Abstract
1,4-Dioxane is a genotoxic carcinogen, and its mutagenic properties were recently observed in the liver of guanine phosphoribosyl transferase (gpt) delta transgenic rats. However, the mechanisms of its genotoxicity remain unclear. We analyzed DNA adduct formation in rat livers following 1,4-dioxane treatment. After administering 1,4-dioxane in drinking water at doses of 0, 20, 200, and 5,000 ppm, liver adduct formation was analyzed by DNA adductome analysis. Adducts in treated rat livers were dose-dependently increased compared with those in the control group. Principal component analysis-discriminant analysis (PCA-DA) clearly revealed two clusters of DNA adducts, associated with 0 ppm and low-dose (20 ppm) 1,4-dioxane-treatment versus middle- and high-dose (200, 5,000 ppm)-treated rats. After confirming the intensity of each adduct, three adducts were screened as characteristic of 1,4-dioxane treatment. Two of the three candidates contained thymine or cytidine/uracil moieties. Another candidate was identified as 8-oxo-dG based on mass fragmentation together with high-resolution accurate-mass (HRAM) mass spectrometry data. Oxidative stress responses may partly explain the mechanisms of increased mutations in the liver of gpt delta rats following 1,4-dioxane treatment.
Keywords: DNA adduct; 1,4-dioxane; gpt delta rat
Introduction
1,4-Dioxane is a synthetic industrial chemical that is widely used as a solvent for organic products, including dyes, waxes, and cosmetics,1–4) hence, it is often found as an impurity in various consumer products, such as deodorants, shampoos, and cosmetics. Moreover, 1,4-dioxane is released into the environment during production, resulting in contamination of drinking water and food, which poses potential health concerns. This compound has been reported to induce liver cancer in rats;5) however, the mechanisms remain unclear. Recently, we evaluated the in vivo genotoxicity of 1,4-dioxane using the liver of guanine phosphoribosyl transferase (gpt) delta transgenic F344 rats, and found that the overall mutation frequency and A:T to G:C transitions and A:T to T:A transversions in the gpt transgene were significantly increased by oral administration of the highest dose (5,000 ppm) of 1,4-dioxane.6) Moreover, the expression level of the DNA repair enzyme methyl guanine methyl transferase (MGMT), was significantly induced at 5,000 ppm 1,4-dioxane, indicating various types of DNA damage (DNA adducts) occurred following 1,4-dioxane exposure, which exceeded the repair capacity of cells in the liver and consequently led to mutations in the liver of gpt delta rats.
DNA adductome analysis is a comprehensive next-generation method involving liquid chromatography-mass spectrometry methods. Recently, we developed an approach using a high-performance liquid chromatography-quadrupole time-of-flight (HPLC-QTOF) mass spectrometer (MS).7,8) One advantage of our approach is that high-resolution accurate-mass (HRAM) analysis provides information on the chemical structures of the detected DNA adducts, which can then be identified by referring to a list of 130 DNA adducts with known m/z [M + H]+ values compiled in our previous study.8) HRAM can be used to acquire accurate spectral data using a mass measurement in the order of 0.001 atomic mass units (amu), which is sufficient for determining the molecular formula of an ion. Moreover, MS/MS fragmentation data obtained in these conditions are used to detect DNA adducts and provide structural confirmation or information. In the present study, we analyzed the DNA adduct formation of 1,4-dioxane through comprehensive analysis of DNA adducts, termed “adductome analysis”, and screened a few characteristic DNA adducts produced following 1,4-dioxane treatment. The potential mechanisms of genotoxicity induced by 1,4-dioxane were also predicted.
Materials and methods
Chemicals.
1,4-Dioxane was purchased from Wako Pure Chemical Industries, Ltd. (Osaka, Japan; purity >99.9%). Nuclease P1 and HPLC-grade acetonitrile were also purchased from Wako Pure Chemical Industries, Ltd. Phosphodiesterase I was purchased from Worthington Biochem (Lakewood, NJ, U.S.A.). Bovine spleen phosphodiesterase II, DNase I, and bacterial alkaline phosphatase Type III (Escherichia coli) were purchased from Sigma-Aldrich (St. Louis, MO, U.S.A.). All other chemicals used were of analytical grade and purchased from Wako.
DNA adductome analysis.
Frozen liver tissue samples from 5 rats per group from the 0 (control), 20, 200, and 5,000 ppm 1,4-dioxane groups from our previous 16-week study were used for DNA adductome analysis.6) In the study, male F344 rats (Charles River Japan, Inc.) at 3 weeks of age were administered 1,4-dioxane in their drinking water for 16 weeks.
DNA from liver tissues was extracted and purified using a Gentra® Puregene™ tissue kit (QIAGEN, Hilden, Germany) in accordance with the manufacturer’s instructions except that desferroxamine (final concentration: 0.1 mM) was added to all solutions to avoid the formation of oxidative adducts during the purification step. The extracted DNA was stored at −80 ℃ until DNA adductome analysis. DNA samples were enzymatically digested as reported previously.7) LC-HRAM analyses were performed using a Shimadzu Prominence LC system (Kyoto, Japan) interfaced with a Triple TOF6600 mass spectrometer (SCIEX, Framingham, MA, U.S.A.) in Information Dependent Acquisition Scanning mode. The HPLC conditions were as follows: column = SynergiTM Fusion-RP (2.5 µm particle size, 2.0 × 100 mm; Phenomenex, Torrance, CA, U.S.A.); flow rate = 0.4 mL/min; and solvent system = a linear gradient from 2.5% to 85% acetonitrile in 10 mM ammonium acetate (pH 5.3) over 30 min, controlled using Analyst TF 1.7.1 software. Sample injection volumes were 10 µL each. MS parameters were as follows: mass range scanned from 50 to 1,000 with a scan duration of 0.5 s (1.0 s total duty cycle), capillary 3.7 kV, sampling cone 40 V, extraction cone 4 V, source temperature 125 ℃, and desolvation temperature 250 ℃. Nitrogen gas was used as the desolvation gas (flow 800 L/h) and cone gas (30 L/h). All data were collected in positive ion mode. A cone voltage of 20 V was used.
The raw data files obtained from the LC-HRAM runs were analyzed using PeakView® 2.1 and MarkerView 1.3 software (SCIEX). These applications detect, integrate, and normalize the intensities of the peaks to the sum of peaks within a specific sample. The resulting multivariate dataset, which consisted of the peak number (based on the retention time and m/z), sample name, and normalized peak intensity, was analyzed using principal component analysis-discriminant analysis (PCA-DA).
Statistical analysis.
Intensity data obtained from the adductome were expressed as the means ± standard deviation (SD) and compared with those of the corresponding solvent control (1,4-dioxane 0 ppm) using the F test followed by Student’s t test.
Results
Comprehensive analysis of DNA adducts induced by 1,4-dioxane treatment.
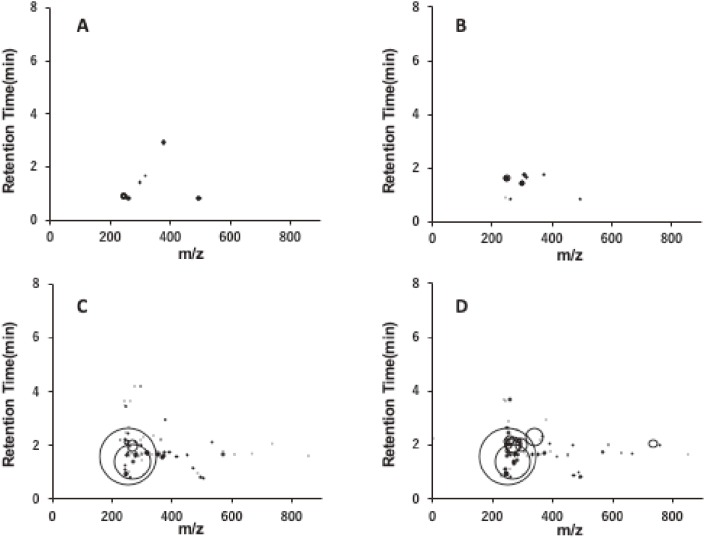
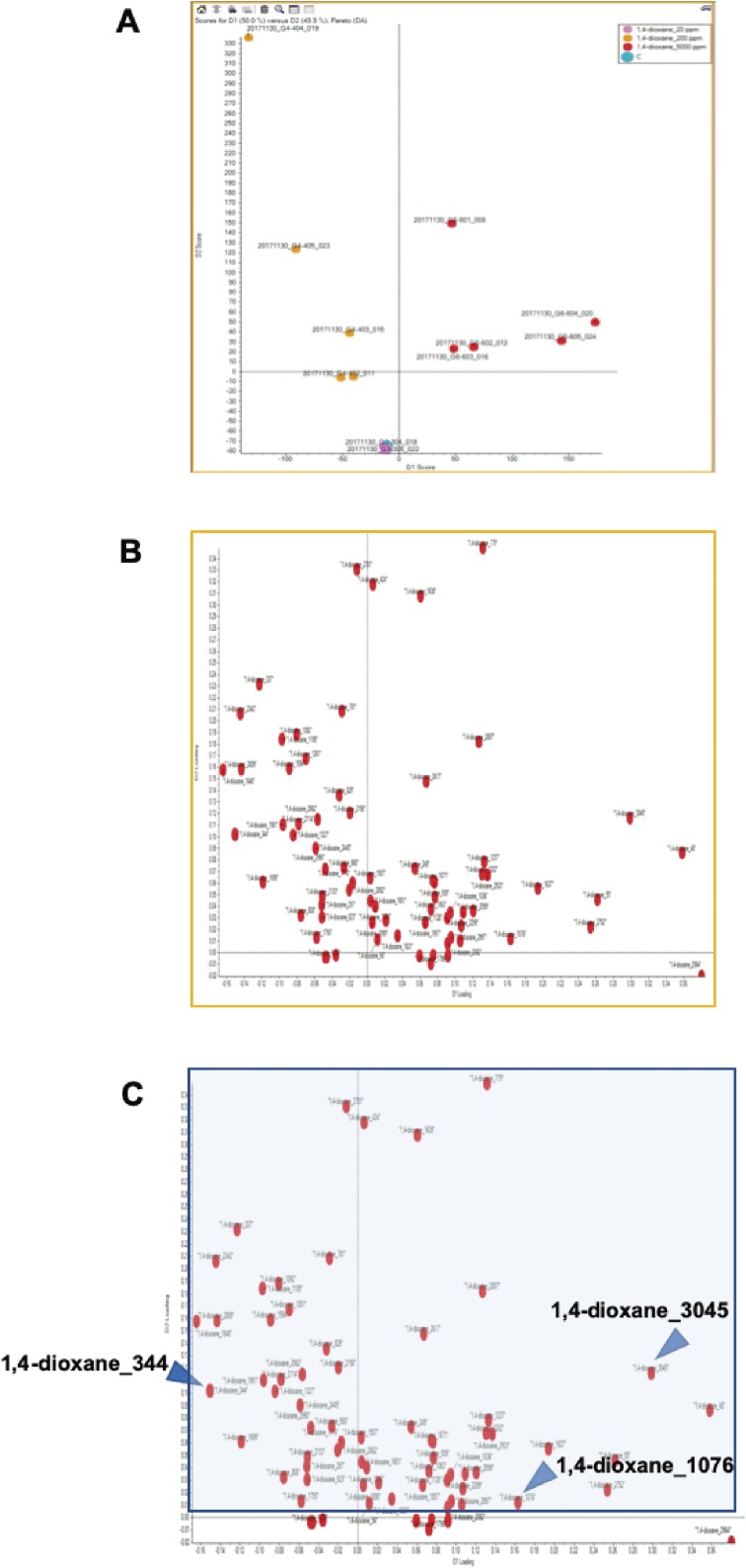
Recently, we reported that 1,4-dioxane was clearly genotoxic in the liver of gpt delta transgenic rats by oral administration at a dose of 5,000 ppm for 16 weeks.6) Mutation spectra analysis showed that most mutations induced by 1,4-dioxane occurred at A/T base pairs, and the prominent mutation types were A:T to G:C transitions followed by A:T to T:A transversions.6) To investigate the mechanisms of the induction of mutations in rat livers by 1,4-dioxane administration, we performed adductome analysis as described previously.7,8) Approximately 140 types of DNA adducts were detected in the control and 1,4-dioxane-treated groups (0, 20, 200, 5,000 ppm) (Fig. 1). Only small numbers of DNA adducts were detected in the control and low-dose (0, 20 ppm) treatment groups, whereas larger numbers of DNA adducts were observed following treatment with middle and high doses of 1,4-dioxane (200 and 5,000 ppm) (Fig. 1). Moreover, DNA adductome map views of the 200 and 5,000 ppm 1,4-dioxane-treated groups showed similar patterns, with a slightly increased number of DNA adducts at the highest dose of 1,4-dioxane treatment (Fig. 1C and D). PCA-DA against a subset of DNA adducts observed in these data sets was applied; the results are shown in the 2D PCA-DA score plot (Fig. 2A) and associated loading plot (Fig. 2B). Clear clustering was observed according to control and low dose (0, 20 ppm) of 1,4-dioxane-treatment versus middle and high dose (200, 5,000 ppm)-treated rats (Fig. 2A). This suggested that the DNA adduct status was altered in the middle- and high-dose groups.
Figure 1.
Comprehensive DNA adducts analysis. Map views of DNA adducts in the liver of rats with or without 1,4-dioxane exposure. (A) 1,4-dioxane 0 ppm (control), (B) 20 ppm, (C) 200 ppm, and (D) 5,000 ppm.
Figure 2.
PCA-DA scores and loading plots. (A) 2D PCA-DA scores of DNA adducts obtained from adductome analysis. Light blue; 1,4-dioxane 0 ppm (control), pink; 1,4-dioxane 20 ppm, yellow; 1,4-dioxane 200 ppm, red; 1,4-dioxane 5,000 ppm. (B) Variable loading plots. Each red spot represents a DNA adduct observed in the DNA adductome analysis. (C) The intensity of each DNA adduct enclosed in a square region was confirmed. The candidate characteristic DNA adducts for 1,4-dioxane exposure are indicated by arrowheads.
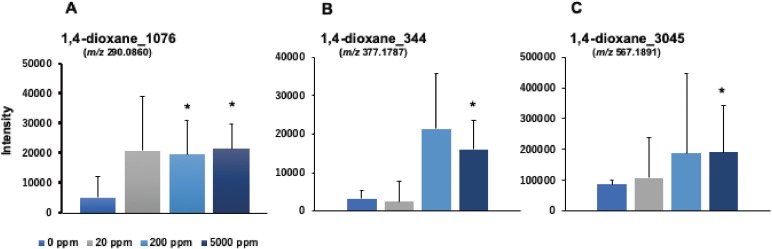
The associated loadings plot demonstrated that the number of DNA adducts made a greater contribution to the middle- and high-dose 1,4-dioxane treatments based on their PCA significance (Fig. 2B). To identify the characteristic DNA adducts formed by 1,4-dioxane treatment, we confirmed the intensity of each DNA adduct plotted in the square region (Fig. 2C). Three DNA adducts were screened as candidate characteristic DNA adducts for 1,4-dioxane exposure because their intensities were significantly higher in the 1,4-dioxane groups treatment rather than in the control group. Figure 3 shows the intensities observed in the control and each treated sample for the three candidates of characteristic DNA adducts following 1,4-dioxane exposure. The DNA adducts, named as 1,4-dioxane_1076 (m/z [M + H]+ 290.0860 at RT 1.98 min), 1,4-dioxane_344 (m/z [M + H]+ 377.1787 at RT 2.95 min), and 1,4-dioxane_3045 (m/z [M + H]+ 567.1891 at RT 1.76 min), showed significantly higher contributions following 1,4-dioxane exposure at the middle- and high-dose treated groups. To confirm the results of PCA-DA analysis, random forest (RF) analysis of the DNA adductome profile data was performed. The DNA adducts that effectively distinguished the groups (control vs. 1,4-dioxane) and are shown in the importance plot (Supplementary Fig. 1). The DNA adduct, 1,4-dioxane_344, was the most important variable causing clustering in both the mean decrease in accuracy and mean decrease in the Gini index (Supplementary Fig. 1). This DNA adduct, named 1,4-dioxane_344, was found in both the PCA-DA and RF analyses to correlate highly with the effects of 1,4-dioxane exposure.
Figure 3.
Quantitative analysis of three DNA adducts screened as candidate characteristic DNA adducts for 1,4-dioxane exposure. The intensity of DNA adducts, 1,4-dioxane_1076 (A), 1,4-dioxane_344 (B) and 1,4-dioxane_3045 (C), screened as characteristic DNA adducts for 1,4-dioxane treatment. Asterisk (*) indicates a significant difference (P < 0.05) from the control by Student’s t-test.
Identification of DNA adducts correlated with 1,4-dioxane treatment using in-house DNA adduct database.
To identify the chemical structures of DNA adducts detected as characteristic to 1,4-dioxane exposure (1,4-dioxane_344, 1076, 3045), we used our in-house DNA adduct database.8) First, the m/z [M + H]+ values were compared with those of known DNA adducts listed in our in-house database. However, these adducts considered as characteristic to 1,4-dioxane exposure were not found in the database. Therefore, these DNA adducts may be novel.
Analysis of chemical structures of characteristic DNA adducts after 1,4-dioxane exposure using HRAM data.
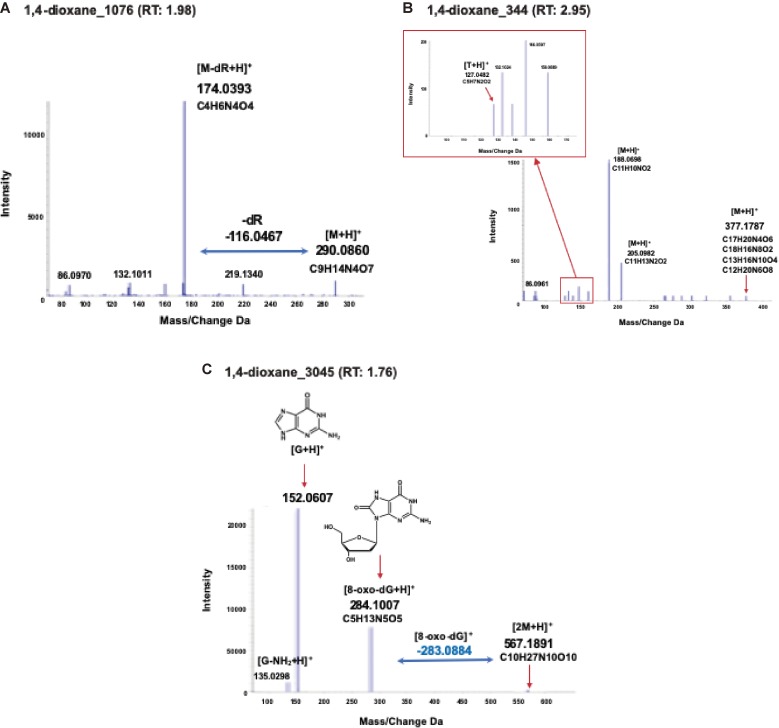
HRAM can be used to obtain accurate spectral data with mass measurements in the order of 0.001 amu, which is sufficient for determining the molecular formula of an ion. Moreover, MS/MS fragmentation data can reveal the presence of a DNA adduct and provide structural confirmation or information. The loss of 116.047 amu, corresponding to a deoxyribose (dR) moiety, is used as a hallmark of deoxyribonucleosides. However, some types of DNA adducts do not show loss of a dR moiety. The N7- and N3-positions of adenine, N7-position of guanine, and O2-position of cytosine and thymine are known to yield unstable nucleoside adducts. Therefore, the adducts formed at these positions and the dR moiety are readily lost upon enzymatic/thermal hydrolysis of the DNA to produce aglycone base adducts. These base adducts show loss of the base moiety during MS analysis.9,10) In the present study, by using accurate m/z values and their fragmentation data, candidate formulas of adducts strongly considered to have resulted from 1,4-dioxane treatment were automatically calculated with PeakView® software (SCIEX), as shown in Fig. 4. In the case of 1,4-dioxane_1076, a fragment ion peak corresponding to the loss of a dR moiety (−116.0467) from the precursor ion peak (m/z 290.0860) was observed (Fig. 4A). Because the candidate formula of [M-dR + H]+ is C4H6N4O4, this adduct contains the cytosine [M + H C4H6N3O] moiety but not the guanine [M + H C5H5N5O], adenine [M + H C5H5N5], and thymine [M + H C5H6N2O2] moieties. Uracil [M + H C4H4N2O2] is another candidate base of this adduct, because deoxyuridine (dU) is produced from spontaneous deamination of deoxycytidine (dC)11) and cytidine deaminase enzymes, such as AID/APOBEC3.12) In contrast, a fragment ion corresponding to loss of a dR moiety was not observed in adduct 1,4-dioxane_344 (Fig. 4B); however, a fragment ion with an m/z 127.0482 [M + H]+ showed a very similar m/z value to thymine with an m/z 127.0500 [T + H]+ (Supplementary Fig. 2). These results suggested that this adduct contains a thymine moiety. Although several candidate formulas of the precursor ion were calculated using the software, precise structural analysis remains difficult. To obtain sufficient information useful for structural identification, further studies are needed. In contrast, the fragment ions observed in 1,4-dioxane_3045 indicate the formation of 8-oxo-dG, an oxidative stress-related adduct.11,13) Indeed, a neutral loss corresponded to 8-oxo-dG (m/z 283.0884 [8-oxo-dG]+) from a precursor ion of m/z 567.1891 and product ion of m/z 284.1007 [M + H]+ (Fig. 4C). The m/z value 284.1007 [M + H]+ was nearly identical to that of 8-oxo-dG (m/z 284.0917 [M + H]+). Based on these results, the precursor ion was considered to indicate a dimer of 8-oxo-dG (m/z 567.1891 [2M + H]+), and we concluded this adduct was an 8-oxo-dG adduct, especially as it has been reported that chemical substances frequently dimerize during ionization in MS analysis.14)
Figure 4.
Product ion scan and MS/MS fragmentation data of candidate DNA adducts. Using HRAM data, a candidate formula of the adduct was automatically calculated using PeakView® software (SCIEX), as described above. (A) MS/MS fragmentation data of 1,4-dioxane_1076 with a retention time of 1.98 min. A peak corresponding to the loss of a dR moiety (−116.0467) from the precursor ion peak (m/z 290.0860) was observed. (B) MS/MS fragmentation data of 1,4-dioxane_344 with a retention time of 2.95 min. A fragment ion corresponding to loss of a dR moiety was not observed; however, a fragment ion with m/z 127.0482 [M + H]+ showing a similar m/z value to thymine with m/z 127.0500 [T + H]+, was observed. (C) MS/MS fragmentation data of 1,4-dioxane_3045 with a retention time of 1.76 min. A neutral loss corresponding to 8-oxo-dG (m/z 283.0884 [8-oxo-dG]+) from the precursor ion with m/z 567.1891, and a product ion with m/z 284.1007 [M + H]+ was observed. The m/z value 284.1007 [M + H]+ was nearly identical to that of 8-oxo-dG (m/z 284.0917 [M + H]+).
Discussion
We demonstrated previously that 1,4-dioxane is a genotoxic hepatocarcinogen in gpt delta transgenic rats.6) The gpt mutation frequency was significantly increased at the high dose administered (5,000 ppm) and showed an increased frequency at the middle dose (1,000 ppm). Mutations induced by 1,4-dioxane treatment included A:T to G:C transitions followed by A:T to T:A transversions. In addition, G:C to T:A transversions were slightly but not significantly increased. However, the detailed genotoxicity mechanisms of 1,4-dioxane remained unclear. It has also been reported that the expression levels of the DNA damage repair enzyme MGMT were increased in the liver of gpt delta rats administered the high dose of 1,4-dioxane, but not in the low-dose groups.6) Therefore, some types of DNA adducts are likely produced by 1,4-dioxane exposure and may be involved in inducing MGMT expression in the liver of rats.
Generally, O6-methyl guanine and O4-methyl thymine are the primary substrates of MGMT.11,15) In fact, mutations at A/T base pairs were greatly increased by the high dose of 1,4-dioxane. However, O4-methyl thymine was not identified as a characteristic DNA adduct in this study. Alternatively, three candidate characteristic DNA adducts for 1,4-dioxane exposure were identified in the DNA adductome by PCA-DA and RF analyses. Because the m/z values of these DNA adducts were not included in our in-house DNA adduct database, these DNA adducts are novel. Although no data are available regarding the precise formulas and chemical structures of these DNA adducts, 1,4-dioxane_344 would include a thymine moiety in the HRAM data. In addition, the intensity of this DNA adduct was statistically increased in the high dose of 1,4-dioxane, it is, therefore, postulated that 1,4-dioxane_344 may contribute to the induction of A:T to G:C and A:T to T:A mutations. Similarly, 1,4-dioxane_1076 is likely a cytidine or uracil adduct. It has been reported that spontaneous deamination of cytidine and/or cytidine deaminase enzymes, such as AID/APOBEC3, produce dU from dC in single-stranded DNA.11,12) Additionally, AID/APOBEC3 was reported to efficiently deaminate dC when commonly damaged bases, such as in alkylation and oxidation, were present in the −2 or −1 positions.16) In addition, 1,4-dioxane_3045 was identified as 8-oxo-dG based on the mass fragmentation and HRAM data. 8-Oxo-dG is produced from reactive oxygen species and is known to be an oxidative stress-related adduct.11,13) Because 8-oxo-dG was found to be involved in G to T transversion in in vitro assays,17,18) it is likely that oxidative stress occurs in the mechanisms that increase mutations in the liver of gpt delta rats after 1,4-dioxane treatment. In contrast to the present results, the level of 8-oxo-dG was similar in the livers of gpt delta rats regardless of the 1,4-dioxane exposure status in our previous study.6) The reason for this discrepancy remains unclear, but differences in sample preparation and detection methods may have influenced the results. However, the degree of increment of 8-oxo-dG in the middle- and high-dose 1,4-dioxane treatment groups was moderate, thus it is suggested that 8-oxo-dG may not be the main contributor to the genotoxicity observed in the liver of F344 rats after 1,4-dioxane treatment. In fact, G to T transversion was slightly increased in the liver of rats at the highest dose of 1,4-dioxane.6) Therefore, based on our results, the host reaction against the genotoxic agent 1,4-dioxane induced the formation of oxidative stress-related DNA adducts, such as 8-oxo-dG, which partly contribute to genotoxicity observed in the liver of F344 rats after 1,4-dioxane treatment. Supporting our hypothesis, Mnaa et al. reported that administration of 1,4-dioxane increased the concentration of the oxidative stress markers of 2-thiobarbituric acid-reactive substances, such as malondialdehyde, in the serum after 42 days of exposure.19) Moreover, oral injection of the antioxidative agent N-acetyl cysteine decreased malondialdehyde levels compared with the vehicle control and diminished liver toxicity induced by 1,4-dioxane.19) Similarly, Qiu et al. reported that high doses of 1,4-dioxane (500 mg/mL) significantly disrupted various metabolic pathways, concomitantly with renal tissue damage and stimulation of the oxidant defense system, and significantly increased glutathione levels in the urine according to transcriptomics and urine metabolomics analyses.20)
Several studies have reported on the genotoxicity of 1,4-dioxane; however, whether 1,4-dioxane is definitively genotoxic remains controversial.21–23) In the present study, the total number of adducts observed in the livers of rats administered middle and high doses of 1,4-dioxane (200 and 5,000 ppm) appeared to increase compared with the control and low-dose (0, 20 ppm) treatment groups. It is, therefore, possible that the apparent threshold existed between low and middle doses. In our previous study, a linear relationship was observed between the carcinogen (2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline; MeIQx) dose and MeIQx-DNA adduct, which formed via direct binding of MeIQx to nucleobases. In contrast, an oxidative stress-DNA adduct (8-OHdG), which is believed to form via secondary responses, such as carcinogen–host interactions, demonstrated a lack of a dose–response relationships.24) Hence, it is suggested that the mechanisms of mutation induction in gpt transgenic rats after 1,4-dioxane treatment may not require direct binding action with DNA.
Finally, three DNA adducts were screened as characteristic adducts for 1,4-dioxane treatment in the present study. However, we could not identify the chemical structures of most adducts, except for the oxidative-DNA damage-related molecule, 8-oxo-dG. Thus, we could not confirm that 1,4-dioxane directly binds to DNA nucleobases and forms DNA adducts. To determine the genotoxic mechanisms of 1,4-dioxane, further studies such as the structural analysis of characteristic DNA adducts for 1,4-dioxane exposure are needed. DNA adductome analysis should be performed to screen and identify the characteristic DNA adducts related to chemical exposure when the m/z values of DNA adducts are present in the in-house database. Additionally, further studies to predict the chemical structure of novel DNA adducts and improve the DNA adduct database are needed.
Acknowledgements
We thank Mr. Shuntaro Akimoto for providing technical assistance. This study was supported by Grants-in-Aid for Research on Risk of Chemical Substances from the Ministry of Health, Labour, and Welfare of Japan.
Abbreviations
- HRAM
high-resolution accurate-mass
- MGMT
methyl guanine methyl transferase
Supplementary materials
Supplementary materials are available at https://doi.org/10.2183/pjab.96.015.
References
- 1).IARC (1999) 1,4-Dioxane, in: re-evaluation of some organic chemicals, hydrazine and hydrogen peroxide. IARC Monogr. Eval. Carcinog. Risks Hum. 7, 589–602. [PMC free article] [PubMed] [Google Scholar]
- 2).Agency for Toxic Substances and Disease Registry (ATSDR) (2012) Toxicological Profile for 1,4-Dioxane. ATSDR, Atlanta, GA. [PubMed] [Google Scholar]
- 3).U.S. EPA (2013) United States Environmental Protection Agency, IRIS Toxicological Review of 1,4-Dioxane (with Inhalation Update) (Final Report) (EPA/635/R-11/003F). https://cfpub.epa.gov/ncea/iris/iris_documents/documents/toxreviews/0326tr.pdf.
- 4).U.S. EPA (2017) United States Environmental Protection Agency, Technical Fact Sheet–1,4-Dioxane (EPA 505-F-17-011). https://www.epa.gov/sites/production/files/2014-03/documents/ffrro_factsheet_contaminant_14-dioxane_january2014_final.pdf.
- 5).Kano H., Umeda Y., Kasai T., Sasaki T., Matsumoto M., Yamazaki K., et al. (2009) Carcinogenicity studies of 1,4-dioxane administered in drinking-water to rats and mice for 2 years. Food Chem. Toxicol. 47, 2776–2784. [DOI] [PubMed] [Google Scholar]
- 6).Gi M., Fujioka M., Kakehashi A., Okuno T., Masumura K., Nohmi T., et al. (2018) In vivo positive mutagenicity of 1,4-dioxane and quantitative analysis of its mutagenicity and carcinogenicity in rats. Arch. Toxicol. 92, 3207–3221. [DOI] [PubMed] [Google Scholar]
- 7).Ishino K., Kato T., Kato M., Shibata T., Watanabe M., Wakabayashi K., et al. (2015) Comprehensive DNA adduct analysis reveals pulmonary inflammatory response contributes to genotoxic action of magnetite nanoparticles. Int. J. Mol. Sci. 16, 3474–3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8).Totsuka Y., Lin Y., He Y., Ishino K., Sato H., Kato M., et al. (2019) DNA adductome analysis identifies N-nitrosopiperidine involved in the etiology of esophageal cancer in Cixian, China. Chem. Res. Toxicol. 32, 1515–1527. [DOI] [PubMed] [Google Scholar]
- 9).Stornetta A., Villalta P.W., Hecht S.S., Sturla S.J., Balbo S. (2015) Screening for DNA alkylation mono and cross-linked adducts with a comprehensive LC-MS3 adductomic approach. Anal. Chem. 87, 11706–11713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10).Villalta P.W., Balbo S. (2017) The future of DNA adductomics analysis. Int. J. Mol. Sci. 18, 1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11).Chatterjee N., Walker G.C. (2017) Mechanisms of DNA damage, repair, and mutagenesis. Environ. Mol. Mutagen. 58, 235–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12).Siriwardena S.U., Chen K., Bhagwat A.S. (2016) Functions and malfunctions of mammalian DNA-cytosine deaminases. Chem. Rev. 116, 12688–12710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13).Kasai H. (2016) What causes human cancer? Approaches from the chemistry of DNA damage. Genes Environ. 38, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14).De Vijlder T., Valkenborg D., Lemière F., Romijn E.P., Laukens K., Cuyckens F. (2018) A tutorial in small molecule identification via electrospray ionization-mass spectrometry: The practical art of structural elucidation. Mass Spectrom. Rev. 37, 607–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15).Sharma S., Salehi F., Scheithauer B.W., Rotondo F., Syro L.V., Kovacs K. (2009) Role of MGMT in tumor development, progression, diagnosis, treatment and prognosis. Anticancer Res. 29, 3759–3768. [PubMed] [Google Scholar]
- 16).Diamond C.P., Im J., Button E.A., Huebert D.N.G., King J.J., Borzooee F., et al. (2019) AID, APOBEC3A and APOBEC3B efficiently deaminate deoxycytidines neighboring DNA damage induced by oxidation or alkylation. Biochim. Biophys. Acta, Gen. Subj. 1863, 129415. [DOI] [PubMed] [Google Scholar]
- 17).Shibutani S., Takeshita M., Grollman A.P. (1991) Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature 349, 431–434. [DOI] [PubMed] [Google Scholar]
- 18).Moriya M. (1993) Single-stranded shuttle phagemid for mutagenesis studies in mammalian cells: 8-Oxoguanine in DNA induces targeted G·C→T·A transversions in simian kidney cells. Proc. Natl. Acad. Sci. U.S.A. 90, 1122–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19).Mnaa S., Shaker E.S., Mahmoud H.I. (2016) Inhibitory activity of protected edible plants on oxidative stress induced by oral 1,4-dioxane. J. Egypt. Soc. Parasitol. 46, 135–143. [DOI] [PubMed] [Google Scholar]
- 20).Qiu J., Cheng J., Xie Y., Jiang L., Shi P., Li X., et al. (2019) 1,4-Dioxane exposure induces kidney damage in mice by perturbing specific renal metabolic pathways: An integrated omics insight into the underlying mechanisms. Chemosphere 228, 149–158. [DOI] [PubMed] [Google Scholar]
- 21).Morita T., Hayashi M. (1998) 1,4-Dioxane is not mutagenic in five in vitro assays and mouse peripheral blood micronucleus assay, but is in mouse liver micronucleus assay. Environ. Mol. Mutagen. 32, 269–280. [DOI] [PubMed] [Google Scholar]
- 22).Roy S.K., Thilagar A.K., Eastmond D.A. (2005) Chromosome breakage is primarily responsible for the micronuclei induced by 1,4-dioxane in the bone marrow and liver of young CD-1 mice. Mutat. Res. 586, 28–37. [DOI] [PubMed] [Google Scholar]
- 23).Itoh S., Hattori C. (2019) In vivo genotoxicity of 1,4-dioxane evaluated by liver and bone marrow micronucleus tests and Pig-a assay in rats. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 837, 8–14. [DOI] [PubMed] [Google Scholar]
- 24).Fukushima S., Wanibuchi H., Morimura K., Wei M., Nakae D., Konishi Y., et al. (2002) Lack of a dose-response relationship for carcinogenicity in the rat liver with low doses of 2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline or N-nitrosodiethylamine. Jpn. J. Cancer Res. 93, 1076–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]