Abstract
Aim
Colon cancer is one of the leading causes of cancer-related mortality. However, specific biomarkers for its diagnosis or treatment are not established well.
Methods
We developed a colon-cancer specific peptide probe using phage display libraries. We validated the specificity of this probe to colon cancer cells with immunohistochemical staining and FACS analysis using one normal cell and five colon cancer cell lines.
Results
This peptide probe maintained binding affinity even after serum incubation. For therapeutic applications, this peptide probe was conjugated to hematoporphyrin, a photosensitizer, which showed a significantly enhanced cellular uptake and high photodynamic effect to kill tumor cells. As another application, we made a nanoparticle modified from the peptide probe. It efficiently delivered SN-38, an anticancer drug, into tumor cells, and its tumor-targeting ability was observed in vivo after intravenous injection to the same xenograft model.
Conclusion
The noble dodecapeptide probe can be a promising candidate for both colon tumor diagnosis and targeted drug delivery.
Introduction
Colorectal cancer (CRC) is one of the major leading causes of cancer-related deaths [1]. Its prognosis is dependent on its stage. Because there are no diagnostic biomarkers approved for clinical use, CRC has been evaluated with colonoscopy [2,3]. Even though white-light colonoscopy is a good modality, missed colon cancer has been a problem. Therefore, more sensitive imaging techniques have been investigated to detect neoplastic lesions efficiently. In the treatment of advanced colon cancers, targeted therapies are not easy due to the absence of specific biomarkers based on tumor-specific biological processes.
Molecular imaging techniques have been studied with the use of different signature of target tissues compared to their correspondence [[4], [5], [6], [7]]. Development of high-affinity antibodies has advanced tumor diagnosis and target therapy via specific binding to target tissues [8]. However, the size, immunogenicity, and pharmacokinetic properties of such antibody products limited their usefulness in solid malignancies [9].
Peptides can bind to a wide range of targets on cell surface and have advantages as a potential target-specific probe. Their pharmacokinetic behaviors including good stability, rapid peak uptake, and quick clearance enable them to be used in a clinical setting to image several malignant diseases including neuroendocrine tumors, lymphomas, and melanomas [[10], [11], [12]]. Peptide probes also can penetrate through deep tissue in tumors while preventing nonspecific uptake by the reticuloendothelial system [13]. They have a low risk to provoke an immune response, which allows for repeated use.
Phage display is a technique providing the selection of specific peptides that bind to target cells [14,15]. To date, numerous studies have been performed to screen peptide-specific binding to cancer cells using phage display methods [[16], [17], [18]]. Several studies have reported peptides targeting colon cancer cells. However, their specificity and sensitivity to colon cancer cells were not validated [19,20].
The aim of this study was to develop a peptide that specifically binds to human colon cancer cells and to validate this peptide in vivo and in vitro. Furthermore, we proved this peptide could be a potential probe as a therapeutic tool after conjugation with drug and nanoparticles (NPs).
Material and Methods
The Ph.D. -12TM phage display peptide library kit containing E. coli host strain ER2738 and M13KE control phage (New England BioLabs, Ipswich, MA) was used. The phage display library contained random peptides constructed at the N-terminus of the coat protein (pIII) of M13 phage. The titer of the library was 0.5 to 2 × 1013 plaque-forming unit. Horseradish peroxidase/anti-M13 monoclonal conjugate antibody (Abcam, Cambridge, UK) was used. Fetal bovine serum and trypsin were obtained from Thermo Fisher Scientific (Waltham, MA). The DNA sequencing primer was synthesized at Cosmogenetech (Seoul, Korea). Bacto-tryptone, bacto-yeast extract, and NaCl were obtained from Sigma-Aldrich (St. Louis, MO). 1, 2-Dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) was purchased from Echelon Biosciences (Salt Lake City, UT). 1,2-Distearoyl-sn-glycero-3-phosphoethanolamine-N-methoxy(polyethylene glycol)(DSPE-mPEG) (molecular weight, 3000) was purchased from Nanosoft Polymers (Winston-Salem, NC). 7-Ethyl-10-hydroxycamptothecin (SN38) was purchased from Carbosynth (San Diego, CA). Hematoporphyrin (HPP), soybean oil, and 1,1′-dioctadecyl-3,3,3′,3′-tetramethyl indocarbo cyanine perchlorate (DiI dye) were purchased from Sigma-Aldrich (St. Louis, MO).
Cell Lines and Cell Culture
Human colon cancer cells (LoVo, HCT116, HT29, SW480, and DLD-1) and colon fibroblast cells (CCD18Co) were used (ATCC, Manassas, VA). Mouse colon carcinoma cell line (CT26) was used for mouse serum stability test (ATCC). Colon cancer cells were cultured in RPMI medium (Thermo Fisher Scientific, Waltham, MA). CCD841 and CCD18Co were cultured in MEM medium (Thermo Fisher Scientific). All cells were incubated at 37°C in a humidified atmosphere with 5% CO2.
Cell-Based Enzyme-Linked Immunosorbent Assay (ELISA)
LoVo and CCD18Co cells were cultured and plated in 96-well plates. The cells were washed with 1× PBS for three times and then fixed with 4% paraformaldehyde for 40 minutes at room temperature (RT). After fixation, the cells were washed with 1× PBS and blocked with 1× PBS containing 3% BSA for 1 hour at RT. Each phage was incubated separately in a 96-well plate in triplicate at RT for 1 hour. The well plate was washed again with 1× PBS, 100 μl of horseradish peroxidase conjugated anti M13 monoclonal antibody (1:500) was added to each well, and the plate was incubated RT for 1 hour with 80 rpm shaking. After washing, 100 μl of TMB substrate solution was added for 15 minutes, and 0.5 mol/l sulfuric acid solution was added for reaction termination. The plate was read on an automated ELISA plate reader at wavelength of 410 nm. Only PBS adding wells without phage were used as negative controls.
Phage Display Biopanning and Tittering
Screening procedures were performed according to the manufacturer's protocol (version 1.2). First, LoVo cell was digested with trypsin and blocked with 1× PBS containing 3% BSA at RT for 1 hour. Ph.D.-12 phage display peptide library was blocked in the same manner, and centrifuged supernatant containing phages was transferred to blocked LoVo cell containing tubes and incubated at RT for 2 hours. After binding, the library was centrifuged at 13,000 rpm for 10 minutes and washed with 1× PBS three times. After intensive washes, 1 ml of elution buffer (0.2 M glycine-HCl, pH 2.2) was added and incubated for 10 minutes. The eluate was immediately neutralized by the addition of 150 μl of 1 M Tris-HCl at pH 9.1 for 5 minutes. An aliquot of output samples was used for titering, and the rest of the phage was amplified for 5 hours using E. coli for the next biopanning. Specific enrichment of LoVo cell bound phages was finished after three rounds of panning, and titers of each round were calculated using a blue plague forming assay on agar plates containing isopropyl b-D-1 thiogalactopyranoside and X-gal selection reagents.
Sequencing of the Selected Phages
After three rounds of panning, 48 random phage colonies were picked and ssDNA was isolated from the phage. After amplifying selected phage, the pellet was suspended thoroughly in 100 μl of iodide buffer by tapping the tube. A total of 250 μl of 100% ethanol was added following incubation for 10- 20 minutes at RT. The pellet was spun in a microfuge at 14,000 rpm for 10 minutes, and the supernatant was discarded. The pellet was washed with 0.5 ml of 70% ethanol and briefly dried in vacuum. The primer used for sequencing was 5′-CCC TCA TAG TTA GCG TAA CG-3′ (−96 gIII sequencing primer) and 5′-GTA TGG GAT TTT GCT AAA CAA C-3′ (−28 gIII sequencing primer). Sequence alignment was performed according to ExPASy translate tool and reverse complement bioinformatics tool.
Peptide Synthesis
The candidate peptide was synthesized and purified by Peptron Co. (Daejeon, Korea). Fluorescein isothiocyanate (FITC) and cyanine 5.5 dye were conjugated with the N-terminal of candidate peptide and purified to a minimum purity of 95% by high-performance liquid chromatography.
Immunocytochemistry and Flow Cytometry
Purified candidate phage and FITC-labeled candidate peptides were used for the peptide-based immunofluorescence assay to confirm the selective binding of the selected peptides to LoVo, SW480, HCT116, HT29, and CCD841 cells. Cells were cultured and plated in eight-well chamber slide (Nunc, Naperville, IL) until they reached 70%-80% confluence. The cells were washed three times with PBS and blocked with 1× PBS containing 3% BSA for 1 hour at RT. Purified phage was added to each well and incubated at 4°C overnight. Slides were washed with 1× PBS and incubated with Ms. mAb to M13 antibody (Abcam) (1/500, v/v) for 2Please check if data here were captured correctly.
hours at RT. Goat anti rabbit IgG-Alexa 488 (1/1000, v/v) was added and incubated for 1 hour at RT. After washing, 4′,6-diamidino-2-phenylindole (DAPI) was added for 1 minute and mounted with Vectashield mounting medium (Vector Laboratories, Burlingame, CA).
FITC-labeled peptide-based immunofluorescence assay was performed, and FITC-labeled peptide (10 μM of final concentration) was added on each well and incubated for 1 hour at RT. Without second antibody incubation, the cells were directly mounted with Vectashield mounting medium after DAPI staining and visualized with a fluorescence microscope (ZEISS, Oberkochen, Germany). Flow cytometry was performed to detect L20 binding affinity on colon cancer cell lines. Five colon cancer and fibroblast cell lines were harvested and blocked with 3% BSA in 1× PBS, then added with FITC-L20 peptide (final concentration of 10 μM) for 1 hour at RT. After being washed with 1× PBS, these cells were suspended with 1% BSA in 1× PBS and observed under a BD FACS Canto II (BD Biosciences, San Jose, CA).
Serum Stability Test
Colon cancer cells were cultured and plated in eight-well chambers (Lab-Tek, Thermo Scientific). The cells were fixed with 4% paraformaldehyde for 40 minutes and blocked with 5% BSA for 1 hour. Human serum was obtained after centrifuge and then treated with 50% volume of FITC-L20 peptide for 30 minutes at RT. CT26 mouse-derived cell was plated in eight-well chambers and treated with mouse serum according to the incubation time point. The other process is the same with above. The well was washed again with 1× PBS and visualized with microscopy after counterstaining with DAPI.
Preparation and Characterization of NPs
L20-PEG-DSPE was synthesized by amide coupling reaction. DSPE-PEG5-NHS (50 mg, 9.2 μmol), amine-modified L20 peptide (16.5 mg, 9.7 μmol), and trimethylamine (1.5 μl, 10.2 μmol) were dissolved in MeOH (60 μl). The mixture was vortexed and mixed at ambient condition in a light-proof setting for 12 hours. The reaction mixture was purified with reverse-phase silica gel chromatography (H2O:ACN = 9:1 to ACN only) to yield L20-PEG-DSPE (12.1 mg, 1.7 μmol, 18.5% yield) white solid. To analyze the product, 1H NMR data were recorded on an JEOL ECZ-600R Magnetic Resonance System (600 MHz) at Ajou University. The NMR solvent used to acquire the spectra was methanol-D4 containing TMS. Using the resulting L20-PEG-DSPE, L20-NPs with different amount of peptide (0-0.5mg) were prepared by a traditional oil-in-water emulsion method via self-assembly. DPPC (0.5mg), DSPE-MPEG, and L20-PEG-DSPE were dissolved in 2 ml 2%(v/v) glycerol in water. The NPs were named as pep x, where x means the milligram weight of the L20-PEG-DSPE used. The sum of DSPE-MPEG and L20-PEG-DSPE was set as 0.5 mg in all NPs. Also, 20 mg of soybean oil was dissolved in 100 μl DMSO (containing 0.2 mg of DiI, 0.2 mg of DiD, or 1 mg of SN38), mixed with phospholipids and peptide solution at 60°C, and dispersed by sonication using C505 (SONIC & MATERIALS INC, Newtown, CT) for 20 minutes. Then, the resulting L20-NPs were purified by dialysis (MWCO: 13kDa) in distilled water for 1 hour. The size and zeta potentials of L20-NP were measured at 25°C in PBS (pH 7.4) using Zetasizer (Nano ZS90; Malvern Instruments, Malvern, UK) and analyzed with the Zetasizer software (version 7.12). In order to confirm L20-NP morphology, we used transmission electron microscopy (TEM) with negative staining using 2%(w/v) uranyl acetate solution.
Cellular Uptake Assay
First, L20-NPs were fabricated with DiI dye (0.2mg, λex/λem = 550/567 nm) for cellular uptake fluorescence imaging. LOVO cells were seeded at 4 × 104 cells/well in 24-well plates and cultured for 1 day. Then, the cells were washed with DPBS and treated with different concentration of NP (200-800 μg/ml) in serum-free medium and incubated for 2 hours. After incubation, LOVO cells were washed with DPBS twice and fixed with 4% paraformaldehyde in 4°C. Cells were stained with Hoechst 33342 (2 μg/ml) for 15 minutes and washed with DPBS. Cell imaging was obtained with a Fluorescence Inverted Microscope IX 71 (Olympus, Tokyo, Japan).
MTT Assay
The LOVO cell viability test was performed by MTT assay. L20-NPs were made with SN38 (1 mg), a chemotherapeutic drug. To measure the concentration of loaded SN38 in NPs, we analyzed the absorbance of SN38 (385 nm) using a synergy H1 Hybrid Multi-Mode Reader (BioTek Instruments, Inc., Winooski, VT) after completely destroying the NPs in detergent solution (DMSO:PBS:DW = 5:4:1, 1% Triton X-100). LOVO cells were seeded at 0.8 × 104 cells per well in 96-well plate and cultured for 1 day. Then, the cells were treated with 100 μl of each L20-NP (0.125-2 μg/ml of SN38) in serum-free medium for 15 minutes. Then, the medium was replaced with serum-containing RPMI medium and incubated for 48 hours, and 20 μl of MTT solution (0.5 mg/ml) was added to each well. After 2 hours of incubation at 37°C, the medium was removed, and 100 μl of DMSO was added. The absorbance was measured at 570 nm with PowerWave HTMicroplate Spectrophotometer (BioTek Inc., Winooski, VT).
In Vivo and Ex Vivo Near Infrared Fluorescence (NIRF) Imaging
The animal study was approved by the Institutional Review Board of our university (approval number: CUMC-2017-0133-02). We used BALB/C nude mice (4 weeks old, OrientBio, Seongnam city, Korea) for imaging. In order to establish tumor-bearing mouse models (n = 3 per each group), 2 × 106 LOVO cells in PBS with Matrigel (1:1, 100 μl) were subcutaneously injected into the left femoral region. For mice whole body imaging, L20-NPs were made with DiD, NIRF dye (0.2 mg). After tumors grew to approximately 150 mm3, each L20-NP sample with DiD Dye (100 μl in physiological saline) was intravenously injected into the mice tail vein. The mice were anesthetized with isofurane by a respiratory route, and whole body images of mice were gained with IVIS Lumina XRMS (PerkinElmer Inc., Waltham, MA) set at excitation 660 nm and emission 710 nm (cyanine 5.5 filter) at 3, 6, and 24 hours postinjection. For ex vivo analysis, 24 hours postinjection of L20-NP, the tumor and other organs (heart, lung, liver, spleen, and kidney) were dissected. The images were obtained by IVIS Lumina XRMS.
The dissected tumor from each mouse was collected with O.C.T compound in mold and stored at −80°C. The tumor tissues were cut into 5-μm–thick slices and washed with PBS. Then, fluorescence imaging was performed using a Fluorescence Inverted Microscope IX 71 (Olympus, Tokyo, Japan).
Model Systems and Energy Minimization for Molecular Modeling
Maestro (Schrödinger, LLC) software and included software packages were used for 3D modeling and calculation. We generated three types of 3D model for structural simulation of L20 peptide. Each model was arbitrarily elongated with fixed dihedral angles. The 3D structures were optimized energetically by the Polak-Ribiere conjugate gradient method and then the truncated Newton conjugate gradient method [21]. At each step, the iteration proceeded under water solvent conditions until the gradient converged using AMBER as the force field (convergence threshold of 0.05).
Molecular Dynamics (MD)
The MD simulation was performed for a total of 50 nanoseconds, three times for each model using the implicit solvent model and AMBER as the force field. For better similarity with the state of the sample at 25°C, the calculation was performed at 295 K without any symmetry restrictions or constraints. The energies for each interaction were calculated by using the generalized Born model. In the Born calculation for aqueous environment, the internal and external dielectric constants are generally set to 1.0 and 78.5, respectively, but we also considered the condition of setting the dielectric constant of the external environment to 5.0 in consideration of the similarity with the cell membrane environment. A volume-based continuum solvation model (GB/SA model) was used for calculation of the polarity component. The lengths of bonds to hydrogens were limited to equilibrium lengths using the SHAKE module [22]. And the cutoff distance parameters for noncovalent interactions such as van der Waals, electrostatic interactions, and hydrogen bonding were set to 8.0, 20.0, and 4.0 Å, respectively, using extended offsets. The time integration step was 1.0 femtosecond, and the intervals were set to 5.0 picoseconds for equilibration in all simulation processes. We selected the most stable structure from the energy point of view among the structures of MD simulation results we performed.
Statistical Analysis
All data are given as means ± standard deviation of N independent measurements. Statistical analysis was performed using a ttest and repeated-measures ANOVA. Statistical significance was assigned for P values <.05. All analyses were performed in SAS version 9.4 (SAS Institute, Inc., Cary, NC).
Results
Identification of Colon Cancer–Specific Peptides
After three rounds of whole cell biopanning on SW480, HCT116, HT29, and LoVo cells, a number of phage clones were randomly selected and amplified in E. coli ER2738. The titers amplified from the first to third round are shown in Supplementary Table 1. After three rounds of biopanning, the number of phages recovered was gradually increased about 100-fold compared to the first round, and positive candidate phage clones were enriched for DNA preparation. The binding ability of these selected phage clones was evaluated on colon cancer cells (SW480, HT29, HCT116, LoVo, and DLD1) as well as on a normal cell line (CCD-18co) by cell phage ELISA (Supplementary Figure 1). The binding affinities (relative OD values) showed more than two- to five-fold higher selectivity for colorectal cells than the negative controls. The phage clone with the highest optical density (450 nm) compared to the negative control cells was selected for DNA sequencing. The encoded peptide ANLNLWTDYIRW was named L20 peptide for further studies.
Binding of L20 Phage to Colon Cancer Cells
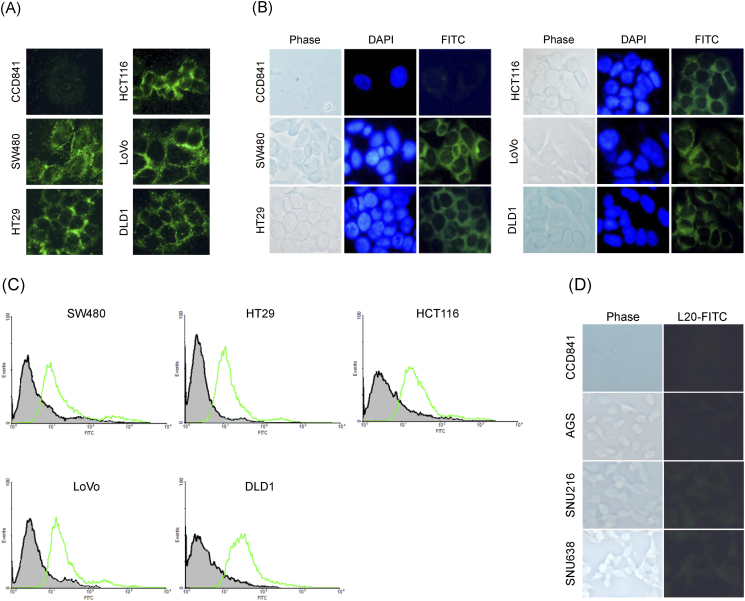
Binding of the L20 phage to five kinds of colon cancer cells as well as a control cell line was examined by immunostaining (Figure 1). Fixed cells were incubated with the L20 phage, the unbound phages were removed by washing the cells with PBS buffer, and the bound phages were detected using fluorescence-conjugated anti-M13 monoclonal antibody. Compared to the control cells, the L20 phage demonstrated a significantly higher binding to colon cancer cells, as shown in Figure 1A. To determine whether the synthetic L20 peptide binds to human colon cancer cells, the L20 peptide was labeled with fluorescein and then incubated with colon cancer cells and control cells. As shown in Figure 1B, a very intense fluorescence was observed on the five colon cancer cells, while the fluorescence on the control cell line was negligible. This result revealed that the synthetic L20 peptide has a significantly higher binding affinity on colon cancer cells in comparison to the control cells tested in this study. The binding affinity was also evaluated by FACS analysis with a control and the L20 peptide in the five colon cancer cell lines. As shown in Figure 1C, this experiment showed the right shift of fluorescence by the L20 peptide compared to the control peptide in all colon cancer cells. To further study the specificity of the L20 peptide, we examined the binding affinity in gastric cancer cells including AGS, SNU216, and SNU638. As shown in Figure 1D, the incubation of the L20 peptide did not show higher fluorescence in gastric cancer cell lines compared to the control cells.
Figure 1.
Binding affinity studies in colon cancer cells. (A) Immunohistochemical staining was performed using Alexa 488 in order to confirm the binding affinity of the phage clone to five colon cancer cells and a normal cell line. (B) Five colon cancer cell lines and CCD841 cells were subjected to immunofluorescence staining to confirm the cell binding ability of the L20 peptide. L20 peptide was labeled with FITC, and cells were stained with DAPI.
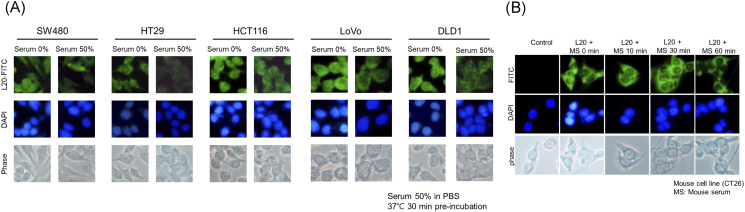
Serum Stability Tests
Before in vivo studies, the stability of L20 peptide was evaluated by incubation in human serum at 37°C for 30 minutes. As shown in Figure 2A, immunohistochemical staining indicated that fluorescence was clearly observed in the five colon cancer cell lines even after incubation of the L20 peptide for 30 minutes in human serum. The percentage of fluorescence reduction in SW480 was 27.1% after incubation of 50% human serum and 40.5% reduction in HT29, 19.5% in HCT116, 49.5% in LoVo, and 45.8% in DLD1 (Image J 1.46r, Bethesda, MD). We also evaluated the stability of L20 peptide in mouse serum (Figure 2B). As shown in Figure 2B, the fluorescence of the L20 peptide was maintained 60 minutes after incubation in mouse serum, showing that the binding ability was preserved.
Figure 2.
Serum stability test of L20 peptide in (A) human and (B) mouse serum. Immunohistochemical staining was performed. (A) Fluorescence was observed even after incubation of the L20 peptide for 30 minutes in human serum. (B) The L20 peptide adherence to mouse colon cancer cells was present even after incubating the L20 peptide for 60 minutes in mouse serum.
L20 Conjugation to HPP
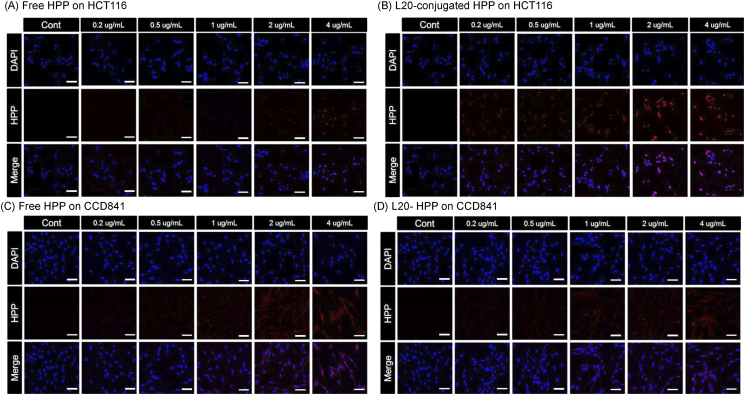
To explore possible therapeutic applications of the L20 peptide, we tried selective photodynamic therapy. For this purpose, HPP, a photosensitizer, was conjugated with L20 peptide (Supplementary Figure 2). Then, the cellular uptake of this conjugate was compared to free HPP in HCT116 cells. Compared to the free HPP (Figure 3A), L20-conjugated HPP showed higher fluorescence in all concentrations of loaded HPP in HCT116 cells (Figure 3B). However, the fluorescence was not significantly different between free and L20-conjugated HPP on CCD841 cells (Figure 3, C and D). These showed that HPP could bind selectively and efficiently to colon cancer cells when it was conjugated with L20 peptide.
Figure 3.
Cell imaging with L20-conjugated HPP. (A) Fluorescence image of HCT116 cells after treatment of free HPP, (B) fluorescence image of HCT116 cells after treatment of L20-conjugated HPP, (C) fluorescence image of CCD841 cells after treatment of free HPP, (D) fluorescence image of CCD841 cells after treatment of L20-conjugated HPP. Scale bar means 20 μm.
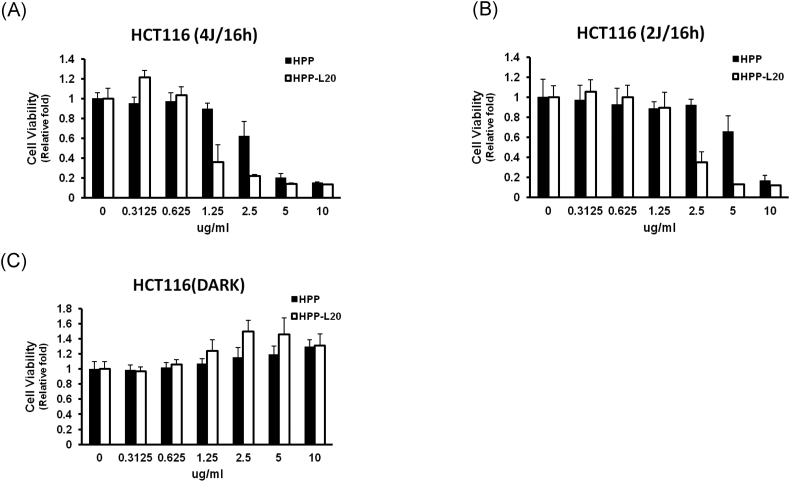
Photodynamic Therapy Using L20-Conjugated HPP
After treatment with free or L20-conjugated HPP, HCT116 cells were irradiated with laser (Figure 4). As shown in Figure 4A, HCT116 cells pretreated with L20-conjugated HPP showed significantly decreased survival at 16 hours after laser irradiation of 4 J/cm2 compared with free HPP pretreatment group in doses of 1.25 and 2.5 μg/ml. This difference was also observed under the weaker laser irradiation of 2 J/cm2 (Figure 4B). These results in photodynamic therapy were consistent with the difference of the cellular uptake of HPP, as shown in Figure 2, A and B.
Figure 4.
Cell survival assay after photodynamic therapy using free and L20-conjugated HP. (A) MTT assay 16 hours after laser (4 J/cm2), (B) MTT assay 16 hours after laser irradiation (2 J/cm2), and (C) dark toxicity.
Preparation and Characterization of NPs
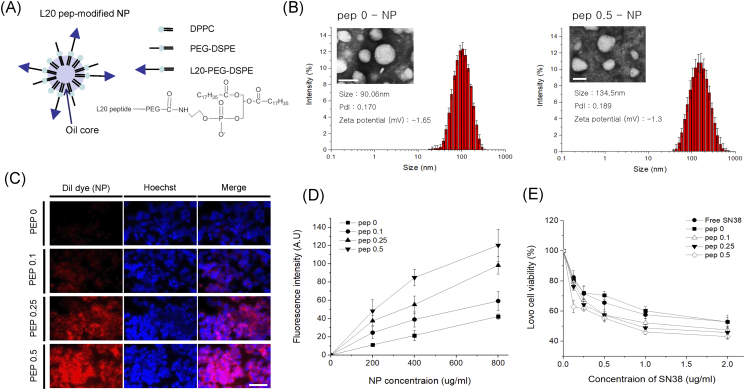
We prepared L20 peptide-modified NPs (L20-NPs) by the traditional oil-in-water emulsion method through self-assembly. We made four different kinds of L20-NPs, in variable L20-DSPE-PEG doses (0, 0.1, 0.25, 0.5 mg). L20-DSPE-PEG was synthesized by amide bond between L20 peptide and NHS-PEG-DSPE (Supplementary Figure 3), which was confirmed by 1HNMR spectroscopy (Supplementary Figure 4). Phospholipid, as a shell, was composed of DPPC, DSPE-PEG, and L20-DSPE-PEG with different ratios (Figure 5A), and the hydrophobic core was filled with soybean oil. The sizes of the L20-NPs were about 100 nm, which is suitable for enhanced permeation and retention effect in vivo. The size of NP with no L20 peptide (pep 0) was about 90.06 nm, while L20-NPs showed slightly increased sizes (Figure 5B and Supplementary Figure 4). Also, the resulting L20-NPs showed near neutral surface charges in zeta-potential measurement, and TEM images showed their spherical morphology.
Figure 5.
L20 peptide-modified nanoparticles and their in vitro test. (A) Illustration and chemical structure (DSPE-PEG-L20), (B) DLS and TEM image, (C) cell binding images, and (D) MTT (SN38-loaded NPs).
In Vitro Cellular Uptake Assay of L20-Conjugated NPs
In order to compare the cellular uptake of L20-NPs, DiI dye with red fluorescence was loaded into NPs. Then, LOVO human colon cells were treated with different concentrations of L20-NPs for 15 minutes (Figure 5C). As expected, pep 0.5-NP–treated cells showed strong fluorescence intensity than the other L20-NPs, indicating excellent targeting ability of the L20 peptide. As the amount of L20 peptide amount in NPs increased, so did fluorescence intensity. Pep 0-NP–treated cells showed weak fluorescence intensity because of the effect of PEG chain, which could inhibit the interaction between cell surface and NPs. All NPs showed dose-dependent increase of cellular uptake at 200-800 μg/ml (Figure 5D).
Cell Viability Test Using a Chemotherapeutic Drug Loaded L20-Conjugated NPs Loading
For a drug delivery in vitro study, we loaded SN38, a hydrophobic anticancer drug, into NPs. In vitro cytotoxicity of L20-NPs loaded with SN38 was measured using MTT assay in variable range of concentration for 15 minutes (0.125-2 μg/ml of SN38). The viability of L20-NP–treated LOVO cells decreased as SN38 concentration increased (Figure 5D). Pep 0.5-NP–treated cells showed poor viability of cells than the other groups in all concentration ranges. This result was correlated with the trend in cellular uptake, meaning L20 peptide could successfully help the uptake of NPs into tumor cells.
In Vivo and Ex Vivo NIRF Imaging in LOVO Tumor-Bearing Mice
To obtain in vivo whole body biodistribution by NIRF imaging, L20-NPs containing DiD dye were intravenously injected into LOVO tumor-bearing mice. All images were obtained at a predetermined postinjection time using the IVIS Lumina XRMS system. The NIRF signal of tumor site in pep 0.1-, 0.25-, and 0.5-NP–treated mice was higher than pep 0-NP–treated mice group, indicating that the L20 peptide increased tumor accumulation of NPs in vivo (Figure 6A). Also, ex vivo NIRF images from dissected tumor showed similar results with whole body image (Figure 6B). In pep 0.25-NP–treated mice, the tumor florescence intensity was the highest in tumor sites compared to mice groups with different in vitro data. Frozen sections of dissected tumor tissue were acquired from each group and observed by fluorescence microscopy (Figure 6C). Pep 0.25-NP–treated tumor exhibited greater fluorescence intensity than pep 0-NP–treated tumor, showing superior tumor targeting ability of the L20 peptide in vivo.
Figure 6.
In vivo biodistribution of L20 peptide-modified nanoparticles. (A) Whole body images of mice after intravenous injection of L20-conjugated NPs and fluorescence intensities in tumor site. (B) Ex vivo images of the major organs and tumors from mice. (C) Fluorescence images of sliced tumor tissues.
Structural Modeling and MD Simulation of L20 Peptide
To understand the mechanism behind the strong cell attachment and colon cancer cell specificity of L20 peptide, we performed computational structure calculation and MD simulation. The result of computational study from Maestro software package shows that the L20 peptide is expected to form an α-helix conformation (Figure 7A). We cross-validated the structure prediction with de novo modeling based on the Robetta protocol[23], and the results also showed that the L20 peptide formed α-helix. A helix wheel diagram representation shows that the amino acid residues of L20 are arranged to form amphipathic α-helix, i.e., the hydrophilic residues are aligned on one face of the helix and the hydrophobic residues on the other (Figure 7B). Because the interior of cell lipid bilayer membrane is hydrophobic, it is common in cell biology that amphipathic helices are readily inserted into cell membrane by forming α-helical bundles, where the hydrophobic faces are oriented outwards. Thus, the strong cell attachment of L20 is likely related to the propensity of the peptide to form an amphipathic α-helix. The cell specificity may be related to the specific lipid composition and outer membrane environment of the colon cancer cells; however, probing the exact mechanism requires further in-depth studies.
Figure 7.
Structural modeling and MD simulation of L20 peptide. (A) Calculated structure of the L20 peptide using Maestro software package. (B) A helix wheel diagram representation of L20 and the plausible model of the helix bundle formation in cell membrane.
Discussion
In this study, we found a 12-mer peptide sequence, named L20, using phage display libraries, which was specific to colon cancer cells. We validated the L20 peptide probe by comparing its binding affinity in five colon cancer cell lines versus a normal cell line with the immunohistochemical staining and FACS analysis. L20 maintained binding affinity after serum incubation. For therapeutic applications, we first conjugated L20 with HPPs, which showed a significantly higher effect in photodynamic therapy compared to free HPPs. As a secondary therapeutic application, we developed NPs modified with L20. L20-NPs delivered a significantly higher tumoricidal effect compared with a free drug. Also, L20 enhanced tumor tissue accumulation of NPs after intravenous injection to mice models.
Molecular imaging techniques have been focused on different signature of target tissues compared to their correspondence. In the present study, we showed solid data for specific binding to colon cancers with the use of five kinds of colon cancer cell lines, all of which showed clear contrast to normal fibroblast cells. Considerable efforts have been made toward the development of peptide-based drug conjugates for cancer targeted imaging and therapy. Peptide probes are advantageous in that they are nonantigenic, are structurally simple, and can be easily synthesized [24]. One of the approaches to find tumor selective peptides is phage display technique. As shown in the present study, peptide probes can be conjugated with diagnostic labels or therapeutic agents [19,25,26]. We proved the specific binding of L20 on colon cancer cells for a diagnostic potential with several kinds of conjugates such as a fluorescence dye, a photosensitizer, and nanoparticle. L20 was also used for a targeted photodynamic therapy after conjugation with a photosensitizer, and as chemotherapy with drug-containing NPs.
Nanomedicine can provide a more effective approach to targeting cancer by focusing on the vascular, tissue, and cellular characteristics that are unique to solid tumors [27]. In particular, NPs have been paid much attention as effective drug carriers [28]. Drug targeting using NPs can decrease the likelihood of resistant clonal populations of cancerous cells. They can encapsulate hydrophobic drugs and increase their water solubility. They also can provide long circulation time after injection into body and increase therapeutic efficacy [29]. We expect that our peptide can be applied to various kinds of NPs containing different drugs and enhance their targeting ability. It will be helpful for developing new drug carriers as well as diagnostic application of the peptide probe.
This study has several strong points. First, we tested the colon cancer specificity in five colon cancer cell lines to increase generalizability and to overcome tumor heterogeneity. Second, we validated tumor specificity not only in vitro but also in vivo. Third, this study showed various possible applications as a theranostic probe by revealing that a chemotherapeutic agent as well as a photosensitizer after conjugation with L20 was delivered efficiently to colon cancer cells, resulting in higher tumoricidal effects compared to their respective free forms.
On the other hand, our study has several limitations. Even though we demonstrated colon cancer specificity with many experiments, we did not perform experiments to find out the binding partners of L20 on the cell surface yet. We did not validate tumor specificity in human colon cancer tissues. To apply L20 into clinical practice, we need further validation processes in human tissues. These experiments were performed in cancer cell lines. Therefore, the results may potentially prove different in the actual human cancer. We need further human trials to confirm our results.
In conclusion, the 12-mer peptide L20 acquired by phage-display technology showed significant specific binding to colon cancer cells and tissues. Based on its high and specific binding to colon cancer cells in vitro and ex vivo, L20-conjugated therapeutics showed a target-specific treatment. Therefore, L20 can be used as a cancer targeting identifier and as a targeted therapy for the treatment of colon cancers.
Acknowledgments
Acknowledgements
We thank Drs. Yoon Jin Roh, Ju Hee Kim, and In-Wook Kim for supporting technical assistance and experimental processes.
Funding
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education, Science, and Technology (2016R1C1B3013951, 2017R1D1A1B03035104, and 2019R1A5A2027588).
Declaration of Competing Interest
The authors declare no potential conflicts of interest.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tranon.2020.100798.
Contributor Information
Heebeom Koo, Email: hbkoo@catholic.ac.kr.
Jae Myung Park, Email: parkjerry@catholic.ac.kr.
Appendix A. Supplementary data
Supplementary material
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2016. CA Cancer J. Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Lieberman D.A. Clinical practice. Screening for colorectal cancer. N. Engl. J. Med. 2009;361:1179–1187. doi: 10.1056/NEJMcp0902176. [DOI] [PubMed] [Google Scholar]
- 3.Zauber A.G., Winawer S.J., O'Brien M.J., Lansdorp-Vogelaar I., van Ballegooijen M., Hankey B.F. Colonoscopic polypectomy and long-term prevention of colorectal-cancer deaths. N. Engl. J. Med. 2012;366:687–696. doi: 10.1056/NEJMoa1100370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rutter M., Bernstein C., Matsumoto T., Kiesslich R., Neurath M. Endoscopic appearance of dysplasia in ulcerative colitis and the role of staining. Endoscopy. 2004;36:1109–1114. doi: 10.1055/s-2004-826049. [DOI] [PubMed] [Google Scholar]
- 5.Kahi C.J., Imperiale T.F., Juliar B.E., Rex D.K. Effect of screening colonoscopy on colorectal cancer incidence and mortality. Clin. Gastroenterol. Hepatol. 2009;7:770–775. doi: 10.1016/j.cgh.2008.12.030. quiz 11. [DOI] [PubMed] [Google Scholar]
- 6.Joshi B.P., Wang T.D. Targeted optical imaging agents in cancer: focus on clinical applications. Contrast Media Mol Imaging. 2018;2018:2015237. doi: 10.1155/2018/2015237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heresbach D., Barrioz T., Lapalus M.G., Coumaros D., Bauret P., Potier P. Miss rate for colorectal neoplastic polyps: a prospective multicenter study of back-to-back video colonoscopies. Endoscopy. 2008;40:284–290. doi: 10.1055/s-2007-995618. [DOI] [PubMed] [Google Scholar]
- 8.Kennedy P.J., Oliveira C., Granja P.L., Sarmento B. Antibodies and associates: partners in targeted drug delivery. Pharmacol. Ther. 2017;177:129–145. doi: 10.1016/j.pharmthera.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 9.Krumpe L.R., Mori T. The use of phage-displayed peptide libraries to develop tumor-targeting drugs. Int. J. Pept. Res. Ther. 2006;12:79–91. doi: 10.1007/s10989-005-9002-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kostenich G., Oron-Herman M., Kimel S., Livnah N., Tsarfaty I., Orenstein A. Diagnostic targeting of colon cancer using a novel fluorescent somatostatin conjugate in a mouse xenograft model. Int. J. Cancer. 2008;122:2044–2049. doi: 10.1002/ijc.23353. [DOI] [PubMed] [Google Scholar]
- 11.Kostenich G., Livnah N., Bonasera T.A., Yechezkel T., Salitra Y., Litman P. Targeting small-cell lung cancer with novel fluorescent analogs of somatostatin. Lung Cancer. 2005;50:319–328. doi: 10.1016/j.lungcan.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 12.Becker A., Hessenius C., Licha K., Ebert B., Sukowski U., Semmler W. Receptor-targeted optical imaging of tumors with near-infrared fluorescent ligands. Nat. Biotechnol. 2001;19:327–331. doi: 10.1038/86707. [DOI] [PubMed] [Google Scholar]
- 13.Hamzeh-Mivehroud M., Alizadeh A.A., Morris M.B., Church W.B., Dastmalchi S. Phage display as a technology delivering on the promise of peptide drug discovery. Drug Discov. Today. 2013;18:1144–1157. doi: 10.1016/j.drudis.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 14.Gray B.P., Brown K.C. Combinatorial peptide libraries: mining for cell-binding peptides. Chem. Rev. 2014;114:1020–1081. doi: 10.1021/cr400166n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bazan J., Calkosinski I., Gamian A. Phage display—a powerful technique for immunotherapy: 1. Introduction and potential of therapeutic applications. Hum Vaccin Immunother. 2012;8:1817–1828. doi: 10.4161/hv.21703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saw P.E., Song E.W. Phage display screening of therapeutic peptide for cancer targeting and therapy. Protein Cell. 2019;10:787–807. doi: 10.1007/s13238-019-0639-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mandelin J., Cardo-Vila M., Driessen W.H., Mathew P., Navone N.M., Lin S.H. Selection and identification of ligand peptides targeting a model of castrate-resistant osteogenic prostate cancer and their receptors. Proc. Natl. Acad. Sci. U. S. A. 2015;112:3776–3781. doi: 10.1073/pnas.1500128112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferreira D., Silva A.P., Nobrega F.L., Martins I.M., Barbosa-Matos C., Granja S. Rational identification of a colorectal cancer targeting peptide through phage display. Sci. Rep. 2019;9:3958. doi: 10.1038/s41598-019-40562-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y., Chen J., Zhang Y., Hu Z., Hu D., Pan Y. Panning and identification of a colon tumor binding peptide from a phage display peptide library. J. Biomol. Screen. 2007;12:429–435. doi: 10.1177/1087057106299164. [DOI] [PubMed] [Google Scholar]
- 20.Liu Z., Gray B.D., Barber C., Bernas M., Cai M., Furenlid L.R. Characterization of TCP-1 probes for molecular imaging of colon cancer. J. Control. Release. 2016;239:223–230. doi: 10.1016/j.jconrel.2016.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Polak E., Ribiere G. Note sur la convergence de méthodes de directions conjuguées. Revue française d'informatique et de recherche opérationnelle Série rouge. 1969;3:35–43. [Google Scholar]
- 22.Ryckaert J.-P., Ciccotti G., Berendsen H.J. Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 23.Chivian D., Kim D.E., Malmstrom L., Bradley P., Robertson T., Murphy P. Automated prediction of CASP-5 structures using the Robetta server. Proteins. 2003;53(Suppl 6):524–533. doi: 10.1002/prot.10529. [DOI] [PubMed] [Google Scholar]
- 24.Gilad Y., Firer M., Gellerman G. Recent innovations in peptide based targeted drug delivery to cancer cells. Biomedicine. 2016;4 doi: 10.3390/biomedicines4020011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelly K.A., Jones D.A. Isolation of a colon tumor specific binding peptide using phage display selection. Neoplasia. 2003;5:437–444. doi: 10.1016/s1476-5586(03)80046-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kelly K., Alencar H., Funovics M., Mahmood U., Weissleder R. Detection of invasive colon cancer using a novel, targeted, library-derived fluorescent peptide. Cancer Res. 2004;64:6247–6251. doi: 10.1158/0008-5472.CAN-04-0817. [DOI] [PubMed] [Google Scholar]
- 27.Lammers T., Aime S., Hennink W.E., Storm G., Kiessling F. Theranostic nanomedicine. Acc. Chem. Res. 2011;44:1029–1038. doi: 10.1021/ar200019c. [DOI] [PubMed] [Google Scholar]
- 28.Ryu J.H., Koo H., Sun I.C., Yuk S.H., Choi K., Kim K. Tumor-targeting multi-functional nanoparticles for theragnosis: new paradigm for cancer therapy. Adv. Drug Deliv. Rev. 2012;64:1447–1458. doi: 10.1016/j.addr.2012.06.012. [DOI] [PubMed] [Google Scholar]
- 29.Koo H., Huh M.S., Sun I.C., Yuk S.H., Choi K., Kim K. In vivo targeted delivery of nanoparticles for theranosis. Acc. Chem. Res. 2011;44:1018–1028. doi: 10.1021/ar2000138. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material