Abstract
The increased proliferation and migration of vascular smooth muscle cells (VSMCs) are critical in the progression of atherosclerosis (AS). Platelet-derived growth factor type BB (PDGF-BB) may induce VSMC proliferation and migration. miR-142-5p plays a critical role in various biological processes, including tumorigenesis, angiogenesis and inflammation. However, whether miR-142-5p is involved in regulating the pathological process of arteriosclerosis remains to be elucidated. Therefore, in this study, the role of miR-142-5p in PDGF-BB-induced human aortic smooth muscle cell (HSAMC) proliferation and migration was investigated. The results revealed that the expression level of miR-142-5p was enhanced in the serum of patients with AS, while that of its target gene, myocardin-like protein 2 (MKL2) was decreased, compared with that in healthy volunteers. Moreover, there was a negative correlation between miR-142-5p and MKL2 expression in the serum of patients with AS. Furthermore, the downregulation of miR-142-5p inhibited PDGF-BB-induced HASMC proliferation and migration; however, the inhibition of HASMC proliferation and migration was reversed by co-transfection with small interfering RNA (siRNA) against MKL2 (siRNA-MKL2). In addition, transfection with miR-142-5p inhibitor significantly increased the expression levels of MKL2, and decreased those of matrix metalloproteinase (MMP)2 and 9, and these effects were reversed by transfection with siRNA-MKL2. Finally, MKL2 was proven to be a target of miR-142-5p. On the whole, the findings of the present study demonstrate that the downregulation of miR-142-5p inhibits human aortic smooth muscle cell (HSAMC) proliferation and migration possibly by targeting MKL2. Hence, miR-142-5p may prove to be a novel therapeutic target in the treatment of AS.
Keywords: microRNA-142-5p, myocardin-like protein 2, atherosclerosis, proliferation and migration, human aortic smooth muscle
Introduction
Atherosclerosis (AS) is a chronic systemic disease in which the inside of an artery narrows due to the build-up of plaque, particularly in the brain, heart and lower extremities (1). Previous studies have reported that the initial step of AS is intimal injury, followed by platelet aggression or leukocyte invasion beneath the endothelial monolayer (2,3). When AS becomes severe, it may result in coronary artery disease, peripheral artery disease or kidney issues, depending on which arteries are affected (4).
The aberrantly increased proliferation and migration of vascular smooth muscle cells (VSMCs) are critical in the pathogenesis and progression of AS (5,6). In addition, previous studies have suggested that the proliferation of VSMCs can be induced or stimulated by cytokines and growth factors (7,8), such as platelet-derived growth factor (PDGF). Platelet-derived growth factor type BB (PDGF-BB), a subunit of PDGF, has been reported to regulate cell growth and division (9,10). In particular, PDGF-BB plays a critical role in initiating numerous biological effects by activating intracellular transduction pathways that are critical in modulating the proliferation and migration of VSMCs (8,11). Collectively, preventing PDGF-modulated VSMC proliferation and migration may contribute to the treatment of AS.
MicroRNAs (miRNAs or miRs) are a class of non-coding, short (22 nucleotides in length) endogenous RNAs that post-transcriptionally regulate gene expression by binding to the 3′-untranslated region (UTR) of their target genes (12,13). miRNAs carry out critical functions in cardiovascular systems (14–16), including AS (17,18). Accumulating evidence has demonstrated that miR-142-5p is upregulated in the plaques of apoE−/− mice with AS (19) and that the overexpression of miR-142-5p may induce VSMC proliferation (20). However, the effect of miR-142-5p on modulating HASMC proliferation and migration remains unclear. Therefore, this study aimed to explore the roles of miR-142-5p in the regulation of HASMC physiology and to elucidate the underlying mechanisms.
Materials and methods
Patient studies
Serum of patients with AS was obtained from 35 patients who had been diagnosed with AS by Doppler ultrasonography (age, 41.9±7.1 years; sex, 17 males and 18 females), and normal serum specimens were obtained from 35 healthy volunteers (age, 42.5±6.9 years; sex, 19 males and 16 females). All the specimens were acquired between January 2016 and June 2017 at the Central Hospital of Wuhan, Tongji Medical College, Huazhong University of Science and Technology. The present study was approved by the Ethics Committee of the Central Hospital of Wuhan, Tongji Medical College, Huazhong University of Science and Technology and prior informed consent was obtained from the patients with AS and the healthy volunteers.
Cells and cell culture
The human artery vascular smooth muscle cell line, HASMC (BeNa Culture Collection), was purchased and incubated in DMEM (Thermo Fisher Scientific, Inc.), supplemented with 10% FBS and 1% penicillin/streptomycin (Gibco; Thermo Fisher Scientific, Inc.) at 37°C in a humidified atmosphere with 5% CO2 for 24 h.
Cell transfection
Cells were seeded in 96-wells with a density of 1×104 cells/well containing 25 ng/ml of PDGF-BB (HEGFP-1601; Cyagen) and cultured for 24 h to reach 60% confluence. Subsequently, the expression levels of miR-142-5p before and after the addition of PDGF-BB were detected accordingly. The cells were then transfected with either 50 nM miR-142-5p inhibitor (5′-CACAAGGUAGAAAGCACUACU-3′) or NC (5′-GUGUAACACGUCUAUACGCCCA-3′; Biomics Biotechnologies Co. Ltd.) using Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) as per the manufacturer's instructions. In order to confirm the transfection efficiency of miR-142-5p, the cells were divided into 3 groups as follows: i) The control group, which were untransfected cells; ii) the NC group, in which cells were transfected with NC (miR-142-5p inhibitor negative control); iii) the inhibitor group, in which cells were transfected with miR-142-5p inhibitor. After transfection, the transfected cells were incubated for 24 h prior to further experimentation.
In addition, in order to confirm the transfection efficiency of myocardin like 2 (MKL2), the cells were divided into 3 groups as follows: i) The control group, which were untransfected cells; ii) the small interfering RNA (siRNA) group, in which cells were transfected with negative control siRNA; iii) the siRNA-MKL2 group, in which cells were transfected with siRNA against MKL2. Briefly, the cells were seeded in 96-wells at a density of 1×104 cells/well and grown for 24 h to reach 60% confluence. The cells were then transfected with either 20 µM siRNA or MKL2 siRNA using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.,) as per the manufacturer's instructions. After transfection, the transfected cells were incubated for 24 h prior to further experimentation. siRNA-MLK2 and negative control siRNA were designed and purchased from Shanghai GenePharma Co., Ltd. The sequences were as follows: Negative control siRNA forward, 5′-CGCCCTCATCAGTGCATACAA-3′ and reverse, 5′-CATAGCAAAGAAAGACTTAAA-3′; siRNA-MKL2 forward, 5′-CGCCATCATCGATGACTACAA-3′ and reverse, 5′-CTACGAGCAGATCAAGATAAA-3′.
Cell proliferation measured by CCK-8 assay
A CCK-8 assay was conducted to measure HASMC proliferation, according to the manufacturer's protocol. Briefly, the cells were seeded into 96-well plates at a density of 2×103 cells/well. The cells were then incubated at 37°C in serum-free DMEM containing 25 ng/ml of PDGF-BB (HEGFP-1601; Cyagen) for 24 h following transfection. Subsequently, 10 µl CCK-8 solution (Dojindo Molecular Technologies, Inc.) was added to each well followed by incubation for a further 4 h at room temperature and the absorbance was measured at 450 nm using Multiscan FC Microplate Photometer (Thermo Fisher Scientific, Inc.).
Cell migration measured by wound scratch assay
The HASMCs were seeded into a 12-well plate at a density of 1×103 cells/well. The cells were then incubated at 37°C in serum-free DMEM containing the 25 ng/ml of PDGF-BB (HEGFP-1601; Cyagen) for 24 h following transfection. A straight scratch wound was then created using a sterilized 10-µl pipette in each well. Following 24 h of incubation at 37°C in a humidified atmosphere with 5% CO2, the wound was visualized and photographed under an inverted microscope at ×200 magnification (BDS500 Trinocular; SCOPE).
Cell migration measured by Transwell migration assay
A total of 200 µl transfected cells were resuspended in serum-free DMEM medium and placed in the upper compartment of 8-µm pore size Transwell chambers without Matrigel (Costar Inc.), while the lower chamber was supplemented with serum-free DMEM containing 5 ng/ml of PDGF-BB. Following incubation for 24 h at 37°C, 5% CO2, non-migrated HASMCs in the upper compartment were removed using cotton swabs, while the migrated cells were stained with 0.1% crystal violet (Sigma-Aldrich; Merck KGaA) for 30 min at room temperature. The cells on the bottom side of the membrane were calculated under a microscope (SZX7-1063; Olympus Corporation) in order to confirm the number of migrated cells.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from the serum of patients with AS and the healthy volunteers or the cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Subsequently, the RNA was reversed transcribed into cDNA using the TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) or TaqMan Gene Expression Assay (Applied Biosystems; Thermo Fisher Scientific, Inc.), respectively as per the manufacturer's instructions. The cDNA was used to template with the SYBR-Green PCR Master Mix kit (Applied Biosystems; Life Technologies) on the CFX96 Real-Time RCR (Bio-Rad) according to the manufacturer's instructions. The PCR reactions were described as follows: An initial denaturation at 95°C for 5 min; 40 thermal cycles of denaturation at 95°C for 30 sec, annealing at 56°C for 30 sec and extension at 72°C for 30 sec; final extension at 60°C for 5 min. For relative quantification, the levels of individual gene mRNA transcripts were normalized to GAPDH, while the expression level of miR-142-5p was normalized to U6. The expression levels of relative genes were calculated using the 2−∆∆Cq method (21). All the experiments were conducted at least in triplicate. The following primers were used: miR-142-5p forward, 5′-AACTCCAGCTGGTCCTTAG-3′ and reverse, 5′-TCTTGAACCCTCATCCTGT-3′; U6 forward, 5′-CTCGCTTCGGCAGAC-3′ and reverse, 5′-AACGCTTACGAATTT-3′; MKL2 forward, 5′-AGATCAGAAGGGTGAGAAGAATG-3′ and reverse, 5′-GGATGGTCTGGTAGTTGTAGTG-3′; matrix metalloproteinase (MMP)2 forward, 5′-TGTGTTCTTTGCAGGGAATGAAT-3′ and reverse, 5′-TGTCTTCTTGTTTTTGCTCCAGTTA-3′; MMP9 forward, 5′-CCTCTGGAGGTTCGACGTGA-3′ and reverse, 5′-TAGGCTTTCTCTCGGTACTGGAA-3′; GAPDH forward, 5′-ACTCCACTCACGGCAAATTC-3′ and reverse, 5′-TCTCCATGGTGGTGAAGACA-3′.
Western blot analysis
Total proteins were lysed with RIPA lysis buffer (Beyotime Institute of Biotechnology) supplemented with a protease inhibitor cocktail (K1010; Apexbio) as per the manufacturer's instructions. The concentration of proteins was measured using a bicinchoninic acid assay kit (Thermo Fisher Scientific, Inc.). The proteins (20 µg/lane) were separated by 10% SDS-PAGE (cat. no. LC26755; Invitrogen; Thermo Fisher Scientific, Inc.) and transferred onto polyvinylidene fluoride membranes (cat. no. abs932; Absin). The membranes were blocked with Tris-buffered saline (TBS; cat. no. SIG-32380-500; Kanglang) containing 5% non-fat milk at 37°C for 2 h. After blocking, the proteins were probed with the following primary antibodies overnight at 4°C: Rabbit anti-MKL2 (cat. no. ab191496; 1:1,000), rabbit anti-MMP2 (cat. no. ab37150; 1:1,000), rabbit anti-MMP9 (cat. no. ab38898; 1:1,000) and rabbit anti-GAPDH (cat. no. ab9485; 1:2,500) (all from Abcam). Subsequently, the blots were washed with Tris-buffered saline/Tween-20 (TBST; T1081-500; Salarbio) three times and incubated with the secondary antibody IgG H&L (cat. no. ab6940; 1:1,000; Abcam) at 37°C for 1 h. After washing, the signals were detected using chemiluminescence HRP Substrate (Clontech), imaged using a GE ImageQuant Las 4000 mini phosphorimager (GE Healthcare Life Sciences) and presented as the density ratio vs. GAPDH. The quantification was performed using ImageJ software version 1.41 (National Institutes of Health). All the procedures above were conducted in triplicate.
Luciferase reporter assay
Using the online tool TargetScan, MKL2 was considered to be a putative target gene of miR-142-5p. In order to confirm the targeted association between miR-142-5p and MKL2, a dual-luciferase reporter assay was performed accordingly. In brief, the MKL2 mRNA 3′-UTR containing the putative or mutated binding sites for miR-142-5p were amplified by PCR and cloned into the pMIR-REPORT Luciferase (BR031; Fenghbio). The HASMC cell line (BeNa Culture Collection) was co-transfected with 20 µM miR-142-5p mimics or NC using Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.,). Luciferase activities were determined at 48 h using the Dual-luciferase Reporter Gene Assay kit (Promega Corp.). Luciferase activity was compared to Renilla luciferase activity as per the manufacturer's protocol. The miR-NC (5′-GUGUAACACGUCUAUACGCCCA-3′) and miR-142-5p mimic (5′-CAUAAAGUAGAAAGCACUACU-3′) were designed and synthesized by Biomics Biotechnologies Co. Ltd.
Statistical analysis
SPSS version 17.0 software (SPSS, Inc.) was used to analyze the data. The data are presented as the mean ± SD in the present study. Pearson's correlation analysis was performed to evaluate the correlation between miR-142-5p and MKL2. The Student's t-test is applied to distinguish differences between 2 groups, while one-way analysis of variance (ANOVA) followed by Newman-Keuls test was used to analyze the data among ≥3 groups. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-142-5p is upregulated in the serum of patients with AS
As demonstrated in Fig. 1A, the expression level of miR-142-5p was significantly upregulated in the serum of patients with AS, compared to that of the healthy volunteers (P<0.01).
Figure 1.
miR-142-5p expression is upregulated, while MKL2 expression is downregulated in the serum of patients with AS. (A) The expression level of miR-142-5p in the serum of 35 patients with AS and 35 healthy volunteers. (B) The mRNA expression level of MKL2 in the serum of 35 patients with AS and 35 healthy volunteers. (C) Results of correlation analysis between the expression of miR-142-5p and the expression of MKL2 in patients with AS. (D) The protein expression level of MKL2 in the patients with AS. (E) The quantified protein expression levels of MKL2 in patients with AS. **P<0.01, AS vs. control group. Control, the serum of 35 healthy volunteers; AS, the serum of 35 patients with AS; AS, atherosclerosis; MKL2, myocardin-like protein 2; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
MKL2 is downregulated in the serum of patients with AS
As demonstrated in Fig. 1B, the mRNA expression level of MKL2 was prominently downregulated in the serum of patients with AS, in contrast with the serum of healthy volunteers (P<0.01). Moreover, the MKL2 protein expression results shown in Fig. 1D and E presented a similar trend of variation (P<0.01) as the mRNA results.
MKL2 expression is negatively regulated by miR-142-5p
As demonstrated in Fig. 1C, the expression of miR-142-5p negatively correlated with the expression level of MKL2 in the serum of patients with AS (r=−0.505; P=0.002).
Transfection efficiency of miR-142-5p and MKL2
Following treatment with PDGF-BB, the expression level of miR-142-5p was determined. As demonstrated in Fig. 2A, the expression level of miR-142-5p significantly increased following treatment with PDGF-BB (P<0.01). Following transfection, the transfection efficiency of miR-142-5p and MKL2 was measured, respectively. As shown in Fig. 2B, the expression level of miR-142-5p markedly decreased in the inhibitor group, as compared with the NC group (P<0.01), while there was no variation between the NC and control group. As demonstrated in Fig. 2C, the mRNA expression level of MKL2 markedly decreased in the siRNA-MKL2 group, as compared with the siRNA group (P<0.01), whereas there was no variation between the siRNA and control group. Moreover, the MKL2 protein expression results shown in Fig. 2D and E presented a similar trend of variation as the mRNA results (P<0.01).
Figure 2.
Transfection efficiency of miR-142-5p and MKL2. (A) The expression level of miR-142-5p before and after treatment with PDGF-BB. **P<0.01, PDGF-BB vs. before group. (B) The relative expression level of miR-142-5p following transfection with miR-142-5p inhibitor. (C) The relative mRNA expression level of MKL2 following transfection with siRNA against MKL2 (siRNA-MKL2). (D) The relative protein expression of MKL2 following transfection with siRNA-MKL2. (E) The protein/GAPDH expression levels of MKL2 following transfection with siRNA-MKL2 are presented. **P<0.01, siRNA-MKL2 vs. siRNA group. Before, cells before treatment with PDGF-BB; PDGF-BB, cells treated with PDGF-BB; Control, untreated cells; NC, miR-142-5p inhibitor negative control-transfected cells; inhibitor, miR-142-5p inhibitor-transfected cells; siRNA, cells transfected with siRNA; siRNA-MKL2, cells transfected with siRNA-MKL2; PDGF-BB, platelet-derived growth factor type BB; MKL2, myocardin-like protein 2; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Downregulation of miR-142-5p inhibits PDGF-BB-induced HASMC proliferation
As demonstrated in Fig. 3, the downregulation of miR-142-5p significantly inhibited HASMC proliferation compared with the NC group (P<0.01). However, the decreased cell viability induced by transfection with miR-142-5p inhibitor was partly reversed by co-transfection with siRNA-MKL2 (P<0.05).
Figure 3.
Downregulation of miR-142-5p inhibits PDGF-BB-induced HASMC proliferation. The viability of HASMCs in different groups was measured. *P<0.05, **P<0.01, inhibitor vs. NC group and inhibitor + siRNA-MKL2 vs. inhibitor group. Control, untransfected cells; NC, miR-142-5p inhibitor negative control-transfected cells; inhibitor, miR-142-5p inhibitor-transfected cells; siRNA-MKL2, cells were transfected with siRNA-MKL2; PDGF-BB, platelet-derived growth factor type BB; HASMC, human aortic smooth muscle cell; MKL2, myocardin-like protein 2.
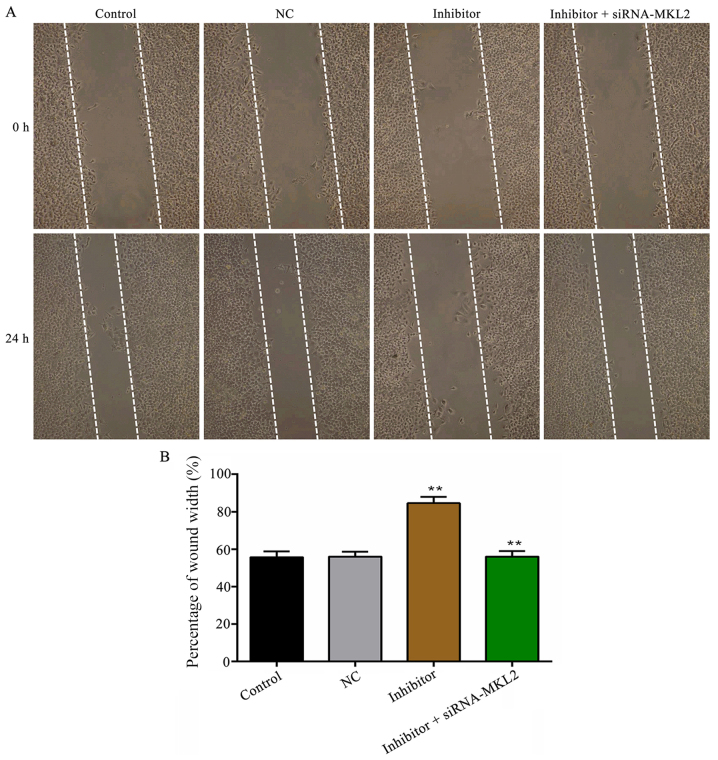
Downregulation of miR-142-5P inhibits the migration of HASMCs following treatment with PDGF-BB
The wound scratch results shown in Fig. 4 demonstrated that the downregulation of miR-142-5p prominently inhibited PDGF-BB-induced HASMC migration in contrast with the NC group (P<0.01). However, the suppressed cell migratory ability induced by transfection with miR-142-5p was partly reversed by co-transfection with siRNA-MKL2 (P<0.01). Similar results were obtained in the Transwell migration assay (P<0.01; Fig. 5).
Figure 4.
Downregulation of miR-142-5p inhibits PDGF-BB-induced HASMC migration measured by wound scratch assay. (A) Wound scratch assay (magnification, ×200). (B) The quantified result of (A) is presented. **P<0.01, inhibitor vs. NC group and inhibitor + siRNA-MKL2 vs. inhibitor group. Control, untransfected cells; NC, miR-142-5p inhibitor negative control-transfected cells; inhibitor, miR-142-5p inhibitor-transfected cells; siRNA-MKL2, cells were transfected with siRNA-MKL2; PDGF-BB, platelet-derived growth factor type BB; HASMC, human aortic smooth muscle cell; MKL2, myocardin-like protein 2.
Figure 5.
Downregulation of miR-142-5p inhibits PDGF-BB-induced HASMC migration measured by Transwell migration assay. (A) Transwell migration assay (scale bar, 100 μm). (B) The quantified result of (A) is presented. **P<0.01, inhibitor vs. NC group and inhibitor + siRNA-MKL2 vs. inhibitor group. Control, untransfected cells; NC, miR-142-5p inhibitor negative control-transfected cells; inhibitor, miR-142-5p inhibitor-transfected cells; siRNA-MKL2, cells were transfected with siRNA-MKL2; PDGF-BB, platelet-derived growth factor type BB; HASMC, human aortic smooth muscle cell; MKL2, myocardin-like protein 2.
Effect of miR-142-5p on the expression levels of MKL2, MMP2 and MMP9
As demonstrated in Fig. 6A-C, as the expression level of miR-142-5p was decreased, the mRNA expression level of MKL2 was increased, while the mRNA expression levels of MMP2 and MMP9 were significantly decreased (P<0.01), compared with the NC group. However, these effects were partly reversed by co-transfection with siRNA-MKL2 (MLK2 and MMP2, P<0.01; MMP9, P<0.05). Furthermore, the protein expression results shown in Fig. 6D and E presented a similar trend of variation as well (P<0.01).
Figure 6.
Effect of miR-142-5p on the mRNA and protein expression levels of MKL2, MMP2 and MMP9. (A) The relative mRNA expression level of MKL2. (B) The relative mRNA expression level of MMP2. (C) The relative expression level of MMP9. (D) Western blot analysis was employed to measure the protein expression levels of MKK2, MMP2 and MMP9. The quantified result of (E) is presented. *P<0.05, inhibitor + siRNA-MKL2 vs. inhibitor group; **P<0.01, inhibitor vs. NC group and inhibitor + siRNA-MKL2 vs. inhibitor group. Control, untransfected cells; NC, miR-142-5p inhibitor negative control-transfected cells; inhibitor, miR-142-5p inhibitor-transfected cells; siRNA-MKL2, cells were transfected with siRNA-MKL2; MKL2, myocardin-like protein 2; MMP2, matrix metalloproteinase-2; MMP9, matrix metalloproteinase-9; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
MKL2 is a target gene of miR-142-5p
The putative seed sequences for miR-142-5p at the 3′-UTR of MKL2 are presented in Fig. 7A. As demonstrated in Fig. 7B, transfection with miR-142-5p mimics significantly decreased the luciferase activity in the WT group, compared with the results in the NC group (P<0.01). However, there was no significant difference observed in the mutant groups.
Figure 7.
MKL2 is identified as a target of miR-142-5p. (A) Target sequences of miR-142-5p in 3′-UTR of the wild-type and mutant MKL2. (B) A dual-luciferase reporter assay was employed to determine whether MKL2 was a target of miR-142-5p. **P<0.01, inhibitor vs. NC group. UTR, untranslated region; WT, wild-type; MKL2, myocardin-like protein 2.
Discussion
miRNAs haven been reported to play critical roles in cardiovascular diseases, including AS (22,23). Moreover, the dysregulation of miRNAs may regulate the proliferation and migration of VSMCs (24,25), which may lead to the development of AS through different stimuli (26,27). miR-142-5p is a member of the miR-142 family, playing a critical role in regulating tumorigenesis (28,29) and immune diseases (30). A previous study demonstrated that miR-142-5p was prominently increased in the plaques of apoE−/− mice with AS (19). Consistent with the findings of this previous study, in the present study, miR-142-5p was significantly upregulated in the serum of patients with AS compared with that of healthy volunteers, as determined by RT-qPCR. To further investigate the detailed role of miR-142-5p in HASMCs in AS, CCK-8, Transwell migration and wound scratch assays were conducted to examine cell proliferation and migration, respectively. According to the results, the downregulation of miR-142-5p was demonstrated to inhibit the proliferation and migration of PDGF-BB-treated HASMCs, which was consistent with the findings of previous study by Kee et al (20), who demonstrated that miR-142-5p promoted VSMC proliferation by targeting B cell translocation gene 3 (BTG3).
As is known, miRNAs negatively regulate the expression of their target genes. After confirming that MKL2 is a likely target gene of miR-142-5p by a dual-luciferase reporter assay, correlation analysis further demonstrated that miR-142-5p negatively regulated MKL2 in AS. MKL2 acts as co-activator, controlling genes of relevance for myogenic differentiation and motile function (31), carrying out critical functions in regulating cell proliferation and migration (32,33). Furthermore, another study revealed that MKL2 functioned as a pro-migratory gene in VSMCs (34). In the present study, MKL2 was found to be underexpressed in the serum of patients with AS compared with that of healthy volunteers, which was consistent with the findings of a previous study (35). In addition, via gain-of-function and loss-of-function approaches, transfection with miR-142-5p inhibitor significantly increased the mRNA and protein expression level of MKL2 in HASMCs; however, transfection with MKL2-siRNA resulted in a decreased MKL2 expression. Moreover, the inhibition of HASMC proliferation and migration induced by transfection with miR-142-5p inhibitor was partly reversed by co-transfection with MKL2-siRNA. Collectively, the results of this study suggested that the downregulation of miR-142-5p participated in the inhibition of HASMC proliferation and migration, which is, at least in part, dependent on targeting MKL2.
MMP2 and MMP9 are crucial enzymes of the MMP family members involved in extracellular matrix remodeling and cell migration (36). Moreover, MMP2 and MMP9 expression levels in VSMCs have been reported to be associated with AS (37–39) by regulating the proliferation and migration of VSMCs (40,41), indicating a pathogenic role for MMP2 and MMP9 in regulating the progression of AS. Furthermore, previous studies have demonstrated that the downregulation of MMP2 and MMP9 inhibit cell migration (42–44). Consistent with previous studies, in the present study, the suppression of MMP2 and MMP9 by transfection with miR-142-5p inhibitor contributed to the decreased migration of HASMCs.
In conclusion, this study found that miR-142-5p expression was markedly upregulated in the serum of patients with AS. Moreover, the downregulation of miR-142-5p may inhibit HASMC proliferation and migration partly by targeting MKL2.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
WW performed the statistical analysis and wrote the manuscript. YS collected the patient data and performed the statistical analysis. SD was involved in the study design, data acquisition and manuscript revision. CY and JW were involved in the study design, discussion and completion of the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of the Central Hospital of Wuhan, Tongji Medical College, Huazhong University of Science and Technology and prior informed consent was obtained from the patients with AS and the healthy volunteers.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Gan WQ, Man SF, Senthilselvan A, Sin DD. Association between chronic obstructive pulmonary disease and systemic inflammation: A systematic review and a meta-analysis. Thorax. 2004;59:574–580. doi: 10.1136/thx.2003.019588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ross R, Agius L. The process of atherogenesis-cellular and molecular interaction: From experimental animal models to humans. Diabetologia. 1992;35(Suppl 2):S34–S40. doi: 10.1007/BF00586277. [DOI] [PubMed] [Google Scholar]
- 3.Ross R. The pathogenesis of atherosclerosis: A perspective for the 1990s. Nature. 1993;362:801–809. doi: 10.1038/362801a0. [DOI] [PubMed] [Google Scholar]
- 4.Seely S. Atherosclerosis or hardening of the arteries? Int J Cardiol. 1989;22:5–12. doi: 10.1016/0167-5273(89)90129-0. [DOI] [PubMed] [Google Scholar]
- 5.Owens GK. Regulation of differentiation of vascular smooth muscle cells. Physiol Rev. 1995;75:487–517. doi: 10.1152/physrev.1995.75.3.487. [DOI] [PubMed] [Google Scholar]
- 6.Ross R. Atherosclerosis-an inflammatory disease. New Engl J Med. 1996;340:115–126. doi: 10.1056/NEJM199901143400207. [DOI] [PubMed] [Google Scholar]
- 7.Kawai-Kowase K, Owens GK. Multiple repressor pathways contribute to phenotypic switching of vascular smooth muscle cells. Am J Physiol Cell Physiol. 2007;292:C59–C69. doi: 10.1152/ajpcell.00394.2006. [DOI] [PubMed] [Google Scholar]
- 8.Heldin CH, Westermark B. Mechanism of action and in vivo role of platelet-derived growth factor. Physiol Rev. 1999;79:1283–1316. doi: 10.1152/physrev.1999.79.4.1283. [DOI] [PubMed] [Google Scholar]
- 9.Ha JM, Yun SJ, Kim KW, Jin SY, Lee HS, Song SH, Shin HK, Bae SS. Platelet-derived growth factor regulates vascular smooth muscle phenotype via mammalian target of rapamycin complex 1. Biochem Biophys Res Commun. 2015;464:57–62. doi: 10.1016/j.bbrc.2015.05.097. [DOI] [PubMed] [Google Scholar]
- 10.Shawky NM, Segar L. Sulforaphane inhibits platelet-derived growth factor-induced vascular smooth muscle cell proliferation by targeting Mtor/P70S6kinase signaling independent of Nrf2 activation. Pharmacol Res. 2017;119:251–264. doi: 10.1016/j.phrs.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dzau VJ, Braun-Dullaeus RC, Sedding DG. Vascular proliferation and atherosclerosis: New perspectives and therapeutic strategies. Nat Med. 2002;8:1249–1256. doi: 10.1038/nm1102-1249. [DOI] [PubMed] [Google Scholar]
- 12.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 13.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 14.Ding S, Huang H, Xu Y, Zhu H, Zhong C. miR-222 in cardiovascular diseases: Physiology and pathology. Biomed Res Int. 2017;2017:4962426. doi: 10.1155/2017/4962426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu H, Liu Z, Zhou L. Roles of miR-17-92 cluster cardiovascular development and common diseases. Biomed Res Int. 2017;2017:9102909. doi: 10.1155/2017/9102909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seeger T, Boon RA. MicroRNAs in cardiovascular aging. J Physiol. 2016;594:2085–2094. doi: 10.1113/JP270557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Price NL, Rotllan N, Canfrán-Duque A, Zhang X, Pati P, Arias N, Moen J, Mayr M, Ford DA, Baldán Á, et al. Genetic dissection of the impact of miR-33a and miR-33b during the progression of atherosclerosis. Cell Rep. 2017;21:1317–1330. doi: 10.1016/j.celrep.2017.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ouimet M, Ediriweera H, Afonso MS, Ramkhelawon B, Singaravelu R, Liao X, Bandler RC, Rahman K, Fisher EA, Rayner KJ, et al. microRNA-33 regulates macrophage autophagy in atherosclerosis. Arterioscler Thromb Vasc Biol. 2017;37:1058–1067. doi: 10.1161/ATVBAHA.116.308916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu RJ, Bi CL, Song JT, Wang L, Ge C, Liu XX, Zhang M. Upregulation of miR-142-5p in atherosclerotic plagues and regulation of oxidized low-density lipoprotein-induced apoptosis in macrophages. Mol Med Rep. 2015;11:3229–3234. doi: 10.3892/mmr.2015.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kee HJ, Park S, Kwon JS, Choe N, Ahn Y, Kook H, Jeong MH. B cell translocation gene, a direct target of miR-142-5p, inhibits vascular smooth muscle cell proliferation by down-regulating cell cycle progression. FEBS Lett. 2013;587:2385–2392. doi: 10.1016/j.febslet.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Han Z, Fan Y, Zhang J, Chen K, Gao L, Zeng H, Cao J, Wang C. MicroRNA-9 inhibits NLRP3 inflammasome activation in human atherosclerosis inflammation cell models through the JAK2/STAT signaling pathway. Cell Physiol Biochem. 2017;41:1555–1571. doi: 10.1159/000470822. [DOI] [PubMed] [Google Scholar]
- 23.Liang X, Xu Z, Yuan M, Zhang Y, Zhao B, Wang J, Zhang A, Li G. MicroRNA-16 suppresses the activation of inflammation microphages in atherosclerosis by targeting PDCD4. Int J Mol Med. 2016;37:967–975. doi: 10.3892/ijmm.2016.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen J, Cui L, Yuan J, Zhang Y, Sang H. Circular RNA WDR77 target FGF-2 to regulate vascular smooth muscle cells proliferation and migration by sponging miR-124. Biochem Biophys Res Commun. 2017;494:126–132. doi: 10.1016/j.bbrc.2017.10.068. [DOI] [PubMed] [Google Scholar]
- 25.Xie B, Zhang C, Kang K, Jiang S. miR-559 inhibits vascular smooth muscle cells proliferation and migration by targeting TGFB2. PLoS One. 2015;10:e0141512. doi: 10.1371/journal.pone.0141512. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Bi R, Ding F, He Y, Jiang L, Jiang Z, Mei J, Liu H. miR-503 inhibits platelet-derived growth factor-induced human aortic vascular smooth muscle cell proliferation and migration through targeting the insulin receptor. Biomed Pharmacother. 2016;84:1711–1716. doi: 10.1016/j.biopha.2016.10.081. [DOI] [PubMed] [Google Scholar]
- 27.Sudo R, Sato F, Azechi T, Wachi H. miR-29-mediated elastin down-regulation contributes to inorganic phosphorus-induced osteoblastic differentiation in vascular smooth muscle cells. Genes Cells. 2015;20:1077–1087. doi: 10.1111/gtc.12311. [DOI] [PubMed] [Google Scholar]
- 28.Islam F, Gopalan V, Vider J, Lu CT, Lam AK. MiR-142-5p act as an oncogenenic microRNA in colorectal cancer: Clinicopathological and functional insights. Exp Mol Pathol. 2018;104:98–107. doi: 10.1016/j.yexmp.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 29.Wang Z, Liu Z, Fang X, Yang H. MiR-142-5p suppresses tumorigenesis by targeting PIK3CA in non-small cell lung cancer. Cell Physiol Biochem. 2017;43:2505–2515. doi: 10.1159/000484459. [DOI] [PubMed] [Google Scholar]
- 30.Kwanhian W, Lenze D, Alles J, Motsch N, Barth S, Döll C, Imig J, Hummel M, Tinguely M, Trivedi P, et al. microRNA-142 is mutated in about 20% of diffuse large B-cell lymphoma. Cancer Med. 2012;1:141–155. doi: 10.1002/cam4.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swärd K, Stenkula KG, Rippe C, Alajbegovic A, Gomez MF, Albinsson S. Emerging roles of the myocardin family of proteins in lipid and glucose metabolism. J Physiol. 2016;594:4741–4752. doi: 10.1113/JP271913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hampl V, Martin C, Aigner A, Hoebel S, Singer S, Frank N, Sarikas A, Ebert O, Prywes R, Gudermann T, Muehlich S. Depletion of the transcriptional coactivators megakaryoblastic leukemia 1 and 2 abolishes hepatocellular carcinoma xenograft growth by inducing oncogene-induced senescence. EMBO Mol Med. 2013;5:1367–1387. doi: 10.1002/emmm.201202406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pipes GC, Creemers EE, Olson EN. The myocardin family of transcriptional coactivators: Versatile regulators of cell growth, migration and myogenesis. Genes Dev. 2006;20:1545–1556. doi: 10.1101/gad.1428006. [DOI] [PubMed] [Google Scholar]
- 34.Smith MC, Hudson CA, Kimura TE, White SJ, Sala-Newby GB, Newby AC, Bond M. Divergent regulation of actin dynamics and megakaryoblastic leukemia-1 and −2 (Mkl1/2) by cAMP in endothelial and smooth muscle cells. Sci Rep. 2017;7:3681. doi: 10.1038/s41598-017-03337-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goikuria H, Freijo MDM, Vega Manrigue R, Sastre M, Elizagaray E, Lorenzo A, Vandenbroeck K, Alloza I. Characterization of carotid smooth muscle cells during phenotypic transition. Cells. 2018;7:E23. doi: 10.3390/cells7030023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seo KW, Lee SJ, Ye BH, Kim YW, Bae SS, Kim CD. Mechanical stretch enhances the expression and activity of osteopointin and MMP2 via the Akt1/AP-1 pathways in VSMC. J Mol Cell Cardiol. 2015;85:13–24. doi: 10.1016/j.yjmcc.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 37.Volcik KA, Campbell S, Chambless LE, Coresh J, Folsom AR, Mosley TH, Ni HY, Wagenknecht LE, Wasserman BA, Boerwinkle E. MMP2 genetic variation is associated with measures of fibrous cap thickness: The atherosclerosis risk in communities carotid MRI study. Atherosclerosis. 2010;210:188–193. doi: 10.1016/j.atherosclerosis.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim J, Ko J. Human sLZIP promotes atherosclerosis via MMP-9 transcription and vascular smooth muscle cell migration. FASEB J. 2014;28:5010–5021. doi: 10.1096/fj.14-259218. [DOI] [PubMed] [Google Scholar]
- 39.Eilenberg W, Stojkovic S, Kaider A, Kozakowski N, Domenig CM, Burghuber C, Nanobachvili J, Huber K, Klinger M, Neumayer C, et al. NGAL and MMP-9/NGAL as biomarkers of plaque vulnerability and targets of statins in patiensts with carotid atherosclerosis. Clin Chem Lab Med. 2017;56:147–156. doi: 10.1515/cclm-2017-0156. [DOI] [PubMed] [Google Scholar]
- 40.Ma L, Zhang L, Wang B, Wei J, Liu J, Zhang L. Berberine inhibts Chlamydia pneumoniae infection-induced vascular smooth muscle cell migration through downregulating MMP3 and MMP9 via PI3K. Eur J Pharmacol. 2015;755:102–109. doi: 10.1016/j.ejphar.2015.02.039. [DOI] [PubMed] [Google Scholar]
- 41.Gan J, Li P, Wang Z, Chen J, Liang X, Liu M, Xie W, Yin R, Huang F. Posuvastatin suppresses platelet-derived growth factor-BB-induced vascular smooth muscle cell proliferation and migration via the MAPK signaling pathway. Exp Ther Med. 2013;6:899–903. doi: 10.3892/etm.2013.1265. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Byun HJ, Darvin P, Kang DY, Sp N, Joung YH, Park JH, Kim SJ, Yang YM. Silibinin downregulates MMP2 expression via Jak2/STAT3 pathway and inhibits the migration and invasive potential in MDA-MB-231 cells. Oncol Rep. 2017;37:3270–3278. doi: 10.3892/or.2017.5588. [DOI] [PubMed] [Google Scholar]
- 43.Zhen Y, Liu J, Huang Y, Wang Y, Li W, Wu J. miR-133b inhibits cell growth, migration, and invasion by targeting MMP9 in non-small cell lung cancer. Oncol Rep. 2017;25:1109–1116. doi: 10.3727/096504016X14800889609439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang C, Wang L, Chen J, Liang J, Xu Y, Li Z, Chen F, Du D. Knockdown of Diaph1 expression inhibits migration and decreases the expression of MMP2 and MMP9 in human glioma cells. Biomed Pharmacother. 2017;96:596–602. doi: 10.1016/j.biopha.2017.10.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.