Abstract
The term ‘extracellular vesicles’ refers to a heterogeneous population of vesicular bodies of cellular origin that derive either from the endosomal compartment (exosomes) or as a result of shedding from the plasma membrane (microvesicles, oncosomes and apoptotic bodies). Extracellular vesicles carry a variety of cargo, including RNAs, proteins, lipids and DNA, which can be taken up by other cells, both in the direct vicinity of the source cell and at distant sites in the body via biofluids, and elicit a variety of phenotypic responses. Owing to their unique biology and roles in cell–cell communication, extracellular vesicles have attracted strong interest, which is further enhanced by their potential clinical utility. Because extracellular vesicles derive their cargo from the contents of the cells that produce them, they are attractive sources of biomarkers for a variety of diseases. Furthermore, studies demonstrating phenotypic effects of specific extracellular vesicle-associated cargo on target cells have stoked interest in extracellular vesicles as therapeutic vehicles. There is particularly strong evidence that the RNA cargo of extracellular vesicles can alter recipient cell gene expression and function. During the past decade, extracellular vesicles and their RNA cargo have become better defined, but many aspects of extracellular vesicle biology remain to be elucidated. These include selective cargo loading resulting in substantial differences between the composition of extracellular vesicles and source cells; heterogeneity in extracellular vesicle size and composition; and undefined mechanisms for the uptake of extracellular vesicles into recipient cells and the fates of their cargo. Further progress in unravelling the basic mechanisms of extracellular vesicle biogenesis, transport, and cargo delivery and function is needed for successful clinical implementation. This Review focuses on the current state of knowledge pertaining to packaging, transport and function of RNAs in extracellular vesicles and outlines the progress made thus far towards their clinical applications.
Subject terms: Extracellular signalling molecules, Organelles, Biologics
Extracellular vesicles transfer a variety of cellular components between cells — including proteins, lipids and nucleic acids. There is now evidence indicating that these cargoes, in particular RNAs, can affect the function of recipient cells. Extracellular vesicles are now being actively tested as biomarkers and delivery vehicles for therapeutic agents.
Introduction
Extracellular vesicles are membrane-enclosed nanoscale particles released from essentially all prokaryotic and eukaryotic cells that carry proteins, lipids, RNA and DNA. They can deliver information between cells through the extracellular space, with strong evidence for functional activity provided by their phenotypic effects on recipient cells. Cargo RNAs of extracellular vesicles include various biotypes that represent a selected portion of the RNA content of the source cell, with a strong bias towards small non-coding RNAs, although fragmented and intact mRNA, ribosomal RNA (rRNA) and long non-coding RNA (lncRNA) molecules can be found. Ample evidence supports the ability of RNAs enclosed in extracellular vesicles to impact the functional properties of cells that take them up (Table 1). For example, extracellular vesicles in plasma promote repair of cardiac cells1, vesicles released by astrocytes contribute to neurodegeneration2 and tumour-derived vesicles create a favourable microenvironment for cancer progression3 and metastases4. However, given that the functions of many non-coding RNAs (ncRNAs) are incompletely understood and that delivery of any given extracellular vesicle-associated RNA is accompanied by delivery of multiple other biomolecules, the complex language of extracellular vesicle-mediated communication remains to be deciphered. Moreover, the cellular mechanisms involved in the trafficking and fate of extracellular RNAs (exRNAs), including the possibility of packaging different RNA sequences into different types of extracellular vesicles, differential uptake into specific recipient cell types and fate on uptake are still being elucidated (Fig. 1).
Table 1.
Functional delivery of miRNAs by extracellular vesicles
| Source cell | RNA | Target cell | Effect | Refs |
|---|---|---|---|---|
| Rhabdomyosarcoma | miR-486-5p | Mouse embryonic fibroblasts, C2C12 immortalized mouse myoblast cell line, immortalized myoblasts | Cell migration, invasion, colony formation (in vitro) | 258 |
| Glioblastoma | miR-9 miR-21 | Brain endothelial cells | Angiogenesis (in vitro) | 3,259 |
| Microglia | Immunosuppression (in vivo) | |||
| Breast cancer cells | Various | Epithelial cells | Promote tumorigenesis, invasion, cell proliferation (in vitro) | 260 |
| Endothelial cells | Promote angiogenesis (in vitro) | |||
| Breast cancer cells | Increase drug resistance (in vitro) | |||
| miR-9, miR-195, miR-203 | Cancer stem cells | Increase expression of stemness genes (in vitro and in vivo) | 261 | |
| Cardiac progenitor cells | miR-210, miR-132, miR-21, miR-451, miR-146a | Various | Inhibit apoptosis, promote angiogenesis, improve cardiac function, inhibit myocardial fibrosis (in vitro and in vivo) | Reviewed in262 |
| Cardiomyocytes | miR-320 | Various cell types | Inhibit angiogenesis (in vivo) | Reviewed in263 |
| miR-30a | Regulate autophagy (in vitro) | |||
| miR-29b, miR-455 | Inhibit fibrosis (in vivo) | |||
| miR-27a, miR-28-3p, miR-34a | Contribute to oxidative stress (in vitro) | |||
| miR-208a | Promote fibrosis (in vivo) | |||
| Neuroprecursor cells | miR-21a | Neural progenitor cells | Promote neurogenesis (in vitro) | 264 |
| Adipose macrophage | miR-155 | Bone marrow mesenchymal stem cells | Increase insulin resistance (in vitro and in vivo) | 265 |
| Adipose mesenchymal stem cells | miR-375 | Bone marrow mesenchymal stem cells | Osteogenic differentiation, enhance bone regeneration (in vitro and in vivo) | 266 |
| Adipocytes (from brown adipose tissue) | miR-99b | Liver | Increase Fgf21 expression, increase glucose tolerance (in vivo) | 267 |
miRNA, microRNA.
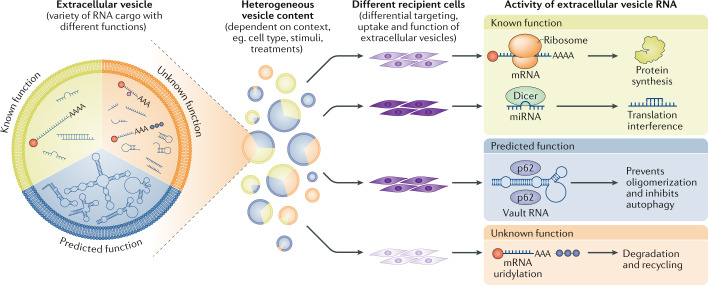
Fig. 1. Principles of functional cell communication by extracellular vesicle RNA.
Extracellular vesicles are generated as highly heterogeneous populations with different types of RNA cargo within them and in different amounts and proportions. Functionally, these RNAs can be divided into those with known functions, for example some mRNA, microRNA (miRNA) and small interfering RNA (green zone), those with predicted functions, for example, some transfer RNA, small nucleolar RNA, small nuclear RNA, Y RNA and vault RNA (blue zone) and those with unknown functions, for example, fragmented and degraded (methylated and uridylidated) RNA species (orange zone). This heterogeneity is further enhanced by the fact that extracellular vesicle cargo content strongly depends on the context (for example, cell type, stimuli and treatments). The effect that different kinds of RNA in vesicles can have on recipient cells is dictated in part by the nature of these cells, which will show differential capability for recognizing specific vesicles, their uptake and ultimately their functional effect.
The RNA contained in extracellular vesicles reflects the type and the physiological/pathological state of the source cells, but differs substantially from the cellular RNA content, in terms of both the types of RNA and the relative concentrations of specific RNA sequences. The extracellular vesicle populations carried in biofluids, tissues and conditioned medium from cultured cells are heterogeneous with respect to size, morphology and composition. Four major subclasses of extracellular vesicles appear to arise from distinct biogenesis pathways and can be distinguished roughly in the basis of size: exosomes (50–150 nm), microvesicles (100–1,000 nm), large oncosomes (1,000–10,000 nm) and apoptotic bodies (100–5,000 nm), but are difficult to distinguish from high-density and low-density lipoproteins, chylomicrons, protein aggregates and cell debris5. Guidelines for standardization of terminology, methods and reporting are being developed to improve experimental reproducibility across studies6,7. The size of most extracellular vesicles (which also limits the number of cargo molecules/vesicles) places them below the resolution and sensitivity thresholds of standard light microscopy and fluorescence-activated sorting techniques. Overlap in the sizes and other biophysical properties among different extracellular vesicle subclasses and lack of known unique markers for each subclass8,9 have made it difficult to define the cargo (including RNAs) of different subclasses with confidence5. Technical factors, including the use of different methods for isolation of extracellular vesicles and their RNA, can strongly influence RNA profiling results (see, for example, refs10–16). Separation of RNA in vesicles from RNAs associated with other exRNA carriers, including lipoproteins17 and ribonucleoproteins18, is also challenging (see refs5,6,10,17,18 and the exRNA Atlas11). A variety of approaches have been used to address these concerns, including culture of cells in serum-free medium (to avoid contamination with serum-derived extracellular vesicles) and separation of extracellular vesicle subclasses and other exRNA carriers by high-resolution density gradient centrifugation10, size-exclusion chromatography19, asymmetric field-flow fractionation20,21 and immunoaffinity purification9.
In addition to serving as a novel mode of communication among cells, RNAs in extracellular vesicles can also serve as biomarkers and therapeutics for a variety of diseases. Since extracellular vesicles have been found in all tested biofluids, many of which can be collected non-invasively, the RNAs within them can be interrogated in real time to evaluate the functional state of a range of cell and tissue types22. The potential of extracellular vesicles to protect nucleic acids and other biological macromolecules from degradation in vivo, and to deliver them to recipient cells in a targeted manner without immune activation, makes them an exciting new modality for delivery of RNA therapeutics, including small interfering RNAs (siRNAs), microRNAs (miRNAs), antisense oligonucleotides, mRNAs, guide RNAs and self-amplifying RNAs.
In this Review, we discuss the biogenesis, biological functions and emerging clinical applications of extracellular vesicle RNAs in mammalian cells. Throughout, we highlight the key open questions and technical challenges that currently limit the broader translational use of extracellular vesicles. We are optimistic that these hurdles will be overcome soon, allowing better understanding and control of extracellular vesicle-mediated delivery of RNAs to specific cellular targets and opening up new opportunities for the detection and treatment of various diseases, including cardiovascular, neurodegenerative and metabolic diseases and cancers.
Extracellular vesicles as RNA carriers
Extracellular vesicles carry within them a wide variety of RNA sequences representing many biotypes of RNA. This diversity of RNAs, as well as the small size and heterogeneity of extracellular vesicles and the overall low concentration of RNA in extracellular vesicles, has complicated characterization of the RNA cargo of extracellular vesicle subclasses (Box 1; Table 2).
Table 2.
Challenges and limitations for extracellular vesicle and exRNA research
| Challenge/limitation | Consequences | Potential solutions |
|---|---|---|
| Heterogeneity of extracellular vesicles and their (RNA) cargo (differences between cell types, influence of external conditions, additional stimuli, etc.) |
Lack of concordance among studies using different cell types or biofluid sources Inability to determine which mechanisms/features of extracellular vesicle biogenesis and cargo selection are universal and which are cell type specific Inability to precisely manipulate the cargo content for improved transfer of that content |
Systematic studies of extracellular vesicle biogenesis and cargo content including multiple diverse cell types |
| Sample-to-sample heterogeneity in extracellular vesicle and other exRNA carriers | High sample-to-sample technical variability that is not related to the biological variable of interest | Develop methods to determine the relative abundance of each carrier subclass in individual samples |
| Differences (efficiency, purity, throughput, etc.) in methods for isolating extracellular vesicle carriers |
Lack of concordance among studies using different extracellular vesicle and exRNA isolation techniques Difficulties in determining exRNA activity on transfer |
Develop methods for unbiased exRNA isolation Establish a comprehensive catalogue of extracellular vesicle and non-extracellular vesicle exRNA carriers Develop methods to isolate each carrier and characterize its cargo, and to perform single-extracellular-vesicle cargo analysis |
| Differences in sensitivity, specificity, reproducibility and bias for different RNA profiling methods | Lack of concordance among studies using different exRNA profiling techniques |
Develop methods for unbiased exRNA profiling Establish standards that can be used to compare sensitivity, specificity and bias among methods |
exRNA, extracellular RNA.
Box 1 Technical considerations in characterization of extracellular vesicle RNA cargo.
Different RNA isolation and measurement methods will produce different extracellular RNA (exRNA) profiles from the same biofluid or RNA sample9,11,16,24,112,285–291 owing to:
unique properties of each exRNA carrier type, such as susceptibility to different chemical disruption methods
biases of each RNA isolation and profiling method
Quantification of the stoichiometry of extracellular vesicle cargo is difficult because:
there are low numbers of cargo molecules per vesicle. On average, only one microRNA per extracellular vesicle35 to one microRNA per 100 extracellular vesicles292 and one intact long RNA molecule (for example, a full-length mRNA) per 1,000 extracellular vesicles112
most sensitive methods (quantitative reverse transcription–PCR and Droplet Digital PCR) are tailored to specific RNA species. However, different RNA sequences can differ greatly in abundance, so it is difficult to accurately extrapolate measurements obtained for one specific RNA to other RNAs
RNA biotypes are not evenly distributed among the different extracellular vesicle subtypes112,135
concordance among existing methods for quantification of extracellular vesicles is poor293–295
single-vesicle characterization methods (for example, through simultaneous visualization of extracellular vesicle protein markers and RNA cargo by super-resolution microscopy296) are needed. Current methods are targeted and of low throughput
Different strategies can be applied to allow profiles from different biological groups (for example, cases and controls for a given disease) to be reliably compared:
use the same method throughout a study. Intramethod reproducibility of many exRNA isolation and analysis methods is quite high, so consistent use of the same method allows differential expression studies to be performed
use computational approaches to separate (or deconvolute) the data from a mixed population of exRNA carriers into profiles of the component carrier subclasses11
Candidate biomarkers and signalling RNAs identified by comprehensive exRNA profiling methods should be validated:
biomarkers should be validated in multiple independent study populations
candidate signalling RNAs should be functionally validated by multiple approaches
Extracellular vesicle RNA content
To understand intercellular communication mediated by extracellular vesicle RNAs, it is important to develop a comprehensive catalogue of the types of RNAs and sequences within them. Initial studies reported that extracellular vesicles contained mRNAs and mature miRNA sequences, as well as ncRNAs, with a peak size of 200 nucleotides, but extending out to 5 kb or more23–25. Since then, more comprehensive studies have found the majority of known ncRNA biotypes, including small nuclear RNAs, small nucleolar RNAs (snoRNAs), rRNAs, lncRNAs, PIWI-interacting RNAs (piRNAs), transfer RNAs (tRNAs), mitochondrial RNAs, Y RNAs and vault RNAs (vtRNAs)26–28. miRNA precursors (pre-miRNAs) have also been reported in tumour microvesicles, along with the RNA interference processing machinery components Dicer and Argonaute29–31. However, the methods used in these studies did not separate extracellular vesicles from free ribonucleoproteins16 or lipoproteins32. Moreover, among the many studies using proteomic methods to analyse the composition of extracellular vesicles, only one has reported the presence of Argonaute 2 (AGO2)33, and none has detected Dicer. One study that used density gradient ultracentrifugation to isolate extracellular vesicles from cell culture supernatant of cancer cells did convincingly demonstrate that overexpression of the KRAS oncogene promotes sorting of AGO2-associated miRNAs into extracellular vesicles34. Thus, whether pre-miRNAs and their processing machinery are transferred via extracellular vesicles in normal cells remains to be established. Finally, extracellular vesicle-associated circular RNAs have also been detected by use of specific data analysis pipelines that can identify back-spliced transcripts35–40.
In general, RNA in extracellular vesicles can be categorized into three types: RNAs that have been established to be functional when carried by extracellular vesicles between cells, such as intact mRNAs and miRNAs; intact RNAs that are predicted to be functional but have not been definitively demonstrated to mediate intercellular communication (for example, piRNAs and vtRNAs); and fragments of RNAs (for example, tRNA fragments and fragments of mRNAs and rRNAs), some of which may be functional but others may be non-functional degradation products (Fig. 1).
Specific enrichment of RNAs in extracellular vesicles
The transcriptomes of different cell types are partially reflected in their extracellular vesicle RNA cargo14,41, but the RNA profiles of extracellular vesicles differ substantially from those of their cells of origin24,42–44, suggesting that some RNA species are selectively incorporated into extracellular vesicles. A recent small RNA-sequencing study focusing on 17–35 nucleotide sequences from extracellular vesicles across five cell lines found that rRNA fragments accounted for 30–94% of the reads and ‘small RNAs’ accounted for 2–40% of the reads, depending on the cell type41. In the ‘small RNA’ category, approximately 15% in some lines and nearly 80% in others mapped to miRNAs, which inversely correlated with the abundance of piRNAs (ranging from 80% to 20%). Notably all of the cell lines included in that study were highly aneuploid, and therefore the results might not reflect the RNA composition of extracellular vesicles in cells with normal genomic composition.
In addition to these differences in RNA biotypes, specific RNA sequences are packaged into extracellular vesicles with different efficiencies. Early studies focused on preferential loading of specific miRNAs into extracellular vesicles45,46, and a recent study found that during T cell activation, tRNA fragments are preferentially enriched in extracellular vesicles compared with miRNAs47. The physiological states of cells can also impact exRNA profiles, including oxidative stress48, pain49 and exercise50–52, all of which alter levels of specific extracellular miRNAs in conditioned medium from cultured cells, as well as in patient serum/plasma.
Packaging of extracellular vesicle RNA
Biogenesis of extracellular vesicles is relatively well studied, and several modes and molecular components that drive the process have been established5,53 (Fig. 2). Exosomes are generated by invagination of the endosomal membrane forming intraluminal vesicles within multivesicular bodies (MVBs). This process involves participation of the endosomal sorting complex required for transport, as well as RAB27A/RAB27B, tumour susceptibility gene 101 protein (TSG101), ALIX, vacuolar protein sorting-associated protein 4 (VPS4)5 and lipid microdomains (lipid rafts)54. When MVBs fuse with the plasma membrane, the intraluminal vesicles are released into the extracellular space as exosomes. Exosome release relies on the cytoskeleton, such that MVBs are transported to the plasma membrane via microtubules55, where branched actin, polymerized by Arp2/3 and stabilized by the actin-binding protein cortactin, supports their docking56. Microvesicles bud out from the plasma membrane directly and share many of the same proteins involved in exosome biogenesis5,20, which makes it challenging to experimentally dissect and manipulate the different extracellular vesicle carriers. In addition to these two basic modes of extracellular vesicle biogenesis, oncosomes form by blebbing off the plasma membrane of tumour cells. These extracellular vesicles are usually larger than microvesicles and are typically associated with cell motility57–60. Release of oncosomes involves recruitment of the actin-bundling protein fascin, which together with the actin–plasma membrane crosslinker ezrin and the cell-surface glycoprotein podocalyxin promotes extrusion and scission of the plasma membrane29. Other large extracellular vesicles, overlapping in size with oncosomes, form during apoptotic cell death when plasma membrane blebs and spike-like protrusions from the plasma membrane (microtubule spikes) break off during apoptosis (forming apoptopodia)5,60. Finally, some extracellular vesicles can be generated with the engagement of proteins of viral origin, such as retroviral-like Gag proteins on the inner plasma membrane that bind RNA and bud off from the cell membrane as virus-like particles.
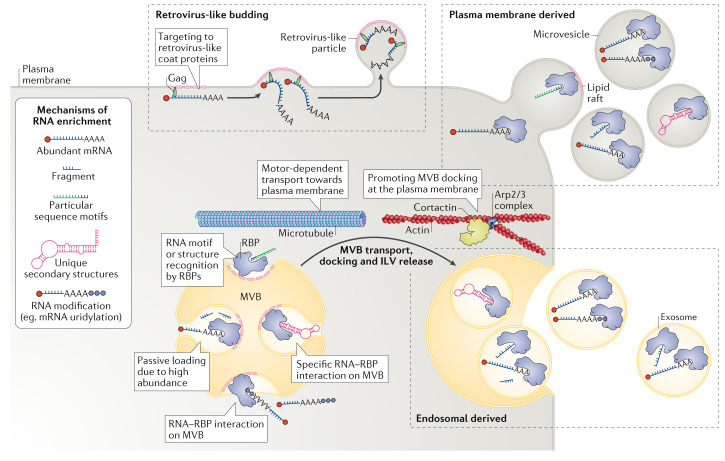
Fig. 2. RNA packaging into extracellular vesicles and their release into the extracellular space.
A variety of different RNA species can be packaged into extracellular vesicles. A number of modalities have been proposed for incorporation of (specific) RNAs into extracellular vesicles. First, RNAs can be targeted to the plasma membrane and released as microvesicles. They can also be targeted to the endosomal compartment and incorporated into intraluminal vesicles (ILVs) of the multivesicular body (MVB), which then can be targeted to the plasma membrane, where it fuses to release ILVs as as exosomes. Both these modes of biogenesis share many factors, and hence the vesicle type and vesicle origin are typically difficult to ascertain and control. Membrane microdomains (lipid rafts) have been strongly associated with the release of extracellular vesicles. Also, cytoskeletal components are implicated in extracellular vesicle biogenesis, in particular for exosomes, which are transported via microtubules, and their docking at the plasma membrane is supported by Arp2/3-generated branched-actin filaments stabilized by the actin-bundling activity of cortactin. Cortical actin remodelling is also an important event in membrane shaping during microvesicle release (not shown). Loading of RNA into extracellular vesicles can occur via multiple routes: passively due to an abundance of the RNA in the cytosol; by recognition via a number of RNA-binding proteins (RBPs), such as Argonaute, annexin A2, major vault protein (MVP), heterogeneous nuclear ribonucleoproteins A2/B1 (HNRNPA2B1),YBX1, SYNCRIP and lupus La protein, that bind particular sequence motifs in the RNA or that recognize unique secondary RNA structures; and through specific modifications, such as uridylation. Packaging of RNA into extracellular vesicles can also be promoted by its recognition by retroviral coat proteins such as Gag (and their silent copies present in animal genomes), which efficiently target RNAs they recognize to the plasma membrane (or the membrane of the MVB; not shown), resulting in virus-like particle release.
During extracellular biogenesis, the cell’s vesiculation machinery packages multiple RNA species into different subclasses of extracellular vesicles, contributing to a substantial portion of the exRNA pool61. Small size, high abundance, ability to associate with membranes and cytoplasmic (versus nuclear) location favour incorporation of a given RNA into extracellular vesicles. Most RNAs transit from the nucleus to specific cellular locations in association with RNA-binding proteins (RBPs), which can coalesce into large ribonucleoprotein particles (containing hundreds of different RNAs and many RBPs) that travel along the cytoskeleton62,63. Of note, there are more than 500 RBPs in mammalian cells64, and RBPs make up about 25% of the protein content of extracellular vesicles41. Information is lacking about the distribution of these RBPs in different extracellular vesicles and their role in loading RNA into these vesicles.
Multiple mechanisms have been implicated in RNA packaging into extracellular vesicles, including specific RNA sequence motifs and/or secondary configuration65,66, differential affinity for membrane lipids67–69 and association with RBPs, such as AGO2 (ref.34), ALIX70, annexin A2 (refs71,72), major vault protein (MVP)73,74, HuR75, heterogeneous nuclear ribonucleoproteins A2/B1 (HNRNPA2B1) (ref.76), YBX1 (refs77–79), SYNCRIP80,81, lupus La protein82 and Arc1 (ref.83) (Fig. 2). Two RBPs, heterogeneous nuclear ribonucleoprotein K (HNRNPK) and scaffold-attachment factor B1 (SAFB), regulate the small non-coding RNA composition of extracellular vesicles via secretory autophagy84. Different RBPs display binding preferences for different RNA sequence motifs, but a full understanding of these binding interactions has yet to be established (see, for example, refs71,76,80,85–87). Other extracellular vesicle sorting signals may include RNA and/or RBP modifications, such as ubiquitylation, sumoylation, phosphorylation and uridylation, which also impact RNA splicing, stability and translation and miRNA biogenesis88. Exosome biogenesis and miRNA-mediated gene silencing may be functionally linked89,90. The small GTPase ARF6 appears to be involved in targeting of pre-miRNAs to oncosomes along with miRNA processing machinery29. Thus, many mechanisms can influence packaging of RNAs into extracellular vesicles, but the extent to which these mechanisms are specific to selected RNA species and how RNA cargo loading is regulated by the cells remain elusive.
Extracellular vesicle RNA delivery
A range of mechanisms have been elucidated for extracellular vesicle uptake by recipient cells; however, what dictates how particular cells respond to specific extracellular vesicles remains unknown5. While uptake of vesicles is relatively well characterized, more work is needed to reveal the mechanisms by which extracellular vesicles deliver functional RNA to recipient cells.
Extracellular vesicle interactions with recipient cells
Interaction of extracellular vesicles with recipient cells depends on the specific proteins, lipids and glycans on their surfaces, as well as their overall negative charge14,91. Uptake of extracellular vesicles can be both selective and non-specific5,53. For example, extracellular vesicles from tumour cells may home to different tissues by virtue of the expression of specific integrins on their surface53, thereby setting up a tissue-specific niche for incoming metastatic tumour cells4. Once they are in contact with recipient cells, multiple routes of extracellular vesicle uptake have been reported, including clathrin/caveolin-mediated endocytosis, macropinocytosis, phagocytosis, lipid raft-mediated uptake or direct membrane fusion92 (Fig. 3). There are also reports of direct cell-to-cell extracellular vesicle transfer via tunnelling nanotubes for RNA delivery93–95. The current focus of extracellular vesicle uptake is on endocytic pathways, which may result in part from the difficulty in assessing fusion events. Extracellular vesicles may also remain bound to cell surfaces without internalization (Fig. 3), engaging in cell signalling and antigen presentation53. It is currently unknown whether cells control the interaction and uptake of extracellular vesicles depending on their subtype and/or physiological state.
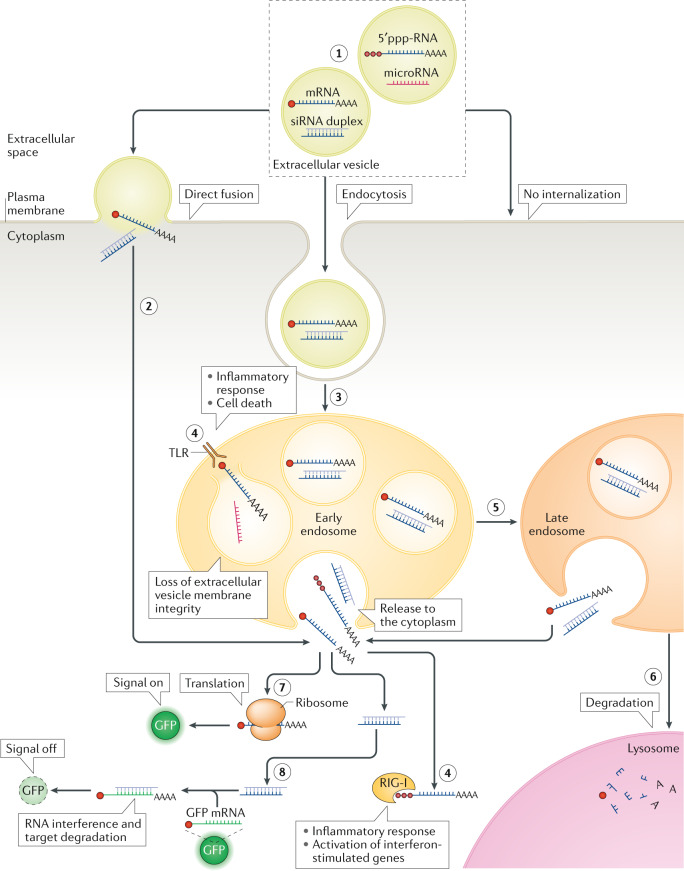
Fig. 3. Extracellular vesicle RNA cargo interaction with recipient cells and its functional delivery.
After encountering the recipient cell, the extracellular vesicle is typically bound to its surface via cell-surface receptors (although extracellular vesicles can also be engulfed from the environment in a process known as macropinocytosis; not shown). After establishing an interaction with the cell surface, the vesicle can remain bound on the surface or can be internalized (1). One possible means of internalization is direct fusion with the plasma membrane (2), but the most common mechanism of internalization involves endocytosis, whereby extracellular vesicles are taken up to endosomes (3). In the endosome, RNA content might be released into the luminal space (if the integrity of the extracellular vesicle membrane is perturbed) or it might be released into the cytoplasm (of note, the frequency of these events is low, and endosomal escape of extracellular vesicle cargo is currently an important bottleneck in functional RNA cargo delivery by extracellular vesicles). In both cases RNAs can be recognized by pattern recognition receptors, such as Toll-like receptors (TLRs) and RIG-I or NOD-like receptors that reside in the endosome and in the cytoplasm, respectively, raising innate immune response signalling (4). Early endosomes will gradually transform into late endosomes with progressive internal acidification and possible release of RNA (stimulated by the decreasing pH) (5). Further down the endocytic pathway, endosomes will mature into lysosomes, in which the cargo that has not been released to the cytoplasm will be degraded (6). RNA cargo that reaches the cytoplasm can elicit its functional effect. For example, mRNA can be translated into a functional protein, such as green fluorescent protein (GFP), and the resulting fluorescence can act as a reporter of functional delivery of extracellular vesicle cargo (7). When small interfering RNA (siRNA) cargo is released into the cytoplasm, it can inhibit translation of specific transcripts, such as those encoding fluorescent proteins. In this case, disappearance of fluorescence will report on functional delivery of extracellular vesicle cargo (8). Extracellular vesicles can be tracked along this route with use of different reagents or labelling strategies (Table 3). 5′ppp-RNA, 5′- triphosphorylated RNA.
Extracellular vesicle uptake and RNA release into recipient cells
For functional activity of RNAs contained in extracellular vesicles within recipient cells, the RNAs need to enter the cytoplasm, and if the RNA enter, through the endosomal pathway, the RNA must ‘escape’ from it (Fig. 3; Supplementary Table 1). Uptake of extracellular vesicles can be promoted at the cell surface. For example, binding to heparan sulfate proteoglycans on the plasma membrane increased uptake of tumour-derived extracellular vesicles via endocytosis96,97, and specific integrin receptors can drive selective uptake of extracellular vesicles by certain primary tumours98. Uptake of extracellular vesicles was also promoted by the interaction of T cell immunoglobulin and mucin domain-containing protein 4 (TIM4) molecules with phosphatidylserine present on extracellular vesicle membranes99. Accordingly, these different types of molecules could be engineered on the surface of extracellular vesicles to promote their uptake99.
Importantly, extracellular vesicle uptake through the endosomal and engulfment (phagocytosis and macropinocytosis) pathways poses challenges to delivery of functional RNA into the cytoplasm, as a prominent role of these pathways is to shuttle cargo to the lysosome for degradation100. Indeed, it has been demonstrated that despite successful uptake, generation of functional protein from extracellular vesicle-derived mRNA in recipient cells was negligible, suggesting that endosomal escape is critical to RNA function101. It is possible that extracellular vesicles might naturally incorporate some mechanisms for endosomal escape, which so far remain unknown. Nevertheless, establishing reliable means of increasing the efficiency of extracellular vesicle cargo escape from the endosomal compartment will likely be essential to widen the application potential of extracellular vesicles (see also later).
Progress on understanding extracellular vesicle cargo delivery to recipient cells has been hampered by the lack of sensitive and specific markers for optical and molecular tracking. Protein delivery can be monitored by use of a GFP–luciferase dual split tag with a fluorescent and bioluminescent readout when extracellular vesicle cargo is united with its other half in the cytosol102. Tools developed for tracking of nanoparticles, liposomes and viruses may prove useful in elucidating these mechanisms (Table 3; Supplementary Table 1). RNA cargo delivery could be facilitated by use of recipient cells null for the RNA being tracked, but this experimental manipulation might alter the cell phenotype, thereby skewing interpretation3,103.
Table 3.
Approaches for extracellular vesicle tracking
| Purpose of labelling | Reagent/approach | Description | Refs |
|---|---|---|---|
| Visualizing extracellular vesicles | |||
| Membrane labelling | DiI, DiD, DiR, DiO | Lipophilic carbocyanines. Incubating donor/recipient cells with DiD labels lipid membrane via lateral diffusion | 268 |
| CellMask | Amphipathic molecule with a lipophilic moiety and a negatively charged hydrophilic dye that anchors the probe in the membrane. Incubation of donor/recipient cells with this dye results in membrane labelling | 248 | |
| FM 4-64, FM 1-43 | Lipophilic styryl compound. Incubating donor/recipient cells with this dye allows the dye to enter the outer leaflet of the surface membrane | 269 | |
| PKH26, PKH67, PKH2 | Aliphatic molecules incorporated via selective partitioning into lipid membrane (of note, PKH26 can aggregate into particles, causing artefacts or false positive signals) | 270 | |
| Antibody-based tracking of extracellular vesicles | Use of Alexa Fluor-conjugated antibodies indirectly binding the extracellular vesicle proteins TSG101, CD63 and HSP70, allowing visualization of extracellular vesicles with fluorescent confocal microscopy or super-resolution microscopy | 271 | |
| Membrane labelling of extracellular vesicles with fluorescent protein | Palmitoylation signal placed in-frame of the N terminus of fluorescent protein | 114 | |
| Reporter tagging of transmembrane extracellular vesicle markers | Extracellular vesicle-associated proteins, such as flotillin 1, flotillin2, RAB5, RAB7 and CD63, can be conjugated to fluorescent proteins such as GFP and mCherry. In this way extracellular vesicles can be imaged intracellularly with time-lapse confocal imaging | 272 | |
| Vesicle interior labelling (cell/extracellular vesicle-encapsulated dyes) | Carboxyfluorescein diacetate succinimidyl ester | A cell-permeable dye that labels intracellular molecules by covalently binding to intracellular lysine residues and other amine sources | 273 |
| Calcein AM | A compound hydrolysed by intracellular esterases resulting in fluorescent signal retained within the cell | 274 | |
| CellTracker CM-Dil | Conjugates to thiol-containing peptides and proteins, retaining the fluorescent signal within the cell | 275 | |
| Labelling RNA cargo | Alexa-labelled RNA | Dye that forms a stable complex with particular nucleic acids | 276 |
| TAMRA NHS | Dye that forms a covalent amide bond with primary amine group | 269 | |
| SYTO RNASelect Green | Cell-permeant nucleic acid stains | 35 | |
| Monitoring extracellular vesicle uptake | |||
| Monitoring fusion | Quenching assays (BODIPY, NBD–rhodamine assays, octadecyl rhodamine B self-quenching) | Dyes with FRET-based self-quenching. Before fusion the probe dimer is self-quenched owing to close proximity. On fusion with a larger membrane (for example, plasma membrane or endosome) the dimer is separated and therefore no longer quenched, releasing a fluorescent signal. Therefore, detection of a fluorescent signal indicates fusion/uptake of extracellular vesicle content | 277–279 |
| Pyrene excimer formation | Pyrene labelling lipids. Lipid probes labelled with pyrene that form dimers emit red-shifted fluorescence but when mixed with non-labelled lipids the dimer fluorescence is replaced by monomer blue-shifted fluorescence | 280 | |
| Monitoring pH changes (indicating endosomal uptake) | pHluorin | pH sensitive variant of GFP | 281 |
| pHRodo | Fluorogenic pH-sensitive dye | 215 | |
| Organelle labelling | Fluorescently fused proteins | Fluorescent protein tagged to organelle-associated proteins EEA1 (early endosome), RAB5 (early/mobile endosome) and RAB7 (maturation to late endosome). Visual tracking of each compartment allows tracking of vesicles within endosomal pathway as well as visualization of escape from the endosomal pathway | 276 |
| LysoTracker | Fluorophore linked to weak base. Labels endolysosomal compartment, allowing tracking of extracellular vesicles to the lysosome or visualization of a separation of extracellular vesicles from the endosomal pathway | 282 | |
| Monitoring functional output | |||
| Visualizing cytosolic release | β-Lactamase assay | CCF2-AM is a FRET substrate that is hydrolysed by the enzyme β-lactamase, producing a coloured product that is directly proportional to enzyme activity. Delivery of the enzyme (via a virus/extracellular vesicle) to a recipient cell expressing CCF2-AM results in a detectable fluorescent signal as a consequence of cytosolic release | 283 |
| Split protein assay (luminescent/fluorescent) | Re-formation of separated protein fragments results in a signal. This confirms uptake of extracellular vesicle content whereby one fragment is present in extracellular vesicles and the complementary fragment is expressed in the recipient cell | 284 | |
| Evidencing mRNA translation with reporter mRNA | Exemplified by the use of reporter mRNA encoding Gaussia luciferase. Translation is evidenced by the signal from the reporter assay, which is prevented by treatment with the translation inhibitor cycloheximide | 114 | |
FRET, fluorescence resonance energy transfer; NBD, 7-nitro-2,1,3-benzoxadiazoI-4-yl.
Biology of extracellular vesicle RNA
Once the extracellular vesicle RNA cargo has been delivered to the recipient cell and has escaped the degradative pathway, it may elicit a functional response. Among the various RNA biotypes present in extracellular vesicles, most is known about miRNAs as mediators of cell–cell communication, with more limited information on the functions of other RNA biotypes. Several difficulties arise in establishing the functionality of RNA in extracellular vesicles. First, overexpression systems that increase the amounts of a particular RNA in extracellular vesicles can result in supraphysiological levels of the RNA in source cells, potentially resulting in changes in the extracellular vesicle release process and/or altering levels of other cargo. Functional effects of the RNA cargo of interest may be affected by the other cargo delivered at the same time. The population of exRNAs — importantly RNA in extracellular vesicles — in conditioned cell culture medium is highly dependent on the culture methods used. Most prominently, bovine serum, which is widely used as a medium supplement, contains bovine RNA in extracellular vesicles and non-vesicular entities. These exRNAs can confound molecular and functional readouts in the extracellular vesicle-recipient cell, particularly as many miRNAs are highly conserved among mammalian species104. Methods to remove exRNAs from bovine serum through ultracentrifugation or affinity purification result in partial, but not complete, depletion12,13. Another confounding problem is mycoplasma infection of cell cultures105, and this must be monitored frequently.
Extracellular vesicle RNA cargoes can be divided on the basis of knowledge of their bioactivity into those with known functions, those with predicted functions and those with unknown functions (Fig. 1), with the third category evoking the early concept of extracellular vesicles as disposal units for some molecules16,106. Nevertheless, even when used as a means for discarding, RNA may have functional effects on both donor and recipient cells. Several prominent examples of functional effects for different species of extracellular vesicle RNA are discussed in the following sections.
miRNAs
Numerous examples of extracellular vesicle-mediated functional transfer of miRNAs have been demonstrated with use of both in vitro and in vivo models for a variety of diseases and physiological states with a broad range of downstream effects. For example, miR-193b is secreted via extracellular vesicles in the central nervous system and decreases levels of amyloid precursor protein in neuronal cells, providing reductions in the pathology of Alzheimer disease, and the content of this miRNA in extracellular vesicles can be used as a biomarker for this disease107. In other examples in the nervous system, neurons produce miR-124a-loaded extracellular vesicles that regulate the glutamate transporter in astrocytes, which is important for synaptic transmission108. In the context of brain cancer, astrocytes secrete extracellular vesicles with miR-19, which inhibits an important tumour suppressor, PTEN, in cancer cells, thereby promoting growth of brain metastases109. As another example, extracellular vesicles from mesenchymal stem cells enriched in miR-375 via its overexpression stimulated bone regeneration by bone marrow stem cells110. Two critical miRNAs that regulate inflammation, miR-155 and miR-146a, are exchanged between dendritic cells in vivo, with the latter inhibiting and the former promoting endotoxin-induced inflammation in mice111. Other recent examples of transfer of functional miRNAs by extracellular vesicles from cancer, cardiac, neural, immune and adipose cells are summarized in Table 1.
mRNAs
Most full-length mRNAs in extracellular vesicles are smaller than 1 kb (ref.112), but clearly, in analogy to retroviruses, longer RNA sequences could be incorporated in a condensed configuration113. mRNAs in extracellular vesicles could serve as a source of novel proteins in recipient cells. Exit from the endosome could be a bottleneck in functional mRNA delivery101 and may depend on the donor and/or recipient cell types. However, active translation of mRNAs transported by extracellular vesicles into recipient cells was reported early on; for example, expression of reporter proteins from mRNA transferred by extracellular vesicles between mast cells23 and from glioblastoma to endothelial cells24. In one study mRNAs transported within extracellular vesicles were translated within 1 hour after extracellular vesicle uptake during bidirectional exchange in co-cultures of glioblastoma and HEK293T cells114. There is also evidence that extracellular vesicle mRNA can generate active proteins, as transfer of Cre recombinase mRNA in extracellular vesicles led to recombination events in the brains of floxed reporter mice115,116. Of note, this study analysed the extracellular vesicles for Cre mRNA and protein, and while the former was detected by reverse transcription–PCR, the latter was not detected by western blot or enzyme-linked immunosorbent assay. But because reverse transcription–PCR is markedly more sensitive than either of the last two protein detection techniques, the contribution of the extracellular vesicle-mediated protein transfer of Cre to this functional effect cannot be ruled out. Extracellular vesicle-mediated transfer of mRNAs between human pulmonary artery smooth muscle cells (stimulated by transforming growth factor-β) and arterial endothelial cells has also been demonstrated with a number of different mRNAs present in these extracellular vesicles, prominently including regulators of the actin cytoskeleton and extracellular matrix remodelling117. Because transforming growth factor-β is a cytokine relevant for the pathology of pulmonary arterial hypertension, this extracellular vesicle-mediated communication could be involved in the disease phenotype117.
Importantly, as extracellular vesicles typically carry both the mRNA and its encoded protein, it is difficult to distinguish newly translated protein in recipient cells. Changing the experimental paradigm revealed that delivery of human erythropoietin mRNA to cells via lipid nanoparticles resulted in release of extracellular vesicles containing the mRNA, which was then translated in mouse cells118. Extracellular vesicles derived from human red blood cells have also been loaded with mRNA for Cas9 by electroporation (see more details later) and used for CRISPR editing in recipient cells119. These studies establish that indeed mRNA delivered by extracellular vesicles can be translated in recipient cells.
lncRNAs
The functions of lncRNAs (longer than 200 bp) are diverse, ranging from negative regulation of miRNAs by serving as miRNA ‘sponges’120,121 to marking of mRNAs for degradation122 and transcriptional regulation of genes123. Several studies implicate the role of extracellular vesicle-transferred lncRNAs in biological processes. One recent study documented that hypoxic cardiomyocytes release extracellular vesicles with high levels of lncRNA NEAT1, which taken up by fibroblasts induces a profibrotic gene expression programme124. In another study, incorporation of hypoxia-inducible factor 1α-stabilizing lncRNA into extracellular vesicles released by tumour-associated macrophages supported breast cancer cell viability125. A future challenge will be determining the mechanism of action of these and other lncRNAs in extracellular vesicles.
tRNA fragments
Next to miRNAs, tRNA fragments are the most abundant small non-coding RNAs in extracellular vesicles. These species function in the regulation of gene expression and epigenetics, particularly in the context of immunomodulation47 and cancer progression126,127. Extracellular vesicles contain different sets of tRNA fragments that can be enriched, as shown for T cells, where 45% of these species were 1.5-fold enriched in extracellular vesicles compared with whole cells47. Specific tRNA fragments derived from the same tRNA are sorted into extracellular vesicles, while others are not. The tRNA fragment content of extracellular vesicles appears to be dependent on the source cell state, as for T cell activation, which results in changes in the types of tRNA fragments incorporated into extracellular vesicles, with some fragments showing activation-dependent sorting into extracellular vesicles47. Silencing of these activation-dependent tRNA fragments promoted T cell activation, suggesting that in this case, secretion of tRNA fragments is a means of removing them from the cell, thereby preventing repression of immune responses47. Whether tRNAs and their fragments in extracellular vesicles have functional effects in recipient cells remains to be established.
snoRNAs
The main function of snoRNAs is to guide chemical modification of other RNA species. snoRNAs are enriched in extracellular vesicles compared with the source cell, and after entry into recipient cells, they are shuttled to the nucleus. The snoRNA content of extracellular vesicles is influenced by the biological state of the source cell and has been shown to change in immune cells, such as dendritic cells treated with immunorepressive or immunostimulatory factors128, and in activated macrophages129. In the latter case, these extracellular vesicles were taken up by distant cells with an associated increase in 2′-O-methylation of RNA — a modification that the snoRNAs released by the macrophages direct on their canonical rRNA targets. Hence, extracellular vesicle-derived snoRNAs can be modulators of inflammation.
ncRNAs signalling via cellular receptors
In addition to their more specific functions, extracellular vesicle-associated ncRNAs can also produce effects via direct interactions with cytoplasmic receptors, in particular immune receptors, such as RIG-I-like receptors, which recognize pathogen-associated molecular patterns (PAMPs), and the NOD-like receptor family, which in addition to PAMPs also recognizes endogenously derived damage-associated molecular patterns (DAMPs). Most of what we know about how exRNA triggers these cytoplasmic receptors has been revealed for extracellular vesicles carrying microbial content130. For example, Mycobacterium tuberculosis RNA is packed into extracellular vesicles released from infected macrophages that are subsequently taken up by other macrophages. This triggers the RIG-I RNA sensing pathway leading to production of type I interferon and accelerated maturation of bacteria-containing phagosomes by recipient macrophages, supporting increased bacterial killing131. A similar mechanism is used during latent infection with Epstein–Barr virus, whereby infected B cells release extracellular vesicles containing viral ncRNAs with the potential to stimulate RIG-I and other related PAMP receptors. These extracellular vesicles act on dendritic cells and stimulate antiviral responses132. There are also hints in the literature that some mammalian RNAs are able to trigger these receptor-mediated pathways. For example, cancer stromal fibroblasts increase the expression of RN7SL1 — an endogenous 5′-triphosphorylated RNA (representing a canonical target for RIG-I) — which is normally shielded by binding to its RBP. This increased expression results in unshielded RN7SL1, which is packaged into extracellular vesicles and when delivered to breast cancer cells induces RIG-I signalling and interferon-stimulated gene expression, resulting in cancer promotion133 and therapy resistance134. However, it is worth noting that in addition to these signalling responses to exRNA, any exogenous RNA may be perceived as ‘foreign’ by the recipient cell, leading to RIG-mediated degradation135. In addition to proactive effects of RIG-I signalling in infection and cancer, recognition of exRNA by immune receptors can also lead to pathological inflammation and rejection of recipient cells by the activation of the immune system.
In addition to immune receptors, some exRNAs are recognized by autophagy receptors in the cytoplasm. For example, vtRNAs present in extracellular vesicles27 as well as in ribonucleoprotein particles136 can serve as important regulators of autophagy. They bind directly to the intracellular autophagy receptor sequestosome 1 (also known as p62), thereby interfering with oligomerization of the receptor and downstream clearance of protein aggregates through the autophagic pathway137. However, the presence of vtRNA in extracellular vesicles has been challenged recently by a study showing that vtRNA was restricted to the non-vesicular exRNA in a human colon cancer cell line and was present only as a minor component in small extracellular vesicles in a human glioma line10. This discrepancy may relate to the way the vesicles were isolated in the different studies or to differences in the cells of origin (non-transformed cells versus cancer cells).
Many of the small RNAs in extracellular vesicles have the potential to influence recipient cells even without reaching the cytoplasm. These exRNAs can potentially trigger Toll-like receptors (TLRs) and thereby elicit an innate immune response106. The most likely TLRs for this task are TLR3, TLR7, TLR8 and TLR9, which are intrinsically capable of sensing PAMPs and DAMPs and operate within the endosomal compartment during its maturation and acidification138. If the lipid layer of extracellular vesicles surrounding the cargo is breached after endocytosis, the RNA cargo could be exposed inside the endosome. As such, TLRs in the endosome of the recipient cells could be triggered by recognizing this single-stranded RNA cargo. This has been demonstrated for miR-21 and miR-29a, which are secreted by lung cancer cells within extracellular vesicles, which are then taken up by surrounding immune cells in vivo and activate endosomal TLR7 and TLR8, thereby inducing a proinflammatory phenotype that supports cancer progression139. Similarly, members of the let-7 miRNA family (let-7a and let-7e) in extracellular vesicles140,141 harbour a specific GU-rich motif (GUUGUGU) that binds to and activates TLR7. TLR7 activation could, in turn, impact cell function in disease-relevant contexts, including neurodegeneration142 and cancer143.
Extracellular vesicles in viral transfer
Nucleic acids of intracellular pathogens can also be packaged into extracellular vesicles in infected cells and trigger protective responses in surrounding cells. In some cases, extracellular vesicles can also be co-opted into facilitating viral RNA transfer and, as such, aid in viral functions. Non-enveloped RNA viruses, such as leishmania RNA virus 1, hepatitis A virus and hepatitis C virus can be packaged into extracellular vesicles, serving as an invisibility cloak to shield them from host immune surveillance144–146. Some viral proteins are specialized in the transmission of viral RNA via extracellular vesicle subspecies that contain elements of both the plasma membrane and the viral envelope. Intriguing reports raise the possibility that even without viral proteins, extracellular vesicles can transport functional hepatitis C virus RNA between cells in a virion-independent manner147,148. Similarly, the retrovirus-derived Gag protein ARC encoded in fly83 and mammalian149 genomes can form and release enveloped capsids that contain their own mRNA (about 1 kb). The trans-synaptic delivery of these extracellular vesicles between neurons and expression of the encoded ARC RNA in postsynaptic neurons were found to be critical in modulating synaptic plasticity.
Modulating RNA cargo and its delivery
Incorporating selected RNA cargo into extracellular vesicles with the potential for targeting to specific cell types in vivo has immense therapeutic potential. One important issue in functional extracellular vesicle transfer is the type of cells the vesicles are obtained from. Depending on the clinical use, cell sources include mesenchymal stem cells, which can be isolated from a number of different tissues and are intrinsically regenerative and non-inflammatory150,151; red blood cells of type O donors, which are easily loaded with cargo152; and dendritic cells, which can present antigens and activate T lymphocytes and thus be used for vaccination153,154. In the last context, extracellular vesicles from tumour cells could be used to elicit antitumour immune responses via presentation of tumour-associated antigens. However, there is concern that vesicles isolated from immortalized cells or tumour cells may carry oncogenic factors and contribute to cancer155.
As mentioned earlier, extracellular vesicles have many similarities to enveloped viruses, which can serve as prototypes to understand the potential for these vesicles to act as vectors for delivery of functional RNAs. Enveloped coronaviruses, for example, have a size similar to that of extracellular vesicles (125 nm) and contain single-stranded RNA of up to 30 kb. This is the largest reported genome size of any RNA virus and may serve as a benchmark for the maximum RNA capacity of these vesicles when packaged in a condensed form. Nevertheless, in addition to RNA, extracellular vesicles contain many other types of cargo, which might have to be displaced to allow full RNA-loading capacity. Different approaches are being explored for loading RNA into extracellular vesicles, targeting strategies and production considerations.
Packaging selected RNA into extracellular vesicles
Loading of the desired cargo for transfer via extracellular vesicles can be achieved either endogenously, by modulation of the donor cell to favour inclusion of the selected cargo into extracellular vesicles, or exogenously, in which case the cargo is loaded into isolated extracellular vesicles in vitro.
In endogenous loading, overexpression of an RNA in the source cell through DNA transfection or lentiviral transduction usually results in enrichment of that RNA, as well as any encoded protein, in the extracellular vesicles generated by those cells156,157. The yield of RNA-loaded extracellular vesicles can be further increased by mechanical extrusion of vesicles through a micrometre-pore-sized filter using cells that express high levels of the desired mRNA158. RNA enrichment can also be achieved by having the donor cells express a protein that both interacts with the plasma membrane (for example, via different lipid anchors)159 or a viral envelope protein (such as vesicular stomatitis virus glycoprotein)101 and has an RNA-binding motif, typically of viral origin (for example, the RNA-binding peptide from the MS2 bacteriophage coat protein)160. In concert, the cargo RNA is equipped with specific sequences recognized by the RNA-binding motif in the protein, allowing targeting of the RNA of interest to the plasma membrane and facilitating its release through budding from the plasma membrane, mimicking retrovirus-like particle release159 (Fig. 2). It is also possible to target RNAs to MVBs, by expressing protein fusions that couple an MVB-enriched protein (such as lysosome-associated membrane protein 2, tetraspanin or CD63) with an RNA-binding motif101,161. One approach for selective enrichment in extracellular vesicles took advantage of a specific subpopulation of microvesicles, referred to as ‘arrestin domain-containing protein 1 (ARRDC1)-mediated microvesicles’. In this approach, ARRDC1 and RNA cargo were engineered to interact — via including the transactivator of transcription (Tat) peptide from HIV in ARRDC1 and its RNA binding-motif, transactivating response (TAR) element, in the RNA — allowing efficient generation of RNA cargo-loaded extracellular vesicles. This strategy was successful in transfer of p53 mRNA, which was translated in recipient cells on uptake (including in vivo delivery into mice)162. Beyond the use of viral components, other approaches have included incorporation of ‘zipcode’ sequences, typically in the 3′ untranslated region of the cargo RNA, which can be recognized by specific RBPs associated with membrane-targeted or MVB-targeted proteins (see, for example, refs85,161). Taking advantage of the lipid biocomposition of extracellular vesicles can also help in loading RNA–RBP complexes into them, by engineering RBPs to bind to extracellular vesicle membranes more efficiently via addition of specific peptides. The different lipids in extracellular vesicles make possible binding with curvature-sensing and lipid composition-specific affinity peptides. Annexin A5 is a well-known example that binds to negative curvature-specific lipid phosphatidylserine on apoptotic bodies and extracellular vesicles, while the effective domain (residues 151–175) of myristoylated alanine-rich C-kinase substrate (MARCKS) protein has an affinity for positive-curvature-associated lipid, phosphatidylinositol 4,5-bisphosphate that is present on plasma membrane-derived vesicles163. Some other proteins, such as bradykinin, can also harbour peptides that bind highly curved membranes164. Other options include incorporation of binding domains of proteins with affinity for intracellular membranes, such as phosphatidylinositol 4,5-bisphosphate or phosphatidylinositol 3,4,5-trisphosphate159.
Common methods to load RNA into isolated extracellular vesicles exogenously include electroporation of siRNAs (see, for example, refs119,165–169), antisense oligonucleotides119 and mRNAs119, as well as use of sonication170 and Lipofectamine169. One of the most promising approaches uses chemically stabilized siRNAs bearing a lipophilic moiety, such as cholesterol, which results in high incorporation onto and through the lipid membrane of the extracellular vesicles with documented functional knockdown of target mRNA in vivo171,172. Still, it is important to keep in mind that isolated extracellular vesicles are inherently already ‘full’ of RNA and proteins, and it is not clear how much more additional cargo can be effectively loaded into them. Nevertheless, the lipid membrane may be extendable, and stabilized siRNAs can be loaded on both the inside and the outside, thereby supporting these in vitro loading strategies. In considering different modes of loading of RNA into isolated extracellular vesicles, one should also keep in mind the integrity of the vesicles and potential antigenic/toxic components included in the loading process, which may limit successive administrations of exogenously loaded vesicles.
Strategies to control extracellular vesicle yield and stability
In addition to cargo loading, to ensure efficient cell–cell communication (especially in a therapeutic setting), it is key to ensure high yields of high-quality extracellular vesicle production173. Generally, obtaining large amounts of pure extracellular vesicle preparations necessitates copious starting material, and enhancing the production of extracellular vesicles is a current focus of many researchers and biotechnology companies. Increasing extracellular vesicle biogenesis in the donor cell is one strategy to increase the overall availability of extracellular vesicle RNA. One possibility is to overexpress regulatory proteins involved in extracellular vesicle biogenesis. A screen with a readout for the numbers of extracellular vesicles released identified factors in the donor cell that promoted exosome biogenesis, including STEAP3 (involved in exosome biogenesis), syndecan 4 (MVB formation) and an increase in the citric acid cycle to boost production of RNA cargo-loaded exosomes161. In another study, overexpression of the actin-bundling protein cortactin to increase MVB docking at the plasma membrane (see Fig. 2) proved efficient in increasing exosome release56. Treatment of source cells with different compounds, such as bafilomycin A1 (a V-ATPase inhibitor174) or heparanase175, which regulates secretion, composition and function of tumour cell-derived exosomes, also increased production of extracellular vesicles. Extracellular vesicle secretion was also increased by elevated levels of intracellular Ca2+ in response to treatment with monesin, a Na+/H+ exchanger176. Natural compounds can influence extracellular vesicle release; for example, microglia exposed to serotonin exhibited increased fusion of MVBs with the plasma membrane, via increase in cytosolic Ca2+ concentration177. Radiation, hypoxia, oxidative stress and low pH can all upregulate extracellular vesicle production178–181. Vesicular stomatitis virus glycoprotein also promotes extracellular vesicle release182,183. Finally, the yield of isolated extracellular vesicles can be increased by in vitro fusion with synthetic lipids, followed by extrusion though small pores (100–400 nm in diameter184). These vesicles could then be loaded with cargo (siRNA) via electroporation and were successfully taken up by recipient cells in culture at a rate that was more than tenfold higher than that of native vesicles184.
Although increased production of extracellular vesicles is generally desired for therapeutic purposes, inhibition of extracellular vesicle release from tumour cells has been proposed as a therapeutic strategy to curtail advancement of cancer. A number of compounds have been identified that reduce secretion of extracellular vesicles. For example, the antihistamine drug ketotifen185 and the antibiotic sulfisoxazole186 curb extracellular vesicle release in multiple cancer cell lines. Screens for modulators of extracellular vesicle secretion have yielded various chemical compounds that both increase secretion (for example, sitafloxacin, chloroindole and fenoterol) and reduce secretion (for example, manumycin A, tipifarnib and neticonazole)187. In a different approach, breast cancer-derived exosomes were removed from the circulation through extracorporeal haemofiltration in an attempt to decrease growth of breast cancer in patients188.
Despite the progress in regulating extracellular vesicle production and loading of cargo, many of the strategies in place so far use treatments that may alter cell phenotypes and extracellular vesicle functionality and/or have a toxic or immunogenic response in vivo. Further work will be needed to expand and evaluate these methods for research and clinical use.
Targeting extracellular vesicles to specific cell types and promoting uptake
Similarly to viral gene therapy, a major challenge in using extracellular vesicles for therapeutic purposes is achieving delivery to diseased cells in living organisms. Extracellular vesicles administered in vivo suffer from rapid clearance mostly by uptake into cells in the liver, kidneys, lungs and spleen189–191 with different routes of administration, including intravascular, intraperitoneal and subcutaneous, altering the distribution only to a minor extent191. Overall, goals include increased targeting of therapeutic extracellular vesicles to specific cell types, facilitation of their translocation through different biological barriers and extending the half-life of vesicles in extracellular spaces in vivo. Some increase in stability in vivo has been achieved by coating extracellular vesicles with the synthetic polymer polyethylene glycol192 or displaying CD47 on their surface, which decreases phagocytosis by macrophages168.
More selective targeting to tissues/cells of interest could be achieved by the incorporation of specific ligands on the surface of extracellular vesicles, including proteins/peptides, antibodies, lipid-associated entities and RNA aptamers193 that will be recognized by the acceptor cell (for a review, see ref.90). In an early attempt to route extracellular vesicles to the brain after intravenous delivery, a peptide from rabies virus glycoprotein that targets acetylcholine receptor-expressing cells in the brain was fused to lysosome-associated membrane protein 2B (which associates with MVBs)165, but its effectiveness differs in different studies. Extracellular vesicles have also been engineered to display a nanobody to the epidermal growth factor receptor on their surface to target tumour cells expressing this receptor194. Tissue-targeting ligands can also be linked to the phosphatidylserine-binding domain of lactadherin, which is exposed on the surface of extracellular vesicles195. In yet another approach, a single-chain variable fragment of an anti-HIV monoclonal antibody was incorporated into an expression vector designed to target recombinant proteins to the surface of mammalian cells to deliver apoptosis-inducing miR-143 (or the drug curcumin) to kill HIV-infected cells196. Finally, a high-throughput screening method (phage display) was used to identify a peptide, CP05, that binds to an extracellular domain of tetraspanin CD63 (ref.197). By linking muscle-targeting peptides and antisense oligonucleotides to CP05, efficient delivery of these antisense RNAs to skeletal muscle in a mouse model of Duchenne muscular dystrophy was achieved197. As an alternative strategy to incorporation of targeting ligands directly in the extracellular vesicle membrane, it is also possible to incorporate artificial lipids into extracellular vesicle membranes, where they can aid in coupling of tissue-targeting antibodies or homing peptides with the extracellular vesicle surface and thus influence the biodistribution of therapeutic extracellular vesicles198.
Besides modification with proteins/peptides, there is growing interest in functionalizing extracellular vesicles with RNA aptamers to control their targeting. In this regard, a bacteriophage Φ29 RNA, which has a three-way junction allowing its self-association, has been engineered to incorporate a membrane-targeting cholesterol moiety, a prostate-specific membrane antigen or RNA aptamer, and an epidermal growth factor receptor RNA aptamer199. This RNA trimer composite incorporated itself spontaneously into membranes of extracellular vesicles ex vivo. These decorated extracellular vesicles could then be loaded with RNA cargo (such as anticancer siRNAs) to inhibit tumour growth in mouse models. Cancer cell-targeting aptamers were also successfully conjugated to a hydrophobic diacyl lipid tail and incorporated in extracellular vesicle membranes following their loading with anticancer drugs200. Extracellular vesicle functionalization has also been performed chemically (via reaction between aptamer-conjugated aldehyde and the amino group of membrane proteins incorporated into extracellular vesicles), whereby extracellular vesicles from bone marrow stromal cells were equipped with a bone marrow-specific RNA aptamer201. These functionalized extracellular vesicles were then efficiently targeted to the bone marrow after intravenous injection in mice and promoted bone regeneration. By contrast, non-functionalized extracellular vesicles primarily accumulated in the liver and lungs and did not have beneficial effects on bone regeneration201.
Overall, a wide variety of specific targeting strategies are being explored for extracellular vesicle delivery. One major caveat of these strategies, however, is that such ligands in the form of non-protected oligonucleotides and peptides/proteins exposed on the surface of the extracellular vesicle may be digested off in biofluids by RNases and proteases, respectively, thereby preventing targeting202,203.
More global approaches for directing tissue accumulation of extracellular vesicles following intravenous administration include chemical modification of their surfaces. For example, treatment with neuraminidase to neutralize the negative charges on extracellular vesicles contributed by polysaccharides and glycosylated phospholipids reduced extracellular vesicle targeting to the liver, with a larger proportion of vesicles taken up by other organs, such as the brain, after intravenous injection in mice204. Engineered glycans can also be displayed on the surface of extracellular vesicles, as these moieties appear to be important for extracellular vesicle recognition and uptake by recipient cells205,206. Increased cellular uptake in general can be achieved by the addition of cationic lipids (for example, in the form of Lipofectamine) to the surface of extracellular vesicles207, by increasing macropinocytosis with an arginine-rich cell-penetrating peptide exposed on the surface of the extracellular vesicles208 or by inducing receptor clustering on recipient cells209.
Increasing cytosolic delivery of the RNA cargo
Cytosolic delivery of extracellular vesicle content can potentially be improved by enhancing the fusogenic properties of extracellular vesicle membranes, which should result in the direct deposition of cargo RNA into the cytoplasm of the cell. Direct fusion of the extracellular vesicles with the plasma membrane can be achieved by incorporating fusion proteins, such as syncytin 1, syncytin 2 and epithelial fusion failure 1 protein210, into their membranes. Furthermore, the gap junction protein connexin 43 is present in the extracellular vesicle membrane and can create a channel in the plasma membrane that allows passage of oligonucleotides into the cytosol211. On the basis of the similarity between extracellular vesicles and viral particles, incorporation of viral proteins, such as vesicular stomatitis virus glycoprotein183, into extracellular vesicles is an appealing strategy to target extracellular vesicles to specific cell types and organs while also improving functional delivery of RNA, albeit at the risk of introducing antigenic proteins113,212. For escape from the endosomal pathway, ideas can be sought from bacteria and viruses. Some enveloped and non-enveloped viruses (for example, adeno-associated virus and influenza virus) are taken up by endocytosis and through pH-sensitive fusion proteins use the acidic pH of the endosomal compartment to promote membrane fusion between the endosomal and vesicular membranes213–215. Many species of pathogenic bacteria produce pore-forming toxins to escape from the degradative route following their uptake into host cell endosomes216–218. Such proteins could be engineered into extracellular vesicle membranes. Addition of compounds such as chloroquine that block acidification of the endosomal compartment and release of viruses paradoxically enhanced extravesicular delivery in one of the studies, possibly through a protein sponge effect leading to swelling and bursting of the endosomes219. Other compound found to increase endosomal escape was small molecule UNC10217832A219.
For liposome-based delivery, increased efficiency of endosomal escape was achieved by inclusion of an amine-rich polymer (polyethylene imine)220. While this may not be directly applicable to extracellular vesicles, it does highlight the existence of compounds that can specifically increase rates of endosomal escape and might improve intercellular delivery of extracellular vesicle content, with the possibility of combining liposome components with extracellular vesicles221.
Potential clinical applications
Extracellular vesicles show promise for clinical applications both as biomarkers and as therapeutic delivery vehicles. Here we focus on uses of their RNA cargo, but proteins, lipids and glycans can all be informative as biomarkers that carry the information about their cellular source, whereby their make-up informs on the physiological state, with changes in composition often associated with a disease state. With their use as biomarkers, the main issue is detecting the subset of extracellular vesicles from a particular tissue in a sea of vesicles from other cell sources in biofluids. For therapy, extracellular vesicles can be used to deliver not only RNA but also proteins and drugs and combinations thereof. Issues remain concerning best practices for isolating large numbers of clinical grade quality vesicles, loading them and directing their biodistribution to target tissues. In most scenarios they would be used to promote delivery of therapeutic cargo — be it functional RNA or protein — to the diseased tissue or tumour and would need to be readministered repeatedly throughout the disease state. Nevertheless, extracellular carriers can also serve as carriers of CRISPR technology to induce pathological error correction, and in this case they might be effective in a single dose.
Use of extracellular vesicle RNA cargo in diagnostics
RNAs in extracellular vesicles are in essence a snapshot of the content and state of the cells that secrete them and are therefore very useful for biomarker discovery61. Extracellular vesicles are found in a wide variety of biofluids, including serum/plasma, urine, saliva, cerebrospinal fluid and breast milk222,223. An advantage of using extracellular vesicle-derived RNAs as biomarkers is that they are protected from degradation by RNases and thus are quite stable, and that the vesicles display on their surface the antigenic markers of the cells from which they were derived, allowing enrichment of vesicles from a particular tissue source (for example, using antibody capture for the enrichment of tumour-derived vesicles in serum224). Many new sensitive technologies, such as Droplet Digital reverse transcription–PCR are being used to enhance detection of specific RNA species in extracellular vesicles225.
RNAs can serve as biomarkers based on the mutations, splice variants and fused mRNAs as well as alterations in levels of specific RNAs associated with various disease states. The first example using extracellular vesicles in biofluids was the detection of a tumour-specific mRNA: mutant epidermal growth factor receptor in serum of patients with glioblastoma24. In other examples, elevated levels of specific miRNAs have been found in cerebrospinal fluid from patients with glioblastoma226, patients with Alzheimer disease and Parkinson disease227, and patients with breast cancer228. Changes in levels of mRNA splice variants are detected in urine from patients with myotonic dystrophy229. lncRNA profiles in blood samples from patients with type 2 diabetes correlate with heart functions230. This biomarker application of extracellular vesicles has already been successfully translated into the clinical setting. Currently, mutated mRNAs in extracellular vesicles and other carriers in plasma are being used as biomarkers for the diagnosis of haematological disorders231 (for example, NeoLAB liquid biopsy from NeoGenomics Laboratories) and for prognostic assessment of prostate, lung and other solid tumours232,233 (for example, ExoDx prostate test from Bio-Techne).
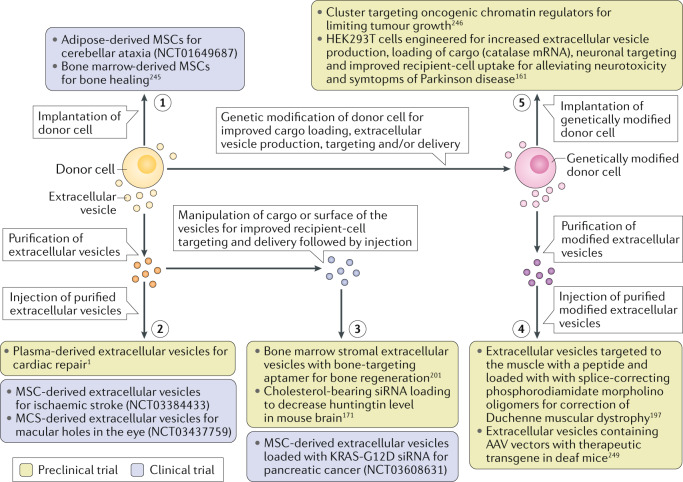
Therapeutic RNA delivery via extracellular vesicles
There is tremendous interest in using extracellular vesicles as vehicles to deliver RNA therapeutics. Extracellular vesicles as therapeutic carriers have several advantages: they can be obtained from a variety of sources, including the patient’s own extracellular vesicles (in which case they would be immune neutral); they protect the cargo within a lipid bilayer; they provide a membrane platform for displaying ligands; and they can carry not only RNA but also other therapeutic agents at the same time. At the present time, these applications are still limited by the inherent heterogeneity of extracellular vesicles and the inability to achieve a fully characterized pure extracellular vesicle population. Extracellular vesicle preparations being used for preclinical trials are more appropriately referred to as ‘secretomes’ as they contain a mixture of extracellular elements234,235. Despite these limitations, therapeutic RNA delivery via extracellular vesicles has already entered clinical investigations (see ref.150 for a review). Preclinical studies have shown that unmodified mesenchymal stem cell-derived extracellular vesicles are distributed broadly after intravenous injection236,237 and can ameliorate the effects of various diseases in the liver, brain, heart, kidneys, lungs, skeletal muscle and immune system238. There are now also strategies in place to obtain extracellular vesicles in considerable amounts from various sources, such as commonly used cells that can be expanded on a large scale (for example, mesenchymal stem cells172,239, red blood cells240 and HEK293 cells241), biofluids that are readily available in large volumes (for example, bovine milk242 and human plasma167) and even plant materials (for example, grapefruit243 and ginger167). Vesicles and therapeutic agents generated from plants and other food items have the advantage that they fall into the category of natural substances and as such are not under the strict guidelines of the US Food and Drug Administration for clinical trials244. Furthermore, as discussed earlier, extracellular vesicles can be further modified to be loaded with cargo, generating ‘designer’ vesicles with increased production, cargo loading and/or targeting for improved therapeutic outcome.
Five basic variations on the theme of using extracellular vesicles as therapeutic vehicles are being tested preclinically or in clinical trials: implantation of donor cells, such as mesenchymal stem cells, which release extracellular vesicles on site; injection of purified, unmodified extracellular vesicles from cultured donor cells; injection of purified extracellular vesicles that have been modified for targeting or loading of cargo in vitro; injection of extracellular vesicles enriched in cargo of interest and/or with enhanced targeting/uptake properties from genetically modified cells; and implantation of genetically modified cells that release modified extracellular vesicles in vivo (Fig. 4). A few examples of these strategies taken from a large literature are provided here. First, mesenchymal stem cell-derived extracellular vesicles with their intrinsic composition of RNAs and proteins are being evaluated for treatment of cerebellar ataxia (ClinicalTrials.gov identifier NCT01649687), bone repair245 and regeneration of macular holes in the eye (NCT03437759), although the role of specific RNAs in therapeutic efficacy has not yet been defined. Extensive effort is directed at loading isolated extracellular vesicles and adding targeting moieties to their surface to improve cargo delivery. For example, mesenchymal stem cell-derived extracellular vesicles transfected with miR-124 — an miRNA most abundantly expressed in the central nervous system and negatively associated with neural progenitor differentiation — are being tested as treatment for ischaemic stroke (NCT03384433), and mesenchymal stem cell-derived extracellular vesicles loaded with an miRNA against mutant KRAS are being tested as a therapy for pancreatic cancer (NCT03608631). In another example, loading of extracellular vesicles with cholesterol-conjugated siRNA to huntingtin mRNA has been shown to be effective in lowering levels of the protein in a mouse model of Huntington disease171. As discussed earlier, including a bone-targeting aptamer on the surface of extracellular vesicles derived from bone marrow stromal cells is an efficient strategy to therapeutically target the bone marrow in a mouse model of osteoporosis201. It is also possible to harvest extracellular vesicles from genetically modified producer cells presenting targeting peptides, load them with desired cargo and administer them in vivo. This strategy alleviated pathology in a mouse model of Duchenne muscular dystrophy, where extracellular vesicles were designed to target muscle and loaded with splice-correcting antisense nucleotides targeting dystrophin (the alteration of which causes pathology of Duchenne muscular dystrophy)197. On the front of using entire cells rather that purified extracellular vesicle administration, preclinical studies have explored genetically modified glioma cells expressing high levels of a cluster of miRNAs (which are incorporated into extracellular vesicles) to suppress growth of non-modified glioma cells in mouse brain246. In another study, increased yields of extracellular vesicles from HEK293T cells loaded with mRNA for the antioxidant enzyme catalase with ligands for neuronal targeting and endosomal escape were effective in combatting neurotoxicity and Parkinson disease-associated neurodegeneration in mice after subcutaneous implantation161.
Fig. 4. Different strategies for using extracellular vesicles for therapeutic applications.
Examples of therapeutic applications of extracellular vesicles that have resulted in either preclinical assessment or clinical trials (references to primary research or clinical trials are given). Generally, there are five strategies that can be used: implantation of donor cells, such as mesenchymal stem cells (MSCs), which release extracellular vesicles on-site (1); injection of purified, unmodified extracellular vesicles from cultured donor cells (2); injection of purified extracellular vesicles that have been modified for targeting or loading of cargo in vitro (3); injection of extracellular vesicles enriched in the cargo of interest and/or with enhanced targeting/uptake properties from genetically modified cells (4); and implantation of genetically modified cells that release modified extracellular vesicles in vivo (5). AAV, adeno-associated virus; siRNA, small interfering RNA.
Potential advantages associated with use of extracellular vesicles compared with other RNA delivery strategies, such as synthetic lipid nanoparticles or naked modified RNA molecules, include lower immunogenicity and toxicity241,247, better ability to cross cell and tissue barriers21,110 and the potential to achieve more efficient delivery into cells248, while maintaining the possibility of incorporating targeting molecules and of loading both endogenous and synthetic RNA cargo. There is also potential for the use of extracellular vesicles in combinatorial therapeutic approaches. An emerging strategy of combining extracellular vesicle-mediated delivery with gene therapy showed improved therapy for hearing loss when adeno-associated viruses harbouring therapeutic transgenes were packaged into extracellular vesicles in a mouse model of hereditary deafness249. Extracellular vesicles250 and exosome–liposome hybrids251 can also be used for delivery of Cas9 and potentially the guide RNA as well for CRISPR gene editing.
Some of the first clinical trials using extracellular vesicles involved their use as vaccines, specifically those targeting tumours. In this case, dendritic-cell-derived extracellular vesicles harbouring on their surface major histocompatibility complexes, which before injection were pulsed with tumour-specific peptides, thereby providing a platform for efficient delivery of immunogenic tumour antigens, were injected into patients (see, for example, ref.252). In further development of extracellular vesicles as vaccines, it is possible that extracellular vesicle delivery of mRNAs for pathogenic proteins into dendritic cells (see, for example, ref.253) might potentiate vaccination without the need to generate and inactivate pathogenic viruses or bacteria. Similarly, dendritic cell-derived extracellular vesicles can be loaded with total tumour RNA or lysate, providing an abundant source of tumour-specific antigens254. In another avenue of using extracellular vesicles in antitumour vaccination, it was shown that tumour-derived extracellular vesicles can promote the antitumorigenic responses in dendritic cells. In this case extracellular vesicles from the tumour can be used directly or could be used for preloading dendritic cells before their implantation into the patient (as dendritic cell-based antitumour vaccines), with the latter showing so far greater therapeutic potential.
Given the promising results from early studies of extracellular vesicles as therapeutics, substantial investment is being made in this area, both from funding sources such as the US National Institutes of Health (with 302 currently funded grants) and from venture capital firms (at least ten biotechnology companies).
Conclusions and perspective
While there is consensus that RNAs in extracellular vesicles mediate communication among cells, and there are demonstrated examples of specific RNAs carried by extracellular vesicles from one cell type mediating defined phenotypic effects on target cells, many questions remain to be resolved before we will be able to fully decipher this complex language. There are a variety of types of extracellular vesicles that differ in size and protein, lipid and RNA cargo. So far, attempts to characterize unique extracellular vesicle subclasses have proved challenging, in large part owing to technical and biological limitations and challenges involved in separation and molecular analysis of these nanoscale structures. Moreover, the RNA content of extracellular vesicles is quite complex, mostly comprising small non-coding RNAs, the function of most of which are just beginning to be elucidated. There is growing evidence that different extracellular vesicle subclasses carry different RNA cargo, but a comprehensive cataloguing of this cargo awaits the development of efficient methods to identify and separate extracellular vesicle subclasses. It is possible that some extracellular vesicles have a higher content of functional RNAs and others serve more as ‘rubbish bags’ for the source cell (still, jettisoning of certain RNAs can have functional consequences for both source cells and recipient cells). In terms of extracellular vesicle-mediated signalling, the modes by which extracellular vesicle subclasses are ‘addressed’ to different recipient cell types await delineation. Although we know miRNAs and mRNAs in extracellular vesicles can be transferred to and be active in recipient cells, it remains to be determined what percentage of the RNA cargo performs functions and what percentage is lost to degradation (or is simply not functional in the recipient cell). A key issue here is the endosomal escape routes of RNA in extracellular vesicles, as this step appears to be the major bottleneck in efficient functional delivery of cargo. Given the apparent roles of extracellular vesicles in physiology and disease, understanding these issues is critical to providing insights into pathogenic mechanisms and potential therapeutic interventions. In this Review, we have provided a platform of the current state of knowledge of RNA transported in extracellular vesicles on which the field can build another level of understanding. In parallel, there is a burgeoning field of nanoparticles that can efficiently carry mRNA and siRNA to cells and escape the endosome (see, for example, refs255,256). It will be important for the extracellular vesicle and nanoparticle/liposome fields to engage in dialogue and combine the best properties of both for therapeutic applications257.
Supplementary information
Acknowledgements
The Breakefield laboratory acknowledges grant support from the US National Institutes of Health National Cancer Institute (P01 CA069246 and R35 CA232103). S.U. acknowledges financial support from Associazione Italiana per la Ricerca sul Cancro (grant IG 20210) to S. Giordano. L.C.L. is supported by US National Institutes of Health grant U01HL126494. The authors thank M. Brennan for insights on mesenchymal stem cells and S. McDavitt for skilled editorial assistance.
Glossary
- Astrocytes
Glial cells of the central nervous system.
- Large oncosomes
Extracellular vesicles between 1,000 nm and 10,000 nm in diameter derived from tumour cells that contain abnormal and transforming macromolecules, in addition to other cargo.
- Lipoproteins
Macromolecular aggregates of proteins (apoproteins) and different classes of hydrophobic and amphipathic biomolecules that range in size from 10 nm to 1 μm. These aggregates have the function of collecting and transporting lipids, triglycerides, free cholesterol (unesterified cholesterol) and esterified cholesterol in blood plasma.
- Ribonucleoproteins
Proteins structurally associated with nucleic acids, the prosthetic group (nucleic acid) of which contains ribose.
- High-resolution density gradient centrifugation
A method in which extracellular vesicles and particles are centrifuged in a gradient medium (for example, iodixanol) and separated primarily on the basis of their density.
- Size-exclusion chromatography
A technique in which a porous stationary phase is used to sort extracellular vesicles from proteins on the basis of the larger size of the former.
- Asymmetric field-flow fractionation
A technique that separates differentially sized particles ranging from 1 nm to 100 μm in a fluid suspension where the rate of laminar flow is not uniform. A force is applied in a perpendicular direction, facilitating the separation of particles on the basis of size.
- Immunoaffinity purification
A method of isolation of extracellular vesicles using antibodies, typically conjugated to magnetic beads, that recognize proteins exposed on the surface of extracellular vesicles.
- Antisense oligonucleotides
Sequences of DNA or RNA roughly 15–25 nucleotides in length that hybridize to complementary RNA targets, resulting in altered splicing or degradation of the targeted RNA.
- Guide RNAs
RNAs with a specific sequence that recognize and aid nucleases, such as Cas9, in cutting the genomic DNA region of interest for targeted genome engineering.
- Self-amplifying RNAs
Engineered viral genomes encoding proteins involved in the replication mechanism and therefore capable of directing their own replication.
- Small nuclear RNAs
Short sequences (about 150 nucleotides) of ribonucleic acid, usually very rich in uracil, involved in the maturation of mRNA in the cell nucleus.
- Small nucleolar RNAs
(snoRNAs). RNA species consisting of 60–300 nucleotides capable of promoting certain chemical modifications and maturation of many cellular RNAs.
- PIWI-interacting RNAs
(piRNAs). RNAs approximately 21–35 nucleotides long representing a class of RNAs that form protein–RNA complexes by interacting with PIWI proteins. These piRNA complexes have been linked to the transcriptional silencing of retrotransposons and other genetic elements in germ line cells.
- Y RNAs
Small non-coding RNAs essential for the initiation of chromosomal DNA replication. When bound to the RO60, they are involved in RNA stability and cellular responses to stress.
- Vault RNAs
(vtRNAs). Small RNA components of the vault ribonucleoprotein complex. Approximately 100 nucleotides in length, they function as the intracellular and nucleocytoplasmic transporters.
- Dicer
The RNase III known as Dicer is involved in the cleavage of double-stranded RNA and microRNA precursor molecules forming short double-stranded fragments called ‘small interfering RNAs’ or ‘microRNAs’.
- Argonaute
The Argonaute protein family has a critical role in the RNA-induced silencing complex. These proteins form complexes with microRNAs, small interfering RNAs and PIWI-interacting RNAs, interfering with mRNA translation and/or degrading target mRNAs.
- Circular RNAs
Single-stranded, non-coding RNAs that may play a role in transcriptional regulation and in mediating protein interactions. They are characterized by a covalently closed loop feature without 5′-end caps or 3′ poly(A) tails.
- tRNA fragments
Transfer RNA (tRNA)-derived fragments are cleaved from either mature or precursor tRNAs. Functionally, they can act through RNA interference pathways, participate in the formation of stress granules, move mRNA from RNA-binding proteins and/or inhibit translation.
- Multivesicular bodies
(MVBs). Represent a late endosomal compartment, which contains luminal vesicles. Vesicles within the MVBs either can be degraded when MVBs fuse with lysosomes or can be secreted as exosomes when MVBs fuse with the plasma membrane.
- Lipid rafts
Subdomains of the plasma membrane represented by accumulations of proteins and lipids, with high concentrations of cholesterol and glycosphingolipids.
- Blebbing
A bulging of the plasma membrane accompanied by membrane decoupling from the underlying cytoskeleton, which has the potential to release vesicles surrounded by the plasma membrane.
- Secretory autophagy
The transportation of proteins via the autophagosome to the plasma membrane, multivesicular bodies or secretory lysosomes resulting in extracellular release of proteins.
- Macropinocytosis
An endocytic process that results in the cellular uptake of fluid and particles less than 0.2 μm in diameter via invaginated membranes that then form intracellular vesicles.
- Tunnelling nanotubes
Dynamic actin-driven protrusions from the plasma membrane of cells that form connections between cells more than 100 μm away to facilitate intercellular communication.
- Proteoglycans
Highly glycosylated proteins that form the major component of the extracellular matrix. Glycosaminoglycans of proteoglycans are divided into seven main classes: hyaluronic acid, chondroitin sulfate A, chondroitin sulfate B, chondroitin sulfate C, keratan sulfate I, keratan sulfate II and heparin. Proteoglycans act as molecular filters, regulating movement of macromolecules, such as albumin and immunoglobulins, throughout the matrix.
- Liposomes
Small spherical nanoparticles that contain an aqueous core solution and at least one phospholipid bilayer. They can be prepared from biological membranes and are often used for drug delivery.
- Amyloid precursor protein
A transmembrane protein that is a precursor of β-amyloid, a protein involved in the pathology of Alzheimer disease.
- Mesenchymal stem cells
Multipotent stromal progenitor cells found in a number of different tissues that are capable of self-renewal and differentiation into a variety of cell types.
- Cre recombinase
A type I topoisomerase from bacteriophage P1 that catalyses cleavage and ligation of DNA at specific loxP recognition sites.
- RIG-I-like receptors
Intracellular pattern recognition receptors that belong to the RNA box helicase family that function as cytoplasmic sensors of pathogen-associated molecular patterns elicited by viral RNA.
- NOD-like receptor family
Cytoplasmic receptors expressed by innate immune and other cells. They are part of the family of intracellular pattern recognition receptors and can recognize structures associated with pathogenic microorganisms (pathogen-associated molecular patterns) or with cell damage (damage-associated molecular patterns), thereby activating the innate immune response and inflammation.
- Type I interferon
A large subgroup of cytokines that help to regulate the activity of the immune system. They act to degrade viral DNA and proteins in infected cells and provide protection to surrounding cells through interaction with and signalling from interferon receptors.
- Toll-like receptors
(TLRs). A class of transmembrane proteins involved in the defence of the organism through innate immunity. They are mainly expressed on the membrane of sentinel cells, such as macrophages and dendritic cells, and recognize certain typical structures of pathogens and microorganisms.
- Lysosome-associated membrane protein 2B
A particular isoform of LAMP2 — a protein that associates with the membrane of multivesicular bodies and lysosomes.
- Tetraspanin
A family of membrane proteins consisting of four transmembrane domains, with intracellular N and C terminals and two extracellular domains. Tetraspanins are present in the membranes of extracellular vesicles.
- ‘Zipcode’ sequences
Sequences typically found in the 3′ untranslated regions of mRNA transcripts that can be recognized by specific RNA-binding proteins.
- Lipofectamine
A transfection reagent, composed of a cationic tail and a lipid region. Lipofectamine binds exogenous DNA and RNA, creating liposomes that, once in contact with the plasma membrane of the cells, merge with it and facilitate entry of nucleic acids into the cytoplasm.
- V-ATPase
Vacuolar-type H-ATPase. A transmembrane protein found mainly in intracellular organelles and the plasma membrane. It catalyses the hydrolysis of ATP, allowing transport of solutes and acidification of organelles.
- Heparanase
Heparanases are a group of enzymes that act both at the cell surface and within the extracellular matrix to degrade polymeric heparan sulfate molecules into shorter chain length oligosaccharides.
- Gene therapy
A variety of methods resulting in alterations in the genome and/or transcriptome of cells in order to correct deficiency or dominant negative effects caused by genetic mutations or infections.
- RNA aptamers
Small oligonucleotides that bind to target molecules with high affinity and specificity through their 3D structures.
- Major histocompatibility complexes
A group of genes encoding plasma membrane proteins that facilitate recognition of foreign peptides by the adaptive immune system.
Author contributions
The authors contributed equally to all aspects of the article, with K.O’B. and K.B. doing the major draft writing.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Biotechnology companies: https://www.evaluate.com/vantage/articles/news/snippets/exosomes-start-deliver-deals
ClinicalTrials.gov: https://www.clinicaltrials.gov/
exRNA Atlas: https://exrna-atlas.org/
Contributor Information
Louise C. Laurent, Email: llaurent@ucsd.edu
Xandra O. Breakefield, Email: breakefield@hms.harvard.edu
Supplementary information
Supplementary information is available for this paper at 10.1038/s41580-020-0251-y.
References
- 1.Danielson KM, et al. Plasma circulating extracellular RNAs in left ventricular remodeling post-myocardial infarction. EBioMedicine. 2018 doi: 10.1016/j.ebiom.2018.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Varcianna A, et al. Micro-RNAs secreted through astrocyte-derived extracellular vesicles cause neuronal network degeneration in C9orf72 ALS. EBioMedicine. 2019 doi: 10.1016/j.ebiom.2018.11.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abels ER, et al. Glioblastoma-associated microglia reprogramming is mediated by functional transfer of extracellular miR-21. Cell Rep. 2019 doi: 10.1016/j.celrep.2019.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoshino A, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015 doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mathieu M, Martin-Jaular L, Lavieu G, Théry C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019 doi: 10.1038/s41556-018-0250-9. [DOI] [PubMed] [Google Scholar]
- 6.Théry C, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the international society for extracellular vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles. 2018 doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Deun J, Hendrix A. Is your article EV-TRACKed? J. Extracell. Vesicles. 2017 doi: 10.1080/20013078.2017.1379835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee K, et al. Multiplexed profiling of single extracellular vesicles. ACS Nano. 2018 doi: 10.1021/acsnano.7b07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kowal J, et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl Acad. Sci. USA. 2016 doi: 10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeppesen DK, et al. Reassessment of exosome composition. Cell. 2019 doi: 10.1016/j.cell.2019.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murillo OD, et al. exRNA atlas analysis reveals distinct extracellular RNA cargo types and their carriers present across human biofluids. Cell. 2019 doi: 10.1016/j.cell.2019.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei Z, Batagov AO, Carter DRF, Krichevsky AM. Fetal bovine serum RNA interferes with the cell culture derived extracellular. RNA. Sci. Rep. 2016 doi: 10.1038/srep31175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lehrich BM, Liang Y, Fiandaca MS. Response to “Technical approaches to reduce interference of fetal calf serum derived RNA in the analysis of extracellular vesicle RNA from cultured cells”. J. Extracell. Vesicles. 2019 doi: 10.1080/20013078.2019.1599681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Srinivasan S, et al. Small RNA sequencing across diverse biofluids identifies optimal methods for exRNA isolation. Cell. 2019 doi: 10.1016/j.cell.2019.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karttunen J, et al. Precipitation-based extracellular vesicle isolation from rat plasma co-precipitate vesicle-free microRNAs. J. Extracell. Vesicles. 2019 doi: 10.1080/20013078.2018.1555410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Deun J, et al. The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling. J. Extracell. Vesicles. 2014 doi: 10.3402/jev.v3.24858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011 doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arroyo JD, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl Acad. Sci. USA. 2011 doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guerreiro EM, et al. Efficient extracellular vesicle isolation by combining cell media modifications, ultrafiltration, and size-exclusion chromatography. PLoS One. 2018 doi: 10.1371/journal.pone.0204276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang H, et al. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat. Cell Biol. 2018 doi: 10.1038/s41556-018-0040-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Q, et al. Transfer of functional cargo in exomeres. Cell Rep. 2019 doi: 10.1016/j.celrep.2019.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shah R, Patel T, Freedman JE. Circulating extracellular vesicles in human disease. N. Engl. J. Med. 2018 doi: 10.1056/NEJMra1704286. [DOI] [PubMed] [Google Scholar]
- 23.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007 doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 24.Skog J, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008 doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ekström K, et al. Characterization of mRNA and microRNA in human mast cell-derived exosomes and their transfer to other mast cells and blood CD34 progenitor cells. J. Extracell. Vesicles. 2012 doi: 10.3402/jev.v1i0.18389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang X, et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics. 2013 doi: 10.1186/1471-2164-14-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Balkom BWM, Eisele AS, Michiel Pegtel D, Bervoets S, Verhaar MC. Quantitative and qualitative analysis of small RNAs in human endothelial cells and exosomes provides insights into localized RNA processing, degradation and sorting. J. Extracell. Vesicles. 2015 doi: 10.3402/jev.v4.26760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chakrabortty SK, Prakash A, Nechooshtan G, Hearn S, Gingeras TR. Extracellular vesicle-mediated transfer of processed and functional RNY5 RNA. RNA. 2015 doi: 10.1261/rna.053629.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clancy JW, Zhang Y, Sheehan C, D’Souza-Schorey C. An ARF6–exportin-5 axis delivers pre-miRNA cargo to tumour microvesicles. Nat. Cell Biol. 2019 doi: 10.1038/s41556-019-0345-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melo SA, et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell. 2014 doi: 10.1016/j.ccell.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Squadrito ML, et al. Endogenous RNAs modulate microRNA sorting to exosomes and transfer to acceptor cells. Cell Rep. 2014 doi: 10.1016/j.celrep.2014.07.035. [DOI] [PubMed] [Google Scholar]
- 32.Karimi N, et al. Detailed analysis of the plasma extracellular vesicle proteome after separation from lipoproteins. Cell. Mol. Life Sci. 2018 doi: 10.1007/s00018-018-2773-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu R, Greening DW, Rai A, Ji H, Simpson RJ. Highly-purified exosomes and shed microvesicles isolated from the human colon cancer cell line LIM1863 by sequential centrifugal ultrafiltration are biochemically and functionally distinct. Methods. 2015 doi: 10.1016/j.ymeth.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 34.McKenzie AJ, et al. KRAS-MEK signaling controls Ago2 sorting into exosomes. Cell Rep. 2016 doi: 10.1016/j.celrep.2016.03.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li M, et al. Analysis of the RNA content of the exosomes derived from blood serum and urine and its potential as biomarkers. Philos. Trans. R. Soc. B Biol. Sci. 2014 doi: 10.1098/rstb.2013.0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang J, et al. Circular RNA expression in exosomes derived from breast cancer cells and patients. Epigenomics. 2019 doi: 10.2217/epi-2018-0111. [DOI] [PubMed] [Google Scholar]
- 37.Zhang H, et al. Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene. 2019 doi: 10.1038/s41388-018-0619-z. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Zhang H, et al. Exosomal circRNA derived from gastric tumor promotes white adipose browning by targeting the miR-133/PRDM16 pathway. Int. J. Cancer. 2019 doi: 10.1002/ijc.31977. [DOI] [PubMed] [Google Scholar]
- 39.He J, et al. Exosomal circular RNA as a biomarker platform for the early diagnosis of immune-mediated demyelinating disease. Front. Genet. 2019 doi: 10.3389/fgene.2019.00860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fanale D, Taverna S, Russo A, Bazan V. Circular RNA in exosomes. Adv. Exp. Med. Biol. 2018 doi: 10.1007/978-981-13-1426-1_9. [DOI] [PubMed] [Google Scholar]
- 41.Sork H, et al. Heterogeneity and interplay of the extracellular vesicle small RNA transcriptome and proteome. Sci. Rep. 2018 doi: 10.1038/s41598-018-28485-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baglio SR, et al. Human bone marrow- and adipose-mesenchymal stem cells secrete exosomes enriched in distinctive miRNA and tRNA species. Stem Cell Res. Ther. 2015 doi: 10.1186/s13287-015-0116-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guduric-Fuchs J, et al. Selective extracellular vesicle-mediated export of an overlapping set of microRNAs from multiple cell types. BMC Genomics. 2012 doi: 10.1186/1471-2164-13-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mittelbrunn M, et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat. Commun. 2011 doi: 10.1038/ncomms1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pigati L, et al. Selective release of microRNA species from normal and malignant mammary epithelial cells. PLoS One. 2010 doi: 10.1371/journal.pone.0013515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nolte’T Hoen ENM, et al. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chiou NT, Kageyama R, Ansel KM. Selective export into extracellular vesicles and function of tRNA fragments during T cell activation. Cell Rep. 2018 doi: 10.1016/j.celrep.2018.11.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsuzaki J, Ochiya T. Extracellular microRNAs and oxidative stress in liver injury: a systematic mini review. J. Clin. Biochem. Nutr. 2018 doi: 10.3164/jcbn.17-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beninson LA, et al. Acute stressor exposure modifies plasma exosome-associated heat shock protein 72 (Hsp72) and microRNA (miR-142-5p and miR-203) PLoS One. 2014 doi: 10.1371/journal.pone.0108748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma C, et al. Moderate exercise enhances endothelial progenitor cell exosomes release and function. Med. Sci. Sports Exerc. 2018 doi: 10.1249/MSS.0000000000001672. [DOI] [PubMed] [Google Scholar]
- 51.D’souza RF, et al. Circulatory exosomal miRNA following intense exercise is unrelated to muscle and plasma miRNA abundances. Am. J. Physiol. Endocrinol. Metab. 2018 doi: 10.1152/ajpendo.00138.2018. [DOI] [PubMed] [Google Scholar]
- 52.Aoi W, et al. Muscle-enriched micro RNA miR-486 decreases in circulation in response to exercise in young men. Front. Physiol. 2013 doi: 10.3389/fphys.2013.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van Niel G, D’Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018 doi: 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- 54.Pollet H, Conrard L, Cloos AS, Tyteca D. Plasma membrane lipid domains as platforms for vesicle biogenesis and shedding? Biomolecules. 2018 doi: 10.3390/biom8030094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mittelbrunn M, Vicente-Manzanares M, Sánchez-Madrid F. Organizing polarized delivery of exosomes at synapses. Traffic. 2015 doi: 10.1111/tra.12258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sinha S, et al. Cortactin promotes exosome secretion by controlling branched actin dynamics. J. Cell Biol. 2016 doi: 10.1083/jcb.201601025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hoshino D, et al. Exosome secretion is enhanced by invadopodia and drives invasive behavior. Cell Rep. 2013 doi: 10.1016/j.celrep.2013.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Minciacchi VR, Freeman MR, Di Vizio D. Extracellular vesicles in cancer: exosomes, microvesicles and the emerging role of large oncosomes. Semin. Cell Dev. Biol. 2015 doi: 10.1016/j.semcdb.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meehan B, Rak J, Di Vizio D. Oncosomes - large and small: what are they, where they came from? J. Extracell. Vesicles. 2016 doi: 10.3402/jev.v5.33109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Caruso S, Poon IKH. Apoptotic cell-derived extracellular vesicles: more than just debris. Front. Immunol. 2018 doi: 10.3389/fimmu.2018.01486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Das S, et al. The Extracellular RNA Communication Consortium: establishing foundational knowledge and technologies for extracellular RNA research. Cell. 2019 doi: 10.1016/j.cell.2019.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Di Liegro CM, Schiera G, Di Liegro I. Regulation of mRNA transport, localizationand translation in the nervous system of mammals (review) Int. J. Mol. Med. 2014 doi: 10.3892/ijmm.2014.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Eliscovich C, Buxbaum AR, Katz ZB, Singer RH. mRNA on the move: the road to its biological destiny. J. Biol. Chem. 2013 doi: 10.1074/jbc.R113.452094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gerstberger S, Hafner M, Ascano M, Tuschl T. Evolutionary conservation and expression of human RNA-binding proteins and their role in human genetic disease. Adv. Exp. Med. Biol. 2014 doi: 10.1007/978-1-4939-1221-6_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ragusa M, et al. Asymmetric RNA distribution among cells and their secreted exosomes: biomedical meaning and considerations on diagnostic applications. Front. Mol. Biosci. 2017 doi: 10.3389/fmolb.2017.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mateescu B, et al. Obstacles and opportunities in the functional analysis of extracellular vesicle RNA - an ISEV position paper. J. Extracell. Vesicles. 2017 doi: 10.1080/20013078.2017.1286095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kosaka N, et al. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J. Biol. Chem. 2010 doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Khvorova A, Kwak YG, Tamkun M, Majerfeld I, Yarus M. RNAs that bind and change the permeability of phospholipid membranes. Proc. Natl Acad. Sci. USA. 1999 doi: 10.1073/pnas.96.19.10649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Janas T, Janas MM, Sapoń K, Janas T. Mechanisms of RNA loading into exosomes. FEBS Lett. 2015 doi: 10.1016/j.febslet.2015.04.036. [DOI] [PubMed] [Google Scholar]
- 70.Iavello A, et al. Role of Alix in miRNA packaging during extracellular vesicle biogenesis. Int. J. Mol. Med. 2016 doi: 10.3892/ijmm.2016.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hagiwara K, Katsuda T, Gailhouste L, Kosaka N, Ochiya T. Commitment of annexin A2 in recruitment of microRNAs into extracellular vesicles. FEBS Lett. 2015 doi: 10.1016/j.febslet.2015.11.036. [DOI] [PubMed] [Google Scholar]
- 72.Otake K, Kamiguchi H, Hirozane Y. Identification of biomarkers for amyotrophic lateral sclerosis by comprehensive analysis of exosomal mRNAs in human cerebrospinal fluid. BMC Med. Genomics. 2019 doi: 10.1186/s12920-019-0473-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Teng Y, et al. MVP-mediated exosomal sorting of miR-193a promotes colon cancer progression. Nat. Commun. 2017 doi: 10.1038/ncomms14448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Statello L, et al. Identification of RNA-binding proteins in exosomes capable of interacting with different types of RNA: RBP-facilitated transport of RNAs into exosomes. PLoS One. 2018 doi: 10.1371/journal.pone.0195969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mukherjee K, et al. Reversible HuR‐micro RNA binding controls extracellular export of miR‐122 and augments stress response. EMBO Rep. 2016 doi: 10.15252/embr.201541930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Villarroya-Beltri C, et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013 doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shurtleff MJ, Temoche-Diaz MM, Karfilis KV, Ri S, Schekman R. Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. Elife. 2016 doi: 10.7554/eLife.19276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yanshina DD, et al. Structural features of the interaction of the 3′-untranslated region of mRNA containing exosomal RNA-specific motifs with YB-1, a potential mediator of mRNA sorting. Biochimie. 2018 doi: 10.1016/j.biochi.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 79.Kossinova OA, et al. Cytosolic YB-1 and NSUN2 are the only proteins recognizing specific motifs present in mRNAs enriched in exosomes. Biochim. Biophys. Acta. 2017 doi: 10.1016/j.bbapap.2017.03.010. [DOI] [PubMed] [Google Scholar]
- 80.Santangelo L, et al. The RNA-binding protein SYNCRIP is a component of the hepatocyte exosomal machinery controlling microRNA sorting. Cell Rep. 2016 doi: 10.1016/j.celrep.2016.09.031. [DOI] [PubMed] [Google Scholar]
- 81.Hobor F, et al. A cryptic RNA-binding domain mediates Syncrip recognition and exosomal partitioning of miRNA targets. Nat. Commun. 2018 doi: 10.1038/s41467-018-03182-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Temoche-Diaz MM, et al. Distinct mechanisms of microRNA sorting into cancer cell-derived extracellular vesicle subtypes. Elife. 2019 doi: 10.7554/eLife.47544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ashley J, et al. Retrovirus-like Gag protein Arc1 binds RNA and traffics across synaptic boutons. Cell. 2018 doi: 10.1016/j.cell.2017.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Leidal AM, et al. The LC3-conjugation machinery specifies the loading of RNA-binding proteins into extracellular vesicles. Nat. Cell Biol. 2020 doi: 10.1038/s41556-019-0450-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bolukbasi MF, et al. miR-1289 and ‘zipcode’-like sequence enrich mRNAs in microvesicles. Mol. Ther. Nucleic Acids. 2012 doi: 10.1038/mtna.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shurtleff MJ, et al. Broad role for YBX1 in defining the small noncoding RNA composition of exosomes. Proc. Natl Acad. Sci. USA. 2017 doi: 10.1073/pnas.1712108114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ahadi A, Brennan S, Kennedy PJ, Hutvagner G, Tran N. Long non-coding RNAs harboring miRNA seed regions are enriched in prostate cancer exosomes. Sci. Rep. 2016 doi: 10.1038/srep24922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu X, et al. A microRNA precursor surveillance system in quality control of microRNA synthesis. Mol. Cell. 2014 doi: 10.1016/j.molcel.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat. Cell Biol. 2009 doi: 10.1038/ncb1929. [DOI] [PubMed] [Google Scholar]
- 90.Murphy DE, et al. Extracellular vesicle-based therapeutics: natural versus engineered targeting and trafficking. Exp. Mol. Med. 2019 doi: 10.1038/s12276-019-0223-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.György B, Hung ME, Breakefield XO, Leonard JN. Therapeutic applications of extracellular vesicles: clinical promise and open questions. Annu. Rev. Pharmacol. Toxicol. 2015 doi: 10.1146/annurev-pharmtox-010814-124630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mulcahy LA, Pink RC, Carter DRF. Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles. 2014 doi: 10.3402/jev.v3.24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Panasiuk M, Rychłowski M, Derewońko N, Bieńkowska-Szewczyk K. Tunneling nanotubes as a novel route of cell-to-cell spread of herpesviruses. J. Virol. 2018 doi: 10.1128/jvi.00090-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rustom A, Saffrich R, Markovic I, Walther P, Gerdes HH. Nanotubular highways for intercellular organelle transport. Science. 2004 doi: 10.1126/science.1093133. [DOI] [PubMed] [Google Scholar]
- 95.Haimovich G, et al. Intercellular mRNA trafficking via membrane nanotube-like extensions in mammalian cells. Proc. Natl Acad. Sci. USA. 2017 doi: 10.1073/pnas.1706365114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shukla D, et al. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell. 1999 doi: 10.1016/S0092-8674(00)80058-6. [DOI] [PubMed] [Google Scholar]
- 97.Christianson HC, Svensson KJ, Van Kuppevelt TH, Li JP, Belting M. Cancer cell exosomes depend on cell-surface heparan sulfate proteoglycans for their internalization and functional activity. Proc. Natl Acad. Sci. USA. 2013 doi: 10.1073/pnas.1304266110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hurwitz SN, Meckes DG. Extracellular vesicle integrins distinguish unique cancers. Proteomes. 2019 doi: 10.3390/proteomes7020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Miyanishi M, et al. Identification of Tim4 as a phosphatidylserine receptor. Nature. 2007 doi: 10.1038/nature06307. [DOI] [PubMed] [Google Scholar]
- 100.Iversen TG, Skotland T, Sandvig K. Endocytosis and intracellular transport of nanoparticles: present knowledge and need for future studies. Nano Today. 2011 doi: 10.1016/j.nantod.2011.02.003. [DOI] [Google Scholar]
- 101.Hung ME, Leonard JN. A platform for actively loading cargo RNA to elucidate limiting steps in EV-mediated delivery. J. Extracell. Vesicles. 2016 doi: 10.3402/jev.v5.31027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Toribio V, et al. Development of a quantitative method to measure EV uptake. Sci. Rep. 2019;9:10522. doi: 10.1038/s41598-019-47023-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bala S, et al. Biodistribution and function of extracellular miRNA-155 in mice. Sci. Rep. 2015 doi: 10.1038/srep10721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mannerström B, et al. Extracellular small non-coding RNA contaminants in fetal bovine serum and serum-free media. Sci. Rep. 2019 doi: 10.1038/s41598-019-41772-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gaurivaud P, et al. Mycoplasmas are no exception to extracellular vesicles release: revisiting old concepts. PLoS One. 2018 doi: 10.1371/journal.pone.0208160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hardy MP, et al. Apoptotic endothelial cells release small extracellular vesicles loaded with immunostimulatory viral-like RNAs. Sci. Rep. 2019 doi: 10.1038/s41598-019-43591-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liu CG, Song J, Zhang YQ, Wang PC. MicroRNA-193b is a regulator of amyloid precursor protein in the blood and cerebrospinal fluid derived exosomal microRNA-193b is a biomarker of Alzheimer’s disease. Mol. Med. Rep. 2014 doi: 10.3892/mmr.2014.2484. [DOI] [PubMed] [Google Scholar]
- 108.Morel L, et al. Neuronal exosomal mirna-dependent translational regulation of astroglial glutamate transporter glt1. J. Biol. Chem. 2013 doi: 10.1074/jbc.M112.410944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhang L, et al. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature. 2015 doi: 10.1038/nature15376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chen CC, et al. Elucidation of exosome migration across the blood–brain barrier model in vitro. Cell. Mol. Bioeng. 2016 doi: 10.1007/s12195-016-0458-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Alexander M, et al. Exosome-delivered microRNAs modulate the inflammatory response to endotoxin. Nat. Commun. 2015 doi: 10.1038/ncomms8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wei Z, et al. Coding and noncoding landscape of extracellular RNA released by human glioma stem cells. Nat. Commun. 2017 doi: 10.1038/s41467-017-01196-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hoen EN, Cremer T, Gallo RC, Margolis LB. Extracellular vesicles and viruses: Are they close relatives? Proc. Natl Acad. Sci. USA. 2016 doi: 10.1073/pnas.1605146113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lai CP, et al. Visualization and tracking of tumour extracellular vesicle delivery and RNA translation using multiplexed reporters. Nat. Commun. 2015 doi: 10.1038/ncomms8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ridder K, et al. Extracellular vesicle-mediated transfer of genetic information between the hematopoietic system and the brain in response to inflammation. PLoS Biol. 2014 doi: 10.1371/journal.pbio.1001874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zomer A, et al. In vivo imaging reveals extracellular vesicle-mediated phenocopying of metastatic behavior. Cell. 2015 doi: 10.1016/j.cell.2015.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.De La Cuesta F, et al. Extracellular vesicle cross-talk between pulmonary artery smooth muscle cells and endothelium during excessive TGF-β signalling: implications for PAH vascular remodelling. Cell Commun. Signal. 2019 doi: 10.1186/s12964-019-0449-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Maugeri M, et al. Linkage between endosomal escape of LNP-mRNA and loading into EVs for transport to other cells. Nat. Commun. 2019 doi: 10.1038/s41467-019-12275-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Usman WM, et al. Efficient RNA drug delivery using red blood cell extracellular vesicles. Nat. Commun. 2018 doi: 10.1038/s41467-018-04791-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang J, et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang Y, et al. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev. Cell. 2013 doi: 10.1016/j.devcel.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 122.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 39 UTRs via Alu elements. Nature. 2011 doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Long Y, Wang X, Youmans DT, Cech TR. How do lncRNAs regulate transcription? Sci. Adv. 2017 doi: 10.1126/sciadv.aao2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kenneweg F, et al. Long noncoding RNA-enriched vesicles secreted by hypoxic cardiomyocytes drive cardiac fibrosis. Mol. Ther. Nucleic Acids. 2019 doi: 10.1016/j.omtn.2019.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chen F, et al. Extracellular vesicle-packaged HIF-1α-stabilizing lncRNA from tumour-associated macrophages regulates aerobic glycolysis of breast cancer cells. Nat. Cell Biol. 2019 doi: 10.1038/s41556-019-0299-0. [DOI] [PubMed] [Google Scholar]
- 126.Maute RL, et al. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc. Natl Acad. Sci. USA. 2013 doi: 10.1073/pnas.1206761110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Goodarzi H, et al. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell. 2015 doi: 10.1016/j.cell.2015.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Driedonks TAP, et al. Immune stimuli shape the small non-coding transcriptome of extracellular vesicles released by dendritic cells. Cell. Mol. Life Sci. 2018 doi: 10.1007/s00018-018-2842-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rimer JM, et al. Long-range function of secreted small nucleolar RNAs that direct 2-O-methylation. J. Biol. Chem. 2018 doi: 10.1074/jbc.RA118.003410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tsatsaronis JA, Franch-Arroyo S, Resch U, Charpentier E. Extracellular vesicle RNA: a universal mediator of microbial communication? Trends Microbiol. 2018 doi: 10.1016/j.tim.2018.02.009. [DOI] [PubMed] [Google Scholar]
- 131.Schorey J, Cheng Y. Extracellular vesicles promote host immunity during an M. tuberculosis infection through RNA sensing. bioRxiv. 2018 doi: 10.1101/346254. [DOI] [Google Scholar]
- 132.Baglio SR, et al. Sensing of latent EBV infection through exosomal transfer of 5’pppRNA. Proc. Natl Acad. Sci. USA. 2016 doi: 10.1073/pnas.1518130113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Nabet BY, et al. Exosome RNA unshielding couples stromal activation to pattern recognition receptor signaling in cancer. Cell. 2017 doi: 10.1016/j.cell.2017.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Boelens MC, et al. Exosome transfer from stromal to breast cancer cells regulates therapy resistance pathways. Cell. 2014 doi: 10.1016/j.cell.2014.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lässer C, et al. Two distinct extracellular RNA signatures released by a single cell type identified by microarray and next-generation sequencing. RNA Biol. 2017 doi: 10.1080/15476286.2016.1249092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Persson H, et al. The non-coding RNA of the multidrug resistance-linked vault particle encodes multiple regulatory small RNAs. Nat. Cell Biol. 2009 doi: 10.1038/ncb1972. [DOI] [PubMed] [Google Scholar]
- 137.Horos R, et al. The small non-coding vault RNA1-1 acts as a riboregulator of autophagy. Cell. 2019 doi: 10.1016/j.cell.2019.01.030. [DOI] [PubMed] [Google Scholar]
- 138.Blasius AL, Beutler B. Intracellular Toll-like receptors. Immunity. 2010 doi: 10.1016/j.immuni.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 139.Fabbri M, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc. Natl Acad. Sci. USA. 2012 doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Joerger-Messerli MS, et al. Extracellular vesicles derived from Wharton’s jelly mesenchymal stem cells prevent and resolve programmed cell death mediated by perinatal hypoxia-ischemia in neuronal cells. Cell Transplant. 2018 doi: 10.1177/0963689717738256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bayraktar R, Bertilaccio MTS, Calin GA. The interaction between two worlds: microRNAs and Toll-like receptors. Front. Immunol. 2019 doi: 10.3389/fimmu.2019.01053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lehmann SM, et al. An unconventional role for miRNA: let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat. Neurosci. 2012 doi: 10.1038/nn.3113. [DOI] [PubMed] [Google Scholar]
- 143.Buonfiglioli A, et al. let-7 microRNAs regulate microglial function and suppress glioma growth through Toll-like receptor 7. Cell Rep. 2019 doi: 10.1016/j.celrep.2019.11.029. [DOI] [PubMed] [Google Scholar]
- 144.Atayde VD, et al. Exploitation of the Leishmania exosomal pathway by Leishmania RNA virus 1. Nat. Microbiol. 2019 doi: 10.1038/s41564-018-0352-y. [DOI] [PubMed] [Google Scholar]
- 145.Ramakrishnaiah V, et al. Exosome-mediated transmission of hepatitis C virus between human hepatoma Huh7.5 cells. Proc. Natl Acad. Sci. USA. 2013 doi: 10.1073/pnas.1221899110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Feng Z, et al. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature. 2013 doi: 10.1038/nature12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Longatti A, Boyd B, Chisari FV. Virion-independent transfer of replication-competent hepatitis C virus RNA between permissive cells. J. Virol. 2015 doi: 10.1128/jvi.02721-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Bukong TN, Momen-Heravi F, Kodys K, Bala S, Szabo G. Exosomes from hepatitis C infected patients transmit HCV infection and contain replication competent viral RNA in complex with Ago2-miR122-HSP90. PLoS Pathog. 2014 doi: 10.1371/journal.ppat.1004424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Pastuzyn ED, et al. The neuronal gene arc encodes a repurposed retrotransposon Gag protein that mediates intercellular RNA transfer. Cell. 2018 doi: 10.1016/j.cell.2017.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wiklander OPB, Brennan M, Lötvall J, Breakefield XO, Andaloussi SEL. Advances in therapeutic applications of extracellular vesicles. Sci. Transl. Med. 2019 doi: 10.1126/scitranslmed.aav8521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Martin-Rufino JD, Espinosa-Lara N, Osugui L, Sanchez-Guijo F. Targeting the Immune system with mesenchymal stromal cell-derived extracellular vesicles: what is the cargo’s mechanism of action? Front. Bioeng. Biotechnol. 2019 doi: 10.3389/fbioe.2019.00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kuo WP, Tigges JC, Toxavidis V, Ghiran I. Red blood cells: a source of extracellular vesicles. Methods Mol. Biol. 2017 doi: 10.1007/978-1-4939-7253-1_2. [DOI] [PubMed] [Google Scholar]
- 153.Tkach M, et al. Qualitative differences in T-cell activation by dendritic cell-derived extracellular vesicle subtypes. EMBO J. 2017 doi: 10.15252/embj.201696003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Poon IKH, Gregory CD, Kaparakis-Liaskos M. Editorial: the immunomodulatory properties of extracellular vesicles from pathogens, immune cells, and non-immune. Cells. Front. Immunol. 2018 doi: 10.3389/fimmu.2018.03024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Choi D, et al. Extracellular vesicle communication pathways as regulatory targets of oncogenic transformation. Semin. Cell Dev. Biol. 2017 doi: 10.1016/j.semcdb.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 156.Ohno SI, et al. Systemically injected exosomes targeted to EGFR deliver antitumor microrna to breast cancer cells. Mol. Ther. 2013 doi: 10.1038/mt.2012.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ridder, K. et al. Extracellular vesicle-mediated transfer of functional RNA in the tumor microenvironment. Oncoimmunology (2015). 10.1080/2162402X2015.1008371 [DOI] [PMC free article] [PubMed]
- 158.Jang SC, et al. Bioinspired exosome-mimetic nanovesicles for targeted delivery of chemotherapeutics to malignant tumors. ACS Nano. 2013 doi: 10.1021/nn402232g. [DOI] [PubMed] [Google Scholar]
- 159.Shen B, Wu N, Yang M, Gould SJ. Protein targeting to exosomes/microvesicles by plasma membrane anchors. J. Biol. Chem. 2011 doi: 10.1074/jbc.M110.208660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Tutucci E, et al. An improved MS2 system for accurate reporting of the mRNA life cycle. Nat. Methods. 2018 doi: 10.1038/nmeth.4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kojima R, et al. Designer exosomes produced by implanted cells intracerebrally deliver therapeutic cargo for Parkinson’s disease treatment. Nat. Commun. 2018 doi: 10.1038/s41467-018-03733-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wang Q, et al. ARMMs as a versatile platform for intracellular delivery of macromolecules. Nat. Commun. 2018 doi: 10.1038/s41467-018-03390-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Flynn AD, Yin H. Lipid-targeting peptide probes for extracellular vesicles. J. Cell. Physiol. 2016 doi: 10.1002/jcp.25354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Gori A, et al. Membrane-binding peptides for extracellular vesicles on-chip analysis. chemRxiv. 2020 doi: 10.26434/chemrxiv.9885167.v3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Alvarez-Erviti L, et al. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011 doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 166.Kooijmans SAA, et al. Electroporation-induced siRNA precipitation obscures the efficiency of siRNA loading into extracellular vesicles. J. Control. Rel. 2013 doi: 10.1016/j.jconrel.2013.08.014. [DOI] [PubMed] [Google Scholar]
- 167.Pomatto MAC, et al. Improved loading of plasma-derived extracellular vesicles to encapsulate antitumor miRNAs. Mol. Ther. Methods Clin. Dev. 2019 doi: 10.1016/j.omtm.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kamerkar S, et al. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017 doi: 10.1038/nature22341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Shtam, T. A. et al. Exosomes are natural carriers of exogenous siRNA to human cells in vitro. Cell Commun. Signal. (2013). 10.1186/1478-811X-11-88 [DOI] [PMC free article] [PubMed]
- 170.Lamichhane TN, et al. Oncogene knockdown via active loading of small RNAs into extracellular vesicles by sonication. Cell. Mol. Bioeng. 2016 doi: 10.1007/s12195-016-0457-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Didiot MC, et al. Exosome-mediated delivery of hydrophobically modified siRNA for huntingtin mRNA silencing. Mol. Ther. 2016 doi: 10.1038/mt.2016.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Haraszti RA, et al. Optimized cholesterol-siRNA chemistry improves productive loading onto extracellular vesicles. Mol. Ther. 2018 doi: 10.1016/j.ymthe.2018.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Gimona M, Pachler K, Laner-Plamberger S, Schallmoser K, Rohde E. Manufacturing of human extracellular vesicle-based therapeutics for clinical use. Int. J. Mol. Sci. 2017 doi: 10.3390/ijms18061190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Edgar JR, Manna PT, Nishimura S, Banting G, Robinson MS. Tetherin is an exosomal tether. Elife. 2016 doi: 10.7554/eLife.17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Thompson CA, Purushothaman A, Ramani VC, Vlodavsky I, Sanderson RD. Heparanase regulates secretion, composition, and function of tumor cell-derived exosomes. J. Biol. Chem. 2013 doi: 10.1074/jbc.C112.444562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Savina A, Furlán M, Vidal M, Colombo MI. Exosome release is regulated by a calcium-dependent mechanism in K562 cells. J. Biol. Chem. 2003 doi: 10.1074/jbc.M301642200. [DOI] [PubMed] [Google Scholar]
- 177.Glebov K, et al. Serotonin stimulates secretion of exosomes from microglia cells. Glia. 2015 doi: 10.1002/glia.22772. [DOI] [PubMed] [Google Scholar]
- 178.Jabbari N, Nawaz M, Rezaie J. Ionizing radiation increases the activity of exosomal secretory pathway in MCF-7 human breast cancer cells: a possible way to communicate resistance against radiotherapy. Int. J. Mol. Sci. 2019 doi: 10.3390/ijms20153649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.King HW, Michael MZ, Gleadle JM. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer. 2012 doi: 10.1186/1471-2407-12-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Atienzar-Aroca S, et al. Oxidative stress in retinal pigment epithelium cells increases exosome secretion and promotes angiogenesis in endothelial cells. J. Cell. Mol. Med. 2016 doi: 10.1111/jcmm.12834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Parolini I, et al. Microenvironmental pH is a key factor for exosome traffic in tumor cells. J. Biol. Chem. 2009 doi: 10.1074/jbc.M109.041152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Mangeot PE, et al. Protein transfer into human cells by VSV-G-induced nanovesicles. Mol. Ther. 2011 doi: 10.1038/mt.2011.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Meyer C, et al. Pseudotyping exosomes for enhanced protein delivery in mammalian cells. Int. J. Nanomed. 2017 doi: 10.2147/IJN.S133430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Jhan YY, et al. Engineered extracellular vesicles with synthetic lipids via membrane fusion to establish efficient gene delivery. Int. J. Pharm. 2020 doi: 10.1016/j.ijpharm.2019.118802. [DOI] [PubMed] [Google Scholar]
- 185.Khan FM, et al. Inhibition of exosome release by ketotifen enhances sensitivity of cancer cells to doxorubicin. Cancer Biol. Ther. 2018 doi: 10.1080/15384047.2017.1394544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Im EJ, et al. Sulfisoxazole inhibits the secretion of small extracellular vesicles by targeting the endothelin receptor A. Nat. Commun. 2019 doi: 10.1038/s41467-019-09387-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Datta A, et al. High-throughput screening identified selective inhibitors of exosome biogenesis and secretion: a drug repurposing strategy for advanced cancer. Sci. Rep. 2018 doi: 10.1038/s41598-018-26411-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Marleau AM, Chen CS, Joyce JA, Tullis RH. Exosome removal as a therapeutic adjuvant in cancer. J. Transl. Med. 2012 doi: 10.1186/1479-5876-10-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Lai CP, et al. Dynamic biodistribution of extracellular vesicles in vivo using a multimodal imaging reporter. ACS Nano. 2014 doi: 10.1021/nn404945r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Gangadaran P, et al. A new bioluminescent reporter system to study the biodistribution of systematically injected tumor-derived bioluminescent extracellular vesicles in mice. Oncotarget. 2017 doi: 10.18632/oncotarget.22493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Wiklander OPB, et al. Extracellular vesicle in vivo biodistribution is determined by cell source, route of administration and targeting. J. Extracell. Vesicles. 2015 doi: 10.3402/jev.v4.26316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kooijmans SAA, et al. PEGylated and targeted extracellular vesicles display enhanced cell specificity and circulation time. J. Control. Rel. 2016 doi: 10.1016/j.jconrel.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 193.Armstrong JPK, Holme MN, Stevens MM. Re-engineering extracellular vesicles as smart nanoscale therapeutics. ACS Nano. 2017 doi: 10.1021/acsnano.6b07607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Kooijmans SAA, et al. Display of GPI-anchored anti-EGFR nanobodies on extracellular vesicles promotes tumour cell targeting. J. Extracell. Vesicles. 2016 doi: 10.3402/jev.v5.31053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Kooijmans SAA, Gitz-Francois JJJM, Schiffelers RM, Vader P. Recombinant phosphatidylserine-binding nanobodies for targeting of extracellular vesicles to tumor cells: a plug-and-play approach. Nanoscale. 2018 doi: 10.1039/c7nr06966a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Zou X, et al. Extracellular vesicles expressing a single-chain variable fragment of an HIV-1 specific antibody selectively target Env+ tissues. Theranostics. 2019 doi: 10.7150/thno.33925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Gao X, et al. Anchor peptide captures, targets, and loads exosomes of diverse origins for diagnostics and therapy. Sci. Transl. Med. 2018 doi: 10.1126/scitranslmed.aat0195. [DOI] [PubMed] [Google Scholar]
- 198.Antes TJ, et al. Targeting extracellular vesicles to injured tissue using membrane cloaking and surface display. J. Nanobiotechnol. 2018 doi: 10.1186/s12951-018-0388-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Pi F, et al. Nanoparticle orientation to control RNA loading and ligand display on extracellular vesicles for cancer regression. Nat. Nanotechnol. 2018 doi: 10.1038/s41565-017-0012-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Zou J, et al. Aptamer-functionalized exosomes: elucidating the cellular uptake mechanism and the potential for cancer-targeted chemotherapy. Anal. Chem. 2019 doi: 10.1021/acs.analchem.8b05204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Luo ZW, et al. Aptamer-functionalized exosomes from bone marrow stromal cells target bone to promote bone regeneration. Nanoscale. 2019 doi: 10.1039/c9nr02791b. [DOI] [PubMed] [Google Scholar]
- 202.Zaborowski MP, et al. Membrane-bound Gaussia luciferase as a tool to track shedding of membrane proteins from the surface of extracellular vesicles. Sci. Rep. 2019 doi: 10.1038/s41598-019-53554-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Rupp AK, et al. Loss of EpCAM expression in breast cancer derived serum exosomes: role of proteolytic cleavage. Gynecol. Oncol. 2011 doi: 10.1016/j.ygyno.2011.04.035. [DOI] [PubMed] [Google Scholar]
- 204.Royo F, Cossío U, Ruiz De Angulo A, Llop J, Falcon-Perez JM. Modification of the glycosylation of extracellular vesicles alters their biodistribution in mice. Nanoscale. 2019 doi: 10.1039/c8nr03900c. [DOI] [PubMed] [Google Scholar]
- 205.Williams C, et al. Assessing the role of surface glycans of extracellular vesicles on cellular uptake. Sci. Rep. 2019 doi: 10.1038/s41598-019-48499-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Williams C, et al. Glycosylation of extracellular vesicles: current knowledge, tools and clinical perspectives. J. Extracell. Vesicles. 2018 doi: 10.1080/20013078.2018.1442985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Nakase I, Futaki S. Combined treatment with a pH-sensitive fusogenic peptide and cationic lipids achieves enhanced cytosolic delivery of exosomes. Sci. Rep. 2015 doi: 10.1038/srep10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Nakase I, et al. Arginine-rich cell-penetrating peptide-modified extracellular vesicles for active macropinocytosis induction and efficient intracellular delivery. Sci. Rep. 2017 doi: 10.1038/s41598-017-02014-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Nakase I, Kobayashi NB, Takatani-Nakase T, Yoshida T. Active macropinocytosis induction by stimulation of epidermal growth factor receptor and oncogenic Ras expression potentiates cellular uptake efficacy of exosomes. Sci. Rep. 2015 doi: 10.1038/srep10300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Prada I, Meldolesi J. Binding and fusion of extracellular vesicles to the plasma membrane of their cell targets. Int. J. Mol. Sci. 2016 doi: 10.3390/ijms17081296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Soares AR, et al. Gap junctional protein Cx43 is involved in the communication between extracellular vesicles and mammalian cells. Sci. Rep. 2015 doi: 10.1038/srep13243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.van Dongen HM, Masoumi N, Witwer KW, Pegtel DM. Extracellular vesicles exploit viral entry routes for cargo delivery. Microbiol. Mol. Biol. Rev. 2016 doi: 10.1128/mmbr.00063-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Grove J, Marsh M. The cell biology of receptor-mediated virus entry. J. Cell Biol. 2011 doi: 10.1083/jcb.201108131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Schnell JR, Chou JJ. Structure and mechanism of the M2 proton channel of influenza A virus. Nature. 2008 doi: 10.1038/nature06531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Montecalvo A, et al. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood. 2012 doi: 10.1182/blood-2011-02-338004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Tweten RK. Cholesterol-dependent cytolysins, a family of versatile pore-forming toxins. Infect. Immun. 2005 doi: 10.1128/IAI.73.10.6199-6209.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Jiang J, Pentelute BL, Collier RJ, Hong Zhou Z. Atomic structure of anthrax protective antigen pore elucidates toxin translocation. Nature. 2015 doi: 10.1038/nature14247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Geoffroy C, Gaillard JL, Alouf JE, Berche P. Purification, characterization, and toxicity of the sulfhydryl-activated hemolysin listeriolysin O from Listeria monocytogenes. Infect. Immun. 1987;55:1641–1646. doi: 10.1128/IAI.55.7.1641-1646.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Heath N, et al. Endosomal escape enhancing compounds facilitate functional delivery of extracellular vesicle cargo. Nanomedicine. 2019 doi: 10.2217/nnm-2019-0061. [DOI] [PubMed] [Google Scholar]
- 220.Ragelle H, et al. Chitosan nanoparticles for siRNA delivery: optimizing formulation to increase stability and efficiency. J. Control. Rel. 2014 doi: 10.1016/j.jconrel.2013.12.026. [DOI] [PubMed] [Google Scholar]
- 221.Antimisiaris SG, Mourtas S, Marazioti A. Exosomes and exosome-inspired vesicles for targeted drug delivery. Pharmaceutics. 2018 doi: 10.3390/pharmaceutics10040218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Quinn JF, et al. Extracellular RNAs: development as biomarkers of human disease. J. Extracell. Vesicles. 2015 doi: 10.3402/jev.v4.27495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Sadik N, et al. Extracellular RNAs: a new awareness of old perspectives. Methods Mol. Biol. 2018 doi: 10.1007/978-1-4939-7652-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Reátegui E, et al. Engineered nanointerfaces for microfluidic isolation and molecular profiling of tumor-specific extracellular vesicles. Nat. Commun. 2018 doi: 10.1038/s41467-017-02261-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Shao H, et al. New technologies for analysis of extracellular vesicles. Chem. Rev. 2018 doi: 10.1021/acs.chemrev.7b00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Akers JC, et al. A cerebrospinal fluid microRNA signature as biomarker for glioblastoma. Oncotarget. 2017 doi: 10.18632/oncotarget.18332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Burgos K, et al. Profiles of extracellular miRNA in cerebrospinal fluid and serum from patients with Alzheimer’s and Parkinson’s diseases correlate with disease status and features of pathology. PLoS One. 2014 doi: 10.1371/journal.pone.0094839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Hannafon BN, et al. Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res. 2016 doi: 10.1186/s13058-016-0753-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Antoury L, et al. Analysis of extracellular mRNA in human urine reveals splice variant biomarkers of muscular dystrophies. Nat. Commun. 2018 doi: 10.1038/s41467-018-06206-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.De Gonzalo-Calvo D, et al. Circulating long-non coding RNAs as biomarkers of left ventricular diastolic function and remodelling in patients with well-controlled type 2 diabetes. Sci. Rep. 2016 doi: 10.1038/srep37354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Giles FJ, Albitar M. Plasma-based testing as a new paradigm for clinical testing in hematologic diseases. Expert. Rev. Mol. Diagnostics. 2007 doi: 10.1586/14737159.7.5.615. [DOI] [PubMed] [Google Scholar]
- 232.McKiernan J, et al. A prospective adaptive utility trial to validate performance of a novel urine exosome gene expression assay to predict high-grade prostate cancer in patients with prostate-specific antigen 2–10 ng/ml at initial biopsy. Eur. Urol. 2018 doi: 10.1016/j.eururo.2018.08.019. [DOI] [PubMed] [Google Scholar]
- 233.Tao Y, et al. Exploration of serum exosomal LncRNA TBILA and AGAP2-AS1 as promising biomarkers for diagnosis of non-small cell lung cancer. Int. J. Biol. Sci. 2020 doi: 10.7150/ijbs.39123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Rohde E, Pachler K, Gimona M. Manufacturing and characterization of extracellular vesicles from umbilical cord–derived mesenchymal stromal cells for clinical testing. Cytotherapy. 2019 doi: 10.1016/j.jcyt.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 235.Witwer KW, et al. Defining mesenchymal stromal cell (MSC)-derived small extracellular vesicles for therapeutic applications. J. Extracell. Vesicles. 2019 doi: 10.1080/20013078.2019.1609206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Otero-Ortega, L. et al. Exosomes promote restoration after an experimental animal model of intracerebral hemorrhage. J. Cereb. Blood Flow. Metab. (2018). 10.1177/0271678X17708917 [DOI] [PMC free article] [PubMed] [Retracted]
- 237.Grange C, et al. Biodistribution of mesenchymal stem cell-derived extracellular vesicles in a model of acute kidney injury monitored by optical imaging. Int. J. Mol. Med. 2014 doi: 10.3892/ijmm.2014.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Yin K, Wang S, Zhao RC. Exosomes from mesenchymal stem/stromal cells: a new therapeutic paradigm. Biomarker Res. 2019 doi: 10.1186/s40364-019-0159-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Mendt M, et al. Generation and testing of clinical-grade exosomes for pancreatic cancer. JCI Insight. 2018 doi: 10.1172/jci.insight.99263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Zhang DX, Kiomourtzis T, Lam CK, Le MTN. The biology and therapeutic applications of red blood cell extracellular vesicles. Erythrocyte. 2019 doi: 10.5772/intechopen.81758. [DOI] [Google Scholar]
- 241.Zhu X, et al. Comprehensive toxicity and immunogenicity studies reveal minimal effects in mice following sustained dosing of extracellular vesicles derived from HEK293T cells. J. Extracell. Vesicles. 2017 doi: 10.1080/20013078.2017.1324730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Munagala R, Aqil F, Jeyabalan J, Gupta RC. Bovine milk-derived exosomes for drug delivery. Cancer Lett. 2016 doi: 10.1016/j.canlet.2015.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Wang Q, et al. Grapefruit-derived nanovectors use an activated leukocyte trafficking pathway to deliver therapeutic agents to inflammatory tumor sites. Cancer Res. 2015 doi: 10.1158/0008-5472.CAN-14-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Quesenberry PJ, et al. Potential functional applications of extracellular vesicles: a report by the NIH common fund extracellular RNA communication consortium. J. Extracell. Vesicles. 2015 doi: 10.3402/jev.v4.27575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Granchi D, et al. Biomarkers of bone healing induced by a regenerative approach based on expanded bone marrow–derived mesenchymal stromal cells. Cytotherapy. 2019 doi: 10.1016/j.jcyt.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 246.Bhaskaran V, et al. The functional synergism of microRNA clustering provides therapeutically relevant epigenetic interference in glioblastoma. Nat. Commun. 2019 doi: 10.1038/s41467-019-08390-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Saleh AF, et al. Extracellular vesicles induce minimal hepatotoxicity and immunogenicity. Nanoscale. 2019 doi: 10.1039/c8nr08720b. [DOI] [PubMed] [Google Scholar]
- 248.Heusermann W, et al. Exosomes surf on filopodia to enter cells at endocytic hot spots, traffic within endosomes, and are targeted to the ER. J. Cell Biol. 2016 doi: 10.1083/jcb.201506084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.György B, et al. Rescue of hearing by gene delivery to inner-ear hair cells using exosome-associated AAV. Mol. Ther. 2017 doi: 10.1016/j.ymthe.2016.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Kim SM, et al. Cancer-derived exosomes as a delivery platform of CRISPR/Cas9 confer cancer cell tropism-dependent targeting. J. Control. Rel. 2017 doi: 10.1016/j.jconrel.2017.09.013. [DOI] [PubMed] [Google Scholar]
- 251.Lin Y, et al. Exosome–liposome hybrid nanoparticles deliver CRISPR/Cas9 system in MSCs. Adv. Sci. 2018 doi: 10.1002/advs.201700611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Escudier B, et al. Vaccination of metastatic melanoma patients with autologous dendritic cell (DC) derived-exosomes: results of the first phase 1 clinical trial. J. Transl. Med. 2005 doi: 10.1186/1479-5876-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Van Craenenbroeck AH, et al. Induction of cytomegalovirus-specific T cell responses in healthy volunteers and allogeneic stem cell recipients using vaccination with messenger RNA-transfected dendritic cells. Transplantation. 2015 doi: 10.1097/TP.0000000000000272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Markov O, Oshchepkova A, Mironova N. Immunotherapy based on dendritic cell-targeted/-derived extracellular vesicles — a novel strategy for enhancement of the anti-tumor immune response. Front. Pharmacol. 2019 doi: 10.3389/fphar.2019.01152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Song P, et al. Lipidoid-siRNA nanoparticle-mediated IL-1β gene silencing for systemic arthritis therapy in a mouse model. Mol. Ther. 2019 doi: 10.1016/j.ymthe.2019.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Liu J, et al. Fast and efficient CRISPR/Cas9 genome editing in vivo enabled by bioreducible lipid and messenger RNA nanoparticles. Adv. Mater. 2019 doi: 10.1002/adma.201902575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Haraszti RA, et al. Serum deprivation of mesenchymal stem cells improves exosome activity and alters lipid and protein composition. iScience. 2019 doi: 10.1016/j.isci.2019.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Ghamloush F, et al. The PAX3-FOXO1 oncogene alters exosome miRNA content and leads to paracrine effects mediated by exosomal miR-486. Sci. Rep. 2019 doi: 10.1038/s41598-019-50592-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Lucero R, et al. Glioma-derived miRNA-containing extracellular vesicles induce angiogenesis by reprogramming brain endothelial cells. Cell Rep. 2020 doi: 10.1016/j.celrep.2020.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Liu Q, Peng F, Chen J. The role of exosomal microRNAs in the tumor microenvironment of breast cancer. Int. J. Mol. Sci. 2019 doi: 10.3390/ijms20163884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Shen M, et al. Chemotherapy-induced extracellular vesicle miRNAs promote breast cancer stemness by targeting OneCUT2. Cancer Res. 2019 doi: 10.1158/0008-5472.CAN-18-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Wang L, et al. Role of cardiac progenitor cell-derived exosome-mediated microRNA-210 in cardiovascular disease. J. Cell. Mol. Med. 2019 doi: 10.1111/jcmm.14562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Yu H, Wang Z. Cardiomyocyte-derived exosomes: biological functions and potential therapeutic implications. Front. Physiol. 2019 doi: 10.3389/fphys.2019.01049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Ma Y, et al. Exosomes released from neural progenitor cells and induced neural progenitor cells regulate neurogenesis through miR-21a. Cell Commun. Signal. 2019 doi: 10.1186/s12964-019-0418-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Ying W, et al. Adipose tissue macrophage-derived exosomal miRNAs can modulate in vivo and in vitro insulin sensitivity. Cell. 2017 doi: 10.1016/j.cell.2017.08.035. [DOI] [PubMed] [Google Scholar]
- 266.Chen S, et al. Exosomes derived from miR-375-overexpressing human adipose mesenchymal stem cells promote bone regeneration. Cell Prolif. 2019 doi: 10.1111/cpr.12669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Thomou T, et al. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature. 2017 doi: 10.1038/nature21365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Miyauchi K, Kim Y, Latinovic O, Morozov V, Melikyan GB. HIV enters cells via endocytosis and dynamin-dependent fusion with endosomes. Cell. 2009 doi: 10.1016/j.cell.2009.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Tian T, Wang Y, Wang H, Zhu Z, Xiao Z. Visualizing of the cellular uptake and intracellular trafficking of exosomes by live-cell microscopy. J. Cell. Biochem. 2010 doi: 10.1002/jcb.22733. [DOI] [PubMed] [Google Scholar]
- 270.Koga K, et al. Purification, characterization and biological significance of tumor-derived exosomes. Anticancer Res. 2005;25:3703–3707. [PubMed] [Google Scholar]
- 271.Mondal A, Ashiq KA, Phulpagar P, Singh DK, Shiras A. Effective visualization and easy tracking of extracellular vesicles in glioma cells. Biol. Proced. Online. 2019 doi: 10.1186/s12575-019-0092-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Planchon D, et al. MT1-MMP targeting to endolysosomes is mediated by upregulation of flotillins. J. Cell Sci. 2018 doi: 10.1242/jcs.218925. [DOI] [PubMed] [Google Scholar]
- 273.Escrevente C, Keller S, Altevogt P, Costa J. Interaction and uptake of exosomes by ovarian cancer cells. BMC Cancer. 2011 doi: 10.1186/1471-2407-11-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Gray WD, Mitchell AJ, Searles CD. An accurate, precise method for general labeling of extracellular vesicles. MethodsX. 2015 doi: 10.1016/j.mex.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Morales-Kastresana A, et al. High-fidelity detection and sorting of nanoscale vesicles in viral disease and cancer. J. Extracell. Vesicles. 2019 doi: 10.1080/20013078.2019.1597603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Wittrup A, et al. Visualizing lipid-formulated siRNA release from endosomes and target gene knockdown. Nat. Biotechnol. 2015 doi: 10.1038/nbt.3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Da Poian AT, André AM, Coelho-Sampaio T. Kinetics of intracellular viral disassembly and processing probed by Bodipy fluorescence dequenching. J. Virol. Methods. 1998 doi: 10.1016/S0166-0934(97)00166-3. [DOI] [PubMed] [Google Scholar]
- 278.Struck DK, Hoekstra D, Pagano RE. Use of resonance energy transfer to monitor membrane fusion. Biochemistry. 1981 doi: 10.1021/bi00517a023. [DOI] [PubMed] [Google Scholar]
- 279.Herrmann A, et al. Effect of erythrocyte transbilayer phospholipid distribution on fusion with vesicular stomatitis virus. Biochemistry. 1990 doi: 10.1021/bi00469a005. [DOI] [PubMed] [Google Scholar]
- 280.Wahlberg JM, Bron R, Wilschut J, Garoff H. Membrane fusion of Semliki Forest virus involves homotrimers of the fusion protein. J. Virol. 1992;66:7309–7318. doi: 10.1128/JVI.66.12.7309-7318.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Sung BH, Pelletier R, Weaver AM. pHluo_M153R-CD63, a bright, versatile live cell reporter of exosome secretion and uptake, reveals pathfinding behavior of migrating cells. bioRxiv. 2019 doi: 10.1101/577346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Yuhua H, et al. Cytosolic delivery of membrane-impermeable molecules in dendritic cells using pH-responsive core-shell nanoparticles. Nano Lett. 2007 doi: 10.1021/nl071542i. [DOI] [PubMed] [Google Scholar]
- 283.Jones DM, Padilla-Parra S. The β-lactamase assay: harnessing a FRET biosensor to analyse viral fusion mechanisms. Sensors. 2016 doi: 10.3390/s16070950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Lönn P, et al. Enhancing endosomal escape for intracellular delivery of macromolecular biologic therapeutics. Sci. Rep. 2016 doi: 10.1038/srep32301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Godoy PM, et al. Large differences in small RNA composition between human biofluids. SSRN Electron. J. 2018 doi: 10.2139/ssrn.3155656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Freedman JE, et al. Diverse human extracellular RNAs are widely detected in human plasma. Nat. Commun. 2016 doi: 10.1038/ncomms11106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Giraldez MD, et al. Comprehensive multi-center assessment of small RNA-seq methods for quantitative miRNA profiling. Nat. Biotechnol. 2018 doi: 10.1038/nbt.4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Godoy PM, et al. Comparison of reproducibility, accuracy, sensitivity, and specificity of miRNA quantification platforms. Cell Rep. 2019 doi: 10.1016/j.celrep.2019.11.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Yeri A, et al. Evaluation of commercially available small RNASeq library preparation kits using low input RNA. BMC Genomics. 2018 doi: 10.1186/s12864-018-4726-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Crescitelli R, et al. Distinct RNA profiles in subpopulations of extracellular vesicles: apoptotic bodies, microvesicles and exosomes. J. Extracell. Vesicles. 2013 doi: 10.3402/jev.v2i0.20677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Gámbaro F, et al. Stable tRNA halves can be sorted into extracellular vesicles and delivered to recipient cells in a concentration-dependent manner. RNA Biol. 2019 doi: 10.1080/15476286.2019.1708548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Chevillet JR, et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc. Natl Acad. Sci. USA. 2014 doi: 10.1073/pnas.1408301111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Grabarek AD, Weinbuch D, Jiskoot W, Hawe A. Critical evaluation of microfluidic resistive pulse sensing for quantification and sizing of nanometer- and micrometer-sized particles in biopharmaceutical products. J. Pharm. Sci. 2019 doi: 10.1016/j.xphs.2018.08.020. [DOI] [PubMed] [Google Scholar]
- 294.Akers JC, et al. Comparative analysis of technologies for quantifying extracellular vesicles (EVs) in clinical cerebrospinal fluids (CSF) PLoS One. 2016 doi: 10.1371/journal.pone.0149866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.van der Pol E, et al. Particle size distribution of exosomes and microvesicles determined by transmission electron microscopy, flow cytometry, nanoparticle tracking analysis, and resistive pulse sensing. J. Thromb. Haemost. 2014 doi: 10.1111/jth.12602. [DOI] [PubMed] [Google Scholar]
- 296.Oleksiuk O, et al. Single-molecule localization microscopy allows for the analysis of cancer metastasis-specific miRNA distribution on the nanoscale. Oncotarget. 2015 doi: 10.18632/oncotarget.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.