Abstract
TYRP1 mRNA is of interest due to its potential non-coding role as a sponge sequestering tumor-suppressive miRs in melanoma. To our knowledge, there is no report on changes in TYRP1 expression in melanomas after development of resistance to targeted therapies. We used patient-derived drug-naïve RASQ61R and BRAFV600E melanoma cell lines. In BRAFV600E melanoma cells, resistance to vemurafenib and trametinib was developed. A time-lapse fluorescence microscope was used to rate proliferation, qRT-PCR and Western blotting were used to assess TYRP1 expression and MITF-M level and activity. A high TYRP1 protein level in RASQ61R cells corresponded with high TYRP1 mRNA level, whereas undetectable TYRP1 protein in BRAFV600E cells was accompanied by medium mRNA level, also in cells carrying NF1R135W variant in addition. TYRP1 expression was MITF-M-independent, since similar transcript status was found in MITF-Mhigh and MITF-Mlow cells. For the first time, we showed that TYRP1 expression remained unaltered in melanoma cells that became resistant to vemurafenib or trametinib, including those cells losing MITF-M. Also drug discontinuation in resistant cells did not substantially affect TYRP1 expression. To verify in vitro results, publicly available microarray data were analyzed. TYRP1 transcript levels stay unaltered in the majority of paired melanoma samples from patients before treatment and after relapse caused by resistance to targeted therapies. As TYRP1 mRNA level remains unaltered in melanoma cells during development of resistance to vemurafenib or trametinib, therapies developed to terminate a sponge activity of TYRP1 transcript may be extended to patients that relapse with resistant disease.
Keywords: TYRP1, MITF, miR sponge, Melanoma, Resistance, Targeted therapy
Introduction
Melanoma is the most deadly form of skin cancer. One of the important factors determining melanoma risk is the epidermal melanin content [45]. TYRP1 (tyrosinase-related protein 1) and two other enzymes, tyrosinase and TYRP2, are active in the melanin production. TYRP1 is a marker of melanocyte differentiation but it is also involved in the survival response to oxidative stress [16]. TYRP1 protein expression is reduced in invasive melanomas [4], while TYRP1 mRNA level is increased [28]. TYRP1 mRNA, in addition to the protein-coding function, has been recognized as contributing to regulation of gene expression by serving as an endogenous miR sponge [17].
In melanocytes, expression of about one hundred genes, including TYRP1, is microphthalmia-associated transcription factor (MITF)-dependent [6, 14, 18, 33, 42]. The M isoform of MITF (MITF-M) is one of the major players affecting melanoma phenotype [5, 20, 26]. Its regulation is complex and involves microenvironmental components [20, 24]. A high MITF-M level is connected with differentiation, a medium level with proliferation, whereas a low level is associated with an invasive and stem-like phenotype [5, 20, 26]. MITF-M also plays a prosurvival role in melanoma cells [21]. The MITF level is enhanced in BRAFV600E melanoma cells upon acute exposure to vemurafenib [35], and MITF inhibition increases sensitivity of cells to this inhibitor [1]. On the contrary, the MITF level and expression of several MITF-dependent genes are markedly reduced in vemurafenib-resistant melanomas resulting in more primitive phenotypes of melanoma cells [40]. Reduced MITF expression followed by the suppression of MITF-dependent pigmentation program were recently reported not only in vemurafenib-resistant cell lines but also in most of trametinib-resistant cell lines [8].
Therefore, we found it interesting to investigate changes of TYRP1 transcript levels in relation to MITF level and its activity shown as transcript levels of other MITF-dependent genes, SLC45A2, BIRC7/livin and BCL2A1, during the development of drug resistance. SLC45A2 (solute carrier family 45), together with TYRP1 belongs to pigmentation-related genes [44], whereas BIRC7 (baculoviral IAP repeat‐containing 7) and BCL2A1 (BCL2-related protein A1) encode prosurvival proteins [9, 32]. We assumed that diminution of MITF-M level during development of resistance would be accompanied with reduced expression of MITF-M-dependent genes. The question was whether TYRP1 mRNA would also be reduced. The answer is important as reduced level of the TYRP1 transcript may limit its function as a miR sponge in resistant cells. We performed our study in drug-naïve MITF-Mhigh and MITF-Mlow patient-derived melanoma cell lines and their vemurafenib- or trametinib-resistant counterparts, also subjected to drug discontinuation (‘drug holiday’).
Materials and methods
Drugs
Vemurafenib and trametinib were purchased from Selleck Chemicals LLC (Houston, TX, USA).
Melanoma cell line generation and culture
Tumor tissues from drug-naïve melanoma patients were processed as described previously [22]. The study was approved by Ethical Commission of Medical University of Lodz and informed consent was obtained from all individual participants included in the study. Melanoma cells were maintained in culture as described previously [37]. To generate lines resistant to vemurafenib or trametinib, cells were cultured for 4–5 months with increasing concentrations of drugs, from 1 to 10 µM and from 1 to 50 nM, respectively. For ‘drug holiday’ experiments, the drug was removed from the medium for 10 days.
A time-lapse fluorescence microscopy
Melanoma cells were grown in 96-well plates at 8 × 103 cells/well. For cell proliferation, a time-lapse fluorescence microscope system (IncuCyte, Essen Bioscience) was used. The data were analyzed using the IncuCyte Zoom original software. Proliferation was assessed as changes in the area occupied by cells (% of confluence) over time. It was expressed as % of confluence of cells at indicated time divided by % of confluence of cells at time 0.
RNA isolation and quantitative real-time PCR (qRT-PCR)
Extraction of RNA, cDNA synthesis and qRT-PCR were described previously [22]. Primer sequences are shown in Table 1. To calculate the relative normalized expression of target genes, a reference gene RPS17 and a mathematical model including an efficiency correction were applied.
Table 1.
Primer sequences, forward (F) and reverse (R) used in the qRT-PCR experiments
| Gene | Sequence | TM (oC) | Amplicon (bp) |
|---|---|---|---|
| BCL2A1 | F: GGATAAGGCAAAACGGAGGCTG | 62 | 183 |
| R: CAGTATTGCTTCAGGAGAGATAGC | 59 | ||
| BIRC7 | F: TGTCCACAGTGTGCAGGAGACT | 64 | 127 |
| R: GGCACTTTCAGACTGGACCTCT | 64 | ||
| MITF-M | F: GCTGGAAATGCTAGAATA | 57 | 379 |
| R: TTCCAGGCTGATGATGTC | 59 | ||
| RPS17 | F: AATCTCCTGATCCAAGGCTG | 60 | 142 |
| R: CAAGATAGCAGGTTATGTCACG | 58 | ||
| SLC45A2 | F: CTTTGCATCAGCCACCTCATTGG | 65 | 153 |
| R: TCCAACCTCGACTCCTCTTTCG | 64 | ||
| TYRP1 | F: GAAAAGAGCCACTTTGTCAGGG | 62 | 104 |
| R: CCATCTGGTCCCAGTATGTCT | 61 |
Cell lysate preparation and Western blotting
Cell lysate preparation and Western blotting were described elsewhere [34]. Antibodies detecting MITF (Cell Signaling, Danvers, MA, USA), TYRP1, GAPDH (Santa Cruz Biotechnology, Santa Cruz, CA, USA), or β-actin (Sigma-Aldrich) were used followed by binding of HRP-conjugated anti-mouse/anti-rabbit antibodies (Santa Cruz Biotechnology). The proteins were visualized using ChemiDoc Imaging System (Bio-Rad).
Analysis of TYRP1 expression reported in data sets from the Gene Expression Omnibus (GEO) database
The publicly available microarray data sets (accession numbers: GSE77940, GSE61992, GSE50509 and GSE99898) were downloaded from the GEO database (https://www.ncb.nlm.nih.gov). The TYRP1 expression profiles were developed from paired BRAFV600 melanoma samples from 31 patients in pretreatment stage and after relapse due to development of resistance to either vemurafenib or dabrafenib, and from paired melanoma samples from 17 patients before treatment and after relapse due to resistance to a combination of dabrafenib and trametinib. Gene expression values were log2 transformed.
Statistical analysis
Graphs represent mean ± SD of three biological replicates. Student’s t test was used to determine significant differences between the mean values. The difference was considered significant if p ≤ 0.05.
Results
TYRP1 expression in patient-derived melanoma cell lines is MITF-M-independent
Eight patient-derived melanoma cell lines were used initially in this study. Six of them harbor a mutation leading to BRAFV600E, and two of them harbor a RAS variant, either NRASQ61R (DMBC22 cell line) or HRASQ61R (DMBC17 cell line) [23]. DMBC28 cell line harbors an NF1R135W variant in addition to BRAFV600E alteration [23]. They exerted different proliferation rates as assessed by time-lapse fluorescence microscopy (Fig. 1a). TYRP1 expression was MITF-M-independent, since in all BRAFV600E melanoma cell lines, both MITF-Mhigh and MITF-Mlow, levels of TYRP1 transcript were similar (Fig. 1b). Interestingly, TYRP1 mRNA levels in cells carrying mutation either in NRAS or in HRAS were much higher than in BRAFV600E melanoma cells. This was well reflected at the protein level as TYRP1 protein was undetectable in BRAFV600E melanoma cells, whereas for RASQ61R cells a strong signal was obtained (Fig. 1c).
Fig. 1.
TYRP1 transcript level in patient-derived melanoma cell lines is MITF-M-independent and not related to proliferation rate. a Proliferation time-courses. Cell proliferation was monitored by analyzing the occupied area of cell images over time using IncuCyte, and it is shown as an increase in cell confluency relative to the confluency at time 0. n = 3 b Expression of TYRP1 and MITF-M was determined by qRT-PCR and normalized to the expression of a reference gene RPS17. Gene expression levels in each melanoma cell line are expressed relative to the median value of all eight cell lines. Bars represent mean values ± SD, n = 3. c Representative Western blot images showing basal levels of TYRP1 protein in tested melanoma cell lines. β-Actin was used as a loading control. d Expression of three MITF-dependent genes, SLC45A2, BIRC7 and BCL2A1, was determined in two MITF-Mlow and two MITF-Mhigh melanoma cell lines by qRT-PCR and normalized to the expression of a reference gene RPS17. Gene expression levels in each melanoma cell line are expressed relative to the median value of all four cell lines. Bars represent mean values ± SD, n = 3
To verify whether only TYRP1 expression was MITF-M-independent, the transcript levels of other genes, BIRC7, BCL2A1 and SLC45A2, previously recognized as regulated by MITF-M, were assessed in MITF-Mlow and MITF-Mhigh cell lines. Transcript levels of these genes in MITF-Mlow cell lines were substantially lower than in MITF-Mhigh cell lines (Fig. 1d). These results indicate that among the genes whose expression was evaluated only TYRP1 was MITF-M-independent.
TYRP1 transcript level is similar in drug-naïve, vemurafenib- and trametinib-resistant melanoma cells, also those on ‘drug holiday’
First, we modeled the vemurafenib resistance in four BRAFV600E melanoma cell lines. During development of resistance, the MITF-M protein level was substantially diminished in MITF-Mhigh cells and remained low/undetectable in MITF-Mlow melanoma cells (Fig. 2a). TYRP1 protein could not be detected in vemurafenib-resistant cells, similarly as in their drug-naïve counterparts (Fig. 2a). Changes in MITF-M level were reflected in its activity shown as downregulation of three MITF-M-dependent genes, SLC45A2, BIRC7 and BCL2A1 (Fig. 2b). On the contrary, TYRP1 transcript level was stable in vemurafenib-resistant cell lines regardless of changes in the MITF-M level (Fig. 2b). This further supports the notion that TYRP1 expression is MITF-M-independent. Interestingly, when vemurafenib-resistant cells were subjected to drug removal (“drug holiday”) for 10 days, MITF-M level and expression of all four tested genes remained unaffected (Fig. 2a, b). TYRP1 transcript levels, which were almost the same in drug-naïve, vemurafenib-resistant and on-drug-holiday BRAFV600E melanoma cells, were not markedly altered also in cells only shortly exposed to vemurafenib (Fig. 2c). Similarly, short treatment with trametinib, a MEK1/2 inhibitor, did not affect substantially the transcript level of TYRP1 in MITF-Mhigh cell lines, DMBC21 and DMBC28 (Fig. 2d). Reduction of MITF expression observed after development of resistance to trametinib in those cell lines was even potentiated after drug cessation (Fig. 2e). While this was associated with a significant downregulation of SLC45A2, BIRC7 and BCL2A1 (Fig. 2f), TYRP1 transcript level was not significantly altered (Fig. 2f).
Fig. 2.
TYRP1 expression is unaltered at the mRNA level and remains undetectable at the protein level during development of resistance to vemurafenib (PLX) and trametinib (TRA) in BRAFV600E melanoma cells regardless of changes in the MITF-M level. a Representative Western blot images showing comparison of TYRP1 and MITF-M protein levels in drug-naïve (–) vs. vemurafenib-resistant (PLXR) cell lines. The levels of TYRP1 and MITF-M in vemurafenib-resistant melanoma cells subjected to drug discontinuation (‘drug holiday’; PLXR DH) are included. GAPDH was used as a loading control. DMBC17 cell lysate was used in parallel as a positive control for TYRP1 and MITF-M staining. b Comparison of expression of TYRP1, SLC45A2, BIRC7 and BCL2A1 in vemurafenib-resistant BRAFV600E melanoma cell lines either exposed to drug or after drug discontinuation (‘drug holiday’), relative to expression in their drug-naïve counterparts. Bars represent mean values ± SD, n = 3, *p ≤ 0.05. c Expression of TYRP1 at the mRNA level in drug-naïve melanoma cells exposed to vemurafenib for 22 h, relative to its expression in untreated cells. Bars represent mean values ± SD, n = 3. d Expression of TYRP1 at the mRNA level in drug-naïve melanoma cells exposed to trametinib for 22 h, relative to its expression in control cells. Bars represent mean values ± SD, n = 3. e Representative Western blot images showing comparison of TYRP1 and MITF-M protein levels in drug-naïve (–) vs. trametinib-resistant (TRAR) cell lines, also subjected to drug discontinuation (‘drug holiday’; TRAR DH) are included. GAPDH was used as a loading control. f Comparison of expression of TYRP1, SLC45A2, BIRC7 and BCL2A1 in TRAR cell lines either exposed to drug or after drug discontinuation (TRAR DH), relative to expression in their drug-naïve counterparts. Bars represent mean values ± SD, n = 3. Expression of indicated genes in b–d and f was assessed by qRT-PCR and normalized to the expression of a reference gene RPS17
TYRP1 expression remains stable in the majority of melanomas during development of resistance to BRAFV600 and MEK1/2 inhibitors
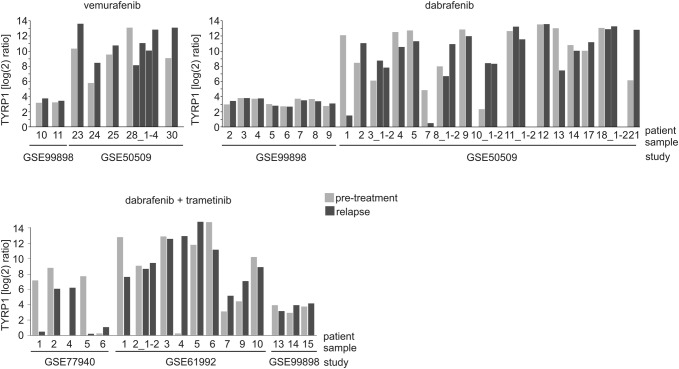
TYRP1 mRNA levels in melanomas from patients who developed resistance to targeted therapeutics were examined using publicly available data sets. The majority of relapsed melanomas did not show any evidence of TYRP1 transcript loss (Fig. 3). Among seven patients who developed resistance to vemurafenib only one exerted reduced TYRP1 expression in three out of four relapsed specimens. Among 24 patients treated with dabrafenib, another BRAFV600 inhibitor, the development of resistance could be linked with reduced TYRP1 expression only in 3 patients that relapsed, when compared with the pretreatment specimens from the same patients. In other relapsed specimens it was either markedly increased (2 specimens) or remained unchanged (19 specimens). Similar trend was observed when TYRP1 transcript level was analyzed in samples from patients treated with a combination of dabrafenib and trametinib. Among 17 patients, diminution of TYRP1 expression was found only in 4 patient specimens, while TYRP1 mRNA level increased in 6 samples and remained unaltered in other 7 specimens (Fig. 3). This analysis further supports the notion that the TYRP1 transcript level is stable also in clinical samples of relapsed melanomas that are resistant to BRAFV600 and BRAFV600 + MEK1/2 inhibitors.
Fig. 3.
TYRP1 expression at the mRNA level is not markedly changed in the majority of the relapsed patients who developed resistance to BRAFV600 inhibitors, vemurafenib or dabrafenib, or combined BRAFV600 and MEK1/2 inhibitors, dabrafenib and trametinib, in comparison to its expression before treatment. Data showing TYRP1 mRNA levels in paired melanoma specimens, pretreatment and post-relapse, are presented as light gray columns and dark gray columns, respectively. For some patients, more than one relapsed samples were examined. Results are expressed as log2 ratios normalized to the mean intensity of pretreatment specimens. Data were obtained from NCBI GEO (https://www.ncbi.nlm.nih.gov/geo/), and study identifiers are indicated.
Discussion
Elevated TYRP1 mRNA levels were detected in metastatic melanoma biopsies [3, 16], and correlated with poor overall patient survival [10–12, 28]. Unlike TYRP1 mRNA, TYRP1 protein did not correlate with overall survival, and TYRP1 protein was not detected in half of the melanoma samples expressing the TYRP1 transcript [11, 12, 28].
Our results indicate that TYRP1 protein is not detectable in the majority of samples that express TYRP1 at the transcript level. Moreover, mutation leading to BRAFV600E might be connected with a medium level of TYRP1 mRNA and undetectable TYRP1 protein, whereas mutation leading to either NRASQ61R or HRASQ61R might be associated with high levels of TYRP1 transcript and protein. It would be interesting to check whether there is any correlation between the molecular type of melanoma (mBRAF, mRAS, mNF1, or triple wild type) [38] and TYRP1 expression.
The high level of TYRP1 mRNA promotes proliferation and tumor growth irrespective of the protein level [17]. In our study, there was no strong association between TYRP1 mRNA level and cell proliferation rate. Melanoma cell lines with the highest proliferation rate were among lines expressing TYRP1 transcript at a medium level, whereas those with the highest expression of TYRP1 at the transcript and protein levels exerted one of the lowest proliferation rates. However, our results are in agreement with the findings of Gilot et al. [17] when cells lacking detectable TYRP1 protein and expressing TYRP1 mRNA at the level below the median value are exclusively considered. This suggests that the proliferation rate may be connected with TYRP1 mRNA level within this narrow range of TYRP1 expression.
The level of TYRP1 mRNA is a consequence of the transcriptional and post-transcriptional regulation [13–15, 18, 31, 41, 43]. It is possible that regulation of TYRP1 expression in MITF-Mlow melanoma cells is stabilized by other transcription factors or regulatory mechanisms. The single-nucleotide polymorphism rs683 located in the 3′-untranslated region of TYRP1 was connected with reduced binding of miR-155 and its mRNA decay activity [10, 31]. miRNA-155 expression is down-regulated in the majority of melanoma cell lines in comparison to melanocytes [30]. Therefore, influence of miR-155 on the TYRP1 transcript stability might be diminished during melanoma development. It has also been shown that miR-16, which interplays with miR-155, participates in the stabilization of TYRP1 mRNA [36].
TYRP1 expression is MITF-M-dependent in melanocytes [14]. A significant correlation between TYRP1 mRNA level and MITF level was found also in a subset of melanoma cell lines [11, 41]. Our results are contradictory to these findings as: (i) TYRP1 expression was similar in MITF-Mlow and MITF-Mhigh melanoma cells and (ii) MITF-M loss accompanying the development of drug resistance in MITF-Mhigh cells, did not influence TYRP1 expression. Interestingly, our results are consonant with an earlier study showing lack of significant changes in TYRP1 expression upon Mitf-transfection of Mitflow SK-MEL-28 melanoma cells [27]. The expression of other genes, BCL2A1, SLC45A2 and BIRC7, was markedly upregulated upon Mitf-transfection in that study [27], indicating that only the TYRP1 promoter was non-responsive to functional exogenous MITF.
The role of RNAs as endogenous miR sponges has been demonstrated [2, 7, 19, 25, 29, 39]. TYRP1 mRNA on its non-canonical miRNA response elements can sequester miR-16 and de-repress targets of miR-16 [17]. For the first time, we have shown that the TYRP1 transcript level is not affected during both acute response and development of resistance to vemurafenib and trametinib, which suggests that the role of TYRP1 mRNA as endogenous ‘miR sponge’ is preserved in resistant cells. Our findings are supported by the analysis of publicly available data on TYRP1 expression in melanoma samples from patients before treatment and at the time of tumor progression due to development of resistance to targeted therapeutics. This analysis confirms stable expression of TYRP1 in melanoma cells irrespective of their sensitivity to BRAFV600 and MEK1/2 inhibitors.
Conclusion
A ‘sponge’ activity of TYRP1 transcript in melanoma cells resistant to BRAFV600E and MEK1/2 inhibitors was not considered until now. Our results proving that TYRP1 expression is MITF-M-independent in melanoma cells suggest that regardless of the mechanisms that are responsible for the stable level of TYRP1 mRNA in melanoma cells, therapies developed to terminate a sponge activity of TYRP1 transcript might be extended to patients that relapse with vemurafenib/trametinib-resistant disease.
Acknowledgements
We thank Dr. Malgorzata Sztiller-Sikorska and Dr. Anna Gajos-Michniewicz for cell culture propagation, Prof. Markus Düchler for stimulating discussion and Ewa Lewandowska for excellent administrative and technical support.
Funding
This work was financially supported by Grant 2014/15/B/NZ7/00947 from National Science Centre (Poland).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Aida S, Sonobe Y, Tanimura H, Oikawa N, Yuhki M, Sakamoto H, Mizuno T. MITF suppression improves the sensitivity of melanoma cells to a BRAF inhibitor. Cancer Lett. 2017;409:116–124. doi: 10.1016/j.canlet.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 2.Babalola O, Mamalis A, Lev-Tov H, Jagdeo J. The role of microRNAs in skin fibrosis. Arch Dermatol Res. 2013;305:763–776. doi: 10.1007/s00403-013-1410-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boissy RE, Sakai C, Zhao H, Kobayashi T, Hearing VJ. Human tyrosinase related protein-1 (TRP-1) does not function as a DHICA oxidase activity in contrast to murine TRP-1. Exp Dermatol. 1998;7:198–204. doi: 10.1111/j.1600-0625.1998.tb00324.x. [DOI] [PubMed] [Google Scholar]
- 4.Bolander A, Agnarsdóttir M, Strömberg S, Ponten F, Hesselius P, Uhlen M, Berggvist M. The protein expression of TRP-1 and galectin-1 in cutaneous malignant melanomas. Cancer Genom Proteom. 2008;5:293–300. [PubMed] [Google Scholar]
- 5.Carreira S, Goodall J, Denat L, Rodriguez M, Nuciforo P, Hoek KS, Testori A, Larue L, Goding CR. Mitf regulation of Dia1 controls melanoma proliferation and invasiveness. Genes Dev. 2006;20:3426–3439. doi: 10.1101/gad.406406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheli Y, Ohanna M, Ballotti R, Bertolotto C. Fifteen-year quest for microphthalmia-associated transcription factor target genes. Pigment Cell Melanoma Res. 2010;23:27–40. doi: 10.1111/j.1755-148X.2009.00653.x. [DOI] [PubMed] [Google Scholar]
- 7.Chi SW, Hannon GJ, Darnell GB. An alternative mode of microRNA target recognition. Nat Struct Mol Biol. 2012;19:321–327. doi: 10.1038/nsmb.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Czyz M, Sztiller-Sikorska M, Gajos-Michniewicz A, Osrodek M, Hartman ML. Plasticity of drug-naïve and vemurafenib- or trametinib-resistant melanoma cells in execution of differentiation/pigmentation program. J Oncol. 2019;2019:1697913. doi: 10.1155/2019/1697913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dynek JN, Chan SM, Liu J, Zha J, Fairbrother WJ, Vucic D. Microphthalmia-associated transcription factor is a critical transcriptional regulator of melanoma inhibitor of apoptosis in melanomas. Cancer Res. 2008;68:3124–3132. doi: 10.1158/0008-5472.CAN-07-6622. [DOI] [PubMed] [Google Scholar]
- 10.El Hajj P, Gilot D, Migault M, Theunis A, Van Kempen LC, Salés F, Fayyad-Kazan H, Badran B, Larsimont D, Awada A, Bachelot L, Galibert MD, Ghanem G, Journe F. SNPs at miR-155 binding sites of TYRP1 explain discrepancy between mRNA and protein and refine TYRP1 prognostic value in melanoma. Br J Cancer. 2015;113:91–98. doi: 10.1038/bjc.2015.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El Hajj P, Journe F, Wiedig M, Laios I, Salès F, Galibert MD, Van Kempen LC, Spatz A, Badran B, Larsimont D, Awada A, Ghanem G. Tyrosinase-related protein 1 mRNA expression in lymph node metastases predicts overall survival in high-risk melanoma patients. Br J Cancer. 2013;108:1641–1647. doi: 10.1038/bjc.2013.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Falkenius J, Lundeberg J, Johansson H, Tuominen R, Frostvik-Stolt M, Hansson J, Egyhazi Brage S. High expression of glycolytic and pigment proteins is associated with worse clinical outcome in stage III melanoma. Melanoma Res. 2013;23:452–460. doi: 10.1097/CMR.0000000000000027. [DOI] [PubMed] [Google Scholar]
- 13.Fang D, Setaluri V. Role of microphthalmia transcription factor in regulation of melanocyte differentiation marker TRP-1. Biochem Biophys Res Commun. 1999;256:657–663. doi: 10.1006/bbrc.1999.0400. [DOI] [PubMed] [Google Scholar]
- 14.Fang D, Tsuji Y, Setaluri V. Selective down-regulation of tyrosinase family gene TYRP1 by inhibition of the activity of melanocyte transcription factor, MITF. Nucleic Acids Res. 2002;30:3096–3106. doi: 10.1093/nar/gkf424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galibert MD, Yavuzer U, Dexter TJ, Goding CR. Pax3 and regulation of the melanocyte-specific tyrosinase-related protein-1 promoter. J Biol Chem. 1999;274:26894–26900. doi: 10.1074/jbc.274.38.26894. [DOI] [PubMed] [Google Scholar]
- 16.Ghanem G, Journe F. Tyrosinase related protein 1 (TYRP1/gp75) in human cutaneous melanoma. Mol Oncol. 2011;5:150–155. doi: 10.1016/j.molonc.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilot D, Migault M, Bachelot L, Journé F, Rogiers A, Donnou-Fournet E, Mogha A, Mouchet N, Pinel-Marie ML, Mari B, Montier T, Corre S, Gautron S, Rambow F, El Hajj P, Ben Jouira R, Tartare-Deckert S, Marine JC, Felden B, Ghanem G, Galibert MD. A non-coding function of TYRP1 mRNA promotes melanoma growth. Nat Cell Biol. 2017;19:1348–1357. doi: 10.1038/ncb3623. [DOI] [PubMed] [Google Scholar]
- 18.Goding CR. Mitf from neural crest to melanoma: signal transduction and transcription in the melanocyte lineage. Genes Dev. 2000;14:1712–1728. [PubMed] [Google Scholar]
- 19.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 20.Hartman ML, Czyz M. MITF in melanoma: mechanisms behind its expression and activity. Cell Mol Life Sci. 2015;72:1249–1260. doi: 10.1007/s00018-014-1791-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartman ML, Czyz M. Pro-survival role of MITF in melanoma. J Invest Dermatol. 2015;135:352–358. doi: 10.1038/jid.2014.319. [DOI] [PubMed] [Google Scholar]
- 22.Hartman ML, Rozanski M, Osrodek M, Zalesna I, Czyz M. Vemurafenib and trametinib reduce expression of CTGF and IL-8 in V600EBRAF melanoma cells. Lab Invest. 2017;97:217–227. doi: 10.1038/labinvest.2016.140. [DOI] [PubMed] [Google Scholar]
- 23.Hartman ML, Sztiller-Sikorska M, Czyz M. Whole-exome sequencing reveals novel genetic variants associated with diverse phenotypes of melanoma cells. Mol Carcinogenesis. 2019;58:588–602. doi: 10.1002/mc.22953. [DOI] [PubMed] [Google Scholar]
- 24.Hartman ML, Talar B, Noman MZ, Gajos-Michniewicz A, Chouaib S, Czyz M. Gene expression profiling identifies microphthalmia-associated transcription factor (MITF) and Dickkopf-1 (DKK1) as regulators of microenvironment-driven alterations in melanoma phenotype. PLoS ONE. 2014;9:e95157. doi: 10.1371/journal.pone.0095157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hausser J, Zavolan M. Identification and consequences of miRNA-target interactions-beyond repression of gene expression. Nat Rev Genet. 2014;15:599–612. doi: 10.1038/nrg3765. [DOI] [PubMed] [Google Scholar]
- 26.Hoek KS, Eichhoff OM, Schlegel NC, Döbbeling U, Kobert N, Schaerer L, Hemmi S, Dummer R. In vivo switching of human melanoma cells between proliferative and invasive states. Cancer Res. 2008;68:650–656. doi: 10.1158/0008-5472.CAN-07-2491. [DOI] [PubMed] [Google Scholar]
- 27.Hoek KS, Schlegel NC, Eichhoff OM, Widmer DS, Praetorius C, Einarsson SO, Valgeirsdottir S, Bergsteinsdottir K, Schepsky A, Dummer R, Steingrimsson E. Novel MITF targets identified using a two-step DNA microarray strategy. Pigment Cell Melanoma Res. 2008;21:665–676. doi: 10.1111/j.1755-148X.2008.00505.x. [DOI] [PubMed] [Google Scholar]
- 28.Journe F, Boufker H, Van Kempen L, Galibert MD, Wiedig M, Salès F, Theunis A, Nonclercq D, Frau A, Laurent G, Awada A, Ghanem G. TYRP1 mRNA expression in melanoma metastases correlates with clinical outcome. Br J Cancer. 2011;105:1726–1732. doi: 10.1038/bjc.2011.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim D, Sung YM, Park J, Kim S, Kim J, Park J, Ha H, Bae JY, Kim S, Baek D. General rules for functional microRNA targeting. Nat Genet. 2016;48:1517–1526. doi: 10.1038/ng.3694. [DOI] [PubMed] [Google Scholar]
- 30.Levati L, Alvino E, Pagani E, Arcelli D, Caporaso P, Bondanza S, Di Leva G, Ferracin M, Volinia S, Bonmassar E, Croce CM, D’Afri S. Altered expression of selected microRNAs in melanoma: antiproliferative and proapoptotic activity of miRNA-155. Int J Oncol. 2009;35:393–400. [PubMed] [Google Scholar]
- 31.Li J, Liu Y, Xin X, Kim TS, Cabeza EA, Ren J, Nielsen R, Wrana JL, Zhang Z. Evidence for positive selection on a number of MicroRNA regulatory interactions during recent human evolution. PLoS Genet. 2012;8:e1002578. doi: 10.1371/journal.pgen.1002578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGill GG, Horstmann M, Widlund HR, Du J, Motyckova G, Nishimura EK, Lin YL, Ramaswamy S, Avery W, Ding HF, Jordan SA, Jackson IJ, Korsmeyer SJ, Golub TR, Fisher DE. Bcl2 regulation by the melanocyte master regulator Mitf modulates lineage survival and melanoma cell viability. Cell. 2002;109:707–718. doi: 10.1016/s0092-8674(02)00762-6. [DOI] [PubMed] [Google Scholar]
- 33.Murisier F, Guichard S, Beermann F. A conserved transcriptional enhancer that specifies Tyrp1 expression to melanocytes. Dev Biol. 2006;298:644–655. doi: 10.1016/j.ydbio.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 34.Osrodek M, Hartman ML, Czyz M. Physiologically relevant oxygen concentration (6% O2) as an important component of the microenvironment impacting melanoma phenotype and melanoma response to targeted therapeutics in vitro. Int J Mol Sci. 2019 doi: 10.3390/ijms20174203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rose AA, Annis MG, Frederick DT, Biondini M, Dong Z, Kwong L, Chin L, Keler T, Hawthorne T, Watson IR, Flaherty KT, Siegel PM. MAPK pathway inhibitors sensitize BRAF-mutant melanoma to an antibody-drug conjugate targeting GPNMB. Clin Cancer Res. 2016;22:6088–6098. doi: 10.1158/1078-0432.CCR-16-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soengas MS, Hernando E. TYRP1 mRNA goes fishing for miRNAs in melanoma. Nat Cell Biol. 2017;19:1311–1312. doi: 10.1038/ncb3637. [DOI] [PubMed] [Google Scholar]
- 37.Sztiller-Sikorska M, Koprowska K, Jakubowska J, Zalesna I, Stasiak M, Duechler M, Czyz ME. Sphere formation and self-renewal capacity of melanoma cells is affected by the microenvironment. Melanoma Res. 2012;22:215–224. doi: 10.1097/CMR.0b013e3283531317. [DOI] [PubMed] [Google Scholar]
- 38.The Cancer Genome Atlas Network Genomic classification of cutaneous melanoma. Cell. 2015;161:1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17:272–283. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- 40.Tsoi J, Robert L, Paraiso K, Galvan C, Sheu KM, Lay J, Wong DJL, Atefi M, Shirazi R, Wang X, Braas D, Grasso CS, Palaskas N, Ribas A, Graeber TG. Multi-stage differentiation defines melanoma subtypes with differential vulnerability to drug-induced iron-dependent oxidative stress. Cancer Cell. 2018;33:890–904. doi: 10.1016/j.ccell.2018.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vachtenheim J, Novotna H, Ghanem G. Transcriptional repression of the microphthalmia gene in melanoma cells correlates with the unresponsiveness of target genes to ectopic microphthalmia-associated transcription factor. J Invest Dermatol. 2001;117:1505–1511. doi: 10.1046/j.0022-202x.2001.01563.x. [DOI] [PubMed] [Google Scholar]
- 42.Widlund HR, Fisher DE. Microphthalamia-associated transcription factor: a critical regulator of pigment cell development and survival. Oncogene. 2003;22:3035–3041. doi: 10.1038/sj.onc.1206443. [DOI] [PubMed] [Google Scholar]
- 43.Wozniak M, Mielczarek A, Czyz M. miRNAs in melanoma: tumor suppressors and oncogenes with prognostic potential. Curr Med Chem. 2016;23:3136–3153. doi: 10.2174/1389557516666160831164544. [DOI] [PubMed] [Google Scholar]
- 44.Yamaguchi Y, Hearing VJ. Physiological factors that regulate skin pigmentation. BioFactors. 2009;35:193–199. doi: 10.1002/biof.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamaguchi Y, Takahashi K, Zmudzka BZ, Kornhauser A, Miller SA, Tadokoro T, Berens W, Beer JZ, Hearing VJ. Human skin responses to UV radiation: pigment in the upper epidermis protects against DNA damage in the lower epidermis and facilitates apoptosis. FASEB J. 2006;20:1486–1488. doi: 10.1096/fj.06-5725fje. [DOI] [PubMed] [Google Scholar]