Abstract
Background:
The role of xeroderma pigmentosum complementation group D (XPD) gene polymorphisms in breast and ovarian cancer development has long been controversial and existing data were inconsistent. Here, we conducted a comprehensive systemic review and meta-analysis to better clarify the association.
Methods:
Relevant case-control studies published in electronic data base from October 1999 to September 2019 were assessed. The statistical analyses of the pooled odds ratios (ORs) and the corresponding 95% confidence intervals (95%CIs) were calculated by using Revman 5.2 software (Cochrane Collaboration, Copenhagen).
Results:
31 articles including 38 case-control studies and 2 XPD polymorphisms (rs1799793 and rs238406) were analyzed. The results showed statistical significance in heterozygous mutants among Asian population for rs1799793 (GA vs GG + AA: OR = 1.38, 95%CI = 1.21–1.56), and Caucasian population for rs238406 (CA vs AA + CC: OR = 0.63, 95%CI = 0.49–0.80), while the rest comparisons including overall groups and subgroups stratified by cancer types and ethnicity failed to indicate any association with breast and ovarian cancer risk.
Conclusions:
The current meta-analysis suggested no concrete correlation of XPD rs1799793(G/A) and rs238406(C/A) polymorphisms with breast cancer or ovarian cancer susceptibility. However, it indicated that heterozygous genotypes might share different pathophysiologic mechanism from not only homozygous wildtypes but also homozygous mutants. More case–control studies with well-adjusted data and diverse populations are essential for validation of our conclusion.
Keywords: breast cancer, ovarian cancer, polymorphism, risk, susceptibility, xeroderma pigmentosum complementation group D, xeroderma pigmentosum complementation group
1. Introduction
Breast and ovarian cancers are 2 leading causes of mortality in women globally. It is reported that 1 in 8 women in the United States will develop breast cancer in her lifetime and 5-year survival rate of ovarian cancer remains in an extremely poor rate of 30% to 40%.[1,2] Although multiple etiologic factors and corresponding therapies have been explored for breast and ovarian cancer risk, the most recent breakthrough in breast and ovarian cancer treatment is the endorsement of PARP inhibitor, which was approved in pts that harbor mutations in either BRCA1 or BRCA2, the 2 most important genes that are involved in homologous recombination triggered by DNA double strand break.[3–5] This implies the importance of detecting inherited DNA repair related genes to prevent carcinogenesis and of developing new therapies that target those genes in breast and ovarian cancer.
Despite homologous recombination repair pathway, there are multiple other pathways to repair different types of DNA damage and maintain genomic integrity. Among these pathways is nucleotide excision repair (NER) pathway that repairs damages including cross-links, oxidative damage and bulky adducts. Xeroderma pigmentosum complementation group D (XPD), also known as ERCC2, plays important roles in the nucleotide NER pathway. The XPD gene is located on chromosome 19q13.3, comprises 23 exons, and spans approximately 54,000 base pairs.[6–8] It encodes an evolutionarily conserved helicase that participates in DNA unwinding and the recognition of bulky adducts and thymidine dimers.
The relation between XPD and multiple cancer types has been recently explored, but with inconsistent results. For instance, Costa et al analyzed DNA samples from 141 ovarian cancer patients and 202 control subjects for XPD polymorphisms using polymerase chain reaction - restriction fragment length polymorphism and observed that XPD rs1799793 genotype carriers have increased susceptibility of ovarian cancer, especially for early stage diseases.[9] However, Bernard-Gallon compared 51 ovarian cancer cases with 1000 controls and conclude that neither homologous mutants nor heterozygous genotypes in rs1799793 had any association with increased risk of ovarian cancer compared with wild genotypes.[10] Gomes-Diaz et al conducted a case-control study to explore the association between the ERCC1 and ERCC2 gene variants and 3 different types of cancer in Mexican patients, but only concluded that rs1799793 was associated with breast cancer.[11] Notably, several meta-analyses were published to clarify the relationship between XPD and various cancer types. For example, one study incorporated 86 articles with 38,848 cases and 48,928 controls including head and neck cancer, gastric cancer, lung cancer, bladder cancer, colorectal cancer as well as hematological malignancies. It concludes that XPD Asp312Asn polymorphism was associated with increased cancer risk, particularly in Asian populations.[12] However, the problem is that not all cancer types share same extent of risks to certain DNA damage genes considering the heterogeneity of different cancer types, thus the conclusion may be confounded by XPD susceptible cancers and is hard to transfer to every cancer type. In consideration of the interactive management of breast and ovarian cancer patients, we conducted a comprehensive systemic review and meta-analysis of relevant case-control studies published in electronic databases, with objective to better clarify the association of XPD polymorphisms in the risk of breast cancer and ovarian cancer.
2. Materials and methods
2.1. Search for eligible literature
A comprehensive electronic search was performed using PubMed, Medline (Ovid), Embase, Weipu, Wanfang and CNKI databases for studies published from October 1999 to September 2019 with the following terms and keywords
“xeroderma pigmentosum complementation group D”, “XPD”, “ovarian cancer”, “breast cancer”, “polymorphism”, “variant” and “mutation”. The search was updated every week until September 25, 2019. No ethical approval and patient consent are required because all analyses were based on previously published studies. The analysis is not a registered study.
2.2. Inclusion and exclusion criteria
Articles fulfilling the following criteria were included:
-
(1)
studied possible XPD polymorphisms in breast and ovarian cancer patients,
-
(2)
provided sufficient data in both case and control groups to calculate the odds ratios (ORs) and the corresponding 95% confidence intervals (95% CIs)
-
(3)
pooled polymorphism should be studied in at least 2 independent studies in order to conduct meta-analysis.
-
(4)
case-control studies.
When duplicate data were present in different articles, only the latest 1 would be taken into consideration. In addition, Newcastle-Ottawa Scale (NOS) was used to assess the quality of the observational studies included. Three aspects of selection, comparability, and exposure (9 scores in total) were carefully assessed. Studies of moderate or high quality were included (score higher than 5).[13] Articles that didn’t fulfill the criteria mentioned above were excluded.
2.3. Data extraction
All potential studies were investigated by 2 independent reviewers from the author list. The following items were extracted: first author, year of publication, ethnicity, cancer type, single nucleotide polymorphisms, control type, genotyping method, source of control. Any discrepancies would be resolved by discussion with a third author until a consensus was reached.
2.4. Statistical analysis
Pooled ORs and corresponding 95% CIs (confidence intervals) were calculated to explore the association of XPD polymorphisms with breast and ovarian cancer risk. Single nucleotide polymorphisms of XPD were considered as binary with dominant allele and mutated allele. Different contrast models were judged:
-
(1)
homozygous mutants contrast (mut/mut vs dom/mut + dom/dom),
-
(2)
homozygous and heterozygous mutants contrast (mut/mut + dom/mut vs dom/dom),
-
(3)
heterozygous mutants contrast (dom/mut vs dom/dom + mut/mut),
-
(4)
homozygous mutants contrast in homozygotes (mut/mut vs dom/dom),
-
(5)
mutant allelic contrast (mutated allele vs dominant allele).
-
(6)
Besides the overall comparisons, we also performed subgroup analyses stratified by cancer type and ethnicity. Heterogeneity assumptions were tested using Higgins I2 test. When the I2 value was less than 50%, a fixed-effects model was used otherwise a random-effects model was applied.[14] The Z test was performed to determine the significance of the pooled ORs where P less than .05 would be considered statistically significant.[15] The presence of publication bias was evaluated by visually inspecting the asymmetry in funnel plots. When the funnel plots showed visible asymmetry, Egger test was performed to further measure the bias, which was considered as existing when P was less than .05.[16] The pooled ORs and corresponding 95% CIs were calculated using the Revman 5.2 software (Cochrane Collaboration, Copenhagen) while the Egger test was performed using STATA 14.0 (StataCorp LP, College Station, TX).
3. Results
3.1. Search results
235 results returned after the primary search. Based on titles and abstracts, 10 articles were duplicates, 92 articles were not related to XPD polymorphisms and cancer risk while 42 articles reported XPD polymorphisms in other cancer types such as lung cancer and bladder cancer thus were excluded. 36 studies were excluded for non-case-control studies such as meta-analysis and lab research. Furthermore, 24 articles were excluded for analyzing different types of mutated alleles that could not be pooled with other independent articles (Fig. 1). For the remaining 31 articles, 23 were of moderate quality (NOS score of 6 or 7) and 8 were of high quality (NOS score of 8 or 9) therefore were all included in this meta-analysis (Table 1).[9–11,17–44]
Figure 1.
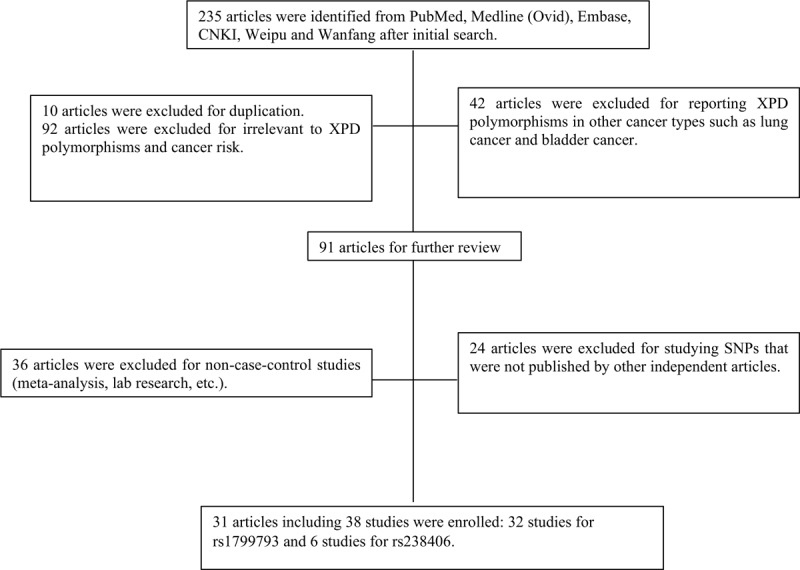
The flow chart of study selection.
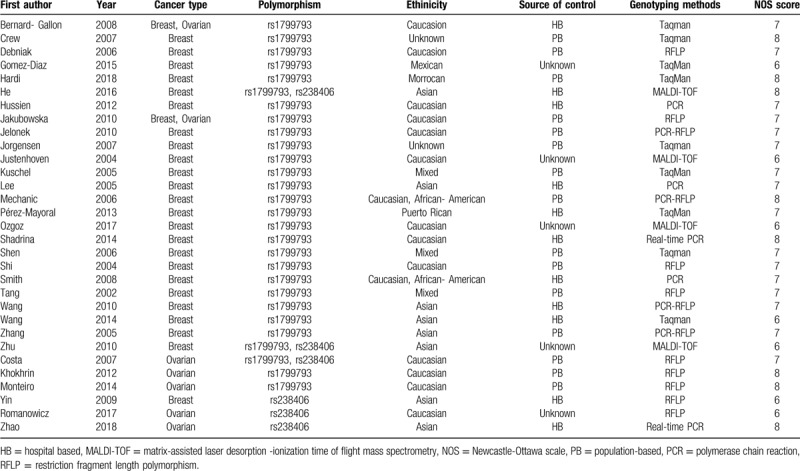
Table 1.
The characteristics of included articles.
3.2. Characteristics of included studies
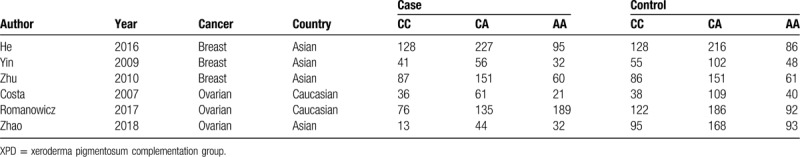
The 31 enrolled articles consisted 38 case-control studies with 2 XPD polymorphisms (rs1799793 in 32 studies and rs238406 in 6 studies). 30 studies focused on breast cancer while 8 studies explored ovarian cancer. Different genotyping methods were utilized including polymerase chain reaction, TaqMan, restriction fragment length polymorphism, and matrix-assisted laser desorption -ionization time of flight mass spectrometry. Ethnicities included Asian, Caucasian, Mexican, Moroccan, Puerto Rican, and mixed. The control sources were either population based or hospital based (Table 1). All studies reported the numbers of corresponding genotypes for both case and control groups as to recessive mutants, heterogeneous mutants, and dominant wild types (Tables 2 and 3).
Table 2.
Genotype distributions in cases and controls for XPD rs1799793(G/A) polymorphisms.
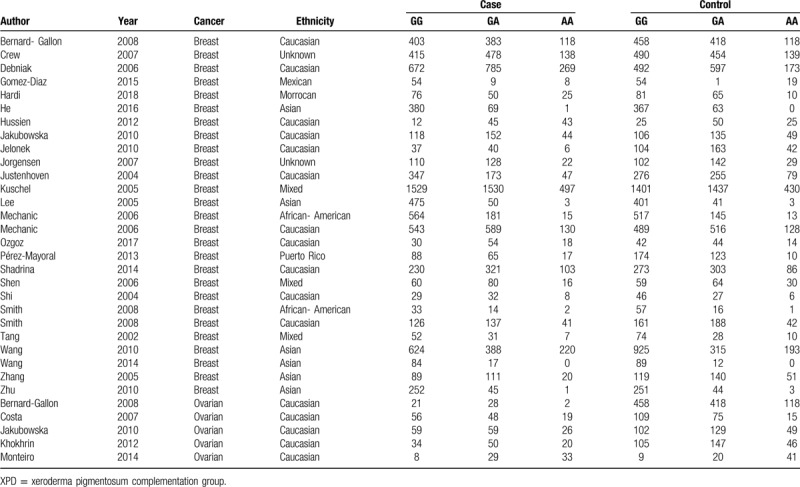
Table 3.
Genotype distributions in cases and controls for XPD rs238406(C/A) polymorphisms.
3.3. XPD rs1799793(G/A) polymorphisms
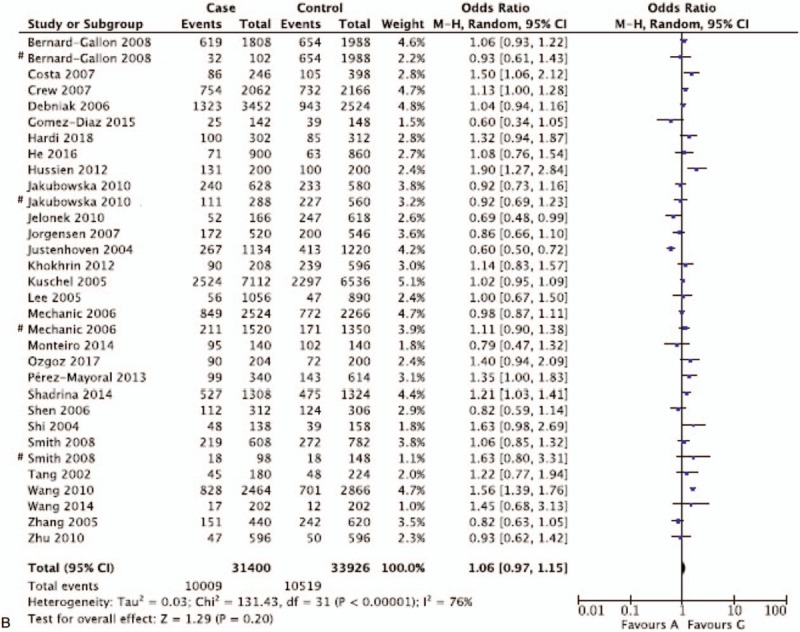
Pooled ORs and corresponding 95%CIs were shown in Table 4. By analyzing 32,663 participants of 32 case-control studies, the meta-analysis displayed no association of XPD rs1799793(G/A) polymorphisms with breast and ovarian cancer risk in the overall group (AA vs GA + GG: OR = 1.04 95%CI = 0.90–1.19; AA + GA vs GG: OR = 1.08, 95%CI = 0.98–1.20; GA vs GG + AA: OR = 1.06, 95%CI = 0.97–1.17; AA vs GG: OR = 1.08, 95%CI = 0.92–1.27; A vs G: OR = 1.06, 95%CI = 0.97–1.15) (Fig. 2 ). The insignificant results were consistent with the outcomes of subgroup analysis for breast cancer and ovarian cancer. However, if stratified by ethnicity, 1 comparison model among Asian population for rs1799793 (GA vs. GG + AA: OR = 1.38, 95%CI = 1.21–1.56) was considered as statistically significant in fixed effect models. Though 5,846 participants were included, the results brought little confidence to conclude that GA was a detrimental factor for breast and ovarian cancer for Asian population. In general, it indicates that G to A variation in the XPD rs1799793 polymorphisms might not correlate with breast and ovarian cancer susceptibility.
Table 4.
Summary of different comparative results for XPD rs1799793 (G/A) polymorphisms.
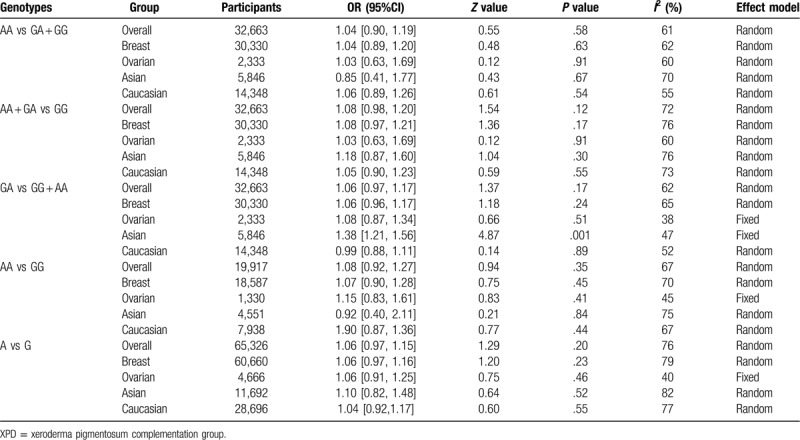
Figure 2.
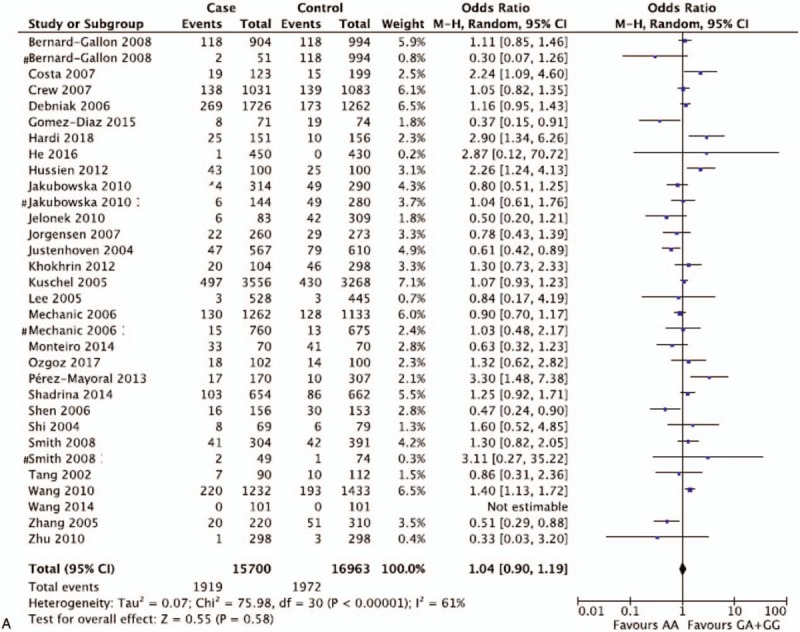
Representative forest plots for XPD rs1799793(G/A) polymorphisms. (A) AA vs GA + GG in overall group analysis. (B) A vs G in overall group analysis. (C) GA vs GG + AA in Asian group analysis. One article was considered as different studies based on ethnicity or cancer type. XPD = xeroderma pigmentosum complementation group.
3.4. XPD rs238406(C/A) polymorphisms
The results of the association of XPD rs238406(C/A) polymorphisms with breast and ovarian cancer risk were shown in Table 5. With 6 studies and 3,360 participants pooled, the results showed a protective trend (OR < 1) of heterozygous mutants for breast and ovarian cancer but statistical significance was only found in Caucasian population (CA vs AA + CC: OR = 0.63, 95%CI = 0.49–0.80). The rest comparisons failed to demonstrate statistically significant ORs, either in overall group analysis (AA vs CA + CC: OR = 1.30, 95%CI = 0.83–2.03; AA + CA vs CC: OR = 1.11, 95%CI = 0.78–1.58, CA vs. AA + CC: OR = 0.85, 95%CI = 0.69–1.05, AA vs CC: OR = 1.30, 95%CI = 0.75–2.24; A vs C: OR = 1.16, 95%CI = 0.85–1.59) or subgroup analysis (either stratified by cancer type and ethnicity) (Fig. 3). The above data suggested that XPD rs238406(C/A) polymorphisms did not pose an increased risk for breast and ovarian cancer, while heterozygous mutants showed a protective trend specifically in Caucasian population, but no solid conclusion should be drawn based on the current statistical derivation.
Table 5.
Summary of different comparative results for XPD rs238406(C/A) polymorphisms.
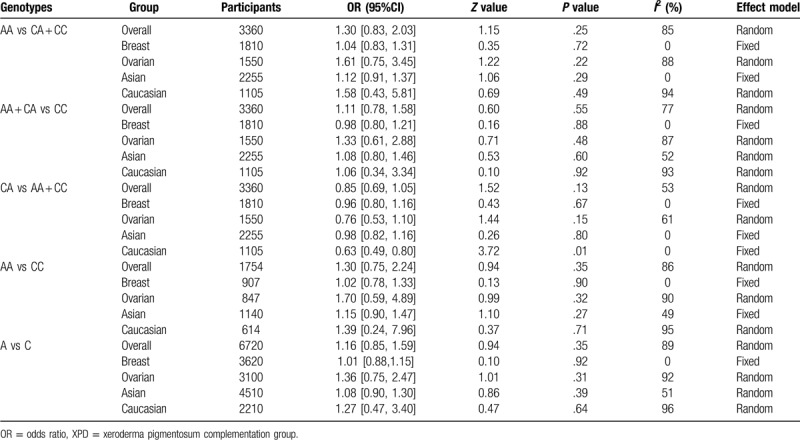
Figure 2 (Continued).
Representative forest plots for XPD rs1799793(G/A) polymorphisms. (A) AA vs GA + GG in overall group analysis. (B) A vs G in overall group analysis. (C) GA vs GG + AA in Asian group analysis. One article was considered as different studies based on ethnicity or cancer type. XPD = xeroderma pigmentosum complementation group.
Figure 3.
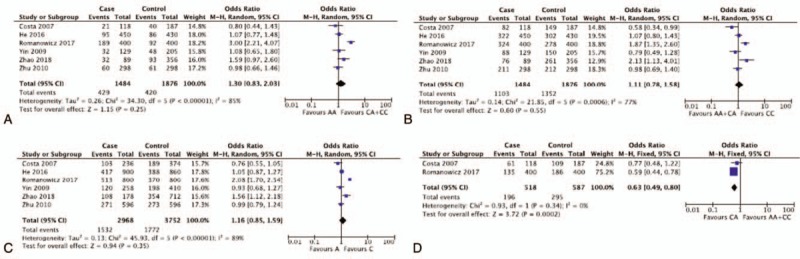
Representative forest plots for XPD rs238406(C/A) polymorphisms. (A) AA vs CA + CC in overall group analysis. (B) AA + CA vs CC in overall group analysis. (C) A vs C in overall group analysis. (D) CA vs AA + CC in Caucasian group analysis. XPD = xeroderma pigmentosum complementation group.
3.5. Publication bias
The publication bias was visually examined on the funnel plots generated by Revman 5.2 software. No obvious asymmetry could be observed (Fig. 4). We further conducted Egger tests in the 3 analyses that indicated significant ORs (2 for rs1799793 polymorphisms and 1 for rs238406 polymorphisms). The results demonstrated no significant publication bias (P > .05, data not shown).
Figure 2 (Continued).
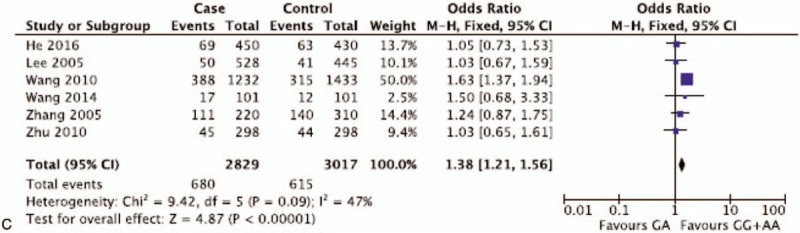
Representative forest plots for XPD rs1799793(G/A) polymorphisms. (A) AA vs GA + GG in overall group analysis. (B) A vs G in overall group analysis. (C) GA vs GG + AA in Asian group analysis. One article was considered as different studies based on ethnicity or cancer type. XPD = xeroderma pigmentosum complementation group.
Figure 4.
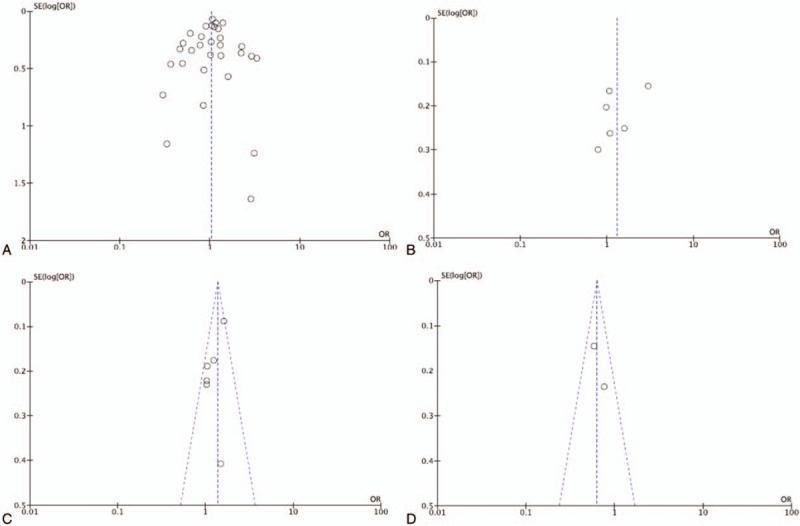
Representative funnel plots for XPD rs1799793(G/A) and rs238406(C/A) polymorphisms. (A) AA vs GA + GG of rs1799793(G/A) in overall group analysis. (B) AA vs CA + CC of rs238406(C/A) in overall group analysis. (C) GA vs GG + AA of rs1799793(G/A) in Asian group analysis. (D) CA vs AA + CC of rs238406(C/A) in Caucasian group analysis. XPD = xeroderma pigmentosum complementation group.
4. Discussion
Since the widely use of poly ADP ribose polymerase (PARP) inhibitors as targeted therapy for BRCA mutated patients and accumulating number of PARPi resistant patients identified, increasing attention has been payed to other gene aberrations involved in DNA repair pathways. XPD genes participate in DNA repair and therefore, when mutated, may contribute to genome instability. Pre-clinical studies have found that XPD aberrance plays a role in activating apoptosis through interaction between p53 and TFIIH to remove damaged cells.[45,46] To date, many publications have shown an association between the XPD polymorphism and risk of cancer. However, the results remain controversial. One meta-analysis in 2014 studied rs1799793 polymorphisms and breast cancer susceptibility. A total of 22 studies with 18,136 cases and 18,351 controls were included. The conclusion was that XPD rs1799793 polymorphisms were not associated with breast cancer.[47] Since then, several new case-control studies were published and no meta-analysis was conducted to see the association between rs1799793 polymorphisms and ovarian cancer, while the correlation between XPD rs238406 polymorphisms with breast and ovarian cancer have not been systemically studied yet. Thus, in order to draw a more concrete conclusion, we searched all related publications and performed a meta-analysis for the 2 XPD polymorphisms by enrolling 38 studies from 31 articles.
The current meta-analysis presented that there was no association of XPD rs1799793(G/A) polymorphisms with breast and ovarian cancer risk in the overall groups and subgroups for breast cancer and ovarian cancer. One comparison model for heterozygous mutants among Asian population was considered as statistically significant. However, the result was hard to transfer to the conclusion that that the heterozygous mutant of GA was a detrimental factor for breast and ovarian cancer for Asian population. This also reflects the complicated role between genes variants and protein functions. The XPD exon 10 rs1799793 polymorphisms were characterized by a G/A nucleotide substitution, causing an Asp/Asn amino acid change at codon 312 of XPD gene.[48] Though the biological function of this amino acid substitution has not yet been elucidated, the fact is that this residue has been highly conserved through evolution.[49] Whether the conservation indicates a protective role in DNA variance against function effect or suggests a strong effect in the enzymatic activity remains to be further studied. The results of XPD rs238406(C/A) polymorphisms showed a protective trend (OR < 1) of heterozygous mutants for breast and ovarian cancer but statistical significance was only found in Caucasian population. The rest comparisons failed to demonstrate statistically significant ORs. This is similar to the result of rs1799793 polymorphisms, suggesting that heterozygous mutants might share different pathophysiologic mechanism from not only homozygous wildtypes but also homozygous mutants, in potentially certain ethnicities. By looking at the separate studies, one case-control study was found to display a strong relationship between XPD rs238406(C/A) polymorphisms and ovarian cancer risk.[43] The variant A allele increased almost 2-fold of the risk of ovarian cancer, which was confirmed in certain histological grades and FIGO staging. The study focused only in Polish population and the authors emphasized that they included only a small group of patients and the obtained results should then be approached as preliminary. Whether ethnicity difference plays a role in the cancer risk brought by the gene variance remains to be solved.
Despite our efforts to include all available publications, several limitations to our meta-analysis should not be ignored. First, most of the original data pooled were unadjusted, not mentioning the reported imbalance of patient characteristics in certain included studies. Those underlying imbalanced risk factors might lead to an inaccurate explanation of pooled data. Whereas, we found no evidence of publication bias thus it convinced us of the reliability of the current meta-analysis. Second, the populations of included studies were limited. It is epidemiologically acknowledged that other ethnicities such as Hispanics and Blacks are also cancer susceptible;[50,51] thus, the lack of data for these populations might affect the overall results, especially when one study suggested a strong relationship between XPD rs238406(C/A) polymorphisms and ovarian cancer risk in Polish population. Third, although the number of pooled participants was so far the largest, the number of several subgroup analyses was still very limited, especially for ovarian cancer patients.
In conclusion, the current meta-analysis suggested no concrete correlation of XPD rs1799793(G/A) and rs238406(C/A) polymorphisms with breast cancer or ovarian cancer susceptibility. However, it indicated that heterozygous genotypes might share different pathophysiologic mechanism from not only homozygous wildtypes but also homozygous mutants. More case–control studies with well-adjusted data and diverse populations are essential for validation of our conclusion.
Author contributions
YT and CB contributed to study concept and design; YT, XL, and FY contributed to literature search and data extractions; YT and JZ contributed to data analysis and the study quality evaluation; YT and KY contributed to the methodology. XL and CB contributed to the supervision of all the study processes. All the authors contributed to writing the manuscript. All authors approved the final version of the manuscript.
Footnotes
Abbreviations: 95%CIs = 95% confidence intervals, NOS = Newcastle-Ottawa scale, OR = odds ratio, XPD = xeroderma pigmentosum complementation group.
How to cite this article: Tian Y, Lin X, Yang F, Zhao J, Yao K, Bian C. Contribution of xeroderma pigmentosum complementation group D gene polymorphisms in breast and ovarian cancer susceptibility: a protocol for systematic review and meta analysis. Medicine. 2020;99:21(e20299).
This study was supported by Key Research Projects of Science and Technology Department Foundation of Sichuan Province (Grant No: 2017SZ0141) and Applied Basic Research Programs of Science and Technology Department Foundation of Sichuan Province (Grant No: 2016JY0122).
The authors have no conflicts of interest to disclose.
The datasets generated during and/or analyzed during the current study are publicly available.
References
- [1].Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin 2015;65:5–29. [DOI] [PubMed] [Google Scholar]
- [2].Ferlay J, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN. Int J Cancer 2012;136:E359–86. [DOI] [PubMed] [Google Scholar]
- [3].Lord CJ, Ashworth A. PARP inhibitors: synthetic lethality in the clinic. Science 2017;355:1152–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Taylor KN, Eskander RN. PARP inhibitors in epithelial ovarian cancer. Recent Pat Anticancer Drug Discov 2018;13:145–58. [DOI] [PubMed] [Google Scholar]
- [5].del Rivero J, Kohn EC. PARP inhibitors: the cornerstone of DNA repair-targeted therapies. Oncology (Williston Park) 2017;31:265–73. [PubMed] [Google Scholar]
- [6].Laine JP, Mocquet V, Bonfanti M, et al. Common XPD (ERCC2) polymorphisms have no measurable effect on nucleotide excision repair and basal transcription. DNA Repair (Amst) 2007;6:1264–70. [DOI] [PubMed] [Google Scholar]
- [7].Lunn RM, Helzlsouer KJ, Parshad R, et al. XPD polymorphisms: effects on DNA repair proficiency. Carcinogenesis 2000;21:551–5. [DOI] [PubMed] [Google Scholar]
- [8].Fontana L, Bosviel R, Delort L, et al. DNA repair gene ERCC2, XPC, XRCC1, XRCC3 polymorphisms and associations with bladder cancer risk in a French cohort. Anticancer Res 2008;28:1853–6. [PubMed] [Google Scholar]
- [9].Costa S, Pinto D, Pereira D, et al. Importance of xeroderma pigmentosum group D polymorphisms in susceptibility to ovarian cancer. Cancer Lett 2007;246:324–30. [DOI] [PubMed] [Google Scholar]
- [10].Bernard-Gallon D, Bosviel R, Delort L, et al. DNA repair gene ERCC2 polymorphisms and associations with breast and ovarian cancer risk. Mol Cancer 2008;7:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Gomez-Diaz B, D.E. Luz Ayala-Madrigal LA, Gutiérrez-Angulo MM, et al. Analysis of ERCC1 and ERCC2 gene variants in osteosarcoma, colorectal and breast cancer. Oncol Lett 2015;9:1657–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Xiao F, Pu J, Wen Q, et al. Association between the ERCC2 Asp312Asn polymorphism and risk of cancer. Oncotarget 2017;8:48488–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Yang F, Mu X, Bian C, et al. Association of excision repair cross-complimentary group 1 gene polymorphisms with breast and ovarian cancer susceptibility. J Cell Biochem 2019;120:15635–47. [DOI] [PubMed] [Google Scholar]
- [14].Chen Y, Qi X, Bian C, et al. The association of FOXP3 gene polymorphisms with cancer susceptibility: a comprehensive systemic review and meta-analysis. Biosci Rep 2019;39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Min L, Mu X, Tong A, et al. The association between HOTAIR polymorphisms and cancer susceptibility: an updated systemic review and meta-analysis. Onco Targets Ther 2018;11:791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Mu X, Zhao J, Yuan X, et al. Gene polymorphisms of toll-like receptor 9 -1486T/C and 2848G/A in cervical cancer risk. Int J Gynecol Cancer 2015;25:1173–8. [DOI] [PubMed] [Google Scholar]
- [17].Crew KD, Gammon MD, Terry M, et al. Polymorphisms in nucleotide excision repair genes, polycyclic aromatic hydrocarbon-DNA adducts, and breast cancer risk. Cancer Epidemiol Biomarkers Prev 2007;16:2033–41. [DOI] [PubMed] [Google Scholar]
- [18].Debniak T, Scott RJ, Huzarski T, et al. XPD common variants and their association with melanoma and breast cancer risk. Breast Cancer Res Treat 2006;98:209–15. [DOI] [PubMed] [Google Scholar]
- [19].Hardi H, Melki R, Boughaleb Z, et al. Significant association between ERCC2 and MTHR polymorphisms and breast cancer susceptibility in Moroccan population: genotype and haplotype analysis in a case-control study. BMC Cancer 2018;18:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].He BS, Xu T, Pan YQ, et al. Nucleotide excision repair pathway gene polymorphisms are linked to breast cancer risk in a Chinese population. Oncotarget 2016;7:84872–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hussien YM, Gharib AF, Awad HA, et al. Impact of DNA repair genes polymorphism (XPD and XRCC1) on the risk of breast cancer in Egyptian female patients. Mol Biol Rep 2012;39:1895–901. [DOI] [PubMed] [Google Scholar]
- [22].Jakubowska A, Gronwald J, Menkiszak J, et al. BRCA1-associated breast and ovarian cancer risks in Poland: no association with commonly studied polymorphisms. Breast Cancer Res Treat 2010;119:201–11. [DOI] [PubMed] [Google Scholar]
- [23].Jelonek K, Gdowicz-Klosok A, Pietrowska M, et al. Association between single-nucleotide polymorphisms of selected genes involved in the response to DNA damage and risk of colon, head and neck, and breast cancers in a polish population. J Appl Genet 2010;51:343–52. [DOI] [PubMed] [Google Scholar]
- [24].Jorgensen TJ, Visvanathan K, Ruczinski I, et al. Breast cancer risk is not associated with polymorphic forms of xeroderma pigmentosum genes in a cohort of women from Washington County, Maryland. Breast Cancer Res Treat 2007;101:65–71. [DOI] [PubMed] [Google Scholar]
- [25].Justenhoven C, Hamann U, Pesch B, et al. ERCC2 genotypes and a corresponding haplotype are linked with breast cancer risk in a German population. Cancer Epidemiol Biomarkers Prev 2004;13:2059–64. [PubMed] [Google Scholar]
- [26].Kuschel B, et al. Common polymorphisms in ERCC2 (Xeroderma pigmentosum D) are not associated with breast cancer risk. Cancer Epidemiol Biomarkers Prev 2005;14:1828–31. [DOI] [PubMed] [Google Scholar]
- [27].Lee SA, Lee KM, Park WY, et al. Obesity and genetic polymorphism of ERCC2 and ERCC4 as modifiers of risk of breast cancer. Exp Mol Med 2005;37:86–90. [DOI] [PubMed] [Google Scholar]
- [28].Mechanic LE, Millikan RC, Player J, et al. Polymorphisms in nucleotide excision repair genes, smoking and breast cancer in African Americans and whites: a population-based case-control study. Carcinogenesis 2006;27:1377–85. [DOI] [PubMed] [Google Scholar]
- [29].Ozgoz A, Hekimler Öztürk K, Yükseltürk A, et al. Genetic variations of DNA repair genes in breast cancer. Pathol Oncol Res 2019;25:107–14. [DOI] [PubMed] [Google Scholar]
- [30].Perez-Mayoral J, Pacheco-Torres AL, Morales L, et al. Genetic polymorphisms in RAD23B and XPC modulate DNA repair capacity and breast cancer risk in Puerto Rican women. Mol Carcinog 2013;52: Suppl 1: E127–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Shadrina AS, Ermolenko NA, Boyarskikh UA, et al. Polymorphisms in DNA repair genes and breast cancer risk in Russian population: a case-control study. Clin Exp Med 2016;16:21–8. [DOI] [PubMed] [Google Scholar]
- [32].Shen J, Desai M, Agrawal M, et al. Polymorphisms in nucleotide excision repair genes and DNA repair capacity phenotype in sisters discordant for breast cancer. Cancer Epidemiol Biomarkers Prev 2006;15:1614–9. [DOI] [PubMed] [Google Scholar]
- [33].Smith TR, Levine EA, Freimanis RI, et al. Polygenic model of DNA repair genetic polymorphisms in human breast cancer risk. Carcinogenesis 2008;29:2132–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wang T, Wang H, Guo H, et al. Polymorphisms in the DNA repair gene ERCC2/XPD and breast cancer risk: a HapMap-based case-control study among Han Women in a Chinese less-developed area. Genet Test Mol Biomarkers 2014;18:703–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zhang L, Zhang Z, Yan W. Single nucleotide polymorphisms for DNA repair genes in breast cancer patients. Clin Chim Acta 2005;359:150–5. [DOI] [PubMed] [Google Scholar]
- [36].Zhu F, et al. Investigations on the association of single nucleotide polymorphisms of DNA repair XPD, XPA genes with the risk of breast cancer. Chin J Clin Lab Sci 2015;33:498–503. [Google Scholar]
- [37].Khokhrin DV, Khrunin AV, Moiseev AA, et al. Association of polymorphisms in glutathione-S-transferase and DNA repair genes with ovarian cancer risk in the Russian population. Genetika 2012;48:901–4. [PubMed] [Google Scholar]
- [38].Monteiro MS, Vilas Boas DB, Gigliotti CB, et al. Association among XRCC1, XRCC3, and BLHX gene polymorphisms and chromosome instability in lymphocytes from patients with endometriosis and ovarian cancer. Genet Mol Res 2014;13:636–48. [DOI] [PubMed] [Google Scholar]
- [39].Shi Q, Wang LE, Bondy ML, et al. Reduced DNA repair of benzo[a]pyrene diol epoxide-induced adducts and common XPD polymorphisms in breast cancer patients. Carcinogenesis 2004;25:1695–700. [DOI] [PubMed] [Google Scholar]
- [40].Tang D, Cho S, Rundle A, et al. Polymorphisms in the DNA repair enzyme XPD are associated with increased levels of PAH-DNA adducts in a case-control study of breast cancer. Breast Cancer Res Treat 2002;75:159–66. [DOI] [PubMed] [Google Scholar]
- [41].Wang HC, Liu CS, Wang CH, et al. Significant association of XPD Asp312Asn polymorphism with breast cancer in Taiwanese patients. Chin J Physiol 2010;53:130–5. [DOI] [PubMed] [Google Scholar]
- [42].Yin J, Liang D, Vogel U, et al. The polymorphism of DNA repair gene ERCC2/XPD Arg156Arg and susceptibility to breast cancer in a Chinese population. Biochem Genet 2009;47:582–90. [DOI] [PubMed] [Google Scholar]
- [43].Romanowicz H, Michalska MM, Samulak D, et al. Association of R156R single nucleotide polymorphism of the ERCC2 gene with the susceptibility to ovarian cancer. Eur J Obstet Gynecol Reprod Biol 2017;208:36–40. [DOI] [PubMed] [Google Scholar]
- [44].Zhao Z, Zhang A, Zhao Y, et al. The association of polymorphisms in nucleotide excision repair genes with ovarian cancer susceptibility. Biosci Rep 2018;38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Yeh YS, Chen YT, Tsai HL, et al. Predictive value of ERCC1, ERCC2, and XRCC expression for patients with locally advanced or metastatic gastric cancer treated with neoadjuvant mFOLFOX-4 chemotherapy. Pathol Oncol Res 2019; 10.1007/s12253-019-00666-5 [DOI] [PubMed] [Google Scholar]
- [46].Lorenzo-Gonzalez M, Ruano-Ravina A2, Torres-Durán M, et al. Residential radon, genetic polymorphisms in DNA damage and repair-related. Lung Cancer 2019;135:10–5. [DOI] [PubMed] [Google Scholar]
- [47].Yan Y, Liang H, Light M, et al. XPD Asp312Asn and Lys751Gln polymorphisms and breast cancer susceptibility: a meta-analysis. Tumour Biol 2014;35:1907–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Fan L, Fuss JO, Cheng Q, et al. XPD helicase structures and activities: insights into the cancer and aging phenotypes from XPD mutations. Cell 2008;133:789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Wolfe KJ, Wickliffe JK, Hill CE, et al. Single nucleotide polymorphisms of the DNA repair gene XPD/ERCC2 alter mRNA expression. Pharmacogenet Genomics 2007;17:897–905. [DOI] [PubMed] [Google Scholar]
- [50].Park HK, Ruterbusch JJ, Cote ML. Recent trends in ovarian cancer incidence and relative survival in the United States by race/ethnicity and histologic subtypes. Cancer Epidemiol Biomarkers Prev 2017;26:1511–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Lee AW, Navajas EE, Liu L. Clear differences in ovarian cancer incidence and trends by ethnicity among Asian Americans. Cancer Epidemiol 2019;61:142–9. [DOI] [PMC free article] [PubMed] [Google Scholar]


