Abstract
Background
Retinitis pigmentosa (RP) is the most common form of inherited retinal degeneration, but genetic defects in nearly half of families remain unresolved. This study aims to identify novel genes associated with RP based on whole exome sequencing (WES) data from 552 probands with RP.
Methods
Biallelic loss-of-function (LoF) variants were selected from the WES data of 552 probands with RP and compared with that of 4728 in-house controls and the gnomAD database. Expression analysis and knockout mice model or knockdown zebrafish model were performed to confirm the association of a few candidate genes with RP.
Findings
Unique biallelic LoF variants in ENSA, DACT2, DDR1, and CCDC188 were identified in four probands with RP, but were absent in 4728 in-house controls and were extremely rare in the gnomAD database. The expression of ENSA was enriched in the rod outer segments of human retina, and significant reduced responses of rods and cones were detected in Ensa knockout mice compared to wild-type mice by electroretinogram. The DACT2 transcript showed the highest expression in human retina and knockdown of dact2 in zebrafish resulted in photoreceptor disc membrane disarrangement.
Interpretation
This study suggests that ENSA is likely a novel gene for RP and DACT2 is a potentially candidate gene for RP. Further studies are expected to evaluate the association between mutations in the other two genes and RP. To our knowledge, mutations in these genes have not been reported to be associated with RP before.
Keywords: Retinitis pigmentosa, ENSA, DACT2, Loss-of-function variant, Knockout mice, Knockdown zebrafish
Research in context.
Evidence before this study
As the most common form of inherited retinal degeneration, retinitis pigmentosa (RP) is one of the major causes of irreversible blindness. To date, mutations in at least 89 genes have been reported to cause RP according to the RetNet database (RetNet, https://sph.uth.edu/retnet/, accessed March 2020). These genes are involved in multiple functional pathways, including phototransduction, ciliary transport, visual cycle, Wnt signaling, ion channels, intermediary metabolism, et al. We have previously performed a comprehensive analysis of mutations in known causative genes in a cohort of Chinese families with RP, and found that mutations in all these genes explain only half of patients with RP.
Added value of this study
Since the genetic defects in nearly half of RP families remain unsolved, we performed whole-exome sequencing in 552 unrelated RP cases and 4728 in-house controls. To our knowledge, this is the first study to reveal the associations between mutations in the four genes (ENSA, DACT2, DDR1, and CCDC188) and RP. The relationship between loss-of-function variants in ENSA or DACT2 and RP was further confirmed by retinal expression analysis, knockout mouse model, or knockdown zebrafish experiment in this study. Among the four novel genes, ENSA is involved in potassium ion channels, DACT2 is involved in Wnt signaling pathway, and DDR1 is involved in intermediary metabolism. In addition, ENSA and DDR1 have potential interaction with genes known to cause retinal degeneration when mutated. Our results are helpful for genetic counseling and potential gene therapy for unexplained patients with RP.
Implications of all the available evidence
This study suggests that ENSA is likely a novel gene for RP and DACT2 is a potentially candidate gene for RP. In addition, our findings demonstrate that the strategy by screening unique biallelic loss-of-function variants in a cohort of probands is feasible for exploring novel RP genes.
Alt-text: Unlabelled box
1. Introduction
Retinitis pigmentosa (RP [MIM: 268000]), the most common form of inherited retinal degeneration (IRD), is a heterogeneous group of hereditary diseases caused by loss of photoreceptor function and subsequent degeneration of retina. It contributes greatly to the etiology of irreversible blindness worldwide with an incidence of approximately one in 3500-5000 individuals [1,2]. Most patients with RP usually initially exhibit night blindness, which is followed by progressive constriction of the visual field and gradual reduction of the visual acuity [3]. The fundus is characterized by a waxy pallor optic disc, attenuated retinal arteries, and pigment abnormalities initially from the midperipheral retina [4]. The rod responses on electroretinogram (ERG) are usually more severely affected than cone responses [5]. RP can be transmitted in autosomal dominant, autosomal recessive, and X-linked patterns, of which autosomal recessive inheritance is the most common [6,7]. Rarely, RP might be transmitted as non-Mendelian inheritance traits, including mitochondrial and digenic inheritances [8,9]. To date, mutations in at least 89 genes have been reported to cause RP (RetNet, https://sph.uth.edu/retnet/, accessed March 2020), but they may explain only half of Chinese families with RP [10]. Identification of additional genes is a prerequisite for genetic counseling and potential gene therapy for undetermined RP patients.
As a part of our ongoing study of the genetic basis for RP based on whole-exome sequencing (WES), a comprehensive analysis of mutations in known causative genes has been performed in 157 unrelated Chinese families with RP [10]. Novel candidate genes were analysed based on undetermined patients with RP from our previously reported cohort and a new RP cohort. In this study, biallelic loss-of-function (LoF) variants were selected if they are rare and exclusively present in any of the 552 families with RP but not in our in-house controls of 4728 unrelated individuals based on WES analysis. A few novel candidate genes, including ENSA, DACT2, DDR1, and CCDC188, were identified. Expression profile, Ensa knockout mice, dact2 knockdown zebrafish, and bioinformatics analysis provided further evidence that biallelic LoF mutations in some of these genes are likely causative for RP.
2. Materials and methods
2.1. Human subjects
Patients with various forms of genetic eye diseases and their available family members were recruited from the Paediatric and Genetic Clinic, Zhongshan Ophthalmic Center. This study was approved by the Institutional Review Board of the Zhongshan Ophthalmic Center and followed the tenets of the Declaration of Helsinki. Written informed consent following the Guidance of Sample Collection of Human Genetic Diseases (863-Plan) by the Ministry of Public Health of China was obtained from the participants or their guardians before clinical data and peripheral blood samples were collected. Medical and ophthalmic histories, visual acuity measurements, slit-lamp examination, and funduscopic examination were performed. Diagnosis of RP was made by a senior ophthalmologist based on night blindness appeared initially, progressive loss of peripheral vision, decreasing visual acuity with age, and fundus changes including waxy pale discs, retinal arteriolar attenuation, tapetoretinal degeneration or pigment deposits in the midperipheral retina. The pedigrees were drawn using questionnaire or oral description.
2.2. WES and bioinformatic analysis
WES was performed on genomic DNA from 552 probands with RP, as well as 4728 unrelated individuals with other ocular condition including 1299 probands with high myopia, 1217 probands with glaucoma, 1720 probands with other genetic eye diseases, and 492 normal controls. The procedures for sequencing, variant calling and annotation, and the filtering of potentially pathogenic variants by multistep bioinformatic analyses were performed as previously described [10]. Samples with pathogenic variants in known RP genes were excluded from this study. Unique biallelic LoF variants were searched in WES data from the 552 probands with RP and then compared with in-house data from 4728 controls and the gnomAD database. Genes with homozygous LoF variants in the gnomAD database were excluded. Genes with homozygous or compound heterozygous variants in one or more samples with RP were selected for further analysis.
2.3. Sanger sequencing
Sanger sequencing was conducted to validate the candidate variants in the probands. Segregation in available family members was further evaluated. Primers (Supplementary Materials, Table S1) were designed using Primer3 (http://primer3.ut.ee/) and used to amplify genomic fragments harboring variants. The procedures used for amplification, sequencing, and target fragment analysis were performed as previously described [10].
2.4. Human ocular tissues
Human ocular tissues were obtained from the Eye Bank of Guangdong Province, which were from an eye donor who died of meningioma. All procedures conformed to the Declaration of Helsinki developed by the World Medical Association and the ethics principles of the International Ethical Guidelines for biomedical research involving human subjects developed by the Council for International Organizations of Medical Sciences. Written informed consent was obtained from the donor prior to the study. This study was approved by the Ethics Committee of Zhongshan Ophthalmic Center, Sun Yat-sen University.
2.5. Reverse transcription PCR (RT-PCR) and quantitative real-time PCR (qPCR)
RNA samples were prepared from 32 human tissues, including 24 extraocular samples purchased from the company (Clontech Laboratories, CA, USA), seven ocular tissues from the eye donor described above, and retinal pigment epithelium (RPE) obtained from the ARPE-19 cell line. The RNA samples from 24 human extraocular tissues were pooled from 12 male and female Caucasian individuals who experienced sudden death at the age of 18 to 54 years. cDNA was synthesized from the total RNA of the 32 tissues using the PrimeScript™ RT Reagent Kit (RR047A, TaKaRa, Japan). All primer sets used for RT-PCR and qPCR (Supplementary Materials, Table S2) were designed using Primer3 (http://primer3.ut.ee/). qPCR was performed with PowerUp SYBR Green Master Mix (A25742, Applied Biosystems), and the fold changes in RNA levels were calculated using the ΔΔCt method, as described in a previous study [11].
2.6. Ensa knockout mice and full-field ERG
All experiments involving animals were conducted according to the Association for Research in Vision and Ophthalmology (ARVO) Statement for the Use of Animals in Ophthalmic and Vision Research. Knockout mice were generated using the CRISPR/Cas9-mediated gene-targeting approach [12]. Single-guide RNAs (sgRNAs, Supplementary Materials, Table S3) were designed to target the upstream and 3’UTR of the Ensa gene respectively (Supplementary Materials, Fig. S1a). In vitro-transcribed Cas9 mRNA and sgRNA were microinjected into the zygotes of C57BL/6J mice, which were then transferred into pseudopregnant females. The newborn mice were genotyped by PCR using tail-derived genomic DNAs (Supplementary Materials, Fig. S1b). The primers used for mouse genotyping were shown in Table S4 in the Supplementary Materials. The homozygous Ensa knockout (Ensa−/−) mice had 6696-bp deletions in both alleles, which resulted in deletion of the entire gene.
ERG of the knockout mice and age-matched wild-type mice were recorded. The mice were adapted to a dark environment overnight and anaesthetized with pentobarbital sodium. The pupils were dilated using tropicamide. Scotopic recordings were obtained at the following increasing light intensities: 0.003, 0.01, 0.03, 0.1, 0.3, 1.0, and 3.0 cd.s/m2. Thereafter, photopic recordings were performed following 5 min light adaptation intervals on a bright green background light intensity of 20 cd.s/m2. Six levels of stimuli (0.3, 1.0, 3.0, 10, 30, and 100 cd.s/m2) were used for the photopic recordings. For each condition (scotopic or photopic), three to ten responses were averaged to the luminance of the flash stimuli, with the stimulus interval varying from 2 to 10 s at low intensities to 1 min at intensities above 3.0 cd.s/m2.
2.7. Morpholino oligonucleotide (MO) injection of zebrafish embryos
The wild-type zebrafish (AB Danio rerio) embryos used in this study were obtained from the Zebrafish Model Animal Facility at the Institute of Clinical and Translational Research of Sun Yat-sen University. MOs targeting the initial translational site (MO TB) of the two genes (dact2 MO TB and extl2 MO TB), and the splicing site at the exon-intron boundary (MO SP) of the two genes (dact2 MO SP [blocking the splice of dact2 exon 1 – intron 1 boundary] and extl2 MO SP [blocking the splice of extl2 exon 2 – intron 2 boundary]), as well as the standard control MO (std MO) were designed by Gene-Tools, LLC (https://www.gene-tools.com, Corvallis, OR, USA) (Supplementary Materials, Table S5). The dact2 MO TB, dact2 MO SP, extl2 MO TB, extl2 MO SP, and std MO were microinjected at dose of 3 ng into the yolk at the 1- cell stage, respectively. For the MO SP efficacy test, 3 days post fertilization (dpf) and 5 dpf larvae from each group were collected and the total RNA was extracted. cDNAs were reverse transcribed by using the PrimeScript™ RT Reagent Kit (RR047A, TaKaRa, Japan), and amplified by using primers in Table S2. The phenotypes were captured by a LEICA microscope (LEICA M205 FA, Germany) at 3 dpf. The eye size across the nasal edge to the temporal edge and the body length were measured by ImageJ software (National Institutes of Health). The larvae with small eyes but without other morphological abnormalities would be cultured until 5 dpf for immunohistochemical staining and transmission electron microscope image when the progenitor cells differentiate into mature cone and rod photoreceptors [13], [14], [15].
2.8. Western blot
The zebrafish embryos (40 for each experimental group) at 24 hours post fertilization (hpf) were collected and washed by 1X PBS. The samples were lysed on ice in RIPA buffer (kingbioR0010, Kingbio, USA) with phenylmethylsulfonyl fluoride (PMSF, P0100-15, Kingbio, USA) for 30 min. Lysates for each group were loaded onto 7.5% and 10% SDS-polyacrylamide gels (PG111 and PG112, EpiZyme scientific, China), and then were transferred onto polyvinylidene fluoride (PVDF) membranes (IPVH00010, Millipore, USA) after electrophoresis. The PVDF membranes were blocked with 5% non-fat dry milk (232100, BD Biosciences, USA) in 1X TBST buffer for 1 hour. For dact2, the membranes were incubated with a mouse anti-β actin loading control (HRP) (1:500, Abcam Cat#ab197277) and a rabbit anti-dact2 antibody (1:100, Sigma Cat#HPA030251, RRID: AB_10611108) together at 4°C overnight. For extl2, the membranes were incubated at 4°C overnight with a mouse anti-β actin (1:500, Abcam Cat#ab197277) and a rabbit anti-extl2 antibody [EPR11130] (1:1000, Abcam Cat#ab168391), respectively. Secondary antibodies included HRP conjugated goat anti-mouse IgG - H&L polyclonal antibody and HRP conjugated goat anti-rabbit IgG - H&L polyclonal antibody (Abcam, Cat#ab6789, RRID: AB_955439, Abcam, Cat#ab6721, AB_955447, both antibodies were used at a final dilution of 1:1000). The band signals were captured using the enhanced chemidoc IMAGING SYSTEM (BIO-RAD, USA). The gray value was analyzed by using Image J.
2.9. Immunohistochemical staining
Paraformaldehyde-fixed human eyes and mouse eyes were embedded in paraffin and cut into 4-μm-thick sections. Paraformaldehyde-fixed zebrafish at 5 dpf were embedded in OCT (Scigen Scientific Gardina, 4583) and cut into 10-μm-thick sections. The sections were subjected to antigen retrieval at 98°C for 30 min and subsequently blocked with 1% bovine serum albumin. Staining for ENSA in human retina was performed with a rabbit anti-ENSA antibody (Novus Cat# NBP2-13963, epitope between residues 1 and 36), a mouse anti-rhodopsin 1D4 antibody (Abcam Cat# ab5417, RRID: AB_304874), and a rabbit anti-opsin red/green antibody (Millipore Cat# AB5405, RRID: AB_177456) at final dilutions of 1:50, 1:2000 and 1:1000, respectively. Serial mouse eye sections were incubated with haematoxylin and eosin, a mouse anti-rhodopsin 1D4 antibody (1:2000, Abcam Cat# ab5417, RRID: AB_304874), and rabbit anti-opsin antibodies (anti-blue, 1:1000, Millipore Cat# AB5407, RRID: AB_177457; anti-green, 1:1000, Thermo Fisher Scientific Cat# OSR00222W, RRID: AB_2178929; anti-red/green, 1:1000, Millipore Cat# AB5405, RRID: AB_177456). Serial zebrafish sections were incubated with a mouse anti-rhodopsin 1D4 antibody (1:2000, Abcam ab5417), a rabbit anti-GNAT2 antibody (1:500, MBL International corporation Cat# PM075), and rabbit anti-opsin antibodies (anti-blue, 1:1000, Millipore Cat# AB5407, RRID: AB_177457; anti-red/green, 1:1000, Millipore Cat# AB5405, RRID: AB_177456). Secondary antibodies included Alexa Fluor 555-conjugated goat anti-rabbit IgG antibodies and Alexa Fluor 488-conjugated goat anti-mouse IgG antibodies (Cell Signaling Technology Cat# 4413, RRID: AB_10694110, Cell Signaling Technology Cat# 4408, RRID: AB_10694704, both antibodies were used at a final dilution of 1:500). Images of stained sections were captured using a confocal microscope (C2, Nikon, Japan). The number of immunofluorescence positive cells was counted using Image J. The opsin red/green+ cells, opsin blue+ cells, and GNAT2+ cells of the ONL were quantified over the whole section by using Image J.
2.10. Images of zebrafish retina by transmission electron microscopy (TEM)
Six groups of larvae at 5 dpf, i.e. wild-type, std MO, dact2 MO TB, dact2 MO SP, extl2 MO TB, and extl2 MO SP, were fixed with 2.5% glutaraldehyde in 0.1 M PBS at 4°C for 2–4 h, respectively. The larvae were washed three times with 0.1 M PBS, and further fixed with 1% osmium tetroxide in 0.1 M PBS buffer at room temperature for 2 h. The larvae were then gradient dehydrated in in 50, 70, 80, 90, 95 and 100% ethanol successively (20 min each) respectively, and then incubated in acetone/812 embedding medium at 37°C overnight. Ultrathin sections were produced using an ultramicrotome (Leica UC7) and stained with 2% uranyl acetate and 2% lead citrate. Images were captured by TEM instrument (HITACHI, HT7700).
2.11. Statistical analysis
Statistical analyses were performed using IBM SPSS Statistics Version 25 (International Business Machines Corporation, New York; USA). Statistical comparisons of ERG responses and cone cell numbers between Ensa knockout mice and wild-type mice were performed using two-tailed Student's t-test. Statistical significance was defined as a p-value less than 0.05 (p < 0.05).
Small eye size or short body length in this study were defined as the eye diameter or the body length less than the mean minus standard deviation (SD) of the eye diameter or the body length of wild-type larvae, whereas an eye size or body length within the mean ± SD of wild-type larvae was considered normal. Statistical differences of eye size and body length from each larvae between targeted gene knockdown zebrafish and wild-type larvae were analysed by One-Way ANOVA (P < 0.01, or 0.05/5, was considered as statistically significant). The proportions of larvae with small eye size and short body length at 3 dpf were compared among different groups by Chi-square test. The corrected significant P value should be less than 0.01 (α = 0.05/5) according to the Bonferroni correction. Statistical differences between the immunofluorescence positive cell numbers of targeted gene knockdown zebrafish and that of the control wild-type larvae were analysed using Chi-square test.
3. Results
3.1. Homozygous LoF variants in ENSA, DACT2, EXTL2, DDR1, and CCDC188 identified in families with RP
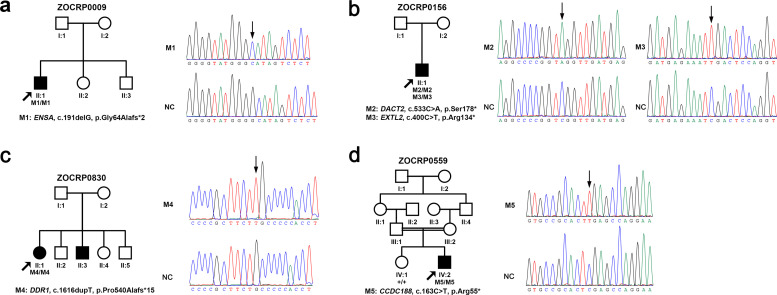
Based on a comparative analysis of WES data, five rare homozygous LoF variants in five genes were exclusively identified in the RP cohort (Table 1). All the five variants were validated by Sanger sequencing (Fig. 1). The five variants in five genes include a homozygous variant in ENSA (endosulfine alpha, HGNC: 3360, MIM 603061) in the proband from family ZOCRP0009, a homozygous variant in DACT2 (dishevelled binding antagonist of beta catenin 2, HGNC: 21231, MIM 608966) as well as a homozygous variant in EXTL2 (exostosin like glycosyltransferase 2, HGNC: 3516, MIM 602411) in the proband from family ZOCRP0156, a homozygous variant in DDR1 (discoidin domain receptor tyrosine kinase 1, HGNC: 2730, MIM 600408) in the proband from family ZOCRP0830, and a homozygous variant in CCDC188 (coiled-coil domain containing 188, HGNC: 51899) in the proband from family ZOCRP0559 (Table 1 and Fig. 1). Potentially pathogenic variants in genes known to cause IRD (listed in RetNet) were not identified in any of the four probands, while several single heterozygous variants were detected in genes responsible for autosomal recessive RP (Supplementary Materials, Table S6). Structural variations or copy number variations involving these genes were not detected by WES in these patients while deep intronic variants of these genes have not been reported before. The presence of these genes in Gene Matcher (https://genematcher.org/) was checked, and no submissions of clinically relevant variants were found for any of these genes. Biallelic LoF variants in the five genes were not present in WES data from 4728 in-house controls. The three variants in DACT2, DDR1, and CCDC188 were novel and not present in the existing databases (gnomAD, 1000 Genomes, or Exome Variant Server), whereas the other two variants, c.191delG in ENSA and c.400C>T in EXTL2, were found in the gnomAD database at frequencies of 1/251460 and 3/247450, respectively (Table 1). Biallelic LoF variants in the five novel genes are all extremely rare according to the gnomAD database (Table 1). Meanwhile, local SNP haplotypes involving the putative disease alleles were generated and a homozygous haplotype involving ENSA, DCAT2, EXTL2, and CCDC188 were detected in the corresponding proband but not in the other three probands (Supplementary Materials, Fig. S2). Besides the five homozygous LoF variants in the four probands, biallelic rare missense variants were also detected in some of the probands (Supplementary Materials, Table S7), which were excluded due to the present of other biallelic variants in controls or relative high frequency in the general population.
Table 1.
Five homozygous rare variants in five novel candidate genes identified in four RP probands
| Genes | Chromosome | Reference | Patient | Variant | Nucleotide | Amino acid | Expression & interaction§ |
Allele frequency in gnomAD |
Allele frequency of other LoF variants |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| position | transcript | ID | number | change | change | Retina | IRD genes | All | EA | control | gnomAD | |
| ENSA | chr01: 150599935 | NM_207168 | ZOCRP0009 | M1 | c.191delG | p.Gly64Alafs*2 | Ninth | OFD1 | 1/251460 | 1/18394 | 2/9456 | 22/282912 |
| DACT2 | chr06: 168710973 | NM_214462 | ZOCRP0156 | M2 | c.533C>A | p.Ser178* | Second | NA | NA | NA | 4/9456 | 144/282912 |
| EXTL2 | chr01: 101343065 | NM_001439 | ZOCRP0156 | M3 | c.400C>T | p.Arg134* | Highest | NA | 3/247450 | 3/18342 | 2/9456 | 37/282912 |
| DDR1 | chr06: 30864446 | NM_001202523 | ZOCRP0830 | M4 | c.1616dupT | p.Pro540Alafs*15 | Fifth | CCT2, BBS10 | NA | NA | 0/9456 | 178/282912 |
| CCDC188 | chr22: 20136745 | NM_001243537 | ZOCRP0559 | M5 | c.163C>T | p.Arg55* | NA | NA | NA | NA | 0/9456 | 63/282912 |
Notes: EA, East Asian; gnomAD, genome aggregation database; IRD, inherited retinal degeneration; LoF, loss-of-function; M, mutation; NA, not available.
§Information based on GeneCards where expression in retina ranked by SAGE.
None of these variants was present in Exome Variant Server or 1000 Genomes.
No homozygous LoF variants in these five genes were found in the gnomAD database.
Fig. 1.
Pedigrees and sequences of the five homozygous loss-of-function variants in ENSA, DACT2, EXTL2, DDR1, and CCDC188. The genotypes of all probands and available family members are shown below each individual. The blackened symbols represent affected individuals. Mx, mutant alleles; +, wild-type allele.
3.2. Expression profile of ENSA and phenotypes of Ensa knockout mice
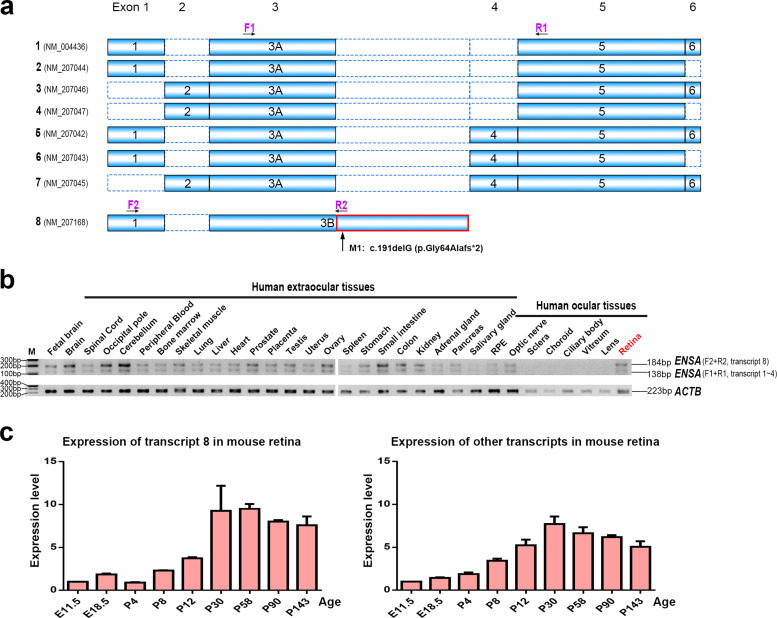
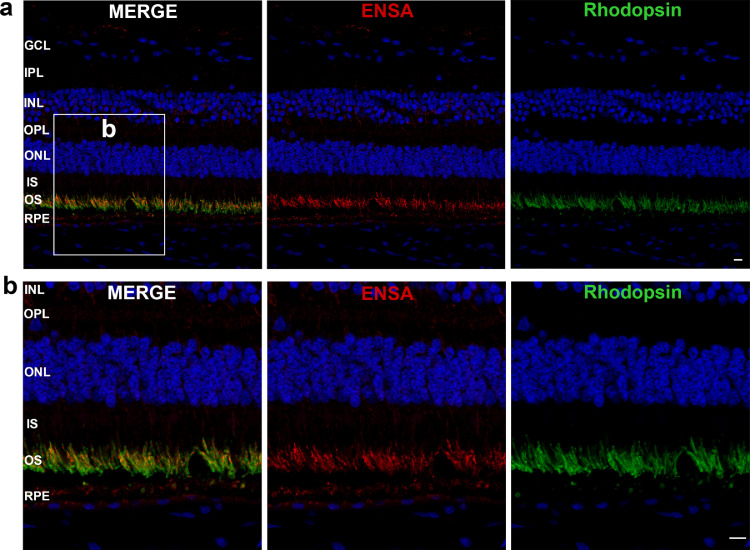
The human ENSA gene, located at 1q21.3, encodes the alpha-endosulfine (UniProt: O43768) [16,17]. Eight different transcripts are found in the NCBI Reference Sequence Database (https://www.ncbi.nlm.nih.gov/refseq/) (Fig. 2a) [18]. The variant identified in the proband from family ZOCRP0009 is located at the second coding exon of transcript 8 (NM_207168), but at 8 bp downstream of an alternative coding exon (i.e., intronic region) in the other seven transcripts (Fig. 2a). To verify the expression of transcript 8 of ENSA in human retina as well as in other human tissues, two primer sets were designed to separately amplify transcript 8 and other transcripts by RT-PCR (Fig. 2a). The results showed that transcript 8 and other transcripts of ENSA were ubiquitously expressed in various human tissues, with transcript 8 having a higher level of expression than other transcripts in the retina as well as in many extraocular tissues (Fig. 2b). Among human ocular tissues, ENSA was predominantly expressed in the retina, optic nerve, and RPE (Fig. 2b), which are the key sites involved in retinopathy. The expression of ENSA in the human retina at the protein level was evaluated by immunostaining, and the results showed that ENSA was predominantly located in the outer segments of rod photoreceptors, with a less extension in the RPE and weak staining in the ganglion cell layer as well as the inner nuclear layer (Fig. 3a). Although the expression pattern was similar to that of rhodopsin, the staining of ENSA appeared relatively stronger at the inner centre of outer segments in rods than that at the outer part (Fig. 3b). The labelling pattern of ENSA in the human retina is compatible with the photoreceptor degeneration of the proband harbouring the ENSA variant. Evaluation of ENSA expression in cones was expected but not confirmed due to unsuccessful co-labelling of ENSA with cone-specific markers in the same section (Supplementary Materials, Fig. S3).
Fig. 2.
Transcripts and expression analysis of ENSA mRNA. (a) The coding scheme of eight alternative transcripts of ENSA. Transcript 8 (NM_207168) is composed of exons 1 and exon 3B. The mutation in ENSA, c.191delG (p.Gly64Alafs*2), is located at exon 3B of transcript 8 (the intronic +8 position of the alternative exon 3A in other transcripts). The differences between exon 3B and exon 3A are indicated in red (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article). Arrows represent the location of primers used for RT-PCR (see also Supplementary Materials, Table S2). (b) The RT-PCR of ENSA in 32 human tissues. Two sets of primers were used to amplify different transcripts of ENSA: set 1 (F1+R1), representing transcripts 1~7, and set 2 (F2+R2), representing transcript 8. ACTB amplification served as a control for cDNA amount in the PCR analysis. It should be noted that transcripts 5, 6, and 7, in which an additional 48 bp exon (exon 4) is encompassed, have been deposited in GenBank, but that we were not able to verify it in any of the analyzed tissues. M, marker. (c) Expression of Ensa in mouse retinas at different developmental stages by qPCR. Ex, embryonic x days. Px, x days after birth. Each of the RNA samples was obtained from three wild-type mice (six retinas). The experiments were repeated 3 times. β-Actin was used for normalization. Error bars represent the standard errors of the relative quantities.
Fig. 3.
Immunostaining for ENSA in human retina. (a) Low-magnification images of the human retina reveal prominent labelling for ENSA in the outer segments of rod photoreceptors and less labelling in the RPE, with weak staining observed in the ganglion cell layer and the inner nuclear layer. (b) Higher-magnification images of the photoreceptor region indicate that the staining for ENSA was mainly located in the inner centre of the rod photoreceptor outer segments, whereas the outer part exhibited markedly lower fluorescence. The staining of rhodopsin indicates the location of rod outer segments. GCL, ganglion cell layer; IPL, inner plexiform layer; INL, inner nuclear layer; OPL, outer plexiform layer; ONL, outer nuclear layer; IS, inner segments of photoreceptors; OS, outer segments of photoreceptors; RPE, retinal pigment epithelium layer. All scale bars represent 10 μm.
A study on mice showed increasing expression of Ensa in the retina after birth. A time-course expression examination of Ensa in mouse retinas at different developmental stages revealed low expression of transcript 8 of Ensa at the embryonic and perinatal stages, followed by a progressive increase from P8 to P30 and sustained high levels at adulthood (Fig. 2c). The expression levels of other Ensa transcripts in the retina increased from E11.5 to P30 and then decreased steadily from P30 to P143 (Fig. 2c). The expression pattern indicated that transcript 8 of ENSA may play an important role in the maintenance of adult retinal function.
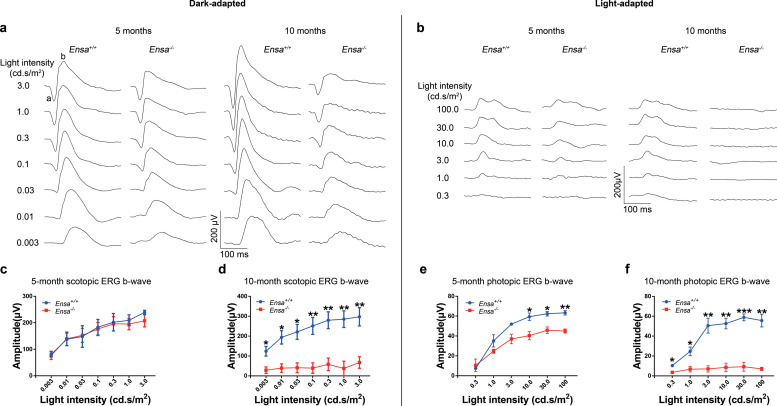
To further confirm the phenotype associated with the loss of ENSA observed in patients with RP, Ensa knockout mice were generated and the photoreceptor function in Ensa knockout mice was assessed by ERG. Rod and cone responses were tested using various stimulus intensities under dark- and light-adapted conditions, respectively. Compared with the wild-type (Ensa+/+) mice, Ensa knockout (Ensa−/−) mice showed a decreased photopic ERG response at 5-month old, especially at higher intensities tested (10, 30, and 100 cd.s/m2), with a slightly decreased scotopic ERG response (Fig. 4 a, b, c, and e). At 10-month old, Ensa−/− mice showed a significantly reduced ERG response under both dark- and light-adapted conditions (Fig. 4 a, b, d, and f). Histologically, a slightly thinner retinal layer and fewer cones, especially green cones, were observed in Ensa−/− mice compared with Ensa+/+ mice by 10 months (Supplementary Materials, Figs. S4 and S5). These findings provide additional evidence showing that loss of Ensa in mice mimics a phenotype of RP in humans.
Fig. 4.
Scotopic and photopic ERG recordings at different flash intensities from wild-type and Ensa knockout mice. (a) Representative ERG waveforms from dark-adapted wild-type (Ensa+/+) and Ensa knockout (Ensa−/−) mice aged 5 and 10 months. The flashlight intensities used to elicit the responses are indicated to the left. (b) Representative ERG waveforms from light-adapted wild-type (Ensa+/+) and Ensa knockout (Ensa−/−) mice aged 5 and 10 months. The flashlight intensities used to elicit the responses are indicated to the left. (c-f) The average amplitudes of scotopic (c-d) and photopic (e-f) ERG b-waves from Ensa+/+ and Ensa−/− mice aged 5 months and 10 months, respectively. Values are shown as the means ± SEMs (n = 3) for all tests. Two-tailed Student's t-test, *P<0.05, **P<0.01, ***P<0.001. SEMs, standard errors of the means.
3.3. Expression profiles of DACT2 and EXTL2, and phenotypes of dact2 or extl2 knockdown zebrafish
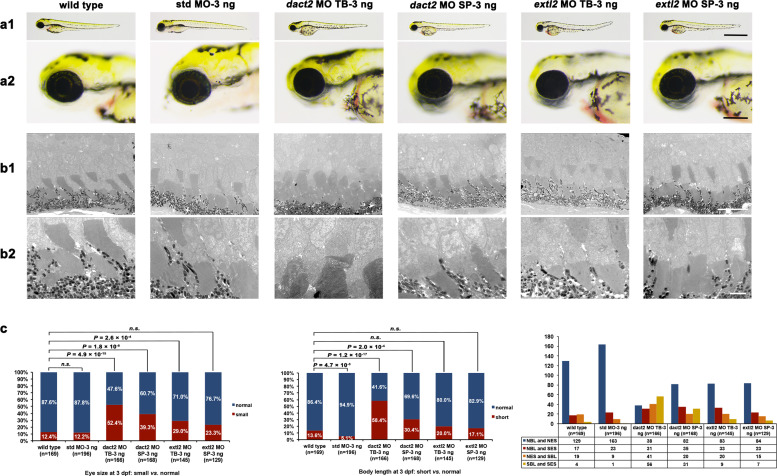
Among the four RP families with rare biallelic LoF variants, only one (ZOCRP0156) exhibited homozygous variants in two genes, DACT2 and EXTL2. To determine whether variants in either DACT2 or EXTL2 contribute to the RP phenotype in ZOCRP0156, expression pattern and knockdown zebrafish of the two genes were performed, respectively. RT-qPCR analysis of the two genes in different human tissues showed that DACT2 expressed at the highest level in retina among the 32 tissues, while the expression of EXTL2 was low in retina (Supplementary Materials, Fig. S6). In addition, MOs targeting either the initial translational site (MO TB) or the splice site at the exon-intron boundary (MO SP) of the two genes, as well as a standard control MO (std MO), were microinjected into zebrafish embryos, respectively. The survival rates of wild-type larvae, std MO, dact2 MO TB, dact2 MO SP, extl2 MO TB, and extl2 MO SP injected larvae were approximately 93.3% (280/300), 87.3% (262/300), 91.0% (273/300), 92.7% (278/300), 88.3% (265/300), and 89.7% (269/300), respectively. The efficacy tests for MO TB and MO SP were analyzed by western blot and RT-PCR. All the four targeted gene MOs, including dact2 MO TB, dact2 MO SP, extl2 MO TB and extl2 MO SP, lead to reduced protein levels compared with wild-type and std MO and the MO SP injection resulted in an intron insertion (Supplementary Materials, Fig. S7). Small eye size was defined as the eye diameters less than 289.3 μm, the eye diameter of mean minus SD for wild-type, while the eye diameters greater than 289.3 μm was classed as normal eye size (Supplementary Materials, Fig. S8). The eye size of the larvae injected with 3 ng of dact2 MO TB (52.4%, n = 166), 3 ng of dact2 MO SP (39.3% n = 168), 3 ng of extl2 MO TB (29.0%, n = 145), and 3 ng of extl2 MO SP (23.3%, n = 129), was significantly smaller than that of the larvae injected with 3 ng std MO (12.2%, n = 196) at 3 dpf (Fig.5). However, the number of photoreceptor cells was not affected in knockdown larvae compared with controls (Supplementary Materials, Fig. S9). The ultrastructure of photoreceptors was further assessed in dact2 or extl2 knockdown larvae using a transmission electron microscopy (TEM), revealing disorganisation of the photoreceptor disc membranes in the dact2 MO TB injected larvae (n = 3) and mild structural alterations such as oedema and minor disarrangement of photoreceptor cells in dact2 MO SP injected larvae (n = 2) (Fig 5). Such TEM change was absent in extl2 MO injected larvae and the control group at 5 dpf (Fig. 5). These results suggest that the homozygous LoF variant in DACT2 is more likely the cause of RP in ZOCRP0156 than the variant in EXTL2.
Fig. 5.
Phenotypes of knockdown dact2 and extl2 in zebrafish. (a1-a2) Eye size and body length in zebrafish at 3 dpf from six groups, i.e. wild-type, std MO-3 ng, dact2 MO TB-3 ng, dact2 MO SP-3 ng, extl2 MO TB-3 ng, and extl2 MO SP-3 ng. a1, the whole body; a2, the eyeballs. The eye diameters (eye size) were 309.5 ± 20.2 μm (n = 169) for wild-type, 308.6 ± 15.7 μm (n = 196) for the std MO group, 285.1 ± 29.2 μm (n = 166) for the dact2 MO TB morphants, 294.2 ± 30.3 μm (n = 168) for the dact2 MO SP morphants, 298.9 ± 25.1 μm (n = 145) for the extl2 MO TB morphants, and 303.8 ± 19.7 μm (n = 129) for the extl2 MO SP morphants. The body lengths were 3345.1 ± 137.7 μm (n = 169) for wild-type, 3431.5 ± 148.5 μm (n = 196) for the std MO group, 3109.5 ± 333.4 μm (n = 166) for the dact2 MO TB morphants, 3318.4 ± 269.1 μm (n = 168) for the dact2 MO SP morphants, 3367.8 ± 271.3 μm (n = 145) for the extl2 MO TB morphants, and 3399.6 ± 257.6 μm (n = 129) for the extl2 MO SP morphants. Scale bar, 1 mm (upper), 200 μm (below). (b1-b2) Under TEM, disorganisation in the membranous disc of the photoreceptor cells was found in the dact2 MO TB injected larvae and mild structural alterations such as oedema and minor disarrangement in photoreceptor cells appeared in dact2 MO SP injected larvae, while the membranous disc of the photoreceptor cells in the extl2 MO TB injected larvae and extl2 MO SP injected larvae were as similar as in wild-type and std MO injected larvae. Scale bar, 10 μm (upper), 2 μm (below). (c) Significant increase proportion of small eye sizes and short bodies were found in dact2 MO TB and dact2 MO SP injected larvae compared with wild-type larvae at 3 dpf, while, in extl2 morphants, significant increase proportion of small eye size was found in extl2 MO TB injected larvae compared with wild-type larvae but insignificant differences between extl2 MO SP injected and wild-type larvae . The left graph showed the quantification of small eye size proportions in zebrafish. The proportions of small eye size were significantly increased in dact2 MO TB morphants, dact2 MO SP morphants, and extl2 MO TB morphants compared with that of the wild-type (P = 4.9 × 10−15, P = 1.8 × 10−8, and P = 2.6 × 10−4, respectively) based on Chi-square test (P < 0.01). The middle graph showed that the proportions of short body length were significantly increased in dact2 MO TB morphants and dact2 MO SP morphants compared with that of the wild-type (P = 1.2 × 10−17 and P = 2.0 × 10−4) but insignificant difference among extl2 MO TB morphants, extl2 MO SP morphants and wild-type (P = 0.1 and P = 0.4), based on Chi-square test (P < 0.01). The right graph showed that the numbers of larvae in the group of NBL and NES, NBL and SES, NES and SBL, and SES and SBL. The number of zebrafish injected in each group is indicated under each column. vs., versus; NBL, normal body length; NES, normal eye size; SES, small eye size; SBL, short body length.
4. Discussion
LoF variants are considered to be deleterious by disrupting the coding-proteins. This kind of variants accounts for approximately 2.4% of protein-coding genetic variants based on the whole exome data from 60706 individuals [19], but accounts for approximately half of known disease-causing mutations according to the Human Gene Mutation Database [20]. Personal genomics study has revealed that LoF variants show a surprising prevalence in a healthy individual, although most of them are common heterozygous variants or even homozygous variants in nonessential genes [21]. Biallelic LoF variants in essential genes are extremely rare in the general population and are frequently associated with inherited diseases. Therefore, a number of novel genes have been identified by screening LoF variants in a cohort of patients [22], [23], [24]. In this study, biallelic LoF variants in five novel candidate genes (ENSA, DACT2, EXTL2, DDR1, and CCDC188) for autosomal recessive RP were identified by comparative analysis of WES data from 552 probands with RP and 4728 in-house controls. According to the gnomAD database, biallelic LoF variants in the five genes were extremely rare. The high expression of ENSA in human photoreceptors and significantly decreased ERG responses obtained from Ensa−/− mice further indicate that ENSA may play an important role in the maintenance of photoreceptor function. The homozygous DACT2 variant and the homozygous EXTL2 variant were found in the same proband, ZOCRP0156. However, the highest expression of DACT2 in the human retina and disorganised photoreceptor disc membranes in dact2 knockdown zebrafish supported that the RP phenotype in ZOCRP0156 was more likely due to the loss of DACT2 rather than that of EXTL2. These lines of evidence not only suggest that biallelic LoF variants in ENSA and DACT2 may cause RP but also suggest that the strategy by screening unique biallelic LoF variants in a cohort of probands is feasible for exploring novel RP genes. This approach has been successfully applied in the identification of biallelic LoF variants in PPP1R21 that cause neurodevelopmental syndrome [22].
ENSA is an endogenous ligand of the sulfonylurea receptor 1 (SUR1), which is one of the regulatory subunits of ATP-sensitive potassium (KATP) channels [16,17]. It has been reported to participate in controlling cell cycle progression [25]. To date, the relationship between variants in ENSA and human Mendelian diseases has not been well elucidated, although a decreased expression level of ENSA has been detected in brains from adult Down syndrome patients [26]. In this study, a homozygous truncated variant, c.191delG (p.Gly64Alafs*2), in ENSA was identified in a proband with RP, and no homozygous LoF variants were detected in 125730 individuals from the gnomAD database. This ubiquitous expression of ENSA in multiple human tissues revealed by RT-PCR is consistent with the results obtained from Northern blot in a set of human tissues that did not include ocular tissues [16]. The specific expression of ENSA in the photoreceptor provides evidence that it could function in photoreceptors, which are specifically affected in patients with RP. A higher expression of Ensa at the postnatal stage than at the embryonic stage in mice is consistent with a previous study's finding that ENSA is not required in early embryonic development [25]. Therefore, the pathogenic mechanism of ENSA-associated retinopathy might involve a disturbance of photoreceptor maintenance by loss of control of cell cycle progression. Furthermore, ENSA might change the cell depolarization and hyperpolarization by perturbing the K+ current in rod photoreceptors, which might not only change the associated signaling but also induce damage to the cells. In addition, ENSA has an interaction with OFD1 [27], which is the causative gene of severe X-linked RP [28].
DACT2, also known as DPR2 or DAPPER2, encodes the disheveled binding antagonist of beta catenin 2. The previous study found that Dpr2 was required for normal convergence extension movements in zebrafish embryos and dpr2-konckdown larvae showed narrow and improperly placed eyes [29]. However, none variants in DACT2 have been reported in patients with eye diseases. In this study, homozygous LoF variant in DACT2 was identified in the proband ZOCRP0156 with RP with another homozygous LoF variant in EXTL2. Functional studies reveal that DACT2 but not EXTL2 were associated with RP for the highest expression in retina and the disorganised photoreceptor disc membranes in dact2 knockdown larvae. In addition, extraocular abnormalities, including short and wide body axes, were found in dact2 knockdown larvae in the current study, which is similar with the aberrant convergent extension movements in the previous study, which were possibly from the functional interaction of dact2 with Stbm [29]. It is interesting that Stbm, a regulator of planar cell polarity (PCP) signaling, participates in the regulation of photoreceptor cell fate as well as cilia and photoreceptor outer segment length in zebrafish in previous studies [30,31]. Therefore, it is supposed that the RP in the patient with DACT2 homozygous LoF variants is from that loss of dact2 possibly leads to the Wnt/PCP inactive by loss of interaction with Stbm, which are awaited for confirmation by further studies.
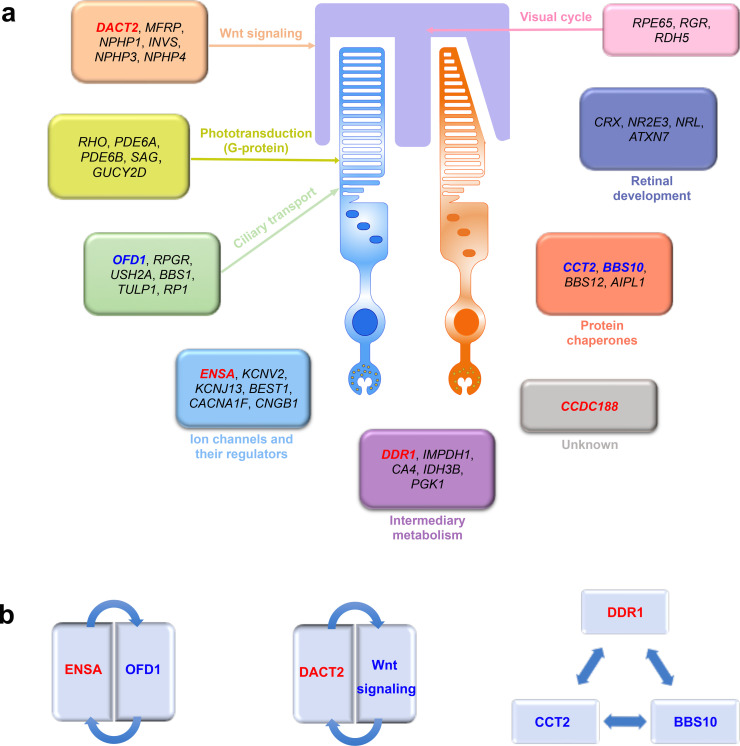
Genes responsible for IRD are involved in a wide range of photoreceptor development, structure, function, and metabolism. Their related functional pathways may include phototransduction, ciliary transport, visual cycle, Wnt signaling, ion channels and their regulators, and so on (Fig. 6a). A web-based search of existing databases revealed that potential pathways related to IRD might be associated with three of the four novel candidate genes, including ENSA, DACT2, and DDR1 (Fig. 6b). However, little is known about the functions of CCDC188 as it is a newly characterized functional gene.
Fig. 6.
Groups and interactions of the novel candidate genes for RP with known genes for inherited retinal degeneration. (a) Three of the four novel candidate genes for RP can be grouped with known genes for inherited retinal degeneration according to the variable functional pathways involving in the photoreceptor degeneration except the newly characterized CCDC188 gene. (b) The potential interactions of three novel candidate genes for RP with known genes for inherited retinal degeneration according to the BioGRID and STRING databases. The four novel candidate genes for RP are highlighted in red, and potential interaction genes with them are highlighted in blue (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
The phenotypes of animal models of ENSA and DACT2 support the association of the two genes with photoreceptor dysfunction or degeneration but are inconsistent with the RP phenotypes in human. A more pronounced decrease of photopic (cone) response than the scotopic (rod) response in ERGs was detected in Ensa−/− mice while the rod responses were more severely affected than the cone responses in RP patients. However, the inconsistent phenotypes between knockout mice and patients were not accidental. For example, mutations in RP2 cause RP in humans but knockout of Rp2 in mice resulted in a more severe effect on photopic response than on the scotopic response [32]. The exact reason why primarily affected photoreceptor cell type (i.e., rods vs. cones) is different between mice and humans is unclear. However, the same situation was also present among different patients with mutations in the same gene, for example, mutations in CERKL, PRPH2, CACNA1F, and others could cause both rod-dominant retinal degeneration and cone-dominant retinal degeneration (RetNet, https://sph.uth.edu/retnet/). These different phenotypes possibly result from the effects of the genetic background or potential modifiers [33,34]. Except for retinal degeneration, the phenotypes of small eye size and short body length identified in zebrafish model are unexpected and not present in patients in humans. However, such additional phenotype was also reported in the RPGR and SLC7A14 genes, in which mutations in human beings cause RP while knockdown of these genes in zebrafish resulted in retinal degeneration as well as small eye size even short body length [35,36].
In summary, four novel candidate genes for RP were identified in the present study by comparative analysis of WES data with the aim of revealing novel rare biallelic LoF variants in a cohort of probands with RP. The correlation of null variants in ENSA with RP was further confirmed by retinal expression analysis and knockout mouse model, while clues were detected about the association of DACT2 with RP from TEM analysis in dact2 knockdown zebrafish model. This approach not only provides a useful strategy to identify novel genes for extremely rare inherited diseases but also expand the list of genes responsible for RP when mutated. Further studies on these genes are expected to validate these findings and help elucidate the molecular mechanism of RP due to LoF of these genes, especially DDR1 and CCDC188.
Author contributions
Xueshan Xiao, Shiqiang Li, and Qingjiong Zhang recruited patients. Zhen Yi, Jiamin Ouyang, Wenmin Sun, Xueshan Xiao, and Shiqiang Li collected the clinical data. Xueshan Xiao and Qingjiong Zhang performed whole exome analysis. Qingjiong Zhang, Zhen Yi, Jiamin Ouyang and Wenmin Sun performed the bioinformatic analysis and designed the study. Zhen Yi performed the expression analysis of ENSA and the mouse experiments. Jiamin Ouyang and Wenmin Sun conducted the expression analysis of DACT2 and EXTL2 and the zebrafish experiments. Zhen Yi, Jiamin Ouyang, Wenmin Sun and Qingjiong Zhang discussed the results and wrote the manuscript. All authors reviewed and approved the manuscript.
Declaration of Competing Interest
The authors declare no conflicts of interest.
Acknowledgments
Acknowledgments
We thank all patients and their relatives for their participation in this study.
Funding sources
This work was supported by grants from the National Natural Science Foundation of China (81970837) and the Fundamental Research Funds of the State Key Laboratory of Ophthalmology.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2020.102792.
Appendix. Supplementary materials
References
- 1.Berger W, Kloeckener-Gruissem B, Neidhardt J. The molecular basis of human retinal and vitreoretinal diseases. Progress Ret Eye Res. 2010;29(5):335–375. doi: 10.1016/j.preteyeres.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 2.Chizzolini M, Galan A, Milan E, Sebastiani A, Costagliola C, Parmeggiani F. Good epidemiologic practice in retinitis pigmentosa: from phenotyping to biobanking. Curr Genomics. 2011;12(4):260–266. doi: 10.2174/138920211795860071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verbakel SK, van Huet RAC, Boon CJF. Non-syndromic retinitis pigmentosa. Progr Ret Eye Res. 2018;66:157–186. doi: 10.1016/j.preteyeres.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 4.Tsang SH, Sharma T. Retinitis Pigmentosa (Non-syndromic) Adv Exp Med Biol. 2018;1085:125–130. doi: 10.1007/978-3-319-95046-4_25. [DOI] [PubMed] [Google Scholar]
- 5.Berson EL. Electroretinographic findings in retinitis pigmentosa. Jpn J Ophthalmol. 1987;31(3):327–348. [PubMed] [Google Scholar]
- 6.Daiger SP. Identifying retinal disease genes: how far have we come, how far do we have to go. Novartis Found Symp. 2004;255:17–27. doi: 10.1002/0470092645.ch3. discussion 27-36, 177-178. doi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daiger SP, Bowne SJ, Sullivan LS. Perspective on genes and mutations causing retinitis pigmentosa. Arch Ophthalmol. 2007;125(2):151–158. doi: 10.1001/archopht.125.2.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beales PL, Badano JL, Ross AJ. Genetic interaction of BBS1 mutations with alleles at other BBS loci can result in non-Mendelian Bardet-Biedl syndrome. Am J Hum Genet. 2003;72(5):1187–1199. doi: 10.1086/375178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kajiwara K, Berson EL, Dryja TP. Digenic retinitis pigmentosa due to mutations at the unlinked peripherin/RDS and ROM1 loci. Science. 1994;264(5165):1604–1608. doi: 10.1126/science.8202715. [DOI] [PubMed] [Google Scholar]
- 10.Xu Y, Guan L, Shen T. Mutations of 60 known causative genes in 157 families with retinitis pigmentosa based on exome sequencing. Hum Genet. 2014;133(10):1255–1271. doi: 10.1007/s00439-014-1460-2. [DOI] [PubMed] [Google Scholar]
- 11.Derveaux S, Vandesompele J, Hellemans J. How to do successful gene expression analysis using real-time PCR. Methods (San Diego, Calif.) 2010;50(4):227–230. doi: 10.1016/j.ymeth.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 12.Birling MC, Herault Y, Pavlovic G. Modeling human disease in rodents by CRISPR/Cas9 genome editing. Mamm Genome. 2017;28(7-8):291–301. doi: 10.1007/s00335-017-9703-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Branchek T, Bremiller R. The development of photoreceptors in the zebrafish, Brachydanio rerio. I. Structure. J Comp Neurol. 1984;224(1):107–115. doi: 10.1002/cne.902240109. [DOI] [PubMed] [Google Scholar]
- 14.Branchek T. The development of photoreceptors in the zebrafish, brachydanio rerio. II. Function. J Comp Neurol. 1984;224(1):116–122. doi: 10.1002/cne.902240110. [DOI] [PubMed] [Google Scholar]
- 15.Wilson ET, Cretekos CJ, Helde KA. Cell mixing during early epiboly in the zebrafish embryo. Dev Genet. 1995;17(1):6–15. doi: 10.1002/dvg.1020170103. [DOI] [PubMed] [Google Scholar]
- 16.Heron L, Virsolvy A, Peyrollier K. Human alpha-endosulfine, a possible regulator of sulfonylurea-sensitive KATP channel: molecular cloning, expression and biological properties. Proc Natl Acad Sci U S A. 1998;95(14):8387–8391. doi: 10.1073/pnas.95.14.8387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heron L, Virsolvy A, Apiou F, Le Cam A, Bataille D. Isolation, characterization, and chromosomal localization of the human ENSA gene that encodes alpha-endosulfine, a regulator of beta-cell K(ATP) channels. Diabetes. 1999;48(9):1873–1876. doi: 10.2337/diabetes.48.9.1873. [DOI] [PubMed] [Google Scholar]
- 18.Thameem F, Farook VS, Yang X. The transcribed endosulfine alpha gene is located within a type 2 diabetes-linked region on 1q: sequence and expression analysis in Pima Indians. Mol Genet Metab. 2004;81(1):16–21. doi: 10.1016/j.ymgme.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 19.Lek M, Karczewski KJ, Minikel EV. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stenson PD, Mort M, Ball EV. The Human Gene Mutation Database: towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum Genet. 2017;136(6):665–677. doi: 10.1007/s00439-017-1779-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacArthur DG, Balasubramanian S, Frankish A. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335(6070):823–828. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rehman AU, Najafi M, Kambouris M. Biallelic loss of function variants in PPP1R21 cause a neurodevelopmental syndrome with impaired endocytic function. Human Mutation. 2019;40(3):267–280. doi: 10.1002/humu.23694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richard EM, Polla DL, Assir MZ. Bi-allelic Variants in METTL5 Cause Autosomal-Recessive Intellectual Disability and Microcephaly. Am J Hum Genet. 2019;105(4):869–878. doi: 10.1016/j.ajhg.2019.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weisz Hubshman M, Broekman S, van Wijk E. Whole-exome sequencing reveals POC5 as a novel gene associated with autosomal recessive retinitis pigmentosa. Hum Mol Genet. 2018;27(4):614–624. doi: 10.1093/hmg/ddx428. [DOI] [PubMed] [Google Scholar]
- 25.Hached K, Goguet P, Charrasse S. ENSA and ARPP19 differentially control cell cycle progression and development. J Cell Biol. 2019;218(2):541–558. doi: 10.1083/jcb.201708105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim SH, Lubec G. Decreased alpha-endosulfine, an endogenous regulator of ATP-sensitive potassium channels, in brains from adult Down syndrome patients. J Neural Transm. Supplement. 2001;(61):1–9. doi: 10.1007/978-3-7091-6262-0_1. [DOI] [PubMed] [Google Scholar]
- 27.Huttlin EL, Bruckner RJ, Paulo JA. Architecture of the human interactome defines protein communities and disease networks. Nature. 2017;545(7655):505–509. doi: 10.1038/nature22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Webb TR, Parfitt DA, Gardner JC. Deep intronic mutation in OFD1, identified by targeted genomic next-generation sequencing, causes a severe form of X-linked retinitis pigmentosa (RP23) Hum Mol Genet. 2012;21(16):3647–3654. doi: 10.1093/hmg/dds194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waxman JS, Hocking AM, Stoick CL, Moon RT. Zebrafish Dapper1 and Dapper2 play distinct roles in Wnt-mediated developmental processes. Development. 2004;131(23):5909–5921. doi: 10.1242/dev.01520. [DOI] [PubMed] [Google Scholar]
- 30.Jenny A, Darken RS, Wilson PA, Mlodzik M. Prickle and Strabismus form a functional complex to generate a correct axis during planar cell polarity signaling. EMBO J. 2003;22(17):4409–4420. doi: 10.1093/emboj/cdg424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song P, Dudinsky L, Fogerty J, Gaivin R, Perkins BD. Arl13b Interacts With Vangl2 to Regulate Cilia and Photoreceptor Outer Segment Length in Zebrafish. Invest Ophthalmol Vis Sci. 2016;57(10):4517–4526. doi: 10.1167/iovs.16-19898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li L, Khan N, Hurd T. Ablation of the X-linked retinitis pigmentosa 2 (Rp2) gene in mice results in opsin mislocalization and photoreceptor degeneration. Invest Ophthalmol Vis Sci. 2013;54(7):4503–4511. doi: 10.1167/iovs.13-12140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brunner S, Skosyrski S, Kirschner-Schwabe R. Cone versus rod disease in a mutant Rpgr mouse caused by different genetic backgrounds. Invest Ophthalmol Vis Sci. 2010;51(2):1106–1115. doi: 10.1167/iovs.08-2742. [DOI] [PubMed] [Google Scholar]
- 34.Samardzija M, Wenzel A, Naash M, Reme CE, Grimm C. Rpe65 as a modifier gene for inherited retinal degeneration. Eur J Neurosci. 2006;23(4):1028–1034. doi: 10.1111/j.1460-9568.2006.04639.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shu X, Zeng Z, Gautier P. Zebrafish Rpgr is required for normal retinal development and plays a role in dynein-based retrograde transport processes. Hum Mol Genet. 2010;19(4):657–670. doi: 10.1093/hmg/ddp533. [DOI] [PubMed] [Google Scholar]
- 36.Jin ZB, Huang XF, Lv JN. SLC7A14 linked to autosomal recessive retinitis pigmentosa. Nat Commun. 2014;5:3517. doi: 10.1038/ncomms4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.