Abstract
Brain protein biomarker clearance to blood in traumatic brain injury (TBI) is not fully understood. The aim of this study was to analyze the effect that a disrupted blood–brain barrier (BBB) had on biomarker clearance. Seventeen severe TBI patients admitted to Karolinska University Hospital were prospectively included. Cerebrospinal fluid (CSF) and blood concentrations of S100 calcium binding protein B (S100B) and neuron-specific enolase (NSE) were analyzed every 6–12 h for ∼1 week. Blood and CSF albumin were analyzed every 12–24 h, and BBB integrity was assessed using the CSF:blood albumin quotient (QA). We found that time-dependent changes in the CSF and blood levels of the two biomarkers were similar, but that the correlation between the biomarkers and QA was lower for NSE (ρ = 0.444) than for S100B (ρ = 0.668). Because data were longitudinal, we also conducted cross correlation analyses, which indicated a directional flow and lag-time of biomarkers from CSF to blood. For S100B, this lag-time could be ascribed to BBB integrity, whereas for NSE it could not. Upon inferential modelling, using generalized least square estimation (S100B) or linear mixed models (NSE), QA (p = 0.045), time from trauma (p < 0.001), time from trauma2 (p = 0.023), and CSF biomarker levels (p = 0.008) were independent predictors of S100B in blood. In contrast, for NSE, only time from trauma was significant (p < 0.001). These findings are novel and important, but must be carefully interpreted because of different characteristics between the two proteins. Nonetheless, we present the first data that indicate that S100B and NSE are cleared differently from the central nervous system, and that both the disrupted BBB and additional alternative pathways, such as the recently described glymphatic system, may play a role. This is of importance both for clinicians aiming to utilize these biomarkers and for the pathophysiological understanding of brain protein clearance, but warrants further examination.
Keywords: albumin quotient, BBB, NSE, S100B, TBI
Introduction
Traumatic brain injury (TBI) is one of the most common causes of death and disability,1 afflicting ∼10,000,000 people annually.2 Unconscious TBI patients deemed to be in need of intracranial monitoring are treated in neurocritical care units (NCCUs).3 Here, different modalities are monitored in order to prevent secondary insults that may lead to irreversible deterioration in the already damaged brain.4–6 Among these modalities, brain-enriched proteins of tissue fate (i.e., “biomarkers”) are increased in body fluids following brain injury and have become increasingly used in the management of TBI.7 The two most studied protein biomarkers include the primarily astrocytic S100 calcium binding protein B (S100B) and the neuronal neuron-specific enolase (NSE). Even though neither is brain specific, both are highly brain enriched and have been associated with a worse intracranial condition and long-term functional outcome following TBI.8–10 In fact, some centers event utilize one or both in clinical routine work,11,12 hence making these two “biomarkers” of particular interest to the neurotrauma translational research field.
The clearance mechanism of protein biomarkers from brain to blood is not fully understood, but is of importance for clinical interpretation and pathophysiological understanding. The adult central nervous system (CNS) comprises three anatomical barriers, namely: (1) the blood–cerebrospinal fluid (CSF) barrier, (2) the blood–brain barrier (BBB), and (3) the arachnoid barrier.13 For simplicity, and to align with the clinical research field, we refer to all of these barriers as “BBB.” In addition, a transport route denoted “the glymphatic system” has recently been described in experimental models.14 Although it has been suggested in pre-clinical models that brain-enriched proteins in CSF passively leave the brain through this perivascular “glymphatic” system,15 others claim that the BBB integrity plays a more decisive role.16–18 In TBI studies, in which the patient likely has concomitant injuries to multiple CNS barriers, the gold-standard strategy for assessing BBB disruption (BBBD) is the CSF to blood albumin quotient (QA).19–25 Previous studies of TBI and subarachnoid hemorrhage assessing how serum levels of S100B are associated with QA have not shown significant correlations.23,26 However, these studies have been limited by a lack of longitudinal analyses and high frequency sampling during constant CSF drainage. The trajectory of secondary pathophysiological mechanisms following TBI likely require a high temporal resolution,3 multi-compartment monitoring, and modeling to elucidate biomarker and QA dynamics. In aggregate, how brain-enriched proteins leave the injured brain warrants further research, as it may increase understanding of brain injury pathophysiology in several conditions and improve the utility of biomarkers in clinical decision making.
Aim
We aimed to assess if, and how, clearance of brain-enriched proteins (S100B and NSE) from brain to blood is affected by BBBD, measured as QA, over time utilizing a high-sampling frequency from multiple compartments in severe TBI patients.
Methods
The included patients were part of a prospective observational study27 undertaken at the NCCU at Karolinska University Hospital (Stockholm, Sweden) between January 1, 2010 and March 1, 2013. Written, informed consent was acquired from next-of-kin. The study was conducted in accordance with the Declaration of Helsinki and Swedish law. Ethical approval was provided by the Stockholm County branch of the Central Ethical Review Board (#2009/1112-31/3), now called the Swedish Ethical Review Authority.
Sample size calculation
Sample size estimation a priori using power analysis was unfeasible because to our knowledge, no assumptions on effect size have been described in the literature using the same approach as in our study. However, using data from our group,26 we could estimate our sample sizes for a cross-sectional rather than a longitudinal data set. Aiming for a correlation coefficient r = 0.60 between S100BCSF and S100Bblood, we found the estimated sample size needed to reach 80% power on the 0.05 significance level (one-sided test) using the R28 package pwr29 to be 15 patients. The effect size estimate was larger than the one previously reported from our group (r = 0.45),26 but smaller than what has more recently been reported30 (r = 0.79), leading us to believe that this was a valid effect size assumption. We therefore set out to include >15 study subjects.
Patient inclusion
Because of the sporadic availability of research personnel, recruitment was made periodically. Patients were not randomized, because no group-specific interventions were conducted. Inclusion criteria for the patients were as follows: (1) age 18–75 years, (2) Glasgow Coma Scale (GCS) score 3–8 (unconscious at admission), and (3) computerized tomography (CT) verified structural intraparenchymal intracranial injury. Exclusion criteria comprised: (1) GCS 3 and bilaterally non-responsive pupils, (2) slit ventricles (because that would preclude the planned patient study management by making external ventricular drain (EVD) insertion impossible), (3) unsurvivable injury, (4) unconsciousness of etiology other than TBI, (5) absence of CT-verified intraparenchymal intracranial injury, (6) other concurrent systemic terminal disorder, and (7) impossibility of follow-up (e.g., being foreign citizens).
Patient management and sample acquisition
The detailed study setup has been described previously.27 Briefly, upon admission, patients received an EVD (Medtronic, Eatontown, NJ). The drain was connected to a four-way stopcock (Multiflo 3, BD, Connecta, Franklin Lakes, NJ), connected to a LiquoGuard® CSF-pump (Möller Medical GmbH, Fulda, Germany) set to a constant drainage velocity of 2 mL CSF per hour. CSF was collected in, and samples were obtained from, the drainage bag in the LiquoGuard system in order to minimize the risk for infections, and freeze blocks were used to keep the CSF cold in order to limit protein degradation. The collected pool of CSF and readily acquired arterial blood were analyzed at 6-and 12-h intervals, for both S100B and NSE. Albumin (plasma and CSF) was analyzed once to twice daily. S100Bblood was analyzed by electrochemiluminescence immunoassay (Elecsys, Roche Diagnostics, Basel, Switzerland), albuminplasma was analyzed by colorometric bromocresol purple (BCP)-binding assay and albuminCSF was analyzed by immunoturbitity on a Roche Cobas/Modular platform (Roche Diagnostics, Basel, Switzerland). NSEblood, NSECSF, and S100BCSF were analyzed by immunoluminometric assay on a LIAISON XL system (DiaSorin, Saluggia, Italy). S100Bblood and S100BCSF were analyzed on different platforms because of local procurements and inability to run CSF samples on the more modern Cobas/Elecsys platforms. Each individual assay has shown robust within- and between-run similarity,31 thus enabling longitudinal sampling as employed in this study. Some claim that the different platforms are not entirely interchangeable,32 whereas others have found excellent associations between the methods (r = 0.932).31 It is noteworthy that, to our knowledge, no studies have examined bicompartmental similarities (i.e., between CSF and blood). This is of importance, because absolute between-assay agreement is of greater importance when identical samples from the same compartment are examined on multiple platforms. However, for the scope of this study, the relative relationship was of greater interest. All laboratory assays were performed at the Karolinska University Laboratory in accordance with local guidelines. Aside from this, patients were treated in accordance with local routine at the NCCU, as previously described.8
Clinical parameters
Baseline clinical data were defined and acquired as follows. Clinical variables comprised GCS,33 (head and non-head) Abbreviated Injury Scale34 (AIS), Injury Severity Score35 (ISS), and significant multitrauma,36 all of which were acquired upon hospital admission. It is of note that an AIS of 6 at admission is impossible, as it denotes an unsurvivable injury. CT variables upon admission were graded using the Marshall CT Classification,37,38 in which classifications V and VI were collapsed into one category (“mass lesion”), as we used only the admission CT. For comparison, we also assessed the Rotterdam39 and Stockholm CT scores.40 Radiological brain injury progression upon subsequent examination was evaluated employing a similar strategy as in previous work.8,41 Glasgow Outcome Scale42 (GOS) was assessed as described previously.27 In short, a neurorehabilitation board-certified physician (P.H.G.) examined patients 6 months following the TBI. GOS comprises 5 categories: (1) dead, (2) persistent vegetative state, (3) severe disability, (4) moderate disability, and (5) low disability.
Definitions
We used “BBBD” as a comprehensive term to denote all types of barrier disruption that can be discerned following TBI, including disruption of the anatomical BBB as well as of the blood–CSF barrier.43 BBBD was defined as the quotient between CSF and blood albumin (Equation 1), because that constitutes the literature gold standard.19,24,43 The reference intervals used for all proteins in the current study were defined using reference intervals stipulated by the Karolinska University Laboratory during the time of patient inclusion or at present (Table 1).44-47
Table 1.
Reference Intervals Used
| Variable | Reference interval |
|---|---|
| AlbuminCSF (mg/L) | 15-29 years: <260 |
| ≥ 50 years: <400 | |
| Albuminblood (g/L) | < 41 years: 36-48 |
| ≥ 71 years: 34-45 | |
| QA (no unit) | 15-29 years: <0.006 |
| ≥ 50 years: <0.009 | |
| S100BCSF (μg/L) | < 5 |
| S100Bblood (μg/L) | < 0.11 |
| NSECSF (μg/L) | < 13 |
| NSEblood (μg/L) | < 18 |
Reference intervals were defined as the reference intervals used by the Karolinska University Laboratory during the time of patient inclusion in the study or at present.44–47
Statistical analysis
Statistical analyses were conducted using R,28 through the interface RStudio®. Continuous data were presented as mean ± standard deviation (SD) if normally distributed, or else median (interquartile range [IQR]). Categorical data were presented as count (%). In graphical depictions and calculations, biomarker values were converted to log10-transformed values, unless otherwise stated. A p value ≤0.05 or a 95% confidence interval (CI), where the range of the CI did not contain the value 0, was considered significant.
Several variables were sampled at different time intervals, resulting in time points without sample overlap. This is demonstrated graphically48 (Fig. S1). Following a comparison between linear and locally weighted scatterplot smoother interpolation (Fig. S2), this was compensated for by linear interpolation. Data rows that still retained “missing” values for a certain time point because of consecutive non-overlapping sampling time points were handled by complete case analysis.
Correlations (regular correlations are here referred to as “momentary” correlations) between biomarkers in different compartments and QA were assessed using repeated measures correlation49 on log10-transformed variables in order to obtain linear relationships. Each model was examined with regard to relevant assumptions. The R packages rmcorr50 and tidyverse51 were used for calculations and graphical depictions. Because biomarker levels in different body compartments might correlate in a time-delayed fashion, we also cross-correlated the original data. We abstained from de-trending or differencing the data because the relationship we examined was the underlying shared time trend. For all, the biomarker sampling time points were subdivided into defined time intervals (“lags”) of 0.5 days (12 h). Multiple measurements occurring within one lag were averaged. Last, cross-correlation was conducted per patient, whereupon results were pooled for all patients and lags. For all cross correlations, original data (i.e., not log10-transformed) were used.
For inferential analysis, we compared a general linear model with correlated errors (general least square estimation, a so-called “marginal model”), and a linear mixed model. For both, the nlme package in R was used.52 Independent of analysis type, we modelled a within-patient variance-covariance matrix, because we used repeated-measures data. The variance-covariance matrix was constructed as a time series model, for which we determined stationarity, autocorrelation, and partial autocorrelation. We deemed that the data were stationary using the Kwiatkowski–Phillips–Schmidt–Shin test, and therefore abstained from autoregressive integrated moving average modeling.53 The variance-covariance matrix hence consisted of an autoregressive-moving average (ARMA) model. We tested 25 different ARMA combinations of varying complexity, and then chose the optimal ARMA structure based on Akaike Information Criterion. For both S100B and NSE we used a first order autoregressive model with a moving average of 1; that is, ARMA(p = 1, q = 1). The resulting models were validated graphically.54 For general linear as well as linear mixed-model selection we employed both a step-up55 and a top-down strategy.56 Random effect structures were evaluated using restricted maximum likelihood-based estimations and fixed effects structure using maximum-likelihood estimations. Model selection was conducted by likelihood ratio tests between nested models and type I F tests. P values of the final models were generated using Satterthwaite approximations in the R package lmerTest.56,57 In all analyses, the dependent variable was the biomarkerblood value (log10 transformed). For S100B, the first 12 h were excluded from analysis, because these time points have been associated with an “extracranial peak.”8,9
In general, Akaike Information Criterion was lower for the mixed-model design in the top-down strategy, whereas they were equal in the step-up strategy. For S100B, the variance-covariance matrix generated in the linear mixed model was not positive-definite, which is why we chose to use the equivalent marginal model for S100B.56 For NSE, we had no such issues, which is why we present results from the linear mixed model. It is noteworthy that results were highly similar independent of linear mixed or marginal model design for each biomarker. Model assumptions were examined graphically with regard to homogeneity of variances, normality of variances, correlation between fitted and observed values, and individual assessment of residual autocorrelation and partial autocorrelation. For NSE, one patient (pat #17) followed a markedly different trajectory than the other patients and also constituted an outlier using the model diagnostics previously described. For validation, the results for NSE were run both including and excluding pat #17, and the results were to a large extent similar, leading us to conclude that the outlier pat #17 did not imply any major alterations with regard to overall conclusions.
Results
Demographics
In total, 17 patients were recruited, of which one patient was excluded because no samples of CSF albumin had been obtained. Among included patients, there was no loss-to-follow up. Demographics are depicted both individually (Table 2) and for the whole cohort (Table S1). Patients were predominantly middle-aged males, with a median GCS of 7 (4–7). Approximately 40% of the patients had sustained a significant multi-trauma. However, the majority of patients had a head AIS score of 5, suggesting that a severe cranial trauma constituted the predominant pathology. Of all patients, 31.3% had progression of CT-verified lesions during the study period. On long-term follow-up, half of the patients had an unfavorable outcome (GOS 1–3).
Table 2.
Individual Patient Demographics
| Patient No. | Age | Gender | GCS | Pupils | |
|
|
ISS | Marshall | Rotterdam | Stockholm | Brain injury progression | GOS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Multi-trauma | HeadAIS | Non-headAIS | |||||||||||
| 1 | 55 | M | 8 | 1 | 0 | 5 | 1 | 26 | VI | 5 | 3.7 | 0 | 4 |
| 2 | 53 | F | 5 | 2 | 0 | 5 | 1 | 25 | VI | 6 | 3.6 | 1 | 3 |
| 4 | 22 | M | 7 | 1 | 1 | 4 | 3 | 29 | VI | 6 | 3.3 | 0 | 3 |
| 5 | 23 | M | 8 | 0 | 0 | 4 | 0 | 16 | III | 4 | 1.5 | 0 | 5 |
| 6 | 20 | M | 8 | 2 | 1 | 3 | 2 | 17 | II | 3 | 1.8 | 1 | 5 |
| 7 | 38 | M | 7 | 0 | 1 | 5 | 3 | 38 | II | 3 | 3.5 | 0 | 3 |
| 8 | 25 | M | 8 | 0 | 0 | 5 | 0 | 25 | IV | 5 | 2 | 0 | 5 |
| 9 | 42 | M | 3 | 1 | 1 | 5 | 3 | 38 | II | 3 | 1.5 | 1 | 3 |
| 10 | 52 | F | 3 | 2 | 1 | 4 | 4 | 29 | III | 4 | 3.5 | 0 | 3 |
| 11 | 59 | M | 7 | 1 | 0 | 5 | 0 | 25 | VI | 4 | 4.9 | 0 | 3 |
| 12 | 62 | M | 3 | 0 | 0 | 5 | 0 | 25 | II | 3 | 2 | 1 | 1 |
| 13 | 49 | M | 3 | 0 | 0 | 4 | 0 | 16 | II | 3 | 1.5 | 0 | 5 |
| 14 | 20 | M | 7 | 0 | 0 | 5 | 0 | 25 | VI | 3 | 2.5 | 0 | 4 |
| 15 | 36 | M | 7 | 0 | 1 | 4 | 3 | 26 | III | 4 | 2.8 | 0 | 4 |
| 16 | 60 | F | 4 | 0 | 0 | 5 | 1 | 25 | VI | 4 | 2.5 | 0 | 4 |
| 17 | 48 | M | 4 | 2 | 0 | 5 | 0 | 25 | VI | 5 | 3.8 | 1 | 1 |
Demographic data of the patient cohort, depicted as each subject's raw data value. One patient (no. 3) was excluded from all analyses because of a lack of albuminCSF samples.
Clinical data upon admission: age (years); gender: M (male), F (female); GCS (Glasgow Coma Scale Score) 3-15; pupils (pupil responsiveness), 0 = bilateral responsive; 1 = unilateral unresponsive; 2 = bilateral unresponsive; multi-trauma, 1 = yes, 0 = no.
Injury scores upon admission: Head and non-head Abbreviated Injury Scale (AIS), (1) minor, (2) moderate, (3) serious, (4) severe, (5) critical, (6) maximum. Injury Severity Score (ISS) (1–75).
Classification upon admission computed tomography (CT): Marshall, (I) no visible pathology,(II) diffuse injury, (III) “swelling,” (IV) shift, V–VI (“mass lesion”). Rotterdam, classes 1–6. Stockholm, tally based. Brain injury progression: 1 = yes, 0 = no.
Outcome data at 6 months follow-up: Glasgow Outcome Scale Score (GOS): (GOS1) dead, (GOS2) persistent vegetative state, (GOS3) severe disability, (GOS4) moderate disability, (GOS5) good recovery.
All patients exhibited profoundly increased values of S100B in CSF and blood following trauma (Fig. 1A, C), all higher than the 0.11 μg/L cutoff used to screen if mild TBI patients have an intracranial lesion. A majority of patients also exhibited NSE CSF and blood concentrations above the upper reference intervals of 13 μg/L (CSF) and 18 μg/L (blood) (Fig. 1B, D). Roughly half of the patients demonstrated a disrupted BBB as defined by QA > 0.006–0.009 (Fig. 1F) upon admission, closely mimicked by CSF levels of albumin >260–400 mg/L (Fig. 1E), but not by blood albumin (Fig. S3). Even though the QA slope showed a groupwise decreasing trend over time, the study period of ∼1 week was too short to observe normalization of QA for several patients.
FIG. 1.
Temporal trajectory of biomarkers and QA in different compartments. S100BCSF was increased above the reference level (A) for all patients following injury and demonstrated a temporal decay. This was reflected in blood (C). For NSECSF (B), one patient demonstrated normal NSECSF values following traumatic brain injury (TBI), and even more exhibited normal NSEblood values (D). AlbuminCSF was increased among roughly half of the patients (E) following the trauma, which was reflected in a pathological QA (F). Over time, there was a decay in the extent of albuminCSF and a decrease in QA, but interestingly, the extent of damage persisted throughout the whole study period for many patients. Dashed line: upper reference interval as used at the Karolinska University Hospital. Upper reference limit: S100BCSF < 5 μg/L, S100Bblood < 0.11 μg/L, NSECSF < 13 μg/L, NSEblood < 18 μg/L. For albuminCSF and QA there is an age-dependent reference limit. Upper reference limits for albuminCSF: 15–29 years, <260 mg/L; ≥ 50 years, <400 mg/L. Upper reference limits for QA: 15–29 years, <0.006; ≥ 50 years: <0.009. Abbreviations: CSF, cerebrospinal fluid; NSE, neuron-specific enolase; QA, albumin quotient. Color image is available online.
Correlations between biomarkers and QA
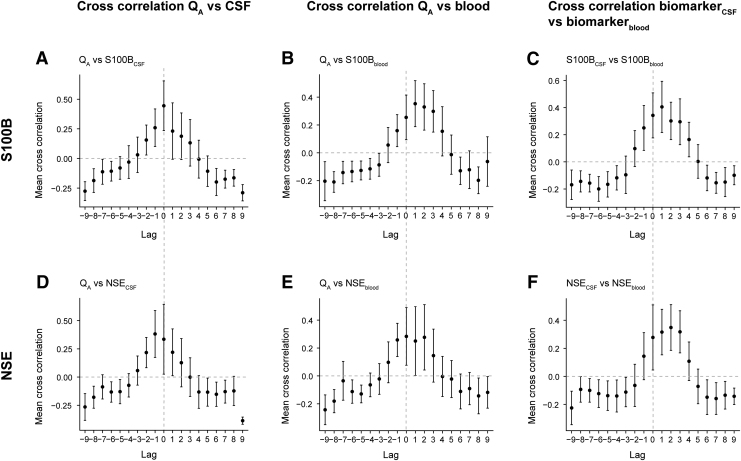
In order to elucidate the relationship between biomarkers in different compartments and QA, we conducted both regular (here denoted “momentary”) correlation and cross-correlation analyses. Cross-correlation analyses were conducted because clearance from CNS to peripheral blood might occur with a time delay. For cross-correlations, we examined three temporal relationships: (1) correlation between biomarkerCSF values and BBBD (Fig. 2A, D); (2) correlation between biomarkerblood values and BBBD (Fig. 2B, E); and (3) cross-correlation between biomarkerCSF and biomarkerblood values (Fig. 2C, F).
FIG. 2.
Cross-correlations of biomarkers between compartments and QA. Cross- correlations between BBBD/biomarkerCSF, BBBD/biomarkerblood, and biomarkerCSF/biomarkerblood are seen for S100 calcium binding protein B (S100B) (A–C) and NSE (D–F). S100BCSF and S100Bblood had peak correlation at lag 1 (C), indicating that S100B is detected in blood ∼12 h later than in CSF. This could be attributed to the delayed clearance through the disrupted BBB (B). For NSECSF and NSEblood, peak correlation occurred at lag 2, equivalent to 24 h (F), but this could not be attributed to either intracranial NSE release (D) or any delayed clearance across the BBB of NSE (E). Overall, both S100B and NSE are detected later in blood than they are in CSF, and NSE is the slower of the two proteins. Each lag corresponds to 0.5 days. Dots correspond to mean cross-correlation across a specific lag. Error bars consist of confidence interval (CI), where CI <0 or CI >0 reflect a significant cross-correlation throughout that lag. Dashed lines harmonize lag 0 across all panels. BBB, blood–brain barrier; BBBD, blood–brain barrier disruption; CSF, cerebrospinal fluid; NSE, neuron-specific enolase; QA, albumin quotient.
There was a strong positive (momentary) correlation between S100BCSF and S100Bblood (ρ = 0.667, CI 0.602–0.723) as well as QA and S100Bblood (ρ = 0.668, CI 0.598–0.728). Cross-correlations for S100B showed that there was an immediate correlation between BBBD and S100BCSF (Fig. 2A), whereas there was a lagged relationship between S100BCSF and S100Bblood (Fig. 2C). The lag was attributed to the identically lagged relationship between BBBD and S100Bblood (Fig. 2B). Hence, S100B exhibited a delayed release to blood, which occurred concomitantly to a delayed leakage across the BBB.
There was also a positive (momentary) correlation between NSECSF and NSEblood (ρ = 0.436, CI 0.343–0.521) as well as QA and NSEblood (ρ = 0.444, CI 0.345–0.533). NSE cross-correlations demonstrated a negative lag between BBBD and NSECSF (Fig. 2D) indicating that NSECSF levels increased more swiftly than the BBBD occurred following an injury. NSECSF and NSEblood (Fig. 2F) were positively lagged against one another, meaning that there is a period of delay before NSE can be detected in blood. This phenomenon could not be attributed to the relationship between BBBD and NSEblood (Fig. 2E), because these exhibited a peak correlation at lag 0; that is, immediate to one another. Hence, there was a discrepancy between NSECSF and NSEblood lags (+2), not accounted for by the other cross correlations (-1 and 0 lags respectively). Further, this means that the delayed NSE clearance did not seem to be a consequence of a delayed intracranial NSE de novo release; that is, higher CSF levels (Fig. 2D), or because of a slower clearance across the disrupted BBB (Fig. 2E, lag 0–2).
In summary, a measured blood biomarker concentration is presumably a reflection of both momentary and delayed brain clearance, as well as the extent of the different disintegrated CNS barrier components. For momentary correlations, there was a stronger correlation coefficient for S100B than for NSE. Generally, there were only small differences between the two biomarkers over the different cross-correlations. However, some noteworthy disparities were found. NSE exhibited fewer lags with significant cross- correlations (Fig. 2D–F) than S100B (Fig. 2A–C). Hence, NSE overall demonstrated a less robust pattern than did S100B, which is why the NSE results should be interpreted cautiously. Moreover, in contrast to S100Bblood, the disintegrated barrier cannot solely explain the temporal pattern of NSE clearance. This is manifested throughout lag 0–2 (equivalent to 0–24 h) (Fig. 2E), where there are, however, small differences in mean cross-correlation and the CIs are very broad, which is why these findings should be interpreted with caution.
Modelling of biomarker clearance from CSF to blood
In order to inferentially determine the importance of QA for blood biomarker levels, we conducted a generalized least square linear model (marginal model) for S100B (Fig. 3A, Table 3) and a linear mixed effects model for NSE (Fig. 3B, Table 3).
FIG. 3.
Graphical depictions of clearance models for S100 calcium binding protein B (S100B) and NSE. A marginal model (general least square estimation) was conducted for S100B (A) and a linear mixed model was conducted for NSE (B). Time from trauma, S100BCSF, and QA predicted S100Bblood in an additive model (A). In contrast, only time from trauma predicted NSEblood (B). For both panels, the size of the circles denotes the biomarkerCSF value, whereas the color gradient represents the value of QA. The black lines are the fitted values in each model. CSF, cerebrospinal fluid; NSE, neuron-specific enolase; QA, albumin quotient. Color image is available online.
Table 3.
Inferential Models of S100B and NSE Clearance from CSF to Blood
| Marginal model of S100B clearance |
Linear mixed model of NSE clearance |
||||
|---|---|---|---|---|---|
| Variable | Estimate | p value | Variable | Estimate | p value |
| Intercept | -1.13 | <0.001 | Intercept | 1.36 | <0.001 |
| Time from trauma | -3.82 | <0.001 | Time from trauma | -0.0511 | <0.001 |
| Time from trauma2 | 0.665 | 0.023 | |||
| S100BCSF | 0.0247 | 0.008 | |||
| QA | -0.0676 | 0.045 | |||
Generalized linear model (marginal model) analysis showing how time from trauma, QA, and S100BCSF were independent predictors of S100Bblood. In all analyses, the dependent variable was S100Bblood. The variables S100Bblood, QA, and S100BCSF were log10-transformed. Time from trauma was used as a polynomial term with a degree of 2. The underlying correlation structure was modelled as an ARMA(1,1) process, for which the error term depends on the AR component ϕ and the MA component θ. These were estimated to be: ϕ = 0.976 (CI: 0.955–0.987) and θ = 0.563 (CI: 0.456–0.654).
Linear mixed model analysis showing how time from trauma was an independent predictor of NSEblood. In all analyses, the dependent variable was NSEblood. The variable NSEblood was log10-transformed. Random effects of the model were a random intercept (patient) and a random slope (time from trauma). The underlying correlation structure was modelled as an ARMA(1,1) process, for which the error term depends on the AR component ϕ and the MA component θ. These were estimated to be: ϕ = 0.637 (CI: 0.454–0.768) and θ = 0.621 (CI: 0.522–0.704).
AR, autoregressive; ARMA, autoregressive moving average; CI, confidence interval; CSF, cerebrospinal fluid; MA, moving average; NSE, neuron-specific enolase; QA, albumin quotient; S100B, S100 calcium binding protein B.
All models used time from trauma and CSF biomarker levels as covariates together with a pre-determined correlation structure, thereby accounting for the longitudinal study design. Interestingly, there were notable differences between the biomarkers. For S100B (Table 3, Fig. 3A), S100Bblood values could be modeled as a function of time from trauma, S100BCSF, and QA, all of which were significant. As can be seen in Figure 1C, the relationship followed a curvilinear slope, making the quadratic time from trauma term significant as well. The finding that QA is a predictor of S100Bblood suggests that the extent of BBBD is related to the levels of S100B in blood.
In contrast, for NSE (Table 3, Fig. 3B) only time from trauma emerged as a significant independent predictor, not NSECSF or QA. These findings were robust even when checked for more complex statistical structures, such as interaction effects. In aggregate, these findings, and the contrast between the biomarkers, indicate that BBBD might affect biomarker clearance from CSF to blood differently for different proteins.
Discussion
We present a high temporal resolution, multi-compartment, prospective biomarker monitoring study conducted after severe TBI. We examined how BBBD, measured as QA, affects brain clearance of the two most studied protein biomarkers in TBI. We found clear correlations between the blood biomarker values and QA, but coefficients were notably larger for S100B. We also found a cross-correlation between the CSF biomarker and the blood biomarker levels for both S100B and NSE, indicative of a slightly delayed/”lagged” clearance from CSF to blood that for S100B, but not NSE, covaried with a delayed clearance across the BBB. Finally, regression modeling demonstrated that for S100B, time from trauma, S100BCSF, and QA were important contributors to S100Bblood. In contrast, only time from trauma emerged as a significant predictor of NSEblood. We suggest that one reason for the discrepancies between the two biomarkers is that they are cleared differently, with S100B being cleared through a disrupted BBB unlike NSE, but this warrants further examination.
The disrupted BBB in TBI
We assessed how a trauma-induced BBBD affects brain biomarker concentrations in blood. Even though some radiological techniques have been proposed to quantify BBBD,58,59 QA remains the gold standard assessment,19–25 because the 67 kDa protein albumin lacks intracranial synthesis,19 is not catabolized in the CNS,60 and has a 200 times lower concentration in CSF compared with serum.19 We found that whereas QA on a group basis had a decreasing trend over the 1st week, patients who had normal QA values at study onset maintained normal values throughout the study period and vice versa. Kleindienst and coworkers also found persistently deranged QA values in a cohort of TBI and subarachnoid hemorrhage (SAH) patients.23 In contrast, Bellander and coworkers found a steep normalization of QA within the 1st week after TBI,26 congruent with data from Blyth and colleagues.20,21 This indicates that the BBB is likely more affected by the primary injury than secondary insults, and that BBBD may persist for at least 7 days, or as some have suggested, even longer.61 Consistently, some patients did not exhibit any signs of BBBD in our study. The underlying reason for this cannot be causally determined by this work because of the small sample size, but we hypothesize that differences in the intracranial injury panorama is a probable explanation. This finding further highlights the necessity of individualizing treatment following TBI. Finally, we noticed that QA and albuminCSF mimicked each other closely. Therefore, albumin levels in CSF are presumably enough to assess the BBBD, although this still necessitates CSF sampling, which is not always feasible in the clinical setting.
Correlation analyses of biomarkers and QA
We found that S100Bblood was momentarily correlated to S100BCSF and QA, in accordance with previous studies.20,21,26,30,59 Others have acquired contrasting results,23,26 possibly because of different analysis strategies. We also noted that momentary NSEblood levels were correlated with NSECSF and QA, something that has been shown in non-TBI studies,60,62 whereas a previous TBI study did not show this association.26 For both biomarkers, we noted similar correlation coefficients between CSF:blood and blood:QA. This implies that a third covariate, most likely time from trauma, inferred these values. We therefore performed cross-correlations, to elucidate the underlying share time trend of the biomarker correlations.
In our cross-correlations, we noted that both S100B and NSE exhibited peak CSF:blood cross correlations at lags >0, suggesting that both biomarkers have a partially delayed clearance from CSF to blood. Previous studies examining S100Bserum and S100BCSF correlation have found a delay varying between 0–24 h30 and 48 h.63 Our S100B CSF:blood cross-correlation exhibited a lag of ∼12 h, corresponding to the delay seen between QA and S100Bblood. Hence, the CSF:blood and the QA:blood pattern mimicked each other, presumably indicating that there is a brief initial accumulation of S100B in CSF before clearance through a disrupted BBB. This constitutes a tentative pathway for S100B clearance to peripheral blood. In contrast, NSE exhibited a more delayed peak in cross-correlations than S100B (∼ 24 h). This is important, because a brain biomarker should confer accurate and timely information if it is to aid in clinical decision making. Moreover, in the three cross- correlations performed on NSE, de novo release of NSECSF seemed to occur more swiftly than BBBD, and BBBD occurred simultaneously to NSEblood detection. Hence, there was a delayed clearance of NSE from CSF to blood, that was not mimicked by either CSF:QA or QA:blood cross-correlations, which was the case for S100B. Data were scarce for all of these observations, but if true, this means that neither delayed BBB clearance nor delayed de novo release of NSE could explain the delayed clearance of NSECSF to blood. Hence, there is a weak but interesting signal suggesting that intracranial NSE accumulates and thereafter is cleared through a route other than the BBB. One tentative clearance pathway is the experimentally described glymphatic system,14 suggested to be a clearance route for both NSE and S100B.15,64 Another possible route includes the recently discovered lymphatic vascular network surrounding dural sinuses, which drains in deep cervical lymph nodes and subsequently blood, with clearance of proteins from the brain interstitium.65,66 The strong momentary correlations we see between CSF and blood suggests a relatively direct pathway in these glymphatic/brain lymphatic routes, which in the case of NSE is seemingly independent of BBBD. However, it should be noted that both the glymphatic and brain lymphatic vasculature have currently only been sufficiently studied in animal models. Some promising work using magnetic resonance imaging favors the existence and importance of the glymphatic system in humans,67,68 but larger human studies and functional characterizations are still warranted.
BBB disruption predicts S100B but not NSE concentrations in blood
For inferential analysis, we employed marginal and linear mixed modelling. For S100Bblood, we found that QA, S100BCSF, and time from trauma were significant independent predictors. This indicates not only that S100B is dependent on the BBB for clearance to peripheral blood, but also that S100Bblood cannot be regarded solely as a marker of BBB integrity as has been suggested.20,69 Whereas some claim that cerebral S100B originates from the astrocytic foot processes that enfolds the BBB,30 our data rather indicate that S100Bblood is a reflection of several factors of which merely one is the extent of BBBD. In a situation in which the BBB is intact,69 it is therefore theoretically possible that the S100Bblood value is falsely low, as this biomarker will then to some extent be “trapped” in the CSF. Applying a similar approach to NSE, we found that time from trauma was retained as the only significant independent variable in the model. This means that neither NSECSF nor QA were predictive of NSEblood. Our findings cohere with one previous study that did not find any increase of NSE following chemical/osmolytic BBBD.70 In summary, there was a clear relationship between QA and S100B, whereas for NSE there was not.
Both our cross-correlation and marginal/linear mixed model analyses hence indicate that S100B and NSE might be cleared differently from CSF to peripheral blood. There are, however, important differences in biochemical, kinetic, and/or pathophysiological properties between the two proteins, potentially confounding our results. Biologically, NSE is larger (39 kDa)71,72 than S100B (9–14 kDa),73 implications of which could be a swifter movement across body compartments for small proteins and a longer serum half-life for larger proteins. Accordingly, NSE has been shown to exhibit an “effective half-life” of up to 48–72 h in serum compared with 24 h for S100B,74 following severe TBI. This would give a slower decline of NSEblood values than S100B values. We compensated for this by employing an ARMA variance-covariance structure to our model; however without finding any predictors of NSEblood other than time from trauma. Next, there are different sources for S100B and NSE throughout the body. S100B is present primarily in astrocytes,75 but extracranially also in muscle, bone, cartilage, adipose tissue, and melanocytes.9,76 The extracranial component is primarily of importance in multi-trauma patients during the first 12 h following the trauma.8,9 We excluded the first 12 h from analysis for S100B and monitored patients for a week, which would result in a very limited contribution of extracranial S100B to our model. In contrast, whereas NSE intracranially locates primarily to neurons,9 extracerebral NSE emanates from, among other sources, erythrocytes.9,77 Hence, extracerebral NSE is a confounding factor following both multi-trauma and hemolysis.9 Importantly, there is no clear period after trauma when this is important, but rather this potential confounder spans the entire data material and contributes a latent data noise. In summary, although S100B and NSE seem to have different clearance routes from the injured brain, there are differences between the proteins, which may have influenced our results.
Limitations
The major limitation in this study relates to the inclusion rate (inclusion was dependent on senior author presence) and the consequently small study sample size. Moreover, there are other factors that may be associated with brain biomarker clearance, such as brain injury severity or type of injury sustained, but these correlations necessitate a larger sample size to study. Conversely, the data was prospectively collected with a uniquely high temporal resolution with constant CSF drainage speed; therefore making this study highly relevant in term of brain protein clearance. Another limitation is that the CSF at time point 0 in our models constituted a pool of CSF collected during the preceding 6 h; therefore, the lag times could be slightly longer than indicated in the cross-correlations, although we believe the current protocol using the CSF pump is the most delicate way to extract CSF for serial sampling using an enclosed system. Further, in animals, it has been shown that acetazolamide treatment, CSF drainage, and sleep deprivation inhibit the glymphatic system.15 Although no patients received acetazolamide, and CSF drainage speed was constant in our study, all patients exhibited different levels of induced sedation, which in theory could affect glymphatic clearance, but how this may affect biomarker levels in humans warrants further research. In spite of being a gold standard metric for BBBD, QA is age dependent,19 which is a source of error across all studies using it. Moreover, for TBI patients, intraventricular hemorrhage26 and administration of large amount of intravenous fluids could both affect QA. For the latter, we propose that CSF albumin alone might be an adequate surrogate for QA (and therefore BBBD). Finally, in the injured brain, multiple factors are in play, and it is possible that CNS half-life is discrepant from blood half-life of certain proteins. We do not account for this in our models, and in order to do so, one needs to employ more extensive pharmacokinetic modeling, which will be the aim for future studies that are warranted. Another amenable avenue is radiological techniques67,68 using biomarker-specific contrast-enhancement combined with sampling techniques similar to ours in order to elucidate discrepancies between BBB and other plausible clearance pathways.
Conclusion
Blood biomarker levels of S100B and NSE correlate momentarily with BBB integrity, measured as QA. However, longitudinally, this relationship was stronger for S100B than for NSE. Upon inferential modeling, blood levels of S100B were dependent on QA, which we could not observe for NSE. Although this could be the result of underlying differences between the two proteins such as hemolysis of NSE, it indicates that S100B and NSE might be cleared differentially from the injured CNS, which is novel and important for brain biomarker interpretation and clinical utility, although future studies are needed to confirm our findings.
Supplementary Material
Acknowledgments
We thank the NCCU staff at the Karolinska University Hospital, Stockholm, Sweden for their hard work throughout all aspects of this study. We also like to thank Per Engzell for statistical advice.
Funding Information
This work was supported by several funding sources. C.L. received salary funding and travel grants from the Karolinska Institute Funds Clinical Scientist Training Programme, Research Internship, Karolinska Institutet Resebidrag (grant numbers K825715022, K825715052, and K825715703). C.L. and E.P.T. received travel grants and salary from the Swedish Society for Medical Research Travel Grant and Post-Doctoral Scholarship (grant numbers SLS-816961, N/A). Equipment was purchased using the Swedish Armed Forces Research and Development Fund (grant number AT.9221010) obtained by M.R.; and Stockholm County ALF (grant number 20180572), obtained by B.M.B. F.A.Z. receives research support from the University of Manitoba Vice-President Research and International (VPRI) Research Investment Fund (RIF), the University of Manitoba Rudy Falk Clinician-Scientist Professorship, the Health Sciences Centre Foundation in Winnipeg, the University of Manitoba Centre on Aging Fellowship, the Canadian Institutes of Health Research (CIHR) (432061), and the National Institute of Neurological Disorders and Stroke (NINDS) (RO3NS114335). None of the other authors received funding. The funders did not participate in study design, data acquisition, data analysis/interpretation, manuscript compilation, or submission decision.
Author Disclosure Statement
No competing financial interests exist.
Supplementary Material
References
- 1. Jennett B. (1996). Epidemiology of head injury. J. Neurol. Neurosurg. Psychiatry 60, 362–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hyder A.A., Wunderlich C.A., Puvanachandra P., Gururaj G., and Kobusingye O.C. (2007). The impact of traumatic brain injuries: a global perspective. NeuroRehabilitation 22, 341–353 [PubMed] [Google Scholar]
- 3. Maas A.I.R., Menon D.K., Adelson P.D., Andelic N., Bell M.J., Belli A., Bragge P., Brazinova A., Buki A., Chesnut R.M., Citerio G., Coburn M., Cooper D.J., Crowder A.T., Czeiter E., Czosnyka M., Diaz-Arrastia R., Dreier J.P., Duhaime A.C., Ercole A., van Essen T.A., Feigin V.L., Gao G., Giacino J., Gonzalez-Lara L.E., Gruen R.L., Gupta D., Hartings J.A., Hill S., Jiang J.Y., Ketharanathan N., Kompanje E.J.O., Lanyon L., Laureys S., Lecky F., Levin H., Lingsma H.F., Maegele M., Majdan M., Manley G., Marsteller J., Mascia L., McFadyen C., Mondello S., Newcombe V., Palotie A., Parizel P.M., Peul W., Piercy J., Polinder S., Puybasset L., Rasmussen T.E., Rossaint R., Smielewski P., Soderberg J., Stanworth S.J., Stein M.B., von Steinbuchel N., Stewart W., Steyerberg E.W., Stocchetti N., Synnot A., Te Ao B., Tenovuo O., Theadom A., Tibboel D., Videtta W., Wang K.K.W., Williams W.H., Wilson L., Yaffe K., and InTBIR Participants and Investigators (2017). Traumatic brain injury: integrated approaches to improve prevention, clinical care, and research. Lancet Neurol. 16, 987–1048 [DOI] [PubMed] [Google Scholar]
- 4. Chesnut R.M., Marshall L.F., Klauber M.R., Blunt B.A., Baldwin N., Eisenberg H.M., Jane J.A., Marmarou A., and Foulkes M.A. (1993). The role of secondary brain injury in determining outcome from severe head injury. J. Trauma 34, 216–222 [DOI] [PubMed] [Google Scholar]
- 5. Jones P.A., Andrews P.J., Midgley S., Anderson S.I., Piper I.R., Tocher J.L., Housley A.M., Corrie J.A., Slattery J., Dearden N.M. and Miller D.J. (1994). Measuring the burden of secondary insults in head-injured patients during intensive care. J. Neurosurg. Anesthesiol. 6, 4–14 [PubMed] [Google Scholar]
- 6. Sarrafzadeh A.S., Peltonen E.E., Kaisers U., Kuchler I., Lanksch W.R., and Unterberg A.W. (2001). Secondary insults in severe head injury––do multiply injured patients do worse? Crit. Care Med. 29, 1116–1123 [DOI] [PubMed] [Google Scholar]
- 7. Thelin E.P., Nelson D.W., and Bellander B.M. (2017). A review of the clinical utility of serum S100B protein levels in the assessment of traumatic brain injury. Acta Neurochir. (Wien) 159, 209–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thelin E.P., Johannesson L., Nelson D., and Bellander B.M. (2013). S100B is an important outcome predictor in traumatic brain injury. J. Neurotrauma 30, 519–528 [DOI] [PubMed] [Google Scholar]
- 9. Thelin E.P., Jeppsson E., Frostell A., Svensson M., Mondello S., Bellander B.M., and Nelson D.W. (2016). Utility of neuron-specific enolase in traumatic brain injury; relations to S100B levels, outcome, and extracranial injury severity. Crit. Care 20, 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Thelin E.P., Al Nimer F., Frostell A., Zetterberg H., Blennow K., Nystrom H., Svensson M., Bellander B.M., Piehl F., and Nelson D.W. (2019). A serum protein biomarker panel improves outcome prediction in human traumatic brain injury. J Neurotrauma 36, 2850–2862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Unden J., Ingebrigtsen T., Romner B., and Scandinavian Neurotrauma C. (2013). Scandinavian guidelines for initial management of minimal, mild and moderate head injuries in adults: an evidence and consensus-based update. BMC Med. 11, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Thelin E.P., Nelson D.W., and Bellander B.M. (2014). Secondary peaks of S100B in serum relate to subsequent radiological pathology in traumatic brain injury. Neurocrit. Care 20, 217–229 [DOI] [PubMed] [Google Scholar]
- 13. Liddelow S.A. (2011). Fluids and barriers of the CNS: a historical viewpoint. Fluids Barriers CNS 8, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Iliff J.J., Wang M., Liao Y., Plogg B.A., Peng W., Gundersen G.A., Benveniste H., Vates G.E., Deane R., Goldman S.A., Nagelhus E.A., and Nedergaard M. (2012). A paravascular pathway facilitates CSF flow through the brain parenchyma and the clearance of interstitial solutes, including amyloid beta. Sci. Transl, Med. 4, 147ra111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Plog B.A., Dashnaw M.L., Hitomi E., Peng W., Liao Y., Lou N., Deane R., and Nedergaard M. (2015). Biomarkers of traumatic injury are transported from brain to blood via the glymphatic system. J. Neurosci. 35, 518–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Marchi N., Bazarian J.J., Puvenna V., Janigro M., Ghosh C., Zhong J., Zhu T., Blackman E., Stewart D., Ellis J., Butler R., and Janigro D. (2013). Consequences of repeated blood–brain barrier disruption in football players. PLoS One 8, e56805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Marchi N., Fazio V., Cucullo L., Kight K., Masaryk T., Barnett G., Vogelbaum M., Kinter M., Rasmussen P., Mayberg M.R., and Janigro D. (2003). Serum transthyretin monomer as a possible marker of blood-to-CSF barrier disruption. J. Neurosci. 23, 1949–1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kanner A.A., Marchi N., Fazio V., Mayberg M.R., Koltz M.T., Siomin V., Stevens G.H., Masaryk T., Aumayr B., Vogelbaum M.A., Barnett G.H., and Janigro D. (2003). Serum S100beta: a noninvasive marker of blood–brain barrier function and brain lesions. Cancer 97, 2806–2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tibbling G., Link H., and Ohman S. (1977). Principles of albumin and IgG analyses in neurological disorders. I. Establishment of reference values. Scand. J. Clin. Lab. Invest. 37, 385–390 [DOI] [PubMed] [Google Scholar]
- 20. Blyth B.J., Farhavar A., Gee C., Hawthorn B., He H., Nayak A., Stocklein V., and Bazarian J.J. (2009). Validation of serum markers for blood–brain barrier disruption in traumatic brain injury. J. Neurotrauma 26, 1497–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Blyth B.J., Farahvar A., He H., Nayak A., Yang C., Shaw G., and Bazarian J.J. (2011). Elevated serum ubiquitin carboxy-terminal hydrolase L1 is associated with abnormal blood–brain barrier function after traumatic brain injury. J. Neurotrauma 28, 2453–2462 [DOI] [PubMed] [Google Scholar]
- 22. Saw M.M., Chamberlain J., Barr M., Morgan M.P., Burnett J.R., and Ho K.M. (2014). Differential disruption of blood–brain barrier in severe traumatic brain injury. Neurocrit. Care 20, 209–216 [DOI] [PubMed] [Google Scholar]
- 23. Kleindienst A., Schmidt C., Parsch H., Emtmann I., Xu Y. and Buchfelder M. (2010). The passage of S100B from brain to blood is not specifically related to the blood–brain barrier integrity. Cardiovasc. Psychiatry Neurol. 2010, 801295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Reiber H. (2003). Proteins in cerebrospinal fluid and blood: barriers, CSF flow rate and source-related dynamics. Restor. Neurol. Neurosci. 21, 79–96 [PubMed] [Google Scholar]
- 25. Nag S.E. (2011). The Blood–Brain and Other Neural Barriers. Reviews and Protocols. Springer Science and Business Media LLC: New York [Google Scholar]
- 26. Bellander B.M., Olafsson I.H., Ghatan P.H., Bro Skejo H.P., Hansson L.O., Wanecek M., and Svensson M.A. (2011). Secondary insults following traumatic brain injury enhance complement activation in the human brain and release of the tissue damage marker S100B. Acta Neurochir. (Wien) 153, 90–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thelin E.P., Nelson D.W., Ghatan P.H., and Bellander B.M. (2014). Microdialysis monitoring of CSF parameters in severe traumatic brain injury patients: a novel approach. Front. Neurol. 5, 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. R Core Team. (2018). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria [Google Scholar]
- 29. Champely S. (2018). pwr: Basic Functions for Power Analysis. R package version 1.2-2. https://CRAN.R-project.org/package=PWR (last accessed July30, 2019)
- 30. Goyal A., Failla M.D., Niyonkuru C., Amin K., Fabio A., Berger R.P., and Wagner A.K. (2013). S100b as a prognostic biomarker in outcome prediction for patients with severe traumatic brain injury. J. Neurotrauma 30, 946–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Smit L.H., Korse C.M., and Bonfrer J.M. (2005). Comparison of four different assays for determination of serum S-100B. Int. J. Biol. Markers 20, 34–42 [DOI] [PubMed] [Google Scholar]
- 32. Muller K., Elverland A., Romner B., Waterloo K., Langbakk B., Unden J., and Ingebrigtsen T. (2006). Analysis of protein S-100B in serum: a methodological study. Clin. Chem. Lab. Med. 44, 1111–1114 [DOI] [PubMed] [Google Scholar]
- 33. Teasdale G., and Jennett B. (1974). Assessment of coma and impaired consciousness – practical scale. Lancet 2, 81–84 [DOI] [PubMed] [Google Scholar]
- 34. Copes W.S., Champion H.R., Sacco W.J., Lawnick M.M., Keast S.L., and Bain L.W. (1988). The injury severity score revisited. J. Trauma 28, 69–77 [DOI] [PubMed] [Google Scholar]
- 35. Baker S.P., O'Neill B., Haddon W. Jr., and Long W.B. (1974). The injury severity score: a method for describing patients with multiple injuries and evaluating emergency care. J. Trauma 14, 187–196 [PubMed] [Google Scholar]
- 36. MRC Crash Trial Collaborators, Perel P., Arango M., Clayton T., Edwards P., Komolafe E., Poccock S., Roberts I., Shakur H., Steyerberg E. and Yutthakasemsunt S. (2008). Predicting outcome after traumatic brain injury: practical prognostic models based on large cohort of international patients. BMJ 336, 425–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Marshall L.F., Marshall S.B., Klauber M.R., Clark M.V., Eisenberg H.M., Jane J.A., Luerssen T.G., Marmarou A., and Foulkes M.A. (1991). A new classification of head-injury based on computerized-tomography. J. Neurosurg. 75, S14–S20 [Google Scholar]
- 38. Marshall L.F., Marshall S.B., Klauber M.R., Van Berkum Clark M., Eisenberg H., Jane J.A., Luerssen T.G., Marmarou A., and Foulkes M.A. (1992). The diagnosis of head injury requires a classification based on computed axial tomography. J. Neurotrauma 9, Suppl. 1, S287–292 [PubMed] [Google Scholar]
- 39. Maas A.I., Hukkelhoven C.W., Marshall L.F., and Steyerberg E.W. (2005). Prediction of outcome in traumatic brain injury with computed tomographic characteristics: a comparison between the computed tomographic classification and combinations of computed tomographic predictors. Neurosurgery 57, 1173–1182 [DOI] [PubMed] [Google Scholar]
- 40. Nelson D.W., Nystrom H., MacCallum R.M., Thornquist B., Lilja A., Bellander B.M., Rudehill A., Wanecek M., and Weitzberg E. (2010). Extended analysis of early computed tomography scans of traumatic brain injured patients and relations to outcome. J. Neurotrauma 27, 51–64 [DOI] [PubMed] [Google Scholar]
- 41. Lindblad C., Thelin E.P., Nekludov M., Frostell A., Nelson D.W., Svensson M., and Bellander B.M. (2018). Assessment of platelet function in traumatic brain injury–a retrospective observational study in the neuro-critical care setting. Front. Neurol. 9, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jennett B., and Bond M. (1975). Assessment of outcome after severe brain damage. Lancet 1, 480–484 [DOI] [PubMed] [Google Scholar]
- 43. Johanson C.E., Stopa E.G., and McMillan P.N. (2011). The blood–cerebrospinal fluid barrier: structure and functional significance. Methods Mol. Biol. 686, 101–131 [DOI] [PubMed] [Google Scholar]
- 44. Klinisk kemi och KUL 24Sju. Albumin, P- [Internet]. Stockholm: Karolinska Universitetssjukhuset; [updated 2018-07-09; cited 2020-01-09]. Available from: https://www.karolinska.se/KUL/Alla-anvisningar/Anvisning/8983
- 45. Klinisk kemi och KUL 24Sju. Albumin, Csv- [Internet]. Stockholm: Karolinska Universitetssjukhuset; [updated 2019-11-25; cited 2020-01-09]. Available from: https://www.karolinska.se/KUL/Alla-anvisningar/Anvisning/8980
- 46. Klinisk kemi och KUL 24Sju. Albuminkvot, Csv/S- [Internet]. Stockholm: Karolinska Universitetssjukhuset; [updated 2019-01-17; cited 2020-01-09]. Available from: https://www.karolinska.se/KUL/Alla-anvisningar/Anvisning/9993
- 47. Klinisk kemi och KUL 24Sju. S100 B, S- [Internet]. Stockholm: Karolinska Universitetssjukhuset; [updated 2019-04-10; cited 2020-01-09]. Available from: https://www.karolinska.se/KUL/Alla-anvisningar/Anvisning/9268
- 48. Tierney Nicholas. ggplot your missing data [Internet]. San Francisco: GitHub, Inc; 2015 [updated 2015-12-01; cited 2019-07-30]. Available from: https://njtierney.github.io/r/missing%20data/rbloggers/2015/12/01/ggplot-missing-data/
- 49. Bakdash J.Z., and Marusich L.R. (2017). Repeated measures correlation. Front. Psychol. 8, 456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bakdash J.Z. and Marusich, L.R. rmcorr: Repeated Measures Correlation, version 0.3.0 [R package]. Vienna: The Comprehensive R Archive Network (CRAN); 2018. [cited 2019-07-30]. Available from: https://CRAN.R-project.org/package=rmcorr
- 51. Wickham H, Averick M, Bryan J, Chang W, D'Agostino McGowan L, François R, Grolemund G, Hayes A, Henry L, Hester J, Kuhn M, Pedersen T.L, Miller E, Bache S.M., Müller K, Ooms J, Robinson D, Seidel D.P, Spinu V, Takahashi K, Vaughan D, Wilke C, Woo K, and Yutani H., (2019). Welcome to the Tidyverse. Journal of Open Source Software, 4(43), 1686, 10.21105/joss.01686 [DOI] [Google Scholar]
- 52. Pinheiro J, Bates D, DebRoy S, Sarkar D, R Core Team. nlme: Linear and Nonlinear Mixed Effects Models, version 3.1-140 [R package]. Vienna: The Comprehensive R Archive Network (CRAN); 2019. [cited 2019-07-30]. Available from: https://CRAN.R-project.org/package=nlme
- 53. Trapletti A., and Hornik K. tseries: Time Series Analysis and Computational Finance, version 0.10-47 [R package]. Vienna: The Comprehensive R Archive Network (CRAN); 2019. [cited 2019-07-30]. Available from: https://CRAN.R-project.org/package=tseries
- 54. Hyndman R.J., and Khandakar Y. (2008). Automatic time series forecasting: the forecast package for R. J, Stat. Softw. 27, 1–22 [Google Scholar]
- 55. Field A., Miles J., and Field Z. (2012). Discovering Statistics Using R. Sage Publications Ltd: London [Google Scholar]
- 56. West B.T., Welch K.B., and Galecki A.T. (2015). Linear Mixed Models A Practical Guide Using Statistical Software, 2nd ed. CRC Press: Boca Raton, FL [Google Scholar]
- 57. Kuznetsova A., Brockhoff P.B., and Christensen R.H.B. (2017). lmerTest Package: tests in linear mixed effects models. J. Stat. Softw. 82, 1–26 [Google Scholar]
- 58. Winter C., Bell C., Whyte T., Cardinal J., Macfarlane D., and Rose S. (2015). Blood–brain barrier dysfunction following traumatic brain injury: correlation of K(trans) (DCE-MRI) and SUVR (99mTc-DTPA SPECT) but not serum S100B. Neurol. Res. 37, 599–606 [DOI] [PubMed] [Google Scholar]
- 59. Dadas A., Washington J., Marchi N., and Janigro D. (2016). Improving the clinical management of traumatic brain injury through the pharmacokinetic modeling of peripheral blood biomarkers. Fluids Barriers CNS 13, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Correale J., Rabinowicz A.L., Heck C.N., Smith T.D., Loskota W.J., and DeGiorgio C.M. (1998). Status epilepticus increases CSF levels of neuron-specific enolase and alters the blood–brain barrier. Neurology 50, 1388–1391 [DOI] [PubMed] [Google Scholar]
- 61. Shlosberg D., Benifla M., Kaufer D., and Friedman A. (2010). Blood–brain barrier breakdown as a therapeutic target in traumatic brain injury. Nat. Rev. Neurol. 6, 393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Martens P., Raabe A., and Johnsson P. (1998). Serum S-100 and neuron-specific enolase for prediction of regaining consciousness after global cerebral ischemia. Stroke 29, 2363–2366 [DOI] [PubMed] [Google Scholar]
- 63. Petzold A., Keir G., Lim D., Smith M., and Thompson E.J. (2003). Cerebrospinal fluid (CSF) and serum S100B: release and wash-out pattern. Brain Res. Bull. 61, 281–285 [DOI] [PubMed] [Google Scholar]
- 64. Kawata K., Liu C.Y., Merkel S.F., Ramirez S.H., Tierney R.T. and Langford D. (2016). Blood biomarkers for brain injury: what are we measuring? Neurosci. Biobehav. Rev. 68, 460–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Aspelund A., Antila S., Proulx S.T., Karlsen T.V., Karaman S., Detmar M., Wiig H., and Alitalo K. (2015). A dural lymphatic vascular system that drains brain interstitial fluid and macromolecules. J. Exp. Med. 212, 991–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Louveau A., Smirnov I., Keyes T.J., Eccles J.D., Rouhani S.J., Peske J.D., Derecki N.C., Castle D., Mandell J.W., Lee K.S., Harris T.H., and Kipnis J. (2015). Structural and functional features of central nervous system lymphatic vessels. Nature 523, 337–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ringstad G., Vatnehol S.A.S., and Eide P.K. (2017). Glymphatic MRI in idiopathic normal pressure hydrocephalus. Brain 140, 2691–2705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Eide P.K., Vatnehol S.A.S., Emblem K.E., and Ringstad G. (2018). Magnetic resonance imaging provides evidence of glymphatic drainage from human brain to cervical lymph nodes. Sci. Rep. 8, 7194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Marchi N., Rasmussen P., Kapural M., Fazio V., Kight K., Mayberg M.R., Kanner A., Ayumar B., Albensi B., Cavaglia M., and Janigro D. (2003). Peripheral markers of brain damage and blood–brain barrier dysfunction. Restor. Neurol. Neurosci. 21, 109–121 [PMC free article] [PubMed] [Google Scholar]
- 70. Kapural M., Krizanac-Bengez L., Barnett G., Perl J., Masaryk T., Apollo D., Rasmussen P., Mayberg M.R., and Janigro D. (2002). Serum S-100beta as a possible marker of blood–brain barrier disruption. Brain Res. 940, 102–104 [DOI] [PubMed] [Google Scholar]
- 71. Kaiser E., Kuzmits R., Pregant P., Burghuber O., and Worofka W. (1989). Clinical biochemistry of neuron specific enolase. Clin. Chim. Acta 183, 13–31 [DOI] [PubMed] [Google Scholar]
- 72. Isgro M.A., Bottoni P. and Scatena R. (2015). Neuron-specific enolase as a biomarker: biochemical and clinical aspects. Adv. Exp. Med. Biol. 867, 125–143 [DOI] [PubMed] [Google Scholar]
- 73. Donato R. (2001). S100: a multigenic family of calcium-modulated proteins of the EF-hand type with intracellular and extracellular functional roles. Int. J. Biochem. Cell Biol. 33, 637–668 [DOI] [PubMed] [Google Scholar]
- 74. Thelin E.P., Zeiler F.A., Ercole A., Mondello S., Buki A., Bellander B.M., Helmy A., Menon D.K., and Nelson D.W. (2017). Serial sampling of serum protein biomarkers for monitoring human traumatic brain injury dynamics: a systematic review. Front. Neurol. 8, 300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Steiner J., Bernstein H.G., Bielau H., Berndt A., Brisch R., Mawrin C., Keilhoff G., and Bogerts B. (2007). Evidence for a wide extra-astrocytic distribution of S100B in human brain. BMC Neurosci. 8, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Anderson R.E., Hansson L.O., Nilsson O., Dijlai-Merzoug R., and Settergren G. (2001). High serum S100B levels for trauma patients without head injuries. Neurosurgery 48, 1255–1260 [DOI] [PubMed] [Google Scholar]
- 77. Johnsson P., Blomquist S., Luhrs C., Malmkvist G., Alling C., Solem J.O., and Stahl E. (2000). Neuron-specific enolase increases in plasma during and immediately after extracorporeal circulation. Ann. Thorac. Surg. 69, 750–754 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.