Abstract
Autophagosome biogenesis is a dynamic membrane event, which is executed by the sequential function of autophagy-related (ATG) proteins. Upon autophagy induction, a cup-shaped membrane structure appears in the cytoplasm, then elongates sequestering cytoplasmic materials, and finally forms a closed double membrane autophagosome. However, how this complex vesicle formation event is strictly controlled and achieved is still enigmatic. Recently, there is accumulating evidence showing that some ATG proteins have the ability to directly interact with membranes, transfer lipids between membranes and regulate lipid metabolism. A novel role for various membrane lipids in autophagosome formation is also emerging. Here, we highlight past and recent key findings on the function of ATG proteins related to autophagosome biogenesis and consider how ATG proteins control this dynamic membrane formation event to organize the autophagosome by collaborating with membrane lipids.
Subject terms: Macroautophagy, Endoplasmic reticulum
Introduction
Under nutrient limited conditions cells survive by degrading their cellular contents to maintain homeostasis, energy levels, and building blocks. Macro-autophagy (hereafter, autophagy) is an intracellular degradation system that delivers cytoplasmic materials to the lysosome/vacuole. In response to autophagy induction, a cup-shaped membrane structure termed the phagophore (also known as the isolation membrane) appears in the cytoplasm. Once the phagophore elongates enough to accommodate its substrates, it closes and seals to form a double membrane structure, called an autophagosome (Fig. 1a). Initially autophagy was mainly analyzed in rat liver hepatocytes by electron microscopy1–3 and biochemical enzyme assays4,5. However, after the discovery of autophagy in the yeast Saccharomyces cerevisiae6, and the isolation of the yeast ATG (autophagy-related) genes7–9, research into autophagy has been transformed from a descriptive phenomenon into a biochemical and molecular field using model organisms, cell culture systems and in vitro reconstitution approaches. Core Atg/ATG proteins are functionally categorized into discrete units: the Atg1/ULK complex, the class III phosphatidylinositol 3-kinase (PI3K) complex, the Atg2-Atg18/WIPI4 complex, Atg9 vesicle, the Atg12 conjugation system, including ATG12–5-16L1 and WIPI2B, and the Atg8/LC3 conjugation system10–12. This molecular understanding of the autophagic machinery has led to the discovery of its importance in tumorigenesis13, mammalian development14,15, lipid metabolism16, degradation of intracellular pathogens17, and neurodegeneration18,19. In recent years, diverse physiological and pathological roles of autophagy have been uncovered20.
Fig. 1. ATG/Atg proteins control dynamic membrane events during autophagosome biogenesis.
a Autophagosome formation can be dissected into five different steps: initiation, nucleation, membrane expansion, closure, and fusion. b, c The intracellular distribution of ATG/Atg proteins under starvation-induced autophagy in mammalian (b)32,34–36,48,56,58,97,127,161,172 and yeast cells (c)33,46,47,55,126,130,154,155. Their localizations are categorized into five groups: -, not detectable; -/+, transient; +, weakly detectable; ++, easily detectable; +++, clearly detectable. Note that ATG/Atg proteins show punctate structures on the ER-related membranes rather than a typical ER-like pattern. ERES ER exit sites, ERGIC ER-Golgi intermediate compartment.
Autophagosome formation is driven by the ATG proteins, but during this dynamic membrane remodeling lipids are major constituents of autophagic membranes. Although the lipid composition of autophagosomes remains obscure, phosphatidylinositol 3-phosphate (PI3P) and phosphatidylethanolamine (PE) are crucial for autophagosome formation. The role of PI3P in autophagy was guided by a study showing the inhibitory activity of 3-methyladenine in rat hepatocytes21. Subsequently, it was shown that class III PI3K is a target of 3-methyladenine22,23. In line with this, Vps34 was identified as a PI3K in yeast24 and mammals25, and a binding partner of Vps30/Atg626. Finally, the Vps34-Vps15-Vps30/Atg6-Atg14 complex was characterized as an autophagy-specific class III PI3K complex26. PI3P is a minor lipid, but its formation is crucial for membrane recruitment of ATG proteins and the early stage of autophagosome formation (Fig. 1a). In contrast, PE is a major phospholipid in eukaryotic cells. Under starved conditions, Atg8/LC3/GABARAP proteins are bound to autophagic membranes through covalent conjugation to PE (Fig. 1a), which is a hallmark of autophagy27. Although Atg8/LC3/GABARAP proteins can be also conjugated to phosphatidylserine (PS) in vitro, PE is the major target of Atg8/LC3/GABARAP conjugation in vivo28,29.
Considering ATG protein recruitment to autophagic membranes is both spatially and temporally controlled, changes in the amount of PI3P and PE on autophagic membranes are unlikely to fully explain how ATG proteins execute their functions and accomplish autophagosome biogenesis. Here, we review key studies and recent findings on the molecular machinery underlying autophagosome formation, and focus on the recent literature investigating the co-operation of ATG proteins with membrane lipids during autophagy, and the emerging roles of membrane lipids in autophagosome formation.
The ULK/Atg1 complex: an initiator of autophagosome formation
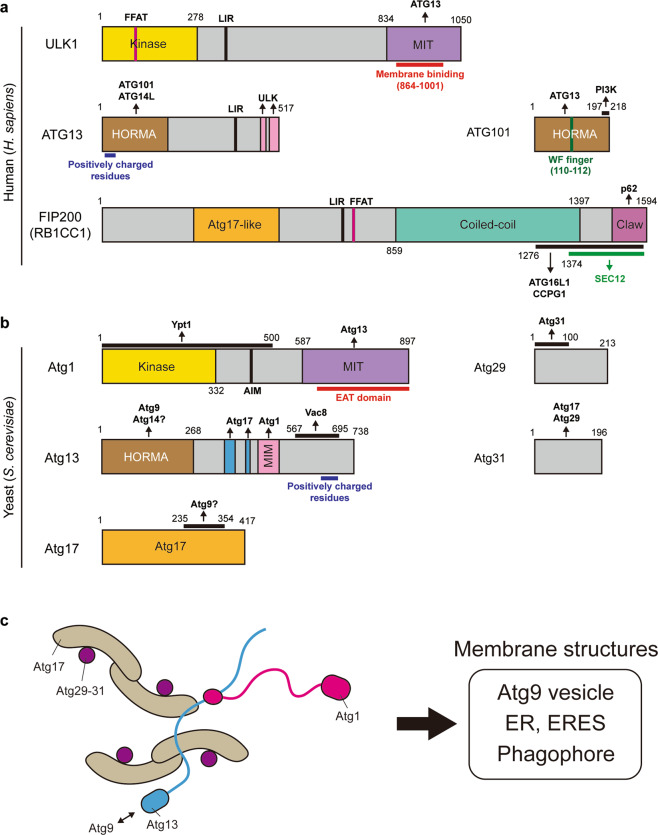
The ULK/Atg1 complex is one of the earliest factors to target to membrane structures at the phagophore formation site (Fig. 1b, c). This complex composed of four components (ULK1/2, ATG13, FIP200/RB1CC1, and ATG101) in mammals (Fig. 2a), and five components (Atg1, Atg13, Atg17, Atg29, and Atg31) in yeast (Fig. 2b), is a pivotal signal transducer of autophagy. So far, the identification of ULK/Atg1 substrates has been largely confined to ones involved in autophagy30,31. There is accumulating evidence showing that membrane recruitment of ULK/Atg1 complex at the initiation stage does not require downstream ATG proteins. In genetic hierarchical analyses, the puncta formation of ULK/Atg1 complex is independent of other Atg complexes in both yeast and mammalian cells32,33. A comprehensive biochemical analysis has shown that ULK complex components are bound to membranes even in the absence of downstream ATG proteins34. Importantly, under starved conditions the formation of the ULK complex puncta can occur during inhibition of PI3P synthesis although the size and lifespan of puncta are reduced34–36. Thus, the ULK/Atg1 complex is first recruited to membrane structures independently of PI3P and downstream ATG proteins, and then its membrane localization is stabilized by PI3P36. Yet, which components of the ULK/Atg1 complex act as determinants for membrane recruitment is unclear.
Fig. 2. The ULK/Atg1 complex is recruited to membrane structures to initiate autophagy.
a The domain structures of H. sapiens ULK complex components. b The domain structures of S. cerevisiae Atg1 complex components. c The proposed structure of the Atg1 complex234. EAT early autophagy targeting and tethering, MIT microtubule interacting and transport, MIM MIT-interacting motif, LIR LC3-interacting region, AIM Atg8 family-interacting motif, FFAT two phenylalanines in an acidic tract, HORMA Hop1/Rev7/Mad2.
To date, there have been several reports on the direct membrane association of the ULK/Atg1 complex components. A putative lipid-binding domain has been identified in ULK/Atg1 proteins. The C-terminus of ULK1 associates with cell membranes (Fig. 2a, red line)37, and in line with this, the Atg1 EAT domain (Fig. 2b, red line), a region that corresponds to the ULK1 C-terminus, binds to liposomes with a preference for small highly curved vesicles38,39, suggesting that the ULK/Atg1 C-terminus serves as a membrane curvature sensor. However, the Atg1 EAT domain needs to interact with Atg13 to induce autophagy40,41, which hinders the membrane association of Atg1 EAT domain38. Therefore, it is unclear if the Atg1 EAT domain in fully assembled Atg1 complex can associate with membranes. A recent paper has proposed that the Atg13-free Atg1 EAT domain is involved in later stages of autophagosome biogenesis42.
ATG13/Atg13 also has a putative lipid-binding region. Membrane binding of human ATG13 is regulated by a cluster of arginine/lysine residues at the N-terminus (Fig. 2a, blue line) via an electrostatic interaction with negatively charged phospholipids, such as phosphatidic acid and phosphatidylinositol phosphates (PIPs)36. Similarly, the C-terminus of yeast Atg13 has positively charged residues (Fig. 2b, blue line) and directly binds to liposomes containing negatively charged lipids in vitro. Thus, the lipid binding ability of Atg13 seems to be conserved from yeast to human, although the domain carrying the cluster of arginine/lysine residues is not conserved39,43.
Secretory pathway proteins can participate in membrane targeting of the ULK/Atg1 complex components. In yeast, a Rab GTPase Ypt1, which is essential for ER-Golgi and Golgi traffic, recruits Atg1 to the preautophagosomal structures (PAS) via an interaction with the N-terminus of Atg1 (Fig. 2b), although other Atg1 complex components, Atg13 and Atg17, are localized to the PAS independently of Ypt144. The initiation step has also been spatially linked to COPII vesicles, which mediate ER-to-Golgi transport45–48. In mammalian cells, SEC12, the activator of COPII assembly, is associated with the FIP200 C-terminus, although this interaction is mainly required for FIP200 function in the remodeling of ER exit sites (ERES), specialized ER regions for COPII vesicle formation48. A recent study has revealed that ULK1 and FIP200 interact with integral ER proteins, VAP proteins (VAPA and VAPB), which can tether the ER to other cell membranes at membrane contact sites49. VAP proteins directly bind FFAT (two phenylalanines in an acidic tract) motif-containing proteins to the ER50. Consistent with this, ULK1 and FIP200 have functional FFAT motifs, suggesting that membrane association of ULK1 and FIP200 is regulated by VAP proteins. Therefore, Ypt1, SEC12, and VAPs facilitate the initiation of autophagy by recruitment of the ULK complex components. However, given that autophagic phenotypes caused by the inhibition of these interactions are partial44,48,49, we suggest the leading players for the recruitment of the Atg1/ULK complex are still missing (Fig. 2c).
Late stage recruitment of ULK/Atg1 complex during autophagosome formation requires LC3/GABARAP/Atg8 proteins. ULK/Atg1 complex components have a LC3/GABARAP/ATG8-interacting (LIR/AIM) motif and bind to ATG8 family proteins (Fig. 2a, b)51–53. These interactions have been proposed to be involved in autophagosome maturation and/or a negative feedback by degrading the ULK/Atg1 complex via autophagy.
Atg9 vesicles: a membrane source for autophagosome formation
ATG9A/Atg9 is a multi-spanning membrane protein essential for the initiation of autophagosome formation (Fig. 1a). ATG9A/Atg9 cycles between different organelle compartments via vesicular transport pathways and delivered to the autophagosome formation site in response to induction of autophagy. In yeast Atg9 localizes to vesicular and tubular structures at the PAS under starved condition54. Atg9-containing vesicles, the diameter of which are 30–60 nm, are highly mobile within the cytoplasm55. As a small part of yeast Atg9 localizes to the autophagosomal outer membrane, it has been proposed that Atg9 vesicles become a seed membrane for phagophore formation in yeast55. In contrast, mammalian ATG9A is not obviously incorporated into autophagosomal membranes. Rather, ATG9A is found on clusters of vesicles and/or tubules in the vicinity of phagophores and transiently associated with the autophagosomal membranes32,56–58. Accordingly, it is thought that ATG9A supplies key components, such as proteins and lipids, to the autophagosomal membranes by transient association. Despite the apparent differences in the localization of ATG9A during autophagy initiation, the two models proposed in yeast and mammalian cells are not mutually exclusive.
Recently, the machinery sorting ATG9A/Atg9 to different locations in the cell has become increasingly clear. Atg9 distribution is partly regulated by the Atg1 complex via its physical interactions with other Atg proteins (Fig. 3a). Cytoplasmic regions of Atg9 interact with Atg1739,59. The N-terminus of Atg9 also binds to Atg13 HORMA domain, which facilitates recruitment of Atg9 vesicles to the PAS60. Atg9 is phosphorylated by Atg1 kinase, and this phosphorylation is required for recruitment of downstream factors such as Atg18 and Atg2, while it does not affect the PAS recruitment of Atg961. As in yeast, the trafficking of ATG9A is dependent on the ULK complex37,62,63 although further analysis is needed to confirm whether ATG9A directly interacts with the ULK complex.
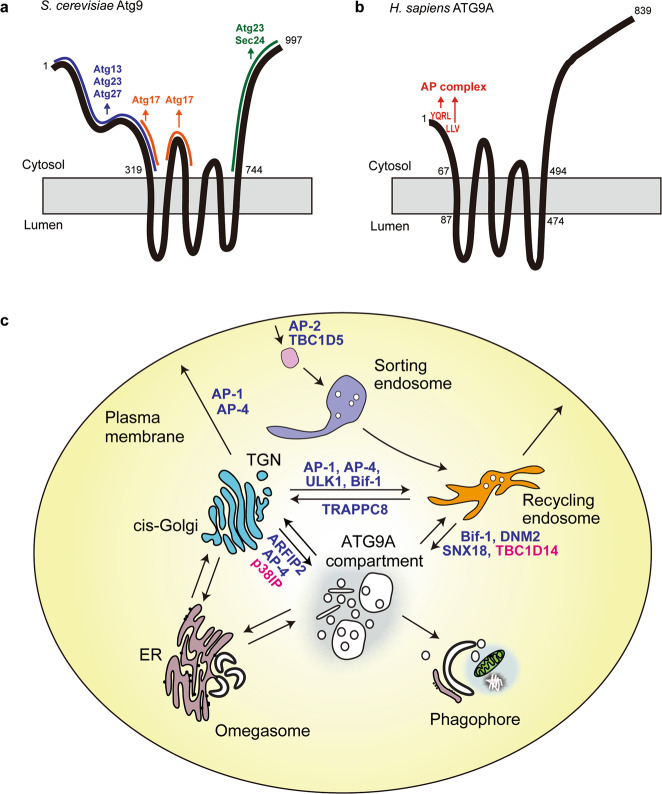
Fig. 3. Atg9/ATG9A vesicles work as a membrane source for autophagosome formation.
a, b The domain structure of S. cerevisiae Atg9 (a) and H. sapiens ATG9A (b) proteins. c Mammalian ATG9A cycles between different organelle compartments via vesicular transport pathways. Positive and negative regulators are shown in blue and red, respectively. AP adaptor protein, ARFIP2 arfaptin-2, SNX18 sorting nexin 18, DNM2 dynamin 2, TBC1D14 TBC1 domain family member 14, TRAPPC8 trafficking protein particle complex 8, p38IP p38-interacting protein.
ATG9A/Atg9 traffic is also controlled by the vesicular transport machinery. The N-terminal tail of ATG9A contains sorting signals recognized by adaptor protein (AP) complexes and interacts with AP-1, AP-2 and AP-4 complex subunits (Fig. 3b, c)63–65. ATG9A distribution is altered in AP-2 knockdown66 and AP-4 KO cells65,67–69: AP-2 likely mediates transport of ATG9A from the plasma membrane and recycling endosomes, while AP-4 regulates exit from the TGN. Yeast Atg9 does not have adaptor complex recognition signals, but instead Atg23 and Atg27 function to export Atg9 into Golgi-derived vesicles70. Rab GTPases, GEFs (guanine nucleotide exchange factor), and GAPs (GTPase-activating protein) also control ATG9A/Atg9 trafficking, mainly between the ERGIC, Golgi, and recycling endosomes. Ypt1 is recruited to Atg9 vesicles in yeast71,72. Similarly, ATG9A is distributed into RAB1- and RAB11-positive compartments73,74. Trs85/TRAPPC8, a subunit of TRAPPIII complex (a GEF for Ypt1/RAB1) and Trs130, a subunit of TRAPPII complex (a GEF for Ypt31/Ypt32) regulate ATG9A/Atg9 traffic presumably by mediating Rab-GTP activity73,75,76. RabGAPs, TBC domain-containing proteins (TBC1D5 and TBC1D14), also contribute to ATG9A cycling66,73. TBC1D14, which has no detectable GAP activity, has been proposed to function as a effector for GTP-loaded RAB1157. TBC1D14 controls RAB1 activity by coupling with RAB11-GTP and the TRAPPIII complex73. Finally, N-BAR-containing proteins BIF-1 and sorting nexin SNX18 are involved in tubulation and fission of ATG9A-positive recycling endosomes by recruiting dynamin 277,78. p38IP, an ATG9A interactor, is required for the redistribution of ATG9A in response to starvation79. Interestingly, phosphatidylinositol 4-phosphate (PI4P) may also regulate Atg9 traffic as a phosphatidylinositol 4-phosphate kinase Pik1 is essential for Atg9 exit from the Golgi80.
The essential role of ATG9A/Atg9 in autophagy remains elusive, but a recent study has revealed a close relationship between ATG9A vesicle and PI4P metabolism81. The BAR-domain-containing protein Arfaptin-2 and two PI4Ks (PI4KIIα and PI4KIIIβ) have been identified as components of ATG9A vesicles. ATG9A vesicles transport PI4KIIIβ to the ER membrane promoting PI4P production at the initiation site, which facilitates recruitment of the ULK complex and initiation of autophagy. In this model, Arfaptin-2 has been proposed to serve as a regulator of ATG9A vesicles by modulating ATG9A exit from the Golgi complex81. PI4KIIα may also be delivered by ATG9A vesicles to provide PI4P for later stages of autophagosome maturation82. Thus, a key role of ATG9A in autophagy may be to supply PI4P to autophagosomal membranes. Very recently, using single-particle cryo-electron microscopy the structure of Arabidopsis thaliana ATG9 was reported at sub-nanometer resolution83. Future structural studies at even higher resolution are needed to reveal some of the unresolved roles of ATG9A/Atg9.
The class III PI3K complex I (PI3KC3-C1): a PI3P generator at the initiation site
The class III PI3K complex I (PI3KC3-C1), that is essential for the nucleation of autophagosomes, consists of Vps34/VPS34, Vps15/p150, Vps30/BECN1, and Atg14/ATG14L (Fig. 4a, b)26,84–86. A fifth subunit Atg38/NRBF2 facilitates the PI3KC3-C1 complex formation and further induces PI3KC3-C1 dimerization87–92. To generate PI3P, the PI3KC3-C1 needs to directly interact with membranes and recognize the substrate lipid, phosphatidylinositol (PI). Several studies have revealed that among the PI3KC3-C1 complex components Vps34/VPS34, Vps30/BECN1, and ATG14L have lipid-binding domains (Fig. 4a, b, red lines)93–96. ATG14L plays a key role in an ER-targeting of the PI3KC3-C197.
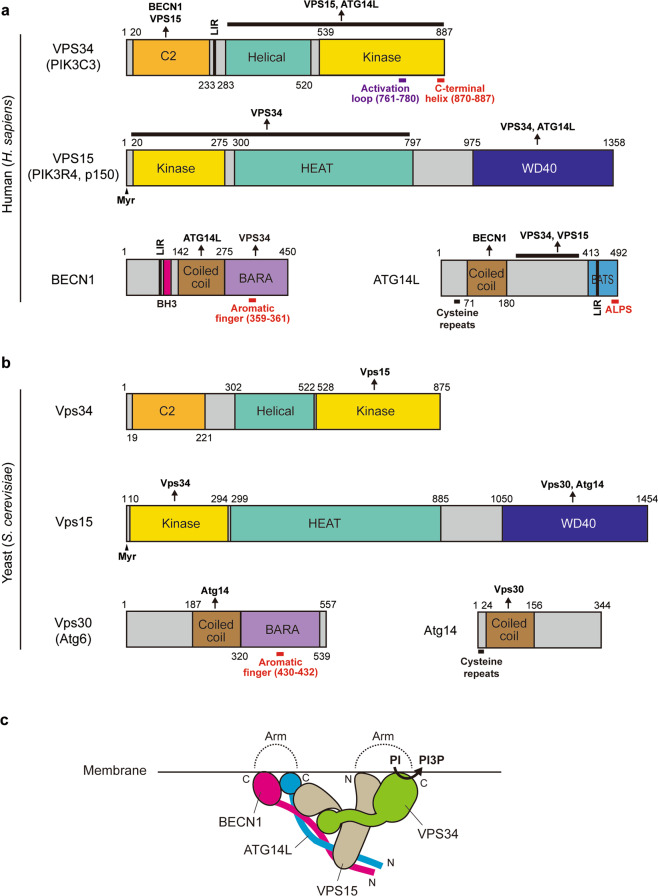
Fig. 4. The class III PI3K complex I (PI3KC3-C1) synthesizes PI3P at the autophagy initiation site.
a, b The domain structures of H. sapiens PI3KC3-C1 components (a) and S. cerevisiae PI3KC3-C1 components (b). c The proposed structure of mammalian PI3KC3-C1 complex. HEAT, Huntingtin, elongation factor 3, the PR65/A subunit of protein phosphatase 2A and the lipid kinase Tor; BH3 Bcl-2 homology 3, LIR LC3-interacting region, BARA β-α repeated autophagy-specific, Myr Myristoylation, BATS BAKOR and ATG14L autophagosome-targeting sequence, ALPS amphipathic lipid packing sensor.
Recent studies by single-particle electron microscopy and crystal structural analysis have provided many insights into how lipid-binding domains individually contribute to the membrane association of the PI3KC3-C1 complex95,98,99. The PI3KC3-C1 complex demonstrates a two-armed V-shaped architecture (Fig. 4c). One arm contains the helical and lipid kinase domains of Vps34/VPS34 and the Vps15/VPS15 N-terminal myristoylation site. The other arm includes the C-terminal domains of Vps30/BECN1 and Atg14/ATG14L, corresponding to BARA and BATS domains, respectively95,98. The crystal structure of Drosophila melanogaster Vps34 revealed a C-terminal loop region can interact with membranes and allow Vps34 activation93. The VPS34 C-terminus is regulated by acetylation at K771, which hinders the affinity of VPS34 for its substrate PI100. Aromatic residues in a surface loop of the BARA domain of Vps30/BECLIN1 serve as a hydrophobic finger to mediate direct association with membranes94,95,101,102. VPS34 catalytic site geometry is strongly influenced by the presence of VPS15, indicating that VPS15 has a central role in scaffolding complex assembly103. Accordingly, it is thought that the PI3KC3-C1 complex is associated with membranes via the tips of the two arms of the PI3KC3-C1 complex carrying the Vps34/VPS34 C-terminus, the myristoylated Vps15/VPS15 N-terminus, the aromatic finger in Vps30/BECN1 and the ATG14L C-terminal BATS domain (Fig. 4c). The BATS domain binds small liposomes containing PI3P or PI(4,5)P2 and senses membrane curvature via an amphipathic helix loop (ALPS motif, Fig. 4a)96. Thus, the VPS34 C-terminus determines the orientation of the PI3KC3-C1 complex, while ATG14L1 BATS domain is critical to sense membrane curvature and mediate the lipid specificity to target membranes95,99.
The PI3KC3-C1 complex components also contain functional LIR motifs and interact preferentially with GABARAP and GABARAPL1 (Fig. 4a). As the LIR motif in ATG14L is in close apposition to its BATS domain, these two motifs might work as a coincidence detector for specific targeting of autophagic membranes104.
Atg18/WIPI proteins: a transmitter of PI3P signals
Atg18/Atg21/WIPI proteins belong to the PROPPINs (β-propellers that bind polyphosphoinositides) family and work as PI3P effectors in autophagy. Atg21 is likely to be restricted to yeast and function mainly in selective autophagy105,106. A major role of Atg18/WIPI proteins is to transmit PI3P signals to downstream ATG proteins (Fig. 5a). In yeast, Atg18 interacts with Atg2 and regulates PAS recruitment of Atg233,107. In mammals, the Atg18 homolog WIPI4 interacts with ATG2 proteins108–110. WIPI2B most prominently functions in LC3 lipidation in mammals111, via its unique interaction with ATG16L110. Thus, individual WIPI proteins have some preferences for their binding partners. This is the case also for C.elegans: the WIPI3/4 ortholog EPG-6 binds to ATG-2, while the WIPI1/2 ortholog ATG-18 does not112. As C. elegans atg-18 and epg-6 mutants show different autophagic defects, it is thought that they act at distinct steps112. These functional differences of Atg18/WIPI proteins in autophagy are presumably caused by differences in their binding partners10,109,112.
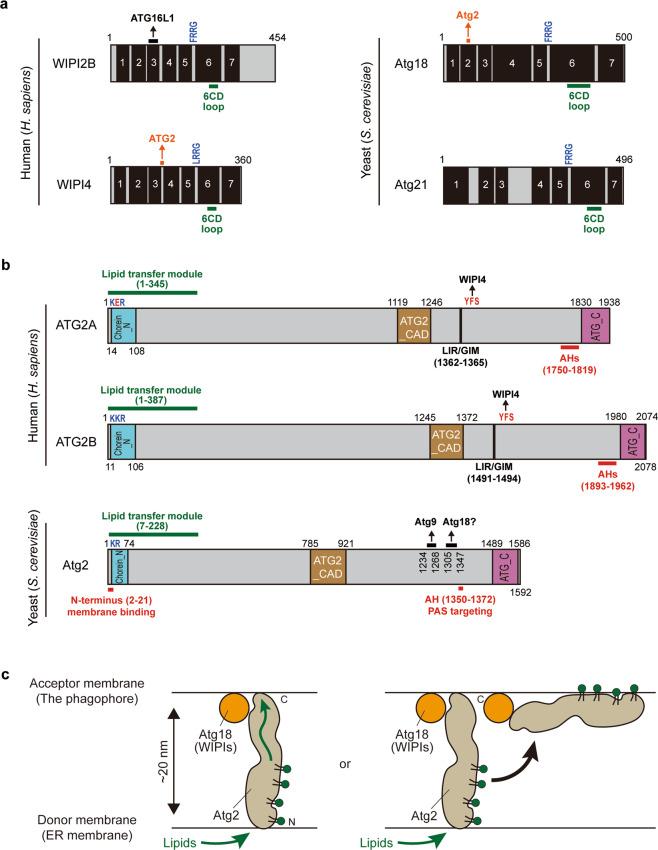
Fig. 5. Atg2/ATG2 functions as a membrane tether and lipid transfer protein in autophagy.
a The domain structures of WIPI2B and WIPI4 in human (left) and Atg18 and Atg21 in S. cerevisiae (right). b The domain structures of Atg2/ATG2 proteins. c Models of Atg2/ATG2-dependent lipid transfer. Chorein_N N-terminal region of Chorein or VPS13, Atg2_CAD autophagy-related protein 2 CAD motif, ATG_C autophagy-related protein C-terminal domain, LIR/GIM LC3/GABARAP-interacting region, AH amphipathic helix.
Atg18/WIPI proteins target to autophagic membranes by directly interacting with PI3P via a conserved FRRG motif (Fig. 5a, FRRG or LRRG)113,114. The PI3P binding of Atg18 proteins is required for full autophagic activity107,115. Structural analyses have revealed that Atg18 proteins fold into a seven-bladed β-propeller and contain two PI3P binding pockets at blades 5 and 6 which are composed of two arginine residues located in the conserved FRRG motif116,117. As Atg2/ATG2-binding and Atg16L1-binding sites are located at the opposite surface from the FRRG motif10,108,110,118,119, this architecture enables Atg18/WIPIs to interact with PI3P and downstream ATG proteins simultaneously.
In addition, a hydrophobic loop in blade 6 serves as a membrane anchor by inserting deeply into the lipid bilayer (Fig. 5a, 6CD loop)117,120. In Komagataella phaffii (previously known as Pichia pastoris), PIPs binding of PpAtg18 is modulated by phosphorylation of the loop region121. Of note, this loop region of yeast Atg18 can fold into an amphipathic α-helix, and is likely to be well conserved among Atg18/WIPI homologs122. The property of the loop might be one of main reasons why Atg18/WIPI proteins preferentially bind to smaller liposomes in vitro120 and localize to the edge of the phagophore in vivo46. Thus, direct membrane association of Atg18/WIPI proteins is controlled by two factors, membrane anchoring via the hydrophobic loop in blade 6 and PI3P-binding via the FRRG motif117,120. Furthermore, Atg2-Atg18 complex formation107,119,123 and Atg18/WIPI oligomerization109,124 can further stabilize the membrane association of Atg18/WIPI proteins.
Atg2/ATG2: a membrane tether and lipid transfer protein
The precise role of Atg2/ATG2 in autophagy has recently been established. Single particle EM analyses have resolved the architecture of Atg2-Atg18 and ATG2A/B-WIPI4 complexes108,110. Atg2/ATG2 and Atg18/WIPI4 demonstrate a rod-shaped and a globular structure, respectively. Atg2 has membrane-binding regions at both ends of the rod structure (Fig. 5b, red lines), the length of which is about ~200 Å (Fig. 5c)110,125,126, and can tether small liposomes in vitro110,126. WIPI4 is stably attached to one end of the ATG2 rod. In a complex with WIPI4, ATG2A can tether a PI3P-containing vesicle to another PI3P-free vesicle110. In line with this, high-resolution microscopy analyses have shown that Atg2/ATG2 localizes to the edge of the phagophore in close apposition to the ERES in yeast46,47 and contact sites between ER and autophagosomal membranes in mammalian cells127. The N-terminal 46 residues of Atg2 localizes to the ER, and the membrane-binding region in the C-terminal region is required for the targeting of the Atg2-Atg18 complex to the PAS126. Furthermore, both N-terminal and C-terminal regions of Atg2 are required to restore autophagy deficiency in atg2△ cells. Thus, Atg2/ATG2 serves as a tether for early autophagic structures to the ER membranes by collaborating with Atg18 in yeast and WIPI4 in mammals110,126. Yet, the contribution of WIPI4 is limited in mammals128,129. Instead, a functional LIR/GIM (LC3/GABARAP-interacting motif) has been found in ATG2, which is indispensable for autophagy129. Atg9 also facilitates Atg2-dependent contact site formation in yeast130. Thus, multiple factors contribute to Atg2/ATG2-dependent tethering activity.
In addition to the tethering function of Atg2/ATG2, it possesses lipid transfer activity. Recently, the crystal structure of the N-terminus of Atg2 has been solved revealing a tubular architecture with a hydrophobic cavity that can harbor tens of glycerophospholipids at once (Fig. 5b, green lines)131, and can transfer phospholipids with little head group specificity using in vitro liposome assays127,131. The Atg2/ATG2-dependent lipid transfer depends on packing defects and negatively charged membranes123,131. In line with these in vitro data, the N-terminal region of Atg2/ATG2 is essential for autophagic flux in both yeast and mammalian cells125–127,131. Surprisingly, overexpression of the Atg2 N-terminus can restore autophagy deficiency in ATG2A/B DKO cells127, suggesting that the tethering function of Atg2/ATG2 can be rescued by overexpression of the lipid transfer domain. Given that the Vps13 N-terminus, a homologous lipid transfer domain132, can be substituted for the corresponding region of Atg2 during autophagy131, the major function of Atg2/ATG2 in autophagy is lipid transport. How Atg2/ATG2 proteins accomplish unidirectional lipid transport and grow the phagophore membranes are key issues to be solved (Fig. 5c).
Atg16/ATG16L1: a determinant of Atg8/LC3 family lipidation sites
The Atg12–Atg5-Atg16/ATG12–ATG5-ATG16L1 complex is composed of the Atg12–Atg5/ATG12–ATG5 conjugate and a dimeric coiled-coil protein Atg16/ATG16L1. Atg12–Atg5/ATG12–ATG5 conjugate acts as an E3-like enzyme in the Atg8/LC3-PE conjugation reaction by facilitating the transfer of Atg8/LC3s/GABARAPs from an E2-like Atg3/ATG3 to PE (Fig. 6a)133–136. Although Atg16/ATG16L1 is dispensable for Atg8/LC3 lipidation reactions in vitro133,135, Atg16 can enhance Atg8 lipidation activity against low-curvature liposomes137 and immobilize Atg8-PE and Atg12–Atg5 complexes on membranes in vitro138. In addition, Atg16/ATG16L1 has a key role in determining the site of Atg8/LC3 lipidation by controlling the targeting of the Atg12–Atg5/ATG12–ATG5 conjugate in vivo139.
Fig. 6. Atg12–5-16/ATG12–5-16L1 and Atg3/ATG3 catalyze lipidation of Atg8/LC3 family proteins.
a The lipidation system of LC3. ATG4 cleaves the C-terminal residues of LC3 to expose glycine (G) residue. Then, LC3 is activated by ATG7 (E1 enzyme) and transferred to ATG3 (E2 enzyme). ATG12–5-16L1 complex facilitates the transfer of LC3 from ATG3 to PE. A PI3P-binding protein WIPI2B controls membrane recruitment of ATG12–5-16L1 complex under starvation condition. b The domain structures of H. sapiens ATG16L1 and S. cerevisiae Atg16 proteins. c The domain structures of H. sapiens ATG3 and S. cerevisiae Atg3 proteins. CC coiled-coil, AH amphipathic helix, FR flexible region, HR handle region, AIM Atg8 family-interacting motif.
The Atg12–Atg5-Atg16/ATG12–ATG5-ATG16L1 complex is recruited to PAS and the phagophore dependent on PI3P140–142. To date, several proteins have been reported as Atg16/Atg16L1-binding proteins (Fig. 6b). ATG16L1 interacts with Rab33B143, FIP200144–146 and WIPI2B10 via distinct amino acid residues in a region between a coiled-coil domain and a WD-repeat domain in ATG16L1. Furthermore, a WD-repeat domain in the C-terminus of mammalian ATG16L1 interacts with ubiquitin146. Similar interactions have been reported in yeast. Atg21, an Atg18 paralog, binds to Atg16147. The Atg16 complex associates with Atg1 complexes via the N-terminal region of Atg12148. Among these interactors, the ATG16L1-WIPI2B binding is the major PI3P-dependent interaction for membrane recruitment of ATG12–ATG5-ATG16L1 complex in starvation-induced autophagy10.
More recently, ATG16L1 was found to have the ability to bind lipids. ATG16L1 has three membrane-binding domains, in the N-terminal region, the coiled-coil domain (CCD) and the β-isoform-specific region. The N-terminal region, which contains an amphipathic helix (Fig. 6b, red lines), is universally required for the lipidation of LC3 family proteins, while the β-isoform-specific region (Fig. 6b, purple lines) is essential for VPS34-independent LC3 lipidation at perturbed endosomes149, while the CCD has an intrinsic ability to bind lipids that is also required for LC3 lipidation (Fig. 6b, green lines)150. In summary, the Atg16/ATG16L1 complex is recruited to the target membranes through multiple-interacting partners, including both proteins and lipids. These dependencies can be changed in response to a physiological context149–151.
Atg3/ATG3: an executor of Atg8/LC3 lipidation
Atg3/ATG3 acts as the E2-like enzyme that catalyzes lipidation of Atg8/LC3 family proteins by localizing to the PAS and the phagophore (Fig. 6a)152–155. An amphipathic helix found in the N-terminus of Atg3 is critical for its association with membranes and Atg8/LC3 lipidation (Fig. 6c, red lines)156–158. This region is involved in the preferential association of Atg3/ATG3 with membranes containing high curvatures and/or conical lipids, such as PE156. On the other hand, lipidation of Atg8/LC3 family proteins is stimulated by acidic phospholipids in vitro29,158. This is probably due to the fact that membrane association of ATG3 relies on membrane charge in addition to lipid packing defects, which lead to cavities for the interaction of peripheral proteins158. Atg3 localization is also regulated by its interaction with Atg8 as a positive feedback to facilitate Atg8 lipidation155. In addition, acetylation at K19 and K48 of Atg3 significantly enhances the membrane association of Atg3 in the presence of physiological level of PE159,160. Overall, these multiple factors mediate Atg3 membrane targeting in vivo.
The ER membrane: a platform for autophagosome formation
The membrane origins and sources for the autophagosome remain under debate, as several membrane sources have been proposed in mammals, including the ER161–163, mitochondria164, the PM165, the ERGIC166, the recycling endosome167,168, and lipid droplets (LD)169. Among these membrane sources it is becoming clear that the ER-related membranes are the primary membrane source and serve as a platform for autophagosome formation. In support of evidence showing the contribution of the ER membranes, omegasomes have been well characterized as an autophagy-related ER structure (ER subdomain): it is a PI3P-enriched membrane labeled by DFCP1, formed in response to starvation and dynamically connected to the ER and the phagophores161. Membrane contacts between the ER and phagophore have been observed by electron microscopy. Rough ER membranes are attached to both the outer and inner surfaces of cup-shaped phagophores162,163. Correlative light and electron microscopy (CLEM) and correlative cryo-fluorescence and cryo-soft X-ray microscopy (cryo-CLXM) have shown that omegasomes are composed of clusters of tubular/vesicular elements, part of which are continuous with the phagophores and/or the ER membranes170,171. These ER subdomains may also form organelle contact sites between the ER and other compartments, such as the mitochondria172, LDs173, and the PM174. It has been also shown that the edge regions of phagophore membranes are attached to the ERES46–48,175 and that COPII coat subunits are required for autophagosome formation45,176,177 and a COPII cargo protein is incorporated into autophagic membranes178, suggesting ER-derived COPII vesicles could contribute to autophagic membranes.
ATG proteins are sequentially recruited to the ER and autophagic membranes in response to autophagy induction (Fig. 1b, c). Their localization is changed depending on different stages of autophagosome formation. In mammalian cells, the ULK complex is first recruited to the ER membrane to initiate autophagy. At middle and late stages of autophagosome formation, the ULK complex also localizes to the omegasome, the phagophore and autophagosome32,34. ATG14L, a component of mammalian PI3KC3-C1, targets to both the ER-related membranes and phagophore to generate PI3P96,97,172, while the lipid transfer protein ATG2 is specifically localized to the ER and phagophore contact sites127. The unique localization of ATG2 presumably reflects its dual membrane targeting and tethering function. WIPI2B and ATG16L1 are mainly localized to the phagophore dependently on PI3P under starvation condition10. Given that it has been proposed that the ERGIC is a key membrane source for LC3 lipidation166, ATG12–5-16L1 and ATG3 are thought to be recruited to the ERGIC in addition to the phagophore membrane to execute LC3 lipidation. Similarly, yeast Atg proteins are also distributed to distinct compartments during autophagosome formation. Atg1 complex, Atg14, Atg12–5-16 complex, and Atg3 localize to the phagophore46,47,154,155, while Atg2, Atg18, and Atg9 are enriched at the edge regions of phagophore membranes, which is close apposition to the ERES46,47,130. On the other hand, there are some differences between yeast and mammalian cells. There are no counterparts to ERGIC and the omegasomes in yeast, but instead the phagophore is formed in the vicinity of the ERES and the vacuole46,47,130. Some Atg proteins such as Atg13, Atg17, and Atg14 are distributed to the vacuole and phagophore contact sites46. Further analyses are needed to reveal how the complex membrane targeting of Atg proteins is achieved in yeast and mammalian cells.
Membrane recruitment of the ULK complex precedes omegasome formation in mammalian cells. In high-resolution analysis using super-resolution microscopy, the initiation of autophagosome formation occurs in regions of the ER, where the ULK complex and the ATG9A vesicles are associated58. Interestingly, ER-localized phospholipid synthesizing enzymes, such as PI synthase and PS synthase, are enriched in close proximity to the autophagosome initiation site34, implying a close relationship between phospholipid synthesis and autophagosome formation. The enrichment of these enzymes might facilitate PI3P generation and ATG2-dependent lipid transfer.
VMP1 and TMEM41B: a key regulator of ion homeostasis during autophagosome formation?
VMP1 (also known as TMEM49) is a multi-spanning membrane protein localized at the ER and required for autophagosome formation. The VMP1 gene is absent in yeast, but conserved in most higher eukaryotes. Given its ER localization, VMP1 has been thought to be a key player in an ER-related event essential for autophagosome formation. Although VMP1-GFP puncta have been observed after overexpression, this puncta formation is not essential for the autophagy function of VMP1179,180. Impaired VMP1 function causes not only accumulation of abnormal autophagic structures181,182, but also pleiotropic effects, such as protein secretion defects183,184, impaired lipoprotein secretion185, abnormal distribution of PI4P and phospholipid metabolizing enzymes186, accumulation of lipid droplets179,187, and enhanced ER-organelle contact sites179,187. Accordingly, VMP1 function is not limited to autophagy, and the molecular mechanism underlying the abnormalities of VMP1-deficient cells remains unclear.
A recent study has revealed a new functional link between VMP1 and a calcium pump SERCA (sarcoendoplasmic reticulum calcium transport ATPase). VMP1 interacts with SERCAs that transport cytosolic Ca2+ into the ER lumen and positively regulates their Ca2+-ATPase activity, suggesting that VMP1 coordinates multiple organelle contact sites by maintaining local Ca2+ levels via SERCA activity regulation179. More detail about VMP1 function has been provided by studies on TMEM41B, a newly identified autophagy-related gene in genome-wide CRISPR screens180,188,189. Interestingly, TMEM41B localizes to the ER and shares a similar protein structure with VMP1. As TMEM41B and VMP1 form a complex, and overexpression of VMP1 restores impaired autophagic flux in TMEM41B KO cells, it has been proposed that they may be half-transporters and function together in autophagy as a full transporter by forming a complex. More work is needed to support this exciting hypothesis and the role of VMP1 and TMEM41B in ion homeostasis during autophagosome formation.
Emerging roles of lipid metabolism in autophagosome biogenesis
There is accumulating evidence that autophagy can be regulated by sphingolipids190–192. Treatment with a short-chain ceramide (C2-ceramide) is able to induce autophagy in various different cell lines, such as human cervical cancer cells, colon cancer cells, and malignant glioma cells193–197. Consistent with these reports, de novo synthesis of ceramide is required for the induction of autophagy in stimulated RAW264.7 macrophage cells198 and the mouse liver199, as well as in yeast200. The pro-autophagic effects by ceramide were proposed to be caused by upregulation of BECN1 activity and/or interfering with class I PI3K/Akt signaling pathway194,197, yet the molecular details remain to be elucidated.
Sphingosine 1-phosphate (S1P) is a simple lysophospholipid known to promote cell survival. Recently, the diverse roles of S1P-metabolizing enzymes in autophagy are being discovered. Overexpression of sphingosine kinase 1 (SK1), which generates S1P from sphingosine, stimulates autophagy in MCF-7 cells and primary neurons201,202. Depletion of S1P phosphohydrolase 1, which mediates degradation of S1P by dephosphorylation, results in the induction of autophagy, suggesting that accumulation of S1P can promote autophagy203. S1P not only binds to S1P receptors at the cell surface204,205, but also acts on intracellular membranes202,203, but how intracellular S1P works on autophagy remains unclear. Notably, S1P is cleaved by SGPL1 (sphingosine phosphate lyase 1) into hexadecenal and ethanolamine phosphate, which can be consumed for the synthesis of PE. Given that autophagosome formation is compromised in SGPL1-deficient brains206, PE generated from the S1P degradation products might play a key role in autophagy in neurons. Sphingomyelin is also involved in autophagy. Overloading of sphingomyelin affects ATG9A transport and induces accumulation of immature autophagosomes207. The increase of ceramide 1-phosphate (C1P) at the Golgi induced by CPTP (C1P transfer protein) knockdown also affects ATG9A distribution208. Neutral sphingomyelinase 2 induces autophagy by increasing ceramide levels in the Golgi209.
The effect of fatty acids on autophagy has drawn some attention in recent years. Saturated fatty acid palmitate induces autophagy in several cell lines210–212, although it has an inhibitory effect in different experimental conditions213. Mono-unsaturated (oleate) and ω-6 poly-unsaturated fatty acids (arachidonic acid) also activate autophagy214–216. Yet, it is reported that the underlying mechanisms of saturated fatty acids- and mono-unsaturated fatty acids-induced autophagy are not the same216. On the other hand, stearoyl-CoA desaturase, an enzyme generating mono-unsaturated fatty acids, is indispensable for efficient autophagosome formation217–219. Therefore, the synthetic pathway of mono-unsaturated fatty acids also contributes to autophagy. Finally, a recent study has shown that trans-unsaturated fatty acids inhibit autophagy induced by saturated fatty acids220, implying complicated connections between autophagic effects caused by different types of fatty acids. Further analyses are needed to elucidate how individual fatty acids act on autophagy.
Autophagosome formation can be affected by changes in phospholipid metabolism. PLD1 hydrolyzes phosphatidylcholine (PC) to generate phosphatidic acid (PA). In mammalian cells, PLD1 localizes to autophagosome-related structures and facilitates autophagy221–223. It has been proposed that its product PA plays a key role in autophagosome formation221,224 and/or lysosomal functions223. However, contrary to these findings, another study has shown that the inhibition of PLD1 results in an enhancement of autophagic flux225. Therefore, PLD1 might function as both positive and negative regulator of autophagy in a context-dependent manner. On the other hand, a role for lipid droplets (LDs) in autophagosome biogenesis has been reported. LDs are made of a hydrophobic core of neutral lipids and a surrounding phospholipid monolayer. In mammalian cells, a neutral lipase PNPLA5 positively regulates autophagy169. In line with this study, autophagy is impaired upon nitrogen starvation in LD-deficient yeast mutant173,226. Interestingly, a recent study has found that the autophagy deficiency in the LD-deficient yeast mutant can be improved by suppressing fatty acid synthesis or by restoring the altered phospholipid composition, indicating that LDs are not necessarily essential for autophagosome biogenesis, and that LDs play a key role in autophagy regulation by relieving metabolic stress caused by excess fatty acid synthesis and altered phospholipid composition227.
Concluding remarks
Thanks to the tremendous development of novel techniques, our knowledge of the molecular machinery underlying autophagosome formation has been expanded. Of note, accumulating evidence suggest that ATG proteins are directly involved in membrane lipid dynamics and organization during autophagy. While these findings have pushed the field forward, our current knowledge of lipid composition and distribution in autophagic membranes is very limited228. Therefore, advanced techniques to detect and evaluate lipid distribution and changes in lipid composition in vivo need to be further developed229,230. In vitro reconstitution231, in silico simulation232 and theoretical analysis233 are also required to obtain deeper insights into the relationship between ATG proteins and membrane lipids. In summary, it will be important to reveal how ATG proteins organize membrane lipids for understanding the detailed mechanisms of autophagosome biogenesis.
Acknowledgements
We thank Hayashi Yamamoto and members of the Tooze lab, especially Harold Jefferies, Javier Hervas, and Wenxin Zhang, for helpful discussions. This project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement No 788708) and the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001187); the UK Medical Research Council (FC001187); and the Welcome Trust (FC001187).
Author contributions
T.N. and S.A.T. wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Taki Nishimura, Email: taki.nishimura@crick.ac.uk.
Sharon A. Tooze, Email: sharon.tooze@crick.ac.uk
References
- 1.Dunn WA., Jr. Studies on the mechanisms of autophagy: formation of the autophagic vacuole. J. Cell Biol. 1990;110:1923–1933. doi: 10.1083/jcb.110.6.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deter RL, Baudhuin P, De Duve C. Participation of lysosomes in cellular autophagy induced in rat liver by glucagon. J. Cell Biol. 1967;35:C11–C16. doi: 10.1083/jcb.35.2.C11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Novikoff AB, Shin WY. Endoplasmic reticulum and autophagy in rat hepatocytes. Proc. Natl Acad. Sci. USA. 1978;75:5039–5042. doi: 10.1073/pnas.75.10.5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kopitz J, Kisen GO, Gordon PB, Bohley P, Seglen PO. Nonselective autophagy of cytosolic enzymes by isolated rat hepatocytes. J. Cell Biol. 1990;111:941–953. doi: 10.1083/jcb.111.3.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stromhaug PE, Berg TO, Fengsrud M, Seglen PO. Purification and characterization of autophagosomes from rat hepatocytes. Biochem. J. 1998;335(Pt 2):217–224. doi: 10.1042/bj3350217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 1992;119:301–311. doi: 10.1083/jcb.119.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-E. [DOI] [PubMed] [Google Scholar]
- 8.Harding TM, Morano KA, Scott SV, Klionsky DJ. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J. Cell Biol. 1995;131:591–602. doi: 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thumm M, et al. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 1994;349:275–280. doi: 10.1016/0014-5793(94)00672-5. [DOI] [PubMed] [Google Scholar]
- 10.Dooley HC, et al. WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1. Mol. Cell. 2014;55:238–252. doi: 10.1016/j.molcel.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 12.Lamb CA, Yoshimori T, Tooze SA. The autophagosome: origins unknown, biogenesis complex. Nat. Rev. Mol. Cell Biol. 2013;14:759–774. doi: 10.1038/nrm3696. [DOI] [PubMed] [Google Scholar]
- 13.Liang XH, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 14.Kuma A, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 15.Tsukamoto S, et al. Autophagy is essential for preimplantation development of mouse embryos. Science. 2008;321:117–120. doi: 10.1126/science.1154822. [DOI] [PubMed] [Google Scholar]
- 16.Singh R, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakagawa I, et al. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- 18.Hara T, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 19.Komatsu M, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 20.Morishita H, Mizushima N. Diverse cellular roles of autophagy. Annu. Rev. Cell. Dev. Biol. 2019;35:453–475. doi: 10.1146/annurev-cellbio-100818-125300. [DOI] [PubMed] [Google Scholar]
- 21.Seglen PO, Gordon PB. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc. Natl Acad. Sci. USA. 1982;79:1889–1892. doi: 10.1073/pnas.79.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blommaart EF, Krause U, Schellens JP, Vreeling-Sindelarova H, Meijer AJ. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur. J. Biochem. 1997;243:240–246. doi: 10.1111/j.1432-1033.1997.0240a.x. [DOI] [PubMed] [Google Scholar]
- 23.Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P. Distinct classes of phosphatidylinositol 3’-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J. Biol. Chem. 2000;275:992–998. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- 24.Schu PV, et al. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science. 1993;260:88–91. doi: 10.1126/science.8385367. [DOI] [PubMed] [Google Scholar]
- 25.Volinia S, et al. A human phosphatidylinositol 3-kinase complex related to the yeast Vps34p-Vps15p protein sorting system. EMBO J. 1995;14:3339–3348. doi: 10.1002/j.1460-2075.1995.tb07340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell Biol. 2001;152:519–530. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell. Dev. Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 28.Sou YS, Tanida I, Komatsu M, Ueno T, Kominami E. Phosphatidylserine in addition to phosphatidylethanolamine is an in vitro target of the mammalian Atg8 modifiers, LC3, GABARAP, and GATE-16. J. Biol. Chem. 2006;281:3017–3024. doi: 10.1074/jbc.M505888200. [DOI] [PubMed] [Google Scholar]
- 29.Oh-oka K, Nakatogawa H, Ohsumi Y. Physiological pH and acidic phospholipids contribute to substrate specificity in lipidation of Atg8. J. Biol. Chem. 2008;283:21847–21852. doi: 10.1074/jbc.M801836200. [DOI] [PubMed] [Google Scholar]
- 30.Mercer TJ, Gubas A, Tooze SA. A molecular perspective of mammalian autophagosome biogenesis. J. Biol. Chem. 2018;293:5386–5395. doi: 10.1074/jbc.R117.810366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Papinski D, Kraft C. Regulation of autophagy by signaling through the Atg1/ULK1 complex. J. Mol. Biol. 2016;428:1725–1741. doi: 10.1016/j.jmb.2016.03.030. [DOI] [PubMed] [Google Scholar]
- 32.Koyama-Honda I, Itakura E, Fujiwara TK, Mizushima N. Temporal analysis of recruitment of mammalian ATG proteins to the autophagosome formation site. Autophagy. 2013;9:1491–1499. doi: 10.4161/auto.25529. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- 34.Nishimura T, et al. Autophagosome formation is initiated at phosphatidylinositol synthase-enriched ER subdomains. EMBO J. 2017;36:1719–1735. doi: 10.15252/embj.201695189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Itakura E, Mizushima N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy. 2010;6:764–776. doi: 10.4161/auto.6.6.12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karanasios E, et al. Dynamic association of the ULK1 complex with omegasomes during autophagy induction. J. Cell Sci. 2013;126:5224–5238. doi: 10.1242/jcs.132415. [DOI] [PubMed] [Google Scholar]
- 37.Chan EY, Longatti A, McKnight NC, Tooze SA. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Mol. Cell Biol. 2009;29:157–171. doi: 10.1128/MCB.01082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ragusa MJ, Stanley RE, Hurley JH. Architecture of the Atg17 complex as a scaffold for autophagosome biogenesis. Cell. 2012;151:1501–1512. doi: 10.1016/j.cell.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rao Y, Perna MG, Hofmann B, Beier V, Wollert T. The Atg1-kinase complex tethers Atg9-vesicles to initiate autophagy. Nat. Commun. 2016;7:10338. doi: 10.1038/ncomms10338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fujioka Y, et al. Structural basis of starvation-induced assembly of the autophagy initiation complex. Nat. Struct. Mol. Biol. 2014;21:513–521. doi: 10.1038/nsmb.2822. [DOI] [PubMed] [Google Scholar]
- 41.Stjepanovic G, et al. Assembly and dynamics of the autophagy-initiating Atg1 complex. Proc. Natl Acad. Sci. USA. 2014;111:12793–12798. doi: 10.1073/pnas.1407214111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin MG, Schoneberg J, Davies CW, Ren X, Hurley JH. The dynamic Atg13-free conformation of the Atg1 EAT domain is required for phagophore expansion. Mol. Biol. Cell. 2018;29:1228–1237. doi: 10.1091/mbc.E17-04-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gatica, D. et al. The carboxy terminus of yeast Atg13 binds phospholipid membrane via motifs that overlap with the Vac8-interacting domain. Autophagy, in press. [DOI] [PMC free article] [PubMed]
- 44.Wang J, et al. Ypt1 recruits the Atg1 kinase to the preautophagosomal structure. Proc. Natl Acad. Sci. USA. 2013;110:9800–9805. doi: 10.1073/pnas.1302337110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ishihara N, et al. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol. Biol. Cell. 2001;12:3690–3702. doi: 10.1091/mbc.12.11.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suzuki K, Akioka M, Kondo-Kakuta C, Yamamoto H, Ohsumi Y. Fine mapping of autophagy-related proteins during autophagosome formation in Saccharomyces cerevisiae. J. Cell Sci. 2013;126:2534–2544. doi: 10.1242/jcs.122960. [DOI] [PubMed] [Google Scholar]
- 47.Graef M, Friedman JR, Graham C, Babu M, Nunnari J. ER exit sites are physical and functional core autophagosome biogenesis components. Mol. Biol. Cell. 2013;24:2918–2931. doi: 10.1091/mbc.e13-07-0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ge L, et al. Remodeling of ER-exit sites initiates a membrane supply pathway for autophagosome biogenesis. EMBO Rep. 2017;18:1586–1603. doi: 10.15252/embr.201744559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao YG, et al. The ER contact proteins VAPA/B interact with multiple autophagy proteins to modulate autophagosome biogenesis. Curr. Biol. 2018;28:1234–1245. doi: 10.1016/j.cub.2018.03.002. [DOI] [PubMed] [Google Scholar]
- 50.Murphy SE, Levine TP. VAP, a versatile access point for the endoplasmic reticulum: review and analysis of FFAT-like motifs in the VAPome. Biochim. Biophys. Acta. 2016;1861:952–961. doi: 10.1016/j.bbalip.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 51.Alemu EA, et al. ATG8 family proteins act as scaffolds for assembly of the ULK complex: sequence requirements for LC3-interacting region (LIR) motifs. J. Biol. Chem. 2012;287:39275–39290. doi: 10.1074/jbc.M112.378109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kraft C, et al. Binding of the Atg1/ULK1 kinase to the ubiquitin-like protein Atg8 regulates autophagy. EMBO J. 2012;31:3691–3703. doi: 10.1038/emboj.2012.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nakatogawa H, et al. The autophagy-related protein kinase Atg1 interacts with the ubiquitin-like protein Atg8 via the Atg8 family interacting motif to facilitate autophagosome formation. J. Biol. Chem. 2012;287:28503–28507. doi: 10.1074/jbc.C112.387514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mari M, et al. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J. Cell Biol. 2010;190:1005–1022. doi: 10.1083/jcb.200912089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamamoto H, et al. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J. Cell Biol. 2012;198:219–233. doi: 10.1083/jcb.201202061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Orsi A, et al. Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol. Biol. Cell. 2012;23:1860–1873. doi: 10.1091/mbc.e11-09-0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Longatti A, et al. TBC1D14 regulates autophagosome formation via Rab11- and ULK1-positive recycling endosomes. J. Cell Biol. 2012;197:659–675. doi: 10.1083/jcb.201111079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Karanasios E, et al. Autophagy initiation by ULK complex assembly on ER tubulovesicular regions marked by ATG9 vesicles. Nat. Commun. 2016;7:12420. doi: 10.1038/ncomms12420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sekito T, Kawamata T, Ichikawa R, Suzuki K, Ohsumi Y. Atg17 recruits Atg9 to organize the pre-autophagosomal structure. Genes Cells. 2009;14:525–538. doi: 10.1111/j.1365-2443.2009.01299.x. [DOI] [PubMed] [Google Scholar]
- 60.Suzuki SW, et al. Atg13 HORMA domain recruits Atg9 vesicles during autophagosome formation. Proc. Natl Acad. Sci. USA. 2015;112:3350–3355. doi: 10.1073/pnas.1421092112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Papinski D, et al. Early steps in autophagy depend on direct phosphorylation of Atg9 by the Atg1 kinase. Mol. Cell. 2014;53:471–483. doi: 10.1016/j.molcel.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Young AR, et al. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J. Cell Sci. 2006;119:3888–3900. doi: 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
- 63.Zhou C, et al. Regulation of mATG9 trafficking by Src- and ULK1-mediated phosphorylation in basal and starvation-induced autophagy. Cell Res. 2017;27:184–201. doi: 10.1038/cr.2016.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Imai K, et al. Atg9A trafficking through the recycling endosomes is required for autophagosome formation. J. Cell Sci. 2016;129:3781–3791. doi: 10.1242/jcs.196196. [DOI] [PubMed] [Google Scholar]
- 65.Mattera R, Park SY, De Pace R, Guardia CM, Bonifacino JS. AP-4 mediates export of ATG9A from the trans-Golgi network to promote autophagosome formation. Proc. Natl Acad. Sci. USA. 2017;114:E10697–E10706. doi: 10.1073/pnas.1717327114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Popovic D, Dikic I. TBC1D5 and the AP2 complex regulate ATG9 trafficking and initiation of autophagy. EMBO Rep. 2014;15:392–401. doi: 10.1002/embr.201337995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Davies AK, et al. AP-4 vesicles contribute to spatial control of autophagy via RUSC-dependent peripheral delivery of ATG9A. Nat. Commun. 2018;9:3958. doi: 10.1038/s41467-018-06172-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.De Pace R, et al. Altered distribution of ATG9A and accumulation of axonal aggregates in neurons from a mouse model of AP-4 deficiency syndrome. PLoS Genet. 2018;14:e1007363. doi: 10.1371/journal.pgen.1007363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ivankovic D, et al. Axonal autophagosome maturation defect through failure of ATG9A sorting underpins pathology in AP-4 deficiency syndrome. Autophagy. 2020;16:391–407. doi: 10.1080/15548627.2019.1615302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Backues SK, et al. Atg23 and Atg27 act at the early stages of Atg9 trafficking in S. cerevisiae. Traffic. 2015;16:172–190. doi: 10.1111/tra.12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lipatova Z, et al. Regulation of selective autophagy onset by a Ypt/Rab GTPase module. Proc. Natl Acad. Sci. USA. 2012;109:6981–6986. doi: 10.1073/pnas.1121299109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kakuta S, et al. Atg9 vesicles recruit vesicle-tethering proteins Trs85 and Ypt1 to the autophagosome formation site. J. Biol. Chem. 2012;287:44261–44269. doi: 10.1074/jbc.M112.411454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lamb CA, et al. TBC1D14 regulates autophagy via the TRAPP complex and ATG9 traffic. EMBO J. 2016;35:281–301. doi: 10.15252/embj.201592695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kakuta S, et al. Small GTPase Rab1B is associated with ATG9A vesicles and regulates autophagosome formation. FASEB J. 2017;31:3757–3773. doi: 10.1096/fj.201601052R. [DOI] [PubMed] [Google Scholar]
- 75.Shirahama-Noda K, Kira S, Yoshimori T, Noda T. TRAPPIII is responsible for vesicular transport from early endosomes to Golgi, facilitating Atg9 cycling in autophagy. J. Cell Sci. 2013;126:4963–4973. doi: 10.1242/jcs.131318. [DOI] [PubMed] [Google Scholar]
- 76.Zou S, et al. Trs130 participates in autophagy through GTPases Ypt31/32 in Saccharomyces cerevisiae. Traffic. 2013;14:233–246. doi: 10.1111/tra.12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Soreng, K. et al. SNX18 regulates ATG9A trafficking from recycling endosomes by recruiting Dynamin-2. EMBO Rep. 19, e44837 (2018). [DOI] [PMC free article] [PubMed]
- 78.Takahashi Y, et al. The Bif-1-Dynamin 2 membrane fission machinery regulates Atg9-containing vesicle generation at the Rab11-positive reservoirs. Oncotarget. 2016;7:20855–20868. doi: 10.18632/oncotarget.8028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Webber JL, Tooze SA. Coordinated regulation of autophagy by p38alpha MAPK through mAtg9 and p38IP. EMBO J. 2010;29:27–40. doi: 10.1038/emboj.2009.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang K, et al. Phosphatidylinositol 4-kinases are required for autophagic membrane trafficking. J. Biol. Chem. 2012;287:37964–37972. doi: 10.1074/jbc.M112.371591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Judith D, et al. ATG9A shapes the forming autophagosome through Arfaptin 2 and phosphatidylinositol 4-kinase IIIbeta. J. Cell Biol. 2019;218:1634–1652. doi: 10.1083/jcb.201901115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang H, et al. GABARAPs regulate PI4P-dependent autophagosome:lysosome fusion. Proc. Natl Acad. Sci. USA. 2015;112:7015–7020. doi: 10.1073/pnas.1507263112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lai LTF, et al. Subnanometer resolution cryo-EM structure of Arabidopsis thaliana ATG9. Autophagy. 2020;16:575–583. doi: 10.1080/15548627.2019.1639300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol. Biol. Cell. 2008;19:5360–5372. doi: 10.1091/mbc.e08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sun Q, et al. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc. Natl Acad. Sci. USA. 2008;105:19211–19216. doi: 10.1073/pnas.0810452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Matsunaga K, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat. Cell Biol. 2009;11:385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 87.Araki Y, et al. Atg38 is required for autophagy-specific phosphatidylinositol 3-kinase complex integrity. J. Cell Biol. 2013;203:299–313. doi: 10.1083/jcb.201304123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cao Y, et al. NRBF2 regulates macroautophagy as a component of Vps34 Complex I. Biochem. J. 2014;461:315–322. doi: 10.1042/BJ20140515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lu J, et al. NRBF2 regulates autophagy and prevents liver injury by modulating Atg14L-linked phosphatidylinositol-3 kinase III activity. Nat. Commun. 2014;5:3920. doi: 10.1038/ncomms4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhong Y, et al. Nrbf2 protein suppresses autophagy by modulating Atg14L protein-containing Beclin 1-Vps34 complex architecture and reducing intracellular phosphatidylinositol-3 phosphate levels. J. Biol. Chem. 2014;289:26021–26037. doi: 10.1074/jbc.M114.561134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ohashi Y, et al. Characterization of Atg38 and NRBF2, a fifth subunit of the autophagic Vps34/PIK3C3 complex. Autophagy. 2016;12:2129–2144. doi: 10.1080/15548627.2016.1226736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Young LN, Cho K, Lawrence R, Zoncu R, Hurley JH. Dynamics and architecture of the NRBF2-containing phosphatidylinositol 3-kinase complex I of autophagy. Proc. Natl Acad. Sci. USA. 2016;113:8224–8229. doi: 10.1073/pnas.1603650113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Miller S, et al. Shaping development of autophagy inhibitors with the structure of the lipid kinase Vps34. Science. 2010;327:1638–1642. doi: 10.1126/science.1184429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Huang W, et al. Crystal structure and biochemical analyses reveal Beclin 1 as a novel membrane binding protein. Cell Res. 2012;22:473–489. doi: 10.1038/cr.2012.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rostislavleva K, et al. Structure and flexibility of the endosomal Vps34 complex reveals the basis of its function on membranes. Science. 2015;350:aac7365. doi: 10.1126/science.aac7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fan W, Nassiri A, Zhong Q. Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L) Proc. Natl Acad. Sci. USA. 2011;108:7769–7774. doi: 10.1073/pnas.1016472108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Matsunaga K, et al. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J. Cell Biol. 2010;190:511–521. doi: 10.1083/jcb.200911141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Baskaran S, et al. Architecture and dynamics of the autophagic phosphatidylinositol 3-kinase complex. Elife. 2014;3:e05115. doi: 10.7554/eLife.05115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ma M, et al. Cryo-EM structure and biochemical analysis reveal the basis of the functional difference between human PI3KC3-C1 and -C2. Cell Res. 2017;27:989–1001. doi: 10.1038/cr.2017.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Su H, et al. VPS34 acetylation controls its lipid kinase activity and the initiation of canonical and non-canonical autophagy. Mol. Cell. 2017;67:907–921. doi: 10.1016/j.molcel.2017.07.024. [DOI] [PubMed] [Google Scholar]
- 101.Noda NN, et al. Structure of the novel C-terminal domain of vacuolar protein sorting 30/autophagy-related protein 6 and its specific role in autophagy. J. Biol. Chem. 2012;287:16256–16266. doi: 10.1074/jbc.M112.348250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chang C, et al. Bidirectional control of autophagy by BECN1 BARA domain dynamics. Mol. Cell. 2019;73:339–353. doi: 10.1016/j.molcel.2018.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Stjepanovic G, Baskaran S, Lin MG, Hurley JH. Vps34 kinase domain dynamics regulate the autophagic PI 3-kinase complex. Mol. Cell. 2017;67:528–534. doi: 10.1016/j.molcel.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Birgisdottir AB, et al. Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs. Autophagy. 2019;15:1333–1355. doi: 10.1080/15548627.2019.1581009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Stromhaug PE, Reggiori F, Guan J, Wang CW, Klionsky DJ. Atg21 is a phosphoinositide binding protein required for efficient lipidation and localization of Atg8 during uptake of aminopeptidase I by selective autophagy. Mol. Biol. Cell. 2004;15:3553–3566. doi: 10.1091/mbc.e04-02-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Meiling-Wesse K, et al. Atg21 is required for effective recruitment of Atg8 to the preautophagosomal structure during the Cvt pathway. J. Biol. Chem. 2004;279:37741–37750. doi: 10.1074/jbc.M401066200. [DOI] [PubMed] [Google Scholar]
- 107.Obara K, Sekito T, Niimi K, Ohsumi Y. The Atg18-Atg2 complex is recruited to autophagic membranes via phosphatidylinositol 3-phosphate and exerts an essential function. J. Biol. Chem. 2008;283:23972–23980. doi: 10.1074/jbc.M803180200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zheng JX, et al. Architecture of the ATG2B-WDR45 complex and an aromatic Y/HF motif crucial for complex formation. Autophagy. 2017;13:1870–1883. doi: 10.1080/15548627.2017.1359381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bakula D, et al. WIPI3 and WIPI4 beta-propellers are scaffolds for LKB1-AMPK-TSC signalling circuits in the control of autophagy. Nat. Commun. 2017;8:15637. doi: 10.1038/ncomms15637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chowdhury S, et al. Insights into autophagosome biogenesis from structural and biochemical analyses of the ATG2A-WIPI4 complex. Proc. Natl Acad. Sci. USA. 2018;115:E9792–E9801. doi: 10.1073/pnas.1811874115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Polson HE, et al. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy. 2010;6:506–522. doi: 10.4161/auto.6.4.11863. [DOI] [PubMed] [Google Scholar]
- 112.Lu Q, et al. The WD40 repeat PtdIns(3)P-binding protein EPG-6 regulates progression of omegasomes to autophagosomes. Dev. Cell. 2011;21:343–357. doi: 10.1016/j.devcel.2011.06.024. [DOI] [PubMed] [Google Scholar]
- 113.Dove SK, et al. Svp1p defines a family of phosphatidylinositol 3,5-bisphosphate effectors. EMBO J. 2004;23:1922–1933. doi: 10.1038/sj.emboj.7600203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jeffries TR, Dove SK, Michell RH, Parker PJ. PtdIns-specific MPR pathway association of a novel WD40 repeat protein, WIPI49. Mol. Biol. Cell. 2004;15:2652–2663. doi: 10.1091/mbc.e03-10-0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nair U, Cao Y, Xie Z, Klionsky DJ. Roles of the lipid-binding motifs of Atg18 and Atg21 in the cytoplasm to vacuole targeting pathway and autophagy. J. Biol. Chem. 2010;285:11476–11488. doi: 10.1074/jbc.M109.080374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Krick R, et al. Structural and functional characterization of the two phosphoinositide binding sites of PROPPINs, a beta-propeller protein family. Proc. Natl Acad. Sci. USA. 2012;109:E2042–E2049. doi: 10.1073/pnas.1205128109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Baskaran S, Ragusa MJ, Boura E, Hurley JH. Two-site recognition of phosphatidylinositol 3-phosphate by PROPPINs in autophagy. Mol. Cell. 2012;47:339–348. doi: 10.1016/j.molcel.2012.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Watanabe Y, et al. Structure-based analyses reveal distinct binding sites for Atg2 and phosphoinositides in Atg18. J. Biol. Chem. 2012;287:31681–31690. doi: 10.1074/jbc.M112.397570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rieter E, et al. Atg18 function in autophagy is regulated by specific sites within its beta-propeller. J. Cell Sci. 2013;126:593–604. doi: 10.1242/jcs.115725. [DOI] [PubMed] [Google Scholar]
- 120.Busse RA, et al. Characterization of PROPPIN-phosphoinositide binding and role of loop 6CD in PROPPIN-membrane binding. Biophys. J. 2015;108:2223–2234. doi: 10.1016/j.bpj.2015.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tamura N, et al. Atg18 phosphoregulation controls organellar dynamics by modulating its phosphoinositide-binding activity. J. Cell Biol. 2013;202:685–698. doi: 10.1083/jcb.201302067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gopaldass N, Fauvet B, Lashuel H, Roux A, Mayer A. Membrane scission driven by the PROPPIN Atg18. EMBO J. 2017;36:3274–3291. doi: 10.15252/embj.201796859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Maeda S, Otomo C, Otomo T. The autophagic membrane tether ATG2A transfers lipids between membranes. Elife. 2019;8:e45777. doi: 10.7554/eLife.45777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Scacioc A, et al. Structure based biophysical characterization of the PROPPIN Atg18 shows Atg18 oligomerization upon membrane binding. Sci. Rep. 2017;7:14008. doi: 10.1038/s41598-017-14337-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tamura N, et al. Differential requirement for ATG2A domains for localization to autophagic membranes and lipid droplets. FEBS Lett. 2017;591:3819–3830. doi: 10.1002/1873-3468.12901. [DOI] [PubMed] [Google Scholar]
- 126.Kotani T, Kirisako H, Koizumi M, Ohsumi Y, Nakatogawa H. The Atg2-Atg18 complex tethers pre-autophagosomal membranes to the endoplasmic reticulum for autophagosome formation. Proc. Natl Acad. Sci. USA. 2018;115:10363–10368. doi: 10.1073/pnas.1806727115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Valverde DP, et al. ATG2 transports lipids to promote autophagosome biogenesis. J. Cell Biol. 2019;218:1787–1798. doi: 10.1083/jcb.201811139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tang Z, et al. TOM40 targets Atg2 to mitochondria-associated ER membranes for phagophore expansion. Cell Rep. 2019;28:1744–1757. doi: 10.1016/j.celrep.2019.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bozic M, et al. A conserved ATG2-GABARAP family interaction is critical for phagophore formation. EMBO Rep. 2020;21:e48412. doi: 10.15252/embr.201948412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gomez-Sanchez R, et al. Atg9 establishes Atg2-dependent contact sites between the endoplasmic reticulum and phagophores. J. Cell Biol. 2018;217:2743–2763. doi: 10.1083/jcb.201710116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Osawa T, et al. Atg2 mediates direct lipid transfer between membranes for autophagosome formation. Nat. Struct. Mol. Biol. 2019;26:281–288. doi: 10.1038/s41594-019-0203-4. [DOI] [PubMed] [Google Scholar]
- 132.Kumar N, et al. VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites. J. Cell Biol. 2018;217:3625–3639. doi: 10.1083/jcb.201807019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hanada T, et al. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J. Biol. Chem. 2007;282:37298–37302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- 134.Otomo C, Metlagel Z, Takaesu G, Otomo T. Structure of the human ATG12~ATG5 conjugate required for LC3 lipidation in autophagy. Nat. Struct. Mol. Biol. 2013;20:59–66. doi: 10.1038/nsmb.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fujioka Y, et al. In vitro reconstitution of plant Atg8 and Atg12 conjugation systems essential for autophagy. J. Biol. Chem. 2008;283:1921–1928. doi: 10.1074/jbc.M706214200. [DOI] [PubMed] [Google Scholar]
- 136.Sakoh-Nakatogawa M, et al. Atg12-Atg5 conjugate enhances E2 activity of Atg3 by rearranging its catalytic site. Nat. Struct. Mol. Biol. 2013;20:433–439. doi: 10.1038/nsmb.2527. [DOI] [PubMed] [Google Scholar]
- 137.Romanov J, et al. Mechanism and functions of membrane binding by the Atg5-Atg12/Atg16 complex during autophagosome formation. EMBO J. 2012;31:4304–4317. doi: 10.1038/emboj.2012.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kaufmann A, Beier V, Franquelim HG, Wollert T. Molecular mechanism of autophagic membrane-scaffold assembly and disassembly. Cell. 2014;156:469–481. doi: 10.1016/j.cell.2013.12.022. [DOI] [PubMed] [Google Scholar]
- 139.Fujita N, et al. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol. Biol. Cell. 2008;19:2092–2100. doi: 10.1091/mbc.e07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Suzuki K, et al. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Mizushima N, et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 2001;152:657–667. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Saitoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 143.Itoh T, et al. Golgi-resident small GTPase Rab33B interacts with Atg16L and modulates autophagosome formation. Mol. Biol. Cell. 2008;19:2916–2925. doi: 10.1091/mbc.e07-12-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gammoh N, Florey O, Overholtzer M, Jiang X. Interaction between FIP200 and ATG16L1 distinguishes ULK1 complex-dependent and -independent autophagy. Nat. Struct. Mol. Biol. 2013;20:144–149. doi: 10.1038/nsmb.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Nishimura T, et al. FIP200 regulates targeting of Atg16L1 to the isolation membrane. EMBO Rep. 2013;14:284–291. doi: 10.1038/embor.2013.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Fujita N, et al. Recruitment of the autophagic machinery to endosomes during infection is mediated by ubiquitin. J. Cell Biol. 2013;203:115–128. doi: 10.1083/jcb.201304188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Juris L, et al. PI3P binding by Atg21 organises Atg8 lipidation. EMBO J. 2015;34:955–973. doi: 10.15252/embj.201488957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Harada K, et al. Two distinct mechanisms target the autophagy-related E3 complex to the pre-autophagosomal structure. Elife. 2019;8:e43088. doi: 10.7554/eLife.43088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lystad AH, et al. Distinct functions of ATG16L1 isoforms in membrane binding and LC3B lipidation in autophagy-related processes. Nat. Cell Biol. 2019;21:372–383. doi: 10.1038/s41556-019-0274-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Dudley LJ, et al. Intrinsic lipid binding activity of ATG16L1 supports efficient membrane anchoring and autophagy. EMBO J. 2019;38:e100554. doi: 10.15252/embj.2018100554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Fletcher K, et al. The WD40 domain of ATG16L1 is required for its non-canonical role in lipidation of LC3 at single membranes. EMBO J. 2018;37:e97840. doi: 10.15252/embj.201797840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ichimura Y, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 153.Ichimura Y, et al. In vivo and in vitro reconstitution of Atg8 conjugation essential for autophagy. J. Biol. Chem. 2004;279:40584–40592. doi: 10.1074/jbc.M405860200. [DOI] [PubMed] [Google Scholar]
- 154.Ngu M, Hirata E, Suzuki K. Visualization of Atg3 during autophagosome formation in Saccharomyces cerevisiae. J. Biol. Chem. 2015;290:8146–8153. doi: 10.1074/jbc.M114.626952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Sakoh-Nakatogawa M, Kirisako H, Nakatogawa H, Ohsumi Y. Localization of Atg3 to autophagy-related membranes and its enhancement by the Atg8-family interacting motif to promote expansion of the membranes. FEBS Lett. 2015;589:744–749. doi: 10.1016/j.febslet.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 156.Nath S, et al. Lipidation of the LC3/GABARAP family of autophagy proteins relies on a membrane-curvature-sensing domain in Atg3. Nat. Cell Biol. 2014;16:415–424. doi: 10.1038/ncb2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hanada T, Satomi Y, Takao T, Ohsumi Y. The amino-terminal region of Atg3 is essential for association with phosphatidylethanolamine in Atg8 lipidation. FEBS Lett. 2009;583:1078–1083. doi: 10.1016/j.febslet.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 158.Hervas JH, et al. Human ATG3 binding to lipid bilayers: role of lipid geometry, and electric charge. Sci. Rep. 2017;7:15614. doi: 10.1038/s41598-017-15057-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Li YT, et al. A semisynthetic Atg3 reveals that acetylation promotes Atg3 membrane binding and Atg8 lipidation. Nat. Commun. 2017;8:14846. doi: 10.1038/ncomms14846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Yi C, et al. Function and molecular mechanism of acetylation in autophagy regulation. Science. 2012;336:474–477. doi: 10.1126/science.1216990. [DOI] [PubMed] [Google Scholar]
- 161.Axe EL, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Hayashi-Nishino M, et al. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 2009;11:1433–1437. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- 163.Yla-Anttila P, Vihinen H, Jokita E, Eskelinen EL. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 2009;5:1180–1185. doi: 10.4161/auto.5.8.10274. [DOI] [PubMed] [Google Scholar]
- 164.Hailey DW, et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat. Cell Biol. 2010;12:747–757. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ge L, Melville D, Zhang M, Schekman R. The ER-Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis. Elife. 2013;2:e00947. doi: 10.7554/eLife.00947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Puri C, Renna M, Bento CF, Moreau K, Rubinsztein DC. Diverse autophagosome membrane sources coalesce in recycling endosomes. Cell. 2013;154:1285–1299. doi: 10.1016/j.cell.2013.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Puri C, et al. The RAB11A-positive compartment is a primary platform for autophagosome assembly mediated by WIPI2 recognition of PI3P-RAB11A. Dev. Cell. 2018;45:114–131. doi: 10.1016/j.devcel.2018.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Dupont N, et al. Neutral lipid stores and lipase PNPLA5 contribute to autophagosome biogenesis. Curr. Biol. 2014;24:609–620. doi: 10.1016/j.cub.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Uemura T, et al. A cluster of thin tubular structures mediates transformation of the endoplasmic reticulum to autophagic isolation membrane. Mol. Cell Biol. 2014;34:1695–1706. doi: 10.1128/MCB.01327-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Duke EM, et al. Imaging endosomes and autophagosomes in whole mammalian cells using correlative cryo-fluorescence and cryo-soft X-ray microscopy (cryo-CLXM) Ultramicroscopy. 2014;143:77–87. doi: 10.1016/j.ultramic.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hamasaki M, et al. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013;495:389–393. doi: 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- 173.Shpilka T, et al. Lipid droplets and their component triglycerides and steryl esters regulate autophagosome biogenesis. EMBO J. 2015;34:2117–2131. doi: 10.15252/embj.201490315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Nascimbeni AC, et al. ER-plasma membrane contact sites contribute to autophagosome biogenesis by regulation of local PI3P synthesis. EMBO J. 2017;36:2018–2033. doi: 10.15252/embj.201797006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Stadel D, et al. TECPR2 cooperates with LC3C to regulate COPII-dependent ER export. Mol. Cell. 2015;60:89–104. doi: 10.1016/j.molcel.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 176.Tan D, et al. The EM structure of the TRAPPIII complex leads to the identification of a requirement for COPII vesicles on the macroautophagy pathway. Proc. Natl Acad. Sci. USA. 2013;110:19432–19437. doi: 10.1073/pnas.1316356110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Davis, S. et al. Sec24 phosphorylation regulates autophagosome abundance during nutrient deprivation. Elife5, e21167 (2016). [DOI] [PMC free article] [PubMed]
- 178.Shima T, Kirisako H, Nakatogawa H. COPII vesicles contribute to autophagosomal membranes. J. Cell Biol. 2019;218:1503–1510. doi: 10.1083/jcb.201809032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Zhao YG, et al. The ER-Localized transmembrane protein EPG-3/VMP1 regulates SERCA activity to control ER-isolation membrane contacts for autophagosome formation. Mol. Cell. 2017;67:974–989. doi: 10.1016/j.molcel.2017.08.005. [DOI] [PubMed] [Google Scholar]
- 180.Morita K, et al. Genome-wide CRISPR screen identifies TMEM41B as a gene required for autophagosome formation. J. Cell Biol. 2018;217:3817–3828. doi: 10.1083/jcb.201804132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Tian Y, et al. C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell. 2010;141:1042–1055. doi: 10.1016/j.cell.2010.04.034. [DOI] [PubMed] [Google Scholar]
- 182.Calvo-Garrido J, King JS, Munoz-Braceras S, Escalante R. Vmp1 regulates PtdIns3P signaling during autophagosome formation in Dictyostelium discoideum. Traffic. 2014;15:1235–1246. doi: 10.1111/tra.12210. [DOI] [PubMed] [Google Scholar]
- 183.Bard F, et al. Functional genomics reveals genes involved in protein secretion and Golgi organization. Nature. 2006;439:604–607. doi: 10.1038/nature04377. [DOI] [PubMed] [Google Scholar]
- 184.Calvo-Garrido J, Carilla-Latorre S, Lazaro-Dieguez F, Egea G, Escalante R. Vacuole membrane protein 1 is an endoplasmic reticulum protein required for organelle biogenesis, protein secretion, and development. Mol. Biol. Cell. 2008;19:3442–3453. doi: 10.1091/mbc.e08-01-0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Morishita, H. et al. A critical role of VMP1 in lipoprotein secretion. Elife8, e48834 (2019). [DOI] [PMC free article] [PubMed]
- 186.Tabara LC, et al. Vacuole membrane protein 1 marks endoplasmic reticulum subdomains enriched in phospholipid synthesizing enzymes and is required for phosphoinositide distribution. Traffic. 2018;19:624–638. doi: 10.1111/tra.12581. [DOI] [PubMed] [Google Scholar]
- 187.Tabara LC, Escalante R. VMP1 establishes ER-microdomains that regulate membrane contact sites and autophagy. PLoS ONE. 2016;11:e0166499. doi: 10.1371/journal.pone.0166499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Moretti, F. et al. TMEM41B is a novel regulator of autophagy and lipid mobilization. EMBO Rep. 19, e45889 (2018). [DOI] [PMC free article] [PubMed]
- 189.Shoemaker CJ, et al. CRISPR screening using an expanded toolkit of autophagy reporters identifies TMEM41B as a novel autophagy factor. PLoS Biol. 2019;17:e2007044. doi: 10.1371/journal.pbio.2007044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Young MM, Wang HG. Sphingolipids as regulators of autophagy and endocytic trafficking. Adv. Cancer Res. 2018;140:27–60. doi: 10.1016/bs.acr.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 191.Harvald EB, Olsen AS, Faergeman NJ. Autophagy in the light of sphingolipid metabolism. Apoptosis. 2015;20:658–670. doi: 10.1007/s10495-015-1108-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Li Y, et al. The pleiotropic roles of sphingolipid signaling in autophagy. Cell Death Dis. 2014;5:e1245. doi: 10.1038/cddis.2014.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Daido S, et al. Pivotal role of the cell death factor BNIP3 in ceramide-induced autophagic cell death in malignant glioma cells. Cancer Res. 2004;64:4286–4293. doi: 10.1158/0008-5472.CAN-03-3084. [DOI] [PubMed] [Google Scholar]
- 194.Scarlatti F, et al. Ceramide-mediated macroautophagy involves inhibition of protein kinase B and up-regulation of beclin 1. J. Biol. Chem. 2004;279:18384–18391. doi: 10.1074/jbc.M313561200. [DOI] [PubMed] [Google Scholar]
- 195.Zeng X, Overmeyer JH, Maltese WA. Functional specificity of the mammalian Beclin-Vps34 PI 3-kinase complex in macroautophagy versus endocytosis and lysosomal enzyme trafficking. J. Cell Sci. 2006;119:259–270. doi: 10.1242/jcs.02735. [DOI] [PubMed] [Google Scholar]
- 196.Guenther GG, et al. Ceramide starves cells to death by downregulating nutrient transporter proteins. Proc. Natl Acad. Sci. USA. 2008;105:17402–17407. doi: 10.1073/pnas.0802781105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Pattingre S, et al. Role of JNK1-dependent Bcl-2 phosphorylation in ceramide-induced macroautophagy. J. Biol. Chem. 2009;284:2719–2728. doi: 10.1074/jbc.M805920200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Sims K, et al. Kdo2-lipid A, a TLR4-specific agonist, induces de novo sphingolipid biosynthesis in RAW264.7 macrophages, which is essential for induction of autophagy. J. Biol. Chem. 2010;285:38568–38579. doi: 10.1074/jbc.M110.170621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Alexaki A, et al. Autophagy regulates sphingolipid levels in the liver. J. Lipid Res. 2014;55:2521–2531. doi: 10.1194/jlr.M051862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Yamagata M, Obara K, Kihara A. Sphingolipid synthesis is involved in autophagy in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 2011;410:786–791. doi: 10.1016/j.bbrc.2011.06.061. [DOI] [PubMed] [Google Scholar]
- 201.Lavieu G, et al. Regulation of autophagy by sphingosine kinase 1 and its role in cell survival during nutrient starvation. J. Biol. Chem. 2006;281:8518–8527. doi: 10.1074/jbc.M506182200. [DOI] [PubMed] [Google Scholar]
- 202.Moruno Manchon JF, et al. Cytoplasmic sphingosine-1-phosphate pathway modulates neuronal autophagy. Sci. Rep. 2015;5:15213. doi: 10.1038/srep15213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Lepine S, et al. Sphingosine-1-phosphate phosphohydrolase-1 regulates ER stress-induced autophagy. Cell Death Differ. 2011;18:350–361. doi: 10.1038/cdd.2010.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Chang CL, et al. S1P(5) is required for sphingosine 1-phosphate-induced autophagy in human prostate cancer PC-3 cells. Am. J. Physiol. Cell Physiol. 2009;297:C451–C458. doi: 10.1152/ajpcell.00586.2008. [DOI] [PubMed] [Google Scholar]
- 205.Taniguchi M, et al. Regulation of autophagy and its associated cell death by “sphingolipid rheostat”: reciprocal role of ceramide and sphingosine 1-phosphate in the mammalian target of rapamycin pathway. J. Biol. Chem. 2012;287:39898–39910. doi: 10.1074/jbc.M112.416552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Mitroi DN, et al. SGPL1 (sphingosine phosphate lyase 1) modulates neuronal autophagy via phosphatidylethanolamine production. Autophagy. 2017;13:885–899. doi: 10.1080/15548627.2017.1291471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Corcelle-Termeau E, et al. Excess sphingomyelin disturbs ATG9A trafficking and autophagosome closure. Autophagy. 2016;12:833–849. doi: 10.1080/15548627.2016.1159378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Mishra SK, et al. CPTP: a sphingolipid transfer protein that regulates autophagy and inflammasome activation. Autophagy. 2018;14:862–879. doi: 10.1080/15548627.2017.1393129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Back MJ, et al. Activation of neutral sphingomyelinase 2 by starvation induces cell-protective autophagy via an increase in Golgi-localized ceramide. Cell Death Dis. 2018;9:670. doi: 10.1038/s41419-018-0709-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Tan SH, et al. Induction of autophagy by palmitic acid via protein kinase C-mediated signaling pathway independent of mTOR (mammalian target of rapamycin) J. Biol. Chem. 2012;287:14364–14376. doi: 10.1074/jbc.M111.294157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Khan MJ, et al. Inhibition of autophagy rescues palmitic acid-induced necroptosis of endothelial cells. J. Biol. Chem. 2012;287:21110–21120. doi: 10.1074/jbc.M111.319129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Martino L, et al. Palmitate activates autophagy in INS-1E beta-cells and in isolated rat and human pancreatic islets. PLoS ONE. 2012;7:e36188. doi: 10.1371/journal.pone.0036188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Las G, Serada SB, Wikstrom JD, Twig G, Shirihai OS. Fatty acids suppress autophagic turnover in beta-cells. J. Biol. Chem. 2011;286:42534–42544. doi: 10.1074/jbc.M111.242412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Mei S, et al. Differential roles of unsaturated and saturated fatty acids on autophagy and apoptosis in hepatocytes. J. Pharmacol. Exp. Ther. 2011;339:487–498. doi: 10.1124/jpet.111.184341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.O’Rourke EJ, Soukas AA, Carr CE, Ruvkun G. C. elegans major fats are stored in vesicles distinct from lysosome-related organelles. Cell Metab. 2009;10:430–435. doi: 10.1016/j.cmet.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Niso-Santano M, et al. Unsaturated fatty acids induce non-canonical autophagy. EMBO J. 2015;34:1025–1041. doi: 10.15252/embj.201489363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Kohler K, et al. A combined proteomic and genetic analysis identifies a role for the lipid desaturase Desat1 in starvation-induced autophagy in Drosophila. Autophagy. 2009;5:980–990. doi: 10.4161/auto.5.7.9325. [DOI] [PubMed] [Google Scholar]
- 218.Ogasawara Y, et al. Stearoyl-CoA desaturase 1 activity is required for autophagosome formation. J. Biol. Chem. 2014;289:23938–23950. doi: 10.1074/jbc.M114.591065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Ogasawara Y, Kira S, Mukai Y, Noda T, Yamamoto A. Ole1, fatty acid desaturase, is required for Atg9 delivery and isolation membrane expansion during autophagy in Saccharomyces cerevisiae. Biol. Open. 2017;6:35–40. doi: 10.1242/bio.022053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Sauvat A, et al. Trans-fats inhibit autophagy induced by saturated fatty acids. EBioMedicine. 2018;30:261–272. doi: 10.1016/j.ebiom.2018.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Dall’Armi C, et al. The phospholipase D1 pathway modulates macroautophagy. Nat. Commun. 2010;1:142. doi: 10.1038/ncomms1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Bae EJ, et al. Phospholipase D1 regulates autophagic flux and clearance of alpha-synuclein aggregates. Cell Death Differ. 2014;21:1132–1141. doi: 10.1038/cdd.2014.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Cai M, et al. Phospholipase D1-regulated autophagy supplies free fatty acids to counter nutrient stress in cancer cells. Cell Death Dis. 2016;7:e2448. doi: 10.1038/cddis.2016.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Holland P, et al. HS1BP3 negatively regulates autophagy by modulation of phosphatidic acid levels. Nat. Commun. 2016;7:13889. doi: 10.1038/ncomms13889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Jang YH, Choi KY, Min DS. Phospholipase D-mediated autophagic regulation is a potential target for cancer therapy. Cell Death Differ. 2014;21:533–546. doi: 10.1038/cdd.2013.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Li D, et al. Storage lipid synthesis is necessary for autophagy induced by nitrogen starvation. FEBS Lett. 2015;589:269–276. doi: 10.1016/j.febslet.2014.11.050. [DOI] [PubMed] [Google Scholar]
- 227.Velazquez AP, Tatsuta T, Ghillebert R, Drescher I, Graef M. Lipid droplet-mediated ER homeostasis regulates autophagy and cell survival during starvation. J. Cell Biol. 2016;212:621–631. doi: 10.1083/jcb.201508102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Shatz O, Holland P, Elazar Z, Simonsen A. Complex relations between phospholipids, autophagy, and neutral lipids. Trends Biochem. Sci. 2016;41:907–923. doi: 10.1016/j.tibs.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 229.Cheng J, et al. Yeast and mammalian autophagosomes exhibit distinct phosphatidylinositol 3-phosphate asymmetries. Nat. Commun. 2014;5:3207. doi: 10.1038/ncomms4207. [DOI] [PubMed] [Google Scholar]
- 230.Baba T, Toth DJ, Sengupta N, Kim YJ, Balla T. Phosphatidylinositol 4,5-bisphosphate controls Rab7 and PLEKHM1 membrane cycling during autophagosome-lysosome fusion. EMBO J. 2019;38:e100312. doi: 10.15252/embj.2019102837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Turco E, Martens S. Insights into autophagosome biogenesis from in vitro reconstitutions. J. Struct. Biol. 2016;196:29–36. doi: 10.1016/j.jsb.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Bahrami AH, Lin MG, Ren X, Hurley JH, Hummer G. Scaffolding the cup-shaped double membrane in autophagy. PLoS Comput. Biol. 2017;13:e1005817. doi: 10.1371/journal.pcbi.1005817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Knorr RL, Dimova R, Lipowsky R. Curvature of double-membrane organelles generated by changes in membrane size and composition. PLoS ONE. 2012;7:e32753. doi: 10.1371/journal.pone.0032753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Yamamoto H, et al. The Intrinsically disordered protein Atg13 mediates supramolecular assembly of autophagy initiation complexes. Dev. Cell. 2016;38:86–99. doi: 10.1016/j.devcel.2016.06.015. [DOI] [PubMed] [Google Scholar]