Abstract
The human sodium iodide symporter (hNIS) is a theranostic reporter gene which concentrates several clinically approved SPECT and PET radiotracers and plays an essential role for the synthesis of thyroid hormones as an iodide transporter in the thyroid gland. Development of hNIS mutants which could enhance translocation of the desired imaging ions is currently underway. Unfortunately, it is hindered by lack of understanding of the 3D organization of hNIS and its relation to anion transport. There are no known crystal structures of hNIS in any of its conformational states. Homology modeling can be very effective in such situations; however, the low sequence identity between hNIS and relevant secondary transporters with available experimental structures makes the choice of a template and the generation of 3D models nontrivial. Here, we report a combined application of homology modeling and molecular dynamics refining of the hNIS structure in its semioccluded state. The modeling was based on templates from the LeuT-fold protein family and was done with emphasis on the refinement of the substrate-ion binding pocket. The consensus model developed in this work is compared to available biophysical and biochemical experimental data for a number of different LeuT-fold proteins. Some functionally important residues contributing to the formation of putative binding sites and permeation pathways for the cotransported Na+ ions and I− substrate were identified. The model predictions were experimentally tested by generation of mutant versions of hNIS and measurement of relative (to WT hNIS) 125I− uptake of 35 hNIS variants.
Graphical Abstract
I. INTRODUCTION
Sodium (Na+) iodide symporter, or NIS, is a secondary transporter from the SLC5 sodium/solute symporter family, which includes several sodium-dependent glucose transporters such as hSGLT1 and hSGLT2.1 NIS is found predominantly in the basolateral membranes of thyrocytes, where it transports the I− ions necessary for the biosynthesis of thyroid hormones against their concentration gradient by coupling to the symport of Na+ ions along their electrochemical gradient. NIS is expressed also in a variety of organs, including the stomach, intestines, salivary glands, placenta, lactating breast, lungs, and testes, where its role is not well understood.2 Iodide transport defects (ITD) in the function of human NIS (hNIS) due to missense pathological mutations and deletions result in iodide deficiency and disorders like goiter and congenital hypothyroidism, which can lead to lifelong cognitive and physical disability. Several mutations related to ITD have been reported in patients.3
Electrophysiology studies performed on rat NIS showed that it enables secondary transport for a number of monovalent anions with remarkable differences in size and molecular shape (e.g., I−, NO3−, SCN−, TcO4−, BF4−).2,4,5 The common environmental pollutant perchlorate (ClO4−), which is also a transported substrate of NIS, serves as a potent competitive inhibitor of the transport of I− and other NIS substrates.6
Since hNIS is expressed and functional in differentiated thyroid cancers, some breast cancers, and cholangiocarcinoma,3 its ability to transport selected positron emission tomography (PET) and single-photon emission computed tomography (SPECT) radiotracers (e.g., 125I−, 99mTcO4−, [18F]-BF4−) can be potentially harnessed for early detection of these cancers.7–10 The transport of 131I− by hNIS has already been used for treatment of Graves disease, toxic nodular goiter, and benign nontoxic goiter and offers potential therapeutic implementations of hNIS in differentiated thyroid tumors, breast cancer, and other malignancies where hNIS is present.3 Importantly, recent advances in gene targeting methods have paved the way for potential use of hNIS for in vitro and in vivo PET and SPECT imaging with selected radiotracers in preclinical models and human subjects.11–14 Maximizing the potential of hNIS for such applications relies on the expression of engineered mutants of hNIS which are transport efficient even in the presence of background ion blockers in the cells of interest (e.g., cancer cells). The administration of the competitive inhibitor ClO4− can effectively suppress radiotracer uptake in all healthy cells expressing wild type hNIS, while cancer cells containing engineered variants of hNIS will accumulate the radiotracer. This procedure would eliminate uptake of radiotracers and cytotoxicity in the healthy tissues expressing wild type hNIS and would prevent background signal interference in the PET/SPECT measurements. Pertechnetate, 99mTcO4−, used in SPECT imaging and tetrafluoroborate, [18F]-BF4− (TFB), used in PET imaging are among the most promising radiotracers of hNIS,15,16 given their short half-life times (6 h for 99mTcO4− and 109.8 min for [18F]-BF4−)10,17 and high quality of the produced imaging signal. Thus, it is desirable that hNIS mutants which are optimized to transport efficiently these two radiotracers in the presence of ClO4− are developed. The rational optimization of hNIS for in vivo or in vitro imaging however is hindered by lack of crystal structures of hNIS and by often-conflicting uptake mechanisms reported for NIS-mediated transport.
Rat NIS, which has a high sequence identity with human NIS, was cloned successfully in 1996 and kickstarted the molecular characterization of the structure and function of NIS.18 Although a crystal structure of NIS is yet to be resolved, a number of studies have elucidated possible structure/activity determinants. Hydropathy analyses, coupled with mutagenesis studies, have shed light on the overall secondary structure of NIS.19 It is accepted that NIS features 13 transmembrane domains (TMs), with an N-terminus in extracellular orientation and a long cytoplasmic C-terminus tail. Glycosylation and phosphorylation positions have been identified in the extracellular loops of TMs 6 and 7 and 12 and 13.19
The transport motif of NIS is composed of TMs 2–11 and likely possesses the 5 + 5 inverse repeat20 found in the crystal structures of the bacterial leucine transporter LeuT21 and various other secondary transporters, including the galactose transporter vSGLT of Vibrio parahemolyticus,22 which is regarded as a homologue of the SLC5 family members hSGLT1 and hSGLT2.1 Moreover, rat NIS is reported to exhibit either electrogenic (Na+:substrate = 2:1) or electroneutral (Na+:substrate = 1:1) transport, depending on the transported anion.2,4–6 Functional mutagenesis studies along with the known pathological ITD mutations have identified several amino acids in NIS as critical for I− transport.23–46 The overall geometry of NIS, the structure of its binding sites, and its transport mechanism, however, remain poorly understood.
Some information about NIS has been obtained from comparison to other proteins which have the same 5 + 5 LeuT-fold as NIS. Among these proteins are two homologues of hNIS, vSGLT, whose structure has been resolved with X-ray diffraction,22 and the proline transporter, PutP, which has been extensively studied with functional mutagenesis.47–61 In the absence of actual crystallographic structures, computational modeling provides an avenue for exploration of structural characteristics and their potential effect on protein function. Validated homology models of PutP, based on vSGLT, have been reported before56,57 and offer the opportunity for structure/activity analysis of PutP in analogy to vSGLT and other LeuT-fold transporters. Homology models of hNIS based on the vSGLT structure have also been reported in recent articles2,3,44–46 without extensive cross-validation. Furthermore, some of the studies did not consider the copermeant Na+ ions that are required for substrate or inhibitor binding.45
Given the potential importance of hNIS as a reporter and therapeutic gene used for early cancer detection and treatment, and the need for better understanding of the transport mechanism in hNIS for construction of engineered strains with enhanced selectivity for I−, 99mTcO4−, and [18F]-BF4−, even in the presence of competitive inhibitors like ClO4−, we report here a thoroughly validated homology model of hNIS, consistent with the available experimental data. Several hNIS models based on different combinations of LeuT-fold templates were constructed, and a combination of Grand Canonical Monte Carlo (GCMC)62 and all-atom molecular dynamics simulations was employed with the aim of identification of putative binding sites for Na+ and I− and other structural elements of relevance to transport. The molecular simulations were used to guide mutagenesis and uptake experiments by mapping residues essential for substrate and copermeant ion binding. The final validated structure of our human NIS model is available upon request.
II. METHODOLOGY
Choice of Templates
It has been suggested that hNIS has a similar architecture as the vSGLT galactose transporter,2 a bacterial orthologue of hSGLT1 and hSGLT2 from the SLC5 protein family to which hNIS belongs.1 The sequence similarity, however, remains relatively low (<35%). vSGLT features the typical 5 + 5 LeuT-fold inverted repeat which is found in a number of proteins transporting a wide range of diverse substrates.20 For sequence alignment, necessary for the construction of the homology models of hNIS, we chose six LeuT-fold proteins crystallized in the occluded or semioccluded state, including vSGLT22 (Na+-coupled galactose transporter, PDB code 3DH4), LeuT21 (Na+-coupled leucine transport, PDB code 2A65), AdiC63 (Na+-independent arginine/agmantine antiport, PDB code 3L1L), Mhp164 (Na+-coupled hydantoin transport, PDB code 4D1B), BetP65 (Na+-coupled betaine transport, PDB code 4DOJ), and dDAT66 (Na+-coupled dopamine transport, PDB code 4XPA). The substrates of these templates are neutral or positively charged organic molecules of different sizes and shapes and have very different physical and chemical properties from iodide. Therefore, for construction of alternative models of hNIS and assessment of its overall 3D structure, we also used transporters involved in selective anion uptake with available high-resolution X-ray or cryo-EM structures such as the bicarbonate transporters (hAE167 and hNBCe168) from the SLC4 family and the chloride/proton antiporter CLC-ec169 from the CLC family of chloride channels and transporters. The sequence identity for hNIS and all of the selected templates is low (20%–30%). The highest sequence identity was observed with the LeuT-fold proteins, especially for the 5 + 5 motifs.
Multiple Sequence Alignment Protocols
Three separate alignments were made for hNIS with respect to each of the available protein folds (LeuT-fold with six templates, SLC4-fold with two templates, and CLC-fold with one template). The multiple sequence alignments were performed with the align tool of Modeller 9.18,70 which overlaps the template crystal structures and uses structural information from this overlap to assign the amino acid sequences to helices and loops. In the case of the LeuT-fold-based models, to improve alignment in the ion/substrate binding domain of hNIS, the putative sequences of TMs 1, 12, and 13 of hNIS were removed, and the remaining 5 + 5 fold of hNIS (residues 50–439) was aligned with the 5 + 5 folds of the six selected templates. TMs 1 and 12 of hNIS were modeled after TMs 1 and 12 of vSGLT (from PDB codes 2XQ2 and 3DH4, respectively), since the rest of the templates showed significant differences in this area. TM13 of hNIS was excluded from modeling with the LeuT-fold templates due to poor sequence identity with all templates. The refined alignment for the LeuT-fold models is presented in Figure S1 of the Supporting Information. For the models based on SLC4 and CLC templates, we prepared alignments using the full hNIS sequence. The sequence identity in both cases was lower than 20%.
3D Threading and Refinement of Models
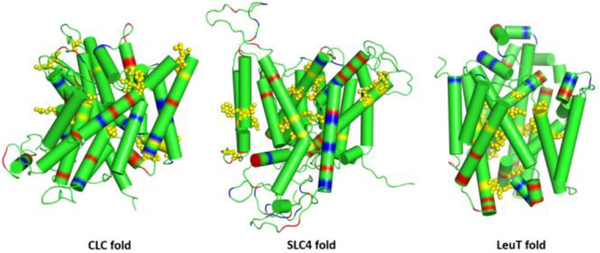
3D models of hNIS with the three different protein folds (CLC, SLC4, and LeuT) were constructed with Modeller 9.1870 and then used to map key transport residues known from functional mutagenesis in NIS. The three models based on the different templates are presented in Figure 1. Only the LeuT-fold model of hNIS satisfied basic considerations of protein structure and provided structural consistency with the mutagenesis data. Therefore, only LeuT-fold templates were used for further modeling of hNIS. 3D structures of hNIS were built using the automodel function in Modeller 9.1870 with alpha helical constraints imposed on residues 85–99, 279–296, 393–401, 409–425, and 428–438 for better reproduction of the helical transmembrane domains. LeuT,21 AdiC,63 BetP,65 and dDAT66 show notable differences from vSGLT22 and Mhp164 in the structure and orientation of TM11 which implies differences in the gating and overall transport mechanism. Such subtle mechanistic differences are also inferred from DEER measurements on Mhp1 and LeuT.20 After assessment of similarities in the 3D structure of the six overlapped LeuT-fold templates, they were divided in two groups: vSGLT-like (including vSGLT and Mhp1) and LeuT-like (including LeuT, AdiC, BetP, and dDAT). Three LeuT-fold models of hNIS were generated afterward: a vSGLT-like model based on vSGLT and Mhp1, a LeuT-like model based on LeuT, AdiC, BetP, and dDAT, and a mixed model based on all six templates. For each model, 500 structures were generated with Modeller 9.18, using automodel, and the quality of the structures was assessed with pdfpdb,70 DOPE,71 and GA34172 scores. Afterward, 5 to 10 structures with the lowest DOPE and highest GA341 scores were selected and overlapped with the templates for assessment of overall backbone structure and position of residues identified as important from experimental data (X-ray diffraction where possible and functional mutagenesis).
Figure 1.
Comparison of three different 3D hNIS models built with protein templates from the chloride transporter CLC-ec1 (PDB: 1KPK), the bicarbonate transporters hAE1 (PDB: 4YZF) and hNBCe1 (PDB: 6CAA) from the SLC4 family, and the LeuT-fold proteins vSGLT (PDB: 3DH4) and Mhp1 (PDB: 4D1B). The position of acidic and basic residues is indicated with red and blue colors, respectively. Key residues for iodide uptake identified in hNIS from functional mutagenesis are shown as yellow spheres.
Comparison of the three different types of hNIS models revealed significant structural differences in the area of extracellular gating (Figure S2), with the LeuT-like and mixed hNIS models adopting helical organization closer to the helical organization in LeuT. Considering the homology between vSGLT and hNIS, only the vSGLT-like model was then used for further structural refinement and MD simulations. The three vSGLT-like structures with the best side-chain overlap with the corresponding residues of interest in the vSGLT and Mhp1 templates were selected for further side-chain relaxation with ROSETTA MP.73
Side-Chain Relaxation with ROSETTA
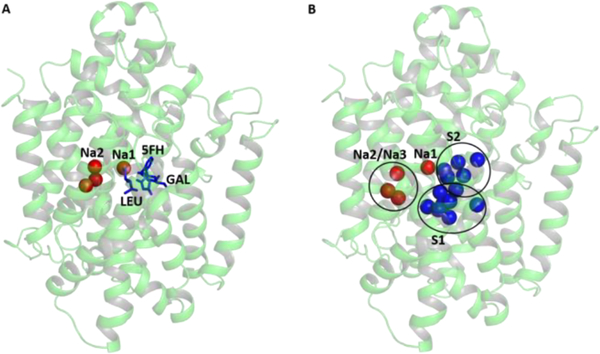
The three vSGLT-like hNIS structures selected from the previous step were subjected to side-chain relaxation with the ROSETTA MP protocols73 in the presence of an implicit membrane. The span files for the membrane were prepared manually with topology based on the initial 3D structures of hNIS. 2000 decoy structures were generated for each of the three individual hNIS models and were scored with the mpframework_smooth_fa_2012 function. The resultant structures were clustered with the cluster tool provided in ROSETTA. The center of the largest lowest energy cluster was chosen as a representative hNIS model for the remaining GCMC and MD simulations and is displayed in Figure 2A.
Figure 2.
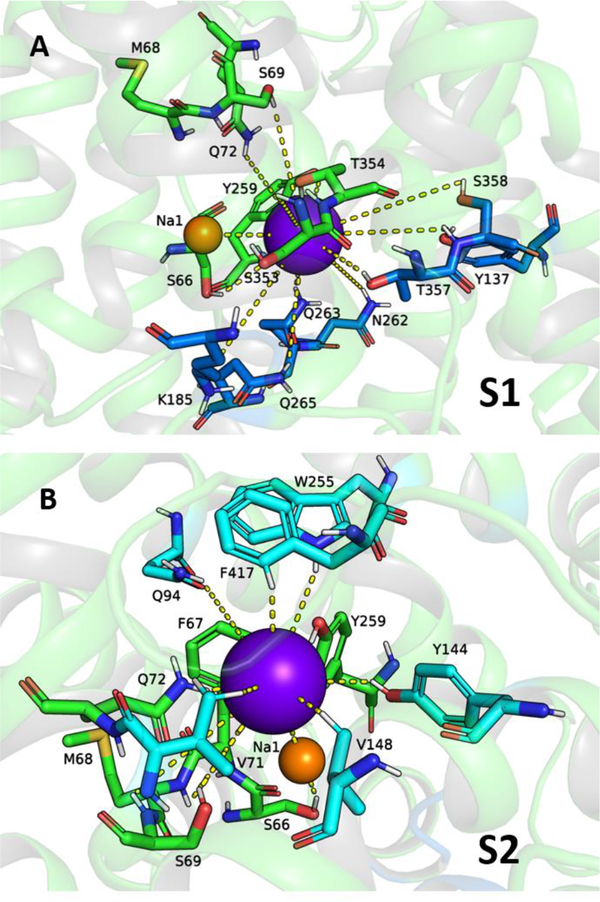
(A) 3D model of hNIS based on the vSGLT and Mhp1 templates. The positions of the bound Na+ ions in the putative sites Na1 and Na2 (red spheres) and substrates (blue sticks) in the available crystal structures of LeuT, vSGLT, and Mhp1 are shown overlapped with the 3D model of hNIS. (B) Putative binding sites for cations (red spheres) and anionic substrates (blue spheres) in hNIS determined from combination of GCMC/PB calculations.
Putative Binding Sites from GCMC and PB Mapping
Identification of binding sites for ions in the protein scaffold is challenging. This work adapts a strategy used previously to map binding sites in the GltPh transporter.74 Briefly, water molecules were used to sample polar cavities in the protein core with a combination of GSBP/GCMC algorithms allowing flexibility of the reduced protein system and representing the membrane/solvent environment implicitly.62,75 For the Grand Canonical Monte Carlo (GCMC) calculations, only the protein residues within a sphere with a 15 Å radius centered at the center of mass for the consensus hNIS model were treated explicitly, with the rest of the system represented as generalized solvent boundary potential (GSBP).75 The GCMC simulation was run for 150 cycles with 5000 MC steps at 300 K with excess chemical potential (μ) for the external water reservoir set to −6.2 kcal/mol. The produced hNIS structures were relaxed with Langevin dynamics for 2 ps per GCMC cycle. Here, 50 relaxed structures from the GCMC calculations were then randomly selected and submitted for electrostatic potential calculations using linear Poisson–Boltzmann (PB) equations and an implicit membrane default setup from the PBEQ solver tool in the CHARMM-GUI server.76,77 The resulting electrostatic density maps were overlapped with the water density map evaluated from the GCMC trajectories, and the areas of highest water density in the putative binding cavity were assigned as either positively charged (putative anion binding area) or negatively charged (putative cation binding area). The position of the putative anion and cation binding sites are shown in Figure 2B as blue or red spheres, respectively.
Refinement of Binding Sites with MD Simulations
The putative binding sites identified from the GCMC/PB calculations were used for further assessment of Na+ and I− binding in hNIS with MD simulations. The MD models included the two proposed transport stoichiometries for rat NIS (1:1 Na+:substrate and 2:1 Na+:substrate)6,78 and were tested with both I− and ClO4− as substrates. In all simulations, one Na+ was placed in a negatively charged area of the protein identified from our GCMC/PB calculations which corresponds to the Na2 binding site in the crystal structure of vSGLT.22 The models with 2:1 Na+:substrate stoichiometry had a second Na+ ion bound at the site with the negative electrostatic potential overlapping with the location of the Na1 site in LeuT21 (Figure 2B). For each anionic substrate (I− or ClO4−) and each tentative transport stoichiometry (1:1 and 2:1 Na+:substrate), 13 different substrate binding positions in the large slightly positive substrate binding cavity of hNIS, inferred from our GCMC calculations, were used as the initial guess for MD simulations (Figure 2B, blue spheres). In addition, all calculations were done with either no water molecules present in the substrate binding cavity at the initial MD step or with 12 water molecules at the remaining 12 out of the predicted 13 GCMC/PB substrate positions (excluding the site for the bound anion). Thus, a total of 104 separate models were constructed for all-atom MD simulations aimed at the refinement of binding sites in hNIS.
The hNIS structures with the respective ions and (where applicable) water molecules were then positioned in a tetragonal periodic box of size 90.44 Å × 90.44 Å × 101.83 Å, containing a POPC bilayer (95 and 92 POPC molecules in upper and lower leaflet, respectively), water (20 Å layers), and 0.15 M KCl solution with the CHARMM-GUI server.76,79,80 The orientation of the hNIS protein in the lipid bilayer was based on the vSGLT membrane orientation evaluated from the OPM server.81 The prepared hNIS models were equilibrated with NAMD 2.1282 following the standard six-step protocol for constrained equilibration.83 The CHARMM36 force field was used for protein, lipids, counterions, and water molecules.84,85 Iodide parameters were taken from ref 86. CHARMM-compatible parameters were developed for ClO4− using the GAAMP protocol87 (Table S1). The 50 ns long MD trajectories for each of the developed hNIS models were performed in the NPaT ensemble at T = 310 K and P = 1 atm. The aggregate sampling time was 5.2 μs. The trajectories were then analyzed with in-house tcl scripts developed for VMD 1.9.3.88
Experimental Protocols
For further validation of our hNIS model, selected residues mapped as important for Na+ or substrate binding or gating modification in hNIS were evaluated experimentally for their roles in transporter uptake efficiency. We focus our experimental studies on positions/substitutions without published data.
Site-Directed Mutagenesis
Targeted mutagenesis of specific residues in human WT hNIS was performed using a QuikChange Lightning Site-Directed Mutagenesis kit (Agilent Technologies, Santa Clara, CA). Nucleotide substitutions were confirmed by sequencing analysis. A human influenza hemagglutinin (HA) tag was added at the C-terminus of hNIS to enable detection of mutant hNIS protein expression using an anti-HA tag antibody. The hNIS mutants were subcloned into a lentiviral expression vector.
Generation of Stable Cell Line
VSV.G pseudotyped lentiviral vectors encoding human NIS WT or human NIS mutants were generated by triple transfection of packaging and vector plasmids with FuGENE HD (Promega, Madison, WI) into 293T cells as previously described.89 Titers of lentiviral vectors were determined by transduction of HeLaH1 cells and quantitated as transducing units/mL using quantitative PCR. HeLaH1 cells stably expressing the various hNIS mutants were generated by transduction of cells at a multiplicity of infection (MOI) of 2. Cells were placed under puromycin selection (0.5 μg/mL). Puromycin-resistant HeLaH1-NIS cells were characterized by flow cytometry analysis with 1:50 dilution of Alexa Fluor 647 conjugated antibodies recognizing the HA Tag 6E2 (Cell Signaling, Danvers, MA). Median fluorescence intensity of Alexa Fluor 647 positive cells was analyzed using the FlowJo (Ashland, OR) software.
In Vitro Uptake Assay
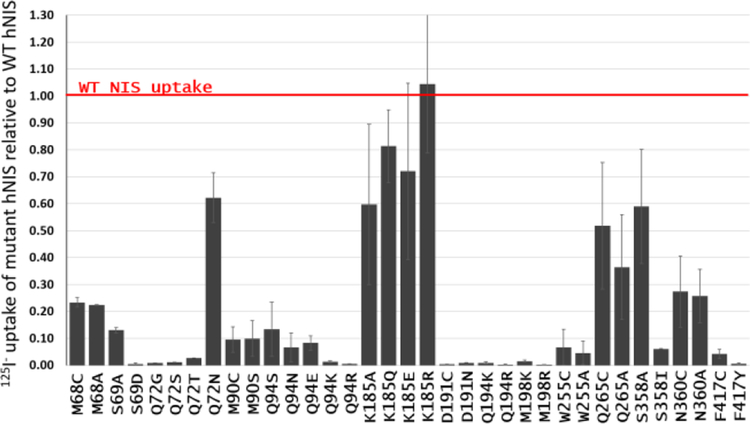
HeLaH1 cells stably expressing human WT hNIS or hNIS mutants were seeded in 12 well plates at a density of 3 × 105 cells/well for 1 day prior to experiments. Cells were washed once in Hanks’ balanced salt solution (HBSS, Gibco) supplemented with 10 mM HEPES (Genesee Scientific, San Diego, CA). The cells were incubated with the 125I− radioisotope (∼100,000 cpm) in a HBSS/HEPES buffer at 37 °C for 1 h. Cells were washed twice with cold HEPES/HBSS and lysed with 1 M NaOH for 10 min. Radioactivity was quantified with a PerkinElmer Wizard-2 2470 gamma counter for 125I−. All the experiments were performed in triplicate and repeated at least three times. To allow comparison between experiments, the uptake of 125I− by the hNIS mutants was normalized with the uptake of 125I− by WT hNIS. Results for the mutants were represented as fractions of the WT hNIS uptake, where the WT hNIS uptake is equivalent to 1 (Figure 3).
Figure 3.
Average 125I− relative uptake of selected hNIS mutants studied in the present work, calculated as fractions of the uptake of cells expressing WT hNIS. Values below 1 signify a decrease in 125I− uptake compared to WT hNIS.
III. RESULTS AND DISCUSSION
Consensus 3D Structural Model of the Semioccluded State of hNIS
Figure 1 displays the 3D hNIS models (cylindrical representation) built with templates from three different protein families: hAE167 and hNBCe168 from the SLC4 family of anion transporters, CLC-ec169 from the CLC family of chloride transporters and channels, and vSGLT22 and Mhp164 from the LeuT-fold transporters. The positions of basic and acidic residues in the models are indicated with blue and red colors, respectively. The hNIS models based on the SLC4 and CLC templates led to regions composed of acidic or basic residues exposed to the hydrophobic core of the membrane. Many of the functionally relevant residues (yellow spheres) mapped from mutagenesis studies appear either in the connecting loops or in the peripheral helices in contradiction to the experimental data reviewed in this work. Only the hNIS model based on the LeuT-fold templates has a structure consistent with the expected membrane protein organization, featuring charged residues concentrated in the hydrophilic loop regions, with the exception of the catalytically relevant D191 which appears to be oriented toward the ion binding cavity. The putative binding residues implied from functional mutagenesis studies are located in the protein core, where the sodium and substrate binding areas have been found in all LeuT-fold transporters with known structures.90 Therefore, hNIS almost certainly has a LeuT protein fold as suggested by previous homology and hydropathy modeling2 instead of a protein fold characteristic for known secondary transporters of anions. The final 3D model of hNIS used in this study (Figure 2) is therefore based on two LeuT-fold proteins (vSGLT and Mhp1), chosen due to their overall structural similarities in the areas of outward-facing and inward-facing gates (which implies similarity of the transport mechanism), the homologous relation to hNIS (in the case of vSGLT),1,2 and the presence of multiple structures in different conformations (in the case of Mhp1).64,91,92
Putative Binding Sites for Cotransported Cations and Substrate in hNIS from Molecular Modeling
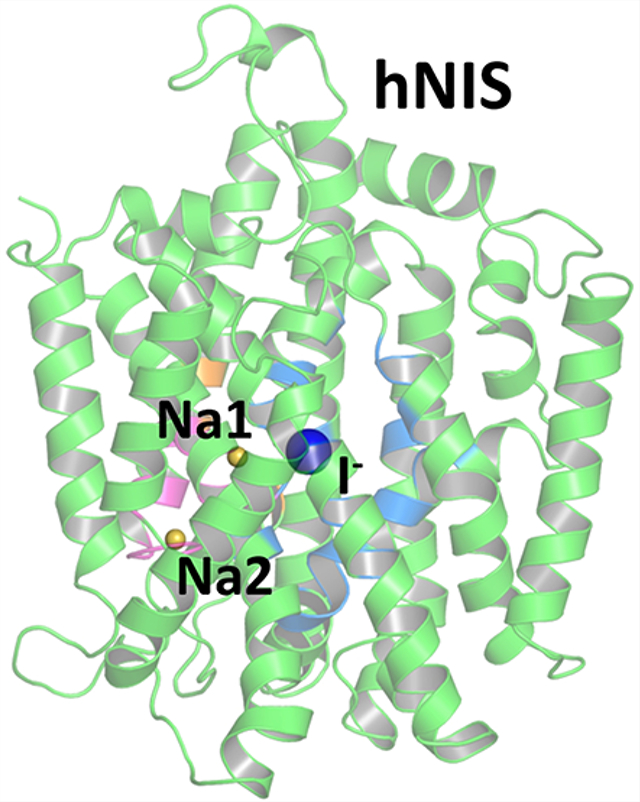
The location of the binding sites in the LeuT-fold family has been well established in several available X-ray structures from various structurally similar proteins1,20,21,63,64,66,90–95 and a plethora of experimental mutagenesis and uptake studies (Table S2). The LeuT-fold proteins feature a variety of transport stoichiometries, where the coupling Na+ ions vary in numbers from n = 0 (e.g., the Na+-independent arginine transporter AdiC)63 to n = 3 (human betaine/GABA transporter).96 In some cases, K+- and Cl−-dependent transport is also observed.66,96–98 Most of the LeuT-fold proteins have at least one Na+ ion bound at a conserved sodium binding site, which corresponds to site Na2 in LeuT.20,99,100 LeuT features also a second sodium ion bound at its Na1 binding site, in immediate proximity to the primary substrate binding site often referred to as the S1 site.21 The hNIS homologues from the human SGLT family use a different number of cations coupled to substrate cotransport depending on the particular family member: hSGLT1 and hSGLT2 couple two or one Na+ ions to glucose transport, respectively.101 Cation binding sites labeled Na1 and Na2 as well as a primary substrate binding site S1 have been identified in several LeuT-fold proteins with available high-resolution X-ray structures.100 A recent structure of an outward-facing open state of the sialic acid transporter, SiaT, which belongs to the LeuT-fold family of proteins, features a new Na+ binding site, labeled as Na3,102 close to site Na2 in vSGLT. The location of the second sodium binding site in hSGLT1 has recently been delegated to the Na3 site.103 The vSGLT and Mhp1 templates used for our hNIS model feature a single sodium ion in the Na2 binding site.22,64 Taken together with the diverse substrates transported by the LeuT-fold proteins, it is evident that the LeuT-fold organization of the transmembrane domains supports a fairly powerful mechanism of secondary transport that can be easily adapted to the chemistry and geometry of many different substrates. The transport stoichiometry for human NIS is as of yet unknown. The available electrophysiological evidence suggests a 2:1 Na+:I− transport stoichiometry in rat NIS and is in support of two cation (Na+) binding sites which we label Na1 and Na2, respectively, in analogy to the Na+ binding sites in LeuT. We have probed both tentative sites Na1 and Na2 in our hNIS model.
While all of the known secondary transporters feature a primary binding site for a substrate (site S1), the existence of a second, presumably allosteric, substrate binding site, S2, has also been proposed in LeuT transporters based on molecular modeling, single-molecule, and functional studies.94,104–107 The LeuT-fold protein, L-Trp transporter MhsT, features two substrate binding sites labeled S1 and S2 using nomenclature introduced for substrate binding sites in LeuT.108 Two substrate binding sites have been identified in the human serotonin transporter hSERT and have been discussed in relation to therapies for mental disorders and addiction.98,109 Existence of a second substrate binding site has been suggested for the galactose transporter, vSGLT, and for the proline transporter, PutP, based on a combination of saturation, mutagenesis, and uptake data.47 vSGLT and PutP are the most likely homologues of the SLC5 protein family and its members hSGLT1, hSGLT2, and hNIS.47 Therefore, hNIS may potentially feature two substrate binding sites for its anionic substrates referred in our work as sites S1 and S2, respectively.
The first step in the elucidation of the potential locations of ion and substrate binding sites in our hNIS model was the collection of excess density maps for a polar molecular probe (water in this work) complemented by ensemble-averaged Poisson–Boltzmann electrostatic potential maps. The combined GCMC/PB data in Figure 2B highlights well-defined binding cavities attractive to cations as well as a well-defined binding pocket for I−. GCMC/PB mapping suggests a potential Na+ binding site in the vicinity of the D191 residue, which corresponds to the Na2 binding site preserved in most known LeuT transporters20,99,100 or the Na3 site in the sialic transporter SiaT.102,103 Another potential sodium binding site, corresponding to site Na1 in LeuT21 was found in the area of S66 and F67. The remaining portion of the cavity is positively charged and expected to attract negative substrates, such as I−. It can be divided into two sections corresponding to the S1 and S2 sites found in vSGLT (Figure 2B).47
To assess in more detail the sodium and substrate binding in hNIS, we ran a number of 50 ns long MD simulation of hNIS where I− or ClO4− were placed in one of the potential substrate binding locations identified from GCMC (13 positions per simulated system, Figure 2B). We constructed models to test transport stoichiometries proposed from the electrophysiological studies in the rat NIS: 2:1 Na+:substrate or 1:1 Na+:substrate.2 In all models, one Na+ ion was placed in the highly preserved Na2 binding site. In the models with the 2:1 Na+:substrate ratio, the second Na+ ion was placed in the putative Na1 binding site, as suggested by our GCMC results (Figure 2B).
The graphs in Figure S3 display the contact frequencies of I−, ClO4−, and Na+ in the Na1 and Na2 sites, with the protein matrix, evaluated from cumulative MD trajectories (e.g., the contact frequencies for iodide are added from all MD trajectories featuring an iodide ion, including those with or without water in the binding cavity at the MD onset and those with 2:1 and 1:1 Na+:I− ratio). The individual contributions for these cumulative graphs are displayed in Figure S4. The protein residues with the highest contact frequencies for each ion are listed in Table S2 of the Supporting Information. The contact frequencies indicate the percent of the MD steps in which an ion of interest can be found within 5 Å from the protein atoms and can be used for identification of protein residues which have frequent contact with the ions (i.e., potential binding sites where the ions tend to reside during the MD simulations). The sodium ion at the Na2 position consistently shows high contact frequencies with a small number of residues indicative of a well-defined binding site and stable ion binding.
Location of the Putative Na2 Site
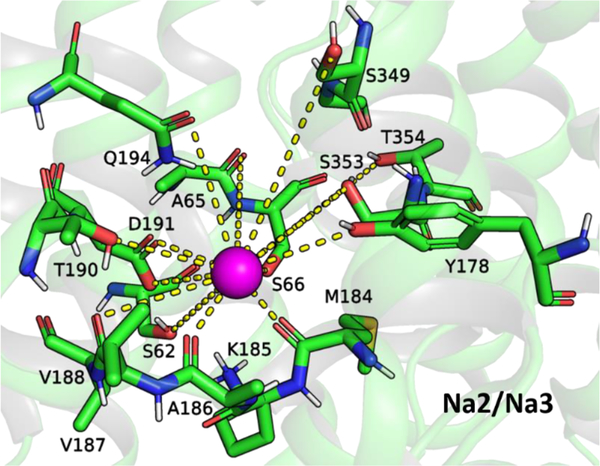
Our findings for the Na2 site in hNIS are mostly consistent with the previously identified Na2 site in a homology model of hNIS based solely on vSGLT.46 The contact frequency patterns for Na+ in site Na2 indicate stable coordination of the Na+ ion by several key residues: S62, A65, S66, M68, Y178 and a nest of carbonyl groups of residues M184–188, T190, D191, Q194, S349, and T354 (Figure 4). Mutations at positions S66, S349, and T354 have proven detrimental to I− uptake.30,38,46 The I− uptake is impacted also by mutations at positions M68, K185, and Q194 (Figure 3).46 The available crystal structures of vSGLT, Mhp1, and LeuT show that their conserved Na+ (site Na2) sodium ion is coordinated by analogues of A65, M68, G350, S353, and T354 in hNIS (Table S2).21,22,64
Figure 4.
Tentative Na2 binding site for Na+ (magenta sphere) in hNIS, based on the contact frequency maps in Figures S3 and S4, the ion density map in Figure S5, and the residues listed in Table S2.
Mutations of D191 in hNIS, which remove the negative charge (D191N, D191C), eliminate transport completely (Figure 3). Residue D191 in hNIS corresponds to D204 in hSGLT1, whose neutralization impairs substrate uptake and leads to the formation of a glucose-gated proton channel.110
Residue D189 in vSGLT, which corresponds to D204 in hSGLT1 and D191 in hNIS, has been shown to interact with the Na+ in site Na2 in MD simulations of the Na+ and substrate exit from the inward-facing vSGLT structure.111–113
Introduction of a positive charge in the vicinity of D191 (Q194R, Q194K, M198R, and M198K shown in Figure 3) leads to drastic decrease in the iodide uptake. It is likely that the positive charge of the substituted amino acid in these cases interferes with Na+ binding at the Na2 binding site and does not adopt the allosteric modulator role of the Na+ ion, as R262 and K158 do in the Na+-independent LeuT-fold transporters CaiT and ApcT, respectively.114,115 Taken together, these results show that a Na+ ion is likely present in hNIS at the areas of TMs 2, 6, and 9, which in our inward-facing open model of hNIS may correspond to either site Na2 in vSGLT or site Na3 in SiaT, and that this presence is determined by the strong electrostatic interaction with a negatively charged residue (conserved in the homologues of hNIS, such as vSGLT, hSGLT1, and PutP) and a number of conserved polar residues (Gln, Ser, Thr) present in many LeuT-fold transporters (Table S2).
Location of the Putative Na1 Site
Among the residues which emerge as potential Na+-coordinating centers in site Na1 are S64, S66, F67, Q72, Y259, L289, and T354, which are also involved in substrate binding (Table S2, Figures S3 and S4, Figure 5). Similar overlap between the substrate and Na1 binding sites has been observed in the crystal structure of LeuT,21 where the Na+ in site Na1 is coordinated by the carboxylate group of the bound Leu substrate and is also inferred from Kd values evaluated from electrophysiology for Na+ and glucose in hSGLT1.113 Unlike the stably bound Na+ (site Na2), the Na+ in site Na1 is fairly mobile, and in the cases of a dehydrated binding cavity often forms an ion pair with the I− anion in the polar substrate binding pocket. Similar behavior has been suggested for the Na+ and CO32− ions bound in the hNBCe1 protein of the SLC4 anion transporter family116 and implies that the Na+ ion in site Na1 may be required for strengthening the anion–protein interactions in the uncharged but slightly polar substrate binding cavity of hNIS, which lacks traditional anion binding residues such as Arg or Lys. Interestingly, the Na+ in site Na1 in LeuT is in proximity to two negative residues (E287 and E290) which are responsible for its coordination and stabilization.21 Molecular dynamics simulations and smFRET studies show that the protonation state of E290 also modulates some of the Na+-coupled conformational changes consistent with allosteric control during the secondary transport in LeuT, which in turns affects the protein affinity for Na+.117–120 The hSGLT1 system, which transports two Na+ ions, has a negatively charged residue (D294) in the vicinity of its putative Na1 site suggested from homology to vSGLT.113 Such residues are missing in vSGLT, PutP, and Mhp1, which have 1:1 Na+:substrate transport stoichiometry.22,56,64 Thus, the I− ion in the hNIS transporter may in turn adopt the role of a negatively charged amino acid residue for stabilization of a second Na+ ion in the polar core of hNIS and may aid in the ion permeation of both ions through the large polar substrate binding cavity after their dehydration. Rehydration of the binding cavity occurs naturally as the inward-facing state opens and is a required step in the ion and substrate exit from the LeuT-fold secondary transporters during the alternate access mechanism.121
Figure 5.
Tentative S1 and S2 binding sites for I− (purple sphere) in hNIS based on the contact frequency maps in Figures S3 and S4, the I− density map in Figure S5, and the residues listed in Table S2. The residues are color coded: green for residues involved in both sites S1 and S2, dark blue for residues unique to site S1, and cyan for residues unique to site S2.
Substrate Binding Sites in hNIS
The I− and ClO4− ions mostly dwell in the positively charged cavity determined from GCMC calculations at least for the duration of the 50 ns long MD simulations performed here. The I− and ClO4− density maps (Figure S5) point at the presence of two major binding regions in the anion-accessible area of the protein. The amino acid composition of these binding sites is presented in Table S2. Comparison to the previously identified hSGLT1 and PutP binding sites (Table S2) established that the two putative substrate binding sites in hNIS are indeed consistent with sites S1 and S2. Figure 5 displays representative coordination of I− within the hNIS core corresponding to the protein areas with the highest anion density. The residues of the binding sites are color coded: dark blue for site S1, cyan for site S2, and green for residues involved in both sites S1 and S2. Coordination of the ClO4− ion is shown in Figure S6 with the same color coding.
The amino acid residues common for both S1 and S2 sites are S66, F67, S69, Q72, Y144, Y259, S353, and T354. Functional mutagenesis and comparison to analogous residues in vSGLT, hSGLT1, PutP, Mhp1, and LeuT show that these residues are essential for substrate uptake. The T354P mutation in hNIS is found in patients with congenital ITDs.25,28 Cysteine or alanine mutations of residues S66 and S353 in rat NIS,38,46,122 as well as S69 in human NIS (Figure 3), lead to a drastic decrease in NIS function. Substitution of S69 with the negatively charged Asp residue eliminates transport. Removal of the amino group at position Q72 (Q72G, Q72S, Q72T) in human NIS is detrimental to I− uptake, even if the substituting residue features a polar OH group, suitable for binding to anions. A Q72N substitution leads to hNIS with lowered but observable I− uptake, hinting at the importance of the presence of an amino group at this position (Figure 3).
PutP mutations to cysteine of the A53-L60 residues, analogous to the A65-Q72 stretch in hNIS, have severe impact on the proline uptake.53 The Cys mutants of PutP are sensitive to inhibition with N-ethylmaleimide (NEM) and are accessible to the fluorescent dye fluorescein-5-maleimide, indicating that they are part of a solvent-accessible cavity, which is likely part of the substrate and ions permeation pathway in PutP. In addition, residues Q72, Y144, S353, Y259, and T354 in hNIS correspond to substrate binding residues found in the substrate-bound crystal structures of vSGLT, Mhp1, and LeuT (Table S2). Residue Y259 is the hNIS analogue of residues Y263 in vSGLT,22 Y290 in hSGLT1,113 Y248 in PutP,56 and F253 in LeuT,21 which have been identified structural components of both sites S1 and S2 in these systems and have been implicated in gating and allosteric control.47 Ala substitution of the Tyr at position 259 in rat NIS drastically decreases iodide uptake, while introduction of a Phe residue recovers some of the transport function, suggesting that a bulky aromatic residue is critical at this position.44 MD simulations in vSGLT reveal that rotation of residue Y263 is coupled to substrate release123 although nongated substrate release is also possible in this system.124 In hSGLT1, the aromatic nature of Y290 and its hydroxide group have been implicated both in substrate and Na+ binding.113 Importantly, Cys and Gly substitutions of Y248 alter the PutP:proline stoichiometry from 2:1 to 1:1 and lead to severe reduction of the proline uptake, regardless of Na+ binding.47
Location of the Putative S1 Site
The S1 binding site contains in addition residues K185, N262, Q263, Q265, T357 and S358. Residue Q263 has been implicated in Na+ (site Na2) binding in a previous homology model of hNIS,46 and substitutions with Asn in rat NIS leads to a drastic decrease of radioiodine uptake and changes in Km for both Na+ and I−. K185A and Q265A/C substitutions in human NIS have significant detrimental impact on I− uptake (Figure 3). The analogue of the hNIS residue N262 in PutP is residue Q251 (Table S2). A Q251C substitution in PutP affects both substrate and Na+ kinetic parameters.49 Q251 is also connected to P252, which is another residue controlling the molar binding ratio in PutP.47 Residue T357 in hNIS corresponds to residues S368 (protein:substrate stoichiometry control) in vSGLT,113 C344 (protein:substrate stoichiometry control) in PutP,113 and N318 (substrate binding residue in the crystal structure) in Mhp1,64 which have proven critical for substrate uptake in these systems. A T357A mutation in hNIS drastically decreases the radioiodine uptake and alters the Km and Vmax parameters for both I− and Na+.38 S358 in hNIS corresponds to residue Q345 in PutP, which is implicated in substrate binding from electrophysiology measurements.50
Location of the Putative S2 Site
The residues which can be considered unique for the S2 binding site are Q94, W255, Y144, V148, F417, and M420. Most of them have analogues in the substrate binding sites established from the available crystal structures of vSGLT, Mhp1, and LeuT (Table S2). W255A/C and F417Y/C mutations in hNIS eliminate transport and substitution of Q94 with other polar or charged residues (Arg, Lys, Asn, Glu) severely decreases I− uptake (Figure 3). Taking into account the low contact frequency of I− and the higher contact frequency for the larger, tetrahedral ClO4− ion, Q94 emerges as a residue from the second coordination sphere of the anions in the vicinity of residues F67 and Q72 which are critical for transport (see above) and are also part of the Na1 putative binding site (Table S2, Figures S3 and S4). Q94 has a higher contact frequency with the Na+ in site Na1 than with I− (Figure S3), which indicates that Na+ may be coordinated by Q94, while the I− ion is in its S2 site. In addition, Q94 is a neighboring residue of G93 (G93R is one of the known pathological mutations found in patients with congenital ITD2,125), which has been implicated in the control of Na+:I− transport stoichiometry.44 It is possible that the alteration of the Gly side chain at the somewhat peripheral position G93 interferes with residue Q94 from the S2 and Na1 binding sites of hNIS, which in turn may impact other Na1 binding residues (e.g., F67, Q72, Y259) leading to the observed changes in Na+:I− stoichiometry in NIS.
Previous homology models based on vSGLT place residue W255 at the periphery of TM7, and the orientation of W255 has been discussed in conjunction with residue G93 as a “ball and socket” structure, relevant to conformational transitions in NIS.44 Theoretical I− binding studies with the ABF method and the Drude force field in this model of hNIS in the absence of Na+ ions have identified residues F67, F87, M90, G93, Q94, W255, and Y259 as a putative binding site for I− and have revealed that the G93T mutation impacts the binding free energy in this site by displacing the W255 side chain into the binding pocket of hNIS.45 In our model, W255 points inward, toward the protein core, and is part of the binding region in hNIS as suggested by the anion density maps from our MD simulations. All transporters listed in Table S2 feature a Trp or a Phe residue at this position, and these residues are involved in substrate binding as seen from the crystal structures of vSGLT, Mhp1, and LeuT.21,22,64 The remaining LeuT-fold proteins used for the multiple sequence alignment (Figure S1) also have a Trp or Phe residue involved in substrate binding in the vicinity of W255. The presence of such aromatic residues in the binding site of anion binding proteins is not unique for the LeuT-fold architecture. Two Trp residues have been identified in the halide binding site of the water-soluble dehalogenases,126 and Tyr is a well-known binding residue in the CLC-ec1 chloride transporter,127 while chloride in the dopamine and serotonin transporters is coordinated with conserved Tyr and Phe residues.66,98 Halide binding in the fluoride ion channels of the Fluc family also involves highly conserved Phe residues.128 The SLC4 family of anion transporters features Phe residues in their putative binding sites, and some of these residues have a strong impact on transport, as suggested by mutagenesis and uptake data.68 The aromatic residues in the substrate binding pockets play a role not only in substrate binding, for which the indole group of the Trp and the hydroxide group of Tyr are well equipped, but also as part of a hydrophobic constriction zone, which aids in the dehydration of the substrate, necessary for stronger binding within the protein.22 The critical role for I− uptake of F417 in hNIS is supported by kinetic modeling in vSGLT and hSGLT1, which shows that the well-established gating residue F424 in vSGLT and its hSGLT1 analogue F453 are involved in coordination of conformational changes and coupling of Na+ and sugar transport in addition to the traditional role of an energy barrier.22,112,113,129,130
A region with small I− density immediately adjacent to sites S1 and S2 can be discerned by our MD simulations in the hNIS systems loaded with I− and 2Na+ ions. This binding site is composed of residues F67, Q72, Q94, and the W255-G260 (Figure S7). Given the small I− density in this area, this site may represent a transient shallow-binding/access region, present during the early production simulations of I− binding to hNIS.
Differences between Iodide and Perchlorate Binding in hNIS
For the most part, the contact frequency and anion density patterns of ClO4− and I− are qualitatively similar and involve residues from sites S1 and S2 as described above (Figures S3–S6). Most of the coordination is achieved through N–H bonds (from the peptide backbone, amino groups in Asn and Gln, or the indole ring in Trp) or O–H bonds (from the hydroxyl groups of Ser, Thr, and Tyr residues), although involvement of the C–H bond from various polar and nonpolar side chains is also possible (Figure 5, Figure S6). Due to the tetrahedral structure of the ClO4− ion and its slightly larger ionic radius (about 0.15 Å larger than the ionic radius of I−),131 it appears to overlap better with the surrounding protein residues, which results in longer duration of anion binding and, consequently, in higher contact frequencies, especially in the area of the S1 binding site (Table S2, Figure S3). The bulk of the ClO4− ion density falls within the S1 site or at the interface between sites S1 and S2 (Figure S5). Perchlorate also forms an ion pair with the Na+ (site Na1) ion more rarely than I− in the MD simulations with a Na+:substrate 2:1 ratio.
The water presence in the substrate binding cavity of hNIS at the onset of the MD simulations generally leads to destabilized protein–ion binding, rapid opening of the cytoplasmic vestibule of hNIS, and frequent exit of at least one ion from the protein, evident from the shorter than 50 ns ion–protein binding duration displayed in Table S3 and the lower percentage of ion–protein contacts in the contact frequency graphs (Figure S4). The most frequent ion exit induced by the water present in the core of the protein is observed in the Na+:ClO4− 2:1 and Na+:I− 1:1 systems, which points to higher protein–ion binding instability for these stoichiometries.
The S1 site is closer to the intracellular exit route for anions (Figure 2B). Due to the inward-facing semiopen conformation of our hNIS model, substrates in this site tend to unbind and move to the intracellular solution in several MD simulations (Table S3). The primary substrate for hNIS (I−) displays more frequent dissociation compared to ClO4− (Table S3). In fact, perchlorate unbinding was observed in only one of the studied hNIS systems, where ClO4− was placed in site S1. No ClO4− exit from site S2 was observed regardless of the protein core hydration. Enhanced protein–anion coordination may explain the less frequent ClO4− dissociation from the substrate binding sites of hNIS and the I− uptake inhibition effect of ClO4− in NIS, where perchlorate may bind stably to one of the available substrate binding sites and prevent iodide binding and translocation. Nevertheless, our MD simulations do not provide conclusive evidence for preference of a 1:1 Na+:ClO4− transport stoichiometry in human NIS. Such evidence would require detailed modeling and experimental studies involving assessment of the thermodynamics of anion binding in hNIS with different ion loads which is outside of the scope of the current work.
While multiple further studies would be necessary to identify the specific roles of the listed residues in the anion uptake function of hNIS, published studies and uptake experiments performed in our work show that the suggested residues from our MD simulations are crucial for hNIS transport and often correspond to residues involved in direct substrate binding and/or regulation of protein:substrate and Na+:substrate stoichiometry in other structurally similar proteins.
Insights for Permeation Pathway and Gating
The governing hypothesis of secondary transport, known as the “alternating access mechanism”, dictates that the binding sites for the substrate and accompanying species are consecutively uncovered to the outside and inside of the cell as the structure of the protein changes.121 The alternating access hypothesis has been backed by crystallographic and modeling studies on a number of secondary transporters, most prominently the bacterial leucine transporter LeuT, which has been crystallized in outward-facing open, inward-facing open, and occluded states in the presence and absence of bound Na+, substrates, and inhibitors.100 These studies have elucidated the function of LeuT and other proteins featuring the LeuT-fold.20,100 Generally, macroscopic motions in the gating regions of the proteins tend to create or destroy salt bridges and hydrophobic plugs which then obstruct or open water filled permeation pathways through the protein cores, necessary for the substrate and ion translocations. These motions are allosterically triggered by binding of Na+ ions and substrates.20,132
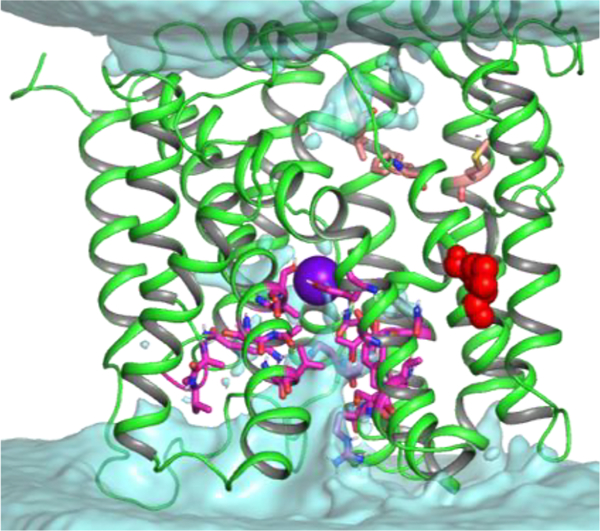
Review of the contact frequency maps (Figures S3 and S4, Table S2) shows that two key for transport residues S66 and T354 (see above) are in contact with all ions (Na+ in sites Na1 and Na2, I−, ClO4−in either S1 or S2 site) present in the hNIS binding cavity. These residues and their neighboring residues from the A65-S69 and T353-S358 stretches therefore may be part of the allosteric interaction network responding to the ion binding and triggering the major conformational changes necessary for ion translocation through the membrane. The plausible permeation pathway in our model of hNIS can be traced with water maps, generated from the MD simulations. A sample water density map for hNIS loaded with two Na+ and one I− ions is shown in Figure 6 and demonstrates a water accessible vestibule at the cytoplasmic side of hNIS. The water density maps of the remaining hNIS models with different ion loads look similar to the map in Figure 6. Such water distribution in our model of hNIS is expected since the crystal structure of vSGLT used in the homology modeling is a dynamic semioccluded or inward-facing open state,112 which relaxes further during the MD simulations. A number of residues line the cytoplasmic water vestibule: S116, T117, Y118, T134, Y137, A180, V181, G182, K185, N262–Q265, Q267, R268, T357, S358, N360, A361, A364,and V365 (Figure 6, magenta sticks). The water molecules permeate the protein to the area of the Na2 and S1 binding sites and provide an exit pathway for the ions bound to them. NMA accessibility studies show that areas of TM2 and TM9 form a hydrophilic and water accessible pore in PutP, consistent with the one observed in hNIS.50,53 MD studies of the gateless galactose exit from vSGLT identify several key residues lining the galactose exit pathway: S368, N371, S372, T375, and R273.124 They correspond to T357, N360, A361, A364, and R268 in the cytoplasmic water vestibule of hNIS, determined from our MD simulations. A Q267E mutation in hNIS, which leads to a decreased turnover rate, is found in patients with congenital ITD.37 Of interest in this area is also residue K185, which belongs to the loop between TM5 and TM6. A positive residue in this region is unique for hNIS and cannot be found in the otherwise hydrophobic corresponding loops in PutP, vSGLT, and hSGLT1. The backbone of residue 185 forms a part of the carbonyl nest, where Na+ (site Na2) is coordinated (Figure 4). The long positive side chain stretches toward the substrate binding site S1 and is involved in direct anion coordination once the anion starts its exit from the protein, implying a role in protein gating. Due to its position on a cytoplasmic loop, Lys demonstrates high flexibility during the MD simulations and may even form a short-lived salt bridge with residue D191, once Na+ is displaced from its Na2 binding site (Figure S8), which may have implications for the inward-facing to outward-facing transition of hNIS once all ions are released in the cytoplasm. A K185R mutation has a negligible effect on iodide uptake, but substitutions which omit the positive charge (K185A, K185Q, and K185E) prove more detrimental to iodide transport. Other residues in the vicinity of K185, which may play a role in the gating process are T117, Y118, Y137, M184, A264, T357, and A361. These residues have bulky hydrophobic methyl or phenyl groups and form an intracellular hydrophobic plug in our semioccluded model of hNIS which opens upon hydration as the MD simulations progress and allows exit of the ions from the protein. At the extracellular site, which in our hNIS model is occluded, residues V76, M90, and F417 (Figure 6, pink sticks) appear in the hydrophobic constriction zone made of residues M73, Y87, and F424, respectively, in the crystal structure of vSGLT.22 An aromatic residue is present at this position in PutP, hSGLT1, Mhp1, and LeuT (Table S2). Mutation of F417 to Cys eliminates I− uptake (Figure 3), proving that residue 417 is critical for anion transport and that hNIS requires the presence of a hydrophobic residue at this position. Similarly, a M90C mutation decreases drastically the I− uptake (Figure 3). The opening of the extracellular gate in vSGLT is attributed to residues P436 and G437, which allow for the necessary structural deformations in TM11.22 Similar Pro-Gly or Gly-Pro motives are found in this area of TM11 in hSGLT1, hNIS, PutP, Mhp1, and the sialic acid transporter SiaT.22,56,64,102 Comparison between the outward-facing open state of SiaT and the occluded structures Mhp1 and vSGLT lends support to the hypothesis that the Pro-Gly pair in TM11 is responsible for the conformational changes of TM11 leading to the opening and closing of the extracellular gate in these transporters. Therefore, it can be expected that the Pro-Gly motif in hNIS (G425-P426, red spheres in Figure 6) has a similar conformation relevant function.
Figure 6.
Water density map calculated from 13 MD simulations of hNIS with 2:1 Na+:I− ion stoichiometry (no water in binding cavity at MD onset). The I− ion is shown as a purple sphere and is located in the S1 site. The S1 site is accessible to water from the intracellular space, consistent with the inward-facing open state of hNIS. The residues shown as magenta sticks line the water permeation pathway. Residues from the hydrophobic constriction zone at the extracellular side of hNIS are shown as pink sticks. The G425-P426 motif, responsible for opening of the extracellular gate is shown as red spheres.
IV. CONCLUSIONS
A combination of homology modeling and molecular dynamics simulations with mutagenesis and uptake experiments led to the development of a cross-validated model for the human NIS transporter. Two putative Na+ binding sites (Na1 and Na2) and a large substrate binding cavity, with two substrate binding sites, S1 and S2, were mapped by Grand Canonical Monte Carlo calculations and assessed further with MD simulations. A number of residues lining the ion and water access pathways to the core of the hNIS transporters and the putative ion binding sites were selected for mutagenesis and uptake experiments, and their functional significance was confirmed by the detrimental impact on uptake following their mutation. The model provides a structural rationale for the substrate uptake and conformational dynamics of hNIS. Taken together, the comparison to a number of LeuT-fold transporters and the results from functional mutagenesis presented in previous studies on hNIS and in our current work afford an extensive validation of our homology model and provide an indispensable structural template for future optimizations of hNIS for enhanced SPECT/PET radiotracer transport.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH 2 R44 TR001191 02A1, the Natural Sciences and Engineering Research Council of Canada (NSERC Grant No. RGPIN-315019), and the Alberta Innovates Technical Futures Strategic Chair in Bio-Molecular Simulations. All calculations were performed on the CFI-supported TNK cluster at the University of Calgary and on the West-Grid/Compute Canada clusters under Research Allocation Award to S.N. H.R.Z. was supported by the Alberta Innovates Health Solution Post-Doctoral Fellowship.
Footnotes
The authors declare the following competing financial interest(s): T.S., R.J, P.J.L, and L.S. are Imanis employees. S.J.R, K.-W.P, and the Mayo Clinic have a financial interest in the NIS technology and Imanis Life Sciences. The other authors declare that they have no competing interests.
Authors will release the atomic coordinates of the hNIS model upon article publication by request sent to the corresponding author.
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.jcim.9b01114
ASSOCIATED CONTENT
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jcim.9b01114.
Multitemplate sequence alignment used for modeling of the LeuT-fold model of NIS. Comparison of LeuT-like, mixed, and vSGLT-like hNIS model. Contact frequencies for Na+, I−, and ClO4−, and perchlorate and iodide densities calculated from the MD simulations of hNIS. Suggested binding sites for ClO4−, suggested allosteric binding site for I−, and role of residue K185 in the hNIS mechanism. Table with ClO4− force field parameters. Comparative table for residues involved in Na+ and substrate binding in hNIS, vSGLT, hSGLT1, PutP, Mhp1, and LeuT. Table with duration of ion binding during all 50 ns long MD simulations. (PDF)
Contributor Information
Hristina R. Zhekova, Centre for Molecular Simulation, Department of Biological Sciences, University of Calgary, Calgary, AB T2N 1N4, Canada.
Toshie Sakuma, Imanis Life Sciences, Rochester, Minnesota 55901, United States; Department of Molecular Medicine, Mayo Clinic, Rochester, Minnesota 55902, United States.
Ryan Johnson, Imanis Life Sciences, Rochester, Minnesota 55901, United States.
Susanna C. Concilio, Department of Molecular Medicine and Mayo Clinic Graduate School of Biomedical Sciences, Mayo Clinic, Rochester, Minnesota 55902, United States
Patrycja J. Lech, Imanis Life Sciences, Rochester, Minnesota 55901, United States
Igor Zdravkovic, Centre for Molecular Simulation, Department of Biological Sciences, University of Calgary, Calgary, AB T2N 1N4, Canada.
Mirna Damergi, Centre for Molecular Simulation, Department of Biological Sciences, University of Calgary, Calgary, AB T2N 1N4, Canada.
Lukkana Suksanpaisan, Imanis Life Sciences, Rochester, Minnesota 55901, United States.
Kah-Whye Peng, Imanis Life Sciences, Rochester, Minnesota 55901, United States; Department of Molecular Medicine, Mayo Clinic, Rochester, Minnesota 55902, United States.
Stephen J. Russell, Imanis Life Sciences, Rochester, Minnesota 55901, United States; Department of Molecular Medicine, Mayo Clinic, Rochester, Minnesota 55902, United States
Sergei Noskov, Centre for Molecular Simulation, Department of Biological Sciences, University of Calgary, Calgary, AB T2N 1N4, Canada.
REFERENCES
- (1).Wright EM; Turk E The Sodium/Glucose Cotransport Family SLC5. Pfluegers Arch. 2004, 447 (5), 510–8. [DOI] [PubMed] [Google Scholar]
- (2).Darrouzet E; Lindenthal S; Marcellin D; Pellequer JL; Pourcher T The Sodium/Iodide Symporter: State of the Art of Its Molecular Characterization. Biochim. Biophys. Acta, Biomembr 2014, 1838 (1), 244–253. [DOI] [PubMed] [Google Scholar]
- (3).Portulano C; Paroder-Belenitsky M; Carrasco N The Na+/I-Symporter (NIS): Mechanism and Medical Impact. Endocr. Rev 2014, 35 (1), 106–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Eskandari S; Loo DD; Dai G; Levy O; Wright EM; Carrasco N Thyroid Na+/I- Symporter. Mechanism, Stoichiometry, and Specificity. J. Biol. Chem 1997, 272 (43), 27230–8. [DOI] [PubMed] [Google Scholar]
- (5).Van Sande J; Massart C; Beauwens R; Schoutens A; Costagliola S; Dumont JE; Wolff J Anion Selectivity by the Sodium Sodide Symporter. Endocrinology 2003, 144 (1), 247–52. [DOI] [PubMed] [Google Scholar]
- (6).Dohan O; Portulano C; Basquin C; Reyna-Neyra A; Amzel LM; Carrasco N The Na+/I Symporter (NIS) Mediates Electroneutral Active Transport of the Environmental Pollutant Perchlorate. Proc. Natl. Acad. Sci. U. S. A 2007, 104 (51), 20250–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Hong H; Yang Y; Zhang Y; Cai W Non-Invasive Cell Tracking in Cancer and Cancer Therapy. Curr. Top. Med. Chem 2010, 10 (12), 1237–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Vandergaast R; Khongwichit S; Jiang H; DeGrado TR; Peng KW; Smith DR; Russell SJ; Suksanpaisan L Enhanced Noninvasive Imaging of Oncology Models Using the NIS Reporter Gene and Bioluminescence Imaging. Cancer Gene Ther. 2019, 2019, na DOI: 10.1038/s41417-019-0081-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Ahn BC Sodium Iodide Symporter for Nuclear Molecular Imaging and Gene Therapy: From Bedside to Bench and Back. Theranostics 2012, 2 (4), 392–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Jiang H; DeGrado TR [(18)F]Tetrafluoroborate ([(18)F]TFB) and Its Analogs for PET Imaging of the Sodium/Iodide Symporter. Theranostics 2018, 8 (14), 3918–3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Miller A; Russell SJ The Use of the NIS Reporter Gene for Optimizing Oncolytic Virotherapy. Expert Opin. Biol. Ther 2016, 16 (1), 15–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Ravera S; Reyna-Neyra A; Ferrandino G; Amzel LM; Carrasco N The Sodium/Iodide Symporter (NIS): Molecular Physiology and Preclinical and Clinical Applications. Annu. Rev. Physiol 2017, 79, 261–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Jiang H; Bansal A; Pandey MK; Peng KW; Suksanpaisan L; Russell SJ; DeGrado TR Synthesis of 18F-Tetrafluoroborate via Radiofluorination of Boron Trifluoride and Evaluation in a Murine C6-Glioma Tumor Model. J. Nucl. Med 2016, 57 (9), 1454–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Domingo-Musibay E; Allen C; Kurokawa C; Hardcastle JJ; Aderca I; Msaouel P; Bansal A; Jiang H; DeGrado TR; Galanis E Measles Edmonston Vaccine Strain Derivatives Have Potent Oncolytic Activity Against Osteosarcoma. Cancer Gene Ther. 2014, 21 (11), 483–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Qaim SM Nuclear Data for Production and Medical Application of Radionuclides: Present Status and Future Needs. Nucl. Med. Biol 2017, 44, 31–49. [DOI] [PubMed] [Google Scholar]
- (16).McConathy J; Goodman MM Non-Natural Amino Acids for Tumor Imaging Using Positron Emission Tomography and Single Photon Emission Computed Tomography. Cancer Metastasis Rev. 2008, 27 (4), 555–73. [DOI] [PubMed] [Google Scholar]
- (17).Nairne J; Iveson PB; Meijer A Imaging in Drug Development. Prog. Med. Chem 2015, 54, 231–80. [DOI] [PubMed] [Google Scholar]
- (18).Dai G; Levy O; Carrasco N Cloning and Characterization of the Thyroid Iodide Transporter. Nature 1996, 379 (6564), 458–60. [DOI] [PubMed] [Google Scholar]
- (19).Levy O; De la Vieja A; Ginter CS; Riedel C; Dai G; Carrasco N N-Linked Glycosylation of the Thyroid Na+/I-Symporter (NIS). Implications for Its Secondary Structure Model. J. Biol. Chem 1998, 273 (35), 22657–63. [DOI] [PubMed] [Google Scholar]
- (20).Kazmier K; Claxton DP; McHaourab HS Alternating Access Mechanisms of LeuT-fold Transporters: Trailblazing Towards the Promised Energy Landscapes. Curr. Opin. Struct. Biol 2017, 45, 100–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Yamashita A; Singh SK; Kawate T; Jin Y; Gouaux E Crystal Structure of a Bacterial Homologue of Na+/Cl–Dependent Neurotransmitter Transporters. Nature 2005, 437 (7056), 215–23. [DOI] [PubMed] [Google Scholar]
- (22).Faham S; Watanabe A; Besserer GM; Cascio D; Specht A; Hirayama BA; Wright EM; Abramson J The Crystal Structure of a Sodium Galactose Transporter Reveals Mechanistic Insights into Na+/Sugar Symport. Science 2008, 321 (5890), 810–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Pohlenz J; Medeiros-Neto G; Gross JL; Silveiro SP; Knobel M; Refetoff S Hypothyroidism in a Brazilian Kindred due to Iodide Trapping Defect Caused by a Homozygous Mutation in the Sodium/Iodide symporter gene. Biochem. Biophys. Res. Commun 1997, 240 (2), 488–91. [DOI] [PubMed] [Google Scholar]
- (24).Fujiwara H; Tatsumi K; Tanaka S; Kimura M; Nose O; Amino N A Novel V59E Missense Mutation in the Sodium Iodide Symporter Gene in a Family with Iodide Transport Defect. Thyroid 2000, 10 (6), 471–4. [DOI] [PubMed] [Google Scholar]
- (25).Fujiwara H; Tatsumi K; Miki K; Harada T; Okada S; Nose O; Kodama S; Amino N Recurrent T354P Mutation of the Na+/I- Symporter in Patients with Iodide Transport Defect. J. Clin. Endocrinol. Metab 1998, 83 (8), 2940–2943. [DOI] [PubMed] [Google Scholar]
- (26).Kosugi S; Bhayana S; Dean HJ A Novel Mutation in the Sodium/Iodide Symporter Gene in the Largest Family with Iodide Transport Defect. J. Clin. Endocrinol. Metab 1999, 84 (9), 3248–3253. [DOI] [PubMed] [Google Scholar]
- (27).Kosugi S; Inoue S; Matsuda A; Jhiang SM Novel, Missense and Loss-of-Function Mutations in the Sodium/Iodide Symporter Gene Causing Iodide Transport Defect in Three Japanese Patients. J. Clin. Endocrinol. Metab 1998, 83 (9), 3365–3368. [DOI] [PubMed] [Google Scholar]
- (28).Kosugi S; Sato Y; Matsuda A; Ohyama Y; Fujieda K; Inomata H; Kameya T; Isozaki O; Jhiang SM High Prevalence of T354P Sodium/Iodide Symporter Gene Mutation in Japanese Patients with Iodide Transport Defect Who Have Heterogeneous Clinical Pictures. J. Clin. Endocrinol. Metab 1998, 83 (11), 4123–4129. [DOI] [PubMed] [Google Scholar]
- (29).Pohlenz J; Rosenthal IM; Weiss RE; Jhiang SM; Burant C; Refetoff S Congenital Hypothyroidism due to Mutations in the Sodium/Iodide Symporter. Identification of a Nonsense Mutation Producing a Downstream Cryptic 3′ Splice Site. J. Clin. Invest 1998, 101 (5), 1028–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Levy O; Ginter CS; De la Vieja A; Levy D; Carrasco N Identification of a Structural Requirement for Thyroid Na+/I-Symporter (NIS) Function From Analysis of a Mutation That Causes Human Congenital Hypothyroidism. FEBS Lett. 1998, 429 (1), 36–40. [DOI] [PubMed] [Google Scholar]
- (31).Paroder V; Nicola JP; Ginter CS; Carrasco N The Iodide-Transport-Defect-Causing Mutation R124H: A Delta-Amino Group at Position 124 is Critical for Maturation and Trafficking of the Na+/I- symporter. J. Cell Sci 2013, 126 (15), 3305–3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Spitzweg C; Morris JC Genetics and Phenomics of Hypothyroidism and Goiter due to NIS Mutations. Mol. Cell. Endocrinol 2010, 322 (1–2), 56–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Fu C; Chen S; Chen R; Fan X; Luo J; Li C; Qian J Mutation Screening of the Sodium Iodide Symporter Gene in a Cohort of 105 China Patients with Congenital Hypothyroidism. Arq. Bras. Endocrinol. Metabol 2014, 58 (8), 828–32. [DOI] [PubMed] [Google Scholar]
- (34).Liang JA; Chen CP; Huang SJ; Ho TY; Hsiang CY; Ding HJ; Wu SL A Novel Loss-of-Function Deletion in Sodium/Iodide Symporter Gene in Follicular Thyroid Adenoma. Cancer Lett. 2005, 230 (1), 65–71. [DOI] [PubMed] [Google Scholar]
- (35).Wu SL; Ho TY; Liang JA; Hsiang CY Histidine Residue at Position 226 is Critical for Iodide Uptake Activity of Human Sodium/Iodide Symporter. J. Endocrinol 2008, 199 (2), 213–9. [DOI] [PubMed] [Google Scholar]
- (36).De la Vieja A; Ginter CS; Carrasco N Molecular Analysis of a Congenital Iodide Transport Defect: G543E Impairs Maturation and Trafficking of the Na+/I- Symporter. Mol. Endocrinol 2005, 19 (11), 2847–58. [DOI] [PubMed] [Google Scholar]
- (37).De La Vieja A; Ginter CS; Carrasco N The Q267E Mutation in the Sodium/Iodide Symporter (NIS) Causes Congenital Iodide Transport Defect (ITD) by Decreasing the NIS Turnover Number. J. Cell Sci 2004, 117 (5), 677–687. [DOI] [PubMed] [Google Scholar]
- (38).De la Vieja A; Reed MD; Ginter CS; Carrasco N Amino Acid Residues in Transmembrane Segment IX of the Na+/I-Symporter Play a Role in its Na+ Dependence and Are Critical for Transport Activity. J. Biol. Chem 2007, 282 (35), 25290–8. [DOI] [PubMed] [Google Scholar]
- (39).Reed-Tsur MD; De la Vieja A; Ginter CS; Carrasco N Molecular Characterization of V59E NIS, a Na+/I- Symporter Mutant that Causes Congenital I- Transport Defect. Endocrinology 2008, 149 (6), 3077–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Dohan O; Gavrielides MV; Ginter C; Amzel LM; Carrasco N Na(+)/I(−) Symporter Activity Requires a Small and Uncharged Amino Acid Residue at Position 395. Mol. Endocrinol 2002, 16 (8), 1893–902. [DOI] [PubMed] [Google Scholar]
- (41).Li CC; Ho TY; Kao CH; Wu SL; Liang JA; Hsiang CY Conserved Charged Amino Acid Residues in the Extracellular Region of Sodium/Iodide Symporter are Critical for Iodide Transport Activity. J. Biomed. Sci 2010, 17, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Li W; Nicola JP; Amzel LM; Carrasco N Asn441 Plays a Key Role in Folding and Function of the Na+/I- Symporter (NIS). FASEB J. 2013, 27 (8), 3229–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Montanelli L; Agretti P; Marco G; Bagattini B; Ceccarelli C; Brozzi F; Lettiero T; Cerbone M; Vitti P; Salerno M; Pinchera A; Tonacchera M Congenital Hypothyroidism and Late-Onset Goiter: Identification and Characterization of a Novel Mutation in the Sodium/Iodide Symporter of the Proband and Family Members. Thyroid 2009, 19 (12), 1419–25. [DOI] [PubMed] [Google Scholar]
- (44).Paroder-Belenitsky M; Maestas MJ; Dohan O; Nicola JP; Reyna-Neyra A; Follenzi A; Dadachova E; Eskandari S; Amzel LM; Carrasco N Mechanism of Anion Selectivity and Stoichiometry of the Na+/I- Symporter (NIS). Proc. Natl. Acad. Sci. U. S. A 2011, 108 (44), 17933–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Vergara-Jaque A; Fong P; Comer J Iodide Binding in Sodium-Coupled Cotransporters. J. Chem. Inf. Model 2017, 57 (12), 3043–3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Ferrandino G; Nicola JP; Sanchez YE; Echeverria I; Liu Y; Amzel LM; Carrasco N Na+ Coordination at the Na2 Site of the Na+/I- Symporter. Proc. Natl. Acad. Sci. U. S. A 2016, 113 (37), E5379–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Li Z; Lee AS; Bracher S; Jung H; Paz A; Kumar JP; Abramson J; Quick M; Shi L Identification of a Second SubstrateBinding site in Solute-Sodium Symporters. J. Biol. Chem 2015, 290 (12), 7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Mazier S; Quick M; Shi L Conserved Tyrosine in the First Transmembrane Segment of Solute:Sodium Symporters is Involved in Na+-Coupled Substrate Co-Transport. J. Biol. Chem 2011, 286 (33), 29347–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Bracher S; Schmidt CC; Dittmer SI; Jung H Core Transmembrane Domain 6 Plays a Pivotal Role in the Transport Cycle of the Sodium/Proline Symporter PutP. J. Biol. Chem 2016, 291 (50), 26208–26215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Raba M; Baumgartner T; Hilger D; Klempahn K; Hartel T; Jung K; Jung H Function of Transmembrane Domain IX in the Na+/Proline Transporter PutP. J. Mol. Biol 2008, 382 (4), 884–93. [DOI] [PubMed] [Google Scholar]
- (51).Raba M; Dunkel S; Hilger D; Lipiszko K; Polyhach Y; Jeschke G; Bracher S; Klare JP; Quick M; Jung H; Steinhoff HJ Extracellular Loop 4 of the Proline Transporter PutP Controls the Periplasmic Entrance to Ligand Binding Sites. Structure 2014, 22 (5), 769–80. [DOI] [PubMed] [Google Scholar]
- (52).Hilger D; Bohm M; Hackmann A; Jung H Role of Ser-340 and Thr-341 in Transmembrane Domain IX of the Na+/Proline Transporter PutP of Escherichia Coli in Ligand Binding and Transport. J. Biol. Chem 2008, 283 (8), 4921–9. [DOI] [PubMed] [Google Scholar]
- (53).Pirch T; Landmeier S; Jung H Transmembrane Domain II of the Na+/Proline Transporter PutP of Escherichia Coli Forms Part of a Conformationally Flexible, Cytoplasmic Exposed Aqueous Cavity Within the Membrane. J. Biol. Chem 2003, 278 (44), 42942–9. [DOI] [PubMed] [Google Scholar]
- (54).Jung H Topology and Function of the Na+/Proline Transporter of Escherichia Coli, a Member of the Na+/Solute Cotransporter Family. Biochim. Biophys. Acta, Bioenerg 1998, 1365 (1–2), 60–4. [DOI] [PubMed] [Google Scholar]
- (55).Jung H; Rubenhagen R; Tebbe S; Leifker K; Tholema N; Quick M; Schmid R Topology of the Na+/Proline Transporter of Escherichia Coli. J. Biol. Chem 1998, 273 (41), 26400–7. [DOI] [PubMed] [Google Scholar]
- (56).Olkhova E; Raba M; Bracher S; Hilger D; Jung H Homology Model of the Na+/Proline Transporter PutP of Escherichia Coli and its Functional Implications. J. Mol. Biol 2011, 406 (1), 59–74. [DOI] [PubMed] [Google Scholar]
- (57).Amin A; Ando T; Saijo S; Yamato I Role of Asp187 and Gln190 in the Na+/Proline Symporter (PutP) of Escherichia Coli. J. Biochem 2011, 150 (4), 395–402. [DOI] [PubMed] [Google Scholar]
- (58).Yamato I; Anraku Y Site-Specific Alteration of Cysteine 281, Cysteine 344, and Cysteine 349 in the Proline Carrier of Escherichia Coli. J. Biol. Chem 1988, 263 (31), 16055–16057. [PubMed] [Google Scholar]
- (59).Quick M; Stolting S; Jung H Role of Conserved Arg40 and Arg117 in the Na+/Proline Transporter of Escherichia Coli. Biochemistry 1999, 38 (41), 13523–9. [DOI] [PubMed] [Google Scholar]
- (60).Yamato I; Ohsawa M; Anraku Y Defective Cation-Coupling Mutants of Escherichia Coli Na+/Proline Symport Carrier. Characterization and Localization of Mutations. J. Biol. Chem 1990, 265 (5), 2450–2455. [PubMed] [Google Scholar]
- (61).Quick M; Jung H A Conserved Aspartate Residue, Asp187, is Important for Na+-Dependent Proline Binding and Transport by the Na+/Proline Transporter of Escherichia Coli. Biochemistry 1998, 37 (39), 13800–6. [DOI] [PubMed] [Google Scholar]
- (62).Woo HJ; Dinner AR; Roux B Grand Canonical Monte Carlo Simulations of Water in Protein Environments. J. Chem. Phys 2004, 121 (13), 6392–400. [DOI] [PubMed] [Google Scholar]
- (63).Gao X; Zhou L; Jiao X; Lu F; Yan C; Zeng X; Wang J; Shi Y Mechanism of Substrate Recognition and Transport by an Amino Acid Antiporter. Nature 2010, 463 (7282), 828–32. [DOI] [PubMed] [Google Scholar]
- (64).Simmons KJ; Jackson SM; Brueckner F; Patching SG; Beckstein O; Ivanova E; Geng T; Weyand S; Drew D; Lanigan J; Sharples DJ; Sansom MS; Iwata S; Fishwick CW; Johnson AP; Cameron AD; Henderson PJ Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1. EMBO J. 2014, 33 (16), 1831–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Perez C; Koshy C; Yildiz O; Ziegler C Alternating-Access Mechanism in Conformationally Asymmetric Trimers of the Betaine Transporter BetP. Nature 2012, 490 (7418), 126–30. [DOI] [PubMed] [Google Scholar]
- (66).Wang KH; Penmatsa A; Gouaux E Neurotransmitter and Psychostimulant Recognition by the Dopamine Transporter. Nature 2015, 521 (7552), 322–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Arakawa T; Kobayashi-Yurugi T; Alguel Y; Iwanari H; Hatae H; Iwata M; Abe Y; Hino T; Ikeda-Suno C; Kuma H; Kang D; Murata T; Hamakubo T; Cameron AD; Kobayashi T; Hamasaki N; Iwata S Crystal Structure of the Anion Exchanger Domain of Human Erythrocyte Band 3. Science 2015, 350 (6261), 680–4. [DOI] [PubMed] [Google Scholar]
- (68).Huynh KW; Jiang J; Abuladze N; Tsirulnikov K; Kao L; Shao X; Newman D; Azimov R; Pushkin A; Zhou ZH; Kurtz I CryoEM Structure of the Human SLC4A4 Sodium-Coupled AcidBase Transporter NBCe1. Nat. Commun 2018, 9 (1), 900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Dutzler R; Campbell EB; Cadene M; Chait BT; MacKinnon R X-ray Structure of a ClC Chloride channel at 3.0 A Reveals the Molecular Basis of Anion Selectivity. Nature 2002, 415 (6869), 287–94. [DOI] [PubMed] [Google Scholar]
- (70).Webb B; Sali A Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc Protein Sci 2016, 86 (1), na DOI: 10.1002/cpps.20. [DOI] [PubMed] [Google Scholar]
- (71).Shen MY; Sali A Statistical Potential for Assessment and Prediction of Protein Structures. Protein Sci. 2006, 15 (11), 2507–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Melo F; Sali A Fold Assessment for Comparative Protein Structure Modeling. Protein Sci. 2007, 16 (11), 2412–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Alford RF; Koehler Leman J; Weitzner BD; Duran AM; Tilley DC; Elazar A; Gray JJ An Integrated Framework Advancing Membrane Protein Modeling and Design. PLoS Comput. Biol 2015, 11 (9), No. e1004398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Larsson HP; Wang X; Lev B; Baconguis I; Caplan DA; Vyleta NP; Koch HP; Diez-Sampedro A; Noskov SY Evidence for a Third Sodium-Binding Site in Glutamate Transporters Suggests an Ion/Substrate Coupling Model. Proc. Natl. Acad. Sci. U. S. A 2010, 107 (31), 13912–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Im W; Berneche S; Roux B Generalized Solvent Boundary Potential for Computer Simulations. J. Chem. Phys 2001, 114 (7), 2924–2937. [Google Scholar]
- (76).Jo S; Kim T; Iyer VG; Im W CHARMM-GUI: A Web-Based Graphical User Interface for CHARMM. J. Comput. Chem 2008, 29 (11), 1859–65. [DOI] [PubMed] [Google Scholar]
- (77).Jo S; Vargyas M; Vasko-Szedlar J; Roux B; Im W PBEQ-Solver for Online Visualization of Electrostatic Potential of Biomolecules. Nucleic Acids Res. 2008, 36, W270–W275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Dohan O; De la Vieja A; Paroder V; Riedel C; Artani M; Reed M; Ginter CS; Carrasco N The Sodium/Iodide Symporter (NIS): Characterization, Regulation, and Medical Significance. Endocr. Rev 2003, 24 (1), 48–77. [DOI] [PubMed] [Google Scholar]
- (79).Jo S; Cheng X; Islam SM; Huang L; Rui H; Zhu A; Lee HS; Qi Y; Han W; Vanommeslaeghe K; MacKerell AD Jr.; Roux B; Im W CHARMM-GUI PDB Manipulator for Advanced Modeling and Simulations of Proteins Containing Nonstandard Residues. Adv. Protein Chem. Struct. Biol 2014, 96, 235–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Wu EL; Cheng X; Jo S; Rui H; Song KC; Davila-Contreras EM; Qi Y; Lee J; Monje-Galvan V; Venable RM; Klauda JB; Im W CHARMM-GUI Membrane Builder Toward Realistic Biological Membrane Simulations. J. Comput. Chem 2014, 35 (27), 1997–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Lomize MA; Pogozheva ID; Joo H; Mosberg HI; Lomize AL OPM Database and PPM Web Server: Resources for Positioning of Proteins in Membranes. Nucleic Acids Res. 2012, 40, D370–D376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Phillips JC; Braun R; Wang W; Gumbart J; Tajkhorshid E; Villa E; Chipot C; Skeel RD; Kale L; Schulten K Scalable Molecular Dynamics With NAMD. J. Comput. Chem 2005, 26 (16), 1781–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Lee J; Cheng X; Swails JM; Yeom MS; Eastman PK; Lemkul JA; Wei S; Buckner J; Jeong JC; Qi Y; Jo S; Pande VS; Case DA; Brooks CL 3rd; MacKerell AD Jr.; Klauda JB; Im W CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field. J. Chem. Theory Comput 2016, 12 (1), 405–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Huang J; MacKerell AD Jr. CHARMM36 All-Atom Additive Protein Force Field: Validation Based on Comparison to NMR Data. J. Comput. Chem 2013, 34 (25), 2135–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Best RB; Zhu X; Shim J; Lopes PE; Mittal J; Feig M; Mackerell AD Jr. Optimization of the Additive CHARMM All-Atom Protein Force Field Targeting Improved Sampling of the Backbone Phi, Psi and Side-Chain Chi(1) and Chi(2) Dihedral Angles. J. Chem. Theory Comput 2012, 8 (9), 3257–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Joung IS; Cheatham TE III Determination of Alkali and Halide Monovalent Ion Parameters for Use in Explicitly Solvated Biomolecular Simulations. J. Phys. Chem. B 2008, 112 (30), 9020–9041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Huang L; Roux B Automated Force Field Parameterization for Non-Polarizable and Polarizable Atomic Models Based on Ab Initio Target Data. J. Chem. Theory Comput 2013, 9 (8), 3543–3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Humphrey W; Dalke A; Schulten K VMD: Visual Molecular Dynamics. J. Mol. Graphics 1996, 14 (1), 33–8 27–8. [DOI] [PubMed] [Google Scholar]
- (89).Pham L; Ye H; Cosset FL; Russell SJ; Peng KW Concentration of Viral Vectors by Co-pPecipitation With Calcium Phosphate. J. Gene Med 2001, 3 (2), 188–94. [DOI] [PubMed] [Google Scholar]
- (90).Shi Y Common Folds and Transport Mechanisms of Secondary Active Transporters. Annu. Rev. Biophys 2013, 42, 51–72. [DOI] [PubMed] [Google Scholar]
- (91).Shimamura T; Weyand S; Beckstein O; Rutherford NG; Hadden JM; Sharples D; Sansom MS; Iwata S; Henderson PJ; Cameron AD Molecular Basis of Alternating Access Membrane Transport by the Sodium-Hydantoin Transporter Mhp1. Science 2010, 328 (5977), 470–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Weyand S; Shimamura T; Yajima S; Suzuki S; Mirza O; Krusong K; Carpenter EP; Rutherford NG; Hadden JM; O’Reilly J; Ma P; Saidijam M; Patching SG; Hope RJ; Norbertczak HT; Roach PC; Iwata S; Henderson PJ; Cameron AD Structure and Molecular Mechanism of a Nucleobase-Cation-Symport-1 Family Transporter. Science 2008, 322 (5902), 709–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (93).LeVine MV; Cuendet MA; Khelashvili G; Weinstein H Allosteric Mechanisms of Molecular Machines at the Membrane: Transport by Sodium-Coupled Symporters. Chem. Rev 2016, 116 (11), 6552–87. [DOI] [PubMed] [Google Scholar]
- (94).Singh SK; Piscitelli CL; Yamashita A; Gouaux E A Competitive Inhibitor Traps LeuT in an Open-to-Out Conformation. Science 2008, 322 (5908), 1655–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (95).de Carvalho FD; Quick M Surprising Substrate Versatility in SLC5A6: Na+-Coupled I- Transport by the Human Na+/Multivitamin Transporter (hSMVT). J. Biol. Chem 2011, 286 (1), 131–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (96).Willford SL; Anderson CM; Spencer SR; Eskandari S Evidence for a Revised Ion/Substrate Coupling Stoichiometry of GABA Transporters. J. Membr. Biol 2015, 248 (4), 795–810. [DOI] [PubMed] [Google Scholar]
- (97).Scimemi A Structure, Function, and Plasticity of GABA transporters. Front. Cell. Neurosci 2014, 8, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).Coleman JA; Green EM; Gouaux E X-Ray Structures and Mechanism of the Human Serotonin Transporter. Nature 2016, 532 (7599), 334–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (99).Perez C; Ziegler C Mechanistic Aspects of Sodium-Binding Sites in LeuT-Like Fold Symporters. Biol. Chem. 2013, 394 (5), 641–8. [DOI] [PubMed] [Google Scholar]
- (100).Penmatsa A; Gouaux E How LeuT Shapes Our Understanding of the Mechanisms of Sodium-Coupled Neurotransmitter Transporters. J. Physiol 2014, 592 (5), 863–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (101).Hummel CS; Lu C; Loo DD; Hirayama BA; Voss AA; Wright EM Glucose Transport by Human Renal Na+/DGlucose Cotransporters SGLT1 and SGLT2. Am. J. Physiol Cell Physiol 2011, 300 (1), C14–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (102).Wahlgren WY; Dunevall E; North RA; Paz A; Scalise M; Bisignano P; Bengtsson-Palme J; Goyal P; Claesson E; CaingCarlsson R; Andersson R; Beis K; Nilsson UJ; Farewell A; Pochini L; Indiveri C; Grabe M; Dobson RCJ; Abramson J; Ramaswamy S; Friemann R Substrate-Bound Outward-Open structure of a Na(+)-coupled Sialic Acid Symporter Reveals a New Na(+) Site. Nat. Commun 2018, 9 (1), 1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (103).Bisignano P; Ghezzi C; Jo H; Polizzi NF; Althoff T; Kalyanaraman C; Friemann R; Jacobson MP; Wright EM; Grabe M Inhibitor Binding Mode and Allosteric Regulation of Na(+)-Glucose Symporters. Nat. Commun 2018, 9 (1), 5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (104).Shi L; Quick M; Zhao Y; Weinstein H; Javitch JA The Mechanism of a Neurotransmitter:Sodium Symporter–Inward Release of Na+ and Substrate Is Triggered by Substrate in a Second Binding Site. Mol. Cell 2008, 30 (6), 667–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (105).Quick M; Winther AM; Shi L; Nissen P; Weinstein H; Javitch JA Binding of an Octylglucoside Detergent Molecule in the Second Substrate (S2) Site of LeuT Establishes an Inhibitor-Bound Conformation. Proc. Natl. Acad. Sci. U. S. A 2009, 106 (14), 5563–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (106).Reyes N; Tavoulari S To be, Or Not To Be Two Sites: That Is The Question About LeuT Substrate Binding. J. Gen. Physiol 2011, 138 (4), 467–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (107).Zhao Y; Terry DS; Shi L; Quick M; Weinstein H; Blanchard SC; Javitch JA Substrate-Modulated Gating Dynamics in a Na+-Coupled Neurotransmitter Transporter Homologue. Nature 2011, 474 (7349), 109–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (108).Quick M; Abramyan AM; Wiriyasermkul P; Weinstein H; Shi L; Javitch JA The LeuT-Fold Neurotransmitter:Sodium Symporter MhsT Has Two Substrate Sites. Proc. Natl. Acad. Sci. U. S. A 2018, 115 (34), E7924–E7931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (109).Joseph D; Pidathala S; Mallela AK; Penmatsa A Structure and Gating Dynamics of Na(+)/Cl(−) Coupled Neurotransmitter Transporters. Front Mol. Biosci 2019, 6, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (110).Quick M; Loo DD; Wright EM Neutralization of a Conserved Amino Acid Residue in the Human Na+/Glucose Transporter (hSGLT1) Generates a Glucose-Gated H+ Channel. J. Biol. Chem. 2001, 276 (3), 1728–34. [DOI] [PubMed] [Google Scholar]
- (111).Li J; Tajkhorshid E Ion-Releasing State of a Secondary Membrane Transporter. Biophys. J 2009, 97 (11), L29–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (112).Zomot E; Bahar I The Sodium/Galactose Symporter Crystal Structure Is a Dynamic, not so Occluded State. Mol. BioSyst 2010, 6 (6), 1040–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (113).Loo DD; Jiang X; Gorraitz E; Hirayama BA; Wright EM Functional Identification and Characterization of Sodium Binding Sites in Na Symporters. Proc. Natl. Acad. Sci. U. S. A 2013, 110 (47), E4557–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (114).Kalayil S; Schulze S; Kuhlbrandt W Arginine Oscillation Explains Na+ Independence in the Substrate/Product Antiporter CaiT. Proc. Natl. Acad. Sci. U. S. A 2013, 110 (43), 17296–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (115).Shaffer PL; Goehring A; Shankaranarayanan A; Gouaux E Structure and Mechanism of a Na+-Independent Amino Acid Transporter. Science 2009, 325 (5943), 1010–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (116).Romero MF; Chen AP; Parker MD; Boron WF The SLC4 Family of Bicarbonate (HCO(3)(−)) Transporters. Mol. Aspects Med 2013, 34 (2–3), 159–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (117).Khelashvili G; Schmidt SG; Shi L; Javitch JA; Gether U; Loland CJ; Weinstein H Conformational Dynamics on the Extracellular Side of LeuT Controlled by Na+ and K+ Ions and the Protonation State of Glu290. J. Biol. Chem 2016, 291 (38), 19786–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (118).Malinauskaite L; Said S; Sahin C; Grouleff J; Shahsavar A; Bjerregaard H; Noer P; Severinsen K; Boesen T; Schiott B; Sinning S; Nissen P A Conserved Leucine Occupies the Empty Substrate Site of LeuT in the Na(+)-Free Return State. Nat. Commun 2016, 7, 11673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (119).Zhao C; Stolzenberg S; Gracia L; Weinstein H; Noskov S; Shi L Ion-Controlled Conformational Dynamics in the Outward-Open Transition From an Occluded State of LeuT. Biophys. J 2012, 103 (5), 878–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (120).Tavoulari S; Margheritis E; Nagarajan A; DeWitt DC; Zhang YW; Rosado E; Ravera S; Rhoades E; Forrest LR; Rudnick G Two Na+ Sites Control Conformational Change in a Neurotransmitter Transporter Homolog. J. Biol. Chem 2016, 291 (3), 1456–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (121).Forrest LR; Zhang YW; Jacobs MT; Gesmonde J; Xie L; Honig BH; Rudnick G Mechanism for Alternating Access in Neurotransmitter Transporters. Proc. Natl. Acad. Sci. U. S. A 2008, 105 (30), 10338–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (122).Ravera S; Quick M; Nicola JP; Carrasco N; Amzel LM Beyond Non-Integer Hill Coefficients: A Novel Approach to Analyzing Binding Data, Applied to Na+-Driven Transporters. J. Gen. Physiol 2015, 145 (6), 555–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (123).Watanabe A; Choe S; Chaptal V; Rosenberg JM; Wright EM; Grabe M; Abramson J The Mechanism of Sodium and Substrate Release from the Binding Pocket of vSGLT. Nature 2010, 468 (7326), 988–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (124).Li J; Tajkhorshid E A Gate-Free Pathway for Substrate Release From the Inward-Facing State of the Na(+)-Galactose Transporter. Biochim. Biophys. Acta, Biomembr 2012, 1818 (2), 263–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (125).Pohlenz J; Refetoff S Mutations in the Sodium/Iodide Symporter (NIS) Gene as a Cause for Iodide Transport Defects and Congenital Hypothyroidism. Biochimie 1999, 81 (5), 469–76. [DOI] [PubMed] [Google Scholar]
- (126).Chaloupkova R; Prudnikova T; Rezacova P; Prokop Z; Koudelakova T; Daniel L; Brezovsky J; Ikeda-Ohtsubo W; Sato Y; Kuty M; Nagata Y; Kuta Smatanova I; Damborsky J Structural and Functional Analysis of a Novel Haloalkane Dehalogenase with Two Halide-Binding Sites. Acta Crystallogr., Sect. D: Biol. Crystallogr 2014, 70 (7), 1884–1897. [DOI] [PubMed] [Google Scholar]
- (127).Accardi A; Picollo A CLC Channels and Transporters: Proteins With Borderline Personalities. Biochim. Biophys. Acta, Biomembr. 2010, 1798 (8), 1457–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (128).Last NB; Kolmakova-Partensky L; Shane T; Miller C Mechanistic Signs of Double-Barreled Structure in a Fluoride Ion Channel. eLife 2016, 5, na DOI: 10.7554/eLife.18767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (129).Adelman JL; Sheng Y; Choe S; Abramson J; Wright EM; Rosenberg JM; Grabe M Structural Determinants of Water Permeation Through the Sodium-Galactose Transporter vSGLT. Biophys. J 2014, 106 (6), 1280–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (130).Sala-Rabanal M; Hirayama BA; Loo DD; Chaptal V; Abramson J; Wright EM Bridging the Gap Between Structure and Kinetics of Human SGLT1. Am. J. Physiol Cell Physiol 2012, 302 (9), C1293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (131).Simoes MC; Hughes KJ; Ingham DB; Ma L; Pourkashanian M Estimation of the Thermochemical Radii and Ionic Volumes of Complex Ions. Inorg. Chem 2017, 56 (13), 7566–7573. [DOI] [PubMed] [Google Scholar]
- (132).Cheng MH; Bahar I Complete Mapping of Substrate Translocation Highlights the Role of LeuT N-Terminal Segment in Regulating Transport Cycle. PLoS Comput. Biol 2014, 10 (10), No. e1003879. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.