Abstract
Acute myeloid leukemia (AML) is among the top four malignancies in Saudi nationals, and it is the top leukemia subtype worldwide. Resistance to available AML drugs requires the identification of new targets and agents. Hsp90 is one of the emerging important targets in AML, which has a central role in the regulation of apoptosis and cell proliferation through client proteins including the growth factor receptors and cyclin dependent kinases. The objective of the first part of this study is to investigate the putative Hsp90 inhibition activity of three novel previously synthesized quinazolines, which showed HL60 cytotoxicity and VEGFR2 and EGFR kinases inhibition activities. Using surface plasmon resonance, compound 1 (HAA2020) showed better Hsp90 inhibition compared to 17-AAG, and a docking study revealed that it fits nicely into the ATPase site. The objective of the second part is to maximize the anti-leukemic activity of HAA2020, which was combined with each of the eleven standard inhibitors. The best resulting synergistic effect in HL60 cells was with the pan cyclin-dependent kinases (CDK) inhibitor dinaciclib, using an MTT assay. Furthermore, the inhibiting effect of the Hsp90α gene by the combination of HAA2020 and dinaciclib was associated with increased caspase-7 and TNF-α, leading to apoptosis in HL60 cells. In addition, the combination upregulated p27 simultaneously with the inhibition of cyclinD3 and CDK2, leading to abolished HL60 proliferation and survival. The actions of HAA2020 propagated the apoptotic and cell cycle control properties of dinaciclib, showing the importance of co-targeting Hsp90 and CDK, which could lead to the better management of leukemia.
Keywords: acute myeloid leukemia (AML), Hsp90, cyclin-dependent kinases (CDK), dinaciclib
1. Introduction
According to the Saudi Cancer Registry (2014), leukemia ranked second and sixth among the top malignancies for Saudi males and females, respectively, while it is the top malignancy for both Saudi sexes below 14 years of age [1]. Moreover, acute myeloid leukemia (AML) ranked fourth, with 11.9% of the total leukemia cases in both Saudi sexes during 1999–2013. The most common subtype of AML in Saudis is M1, with myeloperoxidase, CD13, CD33 and CD117 as the most common reported antigens [2,3]. Globally, AML is considered the top leukemia subtype, with a higher incidence in elderly patients [4].
Reviewing some of the effective targets, FLT3 is regarded as one of the most important mutations in AML, which is prevalent in over a third of its cases [5]. Sunitinib, sorafenib, lestaurtinib and midostaurin are considered the first generation of FLT3 inhibitors, as they have shown a broader effect on kinases VEGFR, PDGFR, KIT, JAK2 and RAS/RAF, beside FLT3 as a main target. Additionally, a number of recently FDA-approved, more FLT3-specific AML inhibitors have offered more promising treatment possibilities including quizartinib, crenolanib and gilteritinib [6]. Moreover, new combinations have shown a prolonged response and lower resistance rate, such as the combination of either midostaurin or glasdegib with cytarabine [4,6]. On another direction of targeting oncogenic mutations, enasidenib and ivosidenib are considered the prototypes of the isocitrate dehydrogenase (IDH) 1 and 2 inhibitors, which may benefit 7–19% of AML patients, yet their clinical significance is pending running clinical trials [4]. However, despite the achievement of complete remission (CR) for some time by many of the established AML inhibitors, multiple AML drug resistance pathways enable a quick recurrence. These resistance pathways include the FLT3, IDH, Bcl2, MDM2, PD1, CD33, CD123, WT1 and RAS family gene alterations [7,8]. Thus, resistance remains a major hurdle, which necessitates the continuation of both new target and drug discoveries [9].
One of the emerging targets for cancer therapeutics are the cyclin-dependent kinases (CDKs), which are important cell cycle regulatory proteins. Many CDK inhibitors have been recently approved for AML [10]. For example, the CDK9 inhibitor alvocidib showed encouraging clinical results for refractory AML through the MCL-1-dependent pathway, while several CDK 4/6 inhibitors are under development to downregulate the FLT3 and KMT2A genes in AML. Additionally, dinaciclib (pan CDK 1, 2, 5 and 9 inhibitor), which showed good tolerability, is being tested as a monotherapy or in combination with venetoclax (Bcl-2 inhibitor) in a relapsed and refractory AML clinical trial phase 1 [11]. Specifically, in HL60 cells, dinaciclib at 4–16 nM (2 h) increases the sub-G1 cell cycle phase, and induces high apoptosis (75% of total cells) [12].
Another attractive emerging target is the Hsp90, which has a central role in the regulation of survival, cell proliferation and apoptosis [13,14]. Cancer can be dependent on many Hsp90 clients like HER2 and EGFR [15]. It was found that the decrease in Hsp90 leads to the activation of caspases, and G1 arrest in leukemia cells [16,17,18]. Moreover, VEGF in HL60 cells were found to increase the expression of Hsp90, leading to cell survival, upregulation of VEGFR and downregulation of the apoptotic machinery [19].
Several Hsp90 inhibitors of natural or synthetic origins displayed potent anticancer activities. As an example, Y306zh exhibited anti-pancreatic cancer cell growth activity through the disruption of the p23, MAPK and Akt signalling pathways. It also distorted ATP binding to Hsp90, and downregulated the Hsp90 client proteins CDK4, Akt and EGFR [20]. Additionally, Hsp90 and surviving antagonist shepherdin induced apoptosis in HL60, K562, U937 and THP-1 patient-derived blast cells [21]. In another report, resistance in melanoma patients was discovered following the use of the combination of the Hsp90 inhibitor XL888 with the BRAF inhibitor vemurafenib, but the use of dinaciclib as a CDK2 inhibitor abolished the resistance of patients to the combination [22].
Moreover, the antibiotic and anticancer drug geldanamycin is known to degrade the Hsp90 substrate [23]. Further, its main derivative 17-N-allylamino-17-demethoxygeldanamycin (17-AAG) was found to inhibit Hsp90 and multiple tyrosine kinases (HER3, AXL, MET and EGFR), and increased p27 and caspases 3/7 in mesothelioma cell lines [24]. 17-AAG was also reported to increase the apoptotic effect of the CDK inhibitor flavopiridol in mantle cell lymphoma (MCL) and in the Kasumi-1 AML cell lines [25,26]. Additionally, the combination of the CDK inhibitor SCH727965 and the Hsp90 inhibitor NVP-AUY922 induced apoptosis and cell cycle disruption in sarcoma cells, while it spared the normal osteoblasts [27].
On the other hand, VEGF/VEGFR-1, -2 and -3 were found to provoke the proliferation of leukemia cells and increase tumorigenesis through the upregulation of hematopoietic growth factors. In addition, VEGF/VEGFR promote Hsp90 expression through binding to Bcl2, all of which builds up resistance against the apoptosis process. It was also found in AML patients that the increase in plasma VEGF levels was connected with a reduction in survival and remission rates and is a strong “independent” indicator of poor prognosis [28].
Recently, our collaborators in King Saud University-KSA synthetized a novel series of 2-[(3-(4-sulfamoylphenethyl)-4(3H)-quinazolinon-2-yl)thio]anilide derivatives. Compounds 1–5 showed cytotoxicity against the HL60, K562, MCF7 and HT29 cell lines, and displayed good selectivity towards the cancer cells compared with the MRC5 normal cells (structures of 1–5 are found in Supplementary Material Table S1). Compounds 1, 4 and 5 displayed a potent inhibitory activity against the VEGFR2, EGFR and HER2 kinases, and their IC50 against HL60 cells were 1.7 µM, 1.5 µM and 0.4 µM, respectively. These three compounds showed better cytotoxicity in HL60 compared with orafenib: 3.14 µM [29].
These previous reports may explain the importance and relationships between CDKs, Hsp90, VEGFR2 and EGFR, which are prevalent in AML. Thus, the first objective of this exploratory drug discovery study is to test the putative Hsp90 inhibitory activity of 1, 4 and 5, and to select the best compound(s) for molecular docking using Hsp90α, as these compounds have previously shown an inhibition of the Hsp90 client proteins VEGFR2, EGFR and HER2. In the second step, a library of eleven inhibitors representing multiple leukemia targets, including CDKs, were tested in combination with the resulting Hsp90 inhibitor, for the possible maximization of its anti-leukemic activity. The main cancer hallmarks, cell cycle and apoptosis, were also investigated to explore the mechanistic features of the resulting compounds and their combinations.
2. Results
2.1. Surface Plasmon Resonance
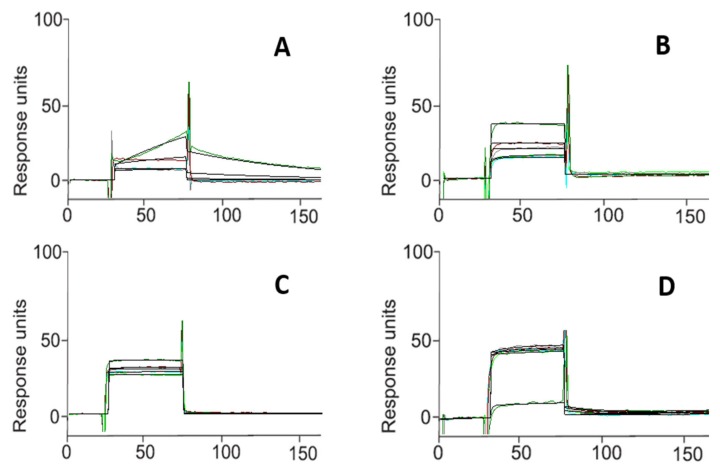
The interaction between each of the three compounds (1, 4, and 5) and Hsp90α was investigated in this study by a surface plasmon resonance (SPR)-based binding assay [30,31,32,33,34]. Only 1 (named HAA2020 in this study) interacted efficiently with the immobilized protein. As a result of fitting the relative sensorgrams to a single-site bimolecular interaction model, the thermodynamic parameters for the resulting complex formation were determined. This approach allowed the measurement of 51.0 ± 2.9 nM KD for the Hsp90α/HAA2020 complex. Interestingly, HAA2020 showed greater affinity towards the chaperone compared with that determined for 17-AAG (Table 1, Figure 1).
Table 1.
Thermodynamic constants measured by surface plasmon resonance (SPR) for the interaction between the tested compounds and immobilized Hsp90α.
| Compound | KD (nM)a |
|---|---|
| 1b | 51.0 ± 2.9 |
| 4 | NBc |
| 5 | NB |
| 17-AAG | 360.0 ± 21.9 |
a: Results were given as the mean ± standard deviation; b: named HAA2020 in this study; c: No binding.
Figure 1.
Surface plasmon resonance sensorgrams acquired for HAA2020 (A), 4 (B), 5 (C) interacting with Hsp90α and for the positive control 17-AAG (D). Each compound was injected onto an Hsp90α modified sensor chip at 6 (n = 3) different concentrations in the range 0.025–4.000 μM.
2.2. Molecular Docking Study
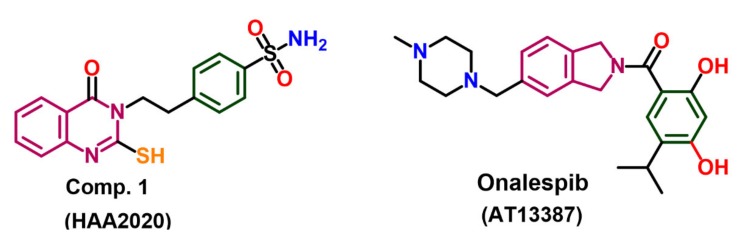
Based on the result of the SPR (Table 1, Figure 1), the binding mode of HAA2020 into the ATPase site of Hsp90 was evaluated in a molecular docking study. The study was performed using AutoDock 4.2. Hsp90 protein with the co-crystallized onalespib (AT13387, Figure 2) as downloaded from Protein Data Bank (http://www.rcsb.org/pdb) [18].
Figure 2.
Chemical structure of HAA2020 and ATI-13387X.
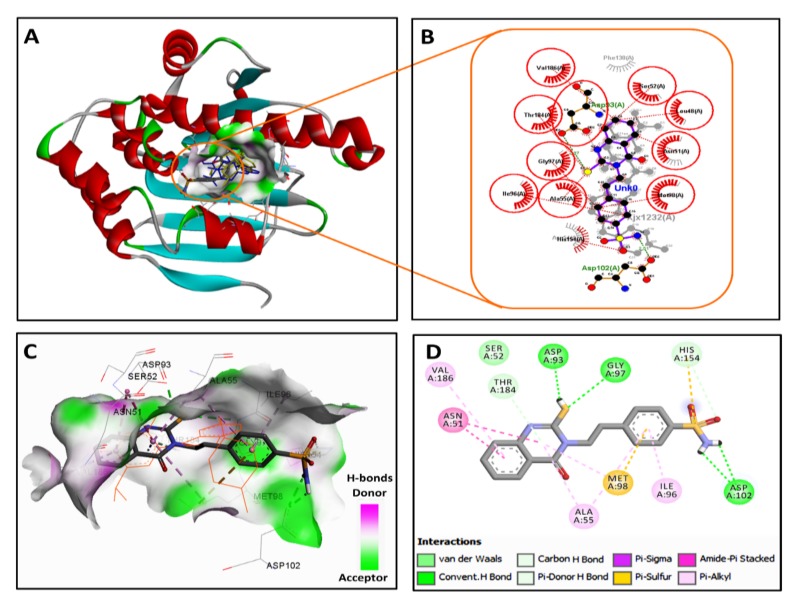
The validation of the docking procedures was done by re-docking the native ligand (onalespib, AT13387) into the active site of Hsp90. The results revealed the superposition of the redocked ligand and the co-crystallized ligand with a root mean square deviation of 2 Å. The results of the docking study of HAA2020 revealed a nice fitting into the ATPase site (Figure. 3), which could count for the inhibition in the catalytic activity of the ATPase. HAA2020 displayed a binding free energy (ΔGb) of −8.62 kcal/mol and an inhibition constant (Ki) of 477.04 nM to Hsp90, compared with a binding free energy of −10.17 kcal/mol and an inhibition constant of 34.93 nM for onalespib. The 2/3D binding modes of HAA2020 and onalespib were generated using a Discovery Studio visualizer [35] and LigPlot+ (v.2.1) [36]. The LigPlot view showed the superposition of HAA2020 with the co-crystallized onalespib with similar binding interactions. HAA2020 showed four conventional hydrogen bonds with Asp93, Gly97 and Asp102 compared with two hydrogen bonds for onalespib. Moreover, HAA2020 exhibited diverse hydrophobic interactions with the hydrophobic residues in Hsp90 (Figure 3).
Figure 3.
2/3D binding modes of onalespib and HAA2020 into the active site of Hsp90 (pdb code: 2XJX): (A) binding modes of the redocked onalespib (coloured in yellow), co-crystallized onalespib (coloured in blue) and HAA2020 (coloured by element) into Hsp90, (B) LigPlot view showing the superposition of HAA2020 and onalespib with two equivalent hydrogen bonds (shown as olive green dotted lines) with Asp93 and Asp102 and several equivalent hydrophobic interactions (shown as brick red dotted lines), (C) 3D binding mode of HAA2020 (shown as sticks coloured by element) overlaid with onalespib (shown as orange lines) into the binding site of Hsp90, receptor shown as the hydrogen bond surface, hydrogen atoms omitted for clarity, and (D) 2D binding mode of HAA2020 showing three conventional hydrogen bonds and different types of hydrophobic interactions.
2.3. MTT Combination Study
HAA2020, which showed good Hsp90 inhibition activity and fitted into the ATPase site of onalespib, was further selected to be tested in combination with a library of eleven compounds for the possible enhancement of its activity against HL60 cells. The eleven compounds represent the multiple-target inhibitor groups for leukemia. The maximum concentration for each of the eleven compounds used in the MTT cytotoxicity assay (HL60, 24 h) was 100 nM, and their resulting IC50 ranged between 7.9–56.5 nM, while the IC50 of HAA2020 was 1814 nM, which was similar to the IC50 of the same compound in a previous study and using the same cell line: 1710 nM [29]. Thus, the eleven compounds were considered for the combination with HAA2020 at 1:50, respectively. The IC50 of the eleven combinations were used with the IC50 of each drug alone including HAA2020, to calculate the combination index (CI) at Fa = 0.9, using the Compusyn software. The combination between HAA2020 and PHA-767491 resulted in a mild synergistic relationship (CI: 0.991), while the combination between HAA2020 and dinaciclib produced stronger synergism (CI: 0.628), and consequently enhanced the activity against HL60 cells to IC50 4.9 nM (Table 2). The rest of the nine compounds resulted in antagonistic activity (CI: >1). The combination parameters for HAA2020 and dinaciclib are shown in Table 3.
Table 2.
Cytotoxic IC50 values of HAA2020, the eleven inhibitors and their combinations in HL60 cells (24 h, nM).
| No. | Inhibitor | Targets | IC50 nM | IC50 (nM) Combination HAA2020/Inhibitor a (50:1) | CI b at Fa = 0.9 |
|---|---|---|---|---|---|
| 1 | PHA-767491 | CDK1/2,CDK5, Cdc7/CDK9, GSK3-β,MK2 PLK1, CHK2 | 26.1 ± 3.8 | 21.8 ± 3.2 | 0.991 |
| 2 | Dinaciclib | CDK1, CDK2, CDK5, CDK9 | 8.2 ± 0.3 | 4.9 ± 1.3 | 0.628 |
| 3 | ON123300 | CDK4, Ark5, PDGFRβ, FGFR RET, Fyn | 37.3 ± 3.5 | 1872.5 ± 45.9 | >1 |
| 4 | AMG 925 | CDK4/FLT3 | 54.4 ± 7.7 | 3355 ± 347.8 | >1 |
| 5 | KW2449 | FLT3, EGFR, FGFR1, Bcr-Abl PDGFRβ, IGF-1R | 56.5 ± 10.8 | 2535.5 ± 318.9 | >1 |
| 6 | AZD2932 | FLT3, c-Kit, VEGFR, PDGFRβ | 15.2 ± 3.1 | 2394 ± 380.4 | >1 |
| 7 | Dasatinib | c-Kit, Abl, Src | 12.1 ± 2.9 | 3036 ± 181.1 | >1 |
| 8 | BGJ398 | Kit, FGFR, VEGFR2, Abl, Fyn, Lck, Lyn, Yes | 16.2 ± 1.7 | 2952.5 ± 170.6 | >1 |
| 9 | Saracatinib | EGFR, Src, Yes, Fyn, Lyn, Blk, Fgr, Lck, Abl | 16.8 ± 1.2 | 1821.5 ± 40.3 | >1 |
| 10 | Lapatinib | EGFR, HER2 | 12.1 ± 2.0 | 2383.5 ± 245.3 | >1 |
| 11 | Taselisib | PI3Kα/δ/γ | 7.9 ± 0.4 | 2647 ± 203.6 | >1 |
| HAA 2020 | VEGFR2, EGFR, HER2 | 1814.5 ± 230.8 | - | - |
a: inhibitors 1–11. b: Combination index. (-): not applicable. Data are represented as mean ± SD (n = 3). Experiment was repeated 3×. IC50 of some combinations is more than 100 nM because the IC50 of HAA2020 alone is more than 100 nM.
Table 3.
The combination index parameters of HAA2020, dinaciclib and their combination.
| Drug/Combination (1:50) | Dm | m | r | CI a at Fa = 0.9 |
|---|---|---|---|---|
| Dinaciclib | 15.985 | –0.358 | –0.911 | - |
| HAA2020 | 0.340 | –1.184 | –0.997 | - |
| HAA2020 + Dinaciclib | 0.552 | –0.722 | –0.998 | 0.628 |
a: Combination index. Definition of Dm, m and r values are found in the Supplementary Material (S2).
2.4. Cell Cycle Analysis
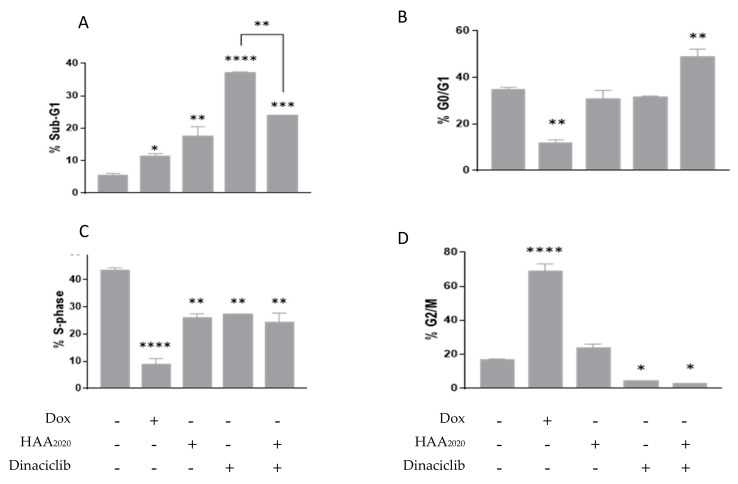
The pan CDK inhibitor dinaciclib was selected as a candidate for further investigations with HAA2020 because it showed the best CI between the eleven inhibitors. The cell cycle distribution was used to examine the anti-proliferative effect of the HAA2020 and dinaciclib combination. Following a 24 h treatment, HAA2020 and dinaciclib (500 nM and 10 nM, respectively) elicited an increase in the sub-G1 phase in HL60 cells, and stronger than that observed for doxorubicin alone as a positive control (100 nM), with the highest increase attributed to dinaciclib. Moreover, the combination treatment showed a significant increase in the sub-G1 phase, but less than the effect caused by each of them alone (Figure 4A). Neither HAA2020 nor dinaciclib had an effect on the G0/G1 phase, however, their combination elicited a significant cell cycle arrest, as observed by an increase of cells in the G0/G1 phase (Figure 4B). However, this was not the case with the addition of doxorubicin, as an opposite observation was apparent, where G0/G1 was reduced. When comparing the differences in the S phase, all three compounds showed significant decreases compared with the untreated control, with the highest decrease attributed to doxorubicin, and little difference between either HAA2020, dinaciclib or their combination (Figure 4C). HAA2020 had no significant effect on the G2/M phase of the cell cycle, in contrast to dinaciclib, where G2/M was significantly reduced both alone or in combination with HAA2020 (Figure 4D). Furthermore, doxorubicin significantly increased cells in the G2/M phase.
Figure 4.
Cell cycle analysis in HL60 cells. Cells were treated (+) for 24 h with either 100 nM doxorubicin (Dox), 500 nM HAA2020, 10 nM dinaciclib or a combination of HAA2020 and dinaciclib (500 nM and 10 nM, respectively). Data are represented as mean ± SEM (n = 2, two independent experiments) for each of the phases of the cell cycle, sub-G1 (A), G0/G1 (B), S (C) and G2/M (D). Statistical differences, compared with the untreated control cells (-), were assessed by a one-way ANOVA with the Tukey’s post-hoc multiple comparison test (GraphPad Prism). p < 0.05 (*), p < 0.01 (**), p < 0.001 (***) and p < 0.0001 (****) were taken as significant.
2.5. Detection of Apoptosis
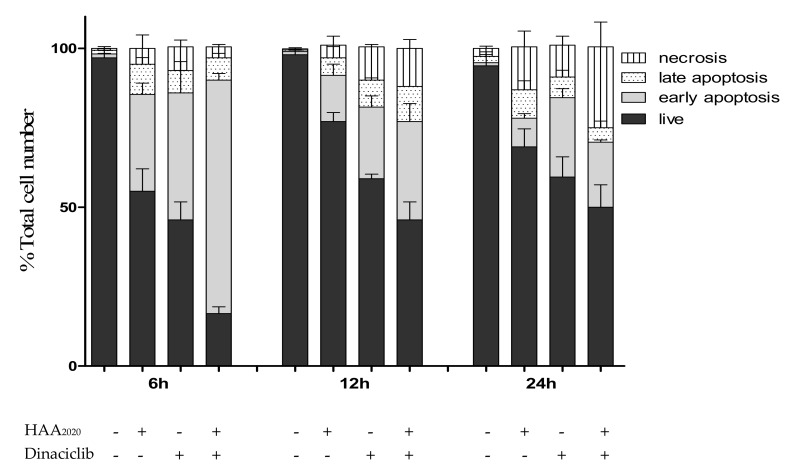
The combination of HAA2020 and dinaciclib showed a synergistic G0/G1 arrest in HL60 cells following the 24 h treatment. Further, each of the two compounds induced significant apoptosis as shown in the sub-G1 phase, but together they induced less apoptosis compared with their effect alone. Thus, the annexin V FITC/PI assay was used at three time points including 24 h to investigate apoptosis in more detail. After 6 h of treatment, each of the two compounds caused an increase in the early apoptotic events compared with the control, and their combination caused synergistic early apoptosis compared with their effect alone in HL60 cells, all with minimal necrosis. At 12 h, each of the two compounds and their combination caused the same effect with a higher percentage of necrosis caused by dinaciclib and the combination, while after 24 h, the necrosis caused by the combination was equivalent to its early and late apoptotic events. The early apoptosis caused by the combination after 6 h, 12 h and 24 h was 70%, 30% and 20%, respectively, which may explain the superiority of HAA2020 and dinaciclib alone compared with their combination in producing a sub-G1 increase in HL60 cells after 24 h (Figure 5). Thus, this three-time point analysis showed that the most effective time point of the combination in HL60 cells is at 6 h.
Figure 5.
Detection of apoptosis in HL60 cells. Cells were treated for 6 h, 12 h and 24 h with either HAA2020 (500 nM), dinaciclib (10 nM) or their combination. Following treatment, the cells were stained with annexin V FITC/PI. A total of 20,000 single-cell events were acquired on a BC-500 flow cytometer and analyzed by the Expo 32 software. Data are represented as mean ± SEM (n = 3, two independent experiments) for each of the cell staining statuses: live cells (annexin V-/PI-), early apoptotic cells (annexin V+/PI-), late apoptotic cells (annexin V+/PI+) and necrotic cells (annexin V-/PI+).
2.6. Real Time PCR
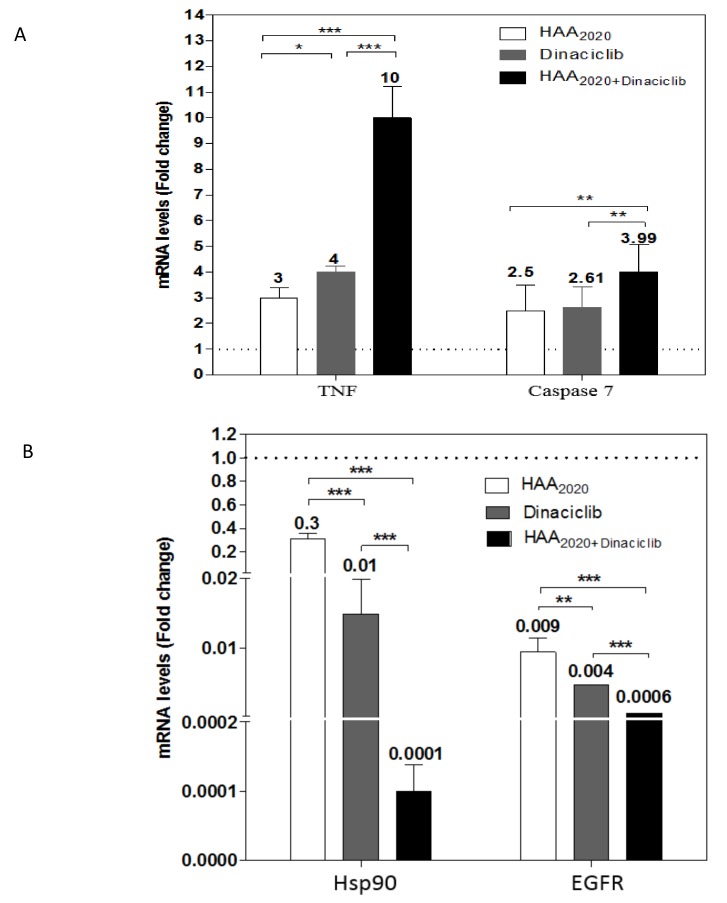
For more details on the apoptotic process of the tested compounds, the mRNA amount of TNF and the caspase-7 genes were evaluated by real-time PCR, following the treatment of HL60 cells at 6 h with either HAA2020 (500 nM), dinaciclib (10 nM) or their combination. Each of the two compounds significantly upregulated the TNF-α and caspase-7 genes in addition to their combination, which showed a synergistic effect, suggesting involvement of both the extrinsic and intrinsic apoptotic pathways. A similar significant effect was exhibited against Hsp90α and its client protein EGFR, but in the decreasing mode, which indicates that the main activities of HAA2020 are maintained when it is combined to the CDK inhibitor dinaciclib (Figure 6).
Figure 6.
mRNA expression levels of TNF-α and caspase-7 (A), Hsp90-α and EGFR (B), compared with the control. HL60 cells were treated for 6 h with either HAA2020 (500 nM), dinaciclib (10 nM) or their combination. Data are represented as mean ± SEM (n = 3, two independent experiments). The results were expressed as fold-change compared with the untreated group. Statistical differences, compared with the untreated control cells (dashed line), were assessed by a one-way ANOVA with the Tukey’s post-hoc multiple comparison test. p < 0.05 (*), p < 0.01 (**) and p < 0.001 (***) were taken as significant.
2.7. Western Blotting
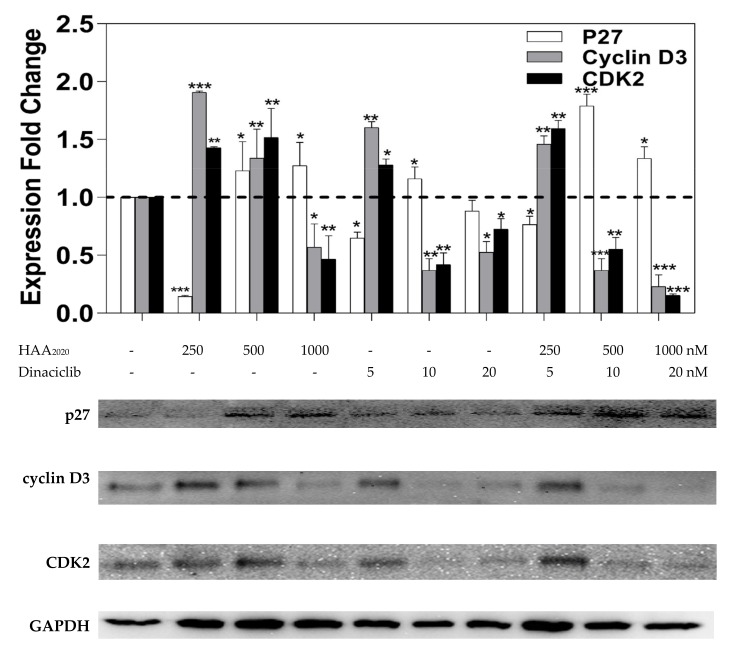
Western blotting was devised to study the effect of HAA2020 and dinaciclib (6 h) on the G1/S cell cycle check point regulatory proteins p27, cyclinD3 and CDK2. The HL60 cells were treated with HAA2020 (250 nM, 500 nM and 1000 nM), dinaciclib (5 nM, 10 nM and 20 nM) and their combination for 6 h. HAA2020 caused a dose-dependent increase in the p27 expression compared with the control GAPDH, while dinaciclib alone, or combined with HAA2020, caused an irregular increase in p27. Each of HAA2020, dinaciclib and their combinations caused a dose-dependent inhibition of cyclin D3 compared with the control, which was more pronounced by the combination. Most importantly, HAA2020 significantly reduced CDK2 at 1000 nM, while dinaciclib caused more inhibition at its IC50 (i.e., 10 nM). The combination of the two compounds caused a dose-dependent and significant decrease in CDK2 compared with GAPDH (Figure 7).
Figure 7.
Expression of p27, cyclin D3 and CDK2 in HL60 cells treated with either HAA2020, dinaciclib or their combination as above for 6 h. Data are represented as mean ± SEM (n = 2, two independent experiments). The result was normalized as fold-change compared with the untreated group. Ratio above 1 (dashed line) shows the amount of mRNA for the three genes in the untreated cells (control = 1). Statistical differences, compared with the untreated control cells, were assessed by a one-way ANOVA with the Tukey’s post-hoc multiple comparison test. p < 0.05 (*), p < 0.01 (**) and p < 0.001 (***) were taken as significant. The Image J software was used for densitometry.
3. Discussion
Many quinazolines which target EGFR and/or HER2, including afatinib, gefitinib, erlotinib and lapatinib, are established anticancer agents. HAA2020 (4-(2-(4-Oxo-2-thioxo-1,4-dihydroquinazolin-3(2H)-yl)ethyl)benzene sulfonamide) is a previously synthetized quinazoline, which showed cytotoxicity against the HL60 and K562 leukemia cell lines and spared the MRC5 normal cells. HAA2020 was two folds stronger against HL60 cells, compared with K562 cells (IC50: 1.71 µM and 3.67 µM, respectively). It also displayed a potent inhibitory activity against the VEGFR2, EGFR and HER2 kinases [29].
In the search for new anticancer agents, Hsp90 poses an attractive target in AML, as it is involved in the regulation of survival, cell proliferation and apoptosis [13,14]. Hsp90 has many important client proteins including the VEGFR2, EGFR and HER2 kinases [15]. A number of previously reported quinazolines have shown Hsp90 inhibition activities including 3-phenyl-2-styryl-3H-quinazolin-4-one [37]. The SPR-based binding assay results in this study showed the efficient interaction of HAA2020 with the Hsp90 protein at 51 nM KD, which was greater towards the chaperone compared with that determined for 17-AAG. This result was supported by a molecular docking study, which showed that HAA2020 fitted nicely into the ATPase site, which could possibly be due to its catalytic activity. Thus, HAA2020 can be considered as a novel Hsp90 inhibitor, among its previously described activities. In a previous study, the treatment of HL60 cells with 17-AAG inhibited Hsp90 [38].
With the purpose of enhancing the activity against AML, which is represented in this study by HL60 cells, HAA2020 was combined with each of the eleven anti-leukemic inhibitors for 24 h. Only two inhibitors resulted in a synergistic effect with HAA2020: PHA-767491 and dinaciclib, both CDK inhibitors. However, dinaciclib showed more synergism with HAA2020 (CI: 0.62) compared with PHA-767491 (CI: 0.99), which was comparable with the synergistic activity of the ganetespib and cytarabine combination (CI: 0.47 [39]). As expected, the combination of HAA2020 and dinaciclib enhanced the cytotoxicity against HL60 (IC50: 4.9 nM) compared with the IC50 of HAA2020 and dinaciclib alone (1814 nM and 8.2 nM, respectively). CDK inhibitors alone have previously shown promising results in the clinic for AML, including alvocidib and dinaciclib [10,11,12]. Moreover, in combination, the CDK inhibitor (SCH727965) and the Hsp90 inhibitor (NVP-AUY922) were found to induce apoptosis and cell cycle disruption in sarcoma cells, while it spared the normal osteoblasts [27]. In another combination study, the resistance in melanoma patients following the use of the Hsp90 inhibitor XL888 with the BRAF inhibitor vemurafenib was reverted by the use of dinaciclib as a CDK2 inhibitor [22]. Thus, dinaciclib was selected for further mechanistic investigations with HAA2020.
Following the 24 h treatment of HL60 cells, the combination of dinaciclib and HAA2020 elicited a significant G0/G1 cell cycle arrest. However, each of the two compounds showed a stronger increase in the sub-G1 phase compared with doxorubicin and with their combination. Thus, a three-time point, more apoptosis-specific assay was performed to investigate the reason behind the fall of the combination to achieve a higher sub-G1 effect compared with HAA2020 and dinaciclib alone. The annexin V FITC/PI assay showed that the most effective early and late apoptosis inducing time point for the combination was at 6 h compared with 12 h and 24 h (combined early and late apoptosis at the three time points: 80%, 45% and 25%, respectively). All of this occurred parallel with the time-dependent increasing necrosis in the HL60 population (necrosis at the three time points: 4%, 12% and 22%, respectively). Similarly, 17-AAG induced 80% apoptosis in HL60 cells at 1 µM (48 h), which was assessed by the annexin V assay, and it caused an increase in the sub-G1 HL60 cells from 3.5% in the untreated cells to 13% at 1 µM, simultaneously with a G0/G1 arrest of 52% (compared with 34% in the untreated cells) at 6 h, all at the expense of reduced S cell cycle phase events (from 52% in the untreated cells down to 7% in the treated ones) [38]. Further, in another previous report, dinaciclib alone increased the sub-G1 phase at 4–16 nM in HL60 cells, and induced high apoptosis following 2 h of treatment, which decreased by two and five folds following 6 h and 24 h, respectively [12].
Previous reports have shown that the stimulation of TNF-α results in the upregulation of caspases 3/7, which leads to apoptosis in various cancer cells [40]. Moreover, Hsp90 is regarded as an anti-apoptotic protein that can be downregulated by the stimulation of TNF-α [41]. In this regard, 17-AAG was previously shown to increase caspases 3/7/9 in HL60 and mesothelioma cell lines [24,38]. Thus, to confirm whether the induced apoptosis in HL60 cells involves this pathway or not, HAA2020 and dinaciclib and their combinations were tested at 6 h, resulting in the significant simultaneous upregulation of TNF-α and caspase-7, confirming the involvement of these multiple apoptotic pathways in the effect of the combination. The combination was also confirmed to show the simultaneous and significant inhibition of Hsp90 and its client protein EGFR in HL60 cells at 6 h, which indicates that the main activities of HAA2020 are maintained when it is combined with the CDK inhibitor. Similarly, AT13387, a drug candidate in clinical trials, showed a dual EGFR and Hsp90 inhibition mechanism of action [42,43].
In cell cycles, the G1/S phases are modulated by the CDK2, CDK4/6 and cyclins A,D checkpoint proteins, while the G2/M phases are modulated by the CDK1 and cyclin B check points [44]. The cell cycle effect of HAA2020 and dinaciclib was tested at 24 h, revealing an increase in the sub-G1 and G0/G1 phases. To confirm the cell cycle effect of the combination at 6 h, which showed more apoptosis in HL60, p27, CDK2 and cyclin D3 were tested. It was found that our combination downregulated p27, cyclin D3 and CDK2, which agrees with previous reports showing that the decrease in Hsp90 is associated with the G1 arrest in leukemia cells [16,21]. Additionally, XL888 synergizes with dinaciclib to inhibit CDK2 [22]. Further, the Hsp90 inhibitor 17-AAG was found to increase p27 and caspases 3/7 in mesothelioma cell lines [24].
Hsp90 inhibitors are important assets in the anticancer armory, as they actively help the control of steroidal hormone receptors and transcription factors, in addition to many kinase pathways in our bodies. In the preclinical settings, Hsp90 inhibitors showed more successful stories compared with the clinical studies. Ganetespib, for example, showed a suppression of the AML-patients’ derived blast cells alone or when combined with cytarabine (CI = 0.47). That activity was mediated by the downregulation of Akt and the induction of apoptosis in HL60 cells [39]. There are currently many Hsp90 inhibitors undergoing clinical trials: , 2 and 1-2. These inhibitors include, among others, SNX-5422 for chronic lymphocytic leukemia (combined with ibrutinib, phase 1) and AT13387 for EGFR-mutant lung cancer (combined with erlotinib hydrochloride, phase 1–2). Moreover, the Hsp90 inhibitors 17-AAG and 17-DMAG showed CDK4 inhibition effects, alone or combined with trastuzumab or sorafineb in separate clinical trials. 17-AAG also exhibited a CDK4 and cyclinD1 inhibition when combined with irinotican. In another clinical trial, AT13387 inhibited CDK4 and caspase-3, while BIIB021 inhibited both CDK4 and HER2 [42,43].
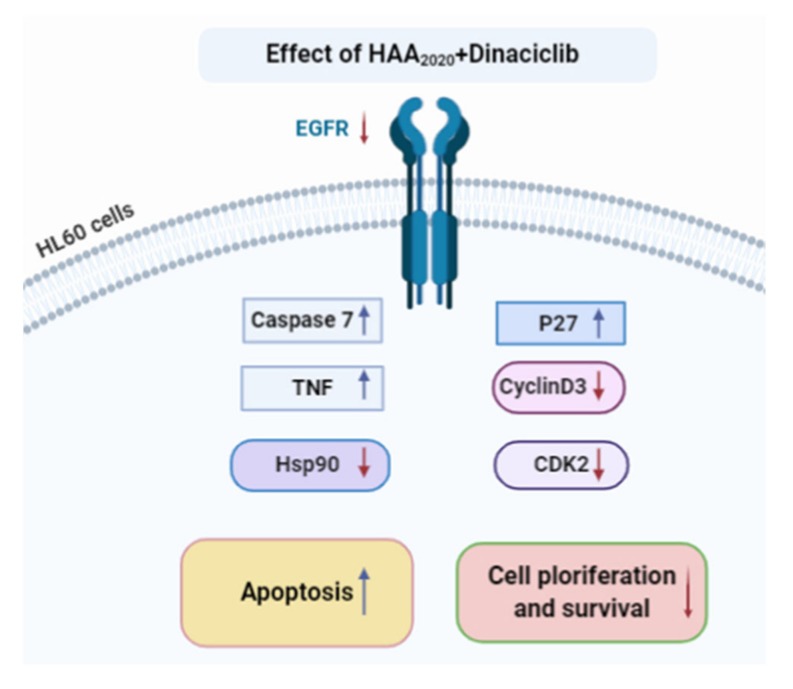
Taking all that together, breast and lung cancers are the only up-to-date clinically successful target organs for Hsp90 inhibitors. Thus, there is a continuous need for mechanistic and combination studies in other cancer platforms, including leukemia, which could help in understanding the role of Hsp90 inhibitors in the clinical stage, and to overcome the efficacy and side effects issues [42,43]. The number of drugs available for leukemia, including AML, are hindered by an array of resistance mechanisms, which necessitates the readiness of more efficient drugs on the front line. The results of this study demonstrate the efficiency of HAA2020 as a Hsp90 inhibitor, which also induced apoptosis and cell cycle effects in HL60 cells, alone or when combined with the pan CDK inhibitor dinaciclib. The combination activity was associated with increased caspase-7 and TNF-α, leading to apoptosis in HL60 cells. In addition, it upregulated p27 simultaneously with the inhibition of cyclinD3 and CDK2, leading to abolished HL60 proliferation and survival (Figure 8). The combination of Hsp90 and CDK inhibitors could pave the way for important emerging drugs in the clinic. Future work may involve in vivo testing of HAA2020, alone or with dinaciclib or other CDK inhibitors, and comparing the activity with AML-gold standard therapies like cytarabine, all of which could better the understanding of the relationship between Hsp90 and CDK inhibition in AML.
Figure 8.
The effect of the HAA2020 and dinaciclib combination on apoptosis, cell proliferation and survival of HL60 cells. Created in BioRender.com.
4. Materials and Methods
4.1. Compounds and Reagents
PHA-767491, dinaciclib, ON123300, AMG925, KW2449, AZD2932, dasatinib, BGJ398, saracatinib, lapatinib and taselisib were purchased from Selleckchem, Houston, TX, USA. Doxorubicin was purchased from Cayman, Ann Arbor, MI, USA. All reagents and kits, including ethanol and dimethyl sulfoxide (DMSO), were purchased from Sigma Chemical Co. Jeddah, MK, KSA, unless otherwise reported.
4.2. Methods
4.2.1. Surface Plasmon Resonance
Surface plasmon resonance (SPR), which is an optical analytic technique for measuring the kinetic and thermodynamic parameters of ligand−protein complexes’ formation and affinity, is widely used to investigate enzyme/inhibitor interactions [30,31,32]. Recently this approach was successfully applied to study the binding of small molecules to Hsp90 [33,45,46,47]. SPR experiments were performed in this study as described elsewhere [33]. Briefly, analyses were carried out using a Biacore 3000 optical biosensor equipped with research-grade CM5 sensor chips (GE Healthcare, Piscataway, NJ, USA). Two separate recombinant Hsp90 surfaces, a BSA surface and an unmodified reference surface, were prepared for simultaneous analyses. The recombinant human Hsp90α (SPP-776, Stress-gen Bioreagents Corporation, Victoria, anada) was dissolved at 100 μg/mL in 10 mM sodium acetate, pH 5.0, then it was immobilized on individual sensor chip surfaces at a flow rate of 5 μL/min using standard amine-coupling protocols to obtain densities of 8−12 kRU. Compounds 1, 4 and 5, [29], as well as 17-AAG used as the positive control [48,49], were dissolved in 100% DMSO to obtain 4 mM solutions and diluted 1:1000 (v/v) in PBS (10 mM NaH2PO4, 150 mM NaCl, pH 7.4) to a final DMSO concentration of 0.1%. A series of concentrations were prepared as 2-fold dilutions into a running buffer: for each sample, the complete binding study was performed using a six-point concentration series, typically spanning 0.025−4 μM, and triplicate aliquots of each test compound were dispensed into single-use vials. Multiple blank samples of the running buffer alone were included in each analysis. Binding experiments were performed at 25 °C, using a flow rate of 50 μL/min, with 60 s monitoring of association and 250 s monitoring of dissociation. Simple interactions were adequately fit to a single-site bimolecular interaction model (A + B = AB), yielding a single KD. Sensorgram elaborations were performed using the BIAevaluation software provided by GE Healthcare (Data files for SPR are available from the Supplementary Material S3, and can be opened by dragging each of the files into https://filext.com/file-extension/BLE).
4.2.2. Molecular Docking Study
A comparative molecular docking study was performed for the best Hsp90 inhibiting compound from the previous analysis, 1 (4-(2-(4-Oxo-2-thioxo-1,4-dihydroquinazolin-3(2H)-yl)ethyl)benzene sulfonamide, named HAA2020 hereafter) and onalespib (AT13387, Figure 2) into the active site of Hsp90. The binding modes, affinities and the binding free energies of HAA2020 were evaluated and compared with those of the co-crystallized ligand (onalespib). The crystal structure of the Hsp90 (pdb code: 2XJX) co-crystallized with onalespib (AT13387) was obtained from Protein Data Bank (http://www.rcsb.org/pdb) in an X-ray resolution of 1.66 Å [18]. Preparations of the protein and ligand files, and the energy minimization of HAA2020 and onalespib were done following the previous report [50]. In addition, the preparation of the grid parameter files was done using AutoGrid according to the same previous report [50]. The 3D grid dimensions were set to the default values with 0.375 Å spacing. The protein structure was set as a rigid file, while HAA2020 and onalespib were docked as flexible molecules. The genetic algorithm was used as the search parameter, and docking parameters were set to the default values. The docking poses were scored and ranked by AutoDock in a decreasing order of their binding free energy. The top ten conformations of the protein–ligand complexes were clustered. The binding modes of the best fit conformations of HAA2020 and onalespib were analyzed. Different types of ligand–protein interactions were vitalized using a Discovery Studio visualizer and LigPlot+. The 2/3D binding modes were generated showing both the hydrogen bonding and hydrophobic interactions (Figure 3).
4.2.3. Cell Culture
HL60 suspension cells (Hematopoietic neoplasm, Human acute myeloid leukemia: AML, Myc+, CLL1, CD33, CD123 and CD135) were obtained from the ATCC. Cells were maintained in RPMI-1640 media (10% FBS, 1% Antibiotic-Antimycotic, Gibco) at 100% humidity, 37 °C and 5% CO2. To keep logarithmic growth, a maximum of 5 × 105 cells/mL were sub-cultured (1–10 passages).
4.2.4. MTT Combination Study
The cytotoxicity of HAA2020 and each of the eleven inhibitors library was assessed by an MTT assay, as previously reported [51,52]. The eleven inhibitors were selected depending on the multiple existing and putative leukemia targets, mainly FLT3, PDGFR, Bcr-Abl, EGFR and CDKs (Table 2) [53,54]. HL60 cells were cultured in 96-well (5 × 104/well). The final HAA2020 concentrations were 0, 312.5, 625, 1250, 2500 and 5000 nM (based on IC50 obtained from previous report: [29]), while the final concentrations of the eleven inhibitors were 0, 6.25, 12.5, 25, 50 and 100 nM (based on IC50 obtained from Sellechem.com). The final DMSO concentration was 0.1%, (n = 3). Depending on the average IC50 of HAA2020 and the eleven inhibitors in HL60 cells (Table 2), the combinations of HAA2020 and each of the eleven inhibitors were performed in a 50:1 ratio. Plates were incubated for 24 h, followed by the addition of MTT (3 h, Life technologies) and centrifugation of the 96-well plates at 400 g/5 min (Eppendorf 5430), resulting in the precipitation of cells at the bottom of the plates. That was followed by suction of the media and the addition of DMSO to each well. The absorbance was read on a multi-plate reader (BIORAD, PR 4100). The optical density of the purple formazan A550 is proportional to the number of viable cells. The compound concentration causing 50% inhibition (IC50) compared with the control cell growth (100%) was determined using GraphPad Prism. The CompuSyn software was used to calculate the combination index (CI) [(D)1/(Dx)1]+ [(D)2/(Dx)2], where (D)1 and (D)2 are concentrations of drug A and drug B, which are employed in the combination study to calculate the IC50. (Dx)1 and (Dx)2 are the IC50 of single drugs A and B. CI <1: synergism, CI = 1: additivity and CI >1: antagonism [55].
4.2.5. Cell Cycle Analysis
Cell cycles are series of changes resulting from the multiple checkpoints and occurring from the initial phase of the cell formation leading to its division. The disruption of the cell cycle causes cancer, which usually takes place in the G1/S (modulated by CDK4/6-cyclinD and CDK2-cyclinA) or the G2/M (modulated by CDK1-cyclinB) check points [44]. Following the treatment of HL60 cells (5 × 105) with each of HAA2020, dinaciclib and their combination (500 nM + 10 nM, respectively) for 24 h, the cells were fixed in 70% ethanol and then processed for a cell cycle analysis using propidium iodide (PI, Santa Cruz, CA, USA), as previously described [56]. A total of 20,000 single-cell events were acquired on a flow cytometer (NovoCyte 3000, ACEA biosciences, San Diego, CA, USA) and analyzed using the cell cycle algorithm of the NovoExpress software (version 1.4.1 ACEA biosciences, San Diego, CA, USA).
4.2.6. Detection of Apoptosis
As a major cell death route, apoptosis is regarded as a non-immunogenic, energy-dependent selective programmed pathway, which cleans dead cells. Apoptosis occur either extrinsically (in the cytoplasm) via the Fas death receptor, a member of tumor necrosis factor (TNF) receptor family, or intrinsically (in the mitochondria), which leads to the release of the cytochrome-c, thus activating cell death signals. Both of the pathways lead to the activation of certain proteases called caspases, ending with cell death [57,58]. In this study, apoptosis was quantified by detecting the cell surface exposure of phosphatidylserine (PS) in apoptotic cells using annexin V FITC/PI, following a previous report [59]. HL60 cells were seeded in 6-well plates at 5 × 105 cells/well for overnight, before the treatment with HAA2020, dinaciclib and their combination (500 nM and 10 nM, respectively) for 6 h, 12 h and 24 h. Cells were inspected microscopically before and after treatment to observe the morphological changes. Then they were collected, centrifuged and the pellets were re-suspended in a binding buffer. Annexin V FITC (Invitrogen, Waltham, MA, USA) was then added to each sample and incubated at room temperature in the dark. A further binding buffer and PI were added. Samples were analyzed by flow cytometry (BC 500, Miami, FL, USA) within 1 h. Viable cells were differentiated from early and late apoptotic/necrotic cells by annexin V (x axis) and PI staining (y axis).
4.2.7. Gene Expression Analysis and Quantitative Real Time PCR
The total RNA resulting from the stimulation of HL60 cells with HAA2020, dinaciclib and their combination was isolated using an RNA isolation mini kit (Thermo Fisher Scientific, Waltham, MA, USA) according to manufacturer’s instructions. The NanoDrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA) was used to determine the concentration and purity of the isolated RNA. The purity of the isolated RNA and its integrity was validated using 1% agarose gel electrophoresis. Subsequently, the complementary DNA (cDNA) was synthesized using the isolated RNA (2 µg) based on the instructions of the manufacturer, using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA, USA). The RT-PCR was carried out in a 96-well plate by the RT-PCR platform using the Applied Biosystems 7500 Fast Real Time PCR System, following a previous report [60]. Briefly, HL60 cells were seeded in 6-well plates (1 × 106 cells/well) and incubated overnight, before treatment with HAA2020, dinaciclib and their combination (500 nM and 10 nM, respectively) for 6 h. The RT-PCR mixture contained 10 µL of 2X SYBR Green I Master mix, 10 ng of cDNA, 0.4 µM of each human primers (TNF-α, caspase-7, Hsp-90α, EGFR and GAPDH as the housekeeping gene, (Table 4), Applied-Biosystems, Thermo Fisher Scientific, Waltham, MA, USA) and PCR-grade water up to a total volume of 20 µL. The RT- PCR program consisted of 45 cycles of denaturation at 95 °C for 15 s followed by annealing/extension at 60 °C for 60 s. All RT-PCR reactions were applied in triplicates and repeated 3 times and included a negative control (no template). Standard comparative methods (ΔCt) were used to evaluate the genes’ expressions. The raw Ct values were converted into relative expression levels (fold-change) using the formula 2−∆∆Ct.
Table 4.
Sequence of Hsp90α, TNF-α, caspase-7, EGFR and GAPDH primers.
| Gene | Sequence |
|---|---|
| Hep90α | (F) TTGGTTACTTCCCCGTGCTG (R) GCCTTTTGCCGTAGGGTTTC |
| TNF-α | (F) CTCTTCTGCCTGCTGCACTTTG (R) ATGGGCTACAGGCTTGTCACTC |
| caspase-7 | (F) GGACCGAGTGCCCACTTATC (R) TCGCTTTGTCGAAGTTCTTGTT |
| EGFR | (F) GCGTCTCTTGCCGGAATGT (R) GGCTCACCCTCCAGAAGGTT |
| GAPDH | (F) AGGTCGGTGTGAACGGATTTG (R) TGTAGACCATGTAGTTGAGGTCA |
4.2.8. Western Blotting
To confirm the cell cycle effect, Western blotting was used to investigate the expression change of the cell cycle proteins p27, cyclinD3 and CDK2. HL60 cells (1 × 106 cells/well of 6-well plates) were treated with HAA2020 (250 nM, 500 nM and 1000 nM), dinaciclib (5 nM, 10 nM and 20 nM) and their combination for 6 h (Figure.7). The total proteins were isolated after the complete lysis of cells by a lysis buffer. The concentration of the total protein was determined by the Bradford method. The loading protein samples were electrophoresed on a polyacrylamide gel and transferred to a membrane. The membrane was incubated with p27, cyclinD3 and CDK2 antibodies (Cell signalling) for 2 h at room temperature and the secondary antibody GAPDH for 1 h at room temperature. The immunoreactivity was visualized by chemiluminescence using horseradish peroxidase (HRP)-conjugated secondary antibodies, and their image was detected by a scanner (GeneGenome, Syngene BioImaging) [61].
4.3. Statistics
Statistical differences of samples, compared with the untreated control cells, were assessed by a one-way ANOVA with the Tukey’s post-hoc multiple comparison test (GraphPad Prism Version 5). p < 0.05 (*), p < 0.01 (**), p < 0.001 (***) and p < 0.0001 (****) were taken as significant.
Abbreviations
| 17-AAG | 17-N-allylamino-17-demethoxygeldanamycin |
| AML | Acute Myeloid Leukaemia |
| CDK | Cyclin dependent kinases |
| CI | Combination index |
| EGFR | Epidermal growth factor receptor |
| Hsp90 | Heat shock protein 90 |
| KSA | Kingdom of Saudi Arabia |
| M1 | AML without maturation |
| PI | Propidium iodide |
| SPR | Surface Plasmon Resonance |
| TNF-α | tumor necrosis factor alfa |
| VEGF | Vascular endothelial growth factor |
| VEGFR | Vascular endothelial growth factor receptor |
Supplementary Materials
The following are available online at https://www.mdpi.com/1420-3049/25/9/2220/s1, Table S1: Structure of compounds 1–5; S2: Definition of Dm, m and r values; Data files S3: Original data files for SPR.
Author Contributions
Conceptualization, design and writing, A.N.A.; Experimental part, M.E.A., A.A. and M.Z.E.-R; Surface plasmon resonance study, A.B., A.V. and N.D.T.; Statistical analysis and proof reading, W.H.M. and M.H.M.; Molecular docking study, A.M.G.; Providing compounds 1, 4 and 5, and revising manuscript, H.M.A., A.A.-M.A.-A., and A.S.E.-A. All authors have read and agreed to the published version of the manuscript.
Funding
The authors would like to thank the Deanship of Scientific Research at Umm Al-Qura University for the continuous support. This work was supported financially by the Deanship of Scientific Research at Umm Al-Qura University to Ashraf N. Abdalla (Grant Code: 15-MED-3-1-0060), and the APC was funded by the Deanship of Scientific Research at Umm Al-Qura University.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: Samples of the compounds are available from the authors. Subject to conditions and terms.
References
- 1.Registry S.C. Saudi Cancer Registry Cancer Incidence Report Saudi Arabia, 2014; 2017, 1–81. [(accessed on 1 April 2020)];2017 Available online: https://nhic.gov.sa/eServices/Documents/2014.pdf.
- 2.Bawazir A., Al-Zamel N., Amen A., Akiel M.A., Alhawiti N.M., AlShehri A. The burden of leukemia in the Kingdom of Saudi Arabia: 15 years period (1999–2013) Bmc Cancer. 2019;19:703. doi: 10.1186/s12885-019-5897-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zahrani M., Al-Quozi A., Alaskar A., Faleh A. Clinical features and outcome of acute myeloid leukemia, a single institution experience in Saudi Arabia. J. Appl. Hematol. 2015;6:6. doi: 10.4103/1658-5127.155171. [DOI] [Google Scholar]
- 4.Lai C., Doucette K., Norsworthy K. Recent drug approvals for acute myeloid leukemia. J. Hematol. Oncol. 2019;12:100–120. doi: 10.1186/s13045-019-0774-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilliland D., Griffin J.D. The roles of FLT3 in hematopoiesis and leukemia. Blood. 2002;100:1532–1542. doi: 10.1182/blood-2002-02-0492. [DOI] [PubMed] [Google Scholar]
- 6.Antar A.I., Otrock Z.K., Jabbour E., Mohty M., Bazarbachi A. FLT3 inhibitors in acute myeloid leukemia: Ten frequently asked questions. Leukemia. 2020;34:682–696. doi: 10.1038/s41375-019-0694-3. [DOI] [PubMed] [Google Scholar]
- 7.Zhang J., Gu Y., Chen B. Mechanisms of drug resistance in acute myeloid leukemia. Oncotargets. 2019;12:1937–1945. doi: 10.2147/OTT.S191621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ball B., Stein E.M. Which are the most promising targets for minimal residual disease-directed therapy in acute myeloid leukemia prior to allogeneic stem cell transplant? Haematol. 2019;104:1521–1531. doi: 10.3324/haematol.2018.208587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mancini M., Yarden Y., Belkacemi L. Resistance to Anti-Cancer Therapeutics Targeting Receptor Tyrosine Kinases and Downstream Pathways. Volume 15. Springer AG; Basel, Switzerland: 2018. pp. 29–49. [Google Scholar]
- 10.Mikhail S., Albanese C., Pishvaian M.J. Cyclin-dependent kinase inhibitors and the treatment of gastrointestinal cancers. Am. J. Pathol. 2015;185:1185–1197. doi: 10.1016/j.ajpath.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee D.J., Zeidner J.F. Cyclin-dependent kinase (CDK) 9 and 4/6 inhibitors in acute myeloid leukemia (AML): A promising therapeutic approach. Expert Opin. Investig. Drugs. 2019;28:989–1001. doi: 10.1080/13543784.2019.1678583. [DOI] [PubMed] [Google Scholar]
- 12.Baker A., Gregory G., Verbrugge I., Kats L.M., Hilton J.J., Vidacs E., Lee E.M., Lock R.B., Zuber J., Shortt J., et al. The CDK9 Inhibitor Dinaciclib Exerts Potent Apoptotic and Antitumor Effects in Preclinical Models of MLL-Rearranged Acute Myeloid Leukemia. Cancer Res. 2015;76:1158–1169. doi: 10.1158/0008-5472.CAN-15-1070. [DOI] [PubMed] [Google Scholar]
- 13.Trepel J., Mollapour M., Giaccone G., Neckers L. Targeting the dynamic HSP90 complex in cancer. Nat. Rev. Cancer. 2010;10:537–549. doi: 10.1038/nrc2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neckers L.M., Workman P. Hsp90 molecular chaperone inhibitors: Are we there yet? Clin. Cancer Res. 2012;18:64–76. doi: 10.1158/1078-0432.CCR-11-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Proia D.A., Bates R.C. Ganetespib and HSP90: Translating Preclinical Hypotheses into Clinical Promise. Cancer Res. 2014;74:1294–1300. doi: 10.1158/0008-5472.CAN-13-3263. [DOI] [PubMed] [Google Scholar]
- 16.Lanneau D., De Thonel A., Maurel S., Mirjolet C., Garrido C. Apoptosis Versus Cell Differentiation. Prion. 2007;1:53–60. doi: 10.4161/pri.1.1.4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimura T., Uesugi M., Takase K., Miyamoto N., Sawada K. Hsp90 inhibitor geldanamycin attenuates the cytotoxicity of sunitinib in cardiomyocytes via inhibition of the autophagy pathway. Toxicol. Appl. Pharm. 2017;329:282–292. doi: 10.1016/j.taap.2017.06.015. [DOI] [PubMed] [Google Scholar]
- 18.Woodhead A.J., Angove H., Carr M.G., Chessari G., Congreve M., Coyle J.E., Cosme J., Graham B., Day P.J., Downham R., et al. Discovery of (2,4-Dihydroxy-5-isopropylphenyl)-[5-(4-methylpiperazin-1-ylmethyl)-1,3-dihydroisoindol-2-yl]methanone (AT13387), a Novel Inhibitor of the Molecular Chaperone Hsp90 by Fragment Based Drug Design. J. Med. Chem. 2010;53:5956–5969. doi: 10.1021/jm100060b. [DOI] [PubMed] [Google Scholar]
- 19.Dias S., Shmelkov S.V., Lam G., Rafii S. VEGF165 promotes survival of leukemic cells by Hsp90-mediated induction of Bcl-2 expression and apoptosis inhibition. Blood. 2002;99:2532–2540. doi: 10.1182/blood.V99.7.2532. [DOI] [PubMed] [Google Scholar]
- 20.Xue N., Jin J., Liu D., Yan R., Zhang S., Yu X., Chen X. Antiproliferative effect of HSP90 inhibitor Y306zh against pancreatic cancer is mediated by interruption of AKT and MAPK signaling pathways. Curr. Cancer Drug Targets. 2014;14:671–683. doi: 10.2174/1568009614666140908101523. [DOI] [PubMed] [Google Scholar]
- 21.Gyurkocza B., Plescia J., Raskett C.M., Garlick D.S., Lowry P.A., Carter B.Z., Andreeff M., Meli M., Colombo G., Altieri D.C. Antileukemic Activity of Shepherdin and Molecular Diversity of Hsp90 Inhibitors. J. Natl. Cancer Inst. 2006;98:1068–1077. doi: 10.1093/jnci/djj300. [DOI] [PubMed] [Google Scholar]
- 22.Azimi A., Caramuta S., Seashore-Ludlow B., Boström J., Robinson J.L., Edfors F., Tuominen R., Kemper K., Krijgsman O., Peeper D.S., et al. Targeting CDK 2 overcomes melanoma resistance against BRAF and Hsp90 inhibitors. Mol. Syst. Boil. 2018;14:e7858. doi: 10.15252/msb.20177858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stebbins E., A Russo A., Schneider C., Rosen N., Hartl F.U., Pavletich N.P. Crystal Structure of an Hsp90–Geldanamycin Complex: Targeting of a Protein Chaperone by an Antitumor Agent. Cell. 1997;89:239–250. doi: 10.1016/S0092-8674(00)80203-2. [DOI] [PubMed] [Google Scholar]
- 24.Ou W.-B., Hubert C., Fletcher J.A., Corson J.M., Bueno R., Flynn D.L., Sugarbaker D.J. Targeted Inhibition of Multiple Receptor Tyrosine Kinases in Mesothelioma. Neoplasia. 2011;13:12–22. doi: 10.1593/neo.101156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee Y.H., Song G.G. 17-AAG enhances the cytotoxicity of flavopiridol in mantle cell lymphoma via autophagy suppression. Neoplasma. 2013;60:607–616. doi: 10.4149/neo_2015_047. [DOI] [PubMed] [Google Scholar]
- 26.Wang L., Harshman S.W., Liu S., Ren C., Xu H., Sallans L., Grever M., Byrd J.C., Marcucci G., Freitas M. Assaying pharmacodynamic endpoints with targeted therapy: Flavopiridol and 17AAG induced dephosphorylation of histone H1.5 in acute myeloid leukemia. Proteomics. 2010;10:4281–4292. doi: 10.1002/pmic.201000080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu W., Sharma S.S., Ma L., Chu B., Bui M.M., Reed D.R., Pledger W.J. Apoptosis of osteosarcoma cultures by the combination of the cyclin-dependent kinase inhibitor SCH727965 and a heat shock protein 90 inhibitor. Cell Death Dis. 2013;4:e566. doi: 10.1038/cddis.2013.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song G., Li Y., Jiang G. Role of VEGF/VEGFR in the pathogenesis of leukemias and as treatment targets (Review) Oncol. Rep. 2012;28:1935–1944. doi: 10.3892/or.2012.2045. [DOI] [PubMed] [Google Scholar]
- 29.Alkahtani H.M., Abdalla A.N., Obaidullah A.J., Alanazi M.M., Almehizia A.A., Alanazi M.G., Ahmed A.Y., Alwassil O.I., Darwish H.W., Abdel-Aziz A.A.-M., et al. Synthesis, cytotoxic evaluation, and molecular docking studies of novel quinazoline derivatives with benzenesulfonamide and anilide tails: Dual inhibitors of EGFR/HER2. Bioorganic Chem. 2020;95:103461. doi: 10.1016/j.bioorg.2019.103461. [DOI] [PubMed] [Google Scholar]
- 30.Caputo M., De Rosa M.C., Rescigno T., Zirpoli H., Vassallo A., De Tommasi N., Torino G., Tecce M.F. Binding of polyunsaturated fatty acids to LXRαand modulation of SREBP-1 interaction with a specific SCD1 promoter element. Cell Biochem. Funct. 2014;32:637–646. doi: 10.1002/cbf.3067. [DOI] [PubMed] [Google Scholar]
- 31.Malafronte N., Vassallo A., Piaz F.D., Bader A., Braca A., De Tommasi N. Biflavonoids from Daphne linearifolia Hart. Phytochem. Lett. 2012;5:621–625. doi: 10.1016/j.phytol.2012.06.008. [DOI] [Google Scholar]
- 32.Dal Piaz F., Ferro P., Vassallo A., Vasaturo M., Forte G., Chini M.G., Bifulco G., Tosco A., De Tommasi N. Identification and mechanism of action analysis of the new PARP-1 inhibitor 2″-hydroxygenkwanol A. Biochim. Et Biophys. Acta Gen. Subj. 2015;1850:1806–1814. doi: 10.1016/j.bbagen.2015.05.014. [DOI] [PubMed] [Google Scholar]
- 33.Terracciano S., Chini M.G., Vaccaro M.C., Strocchia M., Foglia A., Vassallo A., Saturnino C., Riccio R., Bifulco G., Bruno I. Identification of the key structural elements of a dihydropyrimidinone core driving toward more potent Hsp90 C-terminal inhibitors. Chem. Commun. 2016;87:12857–12860. doi: 10.1039/C6CC06379A. [DOI] [PubMed] [Google Scholar]
- 34.Cooper M.A. Label-free screening of bio-molecular interactions. Anal. Bioanal. Chem. 2003;377:834–842. doi: 10.1007/s00216-003-2111-y. [DOI] [PubMed] [Google Scholar]
- 35.Discovery Studio Visualization. Version 2.5.5.9350. [(accessed on 1 March 2020)]; Available online: https://www.3dsbiovia.com/products/collaborative-science/biovia-discovery-studio/visualization-download.php.
- 36.Laskowski R.A., Swindells M.B. LigPlot+: Multiple Ligand–Protein Interaction Diagrams for Drug Discovery. J. Chem. Inf. Model. 2011;51:2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 37.Park H., Kim Y.-J., Hahn J.-S. A novel class of Hsp90 inhibitors isolated by structure-based virtual screening. Bioorganic Med. Chem. Lett. 2007;17:6345–6349. doi: 10.1016/j.bmcl.2007.08.069. [DOI] [PubMed] [Google Scholar]
- 38.Nimmanapalli R., O’Bryan E., Kuhn D., Yamaguchi H., Wang H.G., Bhalla K.N. Regulation of 17-AAG-induced apoptosis: Role of Bcl-2, Bcl-XL, and Bax downstream of 17-AAG-mediated down-regulation of Akt, Raf-1, and Src kinases. Blood. 2003;102:269–275. doi: 10.1182/blood-2002-12-3718. [DOI] [PubMed] [Google Scholar]
- 39.Lazenby M., Hills R.K., Burnett A., Zabkiewicz J. The HSP90 inhibitor ganetespib: A potential effective agent for Acute Myeloid Leukemia in combination with cytarabine. Leuk. Res. 2015;39:617–624. doi: 10.1016/j.leukres.2015.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Totzke J., Gurbani D., Raphemot R., Hughes P.F., Bodoor K., Carlson D.A., Loiselle D.R., Bera A.K., Eibschutz L., Perkins M.M., et al. Takinib, a Selective TAK1 Inhibitor, Broadens the Therapeutic Efficacy of TNF-α Inhibition for Cancer and Autoimmune Disease. Cell Chem. Boil. 2017;24:1029–1039. doi: 10.1016/j.chembiol.2017.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fritsch J., Fickers R., Klawitter J., Särchen V., Zingler P., Adam D., Janssen O., Krause E., Schütze S. TNF induced cleavage of HSP90 by cathepsin D potentiates apoptotic cell death. Oncotarget. 2016;7:75774–75789. doi: 10.18632/oncotarget.12411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yuno A., Lee M.-j., Lee S., Tomita Y., Rekhtman D., Moore B., Trepel J.B. Chaperones Methods and Protocols. Volume 1709. Springer AG; Basel, Switzerland: 2018. Chapter 29: Clinical Evaluation and Biomarker Profiling of Hsp90 Inhibitors. [DOI] [PubMed] [Google Scholar]
- 43.Butler L.M., Ferraldeschi R., Armstrong H., Centenera M., Workman P. Maximizing the Therapeutic Potential of Hsp90 Inhibitors. Mol. Cancer Res. 2015;13:1445–1451. doi: 10.1158/1541-7786.MCR-15-0234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shaltiel I.A., Krenning L., Bruinsma W., Medema R.H. The same, only different—DNA damage checkpoints and their reversal throughout the cell cycle. J. Cell Sci. 2015;128:607–620. doi: 10.1242/jcs.163766. [DOI] [PubMed] [Google Scholar]
- 45.Camero C.M., Vassallo A., De Leo M., Temraz A., De Tommasi N., Braca A. Limonoids from Aphanamixis polystachya Leaves and Their Interaction with Hsp90. Planta Med. 2018;84:964–970. doi: 10.1055/a-0624-9538. [DOI] [PubMed] [Google Scholar]
- 46.Terracciano S., Russo A., Chini M.G., Vaccaro M.C., Potenza M., Vassallo A., Riccio R., Bifulco G., Bruno I. Discovery of new molecular entities able to strongly interfere with Hsp90 C-terminal domain. Sci. Rep. 2018;8:1709. doi: 10.1038/s41598-017-14902-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gualtieri M.J., Malafronte N., Vassallo A., Braca A., Cotugno R., Vasaturo M., De Tommasi N., Piaz F.D. Bioactive Limonoids from the Leaves of Azaridachta indica (Neem) J. Nat. Prod. 2014;77:596–602. doi: 10.1021/np400863d. [DOI] [PubMed] [Google Scholar]
- 48.Schulte T.W., Akinaga S., Soga S., Sullivan W., Stensgard B., Toft D., Neckers L.M. Antibiotic radicicol binds to the N-terminal domain of Hsp90 and shares important biologic activities with geldanamycin. Cell Stress Chaperon. 1998;3:100–108. doi: 10.1379/1466-1268(1998)003<0100:ARBTTN>2.3.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guo W., Reigan P., Siegel D., Zirrolli J., Gustafson D., Ross D. Formation of 17-Allylamino-Demethoxygeldanamycin (17-AAG) Hydroquinone by NAD(P)H:Quinone Oxidoreductase 1: Role of 17-AAG Hydroquinone in Heat Shock Protein 90 Inhibition. Cancer Res. 2005;65:10006–10015. doi: 10.1158/0008-5472.CAN-05-2029. [DOI] [PubMed] [Google Scholar]
- 50.Gouda A.M., Abdelazeem A., Omar H.A., Abdalla A.N., Abourehab M., Ali H. Pyrrolizines: Design, synthesis, anticancer evaluation and investigation of the potential mechanism of action. Bioorganic Med. Chem. 2017;25:5637–5651. doi: 10.1016/j.bmc.2017.08.039. [DOI] [PubMed] [Google Scholar]
- 51.Abdalla A.N., Shaheen U., Abdallah Q.M.A., Flamini G., Bkhaitan M.M., Abdelhady M.I.S., Ascrizzi R., Bader A. Proapoptotic Activity of Achillea membranacea Essential Oil and Its Major Constituent 1,8-Cineole against A2780 Ovarian Cancer Cells. Molecules. 2020;25:1582. doi: 10.3390/molecules25071582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alkahtani H.M., Alanazi M.M., Aleanizy F.S., Alqahtani F.Y., Alhoshani A., Alanazi F.E., Almehizia A., Abdalla A.N., Alanazi M.G., El-Azab A.S., et al. Synthesis, anticancer, apoptosis-inducing activities and EGFR and VEGFR2 assay mechanistic studies of 5,5-diphenylimidazolidine-2,4-dione derivatives: Molecular docking studies. Saudi Pharm. J. 2019;27:682–693. doi: 10.1016/j.jsps.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Y., Wang P.P., Li X.X., Yu C.Y., Yang H., Zhou J., Xue W.W., Tan J., Zhu F. The Human Kinome Targeted by FDA Approved Multi-Target Drugs and Combination Products: A Comparative Study from the Drug-Target Interaction Network Perspective. PLoS ONE. 2016;11:e0165737. doi: 10.1371/journal.pone.0165737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin S.-Y., Hu F.-F., Miao Y.-R., Hu H., Lei Q., Zhang Q., Li Q., Wang H., Chen Z., Guo A.-Y. Identification of STAB1 in Multiple Datasets as a Prognostic Factor for Cytogenetically Normal AML: Mechanism and Drug Indications. Mol. Nucleic Acids. 2019;18:476–484. doi: 10.1016/j.omtn.2019.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chou T.-C. Drug Combination Studies and Their Synergy Quantification Using the Chou-Talalay Method. Cancer Res. 2010;70:440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 56.Shaheen U., Ragab E.A., Abdalla A.N., Bader A. Triterpenoidal saponins from the fruits of Gleditsia caspica with proapoptotic properties. Phytochem. 2018;145:168–178. doi: 10.1016/j.phytochem.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 57.Grothaus P.G., Cragg G.M., Newman D.J. Plant Natural Products in Anticancer Drug Discovery. Curr. Org. Chem. 2010;14:1781–1791. doi: 10.2174/138527210792927708. [DOI] [Google Scholar]
- 58.Ghobrial I.M., Witzig T.E., Adjei A.A. Targeting apoptosis pathways in cancer therapy. Ca A Cancer J. Clin. 2005;55:178–194. doi: 10.3322/canjclin.55.3.178. [DOI] [PubMed] [Google Scholar]
- 59.Malki W.H., Gouda A.M., Ali H.E., Al-Rousan R., Samaha D., Abdalla A.N., Bustamante J., Elmageed Z.Y.A., Ali H. Structural-based design, synthesis, and antitumor activity of novel alloxazine analogues with potential selective kinase inhibition. Eur. J. Med. Chem. 2018;152:31–52. doi: 10.1016/j.ejmech.2018.04.029. [DOI] [PubMed] [Google Scholar]
- 60.El-Boshy M., Basalamah M.A., Ahmad J., Idris S., Mahbub A., Abdelghany A.H., Almaimani R.A., Almasmoum H., Ghaith M.M., Elzubier M., et al. Vitamin D protects against oxidative stress, inflammation and hepatorenal damage induced by acute paracetamol toxicity in rat. Free. Radic. Boil. Med. 2019;141:310–321. doi: 10.1016/j.freeradbiomed.2019.06.030. [DOI] [PubMed] [Google Scholar]
- 61.Burnette W. “Western Blotting”: Electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal. Biochem. 1981;112:195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.