Abstract
Periodontal accelerate osteogenesis orthodontics (PAOO) is an extension of described techniques that surgically alter the alveolar bone; however, the specific mechanism underlying the technique is not completely understood. The aim of the present study was to evaluate the roles of microRNA (miR)-21 during PAOO. Sprague-Dawley rats were divided into the following four groups: i) Group tooth movement (TM), underwent TM and were administered normal saline (NS); ii) Group PAOO, underwent PAOO + TM and were administered NS; iii) Group agomiR-21, underwent PAOO + TM and were administered agomiR-21; and iv) Group antagomiR-21, underwent PAOO + TM and were administered antagomiR-21. To validate the rat model of PAOO, morphological analyses were performed and measurements were collected. Reverse transcription-quantitative PCR, western blotting and immunohistochemical staining were performed to examine the expression levels of programmed cell death 4 (PDCD4), activin A receptor type 2B (ACVR2b), receptor activator of NF-κΒ ligand (RANKL) and C-Fos. Dual-luciferase reporter assays were performed to validate PDCD4 as a target of miR-21 in vitro. Following 7 days of treatment, the TM distance of group PAOO was longer compared with groups TM and antagomiR-21 (P<0.05), but shorter compared with group agomiR-21 (P<0.05). Tartrate-resistant acid phosphatase staining indicated that following treatment with agomiR-21, osteoclast activity was notably increased, whereas the mRNA and protein expression levels of PDCD4 were notably decreased compared with group PAOO. The mRNA and protein expression levels of RANKL and C-Fos in group agomiR-21 were notably increased compared with group PAOO, whereas group antagomiR-21 displayed the opposite pattern (P<0.05). With regard to ACVR2b, no significant differences were observed among the group agomiR-21 and antagomiR-21 compared with group PAOO. Bioinformatics analysis predicted that PDCD4 was a potential target gene of miR-21, and dual-luciferase reporter assays demonstrated that miR-21 directly targeted PDCD4. In conclusion, the present study demonstrated that miR-21 serves an important role during PAOO-mediated orthodontic TM.
Keywords: microRNA-21, periodontal accelerate osteogenesis orthodontics, tooth movement, osteoclasts
Introduction
Tooth movement (TM) is a process in which mechanical force induces alveolar bone resorption on the pressure side and alveolar bone deposition on the tension side (1). As it usually takes ≥2 years to complete orthodontic treatment, it is important that periodontal accelerate osteogenesis orthodontics (PAOO) can increase orthodontic TM and reduce the course of orthodontic treatment (2). The clinical application of PAOO was first introduced by the Wilcko brothers (a periodontist and an orthodontist), and has become a useful modality in the field of surgical orthodontics for the induction of faster TM (3). Traditional orthodontic therapy focuses on applying forces to the teeth, whereas PAOO utilizes the dynamics of bone physiology to enhance TM. PAOO involves controlled surgical damage to cortical bone that accelerates bone metabolism to aid orthodontic TM, resulting in TM that is 2–3 times faster compared with traditional orthodontic therapy (4–6).
MicroRNAs (miRNAs/miRs), which are 22–25 nucleotides in length, post-transcriptionally regulate gene expression in a sequence-specific manner (7,8). A previous study has revealed that certain miRNAs are critical post-transcriptional modulators during osteoblastogenesis and osteoclastogenesis (9). Furthermore, previous studies have demonstrated that certain miRNAs respond to mechanical stimuli in cultured human periodontal ligament cells and periodontal ligament stem cells (10–12). However, whether miRNAs regulate PAOO or mediate alveolar bone remodeling in vivo is not completely understood.
It has been reported that miR-21 mediates stretch-induced osteogenic differentiation of periodontal ligament stem cells in vitro, which supports osteoclast differentiation (10). Although miR-21 is required for the regulation of gene expression under mechanical force in several biological processes, such as bone formation (10,13), its role during PAOO has not been previously reported. Therefore, the present study aimed to investigate the effects of miR-21 expression in vivo and examine its functional roles in the regulation of osteogenesis and osteoclast differentiation during PAOO.
Materials and methods
Study design
A total of 36 male Sprague-Dawley rats [weight, 286–326 g; age, 8 weeks; provided by Beijing Weitong Lihua Experimental Animal Technology Co., Ltd.; certificate no. scxk (Beijing) 2016-0011] were raised under specific pathogen-free conditions. The experiment was performed under sterile conditions with humidity 40–60%, 20–26°C and maximum daily temperature difference of 4°C. The circadian rhythm was changed every 12 h. Rats were randomly divided into four groups (n=9/group) as presented in Table I. The present study was approved by the Experimental Animal Welfare and Ethics Committee of China Medical University (approval no. 16049R).
Table I.
Group assignment.
| Group | Force | Injection |
|---|---|---|
| TM | TM | Normal saline |
| PAOO | PAOO + TM | Normal saline |
| agomiR-21 | PAOO + TM | AgomiR-21 |
| antagomiR-21 | PAOO + TM | AntagomiR-21 |
TM, Tooth movement; PAOO, periodontal accelerate osteogenesis orthodontics.
Overexpression and inhibition of miR-21 in rats
AgomiR-21 and antagomiR-21 were purchased from Guangzhou RiboBio Co., Ltd., which had the following sequences: agomiR-21 forward, 5′-UAGCUUAUCAGACUGAUGUUGA-3′ and reverse, 3′-AUCGAAUAGUCUGACUACAACU-5′; and antagomiR-21, 5′-UCAACAUCAGUCUGAUAAGCUA-3′. Rats were anesthetized by the intraperitoneal injection of sodium pentobarbital (40 mg/kg). Prior to orthodontic loading, rats received an injection of agomiR-21 (3 nmol/time; 20 µl) per day, antagomiR-21 (10 nmol/time; 20 µl) per day or normal saline (20 µl) per day into the buccal, palatal and mesial submucosa of the first maxillary molar continuously for 3 days.
Orthodontic loading
Rats were anesthetized by the intraperitoneal injection of sodium pentobarbital (40 mg/kg). Subsequently, two upper central incisors were drilled horizontally with a 0.25-mm orthodontic ligation wire to penetrate the root hole. The central incisors were ligated together to enhance anchorage and prevent continuous eruption.
Groups PAOO, agomiR-21 and antagomiR-21 were prepared as follows: The labial, palatal and mesial alveolar mucosa of the left maxillary first molar was incised to expose the alveolar bone surface; and subsequently, a high-speed hand piece was used under water irrigation to punch five perforations on the buccal and palate sides, and two perforations on the mesial side (diameter, 0.5 mm; depth; 0.5 mm). Group TM did not undergo the perforation procedure.
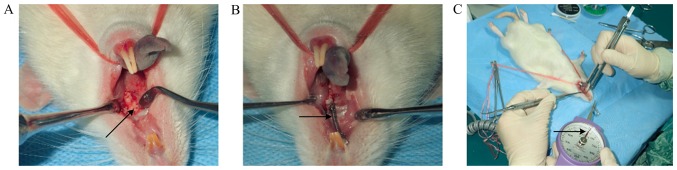
To prevent the tension spring from falling off, a retention groove was grinded into the buccal-palatal surface of the neck of the left maxillary first molars and a ligation wire was inserted into the retention groove of the first molar. The tension spring was fixed between the first maxillary molar and the incisor, and was stretched to pull the first maxillary molar to the mesial position. The orthodontic force was maintained at 25 g using a dynamometer. For the 3 days following surgery, the rats were provided with soft food. The establishment of the PAOO model is presented in Fig. 1.
Figure 1.
Establishment of the rat model of periodontal accelerate osteogenesis orthodontic tooth movement. (A) Full-thickness flaps on the buccal and palatal aspects, and perforations on the corticotomy side were made using a high-speed hand piece. Arrow indicates perforations. (B) Tension spring was fixed between the first maxillary molar and the incisor. Arrow indicates tension spring. (C) Orthodontic force was maintained at 25 g using a dynamometer. Arrow indicates dynamometer.
The distance of TM was measured using an electronic vernier caliper and a stereomicroscope (Motic Instruments). The distance between the first maxillary molar and the upper incisor was measured prior to surgery and following sacrifice to calculate the total distance of TM. The measurements were conducted three times in each rat.
At day 7 post-surgery, the rats were sacrificed by carbon dioxide euthanasia, according to the guidelines (14). The flow rate of CO2 did not displace >30% of the chamber volume/min. Death was verified by monitoring cessation of breathing, heartbeat and deep pain in response to the toe pinch method. The flow rate of CO2 was maintained for at least 1 min after cessation of breath, and absence of heartbeat was assessed for at least 5 min. Subsequently, the mucosal tissues were removed, and the cortical and cancellous bones were collected for further analysis.
Tartrate-resistant acid phosphatase (TRAP) staining
Alveolar bone tissue surrounding the maxillary first molar were fixed for 48 h at 4°C in precooled 4% paraformaldehyde. The decalcification was carried out in 10% EDTA decalcification solution (pH 7.2–3.4). The solution was replaced every 2 days and shaken. The decalcification time was ~3 months until the alveolar bone could be penetrated without resistance under 4°C. Subsequently, the tissue was embedded in paraffin and sectioned into 5-µm thick occlusal serial sections. The sections were stained using an Acid Phosphatase, Leukocyte (TRAP) kit (Sigma-Aldrich; Merck KGaA) according to the manufacturer's protocol. Subsequently, osteoblasts and TRAP+ multinucleated cells that were attached to alveolar bone surfaces were counted. The BX53 Olympus fluorescence microscope (Olympus Corporation) was used under magnification, ×200.
Reverse transcription-quantitative PCR (RT-qPCR)
Dentoalveolar bone tissue from the mesial alveolar bone around the left first maxillary molar, including cortical and cancellous bone tissue, was collected. Total RNA was extracted using QIAzol Lysis Reagent (Qiagen, Inc.). Total RNA was reverse transcribed into cDNA using the TaqMan Reverse Transcription kit (Guangzhou RiboBio Co., Ltd.) according to the manufacturer's protocol. Subsequently, qPCR was performed. QuantiFast® SYBR® Green PCR Master Mix was used (QuantiFast® SYBR® Green PCR kit; Beijing Bulader Technology Development Co., Ltd.). The following thermocycling conditions were used for qPCR: Initial denaturation at 95°C for 5 min; followed by 40 cycles of 95°C for 10 sec and 60°C for 30 sec. mRNA and miRNA expression levels were normalized to the internal reference genes GAPDH and U6, respectively. miRNA expression levels were determined by performing a stem-loop RT-qPCR assay, as previously described (15). The 2−∆∆Cq method was used (16). The following primer sequences were used: Programmed cell death 4 (PDCD4) forward, 5′-CTGTGTTTATGAGACTGTGGTT-3′ and reverse, 5′-CGCGACTTCGTTCGTATC-3′; activin A receptor type 2B (ACVR2b) forward, 5′-TCTCGTACCTGCATGAGGA-3′ and reverse, 5′-TCGCTCTTCAGCAGAACA-3′; C-fos forward, 5′-CAGCTCCCACCAGTGTCTA-3′ and reverse, 5′-CGCGTTGAAACCCGAGAA-3′; receptor activator of NF-κΒ ligand (RANKL) forward, 5′-CGAAGACACAGAAGCACTAC-3′ and reverse, 5′-CACGAACCTTCCATCATAGC-3′; and GAPDH forward, 5′-GCGAGATCCCGCTAACATCA-3′ and reverse, 5′-CTCGTGGTTCACACCCATCA-3′.
Western blotting
Dentoalveolar bone tissues from the mesial alveolar bone around the left first maxillary molar, including cortical and cancellous bone tissues, were collected. The tissues were sonicated in a lysis buffer (Shanghai HuaYi Biology Technology Co., Ltd.) containing 1 mM PMSF. Total protein was extracted and quantified using the bicinchoninic acid method. Total protein (50 µg/lane) was separated via 10% SDS-PAGE and transferred onto nitrocellulose membranes, which were blocked with 5% non-fat milk for 1 h at 37°C. Subsequently, the membranes were incubated with primary antibodies targeted against PDCD4 (1:2,000; cat. no. 12587-1-AP; ProteinTech Group, Inc.), ACVR2b (1:1,000; cat. no. Ag16744; ProteinTech Group, Inc.), RANKL (1:1,000; cat. no. 23408-1-AP; ProteinTech Group, Inc.), C-Fos (1:1,000; cat. no. 15832-1-AP; ProteinTech Group, Inc.) and GAPDH (1:5,000; cat. no. 10494-1-AP; Santa Cruz Biotechnology, Inc.) at 4°C overnight. Subsequently, the membranes were incubated with horseradish peroxidase-conjugated secondary antibodies, goat anti rabbit IgG (1:5,000; cat. no. CW0156S; Kangwei Century Biotechnology Co., Ltd.) at 37°C for 45 min. Protein bands were visualized using Pierce ECL (Pierce; Thermo Fisher Scientific, Inc. GADPH was used as the loading control. Gel Pro analyzer software 4.5 (Beijing Zhongsheng Tiancheng Technology Co., Ltd.) was used to analyze the optical density value of the target strip.
Immunohistochemical staining
To reduce the error and maintain standards between groups, the paraffin sections used for immunohistochemical staining were taken from 1.2 mm away from the root tip. The sections were fixed for 48 h at 4°C in precooled 4% paraformaldehyde. Subsequently, 4-µm thick sagittal mouse maxillae were deparaffinized, treated with 0.25% trypsin for 30 min at 37°C for antigen retrieval and incubated with 3% hydrogen peroxide for 30 min at 37°C. The sections were blocked with 5% bovine serum albumin (Wuhan Boster Biological Technology, Ltd.) in PBS for 2 h at room temperature. Subsequently, the sections were incubated at 4°C overnight with anti-PDCD4 (1:100; cat. no. 12587-1-AP; ProteinTech Group, Inc.) and anti-C-Fos (1:100; cat. no. 15832-1-AP; ProteinTech Group, Inc.) rabbit anti-mouse antibodies. Following primary incubation, the sections were incubated with a horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (1:500; cat. no. CW0103S; Kangwei Century Biotechnology Co., Ltd.) for 2 h at room temperature. The sections were washed three times by PBS for 3 min. The slides were stained with DAB for 4 min at 37°C and counterstained with hematoxylin for 3 min at 37°C. Stained sections were observed under a fluorescence microscope (N-STORM; Nikon Corporation) in ≥3 fields of view under magnification ×200. The number of positive-stained cells over the total periodontal area was quantified using ImageJ software (version 1.47; National Institutes of Health). PDCD4 and C-Fos protein levels were semi-quantitatively detected as the integral optical density.
Cell culture
293T cells (American Type Culture Collection) were cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.) containing 10% FBS (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 50 mg/l streptomycin at 37°C with 5% CO2.
Dual-luciferase reporter assay
Renilla luciferase activities was used as control for normalization. PDCD4 was predicted as a potential target of miR-21 by TargetScan7.2 (http://www.targetscan.org/vert_72/). The 3′-untranslated region (UTR) of PDCD4 was cloned into the pGL3 plasmid (Suzhou GenePharma Co., Ltd.). Subsequently, 293T cells at 50% density were seeded into 24-well plates and co-transfected with PDCD4-3′UTR-wild-type (WT) or the mutant (MUT) sequence and miR-21 mimics or mimics control (20 nM; Suzhou GenePharma Co., Ltd.) using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.; 100 nM) according to the manufacturer's protocol. Following incubation for 48 h at 37°C, luciferase activities were determined using a Dual-Luciferase Reporter assay system (Promega Corporation) according to the manufacturer's protocol. miR-21 mimics forward, 5′-UAGCUUAUCAGACUGAUGUUGAC-3′ and reverse, 3′-CAACAUCAGUCUGAUAAGCUAUU-5′; and mimics control forward, 5′-UUCUCCGAACGUGUCACG-3′ and reverse, 3′-ACGUGACACGUUCGGAGAATT-5′.
Statistical analysis
All statistical analyses were performed using SPSS software (version 13.0; SPSS, Inc.). Data are presented as the mean ± SD. Differences among groups were analyzed using one-way ANOVA followed by Tukey's post hoc test. P<0.05 was considered to indicate a statistically significant difference. Each experiment was repeated three times.
Results
TM in the rat model of PAOO
Following 7 days of treatment, the distance of TM in group PAOO was significantly longer compared with groups TM and antagomiR-21, but was significantly shorter compared with group agomiR-21 (P<0.05; Table II).
Table II.
Mean difference in tooth movement between the four groups.
| Group | Distance (mm) |
|---|---|
| TM | 0.112±0.013a |
| PAOO | 0.251±0.008 |
| agomiR-21 | 0.856±0.012a |
| antagomiR-21 | 0.072±0.010a |
P<0.05 vs. group B.
TRAP staining indicated that group PAOO displayed a higher number of osteoclasts in the alveolar bone surrounding the first molar compared with group TM (Fig. 2A and B). Following treatment with agomiR-21, osteoclast activity was notably increased and a reduced number of osteoclasts were observed in the tissues control compared with group PAOO (Fig. 2C). The osteoclast activity was not notably different in group antagomiR-21 control compared with group PAOO (Fig. 2D). The results indicated that the rat model of PAOO had been successfully established.
Figure 2.
Tartrate-resistant acid phosphatase staining of tissues. (A) Osteoclast in Group TM was small. (B) Osteoclast in Group PAOO was relatively increased. (C) Osteoclast in Group agomiR-21 was notably. (D) Osteoclast in Group antagomiR-21 was small. Arrow indicates osteoclast. PDL, periodontal ligament; AB; alveolar bone.
miR-21 regulates osteoclastogenesis during PAOO-facilitated TM
To assess the association between miR-21 and osteoclastogenesis during PAOO, rats were treated with agomiR-21 or antagomiR-21. To ensure agomir-21 and antagomir-21 penetrated the cortical bone and functioned, the mucosal tissues were removed, and the cortical and cancellous bones were collected. Subsequently, the efficiency of agomiR-21 and antagomiR-21 in local dentoalveolar bone tissues was evaluated by RT-qPCR. Compared with group PAOO, miR-21 expression levels in alveolar bone tissue were significantly decreased in group TM (P<0.05). Furthermore, compared with group PAOO, miR-21 expression levels were significantly increased in rats treated with agomiR-21 (P<0.05), but were significantly decreased in rats treated with antagomiR-21, which suggested that agomiR-21 and antagomiR-21 upregulated and downregulated miR-21 expression, respectively, in vivo (P<0.05; Fig. 3).
Figure 3.

miRNA-21 expression levels in alveolar bone tissues of rats. miRNA-21 expression levels in the alveolar bone tissues of groups TM and antagomiR-21 were significantly decreased compared with group PAOO. By contrast, miRNA-21 expression levels in the alveolar bone tissues of group agomiR-21 were significantly increased compared with group PAOO. *P<0.05 vs. group PAOO. miRNA, microRNA.
To further determine the role of miR-21 during PAOO, the expression of two downstream target genes of miR-21, PDCD4 and ACVR2b, was detected. Compared with group PAOO, rats treated with agomiR-21 displayed decreased mRNA and protein expression levels of PDCD4 notably. By contrast, groups TM and antagomiR-21 displayed significantly increased mRNA and protein expression levels of PDCD4 compared with group PAOO (P<0.05; Fig. 4A). The mRNA and protein expression levels of ACVR2b were significantly increased in group TM compared with group PAOO (P<0.05); however, ACVR2b expression levels were not significantly different between groups PAOO, agomiR-21 and antagomiR-21(P>0.05; Fig. 4B).
Figure 4.
Expression levels of alveolar bone tissue formation-associated factors. mRNA and protein expression levels of (A) PDCD4, (B) ACVR2b, (C) RANKL and (D) C-Fos. *P<0.05 vs. group PAOO. PDCD4, programmed cell death 4; ACVR2b, activin A receptor type 2B; RANKL, receptor activator of NF-κΒ ligand.
RANKL is an important factor that can be used as an indicator of osteoclast activity (17). C-Fos is a key regulator of osteoclast differentiation that is a RANK-activating transcription factor, which induces the expression of various osteoclast-specific downstream target genes (18). To further study the mechanism underlying orthodontic teeth movement, the expression levels of RANKL and C-Fos were examined. The results indicated that, compared with group PAOO, the mRNA and protein expression levels of RANKL and C-Fos were significantly increased in group agomiR-21, but were significantly decreased in groups TM and antagomiR-21 (P<0.05; Fig. 4C and D).
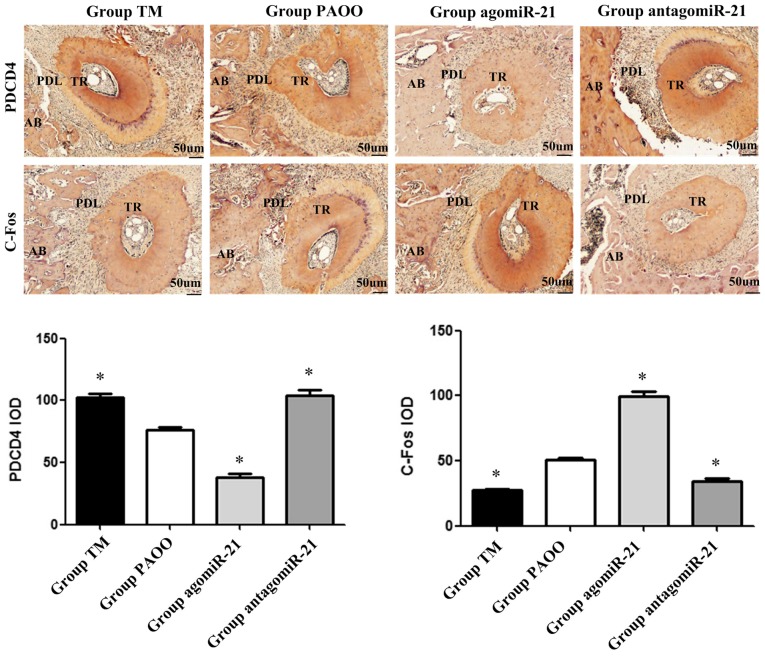
Immunohistochemical staining was used to further examine the relationship between miR-21 and PDCD4 or C-Fos in rat alveolar bone tissues. PDCD4 and C-Fos expression was primarily localized in the cytoplasm and occasionally present in the cell nuclei. The staining intensity of PDCD4 was significantly decreased in group agomiR-21, and was significantly increased in groups TM and antagomiR-21, compared with group PAOO. By contrast, C-Fos staining displayed the opposite trend (P<0.05; Fig. 5).
Figure 5.
PDCD4 and C-Fos expression in alveolar bone tissues. Compared with group PAOO, the staining intensity of PDCD4 and C-Fos was significantly decreased in group agomiR-21, and significantly increased in groups TM and antagomiR-21. *P<0.05 vs. group PAOO. PDCD4, programmed cell death 4; IOD, integral optical density; AB, alveolar bone; PDL, periodontal ligament; TR, tooth root.
PDCD4 is a direct target of miR-21
TargetScan was used to predict the binding site between miR-21 and the 3′UTR of PDCD4 (Fig. 6A). miR-21 overexpression significantly inhibited the luciferase activities of the PDCD4-3′UTR-WT group compared with the NC group. However, miR-21 overexpression did not significantly alter the luciferase activities of the PDCD4-MUT group compared with the NC group, which indicated that miR-21 specifically targeted the 3′UTR of PDCD4 by binding to the predicted sequence (P<0.05; Fig. 6B).
Figure 6.
miR-21 targets the 3′UTR of PDCD4 to suppress PDCD4 expression in 293T cells. (A) The potential binding site between the 3′UTR of PDCD4 and miR-21. (B) miR-21 overexpression inhibited luciferase activities in the WT-PDCD4 group compared with the NC group, whereas luciferase activities in MUT cells were not significantly altered by miR-21 overexpression. *P<0.05 vs. NC. miR, microRNA; UTR, untranslated region; PDCD4, programmed cell death 4; WT, wild-type; MUT, mutant; NC, negative control.
Discussion
PAOO is a clinical procedure that combines selective alveolar corticotomy, particulate bone grafting and the application of orthodontic forces, and is theoretically based on the bone-healing pattern that is known as the regional acceleratory phenomenon (19). PAOO results in increased alveolar bone width, decreased treatment time, increased post-treatment stability and reduced apical root resorption (20,21). To investigate the specific mechanism underlying the clinical procedure, a rat model of PAOO was established in the present study. To validate the rat model, morphological analyses were performed and necessary measurements were taken. Compared with group TM, group PAOO displayed an increased rate of TM, a significant reduction in bone volume and signs of bone resorption, including a higher number of osteoclast cells at 7 days post-surgery.
miRNAs modulate gene expression by binding to target mRNAs, thereby inhibiting their translation or promoting their degradation (22). In a previous study, gene chip technology indicated that miR-21 expression was increased in TM (23). A potential mechanism underlying miR-21 in TM is that the protein that is post-transcriptionally controlled by miR-21 inhibits osteoclastogenesis (23). Although several targets of miR-21 have been identified by bioinformatics analysis, PDCD4 and ACVR2b were the primary genes that were investigated in the present study due to their ability to regulate osteogenic and osteoclastic differentiation (10,24). High miR-21 expression levels are required for PDCD4 downregulation during osteoclastogenesis. PDCD4 suppresses cap-dependent translation of mRNAs with highly structured 5-regions via interaction with the eukaryotic translation initiation factor 4A helicase (25). It has also been reported that PDCD4 can directly alter the activity of the transcription factor activator protein-1 (AP-1), which triggers the transcription ofmiR-21 (26). Several conserved enhancer elements, including the binding site for AP-1, have been identified in the miR-21 promoter region (27). PDCD4 also regulates C-Fos, which is a transcription factor associated with osteoclast production (28,29). As demonstrated by the development of osteopetrosis in mice lacking C-Fos, C-Fos serves an important role during osteoclastogenesis. Therefore, although C-Fos is not required for normal osteoprogenitor development, it is essential for osteoclastogenesis (30,31). Moreover, C-Fos is the principal contributor among PDCD4-induced AP-1 components (32).
In the present study, PDCD4 expression levels were significantly decreased in the agomiR-21 group compared with group PAOO, whereas the antagomir-21 group displayed significantly increased PDCD4 expression levels compared with group PAOO. The results suggested that miR-21 negatively regulated PDCD4 expression during PAOO-mediated TM. Furthermore, the results indicated that C-Fos expression levels increased with agomiR-21 treatment and decreased with antagomiR-21 treatment, as has been reported in previous studies (33).
ACVR2b is a transmembrane serine/threonine receptor kinase that serves a crucial role during the activation of activin, which forms part of the transforming growth factor-β signaling pathway, and acts on cell proliferation and differentiation, as well as several other biological functions (34). Wei et al (10) demonstrated that miR-21 mediated the osteogenic differentiation effect of stretch by directly targeting ACVR2b, which is a key regulator of osteogenic differentiation. In addition, ACVR2b gain- and loss-of-function experiments indicated an association between miR-21 and ACVR2b during stretch-induced periodontal ligament stem cells osteogenic differentiation (10). Moreover, it has been reported that ACVR2b-mediated effects on osteoblasts are due to its interaction with bone morphogenetic protein (35). In the present study, the results indicated that the expression levels of ACVR2b were not significantly altered by miR-21 overexpression or knockdown, which suggested that miR-21 did not serve a biological role via ACVR2b during PAOO-mediated TM.
Several studies have investigated the role of miR-21 during orthodontic TM (10,36); however, to the best of our knowledge, the present study was the first to investigate the role of miR-21 during PAOO. Dual-luciferase reporter assays were performed to verify that miR-21 downregulated PDCD4 expression levels by targeting the 3′UTR of PDCD4. Although the present study investigated the role of miR-21 during PAOO, there were a number of limitations. First, the present study did not perform RNAscope or RNA-fluorescence in situ hybridization experiments. Second, the mechanisms underlying miR-21-mediated regulation of PDCD4, ACVR2b, RANKL and C-Fos expression in vitro were not investigated. Furthermore, due to the absence of in vitro experiments, the dose- and time-dependent relationship between miR-21 and PDCD4, ACVR2b, RANKL and C-Fos was not investigated; therefore, further investigation is required.
In conclusion, the present study suggested a potential mechanism underlying PAOO-mediated acceleration of orthodontic TM. miR-21 overexpression negatively regulated the expression levels of the target gene PDCD4, leading to increased C-Fos expression levels, enhanced RANKL-mediated osteoclast generation, remodeling of the alveolar bone, development of temporary osteopenia and local alveolar osteoporosis, and increased movement of orthodontic teeth.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Department of Education Science and Technology Program of Liaoning Province (grant no. LK201639).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YuZ, YT and YaZ conceived and designed the experiments. XY, ZZ and CF collected and analyzed the imaging and pathology data. YuZ wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Experimental Animal Welfare and Ethics Committee of China Medical University (approval no. 16049R).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Van Schepdael A, Vander Sloten J, Geris L. A mechanobiological model of orthodontic tooth movement. Biomech Model Mechanobiol. 2013;12:249–265. doi: 10.1007/s10237-012-0396-5. [DOI] [PubMed] [Google Scholar]
- 2.Soltani L, Loomer PM, Chaar EE. A novel approach in periodontally accelerated osteogenic orthodontics (PAOO): A case report. Clin Adv Periodontics. 2019;9:110–114. doi: 10.1002/cap.10045. [DOI] [PubMed] [Google Scholar]
- 3.Wilcko WM, Wilcko MT. Accelerating tooth movement: The case for corticotomy-induced orthodontics. Am J Orthod Dentofacial Orthop. 2013;114:4–12. doi: 10.1016/j.ajodo.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 4.Adusumilli S, Yalamanchi L, Yalamanchili PS. Periodontally accelerated osteogenic orthodontics: An interdisciplinary approach for faster orthodontic therapy. J Pharm Bioallied Sci. 2014;6(Suppl 1):S2–S5. doi: 10.4103/0975-7406.137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferguson DJ, Al-Harbi MS, Wilcko WM, Wilcko MT, Erie P. Lower dental arch decrowding comparing non-extraction accelerated osteogenesis and distraction techniques. J Dent Res. 2001;80:181–183. [Google Scholar]
- 6.Yu H, Jiao F, Wang B, Shen SG. Piezoelectric decortication applied in periodontally accelerated osteogenic orthodontics. J Craniofac Surg. 2013;24:1750–1752. doi: 10.1097/SCS.0b013e3182902c5a. [DOI] [PubMed] [Google Scholar]
- 7.He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 8.Bartel D. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lian JB, Stein GS, van Wijnen AJ, Stein JL, Hassan MQ, Gaur T, Zhang Y. MicroRNA control of bone formation and homeostasis. Nat Rev Endocrinol. 2012;8:212–227. doi: 10.1038/nrendo.2011.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei F, Liu D, Feng C, Zhang F, Yang S, Hu Y, Ding G, Wang S. microRNA-21 mediates stretch-induced osteogenic differentiation in human periodontal ligament stem cells. Stem Cells Dev. 2015;24:312–319. doi: 10.1089/scd.2014.0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu Y, Ou Y, Liao C, Liang S, Wang Y. High-throughput sequencing analysis of the expression profile of microRNAs and target genes in mechanical force-induced osteoblastic/cementoblastic differentiation of human periodontal ligament cells. Am J Transl Res. 2019;11:3398–3411. [PMC free article] [PubMed] [Google Scholar]
- 12.Yao S, Zhao W, Ou Q, Liang L, Lin X, Wang Y. MicroRNA-214 suppresses osteogenic differentiation of human periodontal ligament stem cells by targeting ATF4. Stem Cells Int. 2017;2017:3028647. doi: 10.1155/2017/3028647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramanujam D, Sassi Y, Laggerbauer B, Engelhardt S. Viral vector-based targeting of miR-21 in cardiac nonmyocyte cells reduces pathologic remodeling of the heart. Mol Ther. 2016;24:1939–1948. doi: 10.1038/mt.2016.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.NIH ARAC guidelines for euthanasia of rodents using carbon dioxide. NIH Office of Intramural Research and Office of Animal Care and Use, corp-author. https://oacu.oir.nih.gov/oacu-staff 2017 [Google Scholar]
- 15.Feng J, Wang K, Liu X, Chen S, Chen J. The quantification of tomato microRNAs response to viral infection by stem-loop real-time RT-PCR. Gene. 2009;437:14–21. doi: 10.1016/j.gene.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Takegahara N, Kim H, Mizuno H, Sakaue-Sawano A, Miyawaki A, Tomura M, Kanagawa O, Ishii M, Choi Y. Involvement of receptor activator of nuclear factor-κB ligand (RANKL)-induced incomplete cytokinesis in the polyploidization of osteoclasts. J Biol Chem. 2016;291:3439–3454. doi: 10.1074/jbc.M115.677427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Izawa T, Arakaki R, Mori H, Tsunematsu T, Kudo Y, Tanaka E, Ishimaru N. The nuclear receptor AhR controls bone homeostasis by regulating osteoclast differentiation via the RANK/c-Fos signaling axis. J Immunol. 2016;197:4639–4650. doi: 10.4049/jimmunol.1600822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amit G, Jps K, Pankaj B, Suchinder S, Parul B. Periodontally accelerated osteogenic orthodontics (PAOO)-a review. J Clin Exp Dent. 2012;4:e292–e296. doi: 10.4317/jced.50822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma Z, Zheng J, Yang C, Xie Q, Liu X, Abdelrehem A. A new modified bone grafting technique for periodontally accelerated osteogenic orthodontics. Medicine (Baltimore) 2018;97:e12047. doi: 10.1097/MD.0000000000012047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hou HY, Li CH, Chen MC, Lin PY, Liu WC, Cathy Tsai YW, Huang RY. A novel 3D-printed computer-assisted piezocision guide for surgically facilitated orthodontics. Am J Orthod Dentofacial Orthop. 2019;155:584–591. doi: 10.1016/j.ajodo.2018.11.010. [DOI] [PubMed] [Google Scholar]
- 22.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mRNAs are conserved targets of microRNAs. Genome Res. 2019;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pitari MR, Rossi M, Amodio N, Botta C, Morelli E, Federico C, Gullà A, Caracciolo D, Di Martino MT, Arbitrio M, et al. Inhibition of miR-21 restores RANKL/OPG ratio in multiple myeloma-derived bonemarrow stromal cells and impairs the resorbing activity of mature osteoclasts. Oncotarget. 2015;6:27343–27358. doi: 10.18632/oncotarget.4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frankel B, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2008;283:1026–1033. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- 25.Yasuda M, Nishizawa T, Ohigashi H, Tanaka T, Hou DX, Colburn NH, Murakami A. Linoleic acid metabolite suppresses skin inflammation and tumor promotion in mice: Possible roles of programmed cell death 4 induction. Carcinogenesis. 2009;30:1209–1216. doi: 10.1093/carcin/bgp106. [DOI] [PubMed] [Google Scholar]
- 26.Loh PG, Yang HS, Walsh MA, Wang Q, Wang X, Cheng Z, Liu D, Song H. Structural basis for translational inhibition by the tumour suppressor Pdcd4. EMBO J. 2009;28:274–285. doi: 10.1038/emboj.2008.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujita S, Ito T, Mizutani T, Minoguchi S, Yamamichi N, Sakurai K, Iba H. miR-21 Gene expression triggered by AP-1 is sustained through a double-negative feedback mechanism. J Mol Biol. 2008;378:492–504. doi: 10.1016/j.jmb.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 28.Miao W, Gao H, Hou X. Magnesium lithospermate B inhibits titanium particles-induced osteoclast formation by c-fos and inhibiting NFATc1 expression. Connect Tissue Res. 2019;60:487–494. doi: 10.1080/03008207.2019.1593392. [DOI] [PubMed] [Google Scholar]
- 29.Talotta F, Cimmino A, Matarazzo MR, Casalino L, De Vita G, D'Esposito M, Di Lauro R, Verde P. An autoregulatory loop mediated by miR-21 and PDCD4 controls the AP-1 activity in RAS transformation. Oncogene. 2009;28:73–84. doi: 10.1038/onc.2008.370. [DOI] [PubMed] [Google Scholar]
- 30.Friedman AD. Transcriptional control of granulocyte and monocyte development. Oncogene. 2007;26:6816–6828. doi: 10.1038/sj.onc.1210764. [DOI] [PubMed] [Google Scholar]
- 31.Huhe M, Liu S, Zhang Y, Zhang Z, Chen Z. Expression levels of transcription factors c-Fos and c-Jun and transmembrane protein HAb18G/CD147 in urothelial carcinoma of the bladder. Mol Med Rep. 2017;15:2991–3000. doi: 10.3892/mmr.2017.6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haubrock M, Hartmann F, Wingender E. NF-Y binding site architecture defines a C-Fos targeted promoter class. PLoS One. 2016;11:e0160803. doi: 10.1371/journal.pone.0160803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sugatani T, Vacher J, Hruska KA. A microRNA expression signature of osteoclastogenesis. Blood. 2011;117:3648–3657. doi: 10.1182/blood-2010-10-311415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi Y, Massagué J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/S0092-8674(03)00432-X. [DOI] [PubMed] [Google Scholar]
- 35.Kokabu S, Gamer L, Cox K, Lowery J, Tsuji K, Raz R, Economides A, Katagiri T, Rosen V. BMP3 suppresses osteoblast differentiation of bone marrow stromal cells via interaction with Acvr2b. Mol Endocrinol. 2012;26:87–94. doi: 10.1210/me.2011-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen N, Sui BD, Hu CH, Cao J, Zheng CX, Hou R, Yang ZK, Zhao P, Chen Q, Yang QJ, et al. MicroRNA-21 contributes to orthodontic tooth movement. J Dent Res. 2016;95:1425–1433. doi: 10.1177/0022034516657043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.