Abstract
Background:
Ossification of the posterior longitudinal ligament (OPLL) refers to an ectopic ossification disease originating from the posterior longitudinal ligament of the spine. Pressing on the spinal cord or nerve roots can cause limb sensory and motor disorders, significantly reducing the patient's quality of life. At present, the pathogenesis of OPLL is still unclear. The purpose of this study is to integrate microRNA (miRNA)-mRNA biological information data to further analyze the important molecules in the pathogenesis of OPLL, so as to provide targets for future OPLL molecular therapy.
Methods:
miRNA and mRNA expression profiles of GSE69787 were downloaded from Gene Expression Omnibus database and analyzed by edge R package. Funrich software was used to predict the target genes and transcription factors of de-miRNA. Gene ontology and Kyoto Encyclopedia of Genes and Genomes enrichment analysis of differentially expressed genes (DEGs) were carried out based on CLUEGO plug-in in Cytoscape. Using data collected from a search tool for the retrieval of interacting genes online database, a protein-protein interaction (PPI) network was constructed using Cytoscape. The hub gene selection and module analysis of PPI network were carried out by cytoHubba and molecular complex detection, plug-ins of Cytoscape software respectively.
Results:
A total of 346 genes, including 247 up-regulated genes and 99 down-regulated genes were selected as DEGs. SP1 was identified as an upstream transcription factor of de-miRNAs. Notably, gene ontology enrichment analysis shows that up- and down-regulated DEGs are mainly involved in BP, such as skeletal structure morphogenesis, skeletal system development, and animal organ morphogenesis. Kyoto Encyclopedia of Genes and Genomes enrichment analysis indicated that only WNT signaling pathway was associated with osteogenic differentiation. Lymphoid enhancer binding factor 1 and wingless-type MMTV integration site family member 2 Wingless-Type MMTV Integration site family member 2 were identified as hub genes, miR-520d-3p, miR-4782-3p, miR-6766-3p, and miR-199b-5p were identified as key miRNAs. In addition, 2 important network modules were obtained from PPI network.
Conclusions:
In this study, we established a potential miRNA-mRNA regulatory network associated with OPLL, revealing the key molecular mechanism of OPLL and providing targets for future treatment or prevent its occurrence.
Keywords: bioinformatic analysis, lymphoid enhancer binding factor 1, microRNAs, mRNA, ossification of the posterior longitudinal ligament, wingless-type MMTV integration site family member 2, wNT signaling pathway
1. Introduction
Ossification of the posterior longitudinal ligament (OPLL) refers to the abnormal thickening of the posterior longitudinal ligament of the spine and the formation of bone tissue,[1] about 70% occur in the cervical spine and more common in Asian populations.[2,3] Most patients have no clinical symptoms, which are only seen by imaging examination. However, when calcified posterior longitudinal ligament compresses the spinal cord or nerve root, patients will have limb sensory and motor disorders, and even autonomic nerve dysfunction, which will greatly reduce the quality of life of patients.[4,5] Surgery is needed if conservative treatment does not work,[6] although now the improvement of surgical technique, but can’t seem to stop the progress of OPLL.[5] In recent years, many medical researchers on the molecular structure characteristics were studied, the existence of a drug non-surgical treatment method and prevention of OPLL is now the focus of attention.[7,8]
MicroRNA (miRNA) is a kind of endogenous small RNA with a length of about 20 to 24 nucleotides, which has a variety of important and complex regulatory functions in cells.[9] The effect of microRNA on the target gene mRNA mainly depends on the degree of complementarity with the target gene transcribed sequence, which can be expressed in three ways. The first is to cut off the mRNA molecule of the target gene. The miRNA binds to the target gene in a completely complementary manner, the action mode and function are very similar to siRNA (small interfering RNA), and finally cut off the target mRNA, resulting in degradation of the mRNA. The second is to inhibit the translation of target genes. During the action, it does not completely complement and bind to the target gene, thereby inhibiting translation without affecting the stability of the mRNA. The third is binding inhibition. It has the above two modes of action: when it is complementary to the target gene, it directly targets the cleaved mRNA; when it is not completely combined with the target gene, it plays a role in regulating gene expression.[10] It has been reported that miRNA plays a key role in the occurrence and development of OPLL. For example, miR-10a regulates OPLL development by regulating the ID3 / RUNX2 axis.[11] miR-563 can significantly promote osteogenic differentiation of posterior longitudinal ligament cells by down-regulating SMURF1 in vitro.[12] There are significant differences in the expression of MiR-10a-5p, miR-563, and miR-210-3p in OPLL patients and normal population.[13]
In this study, we conducted multiple bioinformatics analysis methods on the data provided by Chen et al,[14] with the purpose of revealing important molecular mechanisms in the pathogenesis of OPLL and providing a possible basis for future targeted and preventive treatment.
2. Materials and methods
2.1. Microarray data
The GSE69787 gene expression data set is derived from GPL17303 Ion Torrent Proton (Homo sapiens) (http://www.ncbi.nlm.nih.gov/geo/) which contains 12 samples in 2 groups of different RNA profiles, mRNA group and miRNA group. In mRNA group, 3 protein–protein interaction (PLL) cell specimens and 3 OPLL cell specimens were included. Meanwhile, there are 3 PLL cell specimens and 3 OPLL cell specimens in miRNA group. This study was based on data from an open database. Ethics and patient consent are not applicable.
2.2. Identification of differentially expressed miRNA and mRNA
The differential expression analysis of mRNA and miRNA was conducted with the edgeR package of R software, an excellent tool for statistical calculation and statistical mapping.[15] The results of the analysis are saved in a file format, the cut-off criteria of differentially expressed genes (DEGs) is set as: FDR < 0.01, |log fold-change |> 2. Venn diagrams were produced by FunRich software, an independent software tool for functional enrichment and interaction network analysis of genes and proteins.[16]
2.3. Prediction potential transcription factors
The upstream transcription factors of DE-miRNAs were predicted using FunRich software. The screened DE-miRNAs were respectively up-regulated and down-regulated into FunRich, and the results showed only the top 10 predictive transcription factors.
2.4. Identification of DEGs
FunRich software was used to identify DEGs. We took the Venn intersection of the up-regulated DE-miRNAs targeted genes predicted by FunRich and the previously obtained down-regulated DE-mRNAs as the down-regulated DEGs. Similarly, the Venn intersection of the down-regulated DE-miRNAs targeted genes predicted by FunRich and the up-regulated DE-mRNAs as the up-regulated DEGs.
2.5. GO and KEGG enrichment analysis
DEGs’ gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis were performed using the plugin ClueGO[17] (version 2.5.4) in the Cytoscape software[18] (3.7.2) and the collated results are then imported into imageGP to adjust the bubble image visualization (http://www.ehbio.com/ImageGP). The standard setting with statistically significant difference was P < .05 and k value was 1.0.
2.6. PPI network construction and molecular complex detection (MCODE)
Integrate the PPI network. The interaction between DEGs was evaluated using the search tool for the retrieval of interacting genes database[19] (version 11.0); a composite score > 0.4 was considered a statistically significant interaction. Subsequently, the analysis results of PPI network were loaded into Cytoscape software for visual adjustment. After that, we detected molecular complex of PPI network by using MCODE (version 1.5.1), which is a plugin of Cytoscape software.
2.7. Identification of hub genes and hub miRNAs
CytoHubba[20] (version 0.1) is a plugin for identifying key genes from PPI network in Cytoscape. Considering the CytoHubba calculation ordering will count the genes for signaling pathways unrelated to osteogenic differentiation that would confuse our purpose and results. We took the top 10 genes after degree calculation method and the osteogenic differentiation pathway related genes to take Venn intersection, and the genes coexisting with the KEGG osteogenic differentiation related signaling pathway were considered as hub genes. Meanwhile, we took Venn intersection between target miRNAs of hub genes predicted by FunRich and De-miRNAs, the result was taken as hub miRNAs.
2.8. miRNA-mRNA network construction
Based on the acquisition of hub genes and hub miRNAs, as well as the differential expression result files analyzed by R software, Cytoscape was used to construct the mRNA-miRNA network according to their fold change values.
3. Results
3.1. Identification of DE-miRNAs, DE-mRNAs, and DEGs
According to the analysis results of edgeR package of R software, there were 344 de-miRNAs and 1631 de-mRNAs, among which 80 miRNAs were up-regulated and 264 miRNAs were down-regulated, 918 mRNAs were up-regulated and 713 mRNAs were down-regulated. According to FunRich prediction results, 2101 targeted genes were predicted by up-regulated de-miRNAs and 4338 targeted genes were predicted by down-regulated de-miRNAs, and 346 DEGs were obtained by Venn intersection, among which 247 DEGs were up-regulated and 99 DEGs were down-regulated (Fig. 1).
Figure 1.

Screen of candidate differentially expressed genes. (A) The intersection of target genes of down-regulated DE-microRNAs and up-regulated DE-mRNAs; (B) The intersection of target genes of up-regulated DE-microRNAs and down-regulated DE-mRNAs; (C) The intersection of genes in osteogenesis related signaling pathways and top 10 genes with degree score.
3.2. Transcription factors of DE-miRNAs predicted
In our study, upstream transcription factors of DE-miRNAs were predicted by FunRich software, and the first 10 predicted results of up-regulated and down-regulated DE-miRNAs were shown in Figure 2, A and B, respectively. Interestingly, it is obvious that SP1 may regulate most DE-miRNAs (Fig. 2).
Figure 2.

Predicted transcription factors of DE-microRNAs. (A) Top 10 transcription factors of up-regulated DE-microRNAs; (B) Top 10 transcription factors of down-regulated DE-miRNAs.
3.3. GO functional enrichment analysis
GO functional enrichment analysis of differentially expressed genes by “ClueGO” (Fig. 3). The results indicate that both up- and down-regulated DEGs are mainly enriched in biological processes. In Figure 3, A and B are the bubble maps of top 5 items of GO functional enrichment analysis of up-regulated and down-regulated genes in “ClueGO”, respectively (standard cut-off P < .05). As is evident from Figure 3, the related biological process of up-regulated DEGs are mainly enriched in skeletal system development, skeletal system morphogenesis, embryonic morphogenesis, regionalization, and embryonic organ morphogenesis. In cellular components analysis, the up-regulated DEGs are mainly enriched in lateral plasma membrane, voltage-gated potassium channel complex, potassium channel complex, presynaptic active zone cytoplasmic component, and cell cortex region. Besides, in molecular function (MF) analysis, up-regulated DEGs are mainly enriched in acetylgalactosaminyltransferase activity, UDP-glycosyltransferase activity, estrogen receptor binding, HMG box domain binding, voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization. However, for down-regulated DEGs, there were no results about CC and MF enrichment analysis, and the main BP enrichment was tertiary alcohol biosynthetic process, cortisol biosynthetic process, aldosterone metabolic process, positive regulation of osteoblast differentiation, animal organ formation.
Figure 3.

Gene ontology function enrichment bubble diagram. (A) Top 5 of up-regulate differentially expressed genes. (B) Top 5 of down-regulate differentially expressed genes (P < .05).
3.4. KEGG pathway enrichment analysis
To more intuitively show the enrichment analysis of DEGs in the KEGG pathways, we concentrated all DEGs together for analysis (P < .05) (Fig. 4). It can be clearly seen that 12 signaling pathways are enriched, and only one of the key pathways associated with osteogenic differentiation is WNT signaling pathway.
Figure 4.
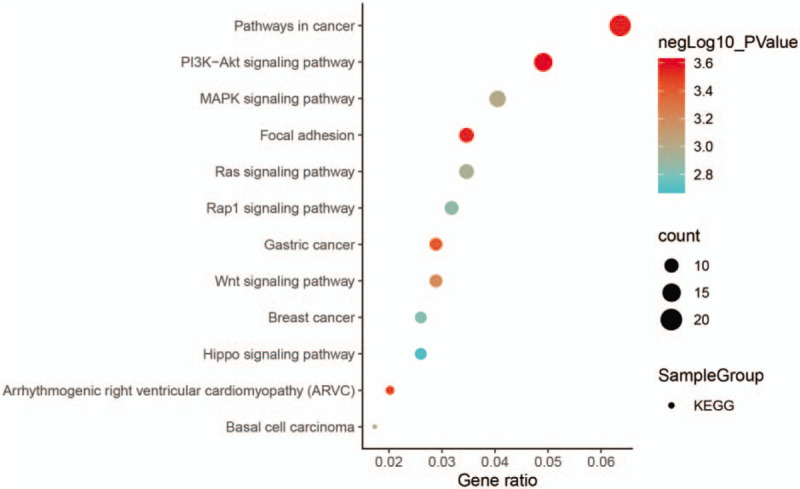
Kyoto Encyclopedia of Genes and Genomes enrichment analysis results of all differentially expressed genes (P < .05).
3.5. 16 Identification of hub genes and hub miRNAs
The top 10 genes were gotten by the CytoHubba (Table 1). Next, we derived the data from the KEGG analysis in Cluego and used the relevant genes in the signaling pathways associated with osteogenic differentiation (Table 1) for the Venn crossover (Fig. 1). As a result, 2 hub genes were identified: wingless-type MMTV integration site family member 2 (Wnt2) and lymphoid enhancer binding factor 1 (LEF1) (Table 2). Based on these 2 hub genes, miR-520d-3p, miR-4782-3p, miR-6766-3p, and miR-199b-5p were identified as hub miRNAs through FunRich miRNA prediction function.
Table 1.
Associated genes in the osteogenesis differentiation pathway and degree top 10 genes.

Table 2.
Function of 2 hub genes.

3.6. PPI network construction and Module analysis
The search tool for the retrieval of interacting genes database is used for all DEGs PPI network construction. After that, the data was entered into Cytoscape to adjust the visualization, which has 266 nodes and 572 edges (Fig. 5). As can be seen in the figure, red represents up-regulated DEGs, blue represents down-regulated DEGs, and the color depth represents the absolute value of fold change. The larger the fold change value is, the darker the color is. Interestingly, the 2 rings in the middle were obtained by analysis of MCODE plug-in from PPI network. However, GO and KEGG enrichment analysis were further performed on these 2 modules (Table 3). Surprisingly, the genes in module 1 are mainly enriched in BP like positive regulation of osteoblast differentiation, positive regulation of animal organ morphogenesis, but KEGG just enriched in Basal cell carcinoma signaling pathway. For module 2, the genes are enriched in collagen fibril organization (BP), extracellular matrix component (CC), and extracellular matrix structural constituent conferring tensile strength (MF).
Figure 5.

Protein–protein interaction network. The red circle represents up-regulated differentially expressed genes, while the blue circle represents down-regulated differentially expressed genes. The larger the absolute value of fold change is, the darker the color is, and the smaller the absolute value of fold change is, the lighter the color is. The middle 2 rings are module 1 and module 2 respectively.
Table 3.
Modules analysis of the PPI network category.

3.7. miRNA-mRNA network construction
Based on the screened hub miRNAs, we constructed the network interacting with miRNAs in DEGs (Fig. 6).
Figure 6.
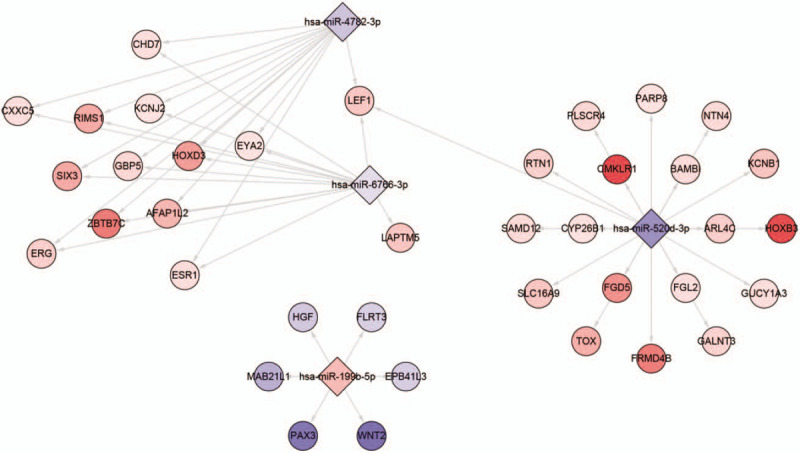
MicroRNA-mRNA network based on hub microRNAs.
4. Discussion
Ossification of the posterior longitudinal ligament of the spine is a degenerative disease. Calcification occurs in normal ligament tissues, which compress the spinal cord and nerve roots, leading to sensory and motor and other autonomic nerve dysfunction.[1–3] This will greatly reduce the quality of life of patients.[21] Currently, there is no other treatment except surgery, but surgical treatment is accompanied by the possibility of complications.[22,23] In addition, the mechanism of OPLL occurrence and development is still poorly understood at present. Exploring the key molecular mechanism of OPLL occurrence provides the possibility of future molecularly targeted therapy, which will greatly improve the quality of life of patients, avoid the possibility of surgery, and even prevent the occurrence of OPLL.
Molecular targeted therapy is a new type of therapy. Targeted therapy drugs inhibit the occurrence of diseases and reverse the progression of diseases by inhibiting specific molecules or signaling pathways.[24] Happily, with the progress of scientific research, more and more mechanisms of tumor occurrence and development have been discovered, and molecular targeted therapy has achieved good efficacy in the field of tumor. Such as targeted therapy in lung cancer[25] and molecular targeted therapy in ovarian cancer.[26]
In our study, we identified 346 differential genes and 344 differentially expressed miRNAs, including 247 up-regulated DEGs and 99 down-regulated DEGs, 80 up-regulated DE-miRNAs and 264 down-regulated DE-miRNAs. SP1 has been identified as the upstream transcription factor of de-miRNAs. Interestingly, recent studies have shown that SP1 regulated mir-545-3p inactivates Wnt/β-catenin signaling by targeting LRP5, thereby acting as an osteo-suppressive factor.[27] GO analysis showed that both up and down regulated DEGs are mainly enriched in biological processes like skeletal structure morphogenesis, skeletal system development and animal organ morphogenesis. KEGG enrichment analysis indicated that pathways related to osteogenic differentiation include MAPK signaling pathway[28] and WNT signaling pathway,[29] while the rest are non-osteogenic pathways such as pathways in cancer, breast cancer signaling pathway, arrhythmogenic right ventricular cardiomyopathy (ARVC) signaling pathway. However, a review of the relevant genes enriched in MAPK signaling pathway suggests that these DEGs-enriched MAPK signaling pathways are related to tumors and not involved in osteogenic differentiation,[30] while the related genes of WNT signaling pathway are involved in osteogenic differentiation. So, we thought that the hub gene was in the WNT signaling pathway. Considering the cytoHubba computational ranking will count the genes of the other non-osteogenic pathways, which will confuse our research. Therefore, we took the top 10 genes in Degree and the genes in WNT signaling pathway to take Venn intersection, and identified 2 Hub genes: Wnt2 and LEF1. Besides, based on the 2 hub genes, we found in our de-miRNAs analysis results that there were 4 miRNAs correlated with them, namely miR-520d-3p, miR-4782-3p, miR-6766-3p, and miR-199b-5p, respectively. Strikingly, in our PPI complex module analysis, GO functional analysis of module 1's genes mainly enriched in the positive regulation of osteoblast differentiation and positive regulation of animal outraged morphogenesis, which are the osteogenesis related functions. However, the KEGG enrichment analysis has only enriched in Basal cell carcinoma, which is considered because the modular analysis is based on the whole PPI network, which includes other non-osteogenic genes, leading to confusion of our results. For module 2, GO functions are enriched in collagen fibril organization, extracellular matrix structural constituent conferring and tensile strength and extracellular matrix component, KEGG no enrichment results. Once again, consider that module 2 comes from the entire PPI network.
Interestingly, the GeneCards database indicates that Wnt2 and LEF1 play important roles in WNT signaling pathway. Wnt2 is a ligand for members of the frizzled family of 7 transmembrane receptors. Functions in the canonical Wnt signaling pathway that results in activation of transcription factors of the TCF/LEF family. Functions as upstream regulator of FGF10 expression. Besides, it plays an important role in embryonic lung development and may contribute to embryonic brain development by regulating the proliferation of dopaminergic precursors and neurons (by similarity).[31] Lymphoid enhancer binding factor 1 (LEF1) participates in the Wnt signaling pathway. Activating transcription of target genes in the presence of CTNNB1 and EP300 and may plays a role in hair cell differentiation and follicle morphogenesis.[32,33] Surprisingly, further investigation showed that Wnt2 and LEF1 regulated the osteogenic differentiation of mesenchymal stem cells through the WNT signaling pathway.[34–36] As shown in Figure 7, SP1 transcription factors down-regulated miR-520d-3p, miR-4782-3p, and miR-6766-3p stimulated LEF1 overexpression, which led to OPLL through WNT signaling pathway. On the other hand, SP1 can also up-regulate mir-199b-5p inactivation of Wnt2, leading to OPLL through the WNT signaling pathway. These 2 mechanisms may work independently or cooperatively to cause OPLL.
Figure 7.
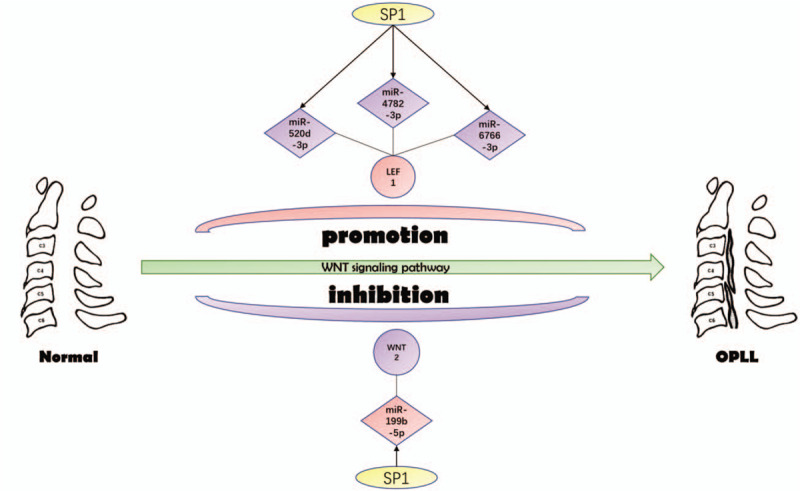
The candidate transcription factor-microRNA-hub gene regulatory network in ossification of the posterior longitudinal ligament.
In this study, although an integrated bioinformatics analysis has been performed and a potential miRNA-mRNA regulatory network has been constructed, there are still some limitations. For example, the samples of gene expression omnibus data set are not large enough, and there is no relevant in vivo and in vitro experimental support that conforms to our conclusion. Although a large amount of evidence can be obtained from the literature review to support our conclusion, further experimental verification is still necessary.
5. Conclusion
In conclusion, we constructed a miRNA-mRNA regulatory network that may be involved in the pathogenesis of OPLL (Fig. 7). Through this study, we will further understand the potential pathogenesis of OPLL, provide targets for future OPLL therapy and improve patients’ quality of life.
Acknowledgments
We are thankful to Dr Xin Li Zhan (Spine and Osteopathy Ward, The First Affiliated Hospital of Guangxi Medical University) for his kindly assistance in all stages of the present study.
Author contributions
Conceptualization: Guoyong Xu, Chong Liu.
Data curation: Guoyong Xu, Tuo Liang, Zhaojie Qin
Formal analysis: Guoyong Xu, Chong Liu.
Funding acquisition: Guoyong Xu, Xinli Zhan.
Investigation: Guoyong Xu, Chong Liu, Chaojie Yu.
Methodology: Guoyong Xu, Chong Liu, Jie Jiang.
Project administration: Guoyong Xu, Chong Liu.
Resources: Guoyong Xu, Zide Zhang.
Software: Guoyong Xu, Tuo Liang, Jie Jiang, Xinli Zhan.
Supervision: Guoyong Xu, Xinli Zhan.
Validation: Guoyong Xu, Chong Liu, Zide Zhang, Jiarui Chen.
Visualization: Guoyong Xu, Tuo Liang, Zhaojie Qin, Chaojie Yu.
Writing – original draft: Guoyong Xu.
Writing – review & editing: Guoyong Xu, Chong Liu.
Footnotes
Abbreviations: DEGs = differentially expressed genes, GO = gene ontology, KEGG = Kyoto Encyclopedia of Genes and Genomes, LEF1 = lymphoid enhancer binding factor 1, miRNA = microRNA, MCODE = molecular complex detection, OPLL = ossification of the posterior longitudinal ligament, PPI = protein–protein interaction, Wnt2 = wingless-type MMTV integration site family member 2.
How to cite this article: Xu GY, Liu C, Liang T, Qin ZJ, Yu CJ, Zhang Z, Jiang J, Chen JR, Zhan XL. Integrated miRNA-mRNA network revealing the key molecular characteristics of ossification of the posterior longitudinal ligament. Medicine. 2020;99:21(e20268).
GX and CL contributed equally to this work and are the co-first authors.
This study was funded by Youth Science Foundation of Guangxi Medical University (Grant/Award Numbers: GXMUYFY 201712), Guangxi Young and Middle aged Teacher's Basic Ability Promoting Project (Grant/Award Number: 2019KY0119), National Natural Science Foundation of China (Grant/Award Numbers: 81560359, 81860393).
The authors have no conflicts of interest to disclose.
The datasets generated during and/or analyzed during the current study are publicly available.
References
- [1].Inamasu J, Guiot BH, Sachs DC. Ossification of the posterior longitudinal ligament: an update on its biology, epidemiology, and natural history. Neurosurgery 2006;58:1027–39. [DOI] [PubMed] [Google Scholar]
- [2].Matsunaga S, Yamaguchi M, Hayashi K, et al. Genetic analysis of ossification of the posterior longitudinal ligament. Spine (Phila Pa 1976) 1999;24:937–8. [DOI] [PubMed] [Google Scholar]
- [3].Saetia K, Cho D, Lee S, Kim DH, Kim SD. Ossification of the posterior longitudinal ligament: a review. Neurosurg Focus. 2011; 30:E1. [DOI] [PubMed] [Google Scholar]
- [4].Iwasaki M, Okuda S, Miyauchi A, et al. Surgical strategy for cervical myelopathy due to ossification of the posterior longitudinal ligament: part 2: advantages of anterior decompression and fusion over laminoplasty. Spine (Phila Pa 1976) 2007;32:654–60. [DOI] [PubMed] [Google Scholar]
- [5].Fargen KM, Cox JB, Hoh DJ. Does ossification of the posterior longitudinal ligament progress after laminoplasty? Radiographic and clinical evidence of ossification of the posterior longitudinal ligament lesion growth and the risk factors for late neurologic deterioration. J Neurosurg Spine 2012;17:512–24. [DOI] [PubMed] [Google Scholar]
- [6].Abiola R, Rubery P, Mesfin A. Ossification of the posterior longitudinal ligament: etiology, diagnosis, and outcomes of nonoperative and operative management. Global Spine J 2016;6:195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kobashi G, Ohta K, Washio M, et al. FokI variant of vitamin D receptor gene and factors related to atherosclerosis associated with ossification of the posterior longitudinal ligament of the spine: a multi-hospital case-control study. Spine (Phila Pa 1976) 2008;33:E553–8. [DOI] [PubMed] [Google Scholar]
- [8].Tsuru M, Ono A, Umeyama H, et al. Ubiquitin-dependent proteolysis of CXCL7 leads to posterior longitudinal ligament ossification. PLoS One 2018;13:e0196204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Mohr AM, Mott JL. Overview of microRNA biology. Semin Liver Dis 2015;35:3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol 2013;14:475–88. [DOI] [PubMed] [Google Scholar]
- [11].Xu C, Zhang H, Gu W, et al. The microRNA-10a/ID3/RUNX2 axis modulates the development of ossification of posterior longitudinal ligament. Sci Rep 2018;8:9225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zhang H, Xu C, Liu Y, et al. MicroRNA-563 promotes the osteogenic differentiation of posterior longitudinal ligament cells by inhibiting SMURF1. Zhonghua Wai Ke Za Zhi 2017;55:203–7. [DOI] [PubMed] [Google Scholar]
- [13].Xu C, Zhang H, Zhou W, et al. MicroRNA-10a, -210, and -563 as circulating biomarkers for ossification of the posterior longitudinal ligament. Spine J 2019;19:735–43. [DOI] [PubMed] [Google Scholar]
- [14].Xu C, Chen Y, Zhang H, et al. Integrated microRNA-mRNA analyses reveal OPLL specific microRNA regulatory network using high-throughput sequencing. Sci Rep 2016;6:21580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Jalal H, Pechlivanoglou P, Krijkamp E, et al. An overview of R in health decision sciences. Med Decis Making 2017;37:735–46. [DOI] [PubMed] [Google Scholar]
- [16].Pathan M, Keerthikumar S, Ang CS, et al. FunRich: an open access standalone functional enrichment and interaction network analysis tool. Proteomics 2015;15:2597–601. [DOI] [PubMed] [Google Scholar]
- [17].Bindea G, Mlecnik B, Hackl H, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009;25:1091–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].von Mering C, Huynen M, Jaeggi D, et al. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res 2003;31:258–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Chin CH, Chen SH, Wu HH, et al. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 2014;8: Suppl 4: S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Fujiwara N, Takeshita K. Updates on ossification of posterior longitudinal ligament. Quality of life (QOL) of patients with OPLL. Clin Calcium 2009;19:1449–56. [PubMed] [Google Scholar]
- [22].Head J, Rymarczuk G, Stricsek G, et al. Ossification of the posterior longitudinal ligament: surgical approaches and associated complications. Neurospine 2019;16:517–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kim DH, Lee CH, Ko YS, et al. The clinical implications and complications of anterior versus posterior surgery for multilevel cervical ossification of the posterior longitudinal ligament, an updated systematic review and meta-analysis. Neurospine 2019;16:530–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lee YT, Tan YJ, Oon CE. Molecular targeted therapy: treating cancer with specificity. Eur J Pharmacol 2018;834:188–96. [DOI] [PubMed] [Google Scholar]
- [25].Mayekar MK, Bivona TG. Current landscape of targeted therapy in lung cancer. Clin Pharmacol Ther 2017;102:757–64. [DOI] [PubMed] [Google Scholar]
- [26].Guan LY, Lu Y. New developments in molecular targeted therapy of ovarian cancer. Discov Med 2018;26:219–29. [PubMed] [Google Scholar]
- [27].Li L, Qiu X, Sun Y, et al. SP1-stimulated miR-545-3p inhibits osteogenesis via targeting LRP5-activated Wnt/beta-catenin signaling. Biochem Biophys Res Commun 2019;517:103–10. [DOI] [PubMed] [Google Scholar]
- [28].Elango J, Robinson J, Zhang J, et al. Collagen peptide upregulates osteoblastogenesis from bone marrow mesenchymal stem cells through MAPK- Runx2. Cells 2019;8: [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Roa LA, Bloemen M, Carels CEL, et al. Retinoic acid disrupts osteogenesis in pre-osteoblasts by down-regulating WNT signaling. Int J Biochem Cell Biol 2019;116:105597. [DOI] [PubMed] [Google Scholar]
- [30].Xie Y, Liu Y, Fan X, et al. MicroRNA-21 promotes progression of breast cancer via inhibition of mitogen-activated protein kinase10 (MAPK10). Biosci Rep 2019;BSR20181000. [DOI] [PubMed] [Google Scholar]
- [31].Gazit A, Yaniv A, Bafico A, et al. Human frizzled 1 interacts with transforming Wnts to transduce a TCF dependent transcriptional response. Oncogene 1999;18:5959–66. [DOI] [PubMed] [Google Scholar]
- [32].Korinek V, Barker N, Willert K, et al. Two members of the Tcf family implicated in Wnt/beta-catenin signaling during embryogenesis in the mouse. Mol Cell Biol 1998;18:1248–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Skokowa J, Klimiankou M, Klimenkova O, et al. Interactions among HCLS1, HAX1, and LEF-1 proteins are essential for G-CSF-triggered granulopoiesis. Nat Med 2012;18:1550–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Park SJ, Bae HS, Park SJC. Osteogenic differentiation and gene expression profile of human dental follicle cells induced by human dental pulp cells. J Mol Histol 2015;46:93–106. [DOI] [PubMed] [Google Scholar]
- [35].Zhou R, Yuan Z, Liu J, et al. Calcitonin gene-related peptide promotes the expression of osteoblastic genes and activates the WNT signal transduction pathway in bone marrow stromal stem cells. Mol Med Rep 2016;13:4689–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hu L, Su P, Yin C, et al. Microtubule actin crosslinking factor 1 promotes osteoblast differentiation by promoting beta-catenin/TCF1/Runx2 signaling axis. J Cell Physiol 2018;233:1574–84. [DOI] [PubMed] [Google Scholar]


