Abstract
Chinese and imported pig breeds differ in fat production potential, which is associated with the polymorphisms in the 5′ proximal regulating region (5′PRR) of thyroid hormone responsive gene (THRSP). In three Chinese breeds (Dingyuan, CDY; Wannanhua, CWH; and Jixi, CJX) and one introduced breed (Yorkshire, YKS), three variant sites were located at T/C-400, A/G-376, and G/A-98 in the 5′PRR. Chinese pig breeds had higher C-400 allele frequencies than YKS. The frequencies of A-376 in CDY and G-376 in CWH were about 0.8. G-98 allele frequencies in CWH and YKS were 0.8617 and 0.8149, respectively. TGG was the dominant haplotype in YKS, CGG in CWH and CJX, and CAA in CDY. According to haplotype frequency, four breeds were clustered into three types, which was consistent with the geographical distribution of the breeds. In CDY, the average backfat thickness (BFT) was the highest with the CC-400 genotype, followed by CT-400 and TT-400 genotypes. In YKS, the pigs with CC-400 or CT-400 genotypes had higher BFT and average daily weight gain, whereas those with CC-400 or TT-400 genotypes had larger lion-eye area. No significant difference was observed in carcass traits among different genotypes at the A/G-376 and G/A-98 loci. The mRNA abundance of THRSP expression for the CCAGAG genotype was significantly higher than that for CTAGAG or TTAGAG genotype. These results indicated that the polymorphisms and genotype distribution of THRSP were closely related to the potential for fat production in pig breeds, which were the result of adaptation to artificial selection and natural selection.
Keywords: Backfat thickness, Carcass trait, Genotype distribution, Pig breed, THRSP gene
Introduction
Owing to high carcass fat content, native Chinese swine breeds are known as fat-type breeds, whereas the leaner, introduced pig breeds are classified as lean-type breeds (Zhao 2003). Differences in carcass fat content lead to differences in product flavors. Fat is an important factor that affects the carcass quality of pigs, and consumer demand for taste, flavor, and juiciness affects the fat content of pig products (Chen et al. 2007). Many genes are involved in the regulation of carcass fat deposition. As gene regulation is complex, it has always been a problem in the field of genetic breeding. THRSP, which encodes thyroid hormone responsive spot 14, is involved in the transcriptional regulation of the gene expression of various lipogenic rate-limiting enzymes (Cunningham et al. 1998; Zhang et al. 2011) and is a candidate gene for fat deposition (Zhou et al. 2011). A previous study showed that nonobese humans downregulate the expression of THRSP in response to fasting, whereas obese subjects do not, and postfasting levels of glucose, insulin, and ketones do not differ between the groups (Kirschner and Mariash 1999). THRSP may affect body adipogenesis via the regulation of its expression through transcriptional elements and polymorphisms in the promoter region (Wang et al. 2004; Shimada et al. 2011). Currently, THRSP polymorphisms have been characterized in chickens (Wang et al. 2004), and SNPs have been reported in cows, rats, mice, and chimpanzees (Kuemmerle and Kinlaw 2011). There are 200 SNPs in the 1000-bp 5′ proximal regulating region (5′PRR) of human THRSP according to the Single Nucleotide Polymorphism Database. Chen et al. (2011) cloned and analyzed the characteristics of the 5′PRR of THRSP, and Rempel et al. (2012) identified 25 SNPs and genotyped 21 of THRSP in which one was related to the weaning-to-estrus interval in swines.
The maintenance of THRSP polymorphisms in animal populations may be an adaptation to selection (Kimura 1983). Although selection is applied to phenotypic traits, the mechanism of traits is based on the function of genes related to the traits; thus, genes are indirectly affected by selection (Ma et al. 2018; Chen et al. 2012). In addition to spontaneous mutation, natural selection and artificial selection are two important factors that affect gene frequency distribution (Kimura 1983). Especially in livestock, artificial selection may cause favorable variation to continuously accumulate and shape gene frequency distribution in a population (Wang et al. 2015; Ma et al. 2019).
The objectives of the present study were to detect the SNPs in the 5′PRR of THRSP in Chinese pig breeds (Dingyuan, CDY; Wannanhua, CWH; and Jixi, CJX) and an introduced pig breed (Yorkshire, YKS) by PCR-sequencing and investigate the association among THRSP, carcass traits, and THRSP expression level in CDY and YKS pigs, in which the polymorphisms are segregating (Caroli et al. 2009).
Materials and methods
Animals and DNA extraction
All procedures that involved animals were approved by the animal care and use committee at the institution in which the experiment was conducted. All procedures that involved animals were also approved and authorized by the Chinese Ministry of Agriculture.
In the present study, three types of Chinese local pig breeds were selected, among which CWH, CJX, and CDY originated from a mountainous area, an area intersecting mountains and swamp, and the Jianghuai hilly area. YKS is an imported breed. The origin of the four pig breeds and the geographical distribution of Chinese pig breeds are shown in Fig. 1. Ear tissue samples of 6-month-old sows were collected from 107 CDY (Dingyuan Swine Farm, Anhui Province, China), 48 CWH (Wannanhua Swine Farm, Anhui Province, China), 65 CJX (Jixi black Swine Farm, Anhui Province, China), and 154 of the introduced breed YKS (Changfeng Animal Husbandry Group, Anhui Province, China). Genomic DNA was extracted by the phenol–chloroform method, dissolved in Tris–EDTA buffer (10 mmol/L Tris–HCl [pH 8.0], 1 mmol/L EDTA [pH 8.0]), and maintained at − 20 °C.
Fig. 1.
Origin of three Chinese pig breeds and their geographical distribution in Anhui Province. The region with red mark name is the origin area of the swine breed. The picture on the right is typical sows of the four pig breeds. Wannanhua, Jixi, and Dingyuan originated from a mountainous area, an area intersecting mountains and swamp, and the Jianghuai hilly area, respectively. Yorkshire is an imported breed
Detection of pig carcass traits
The average backfat thickness (BFT) and lion-eye area (LEA) of YKS at 6 months and the BFT of CDY at 6 months were measured using the EVO II Type Ultrasonic Backfat Instrument (USA). In addition, the average daily weight gain (DWG) of YKS was calculated.
Primers and PCR amplification
Four pairs of primers were designed according to the DNA sequence of the 5′PRR according to the GenBank accession number, NW_003610992, of pig THRSP. The primer P3 sequence, amplified region, product size, and annealing temperature were listed:
F: 5′-CATCCTCACGGATGTTAGTCAG-3′, R: 5′-CCAGACTTCTGTCTGTCATTTCG-3′, product size: 1039 bp, and annealing temperature: 55 °C.
The PCR was carried out using 25-μL samples that contained approximately 1.0 μL of 10 mol/L of each primer, 2.5 μL of 10 × PCR buffer (50 mmol/L KCl, 10 mmol/l Tris–HCl [pH 8.0], 0.1% Triton X-100), 1.5–1.8 μL of 25 mmol/L MgCl2, 2.5 μL of 2.5 mmol/L of each dNTP, 3.0 μL of 50 ng/μL genomic DNA, 1.0 μL of 2.5 U/μL Taq DNA polymerase (Promega, Madison, WI, USA), and ddH2O.
Amplification conditions were: initial denaturation at 94 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 55–60 °C for 30 s, extension at 60 °C for 1 min, and a final extension at 72 ° Cfor 10 min in a Mastercycler@5333 (Eppendorf AG, Hamburg, Germany).
Polymorphism detection and cloning and sequencing
PCR products of P3 were separated on 1.0% agarose gels and recovered using Geneclean II (Promega, Maddison, WI, USA). Each DNA fragment was ligated into pGEM-T Easy Vector (Promega) according to the manufacturer’s instructions at 16 °C overnight. The recombinant plasmid was then transformed into Escherichia coli TOP10 competent cells. Positive clones of transformed cells were identified by restriction enzyme digestion. Three clones of each PCR product of the P3 and each genotype were selected and sequenced. Target DNA fragments in recombinant plasmids were sequenced in both directions using an ABI 3730 automatic sequencer (Perkin Elmer Applied Biosystems, Foster City, CA, USA) by Shanghai Invitrogen Biotechnology Co. Ltd. (Shanghai, China). The SNP haplotypes of three loci were identified by sequencing monoclonal bacteria.
RNA extraction from liver tissue and qPCR
Total RNA was extracted from the livers of CDY and YKS using an RNA Pure Tissue Kit (CWBIO Ltd., Beijing, China). The concentration and integrity of the RNA samples were verified using agarose gel electrophoresis and measured in a Nanodrop 2000 Biophotometer (Thermo Fisher Scientific Inc., West Palm Beach, FL, USA).
Primer sequences for THRSP (NM_001244376.1):
F: 5′-TCTGGCACCACACCTTCT-3′, R: 5′-TGATCTGGGTCATCTTCTCAC-3′, and product size: 114 bp.
β-actin gene (AY_5500069.1) was selected as the internal standard:
F: 5′-AATGCGGTGGGATCGACAAA-3′, R: 5′-CACGCTCACGTTCAGCCTTT-3′, and size: 120 bp.
Quantitative gene expression measurements were performed using the StepOnePlus platform (Life Technologies, Carlsbad, CA, USA) using the comparative ΔCt method. β-actin was selected as the internal reference gene. Each reaction was conducted with a volume of 20 μL that contained 10 μL of SYBR Green Mix, 8.0 μL of diethylpyrocarbonate water, 1.0 μL of cDNA, and 0.5 μL (10 μmol/L) of forward and reverse primers. The amplification conditions were: 95 °C for 10 min, followed by 40 cycles at 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s. The final product was stored at 4 °C.
Statistical analysis
Allelic frequencies and genotype distributions were compared using the χ2 test. The following fixed effects model was employed for the analysis of BFT in all pig breeds, and least squares mean (LSM) was used for multiple comparisons in BFT among different genotypes:
where yijk is the BFT, LEA, or DWG; μ is the population mean; Bi is the fixed effect of the ith breed (i = 1, 2, 3, 4); Gj is the fixed effect of jth genotype (j = 1, 2, …, m); and eijk is the random residual effect of each observation. The analysis was performed using the general linear model procedure of SAS (V. 9.00) (SAS Institute Inc., Cary, NC, USA). The mean separation procedures were performed using a least significant difference test. Statistically significant differences were accepted with a p < 0.05.
Results
Polymorphism detection in pig THRSP gene
The genomic DNA of all pigs was successfully amplified using the primer pairs of P3. The sizes of the amplification fragments were consistent with the target fragments and had sufficient specificity to sequence and analyze. The PCR products that were amplified displayed polymorphisms. Three nucleotide changes were identified in the 5′PRR of THRSP, which were T/C-400, A/G-376, and G/A-98 (Fig. 2).
Fig. 2.
SNP sites in 5′ proximal regulatory region of pig THRSP gene. aatg is the start of coding region of the THRSP, ↑symbol is the SNP site, and negative number is the length (bp) from the start of coding region. The 260 bp sequence is not shown between − 106 and − 386. b The sequence diagram at SNP site
Difference in THRSP genotype distribution among four pig breeds
Polymorphic sequence variations in THRSP in the four pig breeds are shown in Figs. 3,4, 5. At T/C-400, A/G-376, and G/A-98 loci, YKS, CDY, CWH, and CJX were consistent with the Hardy–Weinberg equilibrium (p > 0.05). The genotype distributions of the three mutations among the four pig breeds were significantly different (p < 0.01).
Fig. 3.
Frequency distribution of genetype (a) and allele gene (b) at T/C-400 locus in four pig breeds. Blank column is the theoretical distribution of the genotypes in HW equilibrium. The value at the top of the column is the number of individuals. The χ2 test at the bottom of Fig. 3a is significant difference of genotype distribution between two breeds. YKS is Introduced pig breed, CDY is Chinese Dingyuan, CWH is Chinses Wannanhua and CJX is Chinses Jixi
Fig. 4.
Frequency distribution of genetype (a) and allele gene (b) at A/G-376 locus in four pig breeds. Blank column is the theoretical distribution of the genotypes in HW equilibrium. The value at the top of the column is the number of individuals. The χ2 test at the bottom of Fig. 4a is significant difference of genotype distribution between two breeds. YKS is Introduced breeds, CDY is Chinese Dingyuan, CWH is Chinses Wannanhua and CJX is Chinses Jixi
The genotype distributions at the T/C-400 locus among the pig breeds were significantly different (p < 0.01 or p < 0.05), except for the genotype distribution between CWH and CDY (p > 0.05) (Fig. 3a). The C-400 allele frequency was the highest in CJX, followed by CWH, then CDY, and then YKS (Fig. 3b).
The genotype distributions at the A/G-376 locus among the pig breeds were significantly different (p < 0.01) (Fig. 4a). The A-376 allele frequency in CDY was 0.7944, and the G-376 allele frequency in CWH was 0.8085 (Fig. 4b), which indicated that A-376 and G-376 alleles were evenly distributed in CDY and CWH, respectively, and that this distribution was different from that in YKS and CJX.
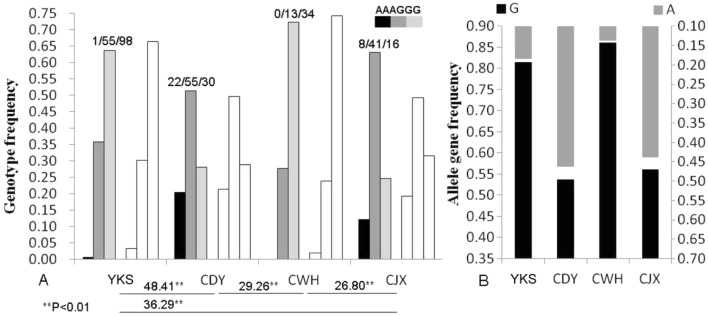
At the G/A-98 locus, there were differences in genotype distributions between YKS and CWH or CDY and CJX (p > 0.05) (Fig. 5a), whereas the difference between other breeds was significant (p < 0.01). The frequencies of the G-98 allele in the four pig breeds were higher than those of the A-98 allele, of which those in CWH and YKS were 0.8617 and 0.8149, respectively (Fig. 5b).
Fig. 5.
Frequency distribution of genetype (a) and allele gene (b) at G/A-98 locus in four pig breeds. Blank column is the theoretical distribution of the genotypes in HW equilibrium. The value at the top of the column is the number of individuals. The χ2 test at the bottom of Fig. 5a is significant difference of genotype distribution between two breeds. YKS is Introduced breeds, CDY is Chinese Dingyuan, CWH is Chinses Wannanhua and CJX is Chinses Jixi
In all SNP haplotypes at the three loci, the frequency of the TGG haplotype in YKS was 0.4310, whereas the frequency of other haplotypes was less than 0.15 (Fig. 6a). The frequencies of CAG, CAA, and CGG haplotypes in Chinese pig breeds were higher than those in the imported breed (YKS) and the frequency of the CGG haplotype in CWH was as high as 0.4245. According to the haplotype frequency, the four pig breeds were divided into three clusters. As shown in Fig. 6b, CWH and CJX were clustered into cluster 1, whereas CDY and YKS were clustered into cluster 2 and cluster 3, respectively.
Fig. 6.
Distribution characteristics of the haplotypes of three loci in four pig breeds. a Frequency distribution of the haplotypes. TGG is the dominant haplotype (high frequency) in YKS, CGG in CWH and CJX, and CAA in CDY. b Cluster of four pig breeds according to the haplotype frequency. The clustering result is consistent with the geographical distribution of the pig breeds
Influence of fixed effects on carcass traits in pig breeds
BFT was significantly influenced by the THRSP genotype. LSMs and standard errors for BFT in different genotypes of THRSP in a Chinese pig breed (CDY) are shown in Table 1. The pigs with CC-400 or CT-400 genotypes had 1.65 cm (p < 0.01) or 1.45 cm (p < 0.05) more BFT than those with the TT-400 genotype. The pigs with the CC-400 genotype had 0.20 cm (p < 0.05) more BFT than those with the CT-400 genotype. The pigs with the AA-376 genotype had significantly more BFT (0.39 cm and 0.35 cm) than those with AG-376 and GG-376 genotypes, respectively (p < 0.05); however, the difference between AG-376 and GG-376 genotypes was not significant. No difference was observed in BFT among pigs at the G/A-98 locus (p > 0.05).
Table 1.
Least squares mean and standard error for backfat thickness of different genotypes of THRSP in CDY
| Locus | Genotype | Number | Backfat thickness (cm) | Sign. test |
|---|---|---|---|---|
| T/C-400 | CC | 33 | 3.16 ± 0.09D | D > E*, D > F**, E > F* |
| CT | 57 | 2.96 ± 0.12E | ||
| TT | 17 | 1.51 ± 0.18F | ||
| A/G-376 | AA | 70 | 3.05 ± 0.064D | D > E* |
| AG | 30 | 2.66 ± 0.166E | ||
| GG | 7 | 2.70 ± 0.335E | ||
| G/A-98 | AA | 22 | 3.01 ± 0.114D | |
| AG | 55 | 2.90 ± 0.095D | ||
| GG | 30 | 2.89 ± 0.120D |
With the same superscript, the difference is not significant
CDY Dingyuan pig
*p < 0.05; **p < 0.01
In YKS, the pigs with CC-400 or CT-400 genotypes had 0.12 cm or 0.07 cm more BFT (p < 0.05), and 21.51 g/d or 22.22 g/d more DWG, than those with the TT-400 genotype, respectively (Table 2). The pigs with CC-400 or TT-400 genotypes had 5.52 cm2 or 2.61 cm2 more LEA than those with the CT-400 genotype (p < 0.05), respectively. No significant difference (p > 0.05) was observed in carcass traits between different genotypes at A/G-376 and G/A-98 loci.
Table 2.
Least squares mean and standard error for carcass traits of different genotypes of THRSP in YKS
| Locus | Genotype | Number | BFT (cm) | LEA (cm2) | DWG (g/d) |
|---|---|---|---|---|---|
| T/C-400 | CC | 7 | 1.09 ± 0.07A | 44.75 ± 2.36A | 560.31 ± 24.62A |
| CT | 89 | 1.04 ± 0.02A | 39.23 ± 0.69B | 561.02 ± 5.95A | |
| TT | 58 | 0.97 ± 0.02B | 41.84 ± 1.05A | 538.80 ± 6.26B | |
| A/G-376 | AA | 7 | 1.09 ± 0.07 | 44.75 ± 2.36 | 560.31 ± 24.62 |
| AG | 101 | 1.03 ± 0.02 | 39.66 ± 0.67 | 557.74 ± 5.54 | |
| GG | 46 | 0.98 ± 0.03 | 41.58 ± 1.21 | 540.21 ± 7.09 | |
| G/A-98 | AA | 1 | 0.98 | 38.32 | 551.11 |
| AG | 55 | 1.03 ± 0.03 | 40.52 ± 1.06 | 561.36 ± 8.60 | |
| GG | 98 | 1.01 ± 0.02 | 40.46 ± 0.70 | 547.72 ± 4.85 |
A > B, P < 0.05; with the same superscript or without superscript, the difference is not significant
BFT Backfat thickness, LEA Lion-eye area, DWG Daily weight gain, YKS Yorkshire
Differential expression among genotypes of THRSP gene in liver tissue
At the T/C-400 locus, the expression of pig THRSP in the liver tissue, with respect to three genotypes, was measured (Fig. 7). The expression of the CCAGAG genotype was significantly higher than that of CTAGAG and TTAGAG genotypes (p < 0.01), whereas there was no difference between CTAGAG and TTAGAG genotypes. The expression of THRSP in CDY was higher than that in YKS (p < 0.01).
Fig. 7.

Expression of THRSP gene in the liver tissue of pigs with three genotypes. The three genotypes are CCAGAG (CC), CTAGAG (CT) or TTAGAG (TT), respectively. CDY is Chinese pig breed. YKS is introduced pig breed. a mRNA abundance of CC type is higher than that of CT- or TT-type whether in CDY or in YKS (p < 0.01). b mRNA abundance of CC-, CT- or TT-type in CDY is highter than that in YKS (p < 0.01)
Discussion
CWH and CJX are distributed in a mountainous area in the southern Anhui Province of China and are exposed to a subtropical humid monsoon climate with an annual precipitation of 1491.3 mm, whereas CDY is distributed in the hilly area of central Anhui, which is in the transition zone between a warm temperate zone and a subtropical zone with an annual precipitation of 988.4 mm (Chen and Jiang 1999; Yuan 2010). Including the introduced breed (YKS), the four breeds have different carcass performances and genetic backgrounds after long-term selection in different habitats (Fig. 1). As our study showed, the frequency of the C-400 allele was higher in CJX, CWH, and CDY, whereas the frequency of the T-400 allele was higher in YKS (Figs. 3b and 6). In contrast, CDY showed higher A-376 and A-98 allele frequencies (Figs. 4b,5b, and 6). The correlation analysis showed that the BFT of pigs with the C-400 allele was always higher.
Subsequently, we detected the differential expression of THRSP among the genotypes at the T/C-400 locus. The expression of the CC-400 genotype was significantly higher than that of TT-400 and CT-400 genotypes in both CDY and YKS (Fig. 7). We infer that the polymorphisms at the T/C-400 locus are associated with the differential expression of THRSP. From the transcription factors (TFs) of the T/C-400 binding site using JASPAR analysis (Fornes et al. 2020), T-400 has the potential binding sites for CCAAT enhancer binding protein delta (CEBPD) and CCAAT enhancer binding protein alpha (CEBPA), whereas C-400 is missing the binding sites of these TFs. CEBPA and CEBPD can activate the transcription of target genes in brown adipocytes by binding to the proximal promoter region (Carmona et al. 2002; Armengol et al. 2012). Therefore, we speculate that the development of brown adipocytes in Chinese pig breeds is not as good as that in introduced breeds; thus, the ability for body fat deposition is significantly higher in Chinese pig breeds with more white adipocytes. The brown adipocytes burn glucose and lipids to maintain thermal homeostasis (Giordano et al. 2014).
In the analysis of the association of genotypes with carcass traits, A/G-376 and G/A-98 loci provided little information, but A-376 is the potential binding site for CTCFL, retinoid X receptor alpha, and hepatocyte nuclear factor 4 gamma, which influence fat content and composition in animal muscles (Goszczynski et al. 2016; Ramayo-Caldas et al. 2014). Our results indicated that the frequency of the A-376 allele in CDY was higher than that in CWH, CJX, and YKS; thus, CDY is higher in intramuscular fat content. In previous studies, CDY had intramuscular fat content of 4.87%, whereas that of YKS was only 1.38% (Wu et al. 2019; Li et al. 2015). G-98 is the potential binding site for nuclear factor 1C (NF1C), estrogen receptor 2 (ESR2), and RFX2, which affect mammary gland development in swines (Verardo et al. 2016; Hurley 2019). NF1C is a proximal promoter of DNA-binding transcription activity and activates the milk genes for carboxyl ester lipase and whey acidic protein, which implies that NF1-C2 participates in the establishment of a functional gland (Johansson et al. 2005; Johansson et al. 2003). ESR2 regulates granulosa cell genes that are essential for follicle maturation and ovulation; thus, it is related to lactation (Khristi et al. 2018). RFX2 is a major transcriptional regulator of spermiogenesis (Kistler et al. 2015). According to the results of the present study, the frequencies of G-98 alleles in CWH (0.8617) and YKS (0.8149) were higher than those in CDY (0.5374) and CJX (0.5615). Whether this is related to the higher lactation performance in CWH and YKS needs further study.
The fact that THRSP mutations are present in fat-type and lean-type pig breeds will enable the development of breeding strategies to maximize the benefits of new varieties of pig for different purposes in these breeds. These results preliminarily demonstrated that the polymorphisms in the 5′PRR of THRSP are related to the potential for fat production in pig breeds. The major gene frequency of influencing characters changes with artificial selection, and can be used as a molecular genetic marker for early auxiliary selection in pig breeding.
Author contributions
HC: conceptualization, funding acquisition, methodology, project administration, supervision, writing—review & editing. XW: investigation, methodology, project administration, roles/writing—original draft, writing—review & editing. JC, WQ: formal analysis, investigation, writing—review & editing. GC, HC: formal analysis, writing—review & editing. XS, MZ, and NB: formal analysis.
Funding
The work was supported by the National Natural Science Foundation of China (NSFC) [Grant No. 31172180] and Project of Anhui Province Scientific Technology Plan (No. 17030701007).
Data availability
Data will be available on request to corresponding or first author.
Compliance with ethical standards
Conflict of interest
All authors declare that they have no conflict of interest..
Ethics approval
All procedures that involved animals were approved by the animal care and use committee at the institution in which the experiment was conducted. All procedures that involved animals were also approved and authorized by the Chinese Ministry of Agriculture.
Footnotes
Xiaohong Wang and Jin Cheng contributed equally to this work.
References
- Armengol J, Villena JA, Hondares E, Carmona MC, Sul HS, Iglesias R, Giralt M, Villarroya F. Pref-1 in brown adipose tissue: specific involvement in brown adipocyte differentiation and regulatory role of C/EBPδ. Biochem J. 2012;443:799–810. doi: 10.1042/BJ20111714. [DOI] [PubMed] [Google Scholar]
- Carmona MC, Iglesias R, Obregón MJ, Darlington GJ, Villarroya F, Giralt M. Mitochondrial biogenesis and thyroid status maturation in brown fat require CCAAT/enhancer-binding protein alpha. J Biol Chem. 2002;277:21489–21498. doi: 10.1074/jbc.M201710200. [DOI] [PubMed] [Google Scholar]
- Caroli AM, Chessa S, Erhardt GJ. Invited review: milk protein polymorphisms in cattle: effect on animal breeding and human nutrition. J Dairy Sci. 2009;92:5335–5352. doi: 10.3168/jds.2009-2461. [DOI] [PubMed] [Google Scholar]
- Chen HQ, Jiang MY. Relationship between native breeding pigs trait and ecological type in Anhui Province. Chinese J Appl Ecol. 1999;10:464–466. doi: 10.13287/j.1001-9332.1999.0120. [DOI] [Google Scholar]
- Chen RS, Zhang WL, Jing RB. Thirty years’ review of pork quality research. Swine Industry Science. 2007;24:90–94. doi: 10.3969/j.issn.1673-5358.2007.07.026. [DOI] [Google Scholar]
- Chen H, Chen HQ, Zhou QQ, Zhang YP, Wei HQ, Li CM, Zhang XY, Peng YL. Identification of transcription regulation activity in 5′ flanking region of pig THRSP gene. Acta Veterinaria et Zootechnica Sinica. 2011;42:329–334. [Google Scholar]
- Chen HQ, Qin J, Zhu YJ, Pan ZT, Xie YN, Jiao MH, Chen GW, Chen H, Chu MX. The polymorphisms of goat THRSP gene associated with ecological factors in Chinese indigenous goat breeds with different lipogenesis ability. Asian J Anim Vet Adv. 2012;7:802–811. doi: 10.3923/ajava.2012.802.811. [DOI] [Google Scholar]
- Cunningham BA, Moncur JT, Huntington JT, Kinlaw WB. “Spot 14” protein: a metabolic integrator in normal and neoplastic cells. Thyroid. 1998;8:815–825. doi: 10.1089/thy.1998.8.815. [DOI] [PubMed] [Google Scholar]
- Fornes O, Castro-Mondragon JA, Khan A, van der Lee R, Zhang X, Richmond PA, Modi BP, Correard S, Gheorghe M, Baranašić D, Santana-Garcia W, Tan G, Chèneby J, Ballester B, Parcy F, Sandelin A, Lenhard B, Wasserman WW, Mathelier A. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2020;48(D1):D87–D92. doi: 10.1093/nar/gkz1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano A, Smorlesi A, Frontini A, Barbatelli G, Cinti S. White, brown and pink adipocytes: the extraordinary plasticity of the adipose organ. Eur J Endocrinol. 2014;170:R159–R171. doi: 10.1530/EJE-13-0945. [DOI] [PubMed] [Google Scholar]
- Goszczynski DE, Mazzucco JP, Ripoli MV, Villarreal EL, Rogberg-Muñoz A, Mezzadra CA, Melucci LM, Giovambattista G. Genetic characterisation of PPARG, CEBPA and RXRA, and their influence on meat quality traits in cattle. J Anim Sci Technol. 2016;58:14. doi: 10.1186/s40781-016-0095-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley WL. Review: mammary gland development in swine: embryo to early lactation. Animal. 2019;13(S1):s11–s19. doi: 10.1017/S1751731119000521. [DOI] [PubMed] [Google Scholar]
- Johansson EM, Kannius-Janson M, Bjursell G, Nilsson J. The p53 tumor suppressor gene is regulated in vivo by nuclear factor 1-C2 in the mouse mammary gland during pregnancy. Oncogene. 2003;22:6061–6070. doi: 10.1038/sj.onc.1206884. [DOI] [PubMed] [Google Scholar]
- Johansson EM, Kannius-Janson M, Gritli-Linde A, Bjursell G, Nilsson J. Nuclear factor 1-C2 is regulated by prolactin and shows a distinct expression pattern in the mouse mammary epithelial cells during development. Mol Endocrinol. 2005;19:992–1003. doi: 10.1210/me.2004-0359. [DOI] [PubMed] [Google Scholar]
- Khristi V, Chakravarthi VP, Singh P, Ghosh S, Pramanik A, Ratri A, Borosha S, Roby KF, Wolfe MW, Rumi MAK. ESR2 regulates granulosa cell genes essential for follicle maturation and ovulation. Mol Cell Endocrinol. 2018;474:214–226. doi: 10.1016/j.mce.2018.03.012. [DOI] [PubMed] [Google Scholar]
- Kimura M. The neutral theory of molecular evolution. Cambridge: Cambridge University Press; 1983. [Google Scholar]
- Kirschner LS, Mariash CN. Adipose S14 mRNA is abnormally regulated in obese subjects. Thyroid. 1999;9:143–148. doi: 10.1089/thy.1999.9.143. [DOI] [PubMed] [Google Scholar]
- Kistler WS, Baas D, Lemeille S, Paschaki M, Seguin-Estevez Q, Barras E, Ma W, Duteyrat JL, Morlé L, Durand B, Reith W. RFX2 is a major transcriptional regulator of spermiogenesis. PLoS Genet. 2015;11:e1005368. doi: 10.1371/journal.pgen.1005368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuemmerle NB, Kinlaw WB. THRSP (thyroid hormone responsive) Atlas Genet Cytogenet Oncol Haematol. 2011;15:480–482. [PMC free article] [PubMed] [Google Scholar]
- Li QG, Wang CL, Xu JY. Slaughter determination and meat quality analysis between the native breed of Weizhu and introduced breed Large white pig. Swine Production. 2015;1:57–58. doi: 10.13257/j.cnki.21-1104/s.2015.01.023. [DOI] [Google Scholar]
- Ma Y, Zhang S, Zhang K, Fang C, Xie S, Du X, Li X, Ni D, Zhao S. Genomic analysis to identify signatures of artificial selection and loci associated with important economic traits in Duroc pigs. G3 (Bethesda) 2018;8:3617–3625. doi: 10.1534/g3.118.200665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Sonstegard TS, Cole JB, VanTassell CP, Wiggans GR, Crooker BA, Tan C, Prakapenka D, Liu GE, Da Y. Genome changes due to artificial selection in U.S. Holstein cattle. BMC Genomics. 2019;20:128. doi: 10.1186/s12864-019-5459-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramayo-Caldas Y, Fortes MR, Hudson NJ, Porto-Neto LR, Bolormaa S, Barendse W, Kelly M, Moore SS, Goddard ME, Lehnert SA, Reverter A. A marker-derived gene network reveals the regulatory role of PPARGC1A, HNF4G, and FOXP3 in intramuscular fat deposition of beef cattle. J Anim Sci. 2014;92:2832–2845. doi: 10.2527/jas.2013-7484. [DOI] [PubMed] [Google Scholar]
- Rempel LA, Nonneman DJ, Rohrer GA. Polymorphism within thyroid hormone responsive (THRSP) associated with weaning-to-oestrus interval in swine. Anim Genet. 2012;43:364–365. doi: 10.1111/j.1365-2052.2011.02303.x. [DOI] [PubMed] [Google Scholar]
- Shimada M, Mochizuki K, Goda T. Feeding rats dietary resistant starch reduces both the binding of ChREBP and the acetylation of histones on the Thrsp gene in the jejunum. J Agric Food Chem. 2011;59:1464–1469. doi: 10.1021/jf103111u. [DOI] [PubMed] [Google Scholar]
- Verardo LL, Silva FF, Lopes MS, Madsen O, Bastiaansen JW, Knol EF, Kelly M, Varona L, Lopes PS, Guimarães SE. Revealing new candidate genes for reproductive traits in pigs: combining Bayesian GWAS and functional pathways. Genet Sel Evol. 2016;48:9. doi: 10.1186/s12711-016-0189-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Carre W, Zhou H, Lamont SJ, Cogburn LA. Duplicated Spot 14 genes in the chicken: characterization and identification of polymorphisms associated with abdominal fat traits. Gene. 2004;332:79–88. doi: 10.1016/j.gene.2004.02.021. [DOI] [PubMed] [Google Scholar]
- Wang C, Wang H, Zhang Y, Tang Z, Li K, Liu B. Genome-wide analysis reveals artificial selection on coat colour and reproductive traits in Chinese domestic pigs. Mol Ecol Resour. 2015;15:414–424. doi: 10.1111/1755-0998.12311. [DOI] [PubMed] [Google Scholar]
- Wu YJ, Zhang P, Fu YR, Su SG, Tian GY, Zhang H, Li QG. Measurement of carcass qulity traits in Dingyuan black pig. Swine Prod. 2019;5:78–80. doi: 10.13257/j.cnki.21-1104/s.2019.05.027. [DOI] [Google Scholar]
- Yuan Q. Investigation and evaluation of cultivated land quality grade in China (Anhui) Beijing: China Land Press; 2010. [Google Scholar]
- Zhang W, Peng W, Zhao M, Lin D, Zeng Z, Zhou W, Bartlam M. Expression, purification and preliminary crystallographic analysis of human thyroid hormone responsive protein. Acta Crystallogr, Sect F: Struct Biol Cryst Commun. 2011;67(Pt 8):941–946. doi: 10.1107/S1744309111021099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao SG. China’s pig industry. Beijing: China Agriculture Press; 2003. [Google Scholar]
- Zhou QQ, Chen HQ, Wei HQ, Qin J, Chen H, Zhang YP. Association of polymorphism in the coding region of THRSP gene with lipogenesis capability in pigs. Prog Biochem Biophys. 2011;38:84–90. doi: 10.3724/SP.J.1206.2010.00419. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be available on request to corresponding or first author.