Abstract
MicroRNAs (miRs) play an important role in the development and progression of spinal cord injury (SCI). The role of miR-138-5p in SCI was investigated in the present study. The anti-inflammatory effects of miR-138-5p and underlying mechanisms were investigated in an SCI rat model and in vitro model. Reverse transcription-quantitative PCR (RT-qPCR) was used to examine the expression of miR-138-5p in the SCI in vivo and in vitro models, as well as patients with SCI; it was found that miR-138-5p was significantly upregulated in SCI. Bioinformatics and dual-luciferase reporter assays were performed to predict and confirm the binding sites between miR-138-5p and the 3′untranslated region of sirtuin 1 (SIRT1). Then, the expression of SIRT1 was detected via RT-qPCR and western blotting, indicating downregulation of SIRT1 in SCI. PC12 cells were transfected with miR-138-5p inhibitor, inhibitor control or miR-138-5p inhibitor + SIRT1 small interfering RNA for 48 h, and then subjected to lipopolysaccharide (100 ng/ml) treatment for 4 h. Then, MTT assay, flow cytometry and ELISA experiments were performed to analyze cell viability, apoptosis, and the levels of tumor necrosis factor-α, interleukin (IL)-1β and IL-6. Findings suggested that downregulation of miR-138-5p increased PC12 cell viability, inhibited cell apoptosis and attenuated proinflammatory responses, which may result in amelioration of SCI. However, all these effects were reversed by SIRT1 knockdown. Finally, it was observed that miR-138-5p altered the related protein expression of the PTEN/AKT pathway. These results indicated that miR-138-5p could regulate inflammatory responses and cell apoptosis in SCI models by modulating the PTEN/AKT signaling pathway via SIRT1, thus playing an important role in the development of SCI. Collectively, the present study demonstrated that miR-138-5p may be a novel therapeutic target for the treatment of SCI.
Keywords: spinal cord injury, microRNA-138-5p, sirtuin 1, apoptosis, inflammation
Introduction
Spinal cord injury (SCI) results from neurological damage in the spinal cord and leads to serious impairment of sensorimotor functions, along with other side effects (1), such as paraplegia and tetraplegia (2,3). Patients with SCI suffer from pain (4). There has been substantial research exploring pathophysiological changes that occur post-SCI (5). However, it is not clear how to most effectively promote spinal cord repair following damage. The systemic inflammatory response has received substantial attention as a major factor contributing to the development of SCI-induced immunological dysfunction (6,7). Moreover, the role of inflammatory response in SCI remains unclear. Therefore, it is important to further clarify the molecular mechanisms of SCI in order to develop novel therapeutic strategies.
MicroRNAs (miRNAs/miRs) are a class of non-coding RNA that regulate gene expression through translational cleavage or repression (8) at the post-transcriptional level (9). miRNAs directly bind to the 3′untranslated region (3′UTR) of mRNAs, resulting in translational repression or mRNA degradation (10). Mounting evidence has revealed that miRNAs are involved in various biological processes (11–13), including cell growth, cell apoptosis and cell differentiation (11,14–19). Altered expression of various miRNAs following traumatic SCI has been observed in adult rats (20). miR-138-5p, a miRNA that has been studied in a variety of tumors (21–23), has also been found to play an important role in Parkinson's disease (24). However, so far, the role of miR-138-5p in SCI has not been reported.
Sirtuin 1 (SIRT1) is a NAD+-dependent deacetylase important in regulating cell apoptosis (25). Studies have also indicated that SIRT1 plays critical roles in the regulation of inflammatory responses (26–29). Notably, potential binding sites between SIRT1 and miR-138-5p were detected in the present study via bioinformatics software analysis. Thus, it was hypothesized that they may interact and in doing so affect SCI progression. As the expression or regulatory mechanisms of miR-138-5p in modulating SCI remain largely unexplored, the purpose of the present study was to determine the role of miR-138-5p in SCI by evaluating its expression and molecular mechanisms.
Materials and methods
Clinical samples
A total of 18 serum specimens from patients with SCI (age range, 27–58 years; 12 male patients and 6 female patients) and 18 serum specimens from healthy controls (age range, 25–60 years; 12 male patients and 6 female patients) were collected at the First People's Hospital of Lianyungang between June 2015 and November 2017. Serum samples were isolated from blood samples by centrifugation at 4°C at 1,000 × g for 15 min. Patients with SCI were diagnosed according to American Spinal Injury Association (30). Written informed consent was obtained from each patient. The present study was approved by the Ethics Committee of the First People's Hospital of Lianyungang.
Animals
A total of 20 adult male Sprague-Dawley rats (6–8 weeks, 200–300 g) were obtained from the Animal Center of Nanjing Medical University, housed in a standard animal room (23±1°C, relative humidity 40–60%, under a 12:12-h light/dark cycle) with free access to standard rodent chow and water. No animals exhibited any neurological disorder prior to the SCI induction protocol. All protocols of animal experiments were performed according to the guidelines for Institutional Animal Care and Use of Laboratory Animals by the National Institutes of Health (31). The present study was approved by the Animal Ethics Committee of the First People's Hospital of Lianyungang.
Establishment of the rat model of SCI
Animals in were randomly divided into two groups: The control group (n=10) and the SCI group (n=10). In the control group, the surgical area was exposed without SCI induction. In the SCI model group, rats were shaved and treated aseptically to construct the SCI model as described previously (32). Briefly, all rats were anesthetized via intraperitoneal injection of pentobarbital (30 mg/kg). After anesthesia, the rat skin was shaved, opened and cleaned with betadine carefully. Then, a 20-mm midline incision was made to expose the vertebral column in the thoracic region. After a thoracic-level (T8-T11) midline skin incision, the paravertebral muscle was dissected and a laminectomy of T10 was undertaken in order to expose the dorsal cord surface without disrupting the dura. Subsequently, SCI was induced by dropping a 10 g rod from a height of 5.0 cm onto the T10 level of the spinal cord. Finally, the incision was sutured and all rats were allowed to recover from anesthesia in warm boxes. At 12 h after SCI induction, animals were anesthetized with pentobarbital (40 mg/kg) through intraperitoneal injection and sacrificed via cervical dislocation, following which specimens were obtained.
Cell lines and cell culture
PC12 cells were obtained from American Type Culture Collection, cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) and 1% penicillin/streptomycin, and maintained at 37°C in a humidified atmosphere with 5% CO2.
PC12 cells (1×106 cell/ml) were transfected with 100 nM miR-138-5p inhibitor (5′-CGGCCUGATTCACAACACCAGCT-3′; Shanghai GenePharma Co., Ltd.), 100 nM inhibitor control (5′-CAGUACUUUUGUGUAGUACAA-3′; Shanghai GenePharma Co., Ltd.), 0.2 µM control-small interfering (si)RNA (cat. no. Sc-36869; Santa Cruz Biotechnology, Inc.), 0.2 µM SIRT1-siRNA (cat. no. Sc-40986; Santa Cruz Biotechnology, Inc.) or 100 nM miR-138-5p inhibitor + 0.2 µM SIRT1-siRNA using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. The efficiency of cell transfection was evaluated via reverse transcription-quantitative PCR (RT-qPCR) and western blot analysis at 48 h after transfection.
An in vitro cell model of SCI in PC12 cells was established according to a previous study (33). In brief, PC12 cells were subjected to lipopolysaccharide (LPS; 100 ng/ml) for 4 h at 37°C. Control cells were left untreated.
miRNA target analysis and dual-luciferase reporter assay
TargetScan (version 7.1; www.targetscan.org/vert_71) was used to predict the potential targets of miR-138-5p. The results showed that SIRT1 was a potential target of miR-138-5p. In order to investigate the direct target binding sites between miR-138-5p and SIRT1, the wild-type 3′UTR (WT-SIRT1) and mutant 3′UTR (MUT-SIRT1) of SIRT1 were cloned into a pMIR-RB-Report™ dual luciferase reporter gene plasmid vector (Guangzhou RiboBio Co., Ltd.) according to the manufacturer's protocols; a QuikChange Site-Directed Mutagenesis kit (Stratagene; Agilent Technologies, Inc.) was used according to the manufacturer's instructions to point-mutate the miR-138-5p binding domain in the 3′UTR of SIRT1. PC12 cells (5×104 cells/well) were co-transfected with 1 ng reporter vector containing WT-SIRT1 or MUT-SIRT1, as well as 50 nM miR-138-5p mimic (5′-AGCUGGUGUUGUGAAUCAGGCCG-3′; Shanghai GenePharma Co., Ltd.) or 50 nM mimic control (5′-CAGUACUUUUGUGUAGUACAA-3′; Shanghai GenePharma Co., Ltd.) using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. The luciferase activity was analyzed at 48 h after co-transfection using a dual-luciferase reporter assay system (Promega Corporation), according to the manufacturer's protocol. Firefly luciferase activities were normalized to Renilla luciferase activities. The experiment was performed at least three times.
RNA extraction and RT-qPCR
Total RNA was isolated from rat spinal cord tissues or PC12 cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocols. Spinal cord specimens isolated from control and SCI model rats were divided into three equal segments to detect gene expression. NanoDrop™ ND-1000 spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific, Inc.) was used to measure the RNA concentrations at 260 and 280 nm. A cDNA Synthesis Kit (Invitrogen; Thermo Fisher Scientific, Inc.) was used to perform RT, according to the manufacturer's protocol. qPCR was performed using a Prism 7000 Real-Time PCR system with Power SYBR Green Master mix (Applied Biosystems; Thermo Fisher Scientific, Inc.). The amplification conditions were as follows: 35 cycles of denaturation at 94°C for 60 sec, annealing at 60°C for 60 sec and chain extension at 72°C for 1 min, followed by a final extension step at 72°C for 10 min. The expression levels of miR-138-5p and SIRT1 were normalized to the expression levels of the control genes U6 and GAPDH, respectively. Primer sequences (Sangon Biotech Co., Ltd.) were as follows: miR-138-5p forward, 5′-AGCTGGTGTTGTGAATCAGGCCG-3′ and reverse, 5′-TGGTGTCGTGGAGTCG-3′; SIRT1 forward, 5′-AATCCAGTCATTAAAGGTCTACAA-3′ and reverse, 5′-TAGGACCATTACTGCCAGAGG-3′; U6 forward, 5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse, 5′-CGCTTCACGAATTTGCGTGTCAT-3′; GAPDH forward, 5′-CTTTGGTATCGTGGAAGGACTC-3′ and reverse, 5′-GTAGAGGCAGGGATGATGTTCT-3′. The relative expression levels of genes were calculated using the 2−∆ΔCq method (34).
Western blot analysis
Tissues and cells were treated with RIPA buffer (Beyotime Institute of Biotechnology) and centrifuged at 1,000 × g for 30 min at 4°C to extract the total protein. Protein concentration was determined with a bicinchoninic acid protein assay kit (Pierce; Thermo Fisher Scientific, Inc.). Then, equal quantities of protein were separated by 10% SDS-PAGE, followed by transferring onto PVDF membranes. The membranes were blocked with 5% skimmed milk for 1 h at room temperature and incubated overnight at 4°C with anti-SIRT1 (1:1,000; cat. no. 9475), anti-PTEN (1:1,000; cat. no. 9188), anti-phosphorylated (p)-AKT (1:1,000; cat. no. 4060), AKT (1:1,000; cat. no. 9272) and anti-β-actin (1:1,000; cat. no. 4970; all Cell Signaling Technology, Inc.) antibodies. After five washes in PBS-0.1% Tween 20, the membranes were incubated with horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (1:2,000; cat. no. 7074) for 1 h at 37°C. Finally, the protein bands were detected using chemiluminescent ECL reagent (EMD Millipore). Protein expression levels were quantified using Image Lab Software (v.6.0; Bio-Rad Laboratories, Inc.).
MTT assay
In order to determine the viability of PC12 cells, cells were seeded (10,000 cells/well) into 96-well plates (BD Biosciences) and cultured for 24 h at 37°C. Then, the culture medium was removed, and miR-138-5p inhibitor, inhibitor control or miR-138-5p inhibitor + SIRT1-siRNA were subsequently transfected into the cells for 48 h at 37°C as previously described. PC12 cells were then subjected to LPS (100 ng/ml) treatment for 4 h. Subsequently, MTT solution (10 µl) was added to the medium and cultivated at 37°C for 4 h according to the manufacturer's instructions. DMSO (100 µl; Nanjing KeyGen Biotech Co., Ltd.) to dissolve the formazan crystals. The 96-well plates were placed in a multifunctional plate reader (VICTOR3™; PerkinElmer, Inc.) to measure the absorbance at 490 nm.
ELISA
After transfection, PC12 cells were treated with LPS (100 ng/ml) for 4 h. Then the cells were collected and centrifuged for measurement of tumor necrosis factor-α (TNF-α; cat. no. 430207), interleukin (IL)-1β (cat. no. 437007) and IL-6 (cat. no. 430507) secretion using ELISA kits according to the manufacturer's instructions (BioLegend, Inc.). The absorbance was measured at 450 nm using a microplate reader (Model 550; Bio-Rad Laboratories, Inc.) and the levels were calculated using standard curves.
Flow cytometric assay
To determine cell apoptosis, PC12 cells were transfected with miR-138-5p inhibitor, inhibitor control or miR-138-5p inhibitor + SIRT1-siRNA for 48 h as aforementioned, and then treated with LPS (100 ng/ml) for 4 h at 37°C. After treatments, the PC12 cells (1×106 cells/well) were trypsinized, washed with PBS, and stained with Annexin V-FITC and propidium iodide (PI) for 30 min at 37°C. An Annexin V-FITC/PI cell apoptosis detection kit (Beyotime Institute of Biotechnology) was used to detect the early and late apoptosis of PC12 cells via flow cytometry using a FACSCalibur flow cytometer (BD Biosciences) and FlowJo software (version 7.6.1; FlowJo LLC).
Statistical analysis
Experiments were repeated at least three times. Data were expressed as the mean ± SD and all statistical analyses were conducted using SPSS 18.0 software (SPSS, Inc.). Differences between groups were determined by two-tailed Student's t-test or one-way ANOVA followed by Tukey's post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of miR-138-5p is increased in SCI tissues and in vitro SCI cell models
miR-138-5p belongs to the miR-138 family that has previously been shown to play an important role in various cancers (21–23). The levels of miR-138-5p in the blood of patients with SCI and the spinal tissues of SCI rats were assessed by RT-qPCR. It was found that the levels of miR-138-5p were upregulated in the blood of patients with SCI compared with healthy controls, and in the spinal cord tissues of the SCI rats compared with the control group (Fig. 1A and B). Next, RT-qPCR was used to measure the levels of miR-138-5p in an in vitro SCI cell model compared with normal control cells (Fig. 1C). It was demonstrated that in LPS-induced PC12 cells, the expression of miR-138-5p was significantly upregulated compared with the control group. These results indicated that miR-138-5p may be involved in the progression of SCI.
Figure 1.
Expression of miR-138-5p in patients with SCI, and rodent and cell SCI models. (A) Expression of miR-138-5p in the blood of patients with SCI and healthy controls. (B) Expression of miR-138-5p in the spinal cord tissues of control and SCI model rats were quantified by RT-qPCR. (C) miR-138-5p expression levels in control and LPS-induced PC12 cells were assessed by RT-qPCR. Data are presented as mean ± SD and the experiments were performed in triplicate. **P<0.01 vs. Control. miR-138-5p, microRNA-138-5p; SCI, spinal cord injury; LPS, lipopolysaccharide; RT-qPCR, reverse transcription-quantitative PCR.
SIRT1 is a direct target of miR-138-5p
To analyze the molecular mechanisms underlying the role of miR-138-5p in PC12 cells, potential targets were predicted using TargetScan. A binding region of miR-138-5p in the 3′UTR of SIRT1 was predicted by, suggesting that SIRT1 was a potential target of miR-138-5p (Fig. 2A). A luciferase reporter assay was performed to validate this prediction. It was demonstrated that miR-138-5p mimic significantly enhanced miR-138-5p levels in PC12 cells (Fig. 2B). Subsequently, PC12 cells were co-transfected with luciferase vector plasmids containing the 3′UTR of SIRT1, along with miR-138-5p mimic or mimic control for 48 h. The results showed that the luciferase activity of luciferase vectors harboring WT-SIRT1 were significantly reduced in the miR-138-5p mimic co-transfection group, whereas no significant inhibition was found in the MUT-SIRT1 + miR-138-5p mimic co-transfection group (Fig. 2C) These results indicated that SIRT1 was a direct target of miR-138-5p.
Figure 2.
(A) miR-138-5p directly targets SIRT1. Predicted binding site of miR-138-5p in the 3′UTR of WT-SIRT1. The sequence of the mutant 3′UTR of SIRT1 is also shown. (B) PC12 cells were transfected with miR-138-5p mimic or mimic control for 48 h, then the level of miR-138-5p in PC12 cells was detected via reverse transcription-quantitative PCR. (C) PC12 cells were co-transfected with WT-SIRT1 or MUT-SIRT1 3′UTR reporter plasmids, along with miR-138-5p mimic or mimic control. After 48 h, luciferase activity were examined by dual-luciferase reporter assays. Data are presented as the mean ± SD. **P<0.01 vs. mimic control. miR-138-5p, microRNA-138-5p; SIRT1, sirtuin 1; 3′UTR, 3′untranslated region; WT, wild-type; MUT, mutant.
Expression of SIRT1 in SCI rat and in vitro SCI cell models is reduced
Based on these observations, it was hypothesized that SIRT1 may be involved in the effects of miR-138-5p in SCI. To test this hypothesis, western blot and RT-qPCR analyses were performed to detect the protein and mRNA expression levels of SIRT1 in the spinal cord tissues of SCI rats and LPS-treated PC12 cells, and it was revealed that SIRT1 expression was downregulated in SCI model spinal tissues compared with the control group (Fig. 3A and B). Additionally, compared with the untreated PC12 cells, SIRT1 expression was significantly downregulated in LPS-treated PC12 cells (Fig. 3C and D).
Figure 3.
Expression of SIRT1 in SCI tissues and cells. (A) Protein and (B) mRNA expression levels of SIRT1 in the spinal cord tissues of control and SCI model rats were analyzed by western blotting and RT-qPCR, respectively. (C) Protein and (D) mRNA expression levels of SIRT1 in control and LPS-induced PC12 cells were detected by western blotting and RT-qPCR, respectively. Data are presented as the mean ± SD. The experiments were performed in triplicate. **P<0.01 vs. Control. miR-138-5p, microRNA-138-5p; SIRT1, sirtuin 1; SCI, spinal cord injury; LPS, lipopolysaccharide; RT-qPCR, reverse transcription-quantitative PCR.
Downregulation of miR-138-5p results in SIRT1 accumulation
In order to evaluate whether miR-138-5p can interfere with SIRT1 expression in an SCI model in vitro, PC12 cells were transfected with control-siRNA, SIRT1-siRNA, miR-138-5p inhibitor, inhibitor control, or SIRT1-siRNA + miR-138-5p inhibitor for 48 h. As presented in Fig. 4A, miR-138-5p levels were significantly decreased in PC12 cells transfected with miR-138-5p inhibitor compared with the inhibitor control. Additionally, SIRT1-siRNA significantly reduced the mRNA levels of SIRT1 in PC12 cells (Fig. 4B). Compared with the control group, the mRNA and protein levels of SIRT1 were significantly enhanced by miR-138-5p inhibitor; this increase was reversed by SIRT1-siRNA (Fig. 4C and D). The findings indicated that miR-138-5p negatively regulates SIRT1 expression in PC12 cells.
Figure 4.
miR-138-5p inhibitor affects the expression of miR-138-5p and SIRT1. (A) RT-qPCR was performed to detect the relative level of miR-138-5p in PC12 cells transfected with miR-138-5p inhibitor or inhibitor control. (B) mRNA expression of SIRT1 in PC12 cells transfected with control-siRNA or SIRT1-siRNA was detected via RT-qPCR. (C) RT-qPCR and (D) western blotting were performed to measure the mRNA and protein expression levels of SIRT1 in PC12 cells transfected with miR-138-5p inhibitor, inhibitor control or miR-138-5p inhibitor + SIRT1-siRNA. Data are presented as the mean ± SD. The experiments were performed in triplicate. **P<0.01 vs. Control; ##P<0.01 vs. inhibitor. miR-138-5p, microRNA-138-5p; SIRT1, sirtuin 1; siRNA, small interfering RNA; RT-qPCR, reverse transcription-quantitative PCR.
Inhibition of miR-138-5p attenuates inflammatory injury in the SCI model in vitro
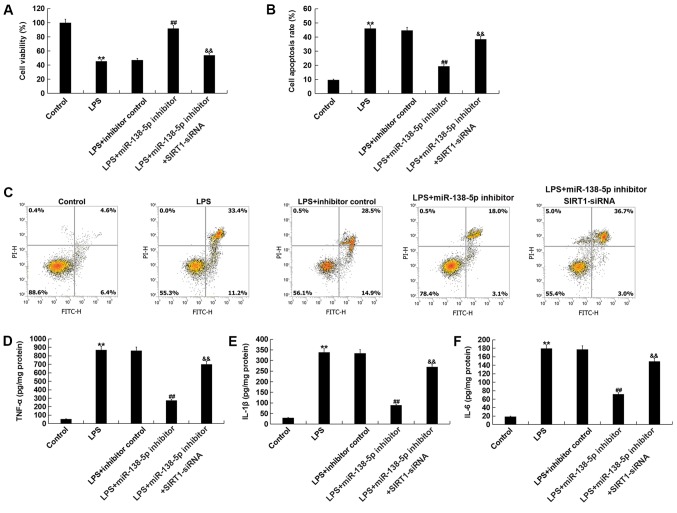
In order to evaluate the function of miR-138-5p in an in vitro SCI model, miR-138-5p inhibitor, inhibitor control, or SIRT1-siRNA + miR-138-5p inhibitor were transfected into PC12 cells for 48 h. Then, the PC12 cells were subjected to 100 ng/ml LPS for 4 h. Subsequently, the viability of cells was assessed using an MTT assay. The results indicated that cell viability was significantly decreased in the LPS treatment group compared with the control group (Fig. 5A). However, compared with LPS treatment alone, miR-138-5p inhibitor significantly promoted PC12 cell viability, which was significantly reversed by SIRT1-siRNA (Fig. 5A). Then, the apoptosis of cells was analyzed via flow cytometry. Compared with the control group, LPS treatment significantly enhanced PC12 cell apoptosis; however, miR-138-5p inhibitor significantly reduced the effect of LPS on apoptosis (Fig. 5B and C). This in turn was reversed by SIRT1-siRNA co-transfection. Additionally, inflammatory factors were detected via ELISA. The results demonstrated that the levels of TNF-α, IL-1β and IL-6 were significantly upregulated in the LPS treatment group compared with the control group. Inhibition of miR-138-5p significantly attenuated the expression of inflammatory factors compared with LPS treatment alone; this reduction was significantly attenuated by SIRT1 silencing (Fig. 5D-F).
Figure 5.
Downregulation of miR-138-5p or SIRT1 knockdown alter cell viability, apoptosis and the expression of inflammatory factors. PC12 cells were transfected with inhibitor control, miR-138-5p inhibitor or miR-138-5p inhibitor + SIRT1-siRNA for 48 h; then, these cells were subjected to LPS treatment (100 ng/ml) for 4 h. (A) Cell viability was measured by MTT assays. (B) Histogram of the percentage of early and late apoptotic cells. (C) Flow cytometric analysis was conducted to evaluate cell apoptosis using Annexin V/PI double staining. Inflammatory factors including (D) TNF-α, (E) IL-1β and (F) IL-6 were detected via ELISA. Data are presented as the mean ± SD. The experiments were performed in triplicate. **P<0.01 vs. Control; ##P<0.01 vs. LPS; &&P<0.01 vs. LPS + inhibitor. miR-138-5p, microRNA-138-5p; SIRT1, sirtuin 1; siRNA, small interfering RNA; LPS, lipopolysaccharide; IL, interleukin; TNF, tumor necrosis factor; PI, propidium iodide.
Role of miR-138-5p in the PTEN/AKT pathway in an in vitro SCI model
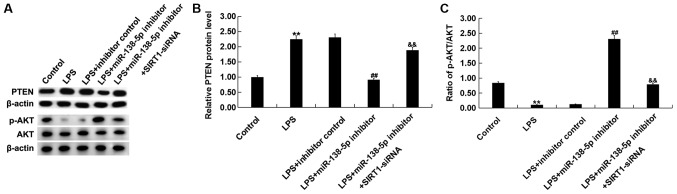
To further investigate the molecular mechanisms of miR-138-5p in SCI, after treatments, the protein levels of PTEN, AKT and p-AKT were measured in PC12 cells via western blotting. Results indicated that compared with the control group, LPS treatment significantly increased PTEN expression (Fig. 6A and B) and decreased AKT phosphorylation (Fig. 6A and C). miR-138-5p inhibitor significantly reversed these effects, which was attenuated by SIRT1 silencing. These results suggested that miR-138-5p modulated the PTEN/AKT signaling pathway by targeting SIRT1 in in vitro SCI cell model.
Figure 6.
Effects of miR-138-5p and SIRT1 on the expression of PTEN/AKT signaling pathway proteins. PC12 cells were transfected with inhibitor control, miR-138-5p inhibitor or miR-138-5p inhibitor + SIRT1-siRNA for 48 h; then, these cells were subjected to LPS treatment (100 ng/ml) for 4 h. (A) Western blot analysis was performed to determine the protein expression of PTEN, AKT and p-AKT in different groups. (B) Protein expression of PTEN was calculated and presented as fold of the control group. (C) p-AKT/AKT was calculated. Data are presented as the mean ± SD. The experiments were performed in triplicate. **P<0.01 vs. Control; ##P<0.01 vs. LPS; &&P<0.01 vs. LPS + inhibitor. miR-138-5p, microRNA-138-5p; SIRT1, sirtuin 1; siRNA, small interfering RNA; LPS, lipopolysaccharide; p, phosphorylated.
Discussion
There has been substantial focus into the importance of miRNAs. Several studies have reported that miRNAs altered the response to SCI by regulating the expression of various key factors in cell growth and apoptosis (35–37). SCI refers to primary mechanical damage in the spinal cord exacerbated by subsequent biological processes, including inflammation, apoptosis and altered gene expression (38). However, an association between miR-138-5p and the proliferation and apoptosis of SCI cells has not previously been identified, to the best of our knowledge. The biological function and underlying mechanisms of miR-138-5p in SCI model rats and cells remain to be further explored.
In the present study, damage was induced at T10 to generate a rodent SCI model. An SCI in vitro cell model was also established by subjecting PC12 cells to LPS exposure for 4 h. Then, RT-qPCR was performed to detect the expression of miR-138-5p in patients with SCI, rats and LPS-induced cells. The results indicated that miR-138-5p was significantly upregulated in SCI. In addition, it was further identified that SIRT1 was a potential target of miR-138-5p. SIRT1 has been implicated as a target of miR-138-5p in various studies (24,39,40). For example, overexpression of miR-138-5p has been reported to suppress manganese-induced autophagy by targeting SIRT1 in SH-SY5Y cells (24). miR-138-5p enhances TNF-α-induced apoptosis in human intervertebral disc degeneration by targeting SIRT1 (39). miR-138-5p has also been found to prevent autophagy in pancreatic cancer by targeting SIRT1 (40).
The present study again demonstrated that SIRT1 was a direct target of miR-138-5p, and further investigated the expression and roles of miR-138-5p and SIRT1 in SCI. In order to further explore the relationship between miR-138-5p and SIRT1, the expression of SIRT1 was detected in an SCI rat model and in vitro cell model. The present study demonstrated high expression of miR-138-5p and low expression of SIRT1 in SCI tissues and cells, suggesting a relationship between the expression levels of miR-138-5p and SIRT1 in the development of SCI. Additionally, it was found that the expression of SIRT1 in PC12 cells was negatively regulated by miR-138-5p. To explore the underlying mechanisms of miR-138-5p in SCI, PC12 cells were transfected with inhibitor control, miR-138-5p inhibitor or SIRT1-siRNA + miR-138-5p inhibitor for 48 h, then the cells were subjected to 100 ng/ml LPS for 4 h. The present findings showed that knockdown of miR-138-5p upregulated SIRT1 and further reduced the apoptosis of SCI model cells.
Results from a previous study indicated that overexpression of proinflammatory factors can promote apoptosis and further aggravate SCI (41). In the present study, the levels of the proinflammatory factors IL-1β, TNF-α and IL-6 were detected via ELISA. The results demonstrated that inhibition of miR-138-5p significantly attenuated the expression of inflammatory factors compared with the inhibitor control group.
Finally, the expression of PTEN and p-AKT was investigated via western blotting. The results indicated that compared with the control group, LPS treatment significantly increased PTEN protein expression and decreased AKT phosphorylation. miR-138-5p inhibitor significantly decreased PTEN protein expression and increased p-AKT protein expression; these changes were attenuated by SIRT1 silencing. Of note, all effects of miR-138-5p inhibitor on LPS-induced PC12 cells were significantly reversed by co-transfection with SIRT1-siRNA. These results suggested that miR-138-5p modulated the PTEN/AKT signaling pathway by targeting SIRT1 in an in vitro model of SCI. Thus, it is hypothesized miR-138-5p inhibitor may suppress SCI-associated biological process by inhibiting the PTEN/AKT signaling pathway, highlighting it as a potential therapeutic target in SCI.
In conclusion, to the best of our knowledge, this was the first study to investigate the relationship between miR-138-5p and SIRT1 following SCI. Further study into miRNAs in SCI is urgently required to develop effective and safe therapeutic strategies for patients with SCI, and to improve the prognosis of SCI. However, the present study is a preliminary study into the role of miR-138-5p in SCI, with further experiments required. For example, the expression of SIRT1, PTEN, p-AKT and AKT in patients with SCI should be detected in order to validate the conclusions of the present study in humans. Additionally, time course studies were not performed for either in vivo or in vitro experiments in the present study; this will be explored in future studies.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JC contributed to designing the study, collecting, analyzing and interpreting the data, and preparing the manuscript. RQ contributed to collecting and analyzing the data, and preparing the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Written informed consent was obtained from each patient. The human study was approved by the Ethics Committee of the First People's Hospital of Lianyungang. The animal study was approved by the Animal Ethics Committee of the First People's Hospital of Lianyungang.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Silva NA, Sousa N, Reis RL, Salgado AJ. From basics to clinical: A comprehensive review on spinal cord injury. Prog Neurobiol. 2014;114:25–57. doi: 10.1016/j.pneurobio.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 2.Brommer B, Engel O, Kopp MA, Watzlawick R, Müller S, Prüss H, Chen Y, DeVivo MJ, Finkenstaedt FW, Dirnagl U, et al. Spinal cord injury-induced immune deficiency syndrome enhances infection susceptibility dependent on lesion level. Brain. 2016;139:692–707. doi: 10.1093/brain/awv375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siddall PJ, McIndoe L, Austin P, Wrigley PJ. The impact of pain on spiritual well-being in people with a spinal cord injury. Spinal Cord. 2017;55:105–111. doi: 10.1038/sc.2016.75. [DOI] [PubMed] [Google Scholar]
- 4.Ahmed MM, King KC, Pearce SM, Ramsey MA, Miranpuri GS, Resnick DK. Novel targets for spinal cord injury related neuropathic pain. Ann Neurosci. 2011;18:162–167. doi: 10.5214/ans.0972.7531.1118413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hooshmand MJ, Galvan MD, Partida E, Anderson AJ. Characterization of recovery, repair, and inflammatory processes following contusion spinal cord injury in old female rats: Is age a limitation? Immun Ageing. 2014;11:15. doi: 10.1186/1742-4933-11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garshick E, Stolzmann KL, Gagnon DR, Morse LR, Brown R. Systemic inflammation and reduced pulmonary function in chronic spinal cord injury. PM R. 2011;3:433–439. doi: 10.1016/j.pmrj.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gris D, Hamilton EF, Weaver LC. The systemic inflammatory response after spinal cord injury damages lungs and kidneys. Exp Neurol. 2008;211:259–270. doi: 10.1016/j.expneurol.2008.01.033. [DOI] [PubMed] [Google Scholar]
- 8.Pillai RS. MicroRNA function: Multiple mechanisms for a tiny RNA? RNA. 2005;11:1753–1761. doi: 10.1261/rna.2248605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hammond SM. An overview of microRNAs. Adv Drug Deliv Rev. 2015;87:3–14. doi: 10.1016/j.addr.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shukla GC, Singh J, Barik S. MicroRNAs: Processing, maturation, target recognition and regulatory functions. Mol Cell Pharmacol. 2011;3:83–92. [PMC free article] [PubMed] [Google Scholar]
- 11.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 12.Feng W, Feng Y. MicroRNAs in neural cell development and brain diseases. Sci China Life Sci. 2011;54:1103–1112. doi: 10.1007/s11427-011-4249-8. [DOI] [PubMed] [Google Scholar]
- 13.Bian S, Sun T. Functions of noncoding RNAs in neural development and neurological diseases. Mol Neurobiol. 2011;44:359–373. doi: 10.1007/s12035-011-8211-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Papagiannakopoulos T, Kosik KS. MicroRNAs: Regulators of oncogenesis and stemness. BMC Med. 2008;6:15. doi: 10.1186/1741-7015-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blandino G, Fazi F, Donzelli S, Kedmi M, Sas-Chen A, Muti P, Strano S, Yarden Y. Tumor suppressor microRNAs: A novel non-coding alliance against cancer. FEBS Lett. 2014;588:2639–2652. doi: 10.1016/j.febslet.2014.03.033. [DOI] [PubMed] [Google Scholar]
- 16.Palanichamy JK, Rao DS. miRNA dysregulation in cancer: Towards a mechanistic understanding. Front Genet. 2014;5:54. doi: 10.3389/fgene.2014.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Kim S, Kim IM. Regulation of metastasis by microRNAs in ovarian cancer. Front Oncol. 2014;4:143. doi: 10.3389/fonc.2014.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Othman N, Nagoor NH. The role of microRNAs in the regulation of apoptosis in lung cancer and its application in cancer treatment. Biomed Res Int. 2014;2014:318030. doi: 10.1155/2014/318030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han C, Yu Z, Duan Z, Kan Q. Role of microRNA-1 in human cancer and its therapeutic potentials. Biomed Res Int. 2014;2014:428371. doi: 10.1155/2014/428371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu NK, Wang XF, Lu QB, Xu XM. Altered microRNA expression following traumatic spinal cord injury. Exp Neurol. 2009;219:424–429. doi: 10.1016/j.expneurol.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roberto GM, Lira RC, Delsin LE, Vieira GM, Silva MO, Hakime RG, Yamashita ME, Engel EE, Scrideli CA, Tone LG, Brassesco MS. microRNA-138-5p as a worse prognosis biomarker in pediatric, adolescent, and young adult osteosarcoma. Pathol Oncol Res. 2019 Mar 12; doi: 10.1007/s12253-019-00633-0. doi: 10.1007/s12253-019-00633-0. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 22.He Z, Ruan X, Liu X, Zheng J, Liu Y, Liu L, Ma J, Shao L, Wang D, Shen S, et al. FUS/circ_002136/miR-138-5p/SOX13 feedback loop regulates angiogenesis in Glioma. J Exp Clin Cancer Res. 2019;38:65. doi: 10.1186/s13046-019-1065-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu D, Gu L, Li Z, Jin W, Lu Q, Ren T. MiR-138-5p suppresses lung adenocarcinoma cell epithelial-mesenchymal transition, proliferation and metastasis by targeting ZEB2. Pathol Res Pract. 2019;215:861–872. doi: 10.1016/j.prp.2019.01.029. [DOI] [PubMed] [Google Scholar]
- 24.Ma J, Zhang Y, Ji H, Chen L, Chen T, Guo C, Zhang S, Jia J, Niu P. Overexpression of miR-138-5p suppresses MnCl2 -induced autophagy by targeting SIRT1 in SH-SY5Y cells. Environ Toxicol. 2019;34:539–547. doi: 10.1002/tox.22708. [DOI] [PubMed] [Google Scholar]
- 25.Nogueiras R, Habegger KM, Chaudhary N, Finan B, Banks AS, Dietrich MO, Horvath TL, Sinclair DA, Pfluger PT, Tschöp MH. Sirtuin 1 and sirtuin 3: Physiological modulators of metabolism. Physiol Rev. 2012;92:1479–1514. doi: 10.1152/physrev.00022.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.da Cunha MSB, Arruda SF. Tucum-do-Cerrado (Bactris setosa Mart.) may promote anti-aging effect by upregulating SIRT1-Nrf2 pathway and attenuating oxidative stress and inflammation. Nutrients. 2017;9:E1243. doi: 10.3390/nu9111243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rada P, Pardo V, Mobasher MA, García-Martínez I, Ruiz L, González-Rodríguez Á, Sanchez-Ramos C, Muntané J, Alemany S, James LP, et al. SIRT1 controls acetaminophen hepatotoxicity by modulating inflammation and oxidative stress. Antioxid Redox Signal. 2018;28:1187–1208. doi: 10.1089/ars.2017.7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan SH, Hung CH, Shih JY, Chu PM, Cheng YH, Lin HC, Tsai KL. SIRT1 inhibition causes oxidative stress and inflammation in patients with coronary artery disease. Redox Biol. 2017;13:301–309. doi: 10.1016/j.redox.2017.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng YY, Kao CL, Ma HI, Hung CH, Wang CT, Liu DH, Chen PY, Tsai KL. SIRT1-related inhibition of pro-inflammatory responses and oxidative stress are involved in the mechanism of nonspecific low back pain relief after exercise through modulation of Toll-like receptor 4. J Biochem. 2015;158:299–308. doi: 10.1093/jb/mvv041. [DOI] [PubMed] [Google Scholar]
- 30.Roberts TT, Leonard GR, Cepela DJ. Classifications in brief: American spinal injury association (ASIA) impairment scale. Clin Orthop Relat Res. 2017;475:1499–1504. doi: 10.1007/s11999-016-5133-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bayne K. Revised guide for the care and use of laboratory animals available. American Physiological Society. Physiologist. 1996;39:199208–199211. [PubMed] [Google Scholar]
- 32.Xin DQ, Hu ZM, Huo HJ, Yang XJ, Han D, Xing WH, Zhao Y, Qiu QH. Schisandrin B attenuates the inflammatory response, oxidative stress and apoptosis induced by traumatic spinal cord injury via inhibition of p53 signaling in adult rats. Mol Med Rep. 2017;16:533–538. doi: 10.3892/mmr.2017.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan Y, Yu L, Zhang C, Chen K, Lu J, Tan L. miRNA-146a attenuates inflammation in an in vitro spinal cord injury model via inhibition of TLR4 signaling. Exp Ther Med. 2018;16:3703–3709. doi: 10.3892/etm.2018.6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 35.Wang C, Pan Y, Cheng B, Chen J, Bai B. Identification of conserved and novel microRNAs in cerebral ischemia-reperfusion injury of rat using deep sequencing. J Mol Neurosci. 2014;54:671–683. doi: 10.1007/s12031-014-0383-7. [DOI] [PubMed] [Google Scholar]
- 36.Wan G, An Y, Tao J, Wang Y, Zhou Q, Yang R, Liang Q. MicroRNA-129-5p alleviates spinal cord injury in mice via suppressing the apoptosis and inflammatory response through HMGB1/TLR4/NF-κB pathway. Biosci Rep. 2020;40:BSR20193315. doi: 10.1042/BSR20193315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun F, Li SG, Zhang HW, Hua FW, Sun GZ, Huang Z. MiRNA-411 attenuates inflammatory damage and apoptosis following spinal cord injury. Eur Rev Med Pharmacol Sci. 2020;24:491–498. doi: 10.26355/eurrev_202001_20022. [DOI] [PubMed] [Google Scholar]
- 38.Weishaupt N, Silasi G, Colbourne F, Fouad K. Secondary damage in the spinal cord after motor cortex injury in rats. J Neurotrauma. 2010;27:1387–1397. doi: 10.1089/neu.2010.1346. [DOI] [PubMed] [Google Scholar]
- 39.Wang B, Wang D, Yan T, Yuan H. MiR-138-5p promotes TNF-α-induced apoptosis in human intervertebral disc degeneration by targeting SIRT1 through PTEN/PI3K/Akt signaling. Exp Cell Res. 2016;345:199–205. doi: 10.1016/j.yexcr.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 40.Tian S, Guo X, Yu C, Sun C, Jiang J. miR-138-5p suppresses autophagy in pancreatic cancer by targeting SIRT1. Oncotarget. 2017;8:11071–11082. doi: 10.18632/oncotarget.14360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bank M, Stein A, Sison C, Glazer A, Jassal N, McCarthy D, Shatzer M, Hahn B, Chugh R, Davies P, Bloom O. Elevated circulating levels of the pro-inflammatory cytokine macrophage migration inhibitory factor in individuals with acute spinal cord injury. Arch Phys Med Rehabil. 2015;96:633–644. doi: 10.1016/j.apmr.2014.10.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.