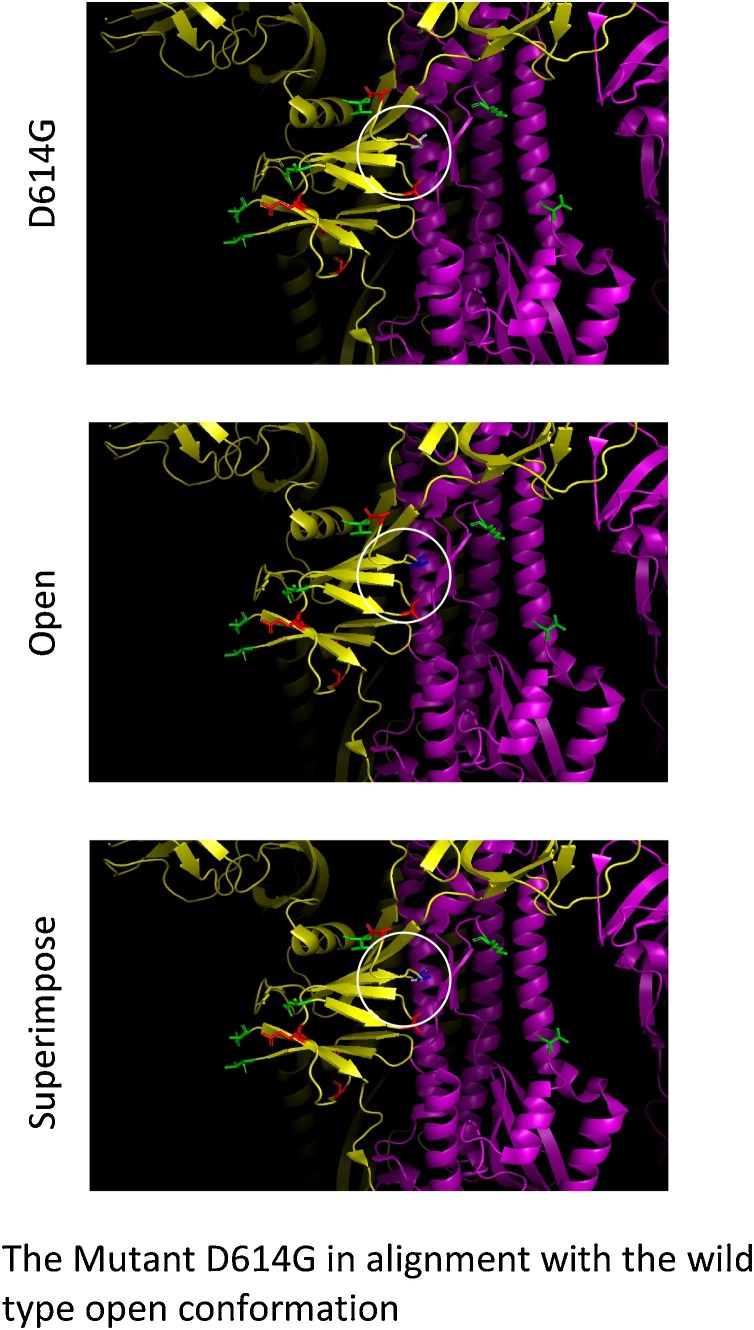
Figure 2.
The mutation observed at the S protein of the SARS-COV-2, D614G in white color, may create conformational changes mimicking the open status and facilitate the cleavage domain’s exposure to proteases FURIN or TMPRSS2 and could be sufficient to speed up the cleavage.
Strikingly, we observed that the variant S-D614G distinguishes the SARS-CoV-2 strains (Figure 1A) that may have caused fatal infections in European populations (Figure 1B). For example, a substantial number of strains with the S-D614G variant are from the countries Belgium, Spain, Italy, France, Netherlands, and Switzerland that top the death toll (Figure 1B) (https://www.worldometers.info/coronavirus/; last accessed May 2, 2020); while Germany and Kuwait, with a lower death toll, constitute most strains with the wild-type 614D at S (Figure 1B). It is also important to note that this scenario in Germany and other European countries remains uncertain as the number of cases is increasing every day. Therefore, continuous assessment and follow-up will be crucial to contain the virus.
Overall, our observation speculates that the S-D614G strains may be more virulent, increasing the severity in infected individuals, especially in Europe where this mutation is prominent. While our evidence is circumstantial, examining the amino acid 614 at S protein site (or other mutations) in mildly ill patients versus critically ill patients may be an asset in our arsenal against COVID-19. In the absence of any experimental studies, our interpretation with limited data will hopefully be helpful for researchers, at this time.
Ethical approval
Not required.
Conflict of interest
All authors have no conflict of interest to declare.
Author contributions
FA-M conceptualized the study. M.E. collected data, analyzed, and wrote the manuscript. AAM proposed protein-related predictions. FA-M critically appraised and edited the manuscript. All authors discussed the hypothesis, critically read and revised the manuscript, and gave final approval for publication.
Acknowledgments
We gratefully acknowledge the authors, originating and submitting laboratories of the sequence data from GISAID on which this research is based. This study was supported by the Kuwait Foundation for the Advancement of Sciences.
References
- Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C. Next strain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34(23):4121–4123. doi: 10.1093/bioinformatics/bty407. Available from: https://nextstrain.org/ncov. [Accessed 2 May 2020] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onder G., Rezza G., Brusaferro S. Case-fatality rate and characteristics of patients dying in relation to COVID-19 in Italy. JAMA. 2020 doi: 10.1001/jama.2020.4683. [DOI] [PubMed] [Google Scholar]
- Phan T. Genetic diversity and evolution of SARS-CoV-2. Infect Genet Evol. 2020;81 doi: 10.1016/j.meegid.2020.104260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y., McCauley J. GISAID: global initiative on sharing all influenza data – from vision to reality. EuroSurveillance. 2017;22(13) doi: 10.2807/1560-7917.ES.2017.22.13.30494. [Accessed 2 May 2020] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Horby P.W., Hayden F.G., Gao G. A novel coronavirus outbreak of global health concern. Lancet. 2020;395(10223):470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . 2020. Coronavirus disease (COVID-2019) situation reports.https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/ [Google Scholar]