Abstract
Case series
Patients: —
Final Diagnosis: Hereditary spastic paraplegia type 4
Symptoms: Progressive psychomotor deterioration • mixed seizure patterns • corneal opacity • dysostotic bones • limb spasticity with extensor plantar responses • axial hypotonia
Medication: —
Clinical Procedure: Phenotype-genotype correlation
Specialty: Genetics • Neurology
Objective:
Rare disease
Background:
Hereditary spastic paraplegia (HSP or SPG) consists of a heterogeneous group of disorders, clinically divided into pure and complex forms. The former is characterized by neurological impairment limited to lower-extremity spasticity. The latter presents additional symptoms such as seizures, psychomotor impairment, cataract, deafness, and peripheral neuropathy. The genetic structure of HSP is diverse, with more than 72 loci and 55 genes identified so far. The most common type is SPG4, accounting for 40% of cases. This case report describes 2 siblings presenting SPG4, one presumptive and one confirmed with a homozygous SPAST variant.
Case Reports:
Two siblings born to third-degree consanguineous and healthy parents presented a SPG4 complex phenotype characterized by progressive psychomotor deterioration, mixed seizure patterns, corneal opacity, dysostotic bones, limb spasticity with extensor plantar responses, and axial hypotonia. After ruling out most inborn errors of metabolism in one of the patients, the complexity of the case derived from exome sequencing. The identification of a homozygous variant in the SPAST gene established a diagnosis for SPG4. The phenotype-genotype did not correlate to classical manifestations, most likely due to the variant’s zygosity. Moreover, 34 patient’s relatives were identified with SPG4 clinical manifestations or asymptomatic with the same genetic variant in heterozygous state.
Conclusions:
We described visual loss and seizures for SPG4 complex phenotype associated with a homozygous variant in the SPAST gene. This diagnosis will lead clinicians to consider it as a differential diagnosis providing adequate genetic counseling.
MeSH Keywords: DNA Mutational Analysis; Exome; Spastic Paraplegia, Hereditary
Background
Hereditary spastic paraplegia (HSP or SPG), also known as familial spastic paraparesis or French settlement disease [1,2], consists of a heterogeneous group of disorders that are mainly characterized by progressive spasticity [2,3]. The main pathologic feature of HSP is degeneration of the longest nerve fibers in the corticospinal tract and posterior column [4].
The clinical presentation of this condition can range from no apparent, subtle, to many symptoms [1]. HSP can be classified as pure or complex. A pure form of HSP is considered if the associated neurological impairment is limited to lower-extremity spasticity, weakness, loss of vibratory sensibility, as well as urinary dysfunction. Additionally, complex forms are accompanied by seizures, cerebellar dysfunction, cataract, deafness, and peripheral neuropathy [5,6].
HSP can be inherited in an autosomal-dominant, autosomal-recessive, X-linked pattern, or via maternal mitochondrial transference [2,7–9]. Its frequency tends to increase with consanguinity [9]. HSP is a very rare disease with an estimated prevalence of 1.8 per 100 000 individuals [10,11], with some other reports indicating ranges from 4.3 to 9.8 per 100 000 individuals [11]. The associated types tend to have a variable penetrance and the age at onset varies greatly even within families [1,12].
The genetic configuration of HSP is diverse, with more than 72 loci and 55 genes actually identified [9,13]. Six genes frequently reported for HSP are: SPAST, ATL1, KIAA1840, CYP7B1, REEP1, and KIF5A [9]. Moreover, 40% of HSP cases are related with SPAST (spastin) pathogenic variants [3], associated to SPG4 (OMIM #182601), more commonly in its pure form [11].
Spastin, localized in chromosome 2p12, encodes an adenosine triphosphatase (ATPase) associated with a variety of cellular activities (AAA) domain that contribute to membrane modeling, as well as intracellular and axonal vesicle transport. This protein is involved in the conformation and stability of micro-tubules in the motor neurons. Its absence causes structural abnormalities and reductions in neuron length and function. Moreover, it can also elicit apoptosis in the absence of proper trophic stimulation [14]. Its intracellular role has been studied in animal models, where mutated spastin has been related to a degenerative process of the central nervous system [15,16].
The objective of this article is to report 2 complex SPG4 cases, one presumptive and one confirmed with a homozygous SPAST variant, associated with more severe clinical symptoms and earlier age at onset. Furthermore, we present the impact of this diagnosis, which led to the identification of 34 patients’ relatives, either with clinical manifestations and / or carrying the same variant in a heterozygous state.
Case Reports
Two siblings were born to third-degree consanguineous and healthy parents with French ancestry. The oldest sister presented severe clinical manifestations at an early age with rapid progression (Table 1). By age 12 years, she presented pneumonia and severe respiratory insufficiency that required tracheal intubation and subsequent artificial respiration via tracheostomy. At the age of 29 years she died due to pneumonia complications that led to respiratory insufficiency.
Table 1.
Index patient’s clinical manifestation and imaging findings in relation to the age at onset.
| Age (years) | 0.5 | 2 | 5 | 8 |
|---|---|---|---|---|
| Clinical manifestations | • Seizures (generalized tonic-clonic seizures and myoclonias) |
|
|
|
| Imaging findings | ||||
| EEG | Poor organization of the basal activity with superimposed theta activity | |||
| CT scan | Third and lateral ventricles dilatation | |||
| MRI | Dandy-Walker variant Cerebellar hypoplasia Asymmetric ventriculomegaly Thinning of the corpus callosum and medulla |
|||
| Visual evoked potentials | Asymmetry in absolute visual latencies | |||
CT scan – computed tomography scan, EEG – electroencephalogram, MRI – magnetic resonance imaging.
The younger brother presented with difficult-to-control, tonic–clonic seizures since infancy. By the age of 15 years, he presented developmental delay, corneal opacity, dysostotic bones in the extremities, and claw-shaped hands. He died due to pneumonia complications that led to a severe respiratory insufficiency.
Both patients had repeated laboratory studies at different institutions, with normal values for blood count, electrolytes, liver function tests, serum and urinary amino acids, urinary organic acids, phytanic acid, serum lactate, pyruvate, and ammonia. As most inborn errors of metabolism were already discarded, and due to the complexity of the case, exome sequencing was recommended. However, the male patient died before a blood sample could be collected, so only the oldest sister was tested.
In brief, genomic DNA was isolated from the female patient’s blood. The sample was prepared using the SureSelect Target Enrichment System (Agilent Technologies, Santa Clara, CA, USA). Data analysis was focused on nonsense and non-synonymous variants, small insertions, and deletions, as well as known splice site alterations. The Ambry Variant Analyzer tool (AVA) was used to annotate nucleotide and amino acid conservation, their biochemical nature, the predicted functional impact, and the population frequency. The previously described genetic variants and polymorphisms were searched in the Single Nucleotide Polymorphism database (dbSNP), 1000 genomes, the Human Gene Mutation Database (HGMD), HapMap data, and online search engines (e.g., PubMed). Variants were further filtered based on possible inheritance models and family history. Co-segregation studies were performed only for the mother.
The female patient’s exome sequencing assay reported a homo-zygous c.1634C>G (rs869312949) variant in the SPAST gene, which produces a nonsense variant (p.S545X) on the encoded protein. These findings supported the likelihood of the homozygous SPAST variant being the cause of the female patient’s clinical symptoms. Additionally, the assay detected a heterozygous c.1165A>G variant in the GABRA1 gene, which produces a protein change in p.I389V (missense variant).
Related to the co-segregation analysis, it was only possible to collect a sample from the mother, in which the same variant in SPAST was confirmed in a heterozygous state without findings within the GABRA1 gene.
Discussion
HSP is a heterogeneous group of neurodegenerative disorders caused by genetic variants in more than 55 genes [17]. Molecular changes within the SPAST gene are the most frequent etiology in the autosomal-dominant form of HSP. This gene encodes spastin, haploinsufficiency of which leads to loss-of-function in microtubule severing [16,18]. Animal model studies concluded that more than one mechanism may also contribute to the pathogenesis, currently demonstrating that the disease is not fully understand [18,19].
Most patients diagnosed with SPG4 have shown clinical symptoms consistent with the HSP pure phenotype [11]. However, our patients presented additional signs such as psychomotor impairment, epilepsy, cataracts, and bone abnormalities. These are considered atypical findings for SPG4. Some authors suggested that the presence of genetic and epigenetic changes, unrelated to SPAST, could explain those manifestations [20,21].
To the best of our knowledge, this is the first case report of a homozygous truncating variant in the SPAST gene, which also presented a complex form for SPG4. The same phenomenon was also observed in mice with homozygous SPAST deletion [15]. The degenerative process, increased in homozygous mice, is characterized by focal axonal swellings and accumulation of cell components. Based on the available evidence to date, the c.1634C>G (p.S545X) alteration in the SPAST gene is expected to be deleterious in nature and a likely pathogenic variant.
In relation to the additional finding, variants in the GABRA1 gene have been associated with an autosomal-dominant form of juvenile myoclonic epilepsy (JME), as well as childhood absence epilepsy (CAE) [22]. Analyses in silico were carried out for the unpublished GABRA1 variant found. SIFT predicted the substitution to be tolerated (score of 0.42); while Polyphen-2 considered it to be possibly damaging (score of 0.956). Previous in silico analyses reported several variants on GABAA receptor subunit genes; even though p.I389V GABRA1 variant was not reported, the variants in the same domain were considered as benign [22]. Moreover, due to the lack of paternal co-segregation, it was not possible to conclude if the variant was de novo or inherited. Thus, the contribution of the index patient’s altered GABRA1 gene to the present clinical course remained uncertain.
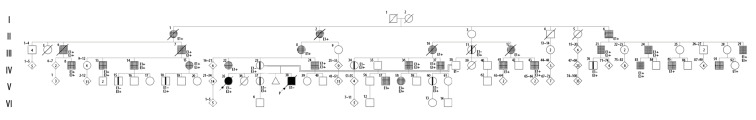
Due to the inheritance pattern, the family received broad genetic counseling, diagnosing 26 affected family members with SPG4 and 8 relatives with no clinical manifestations, but confirmed with the SPAST heterozygous variant (Figure 1). In the nuclear family, the lack of an observable phenotype in the patients’ parents may be due to age-related penetrance (Figure 1: IV–23 and IV–38). Moreover, the sister of the index case (Figure 1: V–37) was confirmed with the heterozygous variant but had no apparent symptoms at the time of evaluation.
Figure 1.
Pedigree of a family with hereditary spastic paraplegia (HSP) according to the standardized human pedigree nomenclature [23].

 Heterozygous SPAST variant with no clinical manifestations (III11; IV23,34,70; V15,18,37,60).
Heterozygous SPAST variant with no clinical manifestations (III11; IV23,34,70; V15,18,37,60).

 Affected individuals with HSP (II1,2,6; III6,7,8,10,12,21,24,29; IV8,13,14,15,22,24,36,37,41,43,83,85,91; V57,58). ● ■ Homozygous SPAST mutation (V35,38). E1 – physical examination for HSP; E2 – clinical exome sequencing (c.1634C>G SPAST); E3 – specific mutation analysis (c.1634C>G SPAST).
Affected individuals with HSP (II1,2,6; III6,7,8,10,12,21,24,29; IV8,13,14,15,22,24,36,37,41,43,83,85,91; V57,58). ● ■ Homozygous SPAST mutation (V35,38). E1 – physical examination for HSP; E2 – clinical exome sequencing (c.1634C>G SPAST); E3 – specific mutation analysis (c.1634C>G SPAST).
In the extended family, only 2 women presented classic clinical manifestations for SPG4 at earlier ages (Figure 1: IV–15 and V–58), including spastic paraparesis and urinary disturbances. However, the patients’ maternal uncle (Figure 1: IV–24) was a confirmed case of SPG4 presenting spastic dysarthria. In general, the affected relatives had late-onset manifestations and were characterized by their variable expression.
A notable feature in this family is that, according to the previously established genealogy, the great-grandparents were of French origin and settled in Mexico. Anecdotally, their great-grandparents presented with gait disturbances (they were referred to as “the ones that limp”), and they may have been part of the French migration to North America and Mexico in the late nineteenth century [24]. In fact, the main manifestation observed in the family is that women “drag their feet” at around 70 years of age. This family history and the SPAST variant finding suggest a founder effect.
The phenotypic variability seen in this family could be caused by unknown epigenetic factors. Furthermore, it seems to be an incomplete penetrance, being important to assess and establish more genotype-phenotype correlations [11]. Previous reports showed that sex and the nature of the SPAST variant act as modifiers of the phenotype. A higher prevalence in males is reported, and estrogens might act as a protective factor, leading to lower penetrance [17]. However, the SPAST variants are correlated with the age at onset, in which missense variants cause a first-decade peak [17,21].
Conclusions
In this article we reported novel clinical manifestations for SPG4 in 2 siblings (one confirmed and one presumptive), possibly associated with a homozygous variant in the SPAST gene. These manifestations were developmental delay, loss of psychomotor abilities, dysostotic bones, and visual disturbances. The homozygous state of this variant had an earlier age at onset and severe phenotypes. The multiple mechanisms of pathogenesis and the impact of other that described for this type of HSP.
Since HSP has a variable penetrance within a variety of inheritance patterns, and given the differences in the precision and accuracy of genetic diagnoses, appropriate genetic counseling is needed. A definitive genetic diagnosis allows testing of other relatives who could be affected, ending the diagnostic odyssey within families. The findings could enable affected individuals to receive appropriate medical follow-up and optimal therapeutic approaches, as well as family planning based on recurrence risks.
This case report helps expand the possible diagnosis by reporting the atypical expression of a known disease. Further molecular studies may allow the identification of other genetic variants that could modify the expression of this disease.
Acknowledgments
We thank the mother of the children presented within this manuscript. English-language editing of this manuscript was provided by Journal Prep.
Footnotes
Conflict of interest
None.
References:
- 1.Meijer IA, Hand CK, Cossette P, et al. Spectrum of SPG4 mutations in a large collection of North American families with hereditary spastic paraplegia. Arch Neurol. 2002;59:281–86. doi: 10.1001/archneur.59.2.281. [DOI] [PubMed] [Google Scholar]
- 2.McDermott C, White K, Bushby K, et al. Hereditary spastic paraparesis: A review of new developments. J Neurol Neurosurg Psychiatry. 2000;69:150–60. doi: 10.1136/jnnp.69.2.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henson BJ, Zhu W, Hardaway K, et al. Transcriptional and post-transcriptional regulation of SPAST, the gene most frequently mutated in hereditary spastic paraplegia. PLoS One. 2012;7:e36505. doi: 10.1371/journal.pone.0036505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salinas S, Proukakis C, Crosby A, et al. Hereditary spastic paraplegia: Clinical features and pathogenetic mechanisms. Lancet Neurol. 2008;7:1127–38. doi: 10.1016/S1474-4422(08)70258-8. [DOI] [PubMed] [Google Scholar]
- 5.Fink JK. Hereditary spastic paraplegia: Clinico-pathologic features and emerging molecular mechanisms. Acta Neuropathol. 2013;126:307–28. doi: 10.1007/s00401-013-1115-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Züchner S, Kail ME, Nance MA, et al. A new locus for dominant hereditary spastic paraplegia maps to chromosome 2p12. Neurogenetics. 2006;7:127–29. doi: 10.1007/s10048-006-0029-1. [DOI] [PubMed] [Google Scholar]
- 7.Faber I, Servelhere KR, Martinez ARM, et al. Clinical features and management of hereditary spastic paraplegia. Arq Neuropsiquiatr. 2014;72:219–26. doi: 10.1590/0004-282x20130248. [DOI] [PubMed] [Google Scholar]
- 8.Finsterer J, Löscher W, Quasthoff S, et al. Hereditary spastic paraplegias with autosomal dominant, recessive, X-linked, or maternal trait of inheritance. J Neurol Sci. 2012;318:1–18. doi: 10.1016/j.jns.2012.03.025. [DOI] [PubMed] [Google Scholar]
- 9.Lo Giudice T, Lombardi F, Santorelli FM, et al. Hereditary spastic paraplegia: Clinical-genetic characteristics and evolving molecular mechanisms. Exp Neurol. 2014;261:518–39. doi: 10.1016/j.expneurol.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 10.Ruano L, Melo C, Silva MC, et al. The global epidemiology of hereditary ataxia and spastic paraplegia: A systematic review of prevalence studies. Neuroepidemiology. 2014;42:174–83. doi: 10.1159/000358801. [DOI] [PubMed] [Google Scholar]
- 11.de Souza PVS, de Rezende Pinto WBV, de Rezende Batistella GN, et al. Cerebellum. Springer; New York LLC: 2017. Hereditary spastic paraplegia: Clinical and genetic hallmarks; pp. 525–51. [DOI] [PubMed] [Google Scholar]
- 12.Fonknechten N, Mavel D, Byrne P, et al. Spectrum of SPG4 mutations in autosomal dominant spastic paraplegia. Hum Mol Genet. 2000;9:637–44. doi: 10.1093/hmg/9.4.637. [DOI] [PubMed] [Google Scholar]
- 13.Novarino G, Fenstermaker AG, Zaki MS, et al. Exome sequencing links corticospinal motor neuron disease to common neurodegenerative disorders. Science. 2014;343:506–11. doi: 10.1126/science.1247363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wood JD. The microtubule-severing protein Spastin is essential for axon outgrowth in the zebrafish embryo. Hum Mol Genet. 2006;15:2763–71. doi: 10.1093/hmg/ddl212. [DOI] [PubMed] [Google Scholar]
- 15.Tarrade A, Fassier C, Courageot S, et al. A mutation of spastin is responsible for swellings and impairment of transport in a region of axon characterized by changes in microtubule composition. Hum Mol Genet. 2006;15:3544–58. doi: 10.1093/hmg/ddl431. [DOI] [PubMed] [Google Scholar]
- 16.Qiang L, Piermarini E, Baas PW. New hypothesis for the etiology of SPAST-based hereditary spastic paraplegia. Cytoskeleton (Hoboken) 2019;76(4):289–97. doi: 10.1002/cm.21528. [DOI] [PubMed] [Google Scholar]
- 17.Parodi L, Fenu S, Barbier M, et al. Spastic paraplegia due to SPAST mutations is modified by the underlying mutation and sex. Brain. 2018;141:3331–42. doi: 10.1093/brain/awy285. [DOI] [PubMed] [Google Scholar]
- 18.Shribman S, Reid E, Crosby AH, et al. Hereditary spastic paraplegia: From diagnosis to emerging therapeutic approaches. Lancet Neurol. 2019;18(12):1136–46. doi: 10.1016/S1474-4422(19)30235-2. [DOI] [PubMed] [Google Scholar]
- 19.Kawarai T, Montecchiani C, Miyamoto R, et al. Spastic paraplegia type 4: A novel SPAST splice site donor mutation and expansion of the phenotype variability. J Neurol Sci. 2017;380:92–97. doi: 10.1016/j.jns.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 20.Guthrie G, Pfeffer G, Bailie M, et al. The neurological and ophthalmological manifestations of SPG4-related hereditary spastic paraplegia. J Neurol. 2013;260(3):906–9. doi: 10.1007/s00415-012-6780-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parodi L, Lynne Rydning S, Tallaksen C, et al. Spastic paraplegia 4. Gene Rev. 2019:19. [Google Scholar]
- 22.Hernandez CC, Klassen TL, Jackson LG, et al. Deleterious rare variants reveal risk for loss of gabaa receptor function in patients with genetic epilepsy and in the general population. PLoS One. 2016;11:e0162883. doi: 10.1371/journal.pone.0162883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bennett RL, French KS, Resta RG, et al. Standardized human pedigree nomenclature: Update and assessment of the recommendations of the National Society of Genetic Counselors. J Genet Couns. 2008;17:424–33. doi: 10.1007/s10897-008-9169-9. [DOI] [PubMed] [Google Scholar]
- 24.Dugast G-A. La tentation mexicaine en France au XIXème siècle: L’image du Mexique et l’Intervention française 1821–1862. 2008. [in French]