Abstract
Introduction
FTND (Fagerstrӧm test for nicotine dependence) and TTFC (time to smoke first cigarette in the morning) are common measures of nicotine dependence (ND). However, genome-wide meta-analysis for these phenotypes has not been reported.
Methods
Genome-wide meta-analyses for FTND (N = 19,431) and TTFC (N = 18,567) phenotypes were conducted for adult smokers of European ancestry from 14 independent cohorts.
Results
We found that SORBS2 on 4q35 (p = 4.05 × 10−8), BG182718 on 11q22 (p = 1.02 × 10−8), and AA333164 on 14q21 (p = 4.11 × 10−9) were associated with TTFC phenotype. We attempted replication of leading candidates with independent samples (FTND, N = 7010 and TTFC, N = 10 061), however, due to limited power of the replication samples, the replication of these new loci did not reach significance. In gene-based analyses, COPB2 was found associated with FTND phenotype, and TFCP2L1, RELN, and INO80C were associated with TTFC phenotype. In pathway and network analyses, we found that the interconnected interactions among the endocytosis, regulation of actin cytoskeleton, axon guidance, MAPK signaling, and chemokine signaling pathways were involved in ND.
Conclusions
Our analyses identified several promising candidates for both FTND and TTFC phenotypes, and further verification of these candidates was necessary. Candidates supported by both FTND and TTFC (CHRNA4, THSD7B, RBFOX1, and ZNF804A) were associated with addiction to alcohol, cocaine, and heroin, and were associated with autism and schizophrenia. We also identified novel pathways involved in cigarette smoking. The pathway interactions highlighted the importance of receptor recycling and internalization in ND.
Implications
Understanding the genetic architecture of cigarette smoking and ND is critical to develop effective prevention and treatment. Our study identified novel candidates and biological pathways involved in FTND and TTFC phenotypes, and this will facilitate further investigation of these candidates and pathways.
Introduction
Cigarette smoking imposes a high health and financial toll on the smokers as well as society at large. Regular cigarette smoking often leads to nicotine dependence (ND). The tendency to develop ND is influenced by both genetic predisposition and environmental factors. In recent years, genetic studies of ND have made significant progress, exemplified by the identification of the CHRNA5-CHRNA3-CHRNB4, CHRNB3-CHRNA6, CHRNA4, and CYP2A6 loci.1–6 However, variants identified in these genes explain only a small proportion of heritability. For example, cigarettes smoked per day (CPD), a common measure used in ND studies, explained about 5.6% of the heritability.7 Many more risk genes remain unidentified.
Cigarette smoking is a complex behavior, and different measures are used to assess ND in both clinical and research settings. The quantity of consumption, typically assessed by self-reported CPD, is a measure used in many studies, including those that identified the CHRNA5-CHRNA3-CHRNB4, CHRNB3-CHRNA6, and CYP2A6 loci.2–4 The Fagerström Test for Nicotine Dependence (FTND)8 is another commonly used measure in genetic studies. CPD is one of the six questions included in the FTND and accounts up to three points in the 10-point FTND scale. Another FTND question, “How soon after you wake up do you smoke your first cigarette?” or time to smoke first cigarette in the morning (TTFC), is a strong predictor of difficulty to quit smoking and smoking relapse,9,10 COPD,11 and lung cancer.12 Although both FTND and TTFC are important measures of ND, comparing with CPD, only a few genome-wide association studies (GWASs)6,13–15 have been conducted using these phenotypes. CHRNA4 and a few intergenic loci were found to be associated with FTND,6,13 but no locus for TTFC was identified.
Here we report results from our GWAS meta-analyses on FTND (N = 19 431) and TTFC (N = 18 567). Our motivation to analyze TTFC separately is to understand what genetic factors contributing to the difficulty of quitting and relapse. Another benefit is that we can compare the results between FTND and TTFC phenotypes and identify convergent candidates. A lesson learned from comprehensive studies of the CHRNA5-CHRNA3-CHRNB4 locus is that a true signal can be detected with many correlated phenotypes.16 Hence, a candidate with corroborative support from both FTND and TTFC would be more credible.
Methods
Datasets
We assembled 14 independent cohorts (FTND, N = 19 431; TTFC, N = 18 567) to examine the association between FTND/TTFC phenotypes and genome-wide genetic variants. A description of the cohorts and the summary of descriptive statistics of the cohorts are provided in Supplementary Material and Supplementary Table S1, respectively.
Genotype Quality Control and Imputation
Genotype quality controls were conducted separately by individual groups for the discovery data sets ISIB, ALSPAC, AUTW, CEDAR, FTC1, FTC2, NTR, and GCD. For discovery data sets obtained from dbGaP (http://www.ncbi.nlm.nih.gov/gap) (SAGE, SC, MGS, COPDGene1, CIDR370v1, CIDR370v3, CIDRc3, CIDRc4, and EAGLE), we used the genotypes provided by the original investigators who conducted quality control procedures following the dbGaP standards. Genotype imputations were also conducted separately by each group using MaCH17,18 or IMPUTE219,20 with the 1000 Genomes reference haplotypes (EUR panel, March 2012 release), using the default settings of the programs. Similarly, genotype quality control for the replication data sets (VTSABD, COPDGene2, S4S, PNAT2, and EAGLE [used for TTFC only]) was conducted by each group separately.
Inclusion Criteria and Phenotypes
Regular smokers, defined as those who smoked daily for at least 1 month or those who smoked at least 100 cigarettes lifetime, of European ancestry were included. FTND scores (0–10) and TTFC scores (0–3) were obtained from self-reported FTND questionnaire, ascertaining the smoking behaviors during the heaviest smoking period of the smokers. Both FTND and TTFC scores were treated as quantitative traits without transformation.
Association and Meta-Analyses
GWAS analyses were performed separately for each data set by individual groups using the PLINK program.21 Assuming a linear mixed effect model, FTND and TTFC were treated as continuous outcomes and genotypes as predictors, whereas sex, age, and the first 10 principal components were included as covariates. Summary statistics from each data set were combined by GWAMA22 program using inverse variance-weighted meta-analysis approach with fixed effects. Because the individual samples were analyzed using the same model, the summary statistics were used directly without further normalization. For the meta-analyses, only bi-allelic single nucleotide polymorphisms (SNPs) with minor allele frequency ≥1% and with high imputation quality (INFO value ≥ 0.4 from IMPUTE2, or r2 ≥ 0.4 from MaCH) were included. p-Values from the meta-analyses were corrected for genomic-control. p-Values below 5 × 10−8 were considered as genome-wide significant, whereas p-values below 5 × 10−5 were considered as suggestive association and this threshold was used for selection of loci for convergence analysis.
SNP Heritability Analyses
To estimate the SNP heritability with genome-wide data, we used the linkage disequilibrium (LD)–adjusted kinship algorithm.23,24 Specifically, we used the control subjects from two large studies25,26 as reference to select LD-adjusted SNP predictors and used the meta-analysis results from the FTND and TTFC to estimate the weights for these SNPs. The heritability was then estimated with these LD-tagged SNPs. The number of SNPs used for FTND heritability estimate was 2 857 113 and that for TTFC was 2 981 471.
Replication Analyses
Six independent data sets of European descent (N = 7010 for FTND and N = 10 061 for TTFC) were used for a replication study of selected SNPs. We conducted replication study for all loci meeting these four criteria: (1) having five or more SNPs with p-value ≤ 5 × 10−5; (2) at least one SNP with p-value ≤ 5 × 10−5 having a minor allele frequency ≥5%; (3) minor allele frequency variation at the locus > 5% between SNPs with p-value ≤ 5 × 10−5; and (4) at least four data sets contributed to the signal at the locus. For each locus, we selected the SNP with smallest p-value for replication testing. The selection of these criteria was based on lessons learned from recent GWASs where true loci have multiple associated SNPs with different frequencies and many loci have multiple independent signals.16,27 The requirement of 5% minor allele frequency was intended to minimize the influence of potential outlier SNPs, and the inclusion of variation of minor allele frequency was to maximize the likelihood that the locus could harbor more than one association signal. p-Values below .0031 (.05/16) were considered as significant replication.
Gene-Based Association Analyses
We performed gene-based association analyses using the results from the GWAS meta-analyses. Specifically, we used the Knowledge-based mining system for Genome-wide Genetic studies (KGG) software,28,29 which uses an extended Simes test to integrate functional information and association evidence to combine the SNP p-values within a gene to obtain an overall p-value for each gene. In these analyses, we filtered out SNPs found in less than four data sets. All analyses were conducted using KGG default settings. We used the Benjamini–Hochberg method30 to correct for multiple testing, and considered FDR q-values below 0.05 as statistically significant.
Pathway and Network Analyses
We conducted pathway enrichment analysis for genes with at least one marker with p ≤ 5 × 10−5 from GWAS meta-analyses of either FTND or TTFC. If a marker was within a gene region, it was assigned to the gene; otherwise, it was mapped to its most proximate gene using the 50-kb flanking regions (both 5′ and 3′ sides). Genes identified using SNPs associated with FTND and TTFC were merged for the pathway enrichment analyses. We used the hypergeometric test implemented in the tool WebGestalt (2014 update)31,32 and the canonical pathways from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. We required each pathway to have at least three genes from our gene list and no more than 300 genes from the reference genome. The p-values from hypergeometric tests were further adjusted by the Benjamini–Hochberg method.30 Only pathways with adjusted p-values < .05 were considered statistically significantly enriched.
We further examined how enriched pathways interacted with each other in function and regulation. Specifically, we applied the Characteristic Sub-Pathway Network (CSPN) algorithm33 to search for significantly interacting pathway pairs.34 CSPN was designed to prioritize pathway pairs with significant interaction of molecules from each pathway pair in the human protein–protein interaction (PPI) network (details in ref. 33). We used the human PPI data from the Protein Interaction Network Analysis (PINA) platform35 as the reference network in this pathway crosstalk analysis. Our working PPI network included a total of 11 318 nodes (protein-coding genes) and 67 936 interactions. We restricted the analysis specifically to the aforementioned merged gene set and their enriched pathways. When running CSPN, a mode “OR” was selected, i.e., we considered all PPIs formed by the supplied genes as well as their one-step extension. In the final step, we selected the significant pathway interaction pairs based on permutation p-values ≤ .05.
Results
FTND and TTFC GWAS Meta-Analyses
In the GWAS meta-analyses of the discovery sample, about 19 000 subjects from 14 independent cohorts were included. For FTND (N = 19 431), only the CHRNA5-CHRNA3-CHRNB4 locus reached genome-wide significance (Table 1). The smallest p-value was observed at rs16969968 (β = −0.21, SE = 0.02, p = 3.96 × 10−19), which is a functional variant in CHRNA5 known to influence smoking behavior, in particular the quantity of cigarettes smoked. Several additional loci (CIB4 on 2p23, BG182718 on 11q22, and DSC3 on 18q12) were promising (Table 1). The meta-analysis had a lambda of 1.03, indicating no significant genomic inflation. The Manhattan and Q-Q plots for FTND are shown in Supplementary Figure S1. In the analyses of TTFC (N = 18 567), four loci (SORBS2 on 4q35, BG182718 on 11q22, AA333164 on 14q21, and CHRNA5-CHRNA3-CHRNB4 on 15q25) reached genome-wide significance (Table 1). A closer examination of the local linkage disequilibrium (LD) indicated that the signals from BG182718 and CHRNA5-CHRNA3-CHRNB4 covered a broad genomic region (~500 kb), whereas the signals from SORBS2 and AA333164 were restricted within small intervals (Figure S2). CHRNB3-CHRNA6 (smallest p = 8.83 × 10−8 for rs11785369) and MIR31HG (smallest p = 5.70 × 10−8 for rs184042824) were approaching genome-wide significance. Several loci meeting our criteria for replication testing are also listed in Table 1. No genomic inflation was observed for TTFC phenotype (λ = 1.01). The Manhattan and Q–Q plots for TTFC are shown in Supplementary Figure S1.
Table 1.
The most significant loci identified in the FTND and TTFC GWAS meta-analyses in the discovery sample
| CHR | BP | SNP | EA | NEA | EAF | β | SE | Z | p | N | Effect* | GENE | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FTND | 2 | 26,860,822 | rs17005545 | T | C | 0.94 | −0.23 | 0.05 | −4.97 | 6.99E-07 | 18621 | ------+-------- | CIB4 |
| 11 | 97,713,579 | rs117029742 | T | C | 0.98 | −0.64 | 0.12 | −5.32 | 1.04E-07 | 12977 | --------??-?-+? | BG182718 | |
| 15 | 78,882,925 | rs16969968 | G | A | 0.64 | −0.21 | 0.02 | −8.83 | 3.96E-19 | 17860 | -----------?-+- | CHRNA5 | |
| 18 | 28,355,510 | rs28510557 | C | T | 0.60 | −0.10 | 0.02 | −4.85 | 1.23E-06 | 18673 | ----++-+----+-- | DSC3 | |
| TTFC | 1 | 40,464,894 | rs34022242 | C | G | 0.94 | −0.13 | 0.02 | −5.16 | 2.48E-07 | 14286 | ----------+?--?? | MFSD2A |
| 1 | 68,862,622 | rs3125927 | G | A | 0.64 | −0.05 | 0.01 | −5.02 | 5.24E-07 | 16091 | ----------+----? | RPE65 | |
| 4 | 103,715,385 | rs223441 | C | T | 0.51 | 0.05 | 0.01 | 4.58 | 4.66E-06 | 18533 | ++++-+++++++-+++ | CISD2 | |
| 4 | 186,514,078 | rs28567706 | C | G | 0.99 | 0.49 | 0.09 | 5.49 | 4.05E-08 | 6533 | +?????+????++??? | SORBS2 | |
| 8 | 42,527,386 | rs11785369 | A | C | 0.89 | 0.09 | 0.02 | 5.35 | 8.83E-08 | 18634 | ++++++++++-++-++ | CHRNB3 | |
| 9 | 21,673,859 | rs184042824 | C | T | 0.98 | −0.31 | 0.06 | −5.43 | 5.70E-08 | 9005 | -+--?+?--?????-? | MIR31HG | |
| 11 | 97,713,579 | rs117029742 | T | C | 0.98 | −0.30 | 0.05 | −5.73 | 1.02E-08 | 13061 | ---------?+-?++? | BG182718 | |
| 14 | 44,608,436 | rs10133756 | C | G | 0.98 | −0.31 | 0.05 | −5.88 | 4.11E-09 | 13283 | ---??----?-??++- | AA333164 | |
| 15 | 78,882,925 | rs16969968 | G | A | 0.66 | −0.06 | 0.01 | −5.82 | 6.21E-09 | 17834 | ----------+-?+-- | CHRNA5 | |
| 17 | 51,940,280 | rs2877510 | C | T | 0.17 | 0.06 | 0.01 | 4.52 | 6.36E-06 | 18647 | ++++--++-+-++-++ | KIF2B | |
| 20 | 15,901,883 | rs6135601 | G | T | 0.97 | −0.13 | 0.03 | −4.55 | 5.53E-06 | 18644 | -----+----+----- | MACROD2 | |
| 22 | 42,058,846 | rs132786 | G | A | 0.19 | −0.07 | 0.01 | −4.61 | 4.07E-06 | 14605 | --?--------?---- | XRCC6 |
EA, effect allele; NEA, non-effect allele; EAF, effect allele frequency; BP, base-pair position according to GRCh37/hg19; FTND, Fagerström Test for Nicotine Dependence; TTFC, time to first cigarette. *, minus and plus signs refer to the direction of effect in each independent dataset (SAGE, SC, MGS, COPDGene1, ISIB, CIDR370v1, CIDR370v3, CIDRc3, CIDRc4, EAGLE, ALSPAC, AUTW, VTSABD, CEDAR, FTC1, FTC2, NTR, GCD), whereas “?” refers to missing data. Genome-wide significant signals are highlighted in bold.
For FTND, four loci were selected for replication in several independent samples (Table 2). Of the four loci tested, only rs16969968 at the CHRNA5-CHRNA3-CHRNB4 locus was successfully replicated (p = 1.19 × 10−11). Rs17005545 in the novel locus CIB4 had a p-value of 2.68 × 10−7 in the combined samples. For TTFC, rs16969968 in CHRNA5 and 11 other loci were selected for replication. Rrs16969968 in CHRNA5 (p = 2.84 × 10−10) was successfully replicated and rs11785369 near the CHRNB3 gene reached genome-wide significance in the combined samples (Table 2).
Table 2.
Replication of candidates for FTND and TTFC
| Marker Info | Discovery | Replication | Combined | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CHR | BP | SNP | EA | NEA | GENE | β | SE | p | β | SE | p | β | SE | p | |
| FTND | 2 | 26,860,822 | rs17005545 | T | C | CIB4 | −0.23 | 0.05 | 6.99E-07 | −0.16 | 0.10 | 0.1078 | −0.22 | 0.04 | 2.68E-07 |
| 11 | 97,713,579 | rs117029742 | T | C | BG182718 | −0.64 | 0.12 | 1.04E-07 | 0.29 | 0.19 | 0.1370 | −0.38 | 0.10 | 1.73E-04 | |
| 15 | 78,882,925 | rs16969968 | G | A | CHRNA5 | −0.21 | 0.02 | 3.96E-19 | −0.29 | 0.04 | 1.19E-11 | −0.23 | 0.02 | 4.24E-28 | |
| 18 | 28,355,510 | rs28510557 | C | T | DSC3 | −0.10 | 0.02 | 1.23E-06 | 0.02 | 0.04 | 0.6672 | −0.08 | 0.02 | 3.19E-05 | |
| TFC | 1 | 40,464,894 | rs34022242 | C | G | MFSD2A | −0.13 | 0.02 | 2.48E-07 | −0.05 | 0.04 | 0.1643 | −0.11 | 0.02 | 3.53E-07 |
| 1 | 68,862,622 | rs3125927 | G | A | RPE65 | −0.05 | 0.01 | 5.24E-07 | 0.02 | 0.02 | 0.2298 | −0.03 | 0.01 | 2.25E-04 | |
| 4 | 103,715,385 | rs223441 | C | T | CISD2 | 0.05 | 0.01 | 4.66E-06 | −0.01 | 0.02 | 0.4071 | 0.03 | 0.01 | 4.42E-04 | |
| 4 | 186,514,078 | rs28567706 | C | G | SORBS2 | 0.49 | 0.09 | 4.05E-08 | 0.08 | 0.07 | 0.2745 | 0.23 | 0.05 | 2.22E-05 | |
| 8 | 42,527,386 | rs11785369 | A | C | CHRNB3 | 0.09 | 0.02 | 8.83E-08 | 0.06 | 0.03 | 2.46E-02 | 0.08 | 0.01 | 7.88E-09 | |
| 9 | 21,673,859 | rs184042824 | C | T | MIR31HG | −0.31 | 0.06 | 5.70E-08 | −0.03 | 0.09 | 0.7136 | −0.23 | 0.05 | 1.53E-06 | |
| 11 | 97,713,579 | rs117029742 | T | C | BG182718 | −0.30 | 0.05 | 1.02E-08 | 0.07 | 0.07 | 0.3574 | −0.17 | 0.04 | 5.28E-05 | |
| 14 | 44,608,436 | rs10133756 | C | G | AA333164 | −0.31 | 0.05 | 4.11E-09 | 0.04 | 0.07 | 0.5277 | −0.18 | 0.04 | 1.26E-05 | |
| 15 | 78,882,925 | rs16969968 | G | A | CHRNA5 | −0.06 | 0.01 | 6.21E-09 | −0.10 | 0.02 | 2.84E-10 | −0.07 | 0.01 | 1.05E-17 | |
| 17 | 51,940,280 | rs2877510 | C | T | KIF2B | 0.06 | 0.01 | 6.36E-06 | 0.01 | 0.02 | 0.5735 | 0.04 | 0.01 | 4.45E-05 | |
| 20 | 15,901,883 | rs6135601 | G | T | MACROD2 | −0.13 | 0.03 | 5.53E-06 | 0.02 | 0.04 | 0.5457 | −0.08 | 0.02 | 8.18E-04 | |
| 22 | 42,058,846 | rs132786 | G | A | XRCC6 | −0.07 | 0.01 | 4.07E-06 | 0.02 | 0.02 | 0.2141 | −0.03 | 0.01 | 5.29E-03 | |
EA, effect allele; NEA, noneffect allele; EAF, effect allele frequency; BP, base-pair position according to GRCh37/hg19; FTND, Fagerström Test for Nicotine Dependence; TTFC, time to first cigarette. Loci reaching GWAS significance (5E-8) were highlighted in bold.
We conducted SNP heritability analyses for both FTND and TTFC using the LD-adjusted kinship algorithm.23,24 The SNP heritability for FTND was estimated at 0.645, and that for TTFC was 0.193.
Convergent Loci Between FTND and TTFC
We compared the results obtained for FTND and TTFC to identify genes showing convergent association signals. We then selected genes/loci showing suggestive association (p ≤ 5 × 10−5) in both FTND and TTFC meta-analyses. A total of 15 genes/loci with convergent association signals were found (Table 3). The CHRNA5-CHRNA3-CHRNB4 locus was the only one reaching genome-wide significance for both FTND and TTFC. Other nicotinic receptors, CHRNB3-CHRNA6 and CHRNA4, also showed convergent association signals. Other highlighted genes included long noncoding RNAs (DA409732 and BG182718) and a RNA-binding gene (RBFOX1), microtubule and actin regulation genes (KIF2B and VAV2), a cAMP/cGMP modulating gene (PDE2A), and an apolipoprotein gene (APOL3). For some genes (THSD7B, PDE2A, KIF2B, and APOL3), the same SNPs showed suggestive association for both phenotypes; for other genes (ZNF804A, VAV2, RBFOX1, and CHRNA4), the association signals for the two phenotypes were from different SNPs.
Table 3.
Genes/loci showing convergent association between FTND and TTFC
| Marker Info | FTND | TTFC | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CHR | BP | SNP | EA | NEA | EAF | β | SE | p | β | SE | p |
GENE |
| 2 | 4,073,151 | rs13425218 | G | C | 0.68 | −0.13 | 0.03 | 3.80E-05 | −0.06 | 0.01 | 2.11E-05 | DA409732 |
| 2 | 137,983,050 | rs17269864 | C | A | 0.67 | −0.10 | 0.02 | 2.77E-06 | −0.04 | 0.01 | 3.17E-05 | THSD7B* |
| 2 | 185,787,483 | rs56321552 | G | C | 0.95 | 0.24 | 0.06 | 2.20E-05 | NL | NL | NL | ZNF804A** |
| 2 | 185,639,136 | rs78011041 | T | G | 0.97 | NL | NL | NL | 0.12 | 0.03 | 3.42E-05 | ZNF804A** |
| 2 | 221,852,642 | rs6709809 | G | A | 0.96 | −0.24 | 0.06 | 4.18E-05 | −0.13 | 0.03 | 2.59E-06 | — |
| 5 | 30,863,015 | rs4502841 | G | A | 0.48 | −0.09 | 0.02 | 1.46E-05 | −0.05 | 0.01 | 2.14E-06 | — |
| 5 | 92,486,203 | rs4526087 | C | G | 0.90 | 0.15 | 0.03 | 1.15E-05 | NL | NL | NL | — |
| 5 | 92,554,821 | rs193049029 | T | C | 0.98 | NL | NL | NL | −0.24 | 0.06 | 2.78E-05 | — |
| 8 | 42,527,386 | rs11785369 | A | C | 0.90 | 0.14 | 0.03 | 2.28E-05 | 0.09 | 0.02 | 8.83E-08 | CHRNB3-CHRNA6 |
| 9 | 136,789,062 | rs7847503 | A | G | 0.97 | −0.23 | 0.06 | 4.32E-05 | NL | NL | NL | VAV2 |
| 9 | 136,768,563 | rs10993850 | C | A | 0.91 | NL | NL | NL | 0.08 | 0.02 | 3.93E-05 | VAV2 |
| 11 | 72,301,503 | rs341047 | G | A | 0.88 | −0.14 | 0.03 | 3.07E-05 | −0.06 | 0.02 | 3.83E-05 | PDE2A |
| 11 | 97,713,579 | rs117029742 | T | C | 0.98 | −0.64 | 0.12 | 1.04E-07 | −0.30 | 0.05 | 1.02E-08 | BG182718 |
| 15 | 78,882,925 | rs16969968 | G | A | 0.64 | −0.21 | 0.02 | 3.96E-19 | −0.06 | 0.01 | 6.21E-09 | CHRNA5-CHRNA3-CHRNB4 |
| 16 | 6,124,714 | rs9927211 | G | C | 0.23 | −0.13 | 0.03 | 1.83E-05 | NL | NL | NL | RBFOX1 *** |
| 16 | 6,071,910 | rs12444143 | T | C | 0.40 | NL | NL | NL | −0.05 | 0.01 | 3.56E-05 | RBFOX1 *** |
| 17 | 51,926,097 | rs3966061 | C | T | 0.17 | 0.12 | 0.03 | 2.14E-05 | 0.06 | 0.01 | 1.46E-05 | KIF2B |
| 20 | 61,989,658 | rs45623037 | G | C | 0.92 | −0.21 | 0.05 | 2.75E-05 | NL | NL | NL | CHRNA4 |
| 20 | 60,522,424 | rs4925328 | T | G | 0.43 | NL | NL | NL | 0.05 | 0.01 | 4.78E-05 | CHRNA4 |
| 22 | 36,503,250 | rs16996373 | G | T | 0.84 | −0.12 | 0.03 | 3.18E-05 | −0.06 | 0.01 | 8.93E-06 | APOL3 |
EA, effect allele; NEA, noneffect allele; EAF, effect allele frequency; BP, base-pair position according to GRCh37/hg19; FTND, Fagerström Test for Nicotine Dependence; TTFC, time to first cigarette; NL, not listed because the p-value > 5 × 10−5; *, linked to alcohol addiction; 43 **, linked to schizophrenia; 58,59 ***, linked to cocaine addiction.42 Genes relevant to addictions or neuropsychiatric co-morbidities are highlighted in bold.
Gene-Based Meta-Analyses
We conducted gene-based analyses using the KGG program. In these analyses, in addition to the CHRNA5 locus and nearby genes, a few novel genes were identified (Table 4). COPB2 and CHRNA4 reached significance for association with FTND, and TTFCP2L1, RELN, and INO80C were significant for the TTFC phenotype. Table 4 also lists other candidates from gene-based analyses (i.e., genes with q-value ≤ 0.1).
Table 4.
Summary of gene-association results
| Gene Information | FTND | TTFC | Functions* | ||||
|---|---|---|---|---|---|---|---|
| Name | Chr | Position | P_value | FDR q_value | P_value | FDR q_value | |
| RPE65 | 1 | 68894506 | 0.49513 | 1.0000 | 0.00004 | 0.0673 | congenital amaurosis |
| CHD1L | 1 | 146714290 | 0.04676 | 0.7808 | 0.00003 | 0.0636 | cancer gene |
| TTFCP2L1 | 2 | 121974163 | 0.89730 | 1.0000 | 0.00001 | 0.0279 | pluripotency, development |
| COPB2 | 3 | 139076432 | 0.00001 | 0.0229 | 0.64899 | 1.0000 | lung cancer |
| LOC102723704 | 4 | 103698193 | 0.08404 | 0.8275 | 0.00004 | 0.0673 | ncRNA |
| UBE2D3 | 4 | 103715539 | 0.12397 | 0.8807 | 0.00005 | 0.0674 | ubiquitination |
| LOC105377348 | 4 | 103750470 | 0.32928 | 1.0000 | 0.00006 | 0.0676 | ncRNA |
| CISD2 | 4 | 103790134 | 0.36281 | 1.0000 | 0.00004 | 0.0673 | intracellular calcium homeostasis |
| SLC9B1 | 4 | 103822084 | 0.51668 | 1.0000 | 0.00004 | 0.0673 | sodium/hydrogen exchanger |
| DNAH8 | 6 | 38683116 | 0.16941 | 0.9251 | 0.00006 | 0.0676 | sperm and respiratory cilia motility |
| AGR3 | 7 | 16899029 | 0.37663 | 1.0000 | 0.00008 | 0.0827 | cancer gene |
| RELN | 7 | 103112230 | 0.31646 | 1.0000 | 4.47E-06 | 0.0223 | schizophrenia, autism, cell adhesion |
| CHRNB3 | 8 | 42552561 | 0.03471 | 0.7313 | 0.00005 | 0.0674 | nicotinic receptor |
| MIR31HG | 9 | 21454266 | 0.03006 | 0.7241 | 0.00005 | 0.0676 | lncRNA, cancer regulation |
| LOC389765 | 9 | 88420916 | 0.06893 | 0.8117 | 0.00006 | 0.0698 | — |
| TMEM132B | 12 | 126106997 | 0.19739 | 0.9368 | 0.00009 | 0.0847 | intracranial aneurysm |
| SLC46A3 | 13 | 29274217 | 0.54791 | 1.0000 | 0.00004 | 0.0673 | Lysosome/cytoplasm transport |
| KLHL1 | 13 | 70274724 | 0.00271 | 0.4838 | 0.00002 | 0.0579 | calcium signaling, cancer |
| IREB2 | 15 | 78730517 | 9.55E-11 | 4.76E-07 | 0.00014 | 0.1254 | lung diseases |
| HYKK | 15 | 78799905 | 3.59E-17 | 8.93E-13 | 1.20E-07 | 9.92E-04 | lung diseases |
| PSMA4 | 15 | 78832746 | 7.80E-11 | 4.76E-07 | 1.76E-05 | 5.48E-02 | lung diseases |
| CHRNA5 | 15 | 78857861 | 1.53E-15 | 1.27E-11 | 9.51E-08 | 9.92E-04 | nicotinic receptor |
| CHRNA3 | 15 | 78887646 | 1.00E-15 | 1.25E-11 | 6.26E-08 | 9.92E-04 | nicotinic receptor |
| CHRNB4 | 15 | 78916635 | 1.47E-10 | 6.12E-07 | 2.60E-06 | 1.62E-02 | nicotinic receptor |
| INO80C | 18 | 33048290 | 0.07345 | 0.8117 | 1.01E-05 | 0.0359 | chromatin remodeling/silencing |
| SSTR4 | 20 | 23016056 | 0.23580 | 0.9667 | 2.32E-05 | 0.0579 | somatostatin receptors |
| CHRNA4 | 20 | 61974661 | 1.39E-06 | 0.0049 | 0.0013 | 0.3773 | nicotinic receptor |
| XRCC6 | 22 | 42017345 | 0.09636 | 0.8438 | 0.0001 | 0.0724 | cancer gene |
| NHP2L1 | 22 | 42069936 | 0.30160 | 0.9951 | 0.0001 | 0.0835 | chromatin remodeling/silencing |
| MEI1 | 22 | 42095517 | 0.42723 | 1.0000 | 0.0001 | 0.0676 | meiotic chromosome synapsis |
FTND, Fagerström Test for Nicotine Dependence; TTFC, time to first cigarette; *, functions relevant to smoking or neuropsychiatric co-morbidities are highlighted in bold. FDR q-values below 0.05 are highlighted in bold.
Pathway and Network Analyses
There were 647 SNPs with p-value ≤ 5 × 10−5 in the FTND meta-analysis and they were mapped to 134 known genes. There were 936 markers with p-value ≤ 5 × 10−5 in the TTFC meta-analysis and they were mapped to 145 genes. Altogether 15 genes were shared between the two phenotypes, yielding a total of 265 unique genes for pathway analyses. In the enrichment analyses using canonical pathways defined in KEGG database, a total of 26 pathways were found to be overrepresented among these genes (Table 5). The neuroactive ligand–receptor interaction pathway was the most significantly enriched pathway, including nicotinic receptors (CHRNA5, CHRNA3, CHRNB4, CHRNB3, CHRNA6, and CHRNA4), gamma-aminobutyric acid receptor (GABRG3), glutamate receptor (GRM5), and somatostatin receptors (SSTR1 and SSTR4). In addition to several pathways known to be involved in ND (cell adhesion molecules, MAPK signaling, tight junction, and axon guidance), our analyses suggested that endocytosis, lysosome, chemokine signaling, and regulation of actin cytoskeleton pathways are also involved in ND.
Table 5.
Summary of pathway enrichment analyses
| Pathway | Genes Found in the Pathway | # Gene in Pathway | # Gene Observed | # Gene Expected | Observed/Expected Ratio | Raw p-Value | Adjusted p-Value |
|---|---|---|---|---|---|---|---|
| Neuroactive ligand- receptor interaction | CHRNA3,CHRNA4,CHRNA5,CHRNA6, CHRNB3,CHRNB4,GABRG3,GRM5, SSTR1,SSTR4,THRB | 272 | 11 | 1.65 | 6.66 | 1.03E-06 | 5.56E-05 |
| Endocytosis | AP2A2,EHD3,FGFR2,IQSEC1,MET,RAB5C | 201 | 6 | 1.22 | 4.91 | 0.0015 | 0.0405 |
| Intestinal immune network for IgA production | HLA-DMB,HLA-DRB5,IL15 | 48 | 3 | 0.29 | 10.29 | 0.0031 | 0.0540 |
| Lysosome | AP1S3,LIPA,MANBA,SUMF1 | 121 | 4 | 0.74 | 5.44 | 0.0065 | 0.0694 |
| Protein export | SPCS3,SRP9 | 23 | 2 | 0.14 | 14.31 | 0.0086 | 0.0694 |
| Cell adhesion molecules (CAMs) | CDH4,HLA-DMB,HLA-DRB5,NRCAM | 133 | 4 | 0.81 | 4.95 | 0.0090 | 0.0694 |
| Arrhythmogenic right ventricular cardiomyopathy (ARVC) | CACNA1D,CACNG4,CTNNA3 | 74 | 3 | 0.45 | 6.67 | 0.0105 | 0.0709 |
| Dilated cardiomyopathy | ADCY4,CACNA1D,CACNG4 | 90 | 3 | 0.55 | 5.49 | 0.0177 | 0.0816 |
| Purine metabolism | ADCY4,PAPSS2,PDE2A,PDE4B | 162 | 4 | 0.98 | 4.06 | 0.0174 | 0.0816 |
| Huntington’s disease | AP2A2,GRM5,PPARG,TAF4 | 183 | 4 | 1.11 | 3.60 | 0.0259 | 0.0816 |
| MAPK signaling pathway | CACNA1D,CACNG4,DUSP16,FGFR2,MRAS | 268 | 5 | 1.63 | 3.07 | 0.0244 | 0.0816 |
| Melanogenesis | ADCY4,FZD8,WNT16 | 101 | 3 | 0.61 | 4.89 | 0.0239 | 0.0816 |
| Allograft rejection | HLA-DMB,HLA-DRB5 | 37 | 2 | 0.22 | 8.90 | 0.0213 | 0.0816 |
| Type I diabetes mellitus | HLA-DMB,HLA-DRB5 | 43 | 2 | 0.26 | 7.66 | 0.0282 | 0.0816 |
| Asthma | HLA-DMB,HLA-DRB5 | 30 | 2 | 0.18 | 10.97 | 0.0143 | 0.0816 |
| Graft-versus-host disease | HLA-DMB,HLA-DRB5 | 41 | 2 | 0.25 | 8.03 | 0.0258 | 0.0816 |
| Rheumatoid arthritis | HLA-DMB,HLA-DRB5,IL15 | 91 | 3 | 0.55 | 5.43 | 0.0182 | 0.0816 |
| Chemokine signaling pathway | ADCY4,TIAM2,VAV2,XCL1 | 189 | 4 | 1.15 | 3.48 | 0.0287 | 0.0816 |
| Regulation of actin cytoskeleton | FGFR2,MRAS,TIAM2,VAV2 | 213 | 4 | 1.29 | 3.09 | 0.0416 | 0.0944 |
| Autoimmune thyroid disease | HLA-DMB,HLA-DRB5 | 52 | 2 | 0.32 | 6.33 | 0.0400 | 0.0944 |
| Hedgehog signaling pathway | RAB23,WNT16 | 56 | 2 | 0.34 | 5.88 | 0.0457 | 0.0944 |
| Basal cell carcinoma | FZD8,WNT16 | 55 | 2 | 0.33 | 5.99 | 0.0442 | 0.0944 |
| Tight junction | CTNNA3,EPB41L1,MRAS | 132 | 3 | 0.8 | 3.74 | 0.0469 | 0.0944 |
| Inositol phosphate metabolism | IMPA1,MINPP1 | 57 | 2 | 0.35 | 5.78 | 0.0472 | 0.0944 |
| Axon guidance | MET,NTNG1,ROBO2 | 129 | 3 | 0.78 | 3.83 | 0.0443 | 0.0944 |
| Staphylococcus aureus infection | HLA-DMB,HLA-DRB5 | 55 | 2 | 0.33 | 5.99 | 0.0442 | 0.0944 |
Statistically significant adjusted p-values are highlighted in bold.
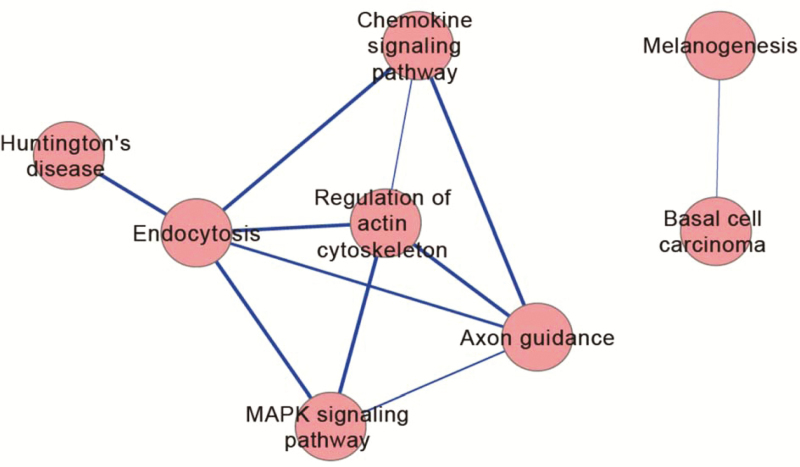
We further tested if and how these pathways interacted with each other. Multiple immune related pathways were identified by two highlighted genes, HLA-DMB and HLA-DRB5; to avoid the extensive network of HLA-related immune responses, we excluded these two genes in our pathway crosstalk analyses. Our analyses revealed interacting networks among the endocytosis, regulation of actin cytoskeleton, MAPK signaling, axon guidance, and chemokine signaling pathways (Figure 1). We also saw that two other pathways, melanogenesis and basal cell carcinoma, interacted and contributed to ND phenotypes.
Figure 1.
Network interaction based on markers with p values ≤5 × 10−5.
Conclusion and Discussion
We conducted GWAS meta-analyses for the FTND and TTFC phenotypes in about 19 000 regular smokers of European ancestry from 14 independent cohorts. We confirmed the known association of the functional variant D398N (rs16969968) in the CHRNA5-CHRNA3-CHRNB4 locus for both FTND and TTFC phenotypes. Although the association between FTND and CHRNA5-CHRNA3-CHRNB4 locus had been reported before, ours is the first to report its association with TTFC. GWAS meta-analysis of FTND identified one potential novel locus (CIB4 on 2p23) with suggestive association in the discovery samples and a trend in the replication samples. CIB4 encodes a calcium binding protein that interacts with integrin.36 It may be involved in the integrin signaling. Our GWAS meta-analyses of TTFC, the largest for this phenotype, identified three novel loci (SORBS2 on 4q35, BG182718 on 11q22, and AA333164 on 14q21) in the discovery samples. SORBS2 encodes an adaptor protein involved in the regulation of actin cytoskeleton37 and is recently suggested to be involved in intellectual disability.38 The 11q22 signal peaks at rs117029742 located 25 kb from the 3′ end of BG182718 (also referred to as RP11-379J13.2). BG182718 is a long noncoding RNA with unknown function. In the interval of 1.5 MB centered at rs117029742, there are no other known genes. Interestingly, ClinVar database (http://www.ncbi.nlm.nih.gov/clinvar/) reports multiple large copy number variants in this region and these variants are reported to be associated with developmental disabilities.39 AA333164 (also referred to as RP11-305B6.3) on 14q21 is another long noncoding RNA with unknown function; it also overlaps with known copy number variants associated with global developmental delay.39 It is not clear how TTFC relates to intellectual disability or developmental delay, or whether there are other functions that account for the association. Independent replication of the three novel loci for TTFC was not evident, presumably due to the lack of power of the replication samples. Statistically significant evidence for independent replication of the observed SNP associations may require even larger sample sizes.
FTND is a commonly used measure in studies of smoking behavior and ND. It consists of six questions, with a sum score ranging from 0 to 10.8 Many studies have used FTND scores to define ND, with varying thresholds (e.g., FTND ≤ 3 as low ND and > 6 as high ND, but also FTND ≥4 as dependent and FTND = 0 as unaffected and those in-between as uncertain), whereas others have treated FTND as a quantitative trait. Hancock and colleagues6 conducted a GWAS using categorized FTND (low-level smoking and mild ND [FTND = 0 to 3]; moderate ND [FTND = 4 to 6]; and severe ND [FTND = 7 to 10]) and discovered that CHRNA4 was associated with FTND at genome-wide significance across GWAS discovery and replication samples. Loukola and colleagues14 used FTND as a binary trait (FTND ≥ 4 as affected) in a study of 1,114 Finns but detected no genome-wide significant associations. Gelernter and colleagues13 conducted a GWAS with FTND as a quantitative trait in a sample of 4117 Caucasians and 3529 African Americans, and detected a genome-wide significant signal in an intergenic region on 7q21. In our analyses, which included the Caucasian subjects from the study of Gelernter and colleagues, only the CHRNA5-CHRNA3-CHRNB4 locus reached genome-wide significance. When the discovery and replication samples were combined, the signal amplified. The signal observed in Gelernter et al.’s study,13 rs13225753, was not significant in our analyses (p = .7188), presumably due to heterogeneity at the locus (Cochran’s Q = 36.86, p = 4.36 × 10−4).
Compared with FTND, the TTFC phenotype is more related to the ability to quitting and relapse.9 Loukola and colleagues14 conducted a GWAS of TTFC in 1114 Finnish subjects, but detected no genome-wide significant signals. In our discovery sample, which also included the subjects from the study of Loukola and colleagues,14 we found four loci reaching genome-wide significance (Table 1). When the discovery and replication samples were combined, the CHRNA5-CHRNA3-CHRNB4 locus remained genome-wide significant for both phenotypes and the CHRNB3-CHRNA6 locus reached genome-wide significance for TTFC, whereas the signals of other loci decreased somewhat due to weaker magnitudes of association in the replication samples (the common “winner’s curse” phenomenon40). The CHRNB3-CHRNA6 locus was originally identified using CPD phenotype,3 and in the current study, a much stronger signal was detected for TTFC (min p = 8.83 × 10−8 for rs11785396) compared with FTND (minimal p = 2.28 × 10−5 for rs11785396; see Table 3). This is consistent with a previous report that the signal at the CHRNB3-CHRNA6 locus is stronger when analyzed with FTND phenotype than that of CPD,15 because the main difference between FTND score and CPD is largely contributed by TTFC score.
TTFC is a component of FTND, and therefore TTFC scores correlate with FTND scores. In the literature, the correlation coefficient was reported to be 0.57.41 In the SC sample, the correlation coefficient was 0.69. However, when we examined the meta-analysis results from these phenotypes, we saw notable differences. For markers reaching genome-wide significance, the CHRNA5-CHRNA3-CHRNB4 locus was the only region showing overlap. Even for this locus, the strength of signal differed substantially between FTND (minimal p = 3.96 × 10−19 for rs16969968) and TTFC (minimal p = 6.21 × 10−9 for rs16969968), despite similar effective sample sizes (Table 1). BG182718 (tagged by rs117029742) also showed signals for both phenotypes, with stronger association with TTFC (p = 1.02 × 10−8) compared with FTND (p = 1.04 × 10−7). On the other hand, there were genes/loci that showed association with one phenotype but not the other. For example, CIB4 (tagged by rs17005545) showed suggestive association with FTND (p = 6.99 × 10−7), but very weak evidence of association with TTFC (p = 2.15 × 10−4). Similarly, rs34022242 located between MFSD2A and CAP1 showed suggestive association with TTFC (p = 2.48 × 10−7), but no association with FTND (p = .024). This suggests that FTND and TTFC measure different aspects of ND. Based on this information, we reasoned that if some genes/loci showed reasonable signals for both phenotypes, it could be seen as a convergent evidence that these genes/loci are likely genuinely associated with ND. Our convergent analyses identified several intriguing genes in addition to those previously highlighted for ND (CHRNA5-CHRNA3-CHRNB4, CHRNB3-CHRNA6, and CHRNA46). These included genes previously associated with diseases for which cigarette smoking is a significant risk factor, such as cocaine addiction (RBFOX1),42 alcohol dependence (THSD7B),43 schizophrenia (ZNF804A)44 and heroin addiction45,46 (ZNF804A), rheumatoid arthritis (PDE2A),47 and prostate cancer (APOL3)48 (Table 3). Furthermore, RBFOX1 was highlighted in a recent study on the genetic relationship between schizophrenia and ND.49 It remains to be elucidated whether these genes are independent risk factors for these diseases among nonsmokers, or is the genetic association arising from mediation or moderation by smoking. For example, the CHRNA5 D398N function variant (rs16969968) is a risk factor for lung cancer but only among smokers.50,51 Our results further reassured that for a true signal, such as the CHRNA5-CHRNA3-CHRNB4 locus, convergent signals are seen with multiple-related phenotypes.
We also noticed the difference in SNP heritability estimates between the FTND and TTFC phenotypes. The SNP heritability estimate of 0.645 for FTND was close to the heritability estimated from twin studies for ND.52 The estimate of 0.193 for TTFC seemed low when compared with twin studies,53 but it was close to the estimate of 0.154 from a study with SNP measure.54 The implication of this difference was not immediately clear. As SNP heritability estimates were influenced by the power of the GWASs, it was likely that our TTFC meta-analysis did not have the power to have a good estimate of heritability.
Gene-based analyses highlighted several other genes besides the CHRNA5-CHRNA3-CHRNB4 and CHRNA4 loci, including a gene involved in pluripotency demethylation and development (TTFCP2L1),55,56 a gene associated with lung cancer (COPB2),57 a gene implicated in schizophrenia (RELN),58,59 and a gene involved in chromatin remodeling (INO80C)60 (Table 4). These were novel genes first reported to be associated with smoking-related phenotypes. It would be interesting to see whether these genes could be replicated in future studies. Our pathway analyses revealed two statistically significantly enriched pathways. Although the involvement of neuroactive ligand-receptor interaction pathway in ND was expected since nicotinic receptors and other surface receptors belong to this pathway, the identification of endocytosis pathway in ND was novel and interesting. Over the years, there has been accumulating evidence that the recycling and internalization of surface receptors, such as nicotinic receptors,61 NMDA receptors,62 glutamate receptors,63 and opioid receptors,64 are involved in drug addiction. Furthermore, it is known that the recycling and internalization of surface proteins are mediated and regulated by endocytosis, actin cytoskeleton, and lysosome functions. Our pathway analyses explicitly identified these pathways in ND (Table 5). Our network analyses indicated that these pathways were interacting with each other, forming an interconnected network. These findings were consistent with the emerging picture in addiction studies.
In summary, our GWAS meta-analyses of FTND and TTFC identified several promising candidates for both phenotypes. Three novel loci (SORBS2, BG182718, and AA333164) were discovered for TTFC. Although we could not validate these novel loci with statistical significance in our replication sample, further investigation is warranted. With supporting information from both FTND and TTFC phonotypes, we also identified promising candidates for ND, including several genes known for association with other psychiatric disorders. Our pathway analyses highlighted the endocytosis pathway, supporting the importance of recycling and internalization of surface receptors in the development of nicotine addiction. In our network analyses, we discovered a multinode network with several interacting pathways known for involvement in substance abuse and psychiatric disorders. This information provides new insights for our understanding of ND and nicotine withdrawal.
Supplementary Material
Acknowledgments
This study was supported, in part, by NIH grants DA032246, DA035825. This work used data from multiple dbGaP studies. We acknowledge the investigators involved in the collection, genotyping, and analysis of these data sets. Detailed description of these samples, their funding agencies, and principal investigators were available in Supplementary Material.
Declaration of Interests
LJB is listed as an inventor on Issued U.S. Patent 8,080,371, “Markers for Addiction” covering the use of certain SNPs in determining the diagnosis, prognosis, and treatment of addiction. HRK has been a consultant, advisory board member, or CME speaker for Indivior, Lundbeck, and Otsuka and is a member of the American Society of Clinical Psychopharmacology’s Alcohol Clinical Trials Initiative (ACTIVE), which was supported in the last 3 years by AbbVie, Alkermes, Ethypharm, Indivior, Lilly, Lundbeck, Otsuka, Pfizer, and XenoPort.
References
- 1. Saccone SF, Hinrichs AL, Saccone NL, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007;16(1):36–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Berrettini W, Yuan X, Tozzi F, et al. Alpha-5/alpha-3 nicotinic receptor subunit alleles increase risk for heavy smoking. Mol Psychiatry. 2008;13(4):368–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Thorgeirsson TE, Geller F, Sulem P, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452(7187):638–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Thorgeirsson TE, Gudbjartsson DF, Surakka I, et al. ; ENGAGE Consortium Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet. 2010;42(5):448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Minicã CC, Mbarek H, Pool R, Dolan CV, Boomsma DI, Vink JM. Pathways to smoking behaviours: Biological insights from the Tobacco and Genetics Consortium meta-analysis. Mol Psychiatry. 2017;22(1):82–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hancock DB, Reginsson GW, Gaddis NC, et al. Genome-wide meta-analysis reveals common splice site acceptor variant in CHRNA4 associated with nicotine dependence. Transl Psychiatry. 2015;5:e651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zheng J, Erzurumluoglu AM, Elsworth BL, et al. ; Early Genetics and Lifecourse Epidemiology (EAGLE) Eczema Consortium LD Hub: A centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics. 2017;33(2):272–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Heatherton TF, Kozlowski LT, Frecker RC, Fagerström KO. The fagerström test for nicotine dependence: A revision of the fagerström tolerance questionnaire. Br J Addict. 1991;86(9):1119–1127. [DOI] [PubMed] [Google Scholar]
- 9. Baker TB, Piper ME, McCarthy DE, et al. ; Transdisciplinary Tobacco Use Research Center (TTURC) Tobacco Dependence Time to first cigarette in the morning as an index of ability to quit smoking: Implications for nicotine dependence. Nicotine Tob Res Off J Soc Res Nicotine Tob. 2007;9 (Suppl 4):S555–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mercincavage M, Branstetter SA, Muscat JE, Horn KA. Time to first cigarette predicts cessation outcomes in adolescent smokers. Nicotine Tob Res. 2013;15(12):1996–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Guertin KA, Gu F, Wacholder S, et al. Time to first morning cigarette and risk of chronic obstructive pulmonary disease: Smokers in the PLCO cancer screening trial. PLoS One. 2015;10(5):e0125973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gu F, Wacholder S, Kovalchik S, et al. Time to smoke first morning cigarette and lung cancer in a case-control study. J Natl Cancer Inst. 2014;106(6):dju118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gelernter J, Kranzler HR, Sherva R, et al. Genome-wide association study of nicotine dependence in American populations: Identification of novel risk loci in both African-Americans and European-Americans. Biol Psychiatry. 2015;77(5):493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Loukola A, Wedenoja J, Keskitalo-Vuokko K, et al. Genome-wide association study on detailed profiles of smoking behavior and nicotine dependence in a twin sample. Mol Psychiatry. 2014;19(5):615–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rice JP, Hartz SM, Agrawal A, et al. ; GENEVA Consortium CHRNB3 is more strongly associated with Fagerström test for cigarette dependence-based nicotine dependence than cigarettes per day: Phenotype definition changes genome-wide association studies results. Addiction. 2012;107(11):2019–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ware JJ, van den Bree M, Munafò MR. From men to mice: CHRNA5/CHRNA3, smoking behavior and disease. Nicotine Tob Res. 2012;14(11):1291–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Willer CJ, Sanna S, Jackson AU, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40(2):161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sanna S, Jackson AU, Nagaraja R, et al. Common variants in the GDF5-UQCC region are associated with variation in human height. Nat Genet. 2008;40(2):198–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39(7):906–913. [DOI] [PubMed] [Google Scholar]
- 20. Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44(8):955–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Purcell S, Neale B, Todd-Brown K, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mägi R, Morris AP. GWAMA: Software for genome-wide association meta-analysis. BMC Bioinformatics. 2010;11:288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Speed D, Hemani G, Johnson MR, Balding DJ. Improved heritability estimation from genome-wide SNPs. Am J Hum Genet. 2012;91(6):1011–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Speed D, Cai N, Johnson MR, Nejentsev S, Balding DJ; UCLEB Consortium Reevaluation of SNP heritability in complex human traits. Nat Genet. 2017;49(7):986–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shi J, Levinson DF, Duan J, et al. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature. 2009;460(7256):753–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bergen SE, O’Dushlaine CT, Ripke S, et al. Genome-wide association study in a Swedish population yields support for greater CNV and MHC involvement in schizophrenia compared with bipolar disorder. Mol Psychiatry. 2012;17(9):880–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Saccone NL, Culverhouse RC, Schwantes-An T-H, et al. Multiple independent loci at chromosome 15q25.1 affect smoking quantity: A meta-analysis and comparison with lung cancer and COPD. PLoS Genet. 2010;6(8):e1001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li MX, Gui HS, Kwan JS, Sham PC. GATES: A rapid and powerful gene-based association test using extended Simes procedure. Am J Hum Genet. 2011;88(3):283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li MX, Kwan JS, Sham PC. HYST: A hybrid set-based test for genome-wide association studies, with application to protein-protein interaction-based association analysis. Am J Hum Genet. 2012;91(3):478–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol. 1995;57(1):289–300. [Google Scholar]
- 31. Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): Update 2013. Nucleic Acids Res. 2013;41(Web Server issue):W77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang B, Kirov S, Snoddy J. WebGestalt: An integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33(Web Server issue):W741–W748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huang Y, Li S. Detection of characteristic sub pathway network for angiogenesis based on the comprehensive pathway network. BMC Bioinformatics. 2010;11(Suppl 1):S32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhao Z, Xu J, Chen J, et al. Transcriptome sequencing and genome-wide association analyses reveal lysosomal function and actin cytoskeleton remodeling in schizophrenia and bipolar disorder. Mol Psychiatry. 2015;20(5):563–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wu J, Vallenius T, Ovaska K, Westermarck J, Mäkelä TP, Hautaniemi S. Integrated network analysis platform for protein-protein interactions. Nat Methods. 2009;6(1):75–77. [DOI] [PubMed] [Google Scholar]
- 36. Huang H, Bogstie JN, Vogel HJ. Biophysical and structural studies of the human calcium- and integrin-binding protein family: Understanding their functional similarities and differences. Biochem Cell Biol. 2012;90(5):646–656. [DOI] [PubMed] [Google Scholar]
- 37. Cestra G, Toomre D, Chang S, De Camilli P. The Abl/Arg substrate ArgBP2/nArgBP2 coordinates the function of multiple regulatory mechanisms converging on the actin cytoskeleton. Proc Natl Acad Sci U S A. 2005;102(5):1731–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang Q, Gao X, Li C, et al. Impaired dendritic development and memory in sorbs2 knock-out mice. J Neurosci Off J Soc Neurosci. 2016;36(7):2247–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Miller DT, Adam MP, Aradhya S, et al. Consensus statement: Chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86(5):749–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kraft P. Curses–winner’s and otherwise–in genetic epidemiology. Epidemiology. 2008;19(5):649–51; discussion 657. [DOI] [PubMed] [Google Scholar]
- 41. Korte KJ, Capron DW, Zvolensky M, Schmidt NB. The Fagerström test for nicotine dependence: Do revisions in the item scoring enhance the psychometric properties? Addict Behav. 2013;38(3):1757–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Feng J, Wilkinson M, Liu X, et al. Chronic cocaine-regulated epigenomic changes in mouse nucleus accumbens. Genome Biol. 2014;15(4):R65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang KS, Liu X, Zhang Q, Pan Y, Aragam N, Zeng M. A meta-analysis of two genome-wide association studies identifies 3 new loci for alcohol dependence. J Psychiatr Res. 2011;45(11):1419–1425. [DOI] [PubMed] [Google Scholar]
- 44. O’Donovan MC, Craddock N, Norton N, et al. ; Molecular Genetics of Schizophrenia Collaboration Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet. 2008;40(9):1053–1055. [DOI] [PubMed] [Google Scholar]
- 45. Sun Y, Zhao L-Y, Wang G-B, et al. ZNF804A variants confer risk for heroin addiction and affect decision making and gray matter volume in heroin abusers. Addict Biol. 2016;21(3):657–666. [DOI] [PubMed] [Google Scholar]
- 46. Hancock DB, Levy JL, Gaddis NC, et al. Replication of ZNF804A gene variant associations with risk of heroin addiction. Genes Brain Behav. 2015;14(8):635–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Okada Y, Terao C, Ikari K, et al. Meta-analysis identifies nine new loci associated with rheumatoid arthritis in the Japanese population. Nat Genet. 2012;44(5):511–516. [DOI] [PubMed] [Google Scholar]
- 48. Johanneson B, McDonnell SK, Karyadi DM, et al. Family-based association analysis of 42 hereditary prostate cancer families identifies the Apolipoprotein L3 region on chromosome 22q12 as a risk locus. Hum Mol Genet. 2010;19(19):3852–3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen J, Bacanu SA, Yu H, et al. ; Cotinine meta-analysis group; FTND meta-analysis group Genetic Relationship between Schizophrenia and Nicotine Dependence. Sci Rep. 2016;6:25671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Timofeeva MN, Hung RJ, Rafnar T, et al. ; Transdisciplinary Research in Cancer of the Lung (TRICL) Research Team Influence of common genetic variation on lung cancer risk: Meta-analysis of 14 900 cases and 29 485 controls. Hum Mol Genet. 2012;21(22):4980–4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gabrielsen ME, Romundstad P, Langhammer A, Krokan HE, Skorpen F. Association between a 15q25 gene variant, nicotine-related habits, lung cancer and COPD among 56,307 individuals from the HUNT study in Norway. Eur J Hum Genet. 2013;21(11):1293–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sullivan PF, Kendler KS. The genetic epidemiology of smoking. Nicotine Tob Res Off J Soc Res Nicotine Tob. 1999;( 1Suppl 2):S51–57; discussion S69-70. [DOI] [PubMed] [Google Scholar]
- 53. Lessov CN, Martin NG, Statham DJ, et al. Defining nicotine dependence for genetic research: Evidence from Australian twins. Psychol Med. 2004;34(5):865–879. [DOI] [PubMed] [Google Scholar]
- 54. Bidwell LC, Palmer RH, Brick L, McGeary JE, Knopik VS. Genome-wide single nucleotide polymorphism heritability of nicotine dependence as a multidimensional phenotype. Psychol Med. 2016;46(10):2059–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tang WW, Dietmann S, Irie N, et al. A Unique Gene Regulatory Network Resets the Human Germline Epigenome for Development. Cell. 2015;161(6):1453–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Takashima Y, Guo G, Loos R, et al. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell. 2014;158(6):1254–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Erdogan E, Klee EW, Thompson EA, Fields AP. Meta-analysis of oncogenic protein kinase Ciota signaling in lung adenocarcinoma. Clin Cancer Res. 2009;15(5):1527–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shifman S, Johannesson M, Bronstein M, et al. Genome-wide association identifies a common variant in the reelin gene that increases the risk of schizophrenia only in women. PLoS Genet. 2008;4(2):e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Greenbaum L, Levin R, Lerer E, et al. Association of reelin (RELN) single nucleotide polymorphism rs7341475 with prepulse inhibition in the Jewish Israeli population. Biol Psychiatry. 2011;69(5):e17–8; author reply e19. [DOI] [PubMed] [Google Scholar]
- 60. Papamichos-Chronakis M, Watanabe S, Rando OJ, Peterson CL. Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell. 2011;144(2):200–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. St John PA, Gordon H. Agonists cause endocytosis of nicotinic acetylcholine receptors on cultured myotubes. J Neurobiol. 2001;49(3):212–223. [DOI] [PubMed] [Google Scholar]
- 62. Lau CG, Zukin RS. NMDA receptor trafficking in synaptic plasticity and neuropsychiatric disorders. Nat Rev Neurosci. 2007;8(6):413–426. [DOI] [PubMed] [Google Scholar]
- 63. Duan JJ, Lozada AF, Gou CY, Xu J, Chen Y, Berg DK. Nicotine recruits glutamate receptors to postsynaptic sites. Mol Cell Neurosci. 2015;68:340–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Whistler JL, Chuang HH, Chu P, Jan LY, von Zastrow M. Functional dissociation of mu opioid receptor signaling and endocytosis: Implications for the biology of opiate tolerance and addiction. Neuron. 1999;23(4):737–746. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.