Abstract
Background:
This study will explore the association between Ki-67 expression and clinical pathological characteristics (CPC) of colorectal cancer (CC).
Methods:
We will search relevant studies from electronic databases (Cochrane Library, PUBMED, EMBASE, Scopus, Cumulative Index to Nursing and Allied Health Literature, China Biology Medicine, and China National Knowledge Infrastructure) from beginning to April 1, 2020 without language and publication time limitations. We will consider all case-controlled studies (CCSs) or randomized controlled studies (RCSs) investigating the association between Ki-67 expression and CPC of CC. We will appraise study quality of CCSs by Newcastle–Ottawa Scale, and RCSs by Cochrane risk of bias tool. Statistical analysis will be carried out by Review Manager 5.3 software.
Results:
The present study will explore the association between Ki-67 expression and CPC of CC.
Conclusion:
Its findings may summarize scientific evidence of the association between Ki-67 expression and CPC of CC, and may provide helpful evidence for clinical practice.
Systematic review registration: PROSPERO CRD42020173795.
Keywords: clinical pathological characteristic, colorectal cancer, Ki-67 expression
1. Introduction
Colorectal cancer (CC), also known as colorectal adenocarcinoma, is the third most commonly diagnosed cancer in males and the second in females.[1–4] It is also one of the most leading reasons of cancer mortality around the world.[5–8] It has been reported that about 1,096,000 new cases of colon cancer and about 704,000 new cases of rectal cancer were diagnosed in 2018.[3] Types of CC include colorectal adenocarcinoma, gastrointestinal carcinoid tumors, primary colorectal lymphomas, gastrointestinal stromal tumors, leiomyosarcomas, and melanomas.[9–14]
Previous studies have reported the association between Ki-67 expression and clinical pathological characteristics (CPC) in patients with CC.[15–31] However, no study has explored the association between Ki-67 expression and CPC in patients with CC at evidenced-medicine level. Thus, this study firstly investigated the association between Ki-67 expression and CPC in patients with CC.
2. Methods
2.1. Study registration
This study has been registered on PROSPERO (CRD42020173795), and has been reported according to the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISRMA) Protocol statement guidelines.[32]
2.2. Eligibility criteria
2.2.1. Types of trials
All potential case-controlled studies (CCSs) or randomized controlled studies (RCSs) that explored the association between Ki-67 expression and CPC of CC will be included. We will exclude any other studies, such as review, comment, and uncontrolled studies.
2.2.2. Types of subjects
We will include all patients who were diagnosed as CC, regarding gender, age, and race.
2.2.3. Types of exposures
In the experimental group, CC tissues from patients with CC were collected.
In the control group, normal tissues adjacent to the CC in patients with CC were harvested.
2.2.4. Types of outcome measurements
Primary outcomes are protein and gene expressions of Ki-67. Gene expression of Ki-67 was measured by real-time polymerase chain reaction. Protein expression of Ki-67 was detected by immunofluorescence or western blot test.
Secondary outcomes are associations between Ki-67 expression and gender, age, clinical stages, tumor sites, pathological features, and lymph node metastasis.
2.3. Literature search and search strategy
The primary sources of literature search will be sought in electronic databases (Cochrane Library, PUBMED, EMBASE, Scopus, Cumulative Index to Nursing and Allied Health Literature, China Biology Medicine, and China National Knowledge Infrastructure) from initiation to April 1, 2020 with no language and publication time restrictions. The CCSs or RCSs that explore the association between Ki-67 expression and CPC of CC will be included. The search strategy details of Cochrane Library are summarized (Table 1). Similar search strategies will be adapted to other electronic databases.
Table 1.
Search strategy used in Cochrane Library database.
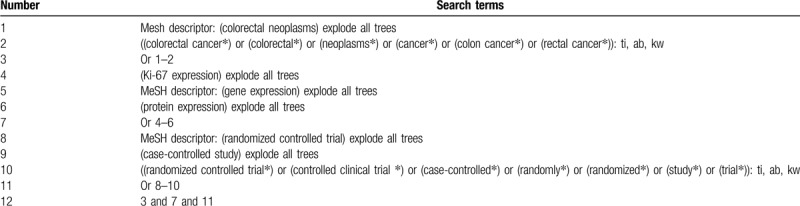
The secondary sources are Google Scholar, conference abstracts, clinical registrations, and reference lists of relevant reviews.
2.4. Study selection
Two researchers will independently identify titles/abstracts of all retrieved studies to remove all irrelevant records. Full-papers of potential studies will be read carefully against eligibility criteria. Any uncertainties between 2 researchers will be solved by a third researcher through discussion. We will record reasons for all excluded literatures. The procedure of study selection will be exerted in a PRISRMA flow chart.
2.5. Data extraction and management
Two researchers will independently extract data using a predefined data extraction sheet. The extracted information comprises of general information (e.g., first author, year of publication, region, gender, ethnicity, disease types, and diagnostic criteria), types of studies (CCSs or RCSs), study quality, outcome measurements (primary and secondary outcomes, and any others), results, and findings. Any differences between 2 researchers will be resolved by a third researcher through discussion.
2.6. Study quality assessment
Two researchers will independently assess study quality for all eligible studies. We will resolve any differences with the help of a third researcher. We will evaluate study quality for CCSs by Newcastle–Ottawa Scale, and that for RCSs by Cochrane risk of bias tool.
2.7. Statistical analysis
RevMan 5.3 software will be used for statistical analysis. We will estimate the treatment effects of dichotomous data as risk ratio and 95% confidence intervals (CIs), and will calculate the treatment effects of continuous data as mean difference or standardized mean difference and 95% CIs. We will employ I2 test to explore statistical heterogeneity across eligible studies. I2 ≤ 50% indicates homogeneity, and a fixed-effects model will be used to pool data. A meta-analysis will be performed if it is necessary. I2 > 50% exerts considerable heterogeneity, and a random-effects model will be utilized to synthesize data, and a subgroup analysis will be carried out to detect the sources of obvious heterogeneity. In addition, we will also report study results by a narrative summary.
2.8. Additional analysis
2.8.1. Subgroup analysis
A subgroup analysis will be performed to identify heterogeneity sources based on the different types of studies, study characteristics, and outcome indicators.
2.8.2. Sensitivity analysis
A sensitivity analysis will be carried out to test robustness of study findings by removing low quality studies.
2.8.3. Reporting bias
Reporting bias will be examined by funnel plot and Egger's regression test if more than 10 studies are included.[33,34]
2.9. Dissemination and ethics
This study will not harvest individual patient data, thus, no ethical approval is required. We expect to publish this study on a peer-reviewed journal or a conference presentation.
3. Discussion
This study will explore the association between Ki-67 expression and CPC in patients with CC. We will collect potential studies from electronic databases and other literature sources. All possible studies will be included to investigate the association between Ki-67 expression and CPC in patients with CC. Although previous studies have reported that Ki-67 expression is associated with CPC of CC, their results are still inconsistent. This study will perform a comprehensive synthesis to assess the association between Ki-67 expression and CPC in patients with CC. We hope that this study can provide a helpful evidence of the association between Ki-67 expression and CPC of CC, which is beneficial to both patients and practitioners.
Author contributions
Conceptualization: Jing Li, Hai-bo Yu, Qing Xue, Hai-tao Yu, Weeks Christina.
Data curation: Jing Li, Zhi-ye Liu, Hai-bo Yu, Xiu-sheng Qu, Qing Xue.
Formal analysis: Jing Li, Zhi-ye Liu, Xiu-sheng Qu, Qing Xue, Hai-tao Yu.
Funding acquisition: Jing Li.
Investigation: Qing Xue.
Methodology: Hai-bo Yu, Xiu-sheng Qu, Qing Xue, Hai-tao Yu.
Resources: Jing Li, Zhi-ye Liu, Hai-bo Yu, Xiu-sheng Qu, Qing Xue, Hai-tao Yu.
Software: Zhi-ye Liu, Xiu-sheng Qu, Qing Xue, Hai-tao Yu.
Validation: Jing Li, Zhi-ye Liu, Hai-bo Yu, Xiu-sheng Qu, Hai-tao Yu, Weeks Christina.
Visualization: Hai-bo Yu, Xiu-sheng Qu, Qing Xue, Hai-tao Yu, Weeks Christina.
Writing – original draft: Jing Li, Zhi-ye Liu, Hai-bo Yu, Xiu-sheng Qu, Qing Xue, Hai-tao Yu, Weeks Christina.
Writing – review & editing: Jing Li, Zhi-ye Liu, Xiu-sheng Qu, Qing Xue, Hai-tao Yu, Weeks Christina.
Footnotes
Abbreviations: CC = colorectal cancer, CCSs = case-controlled studies, CIs = confidence intervals, CPC = clinical pathological characteristics, PRISRMA = Preferred Reporting Items for Systematic Reviews and Meta-Analysis, RCSs = randomized controlled studies.
How to cite this article: Li J, Liu Zy, Yu Hb, Qu Xs, Xue Q, Yu Ht, Weeks C. The association between Ki-67 expression and the clinical pathological characteristics of colorectal cancer: A protocol for a systematic review and meta-analysis. Medicine. 2020;99:21(e19996).
JL and Z-YL contributed equally to this study.
This work is supported by the Scientific Research Project of Heilongjiang Provincial Health and Health Committee (2019-326).
The authors would like to thank Northern medicine and functional food special subject construction project for assistance with some of the experiments.
The authors have no conflicts of interest to disclose.
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
References
- [1].Wong MC, Ding H, Wang J, et al. Prevalence and risk factors of colorectal cancer in Asia. Intest Res 2019;17:317–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Park CH, Kim NH, Park JH, et al. Impact of family history of colorectal cancer on age-specific prevalence of colorectal neoplasia. J Gastroenterol Hepatol 2019;34:537–43. [DOI] [PubMed] [Google Scholar]
- [3].Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- [4].Idrissi Janati A, Karp I, Sabri H, et al. Is a fusobacterium nucleatum infection in the colon a risk factor for colorectal cancer?: a systematic review and meta-analysis protocol. Syst Rev 2019;8:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Matsuda T, Okuyama A. The estimates of 5-year colorectal cancer prevalence in adult population in 2012. Jpn J Clin Oncol 2017;47:669–70. [DOI] [PubMed] [Google Scholar]
- [6].Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, et al. Cancer incidence and mortality patterns in Europe: estimates for 40 countries in 2012. Eur J Cancer 2013;49:1374–403. [DOI] [PubMed] [Google Scholar]
- [7].Delaney J, Cui R, Engel A. Risk of bias judgements and strength of conclusions in meta-evidence from the Cochrane Colorectal Cancer Group. Syst Rev 2019;8:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Goodwin BC, Ireland MJ, March S, et al. Strategies for increasing participation in mail-out colorectal cancer screening programs: a systematic review and meta-analysis. Syst Rev 2019;8:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Stark UA, Frese T, Unverzagt S, et al. What is the effectiveness of various invitation methods to a colonoscopy in the early detection and prevention of colorectal cancer? Protocol of a systematic review. Syst Rev 2020;9:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lombardi N, Bettiol A, Crescioli G, et al. Association between anthraquinone laxatives and colorectal cancer: protocol for a systematic review and meta-analysis. Syst Rev 2020;9:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Silva M, Guerreiro I, Abreu S, et al. Prognostic factors in metastatic colorectal cancer. Ann Oncol 2018;29: Suppl 5: v78–9. [Google Scholar]
- [12].Aliane H, Ghomari-Bezzar S, Belhadj A, et al. Characteristics of colorectal cancer in the elderly patients about 60 cases. Ann Oncol 2018;29: Suppl 5: v80. [Google Scholar]
- [13].Kaidarova D, Smagulova K, Yesentaeva S, et al. Advanced colorectal cancer and risk factors for survival. Ann Oncol 2018;29: Suppl 5: v75–6. [Google Scholar]
- [14].Tao L, Jin L, Dechun L, et al. Galectin-3 expression in colorectal cancer and its correlation with clinical pathological characteristics and prognosis. Open Med (Wars) 2017;12:226–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Aladhraei M, Kassem Al-Thobhani A, Poungvarin N, et al. Association of XPO1 overexpression with NF-κB and Ki67 in colorectal cancer. Asian Pac J Cancer Prev 2019;20:3747–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Woldemeskel M, Hawkins I, Whittington L. Ki-67 protein expression and tumor associated inflammatory cells (macrophages and mast cells) in canine colorectal carcinoma. BMC Vet Res 2017;13:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wang XL, Jin LL, Zhang Y, et al. Expressions of RPN11 and Ki67 in colorectal cancer tissues and their clinicopathological significance. Chin J Histochem Cytochem 2019;28:247–51. [Google Scholar]
- [18].Bao WF, Zhang Q, Cao L. Correlation analysis of intra-rectal ultrasound blood flow resistance index with VEGF, Ki67 expression and pathological factors of rectal cancer. Ningxia Med J 2019;41:193–5. [Google Scholar]
- [19].Xiong F, Chen JG. Clinical effect of tigio combined with oxaliplatin in resectable colorectal cancer and its relationship with tissue Cyclin E and Ki67 protein expression. Mod Med 2017;45:1650–3. [Google Scholar]
- [20].Li Y, Jiang SP, Gao X. The relationship and significance of Ki67, CK20 and CEA expression with clinicopathological characteristics of colorectal cancer. Adv Mod Gen Surg China 2017;20:394–6. [Google Scholar]
- [21].Li P, Xiao ZT, Braciak TA, et al. Association between Ki67 index and clinicopathological features in colorectal cancer. Oncol Res Treat 2016;39:696–702. [DOI] [PubMed] [Google Scholar]
- [22].Yang CK, Guan S, Ying MG. Effects of CO2 pneumoperitoneum on the expression of thymidine kinase 1 and Ki67 in colorectal carcinoma cells. Surg Endosc 2014;28:2863–70. [DOI] [PubMed] [Google Scholar]
- [23].Li W, Zhang G, Wang HL, et al. Expressions and significance of cyclin E, P27kipl and Ki67 in rectal cancer tissues. Chin J Gen Surg (Electronic Edition) 2016;10:422–5. [Google Scholar]
- [24].Zhang ZY, Lei XY, Ji YJ, et al. Expression and clinical significance of HER-2 and Ki-67 in rectal cancer tissues. Hebei Med J 2016;38:2891–4. [Google Scholar]
- [25].Liu P, Xiao XL, Long HA, et al. Clinicopathological significance and correlation of Ki67 and P53 expression in colorectal cancer. J Luzhou Med Coll 2013;36:567–72. [Google Scholar]
- [26].Ye B, Wang XC, Lei L, et al. Expressions of p53 and Ki67 in colorectal cancer and their clinical significance. J Pract Cancer 2013;28:42–4. [Google Scholar]
- [27].Yue ZY, Ma YH, Shen LS, et al. Expressions and significance of Treg, Ki67 and VEGF in colorectal cancer. Int J Lab Med 2012;33:1288–90. [Google Scholar]
- [28].Shi FT, Qin SJ, Kong WM. Expressions of Ki67 and OPN in colorectal cancer and their clinical significance. Chin Commun Phys (Medical Specialty) 2012;14:282. [Google Scholar]
- [29].Jiang GS, Zhang G, Ren WZ. Expressions and pathological significance of Cyclin E, p27kipl and Ki67 in rectal cancer. Hebei Med J 2009;31:2391–2. [Google Scholar]
- [30].Hou H, Xiao MM, Ren QH, et al. Expressions and clinical significance of p53 protein and Ki67 in colorectal cancer. J Pract Oncol 2008;22:424–6. [Google Scholar]
- [31].Yang ZL, Ying WY, Lu XA, et al. Expression of CDX2 and ki67 and DNA ploidy analysis in colorectal cancer. J Oncol 2007;2:103–5. [Google Scholar]
- [32].Shamseer L, Moher D, Clarke M, et al. PRISMA-P Group. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015: elaboration and explanation. BMJ 2015;349:g7647. [DOI] [PubMed] [Google Scholar]
- [33].Sutton AJ, Duval SJ, Tweedie RL, et al. Empirical assessment of effect of publication bias on meta-analyses. BMJ 2000;320:1574–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Egger M, Davey Smith G, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997;315:629–34. [DOI] [PMC free article] [PubMed] [Google Scholar]


