Abstract
Background
Emerging evidence has demonstrated that SPOP functions as an oncoprotein in kidney cancer to promote tumorigenesis by ubiquitination-mediated degradation of multiple regulators of cellular proliferation and apoptosis. However, the detailed molecular mechanism underlying the oncogenic role of SPOP in kidney tumorigenesis remains elusive.
Methods
Multiple approaches such as Co-IP, Transfection, RT-PCR, Western blotting, and animal studies were utilized to explore the role of SPOP in kidney cancer.
Findings
Here we identified LATS1, a critical component of the Hippo tumour suppressor pathway, as a novel ubiquitin substrate of SPOP. We found that LATS1 interacted with Cullin3, and depletion of Cullin 3 upregulated the abundance of LATS1 largely via prolonging LATS1 protein half-life. Mechanistically, SPOP specifically interacted with LATS1, and promoted the poly-ubiquitination and subsequent degradation of LATS1 in a degron-dependent manner. As such, over-expression of SPOP promoted cell proliferation partly through regulating cell cycle distribution in kidney cancer cells. Furthermore, SPOP also promoted kidney cancer cell invasion via degrading LATS1.
Interpretation
Our study provides evidence for a novel mechanism of SPOP in kidney cancer progression in part through promoting degradation of the LATS1 tumour suppressor.
Keywords: Kidney, SPOP, LATS1, Growth, Ubiquitination, Degradation
Research in Context.
Evidence before this study
Kidney cancer is one of the common malignancies in the world.
SPOP plays a critical role in tumorigenesis and progression via ubiquitination and degradation of multiple substrates of SPOP.
SPOP functions as a tumour suppressor in prostate cancer, whereas SPOP acts as an oncoprotein in kidney cancer.
Added value of this study
Cullin3SPOP is the physiological E3 ubiquitin ligase for LATS1 and negatively regulates the protein stability of LATS1.
SPOP mediated ubiquitination and degradation of LATS1 in a degron-dependent manner.
CKΙδ promotes the interaction and degradation of LATS1 by SPOP.
SPOP promotes kidney cancer cell proliferation, invasion and regulates cell cycle in part via promoting the degradation of LATS1. SPOP overexpression accelerated tumour growth in mice.
Implications of all the available evidence
SPOP promotes kidney cancer progression via enhancement of degradation of the LATS1.
Targeting SPOP might be a novel strategy for the treatment of kidney cancer patients.
Alt-text: Unlabelled box
1. Introduction
Kidney cancer is the sixth leading malignancy in the United States, accounting for 5% of all cancer incidents. In 2018, it is expected that approximately 41,730 new cases and nearly 10,010 cancer-related deaths [1]. Clear-cell renal cell carcinoma (ccRCC) is the most common pathological subtype of RCC, which accounts for about 75% of all clinical cases [2]. Nearly 30% of kidney cancer patients present with metastases at the time of diagnosis and almost half of the rest will subsequently develop metastases in their disease progression [3]. As metastatic kidney cancer has been shown to be highly resistant to cytotoxic agents, the first-line treatment of metastatic disease in the 1990s and 2000s largely relied on interferon-alpha [4, 5] and high-dose interleukin-2, despite the response rate was only 5–20 % [6, 7]. Since 2005, several agents targeting mTOR and VEGFR/TKI pathways have been approved for the treatment of metastatic kidney cancer [8]. Unfortunately, most patients still inevitably develop resistance to targeted therapies, leading to tumour progression and eventual death [8]. Thus, it is urgent to comprehensively explore the molecular mechanisms of how kidney cancer develops and progresses and discover promising therapeutic approaches for this deadly disease.
The large tumour suppressor 1 (LATS1) acting as a tumour suppressor is a serine/threonine kinase of the AGC kinase family and is found to be down-regulated in various types of human cancers [9]. Recently, LATS1 has been identified as a central player of the emerging tumour suppressive Hippo signaling pathway. The Hippo pathway was originally discovered in Drosophila, which is a highly conserved pathway and plays a crucial role in regulation of biological processes such as organ size control, stem cell differentiation and renewal, drug resistance, and tumorigenesis [10, 11]. In this pathway, the MST1/2 (mammalian homolog of Drosophila Hippo) kinase phosphorylates activates LATS1/2 which, together with its co-factor MOB1, subsequently phosphorylates and inhibits the YAP (or TAZ, depending on cell context) transcriptional factor by preventing them from translocating to the nucleus [[12], [13]–14]. In addition to the critical role in the Hippo pathway, LATS1 also inhibits in vitro cell proliferation in part by regulating cell apoptosis and cell cycle progression. Ectopic expression of LATS1 induces cell apoptosis by promoting the BAX protein level. Furthermore, ectopic expression of LATS1 also down-regulates Cyclin A and Cyclin B protein levels and inhibits the kinase activity of CDC2, leading to a G2/M blockade [15]. Additionally, LATS1 is localized to the centrosome regulating actin that is necessary for efficient cell migration. As such, knockdown of LATS1 induces cell migration [9]. Thus, recent studies reveal that LATS1 functions as a tumour suppressor through several different mechanisms that negatively regulate tumour development.
Ubiquitin signaling regulates diverse cellular processes through controlling protein ubiquitination and degradation [16]. The protein ubiquitination process involves multistep enzymatic reactions catalyzed by a cascade of enzymes, including the ubiquitin-activating enzyme E1, the ubiquitin-conjugating enzyme E2, and the ubiquitin ligase E3. Ubiquitin ligase recognizes and catalyzes the ubiquitination of substrate proteins for targeted degradation through the 26S proteasome [17, 18]. Recently, it has been reported that Speckle-type POZ (pox virus and zinc finger protein) protein (SPOP) is an adaptor for Cullin 3-based E3 ligases (CRL3). Structurally, SPOP contains MATH and BTB domains: the C-terminal BTB domain that binds Cullin 3, and the N-terminal MATH domain that recruits substrates for ubiquitination [19].
Almost in all ccRCCs, it has been shown that SPOP is overexpressed and accumulated in the cytoplasm of ccRCC cells, whereas SPOP is largely a nucleoprotein in other cell types [20]. The ongoing list of SPOP substrates includes death domain–associated protein (Daxx) [21], the polycomb group protein BMI-1, and the histone variant MacroH2A [22]. SPOP plays a critical role in regulating cell apoptosis, proliferation and animal development. A more recent study showed that SPOP promotes tumorigenesis by ubiquitination and degradation of multiple regulators of cellular proliferation and apoptosis in kidney cancer [23]. However, in other cancer settings including prostate and endometrial cancers, SPOP probably functions largely as a tumour suppressor by ubiquitination and degradation of oncoproteins such as ERG [24, 25], PD-L1 [26], and BRD4 [27]. Recent deep sequencing studies found that SPOP is frequently mutated in prostate cancer with up to 15% mutation rate [28]. However, no SPOP mutation has been detected in kidney cancers thus far [20, 29]. Thus, the physiological role and expression level of SPOP in tumorigenesis are rather context dependent. Hence, we aim to elucidate the tumour promoting mechanism of SPOP in kidney cancer progression.
2. Material and methods
2.1. Cell culture
293T, T98G, and Hela cells were cultured in Dulbecco's Modification of Eagle's Medium (DMEM) (Corning, USA); U2OS and two ccRCC cell lines, 786-O, and A498, were grown in RPMI medium 1640 (Corning). All mediums were supplemented with 10% fetal bovine serum (FBS; Gibco) and 1% Penicillin/Streptomycin. All cells were incubated at 37°C and 5% CO2.
2.2. Antibodies
All antibodies were used at 1:1000 dilution in 5% non-fat milk for Western blot. Anti-SPOP antibody (16750-1-AP) was purchased from Proteintech. Anti-Cul3(2759), anti-LATS1(3477) and anti-CK1(12417) antibodies were purchased from Cell Signaling. Anti-Tubulin(T9028), anti-Actin-Peroxidase(A3854), anti-Flag(F1804) and anti-C-Myc(A5598) antibodies were purchased from Sigma. Peroxidase-conjugated anti-mouse secondary antibody (32430) and peroxides-conjugated anti-rabbit secondary antibody(31462) were purchased from Thermo. Anti-HA antibody (sc-805) was purchased from Santa Cruz Biotechnology.
2.3. Reagents
MG132 and cycloheximide (CHX) were purchased from Sigma. CK1 inhibitor IC261 (SC-3561) and D4476 (SC-202522) were purchased from Santa Cruz Biotechnology. The kidney cancer tissue microarray slides (HKid-CRC180Sur-01) was purchased from Shanghai Outdo Biotech Co., Ltd (Shanghai, China) for measuring the expression of SPOP and LATS1 by IHC staining.
2.4. Plasmids
Myc-tagged Cullins, Myc- tagged SPOP, pLenti-HA-SPOP WT, Myc-tagged CK1δ, CK1α1, CK1α2, CK1γ1, CK1γ2, CK1γ3, CK1ε, Flag-tagged LATS1, and His-tagged Ub were kindly offered by Dr. Wenyi Wei (Harvard Medical School). Various LATS1 mutants were generated in this study. Negative control siRNA and gene-specific siRNAs for human LATS1, CK1δ, Cullin3 were purchased from GenePharma (Shanghai, China). The siRNA transfection of cells was performed according to the manufacturer's instructions. The sequences of the siRNA oligonucleotides are as follows: siLATS1, sense 5′-GAG CUG GAA AGG UUC UAA ATT-3′, antisense 5′-UUU AGA ACC UUU CCA GCU CTT-3′; siCK1δ_1, sense 5′-GCU GCU UGC UGA CCA AAU GTT-3′, antisense 5′-CAU UUG GUC AGC AAG CAG CTT-3′; siCK1δ_2, sense 5′-GCA CCU UGG AAU UGA ACA ATT-3′, antisense 5′-UUG UUC AAU UCC AAG GUG CTT-3′; siCUL3_1, sense 5′-GCU UGG AAU GAU CAU CAA ATT-3′, antisense 5′-UUU GAU GAU CAU UCC AAG CTT-3′; siCUL3_2, sense 5′-CCA AGC ACA UGA AGA CUA UTT-3′, antisense 5′-AUAGUCUUCAUGUGCUUGGTT-3′; siCUL3_3, sense 5′-GGA GCA AGG UAA AGC UCU UTT-3′, antisense 5′-AAG AGC UUU ACC UUG CUC CTT-3′; Negative control (NC) siRNA, sense 5′-UUC UCC GAA CGU GUC ACG UTT3′, antisense 5′-ACG UGA CAC GUU CGG AGA ATT -3′. shRNA to deplete endogenous SPOP was purchased from Genechem Co., Ltd. (Shanghai, China).
2.5. qRT-PCR
The total RNA was extracted with Trizol (Invitrogen, Carlsbad, CA) and reversed-transcribed into cDNA by RevertAid First Strand cDNA Synthesis Kit (Thermo). qPCR was undertaken using Power SYBR Green PCR Master Mix and the results were quantified by 2−ΔΔCt method. The primers used in the PCR are as follows: SPOP, forward primer (5′-GCC CTC TGC AGT AAC CTG TC-3′) and reverse primer (5′-GTC TCC AAG ACA TCC GAA GC-3′); β-actin, forward primer (5′- GGA GAT TAC TGC CCT GGC TCC TA-3′) and reverse primer (5′-GAC TCA TCG TAC TCC TGC TTG CTG -3′); LATS1, forward primer (5′-AAA CCA GGG AAT GTG CAG CAA -3′) and reverse primer (5′-CAT GCC TCT GAG GAA CTA AGG A -3′).
2.6. Protein half-life assays
Cells were treated with indicated condition. For half-life studies, cycloheximide(CHX) was added to the medium with the concentration of 100 μg/ml. At indicated time points thereafter, cells were harvested and protein abundances were measured by Western blotting analysis.
2.7. Immunoblot and immunoprecipitation
Harvested cells were washed by PBS and lysed in protein lysis buffer supplemented with protease inhibitors tablets (Thermo) and phosphatase inhibitors (phosphatase inhibitors cocktail 1, Sigma). Protein concentration was determined using BCA reagent (Thermo Fisher). Same amount of protein samples were separated by electrophoresis in sodium dodecyl sulfonate (SDS)-polyacrylamide gel and transferred onto a nitrocellulose filter (NC) membrane (Amersham). The membrane was incubated in 5% nonfat dry milk/TBST for 1 h; and then incubated with the primary antibody at 4°C overnight. The membrane was washed with TBST for three times, followed by incubated with second antibody for 1 h at room temperature. Proteins of interest were measured by electrochemiluminescence (ECL) assay. For immunoprecipitation assay, 293T cells were transfected with indicated plasmids for 20 h and then treated with MG132 at concentration of 10μM for 10 h. Cells were washed with PBS and lysed in the IP lysis buffer (25mM Tris•HCL pH7.4, 150mM NaCl, 1mM EDTA, 1% NP-40, 5% glycerol, 1 × Thermo protease inhibitor). Protein concentration was determined using BCA reagent (Thermo Fisher) and then incubated 1000μg of cell lysate with the primary antibody-conjugated beads at 4°C for 4 h. The immunocomplexes were washed 3 times with IP lysis buffer before being resolved by SDS-PAGE and immunoblotted with the indicated antibodies.
2.8. In vivo ubiquitination
For the In vivo ubiquitination assay, 293T cells were transfected with plasmids for His-Ub, Flag-LATS1 and other indicated proteins. Cells were treated with 10μM MG132 for 10 h before they were harvested. Cells were washed with PBS and lysed with IP lysis buffer (25mM Tris·HCL pH7.4, 150mM NaCl, 1mM EDTA, 1% NP-40, 5% glycerol, 1 × Thermo protease inhibitor). 1000μg of cell lysate were incubated with 2μg LATS1 antibody at 4°C for 4 h and then incubated with Protein A/G plus agarose overnight. Beads were washed with lysis buffer for 3 times and detected by SDS-PAGE.
2.9. In vitro ubiquitination
This assay was carried out via incubation of the purified proteins including E1, E2 (UbcH5b), ubiquitin, GST-LATS1, SPOP-Cul3-ROC1 complex in ubiquitination buffer for reactions at 37°C for 1 h. The reactions were stopped by the addition of 1% SDS and the reaction products were resolved by SDS-PAGE and probed with the indicated antibodies.
2.10. Immunohistochemistry
Renal cancer tumour microarray slides (HKid-CRC180Sur-01) or 786-O xenograft tumours were deparaffinized, dehydrate and incubated in heat-mediated antigen retrieval. Subsequently, slides were cooled to RT and incubated with 3% H2O2 for 10 min to block endogenous peroxidase activity. After washing, slides were incubated in normal bovine serum (biosharp) to block non-specific binding of IgG. Then, slides were treated with primary antibody LATS1 (abcam, ab234820, 1:500) and SPOP (protein-tech, 16750-1-AP, 1:500) at 4°C overnight. Slides were washed and incubated with streptavidin-conjugated horseradish peroxide (Gene Tech) in PBS for 1 hour at RT. After washing with PBS for 3 times, slides were treated with DAB (Gene Tech) for 5 min. Images were acquired by the Olympus camera and matched software. IHC straining was scored by two independent pathologists on the basis of the “most common” criteria.
2.11. Cell proliferation assays
Cells were seeded in a 96-well plate (5 × 103 cells/well). At the indicated time points, cell proliferation was performed by the CellTiter-Glo Luminescent Cell Viability Assay (CTG, Promega) according to the manufacture's instruction. Independent experiments were repeated in triplicate.
2.12. Cell cycle analysis
Renal cancer cells were seeded in a 6-well plate (3 × 105 cells/well). After 48h, cells were harvested and suspended with 70% cold alcohol for overnight at 4°C. Thereafter, cells were washed with PBS and re-suspended with PBS mixing with 0.1mg/ml RNase I and 50 mg/ml PI at 37°C in the dark room for 30 min. The cell cycle distribution was measured using a FACScalibur flow cytometer (BD, USA).
2.13. Cell invasion assay
For cell invasion assay, 1 × 104- 1 × 105 cells were placed on each upper chamber with 200 μL of serum-free medium and 500μl of medium containing 10% FBS at the bottom. Cells were incubated for 24 h at 37°C in 5% CO2. After washed with PBS, cells in the upper surface of the insert were removed with cotton swabs. Cells attached on the bottom were stained with Wright's-Giemsa for 5 min and washed with ddH2O. The stained cells were photographed and counted under a light microscope in 5 fields with random choice.
2.14. Bioinformatics analysis
The correlation of SPOP expression with the overall survival of KIPR (Kidney renal papillary cell carcinoma) patients initially assessed using the OncoLnc database (www.oncolnc.org).
2.15. Mouse xenograft assay
For mouse xenograft assay, six-week-old BALB/c-nu/nu mice were randomly divided into two different experimental groups. Renal cancer cells were collected and suspended in 100μl PBS mixing with Matrigel (BD 356234, 2:1) and injected into the nude mouse (5 mice for each group). The volume of tumour was measured with a vernier caliper every 4d and calculated by the formula: L × W2 × 0.52 (L: the longest diameter of the tumour, W: the shortest diameter of the tumour). After 40 days, mice were killed and the tumours were dissected and weighted. The animal studies were conducted in accordance with guidelines and approved by Animal Experimentation Ethical Committee of Soochow University (Suzhou, Jiangsu, China).
2.16. Statistical analysis
GraphPad Prism 5 was used for analysis. All results were shown as the means ± S.D. A two-tailed independent student′s t-test and ANOVA were performed to analyze statistical associations between two groups and multiple groups, respectively. P < 0.05 was considered to be statistically significant.
3. Results
3.1. Cullin3SPOP is the physiological E3 ubiquitin ligase for LATS1
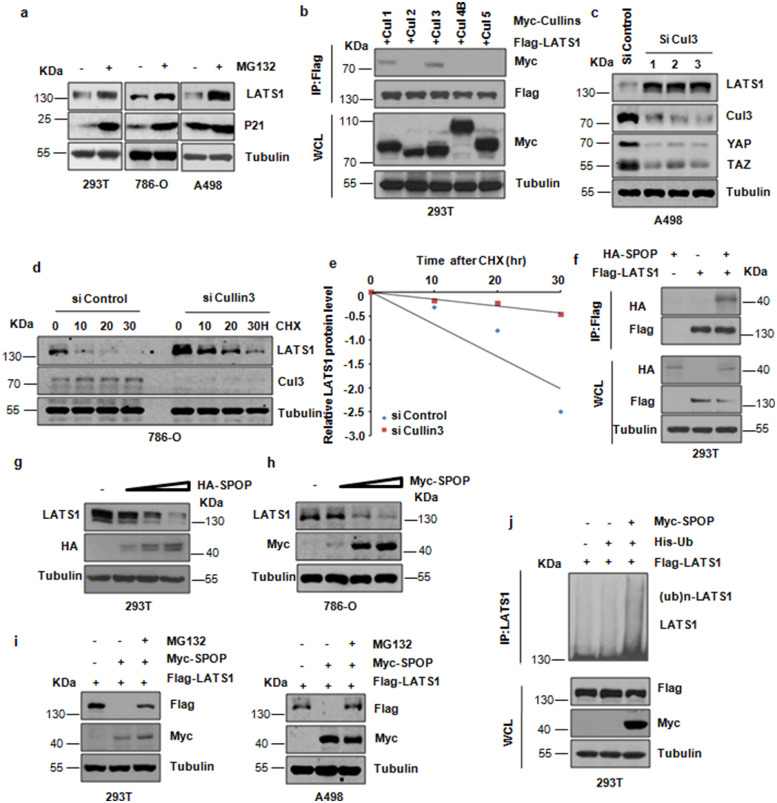
Given the important role of LATS1 in kidney cancer progression [30], it is crucial to explore how LATS1 protein level is regulated at physiological conditions and misregulated in kidney cancer setting. Firstly, we detected that proteasome inhibitor MG132 could stabilize the endogenous LATS1 protein level (Fig. 1a and Supplementary Fig. 1a). In keeping with this notion, we found that LATS1 interacted specifically with Cullin 3 (CUL3) and Cullin 1 (CUL1), but not other members of Cullin family (Fig. 1b), suggesting that CUL3 and CUL1 E3 liagses might be involved in LATS1 ubiquitination and degradation. The role of CUL1 E3 ligase in regulation of LATS1 stability needs further investigation. In keeping with a critical role of Cullin 3 in regulating LATS1 protein stability, depletion of Cullin 3 promoted the abundance of LATS1 (Fig. 1c and Supplementary Fig. 1b). Moreover, depletion of endogenous CUL3 dramatically extended protein half-life of LATS1 (Fig. 1d,e and Supplementary Fig, 1c,d). On the other hand, overexpression of CUL3 reduced the protein half-life of LATS1 (Supplementary Fig, 1e,f). Notably, we observed that LATS1 interacted specifically with SPOP in cells (Fig. 1f and Supplementary Fig. 1g and 1j). In keeping with this notion, we confirmed that overexpression of SPOP down-regulated the LATS1 expression in a dose-dependent manner (Fig. 1g,h and Supplementary Fig. 1h), a process that can be blocked by MG132 treatment (Fig. 1i and Supplementary Fig. 1i). We further demonstrated that SPOP promoted the ubiquitination of LATS1 in cells (Fig. 1j). Together, these results suggest that LATS1 is a potential substrate of the Cullin3/SPOP E3 ligase.
Fig. 1.
Cullin3SPOP is the physiological E3 ubiquitin ligase for LATS1. (a) IB analysis of WCLs derived from 293T, 786-O and A498 cells treated with 10μM MG132 for 10 hr. (b) IB analysis of immunoprecipitates (IPs) and WCLs derived from 293T kidney cells transfected with indicated constructs. Cells were treated with MG132 (10 μM) before harvesting. (c) IB analysis of WCLs derived from A498 cells transfected with Cullin3 siRNA. (d) IB analysis of WCLs derived from 786-O cells after the specified duration of 100 μg/ml cycloheximide (CHX) transfected with Cullin3 siRNA. (e) The abundance of LATS1 protein in (d) was quantified and plotted. (f) IB analysis of WCLs and immunoprecipitates (IPs) derived from 293T cells transfected with indicated constructs. Cells were treated with MG132 (10μM) before harvesting. (g) IB analysis of WCLs derived from 293T kidney cells transfected with increasing doses of plasmid encoding SPOP. (h) IB analysis of WCLs derived from 786-O cells transfected with increasing doses of plasmid encoding SPOP. (i) IB analysis of WCLs derived from 293T kidney cells or A498 cells transfected with indicated constructs. Where indicated, cells were treated with 10μM MG132 before harvesting. (j) IB analysis of WCLs and products of ubiquitination derived from 293T cells transfected with indicated constructs.
3.2. Cullin3SPOP E3 ubiquitin ligase negatively regulates the protein stability of LATS1
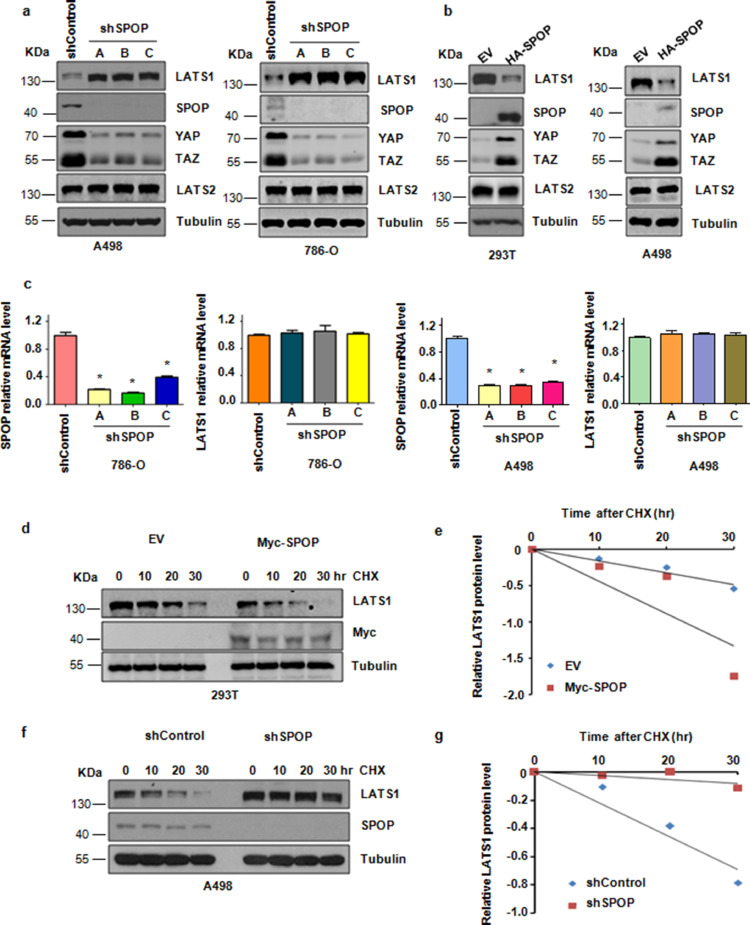
Consistent with an important role for SPOP in modulating the stability of LATS1, we demonstrated that depletion of endogenous SPOP with several different small hairpin RNA (shRNA) markedly elevated the protein abundance of LATS1 in multiple cell lines (Fig. 2a). On the other hand, overexpression of SPOP led to a noticeable decrease in the abundance of LATS1 (Fig. 2b). Notably, we confirmed that in multiple cell lines, depletion of endogenous SPOP with shRNA barely changed the mRNA levels of LATS1 (Fig. 2c). Moreover, the half-life of LATS1 was markedly extended with depleting of endogenous SPOP protein, whereas overexpression of SPOP reduced the protein half-life of LATS1 (Fig. 2d–g). These data collectively support the notion that SPOP regulates the protein stability of LATS1 mainly through a post-translational mechanism.
Fig. 2.
Cullin3SPOP E3 ubiquitin ligase negatively regulates the protein stability of LATS1. (a) IB analysis of WCLs derived from A498 or 786-O cells infected with the indicated lentiviral shRNA vectors. (b) IB analysis of WCLs derived from 293T or A498 cells transfected with HA-tagged SPOP plasmid. (c) Quantitative real-time PCR (qRT-PCR) analysis to detect LATS1 and SPOP mRNA levels derived from 786-O or A498 cells after depletion of SPOP. Data are shown as mean ±SD of three independent experiments. *P<0.05, Student's t test. (d) IB analysis of WCLs derived from 293T cells after the specified duration of cycloheximide (CHX) transfected with Myc-tagged SPOP plasmid. (e) The abundance of LATS1 protein in (d) was quantified and plotted. (f) IB analysis of WCLs derived from A498 cells after the specified duration of 100μg/ml cycloheximide (CHX) infected with indicated lentiviral shRNA vectors. (g) The abundance of LATS1 protein in (f) was quantified and plotted.
3.3. SPOP-mediated ubiquitination and degradation of LATS1 depends on the degron motif
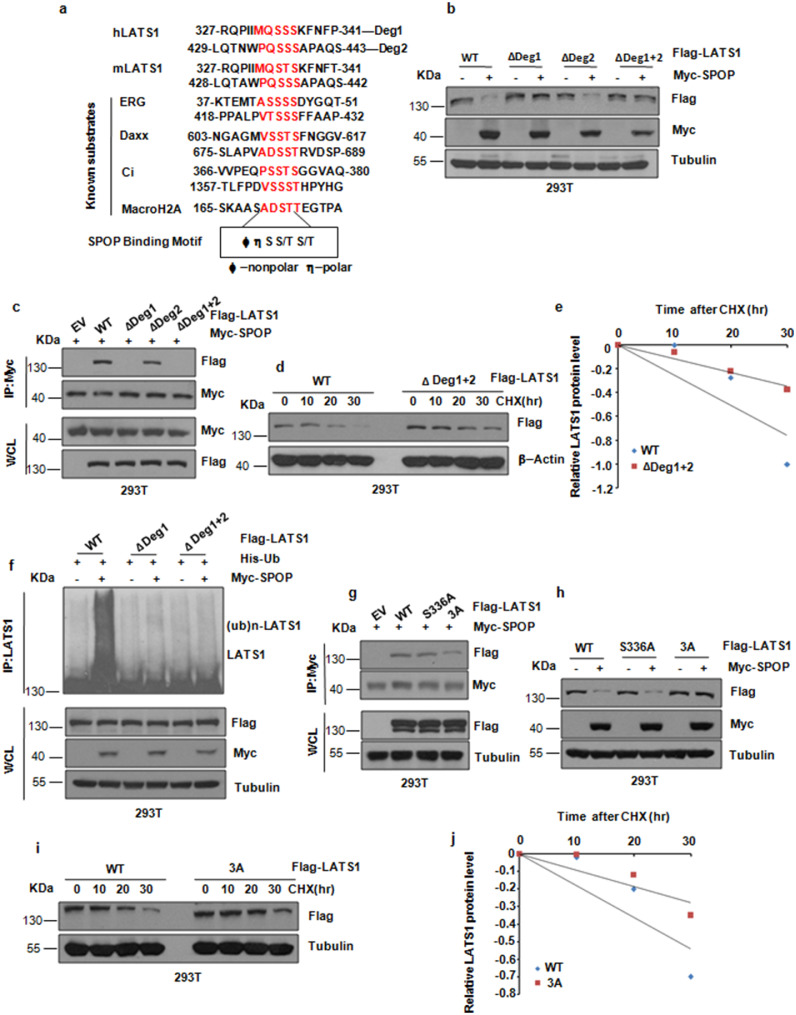
As SPOP contains two conserved domains: the N-terminal conserved MATH domain could recognize its ubiquitin substrate, and the C-terminal BTB domain could bind CRL scaffold protein Cullin3[19]. Notably, we demonstrated that the SPOP mutant with deletion of the MATH domain failed to bind to LATS1 (Supplementary Fig. 2a,b). Moreover, either loss of MATH or BTB domain of SPOP prevented the degradation of LATS1 (Supplementary Fig. 2c). Consistent with previous reports that most of SPOP substrates share a SPOP-binding consensus motif Φ-Π-S-S/T-S/T (Φ-nonpolar; Π, polar), we analyzed LATS1 protein sequence and found two putative motifs, or “degrons” located in the N-terminus of LATS1 (Fig. 3a). Notably, deletion of degron1 (ΔDeg1), and to a lesser extent of degron2 (ΔDeg2), largely blocked SPOP-mediated degradation of LATS1, whereas deletion of both degron1 and 2 (ΔDeg1+2) nearly abolished the LATS1 degradation of SPOP-mediated (Fig. 3b). Consistently, deletion of degron 1 or both degrons dramatically attenuated the interaction of LATS1 with SPOP in cells (Fig. 3c). Moreover, ΔDeg1+2 and ΔDeg1 mutation exhibited resistance to SPOP-mediated destruction (Fig. 3d,e and Supplementary Fig. 2d,e) and poly-ubiquitination (Fig. 3f) in cells. Mutagenesis studies demonstrated that the serine 336-to alanine mutation (S336A) in ΔDeg1 rarely attenuated the interaction of LATS1 with SPOP, but mutating Ser334, Ser335, Ser336 to alanine (LATS1-3A) dramatically attenuated the interaction of LATS1 with SPOP in cells (Fig. 3g). Consistently, the LATS1-3A mutation exhibited resistance to SPOP-mediated degradation in cells (Fig. 3h,j and Supplementary Fig. 2f). Notably, our in vitro ubiquitylation assay result confirmed that LATS1 is a direct ubiquitylation substrate of SPOP (Supplementary Fig. 2g).
Fig. 3.
SPOP-mediated ubiquitination and degradation of LATS1 depends on the degron motif. (a) Amino acid sequence alignment of LATS1 with the SPOP–binding motif (degron) in known substrates of SPOP. (b) IB analysis of WCLs derived from 293T cells transfected with indicated plasmids. (c) IB analysis of WCLs and immunoprecipitates (IPs) derived from 293T cells transfected with indicated plasmids and treated with 10μM MG132 for 10 h before harvesting. (d) IB analysis of WCLs derived from 293T cells after the specified duration of 100μg/ml cycloheximide (CHX) transfected with indicated Flag-tagged LATS1 plasmids. (e) The abundance of LATS1 protein in (d) was quantified and plotted. (f) IB analysis of WCLs and immunoprecipitates (IPs) derived from 293T cells transfected with indicated plasmids and treated with 10μM MG132 for 10 hr before harvesting. (g) IB analysis of WCLs and immunoprecipitates (IPs) derived from 293T cells transfected with indicated plasmids and treated with 10μM MG132 for 10 h before harvesting. (h) IB analysis of WCLs derived from 293T cells transfected with indicated plasmids. (i) IB analysis of WCLs derived from 293T cells after the specified duration of 100μg/ml cycloheximide (CHX) transfected with indicated Flag-tagged LATS1 plasmids. (j) The abundance of LATS1 protein in (i) was quantified and plotted.
3.4. CKΙδ promotes the interaction and degradation of LATS1 by SPOP
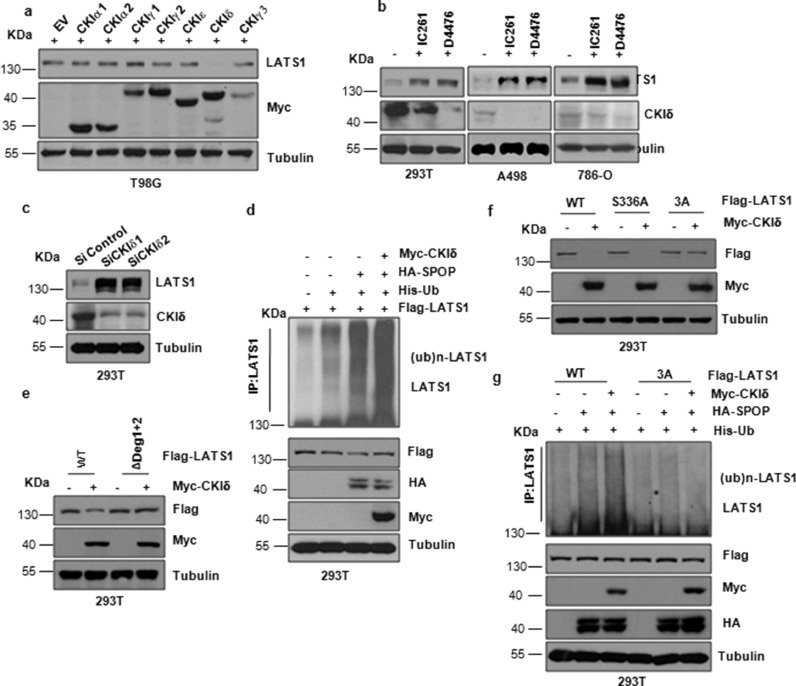
It has been previously reported that proper substrate phosphorylation is necessary before substrate ubiquitination and degradation by SCF type of E3 ligases including FBW7 and β-TRCP. Consistently, recent studies have identified that SPOP promoted SRC-3 and ERG degradation in a casein kinase Ι ε (CKΙε) -dependent and casein kinase Ι δ (CKΙδ)-dependent manner [25, 31]. Given that the SPOP recognizable degron in LATS1 contains several putative CKI phosphorylation sites, we aimed to detect whether CKΙ also involves in SPOP-mediated degradation of LATS1. Notably, we observed that only CKΙδ, but not other CKΙ, could promote LATS1 degradation under overexpression conditions (Fig. 4a). Moreover, depletion of CKΙδ with its siRNA or inhibitors, D4476 and IC261, dramatically elevated the protein level of LATS1 in cells (Fig. 4b,c). More importantly, co-overexpression of CKΙδ in cells enhanced the association of LATS1 with SPOP to promote the ubiquitination of LATS1 (Fig. 4d). Moreover, the LATS1-ΔDeg1+2 and LATS1-3A mutant, but not the LATS1-S336A, exhibited resistance to CKΙδ-mediated LATS1 degradation (Fig. 4e,f). In support of this notion, the ubiquitination of LATS1-3A mutant was reduced in cells (Fig. 4g). In addition, we found that MTS1 kinase that phosphorylates LATS1 did not involve in SPOP-mediated degradation of LATS1 (Fig. S2h).
Fig. 4.
CKΙδ promotes the interaction and degradation of LATS1 by SPOP. (a) IB analysis of WCLs derived from 293T cells transfected with indicated plasmids. (b) IB analysis of WCLs derived from 293T, A498 and 786-O cells incubated with CKΙ inhibitor IC261 (50μM) or D4476 (20μM) before harvesting. (c) IB analysis of WCLs derived from kidney derived 293T cells transfected with CKΙδ siRNA. (d) IB analysis of WCLs and immunoprecipitates (IPs) derived from 293T kidney cells transfected with indicated plasmids and treated with 10μM MG132 for 10 hr before harvesting. (e) IB analysis of WCLs derived from 293T cells transfected with indicated plasmids. (f) IB analysis of WCLs derived from 293T cells transfected with indicated plasmids. (g) IB analysis of WCLs and immunoprecipitates (IPs) derived from 293T cells transfected with indicated plasmids and treated with 10μM MG132 for 10 h before harvesting.
3.5. The biological function of SPOP in kidney cancer cells
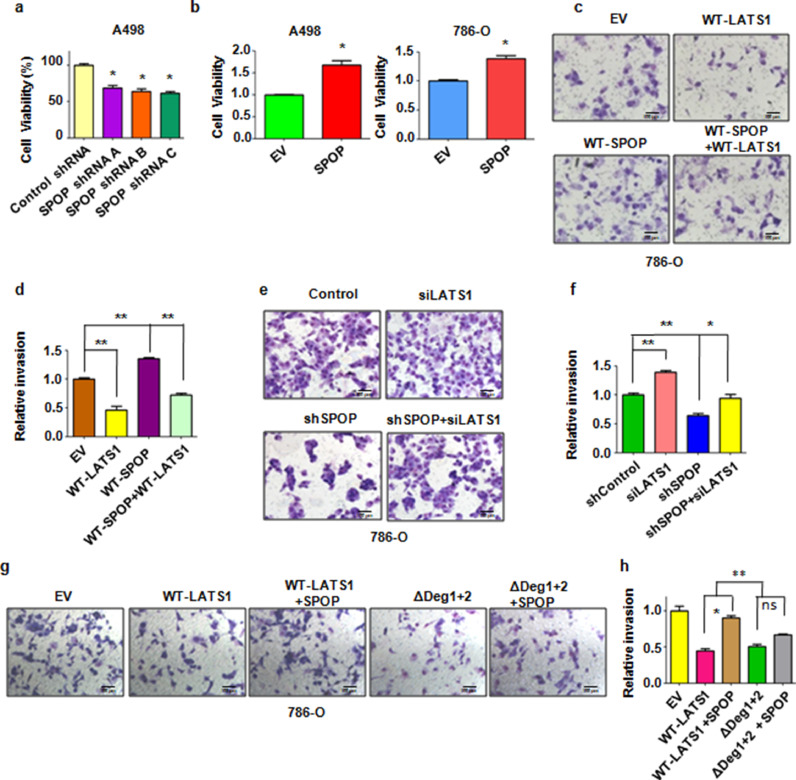
It has been previously reported that SPOP plays a critical role in regulating cell apoptosis, proliferation and animal development through mediating Daxx, BMI-1, and MacroH2A [21, 22]. On the other hand, SPOP could promote tumorigenesis by ubiquitination and degradation of multiple regulators of cellular proliferation and apoptosis in kidney cancer [23]. One recent study demonstrated that SPOP promoted renal cell carcinoma cell epithelial-mesenchymal transition (EMT) and enhanced cell invasion via activation of β-catenin/TCF4 complex [32]. Therefore, we intended to further explore the biological function of SPOP in kidney cancer cells. To this end, we observed that depletion of SPOP decreased cell proliferation in A498 kidney cancer cells (Fig. 5a). On the other hand, overexpression of SPOP promoted cell proliferation in A498 cells and 786-O cells (Fig. 5b). Moreover, we found that overexpression of SPOP enhanced cell invasion (Fig. 5c,d). In contrast, overexpression of LATS1 reduced cell invasion (Fig. 5c,d). Notably, overexpression of SPOP rescued cell invasion inhibition induced by overexpression of LATS1 (Fig. 5c,d). On the other hand, we observed that depletion of SPOP reduced cell invasion which can be rescued by additional depletion of LATS1 (Fig. 5e,f). These results suggest that SPOP downregulates cell invasion partly through modulating LATS1 protein abundance. Moreover, co-expression with SPOP suppressed LATS1-WT but not ΔDeg1+2-mediated inhibition of cell invasion (Fig. 5g,h). Since ShSPOP infection still decreased cell invasion after downregulation of LATS1, indicating that LATS1 might not be the main downstream of SPOP in regulation of cell invasion in ccRCC.
Fig. 5.
SPOP promote the proliferation and invasion of kidney cancer cells. (a) The CTG assay was used to detect the proliferation capability of A498 kidney cancer cells infected with SPOP lentiviral shRNA. Data are shown as mean ±SD of three independent experiments. *P<0.05. (b) The CTG assay was used to detect the proliferation capability of A498 and 786-O kidney cancer cells transfected with SPOP plasmids. Data are shown as mean ±SD of three independent experiments. *P<0.05. (c) Transwell chambers assay was performed to detect the invasion capability of 786-O kidney cancer cells transfected with the indicated plasmids. (d) The quantification of invasion cells in (c). Data are shown as mean ±SD of three independent experiments. **P<0.01. (e) Transwell chambers assay was performed to detect the invasion capability of 786-O kidney cancer cells infected with the indicated lentiviral shRNA or transfected with the indicated siRNA. (f) The quantification of invasion cells in (e). Data are shown as mean ±SD of three independent experiments. *P<0.05, **P<0.01. (g) Transwell chambers assay was performed to detect the invasion capability of 786-O kidney cancer cells transfected with the indicated plasmids. (h) The quantification of invasion cells in (g). Data are shown as mean ±SD of three independent experiments. *P<0.05, **P<0.01. ns: no significant difference.
It has been previously reported that ectopic expression of LATS1 induced cells at G2/M arrest. Thus, we also detected the biological function of SPOP on cell cycle. We found that depletion of SPOP induced cells at G2/M arrest. Notably, simultaneous depletion of SPOP and LATS1 reduced cells populations at G2/M phase compared with single depletion of LATS1 (Supplementary Fig. 3a). In keeping with a possible role of SPOP in regulating cell cycle via LATS1, we found that co-expression with CKΙδ suppressed LATS1-WT-mediated G2/M arrest (Supplementary Fig. 3b). Moreover, co-expression with SPOP suppressed LATS1-WT, but not ΔDeg1+2, -mediated G2/M arrest (Supplementary Fig. 3c). These findings are consistent with the expression of LATS1 and its downstream targets, YAP and TAZ in cells after dysregulation of SPOP and LATS1 (Supplementary Fig. 3d–f). Together, these results suggest that SPOP promotes kidney cancer cell proliferation, invasion and regulates cell cycle in part via promoting the degradation of LATS1.
3.6. SPOP promotes tumorigenesis
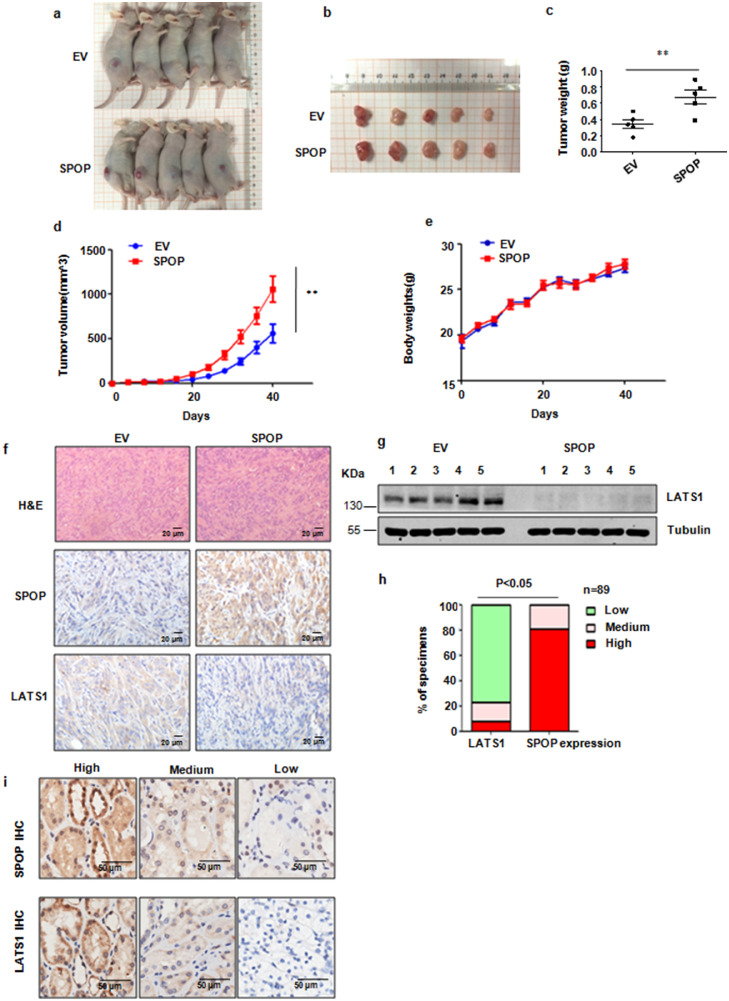
It has been shown that SPOP could promote kidney tumorigenesis [23]. In agreement with this study, we observed that overexpression of SPOP in 786-O cells dramatically elevated the growth of tumour xenografts in mouse models (Fig. 6a–e). We performed Western blotting of LATS1 and immunohistochemistry (IHC) of SPOP and LATS1 on the xenografted tumours and found that LATS1 was down-regulation in the SPOP overexpression xenografted tumours (Fig. 6f,g). On the other hand, depletion of SPOP by shRNA-mediated knockdown in 786-O cells suppressed the growth of tumour xenografts in mouse models (Supplementary Fig. 4a–e). To further investigate the clinical relevance between SPOP and LATS1 in kidney cancer, we monitored SPOP and LATS1 IHC signals in kidney cancer tissues. We found that SPOP was overexpressed in most of kidney cancer clinical tissues. Moreover, we identified an inverse correlation between SPOP and LATS1 expression in kidney cancer tissue cytoplasm (Fig. 6h,i). Therefore, it is necessary to further investigate the associations between the expression levels of SPOP, LATS1, Cullin 3 and survival in renal cancer patients.
Fig. 6.
SPOP promotes tumorigenesis. (a) 786-O-pcDNA3.1, 786-O-SPOP polyclonal stable kidney cancer cell lines were injected subcutaneously into the BALB/c-nu/nu mice. 40 days later, mice were anesthetic and taken a picture. (b) The tumours were dissected and taken a picture. (c) The weights of the dissected tumours in (b). (d) In vivo tumour growth was measured over the indicated time period. (e) The body weights of the BALB/c-nu/nu mice are measured over the indicated time period. (f) Immunohistochemistry staining of SPOP and LATS1 in tissue sections of xenografted tumours in BALB/c-nu/nu mice injected with the indicated cell lines. Scale bars, 20 μm. (g) IB analysis of the LATS1 protein levels in the dissected tumours. (h) Statistical analyses of the correlation between SPOP and LATS1 expression in cytoplasm of the kidney cancer tissue microarray. (i) Representative images of SPOP and LATS1 IHC from 89 cases of kidney cancer.
4. Discussion
Here we identified that the Cullin3/SPOP E3 ligase mediates the abundance of LATS1 protein through promoting its ubiquitination and subsequent destruction. Moreover, previous studies have shown that CKΙ was involved in SRC-3 and ERG degradation mediated by SPOP [25, 31]. In line with those studies, our results demonstrated that only CKΙδ, but not other CKΙ, could promote LATS1 degradation under overexpression conditions. One study showed that long non-coding RNA cytoskeleton regulator RNA (CYTOR) could bind to cytoplasmic β-catenin and prevent CK1-mediated β-catenin phosphorylation, leading to its translocation to the nucleus [33]. Another study revealed that lncRNA01638 impeded SPOP-involved c-Myc degradation in breast cancer [34]. These two studies indicate that lncRNAs could be involved in SPOP-mediated tumorigenesis, which need to be further investigated.
Previous studies demonstrate that SPOP functions as either a tumour suppressor or an oncogene in context-depending [35]. It is implicated that SPOP functions as a tumour suppressor in the prostate cancer because there is a high mutation rate in prostate cancer and mutations is likely inactive [36], [37], [38], [39]. However, there is no detected mutation of SPOP in kidney cancers so far [20, 29]. One study has revealed that SPOP protein and mRNA level were highly expressed in ccRCC compared to normal kidney tissues [32]. Moreover, SPOP was associated with tumour metastasis, tumour recurrence-free survival of ccRCC [32]. Similarly, another study confirmed that positive expression of SPOP and ZEB1 in addition to negative E- cadherin was associated with poor prognosis in ccRCC patients [40].
Importantly, SPOP is a direct transcriptional target of HIFs (hypoxia-inducible factors) in ccRCC. Hypoxia led to cytoplasmic accumulation of SPOP and subsequently caused tumorigenesis, which is through targeting PTEN, ERK phosphatases, Daxx, and Gli2 in ccRCC [23]. RCC cell lines express either both HIF1α and HIF2α or HIF2α that affect the expression of SPOP. For instance, both 786-O and A498 cells predominantly express HIF2α, while Caki-2 cells express both HIF1α and HIF2α [23]. Knockdown of HIF1α in Caki-2 cells, or knockdown of HIF2α in A498 cells, or double-knockdown of both HIF1α and HIF2α in Caki-2, leads to a reduction of SPOP expression at mRNA and protein levels, suggesting that both HIF1α and HIF2α regulate SPOP expression [23]. Mechanistically, SPOP drove EMT and promoted cell invasion via enhancement of β-catenin protein expression and its nuclear translocation and upregulation of TCF4 in ccRCC [32]. Both TCF4 and β-catenin could regulate the ZEB1 expression in ccRCC cells [32]. In addition, PTEN and ERK governed the expression of β-catenin, and mediated EMT in human cancer [41, 42]. Thus, these studies indicate that SPOP mediated cell invasion in part via targeting PTEN and ERK, and activation of β-catenin/TCF4 and subsequent upregulation of ZEB1. Moreover, previous studies revealed that SPOP always misallocated to the cytoplasm which is a nucleoprotein originally [23, 43]. Our results also revealed that SPOP is overexpressed in the cytoplasm in most of the ccRCC clinical tissues (Fig. 6h,i). It has been reported previously that over-expression of SPOP is an unfavorable prognostic indicator in ccRCC patients according to survival regression estimates [32]. Thus, our findings identify that SPOP function as an oncogene in kidney cancer.
It is generally accepted that as a tumour suppressor, LATS1 suppresses cell proliferation through inducing cell apoptosis and G2/M phase arrest [44]. Several E3 ubiquitin ligases including ITCH, WWP1, DCAF1, and NEDD4 have been reported to regulate stability of LATS1 [[45], [46], [47]–48]. Here, we identified SPOP is a new E3 ligase of LATS1 in ccRCC cells. Moreover, we identified that depletion of endogenous SPOP by shRNA induced G2/M arrest via elevating LATS1 protein abundance. In line with this, we observed that overexpression of SPOP could promote cell proliferation partly through regulating cell cycle distribution in A498 cells and 786-O cells (Fig. 5b). On the other hand, SPOP could promote kidney cancer cell invasion in part via regulating LATS1. Therefore, our studies identified E3 ubiquitin ligase Cullin3/SPOP mediated the stability of the tumour suppressor LATS1 through poly-ubiquitination and subsequent degradation of LATS1 in kidney cancer in a degron-dependent manner. It is worth noting that SPOP also targets PTEN, DUSP7, Daxx, and Gli2 in ccRCC [23], thereby suggesting that SPOP could exert its oncogenic function in part via LATS1 pathway.
Together, in this study we demonstrate that SPOP acts as an oncoprotein in kidney cancer. It is worthy to mention that there are several limitations in this study. The basal level of SPOP expression in multiple ccRCC cell lines needs to be measured by Western blotting analysis. The effects of CKIδ inhibitors on the viability and invasion potential of ccRCC cells should be determined. In addition, the expression levels of SPOP in ccRCC cells with different treatments in various control groups, including shRNA control infection and empty vector transfection, are variable. This could be in part due to different exposure times in Western blotting to show the remarkably differences between control and treatment groups. Since LATS1 is a tumour suppressor in ccRCC, it is better to target its upstream regulators such as SPOP to upregulate LATS1 expression. Thus, targeting SPOP in ccRCC is an important strategy for the treatment of ccRCC patients. In fact, several small molecules yield by structure-based design and subsequent hit optimization have been identified to inhibit the SPOP-substrate protein interaction and to suppress oncogenic SPOP pathways [49]. Further investigation is required to discover the novel inhibitors of SPOP including natural compounds as well as its anticancer potential, at least in kidney cancer with elevated SPOP expression. Taken together, our study provided new insight into the molecular mechanisms of SPOP-driven ccRCC progression, in which SPOP promotes that ubiquitination and subsequent degradation of the LATS1 tumour suppressor protein.
Author contributions
L.W. conceived the work, designed and performed the experiments, analyzed the data, and wrote the manuscript. M.L., M.C., and Y.L. performed the experiments and analyzed the data. J.M and Y.H. analyzed the data and critically viewed. ZW.W. wrote the manuscript, and critically viewed and supervised the study. All authors read and approved the final manuscript.
Declaration of Competing Interest
The authors declare that they have no conflict of interest.
Funding sources
This work was supported by grant from National Natural Science Foundation of China (NSFC 81773186) and the major project from the Natural Science Foundation of Education Department of Anhui Province (KJ2019ZD27). The funders play no role in study design, data collection, data analysis, interpretation, and writing of the report.
Footnotes
Abbreviations: BRD4, bromodomain containing 4; ccRCC, Clear-cell renal cell carcinoma; CDC2, cell division cycle 2; CKI, casein kinase I; CRL3, Cullin 3-based E3 ligases; CYTOR, cytoskeleton regulator RNA; Daxx, death domain-associated protein; DMEM, Dulbecco's Modification of Eagle's Medium; EMT, epithelial-mesenchymal transition; KIPR, Kidney renal papillary cell carcinoma; LATS1, Large tumour suppressor 1; mTOR, mammalian target of rapamycin; PD-L1, programmed death ligand 1; SPOP, Speckle-type pox virus and zinc finger protein; TAZ, tafazzin; transcriptional co-activator with a PDZ-binding domain; VEGFR, vascular endothelial growth factor receptor; YAP, Yes-associated protein.
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2020.102795.
Contributor Information
Jia Ma, Email: majiamj10@126.com.
Youhua He, Email: feymnwk@126.com.
Zhi-wei Wang, Email: zwang6@bidmc.harvard.edu, zhiweichina@126.com.
Appendix. Supplementary materials
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Lopez-Beltran A, Scarpelli M, Montironi R. WHO classification of the renal tumors of the adults. Eur Urol. 2006;49(5):798–805. doi: 10.1016/j.eururo.2005.11.035. [DOI] [PubMed] [Google Scholar]
- 3.Hollingsworth JM, Miller DC, Daignault S. Five-year survival after surgical treatment for kidney cancer: a population-based competing risk analysis. Cancer. 2007;109(9):1763–1768. doi: 10.1002/cncr.22600. [DOI] [PubMed] [Google Scholar]
- 4.Interferon-alpha and survival in metastatic renal carcinoma: early results of a randomised controlled trial. Med Res Council Renal Cancer Collab Lancet. 1999;353(9146):14–17. [PubMed] [Google Scholar]
- 5.Pyrhonen S, Salminen E, Ruutu M. Prospective randomized trial of interferon alfa-2a plus vinblastine versus vinblastine alone in patients with advanced renal cell cancer. J Clin Oncol. 1999;17(9):2859–2867. doi: 10.1200/JCO.1999.17.9.2859. [DOI] [PubMed] [Google Scholar]
- 6.Fyfe G, Fisher RI, Rosenberg SA. Results of treatment of 255 patients with metastatic renal cell carcinoma who received high-dose recombinant interleukin-2 therapy. J Clin Oncol. 1995;13(3):688–696. doi: 10.1200/JCO.1995.13.3.688. [DOI] [PubMed] [Google Scholar]
- 7.McDermott DF, Regan MM, Clark JI. Randomized phase III trial of high-dose interleukin-2 versus subcutaneous interleukin-2 and interferon in patients with metastatic renal cell carcinoma. J Clin Oncol. 2005;23(1):133–141. doi: 10.1200/JCO.2005.03.206. [DOI] [PubMed] [Google Scholar]
- 8.Koshkin VS, Rini BI. Emerging therapeutics in refractory renal cell carcinoma. Expert Opin Pharmacother. 2016;17(9):1225–1232. doi: 10.1080/14656566.2016.1182987. [DOI] [PubMed] [Google Scholar]
- 9.Visser S, Yang X. LATS tumor suppressor: a new governor of cellular homeostasis. Cell Cycle. 2010;9(19):3892–3903. doi: 10.4161/cc.9.19.13386. [DOI] [PubMed] [Google Scholar]
- 10.Tremblay AM, Camargo FD. Hippo signaling in mammalian stem cells. Semin Cell Dev Biol. 2012;23(7):818–826. doi: 10.1016/j.semcdb.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 11.Zhao B, Tumaneng K, Guan KL. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat Cell Biol. 2011;13(8):877–883. doi: 10.1038/ncb2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bao Y, Hata Y, Ikeda M. Mammalian Hippo pathway: from development to cancer and beyond. J Biochem. 2011;149(4):361–379. doi: 10.1093/jb/mvr021. [DOI] [PubMed] [Google Scholar]
- 13.Zhao B, Li L, Lei QY. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Gene Dev. 2010;24(9):862–874. doi: 10.1101/gad.1909210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu AM, Wong KF, Jiang X. Regulators of mammalian Hippo pathway in cancer. Biochim Biophys Acta. 2012;1826(2):357–364. doi: 10.1016/j.bbcan.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 15.Xia H, Qi H, Li Y. LATS1 tumor suppressor regulates G2/M transition and apoptosis. Oncogene. 2002;21(8):1233–1241. doi: 10.1038/sj.onc.1205174. [DOI] [PubMed] [Google Scholar]
- 16.Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer. 2006;6(5):369–381. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
- 17.Sun Y. Targeting E3 ubiquitin ligases for cancer therapy. Cancer Biol Ther. 2003;2(6):623–629. [PubMed] [Google Scholar]
- 18.Ciechanover A. The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J. 1998;17(24):7151–7160. doi: 10.1093/emboj/17.24.7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhuang M, Calabrese MF, Liu J. Structures of SPOP-substrate complexes: insights into molecular architectures of BTB-Cul3 ubiquitin ligases. Mol Cell. 2009;36(1):39–50. doi: 10.1016/j.molcel.2009.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J, Ghanim M, Xue L. Analysis of Drosophila segmentation network identifies a JNK pathway factor overexpressed in kidney cancer. Science. 2009;323(5918):1218–1222. doi: 10.1126/science.1157669. [Research Support, Non-U.S. Gov't]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwon JE, La M, Oh KH. BTB domain-containing speckle-type POZ protein (SPOP) serves as an adaptor of Daxx for ubiquitination by Cul3-based ubiquitin ligase. J Biol Chem. 2006;281(18):12664–12672. doi: 10.1074/jbc.M600204200. [DOI] [PubMed] [Google Scholar]
- 22.Hernandez-Munoz I, Lund AH, van der Stoop P. Stable X chromosome inactivation involves the PRC1 Polycomb complex and requires histone MACROH2A1 and the CULLIN3/SPOP ubiquitin E3 ligase. Proc Natl Acad Sci U S A. 2005;102(21):7635–7640. doi: 10.1073/pnas.0408918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li G, Ci W, Karmakar S. SPOP promotes tumorigenesis by acting as a key regulatory hub in kidney cancer. Cancer Cell. 2014;25(4):455–468. doi: 10.1016/j.ccr.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arzuman L, Beale P, Proschogo N. Combination of Genistein and Cisplatin with two designed monofunctional platinum agents in human ovarian tumour models. Anticancer Res. 2015;35(11):6027–6039. [PubMed] [Google Scholar]
- 25.Gan W, Dai X, Lunardi A. SPOP promotes ubiquitination and degradation of the erg oncoprotein to suppress prostate cancer progression. Mol Cell. 2015;59(6):917–930. doi: 10.1016/j.molcel.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J, Bu X, Wang H. Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature. 2018;553(7686):91–95. doi: 10.1038/nature25015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dai X, Gan W, Li X. Prostate cancer-associated SPOP mutations confer resistance to BET inhibitors through stabilization of BRD4. Nat Med. 2017;23(9):1063–1071. doi: 10.1038/nm.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berger MF, Lawrence MS, Demichelis F. The genomic complexity of primary human prostate cancer. Nature. 2011;470(7333):214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cancer Genome Atlas Research N. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499(7456):43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen KH, He J, Wang DL. Methylationassociated inactivation of LATS1 and its effect on demethylation or overexpression on YAP and cell biological function in human renal cell carcinoma. Int J Oncol. 2014;45(6):2511–2521. doi: 10.3892/ijo.2014.2687. [DOI] [PubMed] [Google Scholar]
- 31.Li C, Ao J, Fu J. Tumor-suppressor role for the SPOP ubiquitin ligase in signal-dependent proteolysis of the oncogenic co-activator SRC-3/AIB1. Oncogene. 2011;30(42):4350–4364. doi: 10.1038/onc.2011.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao W, Zhou J, Deng Z. SPOP promotes tumor progression via activation of beta-catenin/TCF4 complex in clear cell renal cell carcinoma. Int J Oncol. 2016;49(3):1001–1008. doi: 10.3892/ijo.2016.3609. [DOI] [PubMed] [Google Scholar]
- 33.Yue B, Liu C, Sun H. A Positive Feed-Forward Loop between LncRNA-CYTOR and Wnt/beta-Catenin Signaling Promotes Metastasis of Colon Cancer. Molecular Therapy. 2018;26(5):1287–1298. doi: 10.1016/j.ymthe.2018.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo L, Tang H, Ling L. LINC01638 lncRNA activates MTDH-Twist1 signaling by preventing SPOP-mediated c-Myc degradation in triple-negative breast cancer. Oncogene. 2018;37(47):6166–6179. doi: 10.1038/s41388-018-0396-8. [DOI] [PubMed] [Google Scholar]
- 35.Mani RS. The emerging role of speckle-type POZ protein (SPOP) in cancer development. Drug Discov Today. 2014;19(9):1498–1502. doi: 10.1016/j.drudis.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.An J, Wang C, Deng Y. Destruction of full-length androgen receptor by wild-type SPOP, but not prostate-cancer-associated mutants. Cell Rep. 2014;6(4):657–669. doi: 10.1016/j.celrep.2014.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barbieri CE, Baca SC, Lawrence MS. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44(6):685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Geng C, He B, Xu L. Prostate cancer-associated mutations in speckle-type POZ protein (SPOP) regulate steroid receptor coactivator 3 protein turnover. Proc Natl Acad Sci U S A. 2013;110(17):6997–7002. doi: 10.1073/pnas.1304502110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Theurillat JP, Udeshi ND, Errington WJ. Prostate cancer. Ubiquitylome analysis identifies dysregulation of effector substrates in SPOP-mutant prostate cancer. Science. 2014;346(6205):85–89. doi: 10.1126/science.1250255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harb OA, Elfeky MA, El Shafaay BS. SPOP, ZEB-1 and E-cadherin expression in clear cell renal cell carcinoma (cc-RCC): Clinicopathological and prognostic significance. Pathophysiology. 2018 doi: 10.1016/j.pathophys.2018.05.004. [DOI] [PubMed] [Google Scholar]
- 41.Ding Q, Xia W, Liu JC. Erk associates with and primes GSK-3beta for its inactivation resulting in upregulation of beta-catenin. Mol Cell. 2005;19(2):159–170. doi: 10.1016/j.molcel.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 42.Perumal E, So Youn K, Sun S. PTEN inactivation induces epithelial-mesenchymal transition and metastasis by intranuclear translocation of beta-catenin and snail/slug in non-small cell lung carcinoma cells. Lung Cancer. 2019;130:25–34. doi: 10.1016/j.lungcan.2019.01.013. [DOI] [PubMed] [Google Scholar]
- 43.Nagai Y, Kojima T, Muro Y. Identification of a novel nuclear speckle-type protein. SPOP. FEBS Lett. 1997;418(1-2):23–26. doi: 10.1016/s0014-5793(97)01340-9. [DOI] [PubMed] [Google Scholar]
- 44.Yang X, Li DM, Chen W. Human homologue of Drosophila lats, LATS1, negatively regulate growth by inducing G(2)/M arrest or apoptosis. Oncogene. 2001;20(45):6516–6523. doi: 10.1038/sj.onc.1204817. [DOI] [PubMed] [Google Scholar]
- 45.Salah Z, Melino G, Aqeilan RI. Negative regulation of the Hippo pathway by E3 ubiquitin ligase ITCH is sufficient to promote tumorigenicity. Cancer Res. 2011;71(5):2010–2020. doi: 10.1158/0008-5472.CAN-10-3516. [DOI] [PubMed] [Google Scholar]
- 46.Li W, Cooper J, Zhou L. Merlin/NF2 loss-driven tumorigenesis linked to CRL4(DCAF1)-mediated inhibition of the hippo pathway kinases Lats1 and 2 in the nucleus. Cancer Cell. 2014;26(1):48–60. doi: 10.1016/j.ccr.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yeung B, Ho KC, Yang X. WWP1 E3 ligase targets LATS1 for ubiquitin-mediated degradation in breast cancer cells. PLoS One. 2013;8(4):e61027. doi: 10.1371/journal.pone.0061027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salah Z, Cohen S, Itzhaki E. NEDD4 E3 ligase inhibits the activity of the Hippo pathway by targeting LATS1 for degradation. Cell Cycle. 2013;12(24):3817–3823. doi: 10.4161/cc.26672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guo ZQ, Zheng T, Chen B. Small-molecule targeting of E3 ligase adaptor SPOP in kidney cancer. Cancer Cell. 2016;30(3):474–484. doi: 10.1016/j.ccell.2016.08.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.