Abstract
Proteins with a functionalized C-terminus such as a C-terminal thioester are key to the synthesis of larger proteins via expressed protein ligation. They are usually made by recombinant fusion to intein. Although powerful, the intein fusion approach suffers from premature hydrolysis and low compatibility with denatured conditions. To totally bypass the involvement of an enzyme for expressed protein ligation, here we showed that a cysteine in a recombinant protein was chemically activated by a small molecule cyanylating reagent at its N-side amide for undergoing nucleophilic acyl substitution with amines including a number of L- and D-amino acids and hydrazine. The afforded protein hydrazides could be used further for expressed protein ligation. We demonstrated the versatility of this activated cysteine-directed protein ligation (ACPL) approach with the successful synthesis of ubiquitin conjugates, ubiquitin-like protein conjugates, histone H2A with a C-terminal posttranslational modification, RNAse H that actively hydrolyzed RNA, and exenatide that is a commercial therapeutic peptide. The technique, which is exceedingly simple but highly useful, expands to a great extent the synthetic capacity of protein chemistry and will therefore make a large avenue of new research possible.
Graphical Abstract
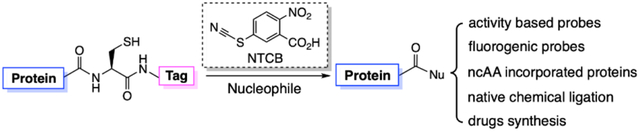
INTRODUCTION
The native chemical ligation concept was first developed by Dawson et al. in 1994, in which one protein or peptide with a C-terminal thioester and the other with a N-terminal cysteine selectively undergo thiol-thioester exchange and then S-to-N acyl transfer to form a larger protein or peptide (Figure 1A)1. Given that a protein with an N-terminal cysteine can be recombinantly produced, the development of the concept made it feasible to synthesize large proteins with a functionalized N-terminus to include either posttranslational or purely chemical modifications. To expand the synthetic scope of native chemical ligation, a related technique termed expressed protein ligation in which a recombinant fusion protein with a C-terminal intein is used to generate a protein thioester was also developed for the synthesis of a protein with a functionalized C-terminus2. Another notable related technique is peptide hydrazide ligation that uses nitrous acid or acetyl acetone to convert a chemically stable peptide hydrazide to a peptide acyl azide or a peptide acyl pyrazole respectively and then a peptide thioester for further native chemical or expressed protein ligation (Figure 1A)3–4. Evident by the original publication garnering more than 2700 citations so far, the advent of native chemical and expressed protein ligation techniques has revolutionized the protein and peptide chemistry field. Groundbreaking applications include the synthesis of a large variety of proteins such as histones, kinases, and RAS proteins with posttranslational modifications for driving basic research advances and the production of many proteins or enzymes for therapeutic and biotechnological purposes5–12. Although developed extensively, further technological improvement in protein ligation is still necessary. The production of a protein thioester using the intein fusion approach is not guaranteed for a lot of proteins. The stringent requirement for the intein catalysis to generate a protein thioester prevents the processing of many fusion proteins that are expressed insolubly and hard to fold13. The C-terminal residue of a targeted protein that is immediate to the intein N-terminus also significantly impacts the protein splicing efficiency, which leads to low splicing efficiency for residues such as proline at this site14–15. The purification of an intein fusion also requires significant caution for avoiding premature hydrolysis2, 16. A split intein may be used to prevent premature hydrolysis but adds more procedural complexity5, 17. Using a protein ligase for expressed protein ligation resolves some issues related to the intein fusion approach but requires a specific amino acid sequence context at the ligation site18. Therefore, a simple method to functionalize a recombinant protein at its C-terminus for expressed protein ligation that requires no enzymatic catalysis, can be broadly applied, and maintains high efficiency in different protein C-terminal sequence contexts is highly desired. In this work, we report such a method and its application in the synthesis of a number of proteins or peptides that can be used in both basic research and therapy.
Figure 1. Protein synthesis by ligation techniques.
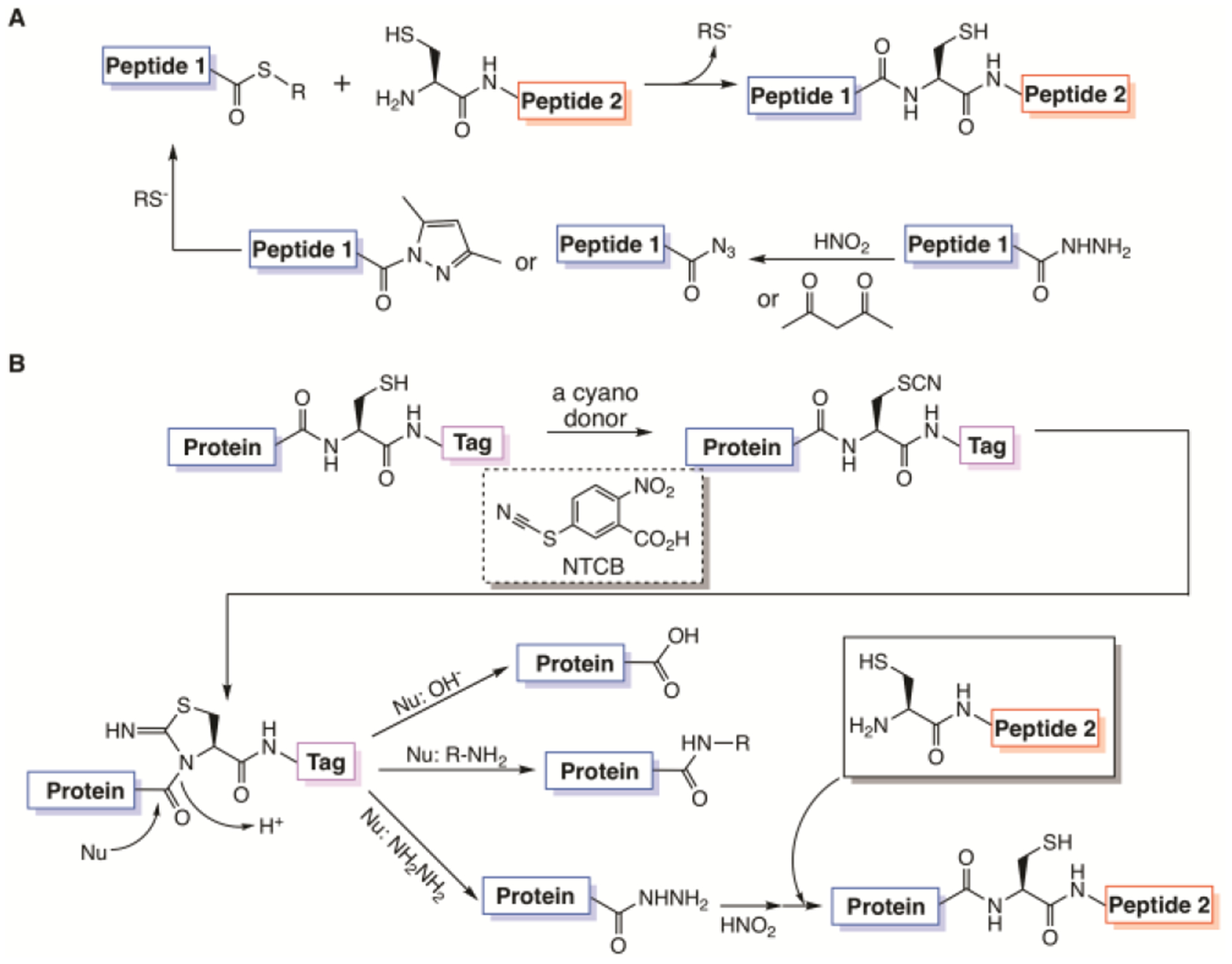
(A) Native chemical ligation and a derivative technique, peptide hydrazide ligation; (B) A proposed ACPL technique based on nucleophilic acyl substitution of an activated cysteine residue in a recombinant protein by a nucleophilic amine. Without a nucleophilic amine, the protein undergoes hydrolysis. When the nucleophile is hydrazine, the afforded protein hydrazide can then undergo peptide hydrazide ligation to form a larger protein.
RESULTS AND DISCUSSION
The Rationale
Among 20 proteinaceous amino acids, cysteine is the most nucleophilic. Its direct activation for generating a peptide thioester was previously explored by Kajihara, Otaka, and their coworkers19–20. To develop a more general, cysteine-based approach that could be applied to proteins for their C-terminal functionalization, we followed a century old industrial chemical process, leather tanning by cyanides. Cyanide salts that reduce disulfide bonds in proteins were used in the early 20th century to treat animal hides and wools21–22. During the process, a cyanide covalently attached to a protein cysteine to form a thiocyanate that underwent reversible intramolecular addition with the cysteine N-amide to generate a 1-acyl-2-iminothiazolidine intermediate. The amide bond in this intermediate was significantly weakened in comparison to a regular protein amide and therefore slowly hydrolyzed to split the protein (Figure 1B)23–24. Early protein chemists exploited this reaction for mapping protein sequences and replaced cyanide salts with other cyanylating reagents such as 2-nitro-5-thiocyanatobenzoic acid (NTCB) that transfers the cyano group directly to a reduced protein cysteine for avoiding the formation of highly toxic cyanide wastes25–26. According to this reaction mechanism, providing a strongly nucleophilic amine in the reaction will trigger nucleophilic acyl substitution with the 1-acyl-2-iminothiazolidine intermediate to replace 2-iminothiazolidine and potentially circumvent the hydrolysis process. The afforded small molecule amine-ligated product can be used for a further protein ligation process. Since this proposed expressed protein ligation that we termed as activated cysteine-based protein ligation (ACPL) doesn’t involve an enzyme and is purely chemically based, it can be highly controllable, selective, and versatile such as functioning for proteins both soluble and insoluble.
Feasibility of ACPL
To demonstrate the feasibility of ACPL, we synthesized Boc-Xxx-Cys-OMe dipeptides in which the Xxx identity varied between seven native amino acids including proline and carried out their reactions with an equivalent amount of NTCB and then ligation with propargylamine (Pa) in dichloromethane (DCM). Our result showed that all dipeptides reacted with Pa to form desired products with varied yields (Table S1). Since the reactions were performed in DCM, amino acids with a hydrophobic and large side chain tended to have high yields, while amino acids with a relatively small and hydrophilic side chain resulted in relatively low yields. To further characterize the effect of the reaction on the chirality of the amino acid at the N-terminal side of cysteine, we synthesized the Boc-L-isoleucine-Pa using ACPL and compared it with both Boc-L-isoleucine-Pa and Boc-DL-isoleucine-Pa that were synthesized using the standard amidation approach as controls. As shown in the 1H NMR spectra (Figure S1), no diastereomeric CH3 peak was observed for the ligated product indicating that this reaction did not lead to the racemization of the amino acid before cysteine.
Versality of ACPL
Encouraged by our small molecule results, we tested ACPL further with recombinant proteins. Ubiquitin (Ub) is natively devoid of cysteine27. We chose it as a model protein for our demonstration. We produced recombinant native Ub and Ub with both a G76C mutation and a C-terminal 6×His tag (Ub-G76C-6H) in E. coli and purified them to homogeneity. We then ligated Ub-G76C-6H with 12 small molecule amines including Pa, allylamine (Aa), hydrazine (Ha), and L- and D-amino acids (Figure 2A) by adding 5 mM NTCB and a 50–1000 mM amine simultaneously to a 2 mg/mL Ub-G76C-6H solution at pH 9 for an overnight incubation at 37 °C. We carefully selected seven L-amino acids for our reactions to represent amino acids in different chemical categories and also different sizes. For all tested compounds including proline that has a secondary amine and two D-amino acids, we obtained ligation products with 50–90% yields that were estimated by the SDS-PAGE analysis of reaction mixtures (Figure S2, Table S2). After using Ni2+ charged resins to simply remove unreacted intermediates, we analyzed all 12 ligation products and the two original Ub and Ub-G76C-6H proteins by electrospray ionization mass spectrometry (ESI-MS) analysis. For all analyzed proteins, their deconvoluted ESI-MS spectra displayed clearly observable monoisotopic peaks. Since there is no commercial software for calculating protein monoisotopic peaks, we wrote a Python script to calculate all theoretical monoisotopic masses and their relative intensities for all proteins and compared them to the determined ESI-MS spectra. Our results showed that determined monoisotopic masses for all proteins agreed very well with their theoretic values in terms of both molecular weight and intensity (Figures S3–S16). Hydrolysis products were either non-detectable or at very low levels. To simplify the comparison, we wrote another Python script to integrate deconvoluted monoisotopic peaks and then calculate the average molecular weights and intensities for all detected protein species in a particular spectrum. The final results are presented in Figures 2B and 2C. For all determined average molecular weights, they matched their theoretical values with a deviation of ± 0.3 Da (Table S2). For all 12 ligation products, we detected very few minor peaks in their ESI-MS spectra indicating that all reactions were very selective. One ligation product Ub-G76G is native Ub itself. Its ESI-MS spectrum in Figure 2C matched that of recombinantly expressed native Ub in Figure 2B. So far, our data demonstrated that ACPL works exactly according to what we proposed on a recombinant protein and this reaction is effective for amines that are primary, secondary, Ha, and amino acids with different configurations, characteristics, and sizes. The ligation with Ha was done in both native and denatured conditions. The results from two conditions showed minimal differences (Figures S16 and S17). Since conditions used for ACPL so far were at 37 °C with a long incubation time that might be a concern for some proteins, we also carried out reactions under lower temperatures and shorter times. We performed the reaction between Ub-G76C-6H and Pa under three different temperatures (8, 23, and 37 °C) as well as four different reaction times (4, 8, 12, and 16 h). The SDS-PAGE analysis of the final reaction mixtures exhibited no clear differences among all conditions. All these conditions yielded the desired product as 50% (Figure S2E). Therefore, a reaction at 8 °C for 4 h is well enough for ACPL.
Figure 2. The synthesis of Ub conjugates by ACPL.
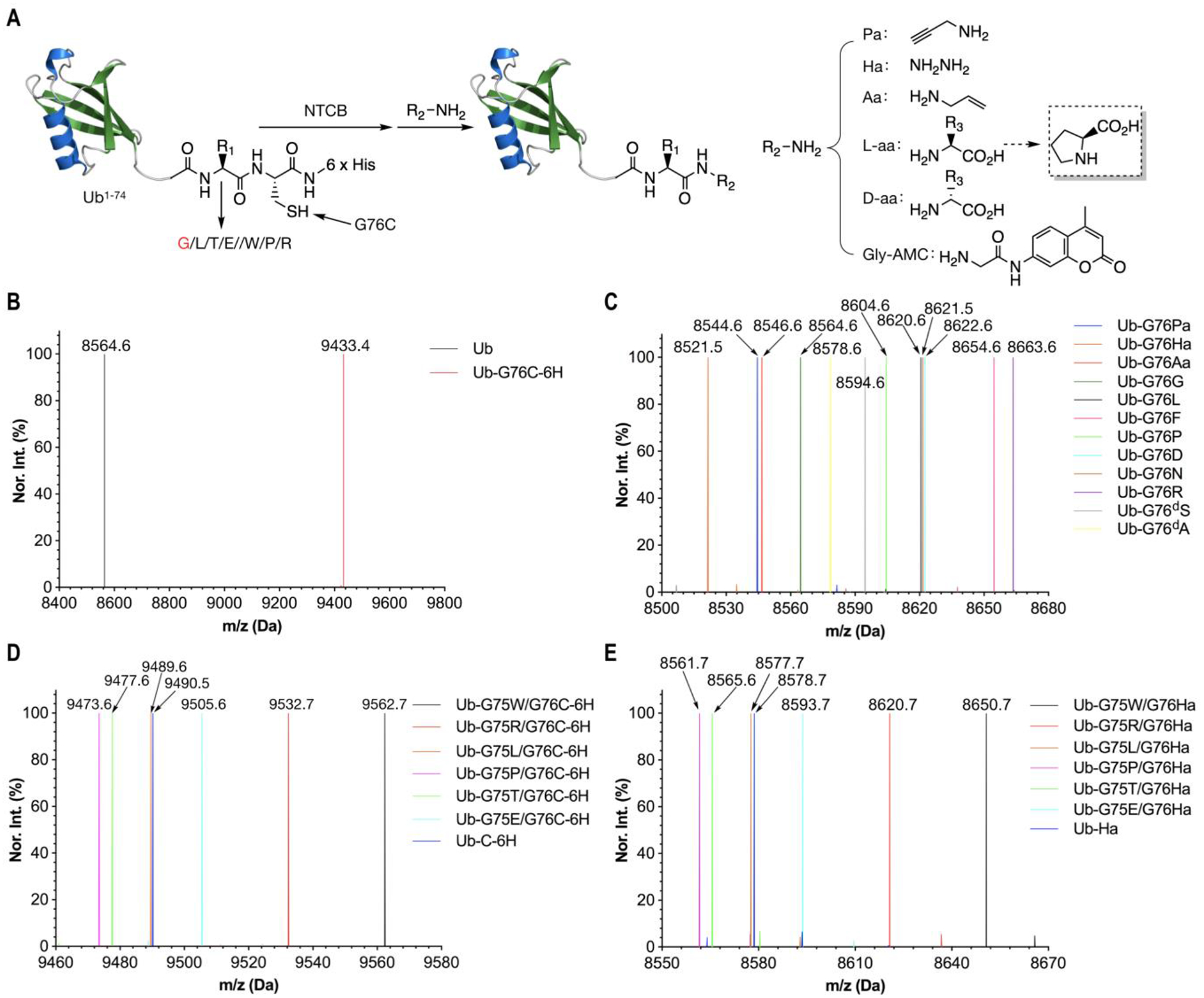
(A) A schematic diagram to show the activation of recombinant Ub proteins containing a cysteine by NTCB followed by nucleophilic acyl substitution with amines, both primary and secondary, to generate different Ub conjugates. The native Ub has 76 residues and glycine at the 75th and 76th positions. (B) The deconvoluted and integrated ESI-MS spectra of wild type Ub and Ub-G76C-6H. 6H represents a 6×His tag. (C) The deconvoluted and integrated ESI-MS spectra of Ub conjugates that were converted from Ub-G76C-6H and had different ligated molecules at the G76 position. Pa, Ha, and Aa are three small molecule amines shown in A. All other ligated molecules are amino acids whose one letter codes are used for labeling. All amino acids are in the L-configuration except two D-amino acids with a footnote d. (D-E) The deconvoluted and integrated ESI-MS spectra of 7 recombinant Ub proteins and products of their reactions with NTCB and Ha. C in Ub-C-6H represents cysteine. All detected molecular weights agreed well with theoretic values in a deviation range of ± 0.3 Da.
Ubiquitin natively has a G75 residue that has the lowest steric hindrance among all amino acids. In Ub-G76C-6H, the glycine immediately N-terminal to G76C might have permitted easy processing of the ligation. Other residues that have different chemical properties and/or are sterically hindered might impede the ligation. To resolve this concern, we mutated G75 in Ub-G76C-6H to six other residues that are large in size, charged, and/or having a secondary amine, recombinantly expressed them, analyzed them with ESI-MS (Figures 2D and S18–S23), and then reacted them in a one-pot fashion with NTCB and Ha. We chose Ha in our demonstration since its ligation products are protein hydrazides that can undergo further peptide hydrazide ligation for making even larger proteins. All reactions progressed exceedingly well and their reaction products displayed average molecular weights matching well to their theoretic values (Figures 2E and S24–S29; Table S3), demonstrating that the residue immediately N-terminal to the targeted cysteine has little detrimental effect on the ligation process. However, large amino acids such as tryptophan did lead to lower yields in comparison to small amino acids such as threonine (Figure S2, Table S3). Putting a cysteine residue right after Ub G76 led to similar ligation results with Ha, Aa, Pa and glycine with an at least 50 % conversion rate (Figures 2D, 2E, and S30–S34 and Table S3). Ub has a flexible C-terminus that may facilitate the ligation. To show that the ligation may work in a more structurally constrained environment, we introduced a cysteine mutation at K48 and K63, two residues in the globular region of Ub and used the two afforded Ub mutants (Figures S35–S36 and Table S3) to undergo ACPL with Ha. ESI-MS of reaction mixtures showed successful formation of two desired protein hydrazides (Figures S37–S38 and Table S3) indicating that ACPL works well in a structurally constrained protein region. Ligation both in a structurally constrained protein region and under a denatured condition is something that the traditional intein and ligase-based methods cannot perform well. Collectively our data strongly demonstrates the versatility of the ACPL technique.
The Use of ACPL to synthesize Ub and Ub-Like Protein (Ubl) Probes
In eukaryotic cells, Ub and Ubls can be posttranslationally attached to proteins for their functional regulation28–30. It has been shown that replacing the C-terminal glycine in Ub and Ubl proteins including SUMO1–3, NEED8, and ISG15 with Pa using either the intein based approach or total synthesis afforded excellent probes to conjugate covalently to deubiquitinases (DUBs) or ubiquitin-like proteases (ULPs) that catalytically remove Ub or Ubls from their conjugated proteins in cells31–35. To recapitulate these results and demonstrate the broad application scope of our ACPL technique in the probe synthesis, we recombinantly expressed Ub and a number of Ubl proteins including SUMO1–4, NEDD8, ISG15, GABARAP, GABARAPL2, UFM1, URM1, and MNSFβ (FLAG-Ub/Ubl-GxC-6H: x denotes the terminal glycine position) that all contained a C-terminal Gly-to-Cys mutation and were also fused with a N-terminal FLAG tag and a C-terminal 6×His tag, purified them to homogeneity, and then carried out their reactions with Pa in the presence of NTCB to afford their Pa-conjugated products. ISG15, SUMO1–4, and MNSFβ natively contain a cysteine residue. This cysteine was mutated to alanine or serine to avoid non-targeted reaction at its location. The yields of all 12 reactions varied between 25% and 80% as indicated by their SDS-PAGE analysis (Figure S2, Table S4). ESI-MS analysis of all 12 products showed their successful and efficient synthesis (Figures 3A and S39–S62; Table S4). The final results are presented in Figures 3A that displayed very little side products for all 12 Pa-conjugated products. In comparison to both intein based and total synthesis approach, our method for the synthesis of these Pa conjugates is much simpler and easier to control. To reproduce some literature results, we used our synthesized Pa-conjugated FLAG-Ub (FLAG-Ub-G76Pa) to react with seven DUBs and observed efficient covalent adduct formation for all tested enzymes in both SDS-PAGE analysis and Western blotting (Figures 3B and S63). We also performed similar tests for seven Pa-conjugated FLAG-Ubl probes and observed their covalent binding to a number of ULPs as shown in Figure 3C. Some ULPs such as SENP1 have only been vaguely confirmed in previous work to deconjugate corresponding Ubls such as SUMO436. All synthesized FLAG-Ub/Ubl conjugates, of which six are synthesized for the first time, are activity-based probes that can be potentially used to profile DUB and ULP proteomes in different tissues or cells. As a demonstration, we incubated the HEK293T cell lysate with FLAG-Ub-G76Pa and then probed the FLAG-Ub-conjugated proteins by an anti-FLAG antibody in Western blotting. The result showed the formation of a number of higher molecular weight species compared to the original FLAG-Ub-G76Pa, indicating conjugation with other proteins in the cell lysate (Figure S64). However, a control reaction using FLAG-Ub-G76C-6H showed no covalent conjugation with any other proteins in the HEK293T cell lysate.
Figure 3. The synthesis of FLAG-Ub/Ubl-Pa and Ub/FLAG-SUMO1–3-AMC probes by ACPL and their applications in covalent conjugation or activity assays of DUB/ULPs.
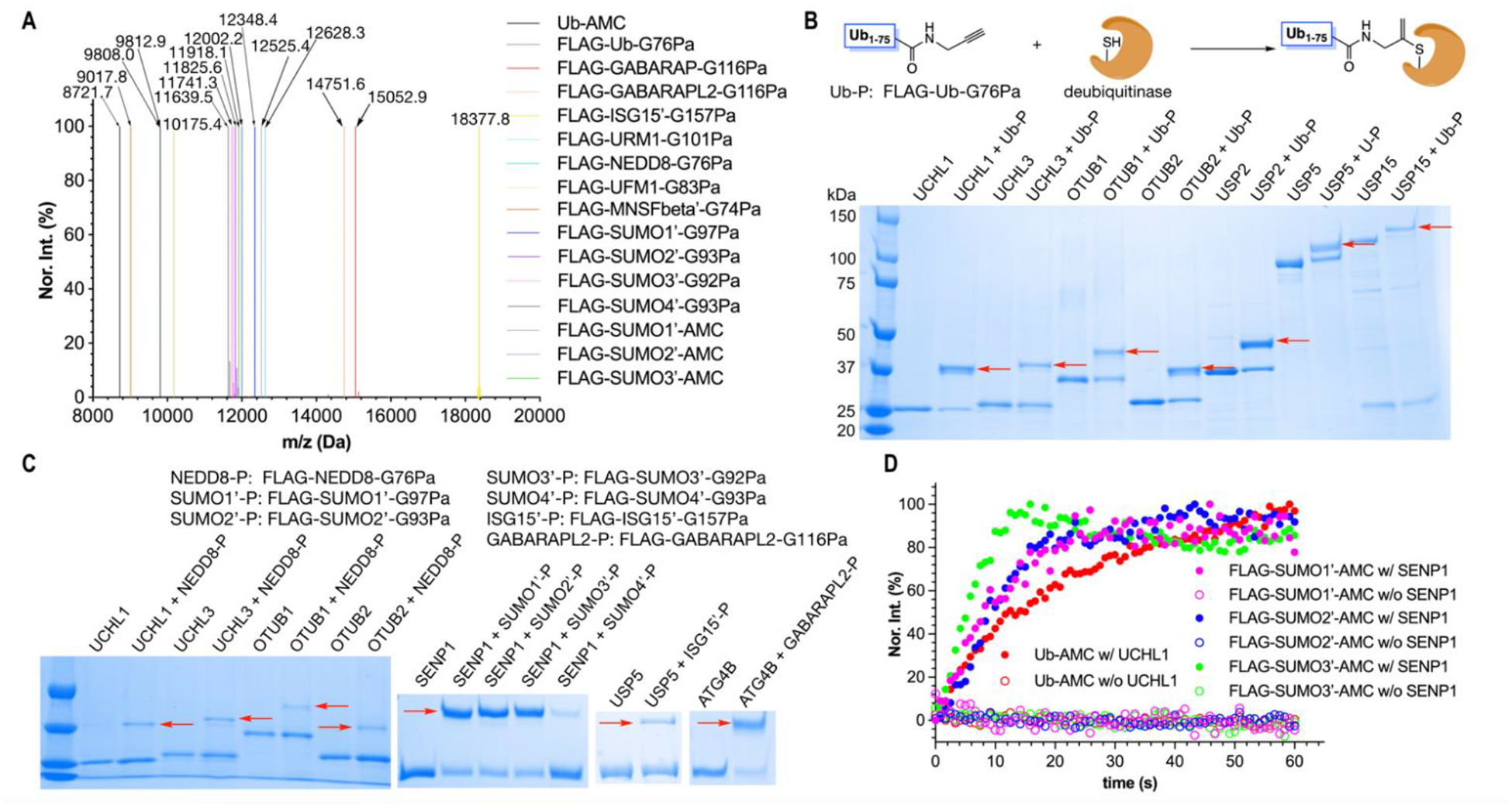
(A) The deconvoluted and integrated ESI-MS of FLAG-Ub/Ubl-Pa and Ub/FLAG-SUMO1–3-AMC probes. Ub-AMC was synthesized from Ub-G76C-6H. All other Pa- and AMC-conjugated Ub/Ubls were generated from FLAG-tagged proteins. Ub/Ubls with their C-terminal glycine mutated to cysteine were expressed and purified as a protein fused with a N-terminal FLAG tag and a C-terminal 6×His tag. ISG15, SUMO1–4, and MNSF have a native cysteine residue. This cysteine was mutated to alanine or serine in all six expressed proteins for avoiding side reactions. The label “′” indicates this mutation. All detected molecular weights agreed well with their theoretic values with a deviation range of 0.5 Da. (B) The formation of covalent adducts between FLAG-Ub-G76Pa and a number of DUBs. Red arrows point to the generated adducts. (C) The formation of covalent adducts, indicated by red arrows, between different FLAG-Ubl-GxPa probes and DUB/ULPs. (D) The DUB/ULP-catalyzed AMC release from Ub-AMC and three FLAG-SUMO-AMC conjugates.
Ub and Ubls conjugated directly to 7-amido-4-methylcoumarin (AMC) at their C-terminus are useful fluorogenic substrates of DUBs and ULPs37. To demonstrate the synthesis of Ub/Ubl-AMC conjugates using our ACPL technique, we made Ub-AMC and FLAG-SUMO1–3-AMC by reacting recombinantly produced Ub-G76C-6H and FLAG-SUMO1–3-GxC-6H proteins with Gly-AMC in the presence of NTCB. The ESI-MS analysis of all four products confirmed their successful formation (Figures 3A and S65–S68) and the following activity assays showed that they served as active substrates for cysteine proteases UCHL1 and SENP1, respectively (Figure 3D). Overall, our combined data of Ub/Ubl probe synthesis establish the broad application scope of the ACPL technique and this technique can make Ub/Ubl probes readily available in a manner that can be performed in almost any biology lab for advancing Ub and Ubl biology studies.
The Use of ACPL to Synthesize H2AK129ac
To further demonstrate its broad application scope, we also carried out ACPL to make non-ubiquitin or non-ubiquitin-like proteins. In human cells, histone H2A can undergo posttranslational acetylation at its terminal lysine K12938. The functional investigation of this acetylation such as how it influences the structure and dynamics of the nucleosome will require the synthesis of the corresponding acetyl-histone, H2AK129ac. We chose to synthesize H2AK129ac to demonstrate that our method can be applied to the synthesis of histones with C-terminal modifications. As a matter of fact, except H3 that natively has a cysteine, all other core histones are lack of cysteine residues. We first recombinantly produced H2A-K129C-6H, an H2A protein with a K129C mutation and a C-terminal 6×His tag and then ligated it to Nε-acetyl-lysine with the assistance of NTCB. The ESI-MS spectrum of the reaction product showed the formation of H2AK129ac (Figures 4A and S69–S70; Table S5) with an at least 60% yield. We folded successfully H2AK129ac into a dimer with H2B and subsequently into a nucleosome (Figure 4B) making it possible to study effects of H2AK129ac on the nucleosome structure and function. In the ESI-MS spectrum of H2AK129ac in Figure 4A, we noticed a minor peak at 13813.6 Da. H2A has two lysine residues, K125 and K127 that are adjacent to K129. Both crystal and cryo-EM structures have indicated all three lysines are located at a flexible C-terminal region of H2A39–40. Potentially either K125 or K127 can undergo intramolecular cyclization with an activated K129C. These intramolecular cyclization products will have a theoretical molecular weight (13813.9 Da) that matches the minor peak in the ESI-MS spectrum. Based on the determined intensities of peaks in the ESI-MS spectrum, this minor peak was lower than 15% of the desired product demonstrating the selectivity of ACPL. By tuning the reaction conditions, this minor peak might be further reduced.
Figure 4. The synthesis of H2AK129ac and RNase H by ACPL.
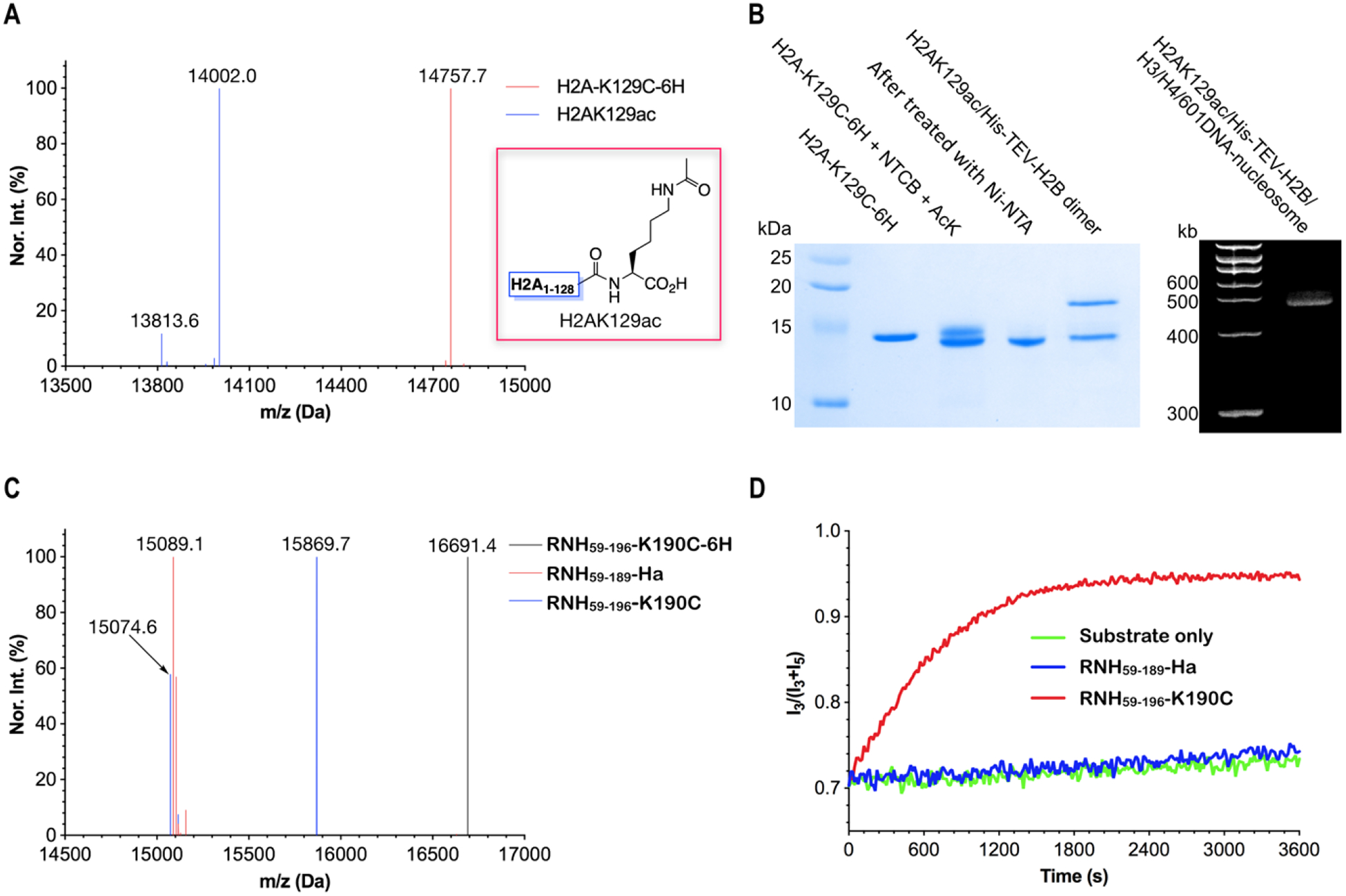
(A) The deconvoluted and integrated ESI-MS spectra of H2A-K129C-6H and H2AK129ac. H2A-K129C-6H was recombinantly expressed and then reacted with NTCB and Nε-acetyl-lysine to afford H2AK129ac. (B) The synthesis of H2AK129ac, its isolation, and folding into an H2AK129ac/H2B dimer and then a nucleosome. The purification of H2AK129ac was achieved by extracting the unreacted intermediate using Ni charged resins. (C) The deconvoluted and integrated ESI-MS spectra of RNH59–196-K190C-6H, RNH59–189-Ha, and RNH59–196-K190C. RNH59–196-K190C-6H was recombinantly expressed in E. coli. It was reacted with NTCB and Ha to afford RNH59–189-Ha that then underwent peptide hydrazide ligation with a 7-mer NH2-CADYGRK-OH peptide to form a catalytic active RNH59–196-K190C. (D) The catalytic hydrolysis of an RNA substrate by RNH59–196-K190C. The RNA substrate had a sequence 5’-Cy3-GACACCUGAUUC-Cy5–3’. A DNA fragment 5’-GAATCAGGTGTC-3’ was used to form a double strand with the RNA substrate for binding to RNH59–196-K190C. The hydrolysis led to improved Cy3 (I3) and decrease Cy5 (I5) emission.
The Use of ACPL in Combination with Peptide Hydrazide Ligation to Synthesize an Active RNase H
For all ligation reactions that we performed thus far, they involved small molecules with only one amino group for avoiding side product formation. For ligation with larger molecules or peptides that have more than one amino group, one can couple ACPL with peptide hydrazide ligation to resolve non-specificity issues. To demonstrate this prospect, we recombinantly produced a B. halodurans RNase H region with a C-terminal Cys-6×His tag (RNH59–196-K190C-6H). Its ligation with Ha in the presence of NTCB led to the synthesis of RNH59–189-Ha, a protein hydrazide that we proceeded further to undergo peptide hydrazide ligation with a 7-mer peptide, NH2-CADYGRK-OH to afford a ligated product RNH59–196-K190C41. ESI-MS analysis showed the successful synthesis of both RNH59–189-Ha and RNH59–196-K190C (Figures 4C and S71–S73; Table S5). Similar to what has been found in previous peptide hydrazide ligation reactions, we also detected a minor hydrolysis product at 15074.6 Da42. The ligated product RNH59–196-K190C was catalytically active to hydrolyze an RNA substrate as shown in Figure 4D. In the contrary, RNH59–189-Ha was completely inactive toward this substrate. Our data related to the synthesis of RNase H demonstrated that ACPL can couple to peptide hydrazide ligation for conjugation with large peptides or even protein fragments.
The Use of ACPL to Synthesize Exenatide, an Anti-Diabetic Medication from a Recombinant Precursor
Exenatide is a 39-mer commercial anti-diabetic peptide drug that has a C-terminal amide. The presence of this C-terminal amide makes it difficult to produce exenatide using the recombinant expression technique43. This bottleneck can be presumably resolved using our ACPL technique by recombinant expression of a precursor protein and then an ACPL reaction with serinamide. To demonstrate this potential, we expressed a 6×His-SUMO-exenatide-S39C-SA-Strep fusion protein that can be largely produced in E. coli followed by the treatment with SUMO protease to obtain the exenatide-S39C-SA-Strep peptide. We then proceeded to carry out its reaction with L-serinamide in the presence of NTCB, purified the final product with a Strep-Tactin column, and analyzed it by ESI-MS. Our results showed that exenatide can be easily procured using this approach (Figures S74–S75).
CONCLUSION
In summary, we have developed a novel expressed protein ligation technique that uses a cyanylating reagent to directly activate a cysteine in a recombinant protein for ligation with small molecule amines and large peptide or protein fragments when coupling with peptide hydrazide ligation. These small molecule amines are primary or secondary and include a number of L- and D-amino acids and functional amines such as Ha, Aa, Pa, and Gly-AMC. The technique that is termed ACPL requires no enzymatic catalysis and is controllable, versatile, specific, and very simple to process. Therefore, it can be broadly applied to synthesize a large variety of proteins with unique functionalities for advanced applications in both basic and applied research. One potential industrial application of the technique is to synthesize therapeutic peptides or proteins like exenatide. ACPL requires the activation of cysteine, one of the two lowest occurring amino acids in proteins. Non-targeted cysteines need to be mutated. For proteins with essential cysteines, one solution for using ACPL is to couple it with the noncanonical amino acid mutagenesis technique. Photocaged cysteines have been genetically incorporated into proteins by amber suppression44–45. The incorporation of a photocaged cysteine to essential cysteine sites in a protein followed by ACPL and then decaging to release protected essential cysteines will allow the processing of proteins with non-targeted cysteines. Overall, our ACPL technique expands to a large extent the synthetic capacity of protein chemistry and will energize the whole field. We anticipate its broad applications in a large variety of research fields and industrial processing of proteins and peptides.
Supplementary Material
ACKNOWLEDGMENT
We thank Dr. Yohannes H. Rezenom who is the director of Texas A&M University Chemistry Mass Spectrometry Facility for helping us on the mass spectrometry analysis. This work was supported by National Institutes of Health (grants R01GM127575 and R01GM121584) and Welch Foundation (A-1715).
Footnotes
The authors declare no competing financial interests. All data supporting the findings in this article are available in the main text or the supplementary information. The two Python scripts are available upon request.
REFERENCES
- 1.Dawson PE; Muir TW; Clark-Lewis I; Kent S, Synthesis of proteins by native chemical ligation. Science 1994, 266 (5186), 776–779. [DOI] [PubMed] [Google Scholar]
- 2.Muir TW; Sondhi D; Cole PA, Expressed protein ligation: a general method for protein engineering. Proc. Natl. Acad. Sci. U. S. A 1998, 95 (12), 6705–6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fang GM; Li YM; Shen F; Huang YC; Li JB; Lin Y; Cui HK; Liu L, Protein chemical synthesis by ligation of peptide hydrazides. Angew. Chem. Int. Ed. Engl 2011, 50 (33), 7645–7649. [DOI] [PubMed] [Google Scholar]
- 4.Flood DT; Hintzen JC; Bird MJ; Cistrone PA; Chen JS; Dawson PE, Leveraging the Knorr Pyrazole Synthesis for the Facile Generation of Thioester Surrogates for use in Native Chemical Ligation. Angew. Chem. Int. Ed. Engl 2018, 130 (36), 11808–11813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muralidharan V; Muir TW, Protein ligation: an enabling technology for the biophysical analysis of proteins. Nat. Method 2006, 3 (6), 429. [DOI] [PubMed] [Google Scholar]
- 6.Shogren-Knaak M; Ishii H; Sun JM; Pazin MJ; Davie JR; Peterson CL, Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 2006, 311 (5762), 844–7. [DOI] [PubMed] [Google Scholar]
- 7.McGinty RK; Kim J; Chatterjee C; Roeder RG; Muir TW, Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature 2008, 453 (7196), 812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chu N; Salguero AL; Liu AZ; Chen Z; Dempsey DR; Ficarro SB; Alexander WM; Marto JA; Li Y; Amzel LM, Akt kinase activation mechanisms revealed using protein semisynthesis. Cell 2018, 174 (4), 897–907. e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwarzer D; Cole PA, Protein semisynthesis and expressed protein ligation: chasing a protein’s tail. Curr. Opin. Chem. Biol 2005, 9 (6), 561–569. [DOI] [PubMed] [Google Scholar]
- 10.Pickin KA; Chaudhury S; Dancy BC; Gray JJ; Cole PA, Analysis of protein kinase autophosphorylation using expressed protein ligation and computational modeling. J. Am. Chem. Soc 2008, 130 (17), 5667–5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dann GP; Liszczak GP; Bagert JD; Müller MM; Nguyen UT; Wojcik F; Brown ZZ; Bos J; Panchenko T; Pihl R, ISWI chromatin remodellers sense nucleosome modifications to determine substrate preference. Nature 2017, 548 (7669), 607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rak A; Pylypenko O; Durek T; Watzke A; Kushnir S; Brunsveld L; Waldmann H; Goody RS; Alexandrov K, Structure of Rab GDP-dissociation inhibitor in complex with prenylated YPT1 GTPase. Science 2003, 302 (5645), 646–650. [DOI] [PubMed] [Google Scholar]
- 13.Stevens AJ; Sekar G; Shah NH; Mostafavi AZ; Cowburn D; Muir TW, A promiscuous split intein with expanded protein engineering applications. Proc. Natl. Acad. Sci. U. S. A 2017, 114 (32), 8538–8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amitai G; Callahan BP; Stanger MJ; Belfort G; Belfort M, Modulation of intein activity by its neighboring extein substrates. Proc. Natl. Acad. Sci. U. S. A 2009, 106 (27), 11005–11010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oeemig JS; Zhou D; Kajander T; Wlodawer A; Iwaï H, NMR and crystal structures of the Pyrococcus horikoshii RadA intein guide a strategy for engineering a highly efficient and promiscuous intein. J. Mol. Biol 2012, 421 (1), 85–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evans TC; Benner J; Xu M-Q, The in vitro ligation of bacterially expressed proteins using an intein from Methanobacterium thermoautotrophicum. J. Biol. Chem 1999, 274 (7), 3923–3926. [DOI] [PubMed] [Google Scholar]
- 17.Vila-Perelló M; Liu Z; Shah NH; Willis JA; Idoyaga J; Muir TW, Streamlined expressed protein ligation using split inteins. J. Am. Chem. Soc 2012, 135 (1), 286–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henager SH; Chu N; Chen Z; Bolduc D; Dempsey DR; Hwang Y; Wells J; Cole PA, Enzyme-catalyzed expressed protein ligation. Nat. Method 2016, 13 (11), 925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okamoto R; Morooka K; Kajihara Y, A Synthetic Approach to a Peptide α‐Thioester from an Unprotected Peptide through Cleavage and Activation of a Specific Peptide Bond by N‐Acetylguanidine. Angew. Chem. Int. Ed. Engl 2012, 51 (1), 191–196. [DOI] [PubMed] [Google Scholar]
- 20.Miyajima R; Tsuda Y; Inokuma T; Shigenaga A; Imanishi M; Futaki S; Otaka A, Preparation of peptide thioesters from naturally occurring sequences using reaction sequence consisting of regioselective S‐cyanylation and hydrazinolysis. Peptide Science 2016, 106 (4), 531–546. [DOI] [PubMed] [Google Scholar]
- 21.Rimington C, The relation between cystine yield and total sulphur in wool. Biochem. J 1929, 23 (1), 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farnworth A; Speakman J, Reactivity of the Sulphur Linkage in Wool. Nature 1949, 163 (4151), 798.18128456 [Google Scholar]
- 23.Wood JL; Catsimpoolas N, Cleavage of the peptide bond at the cystine amino group by the action of cyanide. J. Biol. Chem 1963, 238 (8), PC2887–PC2888. [PubMed] [Google Scholar]
- 24.Catsimpoolas N; Wood JL, Specific cleavage of cystine peptides by cyanide. J. Biol. Chem 1966, 241 (8), 1790–1796. [PubMed] [Google Scholar]
- 25.Patchornik A; Degani Y; Neumann H, Selective cyanylation of sulfhydryl groups. J. Am. Chem. Soc 1970, 92 (23), 6969–6971. [DOI] [PubMed] [Google Scholar]
- 26.Lees A; Nelson BL; Mond JJ, Activation of soluble polysaccharides with 1-cyano-4-dimethylaminopyridinium tetrafluoroborate for use in protein—polysaccharide conjugate vaccines and immunological reagents. Vaccine 1996, 14 (3), 190–198. [DOI] [PubMed] [Google Scholar]
- 27.Komander D; Rape M, The ubiquitin code. Annu. Rev. Biochem 2012, 81, 203–229. [DOI] [PubMed] [Google Scholar]
- 28.Swatek KN; Komander D, Ubiquitin modifications. Cell Res 2016, 26 (4), 399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cappadocia L; Lima CD, Ubiquitin-like protein conjugation: structures, chemistry, and mechanism. Chem. Rev 2017, 118 (3), 889–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van der Veen AG; Ploegh HL, Ubiquitin-like proteins. Annu. Rev. Biochem 2012, 81, 323–357. [DOI] [PubMed] [Google Scholar]
- 31.Ekkebus R; van Kasteren SI; Kulathu Y; Scholten A; Berlin I; Geurink PP; de Jong A; Goerdayal S; Neefjes J; Heck AJ, On terminal alkynes that can react with active-site cysteine nucleophiles in proteases. J. Am. Chem. Soc 2013, 135 (8), 2867–2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pruneda JN; Durkin CH; Geurink PP; Ovaa H; Santhanam B; Holden DW; Komander D, The molecular basis for ubiquitin and ubiquitin-like specificities in bacterial effector proteases. Mol. Cell 2016, 63 (2), 261–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sommer S; Weikart ND; Linne U; Mootz HD, Covalent inhibition of SUMO and ubiquitin-specific cysteine proteases by an in situ thiol–alkyne addition. Bioorg. Med. Chem 2013, 21 (9), 2511–2517. [DOI] [PubMed] [Google Scholar]
- 34.Paudel P; Zhang Q; Leung C; Greenberg HC; Guo Y; Chern Y-H; Dong A; Li Y; Vedadi M; Zhuang Z, Crystal structure and activity-based labeling reveal the mechanisms for linkage-specific substrate recognition by deubiquitinase USP9X. Proc. Natl. Acad. Sci. U. S. A 2019, 116 (15), 7288–7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Basters A; Knobeloch KP; Fritz G, How USP 18 deals with ISG 15‐modified proteins: structural basis for the specificity of the protease. FEBS J. 2018, 285 (6), 1024–1029. [DOI] [PubMed] [Google Scholar]
- 36.Catic A; Fiebiger E; Korbel GA; Blom D; Galardy PJ; Ploegh HL, Screen for ISG15-crossreactive deubiquitinases. PLoS One 2007, 2 (7), e679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dang LC; Melandri FD; Stein RL, Kinetic and mechanistic studies on the hydrolysis of ubiquitin C-terminal 7-amido-4-methylcoumarin by deubiquitinating enzymes. Biochemistry 1998, 37 (7), 1868–1879. [DOI] [PubMed] [Google Scholar]
- 38.Basu A; Rose KL; Zhang J; Beavis RC; Ueberheide B; Garcia BA; Chait B; Zhao Y; Hunt DF; Segal E, Proteome-wide prediction of acetylation substrates. Proc. Natl. Acad. Sci. U. S. A 2009, 106 (33), 13785–13790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frouws TD; Barth PD; Richmond TJ, Site-Specific disulfide crosslinked nucleosomes with enhanced stability. J. Mol. Biol 2018, 430 (1), 45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsumoto S; Cavadini S; Bunker RD; Grand RS; Potenza A; Rabl J; Yamamoto J; Schenk AD; Schübeler D; Iwai S, DNA damage detection in nucleosomes involves DNA register shifting. Nature 2019, 571 (7763), 79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nowotny M; Gaidamakov SA; Crouch RJ; Yang W, Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell 2005, 121 (7), 1005–1016. [DOI] [PubMed] [Google Scholar]
- 42.Zheng JS; Tang S; Qi YK; Wang ZP; Liu L, Chemical synthesis of proteins using peptide hydrazides as thioester surrogates. Nat Protoc 2013, 8 (12), 2483–95. [DOI] [PubMed] [Google Scholar]
- 43.Raufman J-P, Bioactive peptides from lizard venoms. Regul. Pept 1996, 61 (1), 1–18. [DOI] [PubMed] [Google Scholar]
- 44.Wu N; Deiters A; Cropp TA; King D; Schultz PG, A genetically encoded photocaged amino acid. J. Am. Chem. Soc 2004, 126 (44), 14306–14307. [DOI] [PubMed] [Google Scholar]
- 45.Nguyen DP; Mahesh M; Elsässer SJ; Hancock SM; Uttamapinant C; Chin JW, Genetic encoding of photocaged cysteine allows photoactivation of TEV protease in live mammalian cells. J. Am. Chem. Soc 2014, 136 (6), 2240–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


