Abstract
Periostin (POSTN) is an extracellular matrix protein associated with tumor progression and shorter survival in prostate cancer (PCa). Here, we performed an integrative analysis of POSTN’s role in patients with PCa. Clinical and POSTN data from large-scale datasets were analyzed. POSTN cutoffs were identified with X-Tile, and STRING was used for protein-protein interaction analysis. In a cohort of 48 patients with metastatic castration-resistant prostate cancer (mCRPC), we used the AdnaTest platform to isolate circulating tumor cells and extract POSTN mRNA. Plasma samples were also tested for POSTN protein expression by dot blot assay. Data from large-scale datasets did not reveal any association between POSTN genetic alterations and outcome. In primary tumors, we found a significant correlation between POSTN mRNA overexpression, worse baseline prognostic features, and shorter disease-free survival. POSTN was overexpressed in mCRPC and correlated with aggressive features. In our cohort of mCRPC patients, we found a positive correlation between POSTN plasma levels and androgen-receptor variant 7 positivity and an association with shorter overall survival. Our integrative analysis shows that POSTN is associated with poor clinical features and worse outcome in patients with PCa. Further studies are warranted to uncover the function of POSTN in PCa progression and to validate the prognostic significance of POSTN in mCRPC.
Introduction
Periostin (POSTN) is a multifaced extracellular matrix protein involved in physiologic functions such as bone regeneration, cardiac remodeling, skin response to damages, and kidney development [1]. POSTN also participates in many fibrovascular and inflammatory processes that are characteristic of pulmonary and heart fibrosis, chronic kidney and liver diseases, asthma, glaucoma, and retinopathies [1]. This protein functions as a scaffold for many other proteins and appears to be a mediator of cell-to-matrix signaling and epithelial-mesenchymal transition, thus favoring cancer progression [[2], [3], [4], [5]]. Several studies showed that POSTN is overexpressed in various types of cancers, and its overexpression is usually associated with aggressive clinical features and poor outcomes [[6], [7], [8], [9]]. In prostate cancer (PCa), we and other authors have showed that POSTN protein overexpression in primary tumors is associated with worse baseline clinical features, shorter disease-free survival (DFS), and reduced overall survival (OS) [[10], [11], [12], [13], [14]].
In the present integrative analysis, we attempted to identify potential associations between clinical data and POSTN expression at genetic, transcriptional, and proteomic level. In a cohort of patients with metastatic castration-resistant prostate cancer (mCRPC), we also investigated the feasibility to detect POSTN mRNA in circulating tumor cells (CTCs) and POSTN protein in plasma samples in order to explore the association of POSTN expression with clinical outcome in this specific setting.
Results
Periostin in Patients with Prostate Cancer: Results from Large-Scale Datasets
Periostin Genetic Alterations
We explored the genetic landscape of POSTN in cBioPortal [15,16] in order to investigate the prevalence and possible correlation or association of POSTN alterations with clinical features and outcomes. The most common alteration in POSTN gene was deep deletion (up to 12% of samples in the TCGA cohort). POSTN amplifications or gains were also detected (Figure 1). We did not find any significant correlation or association between POSTN genetic alterations and clinical features, DFS, or OS (when available) in PCa patients.
Figure 1.
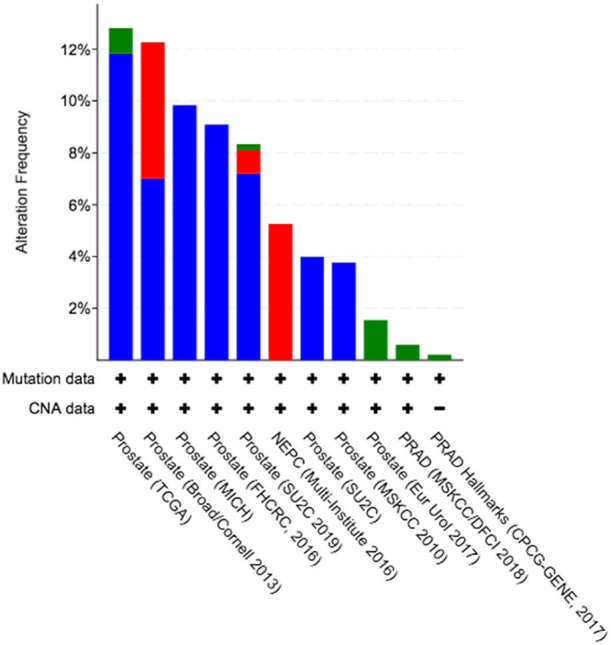
Frequency of POSTN genetic alterations across PCa datasets analyzed by cBioPortal. Deep deletion (blue), amplification (red), and mutation (green). Full description of PCa studies included in the figure is found on https://www.cbioportal.org/.
Periostin mRNA Expression
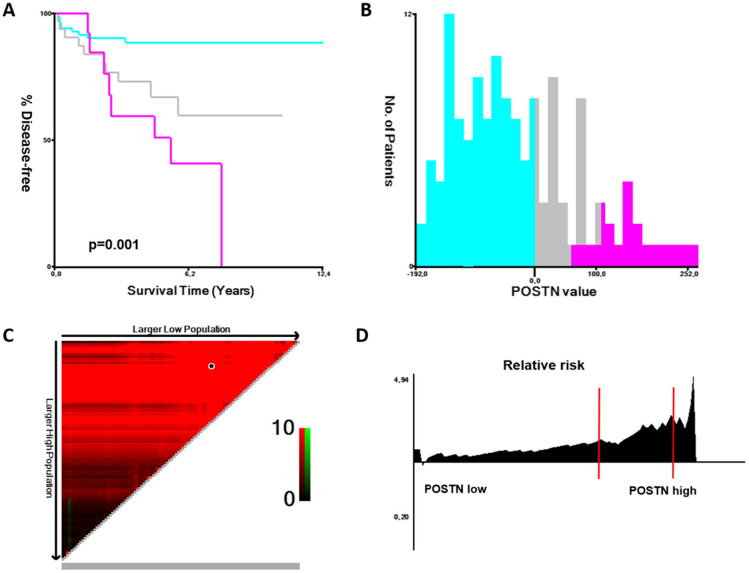
We also investigated the potential correlation and association of POSTN mRNA overexpression with clinical features and outcomes in patients with primary and metastatic PCa. We identified two cohorts of primary PCa with available clinical data (TCGA Firehose Legacy [17] and MSKCC cohorts [18]). Using X-tile [19], we selected a threshold of +1 z-scores to identify patients with POSTN overexpression in cBioPortal (Figure 2). In these two cohorts, 6% and 10% of patients, respectively, had POSTN values above +1 z-scores. We found a significant association between POSTN mRNA expression and DFS in patients enrolled in the MSKCC cohort.
Figure 2.
Association of POSTN mRNA with DFS in the MSKCC cohort of primary PCa. (A) Kaplan-Meier estimations in patients with POSTN >1 (purple), >0-≤1 (gray), and ≤0 (light blue) z-scores; (B) X-tile histogram of POSTN values (expressed as z-scores ×10) according to number of patients; (C) triangular X-tile plot with the choice of POSTN cutoff (black point); (D) continuous relative risk of disease progression (expressed as hazard ratio) according to POSTN values (z-scores cutoffs of 0 and 1 are shown in red lines).
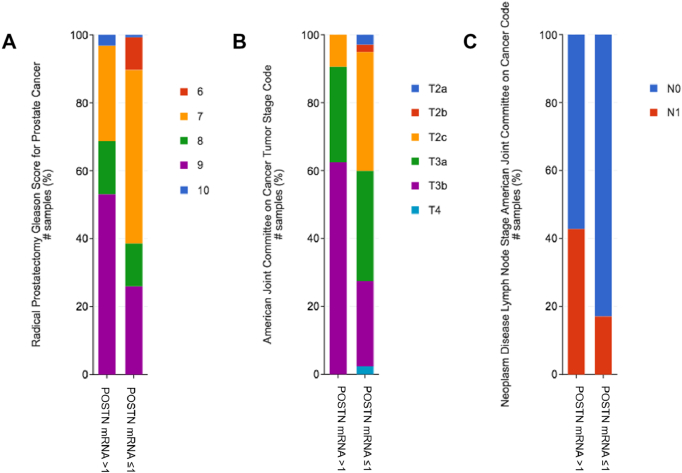
Similar results were found in the provisional data of the TCGA Firehose Legacy cohort (Supplementary Figure 1). In the TCGA cohort, we also found a positive correlation of POSTN expression with Gleason score, tumor stage, nodes positivity, and fraction of genome altered (all P < .001) (Figure 3).
Figure 3.
Correlation of POSTN mRNA expression (z-scores) with Gleason score (A), tumor stage (B), and nodes positivity (C) in the Firehose Legacy TCGA cohort of primary PCa.
In the metastatic samples of SUC2C/PCF cohort (12% of patients with POSTN z-scores >1) [20], we found a positive correlation of POSTN mRNA with neuroendocrine prostate cancer (NEPC) score (P = .016) and an inverse correlation with androgen-receptor (AR) score (P < .001). No data were available for disease progression in this cohort. No association was found between POSTN RNA expression and OS in primary or in metastatic cohorts, but comparisons were inappropriate given the low number of events.
In order to explore the possible differential expression of POSTN mRNA among nontumoral prostate tissue (NT), primary PCa, and mCRPC, we analyzed the GSE32269, GSE70768, and GSE101607 [[21], [22], [23]] datasets by GEO2R tool. POSTN mRNA was significantly overexpressed in mCRPC compared to primary PCa (GSE32269, GSE70768) and, based on AR activity, in non–AR-driven mCRPC compared to AR-driven mCRPC (GSE101607). POSTN mRNA expression decrease was observed in NT tissues in comparison with mCRPC, whereas no significant differences were observed between NT and primary PCa (GSE70768) (Figure 4).
Figure 4.
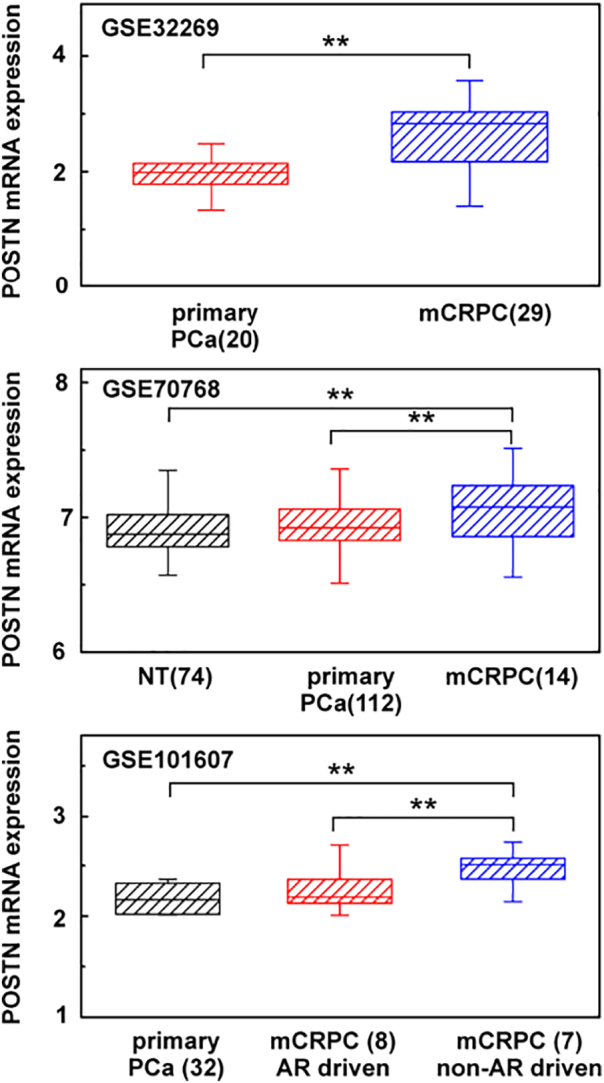
POSTN mRNA expression in NT, primary PCa, and mCRPC using GSE32269, GSE101607, and GSE70768 datasets. Numbers in brackets are sample analyzed. ** P < .01.
Biological Processes Associated with Higher Periostin mRNA Expression in Primary and Metastatic Prostate Cancer
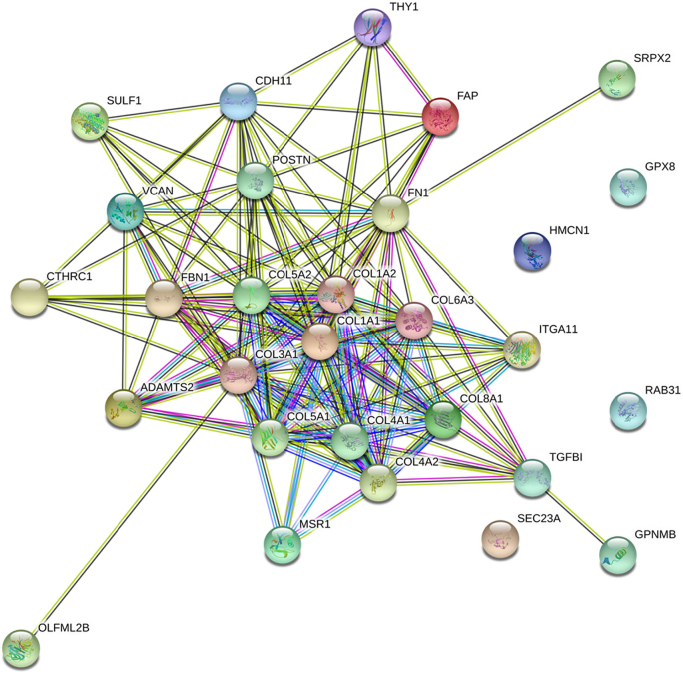
Using the cBioPortal tool, we investigated possible mRNA enrichments in patients with high POSTN mRNA expression in the TCGA and SUC2C/PCF datasets [17,20]. Separately for each cohort of datasets, we selected the first 100 mRNA with the highest statistically significant enrichment in patients with high POSTN mRNA (>1 z-scores) compared to patients with lower POSTN mRNA expression (≤1 z-scores). In patients with high POSTN mRNA expression, we identified an interaction network of 28 mRNAs that were significantly co-expressed with POSTN in both primary and metastatic datasets. This network was visualized by STRING database (Figure 5). The particularly low protein-protein interaction enrichment P value (<1.0e-16) showed that these proteins shared common functions. Gene ontology enrichment analysis revealed that this protein set was tightly involved in extracellular matrix organization, cell adhesion, cell migration, blood vessel development, and morphogenesis, such as collagen, proteoglycans, integrins, and proteases genes.
Figure 5.
STRING protein-protein interaction network of 28 mRNA enriched in patients with high POSTN mRNA expression both in primary TCGA and in metastatic SU2CF/PCF cohorts.
Periostin Protein Expression
To our knowledge, there are no available datasets on POSTN protein expression in PCa patients. However, our group and other authors have investigated the prognostic significance of POSTN protein expression in primary PCa by immunohistochemistry [10,11,13,14]. Most studies report that POSTN overexpression correlates with worse clinical features (higher Gleason score and tumor stage), and it is associated with poorer clinical outcome (shorter time to recurrence and OS).
Noninvasive Detection of Periostin in Patients with Advanced Prostate Cancer
Forty-eight patients with mCRPC were selected for this exploratory study on patients with mCRPC. The main characteristics of cohort patients are summarized in Supplementary Table 1. The median age was 74 (range 56-84), and the median prostate-specific antigen (PSA) was 33 (0.33-4688). In this explorative cohort, 60.4% of patients have not received prior therapy for mCRPC, whereas 39.6% have been previously treated with two or more lines of treatment for mCRPC, including chemo- and hormone therapy.
Feasibility of CTC POSTN Evaluation
We investigated the potential detection of POSTN mRNA in CTC from these patients. Of patients analyzed, only two showed detectable levels of POSTN RNA in CTC, and the values were below the threshold of detection method (<10 cp/ml blood). Therefore, we did not proceed to any further evaluation.
Prognostic and Predictive Value of Plasma POSTN
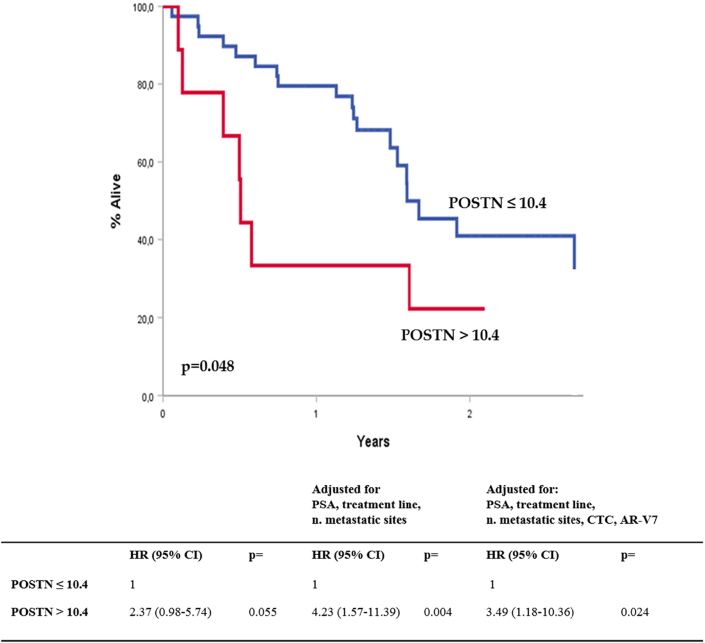
We investigated the potential detection of POSTN protein in plasma of mCRPC patients by dot blot technique. The median POSTN optical density (OD) value in the whole cohort was 4.24 (0.11-18.22). The mean value was 5.56 (SE 0.76). We found a positive correlation between plasma POSTN and androgen-receptor variant 7 (AR-V7) positivity (median POSTN OD: 4.45 vs 9.57 in AR-V7- and AR-V7+; P = .002). Using X-tile with two cut points, we distinguished three prognostic groups (high, medium, low), and we observed a significant difference in the OS of patients with high versus low POSTN plasma values (P = .018) (Supplementary Figure 2). Using one cut point (>10.4 OD), we also found a significant association with shorter OS in patients with high versus low POSTN plasma values that was confirmed after multivariable analyses, adjusting for significant confounding variables (PSA, treatment line, number of metastatic sites, CTC, and AR-V7 status) (Figure 6). Regarding PSA response to treatments, of 9 patients with high POSTN expression (>10.4 OD), only 3 (33.3%) reached the PSA50 compared to 23 (59.0%) of those with lower expression (difference not statistically significant).
Figure 6.
Association of POSTN protein plasma values with OS in our cohort of mCRPC patients. Curves are constructed using the Kaplan-Meier estimations. HR, hazard ratios resulting from the Cox regression models.
Discussion
The most relevant molecular studies on the characterization of primary and advanced PCa have not reported that POSTN gene is a significant determinant of PCa development or progression [24,25]. In addition, POSTN genetic alterations have not emerged as potential predictors of survival or response to therapies in patients with advanced PCa [20,26]. A nonlinear relationship often occurs among genetic alterations, mRNA transcripts, and protein expression [27]. Given the complex role of POSTN within the tumor microenvironment, the adaptive mechanisms of cancer stroma might be involved in POSTN-mediated cancer progression and might not be related with genetic alterations of POSTN gene.
Our analysis of large-scale datasets supports this assumption. POSTN gene deep deletions were found in up to 12% of patients with primary PCa, but we did not find any correlation or association with clinical data (Figure 1). This observation suggests that genetic alterations in POSTN gene might be secondary events within the context of the tumor mutational burden.
Conversely, we found significant correlations and associations between POSTN mRNA expression and clinical data (Figure 2, Figure 3). Patients with POSTN-overexpressing primary tumors had higher Gleason score and tumor stage at baseline. These patients also showed significantly shorter DFS compared to patients with lower POSTN mRNA expression. These results are consistent with the data obtained previously at proteomic level in other cohorts by using immunohistochemistry [[10], [11], [12], [13], [14]]. Our datasets analysis revealed that POSTN is overexpressed in mCRPC compared to primary PCa (Figure 4), and the analysis of metastatic patients included in the SU2C/PCF cohort also showed a positive correlation between POSTN mRNA expression and NEPC score and an inverse correlation with AR score. These observations suggest that POSTN might be overexpressed with the advanced stages of mCRPC when the selective pressure of treatments drives the progression towards a neuroendocrine phenotype [28]. Unfortunately, given the low statistical power of available samples with mRNA data, we were not able to explore the association of POSTN with OS.
Our functional enrichment analysis also uncovered an unexpected result (Figure 5). We found that patients with higher POSTN expression — both in primary and in metastatic cohorts — shared a common mRNA signature that included several other genes involved in extracellular matrix remodeling, cell adhesion, and morphogenesis. This signature might be important in order to identify early aggressive tumors with upregulation of extracellular-matrix proteins that favor metastases and tumor progression in order to select the appropriate treatment choice [29,30]. Several preclinical data support the role of POSTN in the epithelial-mesenchymal transition processes and metastatic potential [2,4,31].
In addition to this integrative analysis, we explored the role of POSTN in a cohort of mCRPC patients treated at our Institute. Firstly, we investigated the feasibility to detect the POSTN RNA in CTC isolated with the AdnaTest system. Only two patients in our cohort showed POSTN mRNA expression in CTC (below the threshold of detection method). Although it has been shown that epithelial tumor cells can express POSTN [10,14], the predominant role of POSTN in the stromal compartment might explain this disappointing result, and our data support the notion that CTCs express low or no levels of POSTN.
Subsequently, the plasma samples from this same cohort of mCRPC patients were analyzed to investigate the feasibility to detect POSTN plasma protein by dot blot technique. Konac and colleagues have recently shown that plasma levels of POSTN are significantly enriched in mCRPC patients compared with healthy controls or with patients affected by benign prostatic hyperplasia [32]. In addition, the negative prognostic value of POSTN plasma levels has been also reported in other neoplasms [[33], [34], [35]]. In our cohort of mCRPC patients, we found a positive correlation between POSTN levels and AR-V7 positivity. AR-V7 positivity is known to be associated with poor prognosis and with poor response to AR signaling inhibitors in mCRPC [36,37]. Patients with higher values of plasma POSTN also showed a significantly higher risk of death, which was confirmed after multivariable analyses including CTC and AR-V7 (Figure 6). Our data also provided evidence that POSTN might be a predictor of response to therapies given that only 33% of patients with high POSTN expression reached the PSA50 compared to 59% of those with lower expression. We acknowledge that POSTN is not prostate specific and that these explorative results need validation in a wider and homogeneous cohort. However, we demonstrated that POSTN protein detection in plasma sample is cheap, is feasible, and could have significant prognostic and predictive implications in the advanced stages of PCa.
Conclusion
Our integrative analysis highlights the relevance of POSTN in primary and advanced PCa. The datasets analysis supports the notion that POSTN-overexpressing tumors share a common mRNA signature that includes genes involved in the extracellular matrix remodeling and cell migration. All available data support a strong correlation between POSTN expression and baseline adverse features in primary PCa, namely, high Gleason score and tumor stage. In addition, our results support an association between high POSTN RNA expression and shorter DFS. Although validation studies are needed, our exploratory cohort on mCRPC patients shows that POSTN levels might be useful to predict prognosis and response to therapies also in the setting of mCRPC.
Materials and Methods
Analysis of POSTN in Large-Scale Datasets
We explored the association of POSTN with clinical features and outcomes of PCa patients using data available on the cBioPortal (http://cbioportal.org) [15,16]. POSTN mRNA cutoffs were identified using X-Tile v3.6.1 [19]. GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/) was used to evaluate differential expression of POSTN mRNA in normal tissue, primary PCa, and mCRPC using GSE32269, GSE101607, and GSE70768 datasets. Functional clustering and GO enrichment were performed using STRING v11 [38].
Analysis of POSTN in Patients with mCRPC
We explored the role of POSTN in a cohort of mCRPC patients who were also tested for the presence of CTC, AR-V7, and AR full-length. The inclusion criteria of the patient population and methodology to detect CTC, AR-V7, and AR full-length have been fully described in our previous paper [36]. Patients' blood samples were collected before the start of the first line for mCRPC or after two or more lines for mCRPC. Study protocol and informed consent were approved by the local Ethical Committee (RP505REG2015).
POSTN mRNA Analysis in CTC
As reported in our prior study [36], CTCs were isolated using the AdnaTest Prostate Cancer Select, and mRNA was retrotranscribed using the AdnaTest Prostate Cancer Detect and SensiScript RT kits (Qiagen, Germany), respectively. Polymerase chain reaction products were analyzed on Agilent chip by Bioanalyzer electrophoresis (Agilent Technologies, USA). Custom primers for POSTN mRNA detection in CTC were developed by Bird srl (Rezzato, Italy), whereas primers specific for epidermal growth factor receptor, prostate-specific membrane antigen, PSA, and actin as internal polymerase chain reaction control were used for CTC positivity.
POSTN Protein Detection in Plasma Samples
Plasma samples from the same patients were tested for POSTN by dot blot assay. For this purpose, 2 ml of blood collected in vacuum tubes containing EDTA was centrifuged at 3000g for 10 minutes. Plasma, obtained by supernatant centrifugation at 15,000g for 10 minutes to eliminate cells and debris, was aliquoted and stored at −80°C. For dot blot assay, 2 μl of plasma was spotted onto a nitrocellulose membrane; dried in air for 1 hour at ambient temperature; and blocked with 0.15 M NaCl, 10 mM Tris–HCl (pH 7.4), and 0.1% Tween-20 solution containing 3% nonfatty milk. After overnight incubation at 4°C with anti-POSTN (1:1000, Acris, Germany), membrane was washed and incubated with HRP-conjugated anti rabbit IgG antibody (1:2000, Cell Signaling Technology Inc., USA). The signal was revealed using the Immobilon Western Chemiluminescent kit (Millipore Corp., USA). After x-ray scanning with GS800 scanner (BioRad, USA), background subtraction, normalization with positive control spots, and densitometric evaluation of each spot were carried out with Quantity One software (BioRad, USA). Comparison of signal intensity, expressed as OD values, was used to determine relative differences in plasmatic levels of POSTN. More detailed information about the POSTN dot blot assay is provided in the supplementary file.
Statistical Analysis
OS and DFS curves were constructed using the Kaplan-Meier method and compared using the log-rank test [39]. Cox proportional hazard models were applied to OS data from the cohort of mCRPC [40]. A stepwise procedure was used with a significance level of P = .05 to retain variables in the model. Hazard ratio (HR) estimates and their 95% confidence intervals were also calculated. All P values were two-tailed. PSA50 was defined as a decline of at least 50% in PSA values from treatment start. Student t test, and Kruskal-Wallis test were applied to compare groups. The Benjamini-Hochberg FDR correction procedure was used to adjust multiple comparisons in the datasets. The IBM software Statistical Package for Social Sciences version 25.0 for Windows (SPSS Inc., Chicago, IL) was used for data analysis.
Authors' Contributions
F. B. conceived and designed the study. C. C. and P. B. performed the datasets analysis. F. B., C. C., L. C., and E. Z. collected patients' data and blood samples. P. B. and M. C. analyzed the blood samples for CTC and POSTN levels. A. R. and L. Z. performed the statistical analyses. F. B., C. C., and P. B. were major contributors in writing the manuscript. All authors read and approved the final manuscript.
Ethics Approval and Consent to Participate
Informed written consent to blood collection and use for experimental purposes was obtained by all patients following study-protocol approval by the local Ethical Committee.
Funding
Supported by 5X1000 and “Ricerca corrente” funds of IRCCS San Martino Polyclinic Hospital, Genoa, Italy. The authors are extremely grateful to Dr. Claudia Casella (Epidemiology Unit-Tumor Registry, IRCCS San Martino Polyclinic Hospital, Genoa, Italy) for helping in data collection. C. C. is supported by a European Society for Medical Oncology Clinical Research Fellowship (2019-2020).
Conflicts of Interest
The authors declare that they do not have any conflict of interest.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tranon.2020.100789.
Contributor Information
Carlo Cattrini, Email: carlo.cattrini@gmail.com.
Francesco Boccardo, Email: fboccardo@unige.it.
Appendix A. Supplementary Data
Supplementary Materials The following are available online: Supplementary Figure 1: Association of POSTN mRNA with DFS in the Provisional Firehose Legacy cohort of primary PCa. (A) Kaplan-Meier estimations in patients with POSTN >1 (gray) and ≤0 (light blue) z-scores; (B) X-tile histogram of POSTN values (expressed as z-scores ×10) according to number of patients; (C) triangular X-tile plot with the choice of POSTN cutoff (black point); (D) continuous relative risk of disease progression (expressed as HR) according to POSTN values (z-score cutoff of 1 is shown in red line). Supplementary Figure 2: Association of POSTN plasma values with OS in our cohort of patients with mCRPC. (A) Kaplan-Meier estimations in patients with POSTN >10.4 (purple), >3.2 ≤ 10.4 (gray) and ≤3.2 (light blue) z-scores; (B) X-tile histogram of POSTN values (expressed as z-scores ×10) according to number of patients; (C) triangular X-tile plot with the choice of POSTN cut-off (black point); (D) continuous relative risk of disease progression (expressed as HR) according to POSTN values (z-scores cutoffs of 10.4 and 3.2 are shown in red lines). Supplementary Table 1: Patient Characteristics. Supplementary File 1: Periostin dot blot assay characterization.
References
- 1.Kudo A. (eds) Periostin. Advances in experimental medicine and biology, vol 1132. Springer, Dermatol. Sin.
- 2.Malanchi I., Santamaria-Martinez A., Susanto E., Peng H., Lehr H.A., Delaloye J.F., Huelsken J. Interactions between cancer stem cells and their niche govern metastatic colonization. Nature. 2012;481(7379):85–89. doi: 10.1038/nature10694. [DOI] [PubMed] [Google Scholar]
- 3.Mosher D.F., Johansson M.W., Gillis M.E., Annis D.S. Periostin and TGF-beta-induced protein: two peas in a pod? Crit. Rev. Biochem. Mol. Biol. 2015;50(5):427–439. doi: 10.3109/10409238.2015.1069791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui D., Huang Z., Liu Y., Ouyang G. The multifaceted role of periostin in priming the tumor microenvironments for tumor progression. Cellular and molecular life sciences : CMLS. 2017;74(23):4287–4291. doi: 10.1007/s00018-017-2646-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y., Huang Z., Cui D., Ouyang G. The multiaspect functions of periostin in tumor progression. Adv. Exp. Med. Biol. 2019;1132:125–136. doi: 10.1007/978-981-13-6657-4_13. [DOI] [PubMed] [Google Scholar]
- 6.Ratajczak-Wielgomas K., Dziegiel P. The role of periostin in neoplastic processes. Folia Histochem. Cytobiol. 2015;53(2):120–132. doi: 10.5603/FHC.a2015.0014. [DOI] [PubMed] [Google Scholar]
- 7.Nuzzo P.V., Buzzatti G., Ricci F., Rubagotti A., Argellati F., Zinoli L., Boccardo F. Periostin: a novel prognostic and therapeutic target for genitourinary cancer? Clinical genitourinary cancer. 2014;12(5):301–311. doi: 10.1016/j.clgc.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 8.Gonzalez-Gonzalez L., Alonso J. Periostin: a matricellular protein with multiple functions in cancer development and progression. Front. Oncol. 2018;8:225. doi: 10.3389/fonc.2018.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moniuszko T., Wincewicz A., Koda M., Domyslawska I., Sulkowski S. Role of periostin in esophageal, gastric and colon cancer. Oncol. Lett. 2016;12(2):783–787. doi: 10.3892/ol.2016.4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nuzzo P.V., Rubagotti A., Zinoli L., Ricci F., Salvi S., Boccardo S., Boccardo F. Prognostic value of stromal and epithelial periostin expression in human prostate cancer: correlation with clinical pathological features and the risk of biochemical relapse or death. BMC Cancer. 2012;12:625. doi: 10.1186/1471-2407-12-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cattrini C., Rubagotti A., Nuzzo P.V., Zinoli L., Salvi S., Boccardo S., Perachino M., Cerbone L., Vallome G., Latocca M.M., Zanardi E., Boccardo F. Overexpression of periostin in tumor biopsy samples is associated with prostate cancer phenotype and clinical outcome. Clinical genitourinary cancer. 2018;16(6):e1257–e1265. doi: 10.1016/j.clgc.2018.07.019. [DOI] [PubMed] [Google Scholar]
- 12.Tsunoda T., Furusato B., Takashima Y., Ravulapalli S., Dobi A., Srivastava S., McLeod D.G., Sesterhenn I.A., Ornstein D.K., Shirasawa S. The increased expression of periostin during early stages of prostate cancer and advanced stages of cancer stroma. Prostate. 2009;69(13):1398–1403. doi: 10.1002/pros.20988. [DOI] [PubMed] [Google Scholar]
- 13.Tischler V., Fritzsche F.R., Wild P.J., Stephan C., Seifert H.H., Riener M.O., Hermanns T., Mortezavi A., Gerhardt J., Schraml P., Jung K., Moch H., Soltermann A., Kristiansen G. Periostin is up-regulated in high grade and high stage prostate cancer. BMC Cancer. 2010;10:273. doi: 10.1186/1471-2407-10-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tian, Y.; Choi, C. H.; Li, Q. K.; Rahmatpanah, F. B.; Chen, X.; Kim, S. R.; Veltri, R.; Chia, D.; Zhang, Z.; Mercola, D.; Zhang, H., Overexpression of periostin in stroma positively associated with aggressive prostate cancer. PloS one2015, 10, (3), e0121502. [DOI] [PMC free article] [PubMed]
- 15.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., Antipin Y., Reva B., Goldberg A.P., Sander C., Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao, J.; Aksoy, B. A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S. O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; Cerami, E.; Sander, C.; Schultz, N., Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling2013, 6, (269), pl1. [DOI] [PMC free article] [PubMed]
- 17.cBioportal. TCGA prostate adenocarcinoma; raw data at the NCI; source mutation data from GDAC Firehose. Last access: 22.01.2020.
- 18.Taylor B.S., Schultz N., Hieronymus H., Gopalan A., Xiao Y., Carver B.S., Arora V.K., Kaushik P., Cerami E., Reva B., Antipin Y., Mitsiades N., Landers T., Dolgalev I., Major J.E., Wilson M., Socci N.D., Lash A.E., Heguy A., Eastham J.A., Scher H.I., Reuter V.E., Scardino P.T., Sander C., Sawyers C.L., Gerald W.L. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18(1):11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Camp R.L., Dolled-Filhart M., Rimm D.L. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clinical cancer research: an official journal of the American Association for Cancer Research. 2004;10(21):7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- 20.Abida, W.; Cyrta, J.; Heller, G.; Prandi, D.; Armenia, J.; Coleman, I.; Cieslik, M.; Benelli, M.; Robinson, D.; Van Allen, E. M.; Sboner, A.; Fedrizzi, T.; Mosquera, J. M.; Robinson, B. D.; De Sarkar, N.; Kunju, L. P.; Tomlins, S.; Wu, Y. M.; Nava Rodrigues, D.; Loda, M.; Gopalan, A.; Reuter, V. E.; Pritchard, C. C.; Mateo, J.; Bianchini, D.; Miranda, S.; Carreira, S.; Rescigno, P.; Filipenko, J.; Vinson, J.; Montgomery, R. B.; Beltran, H.; Heath, E. I.; Scher, H. I.; Kantoff, P. W.; Taplin, M. E.; Schultz, N.; deBono, J. S.; Demichelis, F.; Nelson, P. S.; Rubin, M. A.; Chinnaiyan, A. M.; Sawyers, C. L., Genomic correlates of clinical outcome in advanced prostate cancer. Proceedings of the National Academy of Sciences of the United States of America 2019, 116, (23), 11428–11436. [DOI] [PMC free article] [PubMed]
- 21.[dataset] Changmeng Cai (2011). Expression data for primary localized prostate cancer versus castration-resistant bone metastatic prostate cancer. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE32269.
- 22.[dataset] Ross-Adams H (2015). Prostate cancer stratification using molecular profiles [CamCap ExpressionArray]. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE70768.
- 23.[dataset] Bovinder Ylitalo E (2017). Whole genome expression analysis of clinical bone metastasis samples. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE101607.
- 24.Robinson, D.; Van Allen, E. M.; Wu, Y. M.; Schultz, N.; Lonigro, R. J.; Mosquera, J. M.; Montgomery, B.; Taplin, M. E.; Pritchard, C. C.; Attard, G.; Beltran, H.; Abida, W.; Bradley, R. K.; Vinson, J.; Cao, X.; Vats, P.; Kunju, L. P.; Hussain, M.; Feng, F. Y.; Tomlins, S. A.; Cooney, K. A.; Smith, D. C.; Brennan, C.; Siddiqui, J.; Mehra, R.; Chen, Y.; Rathkopf, D. E.; Morris, M. J.; Solomon, S. B.; Durack, J. C.; Reuter, V. E.; Gopalan, A.; Gao, J.; Loda, M.; Lis, R. T.; Bowden, M.; Balk, S. P.; Gaviola, G.; Sougnez, C.; Gupta, M.; Yu, E. Y.; Mostaghel, E. A.; Cheng, H. H.; Mulcahy, H.; True, L. D.; Plymate, S. R.; Dvinge, H.; Ferraldeschi, R.; Flohr, P.; Miranda, S.; Zafeiriou, Z.; Tunariu, N.; Mateo, J.; Perez-Lopez, R.; Demichelis, F.; Robinson, B. D.; Sboner, A.; Schiffman, M.; Nanus, D. M.; Tagawa, S. T.; Sigaras, A.; Eng, K. W.; Elemento, O.; Sboner, A.; Heath, E. I.; Scher, H. I.; Pienta, K. J.; Kantoff, P.; de Bono, J. S.; Rubin, M. A.; Nelson, P. S.; Garraway, L. A.; Sawyers, C. L.; Chinnaiyan, A. M., Integrative clinical genomics of advanced prostate cancer. Cell 2015, 162, (2), 454. [DOI] [PubMed]
- 25.Network C.G.A.R. The molecular taxonomy of primary prostate cancer. Cell. 2015;163(4):1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mateo, J.; Seed, G.; Bertan, C.; Rescigno, P.; Dolling, D.; Figueiredo, I.; Miranda, S.; Nava Rodrigues, D.; Gurel, B.; Clarke, M.; Atkin, M.; Chandler, R.; Messina, C.; Sumanasuriya, S.; Bianchini, D.; Barrero, M.; Petremolo, A.; Zafeiriou, Z.; Fontes, M. S.; Perez-Lopez, R.; Tunariu, N.; Fulton, B. A.; Jones, R.; McGovern, U. B.; Ralph, C.; Varughese, M.; Parikh, O.; Jain, S.; Elliott, T.; Sandhu, S.; Porta, N.; Hall, E.; Yuan, W.; Carreira, S.; de Bono, J. S., Genomics of lethal prostate cancer at diagnosis and castration-resistance. The Journal of clinical investigation2019. [DOI] [PMC free article] [PubMed]
- 27.Vogel C., Marcotte E.M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nature reviews. Genetics. 2012;13(4):227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davies A.H., Beltran H., Zoubeidi A. Cellular plasticity and the neuroendocrine phenotype in prostate cancer. Nature reviews. Urology. 2018;15(5):271–286. doi: 10.1038/nrurol.2018.22. [DOI] [PubMed] [Google Scholar]
- 29.Kolb, A. D.; Bussard, K. M., The bone extracellular matrix as an ideal milieu for cancer cell metastases. Cancers (Basel)2019, 11, (7), 1020. [DOI] [PMC free article] [PubMed]
- 30.Cattrini C., Zanardi E., Boccardo F. Androgen-deprivation therapy is more than palliation in oligometastatic prostate cancer. J. Clin. Oncol. 2018;36(22):2350. doi: 10.1200/JCO.2018.78.0031. [DOI] [PubMed] [Google Scholar]
- 31.Wang Z., Ouyang G. Periostin: a bridge between cancer stem cells and their metastatic niche. Cell Stem Cell. 2012;10(2):111–112. doi: 10.1016/j.stem.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 32.Konac, E.; Kiliccioglu, I.; Sogutdelen, E.; Dikmen, A. U.; Albayrak, G.; Bilen, C. Y., Do the expressions of epithelial-mesenchymal transition proteins, periostin, integrin-alpha4 and fibronectin correlate with clinico-pathological features and prognosis of metastatic castration-resistant prostate cancer? Experimental biology and medicine (Maywood, N.J.)2017, 242, (18), 1795-1801. [DOI] [PMC free article] [PubMed]
- 33.Zhang, Y.; Yuan, D.; Yao, Y.; Sun, W.; Shi, Y.; Su, X., Predictive and prognostic value of serum periostin in advanced non–small cell lung cancer patients receiving chemotherapy. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine2017, 39, (5), 1010428317698367. [DOI] [PubMed]
- 34.Thuwajit C., Thuwajit P., Jamjantra P., Pairojkul C., Wongkham S., Bhudhisawasdi V., Ono J., Ohta S., Fujimoto K., Izuhara K. Clustering of patients with intrahepatic cholangiocarcinoma based on serum periostin may be predictive of prognosis. Oncol. Lett. 2017;14(1):623–634. doi: 10.3892/ol.2017.6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nuzzo P.V., Rubagotti A., Argellati F., Di Meglio A., Zanardi E., Zinoli L., Comite P., Mussap M., Boccardo F. Prognostic value of preoperative serum levels of periostin (PN) in early breast cancer (BCa) Int. J. Mol. Sci. 2015;16(8):17181–17192. doi: 10.3390/ijms160817181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cattrini, C.; Rubagotti, A.; Zinoli, L.; Cerbone, L.; Zanardi, E.; Capaia, M.; Barboro, P.; Boccardo, F., Role of circulating tumor cells (CTC), androgen receptor full length (AR-FL) and androgen receptor splice variant 7 (AR-V7) in a prospective cohort of castration-resistant metastatic prostate cancer patients. Cancers (Basel)2019, 11, (9). [DOI] [PMC free article] [PubMed]
- 37.Sepe, P.; Verzoni, E.; Miodini, P.; Claps, M.; Ratta, R.; Martinetti, A.; Mennitto, R.; Sottotetti, E.; Procopio, G.; Cappelletti, V.; Daidone, M. G., Could circulating tumor cells and ARV7 detection improve clinical decisions in metastatic castration-resistant prostate cancer? The Istituto Nazionale dei Tumori (INT) experience. Cancers (Basel)2019, 11, (7). [DOI] [PMC free article] [PubMed]
- 38.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P., Jensen L.J., Mering C.V. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–d613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaplan E.L., Meier P. Nonparametric estimation from incomplete observations. J. Am. Stat. Assoc. 1958;53:457–481. [Google Scholar]
- 40.Cox D.R. Regression models and life-tables. J. R. Stat. Soc. Ser. B. 1972;34:187–220. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials The following are available online: Supplementary Figure 1: Association of POSTN mRNA with DFS in the Provisional Firehose Legacy cohort of primary PCa. (A) Kaplan-Meier estimations in patients with POSTN >1 (gray) and ≤0 (light blue) z-scores; (B) X-tile histogram of POSTN values (expressed as z-scores ×10) according to number of patients; (C) triangular X-tile plot with the choice of POSTN cutoff (black point); (D) continuous relative risk of disease progression (expressed as HR) according to POSTN values (z-score cutoff of 1 is shown in red line). Supplementary Figure 2: Association of POSTN plasma values with OS in our cohort of patients with mCRPC. (A) Kaplan-Meier estimations in patients with POSTN >10.4 (purple), >3.2 ≤ 10.4 (gray) and ≤3.2 (light blue) z-scores; (B) X-tile histogram of POSTN values (expressed as z-scores ×10) according to number of patients; (C) triangular X-tile plot with the choice of POSTN cut-off (black point); (D) continuous relative risk of disease progression (expressed as HR) according to POSTN values (z-scores cutoffs of 10.4 and 3.2 are shown in red lines). Supplementary Table 1: Patient Characteristics. Supplementary File 1: Periostin dot blot assay characterization.