Abstract
Background:
While there is strong evidence that genetic risk factors play an important role in the etiologies of structural birth defects, compared to other diseases, there have been relatively few genome-wide association studies (GWAS) of these conditions. We reviewed the current landscape of GWAS conducted for birth defects, noting novel insights, and future directions.
Methods:
This article reviews the literature with regard to GWAS of structural birth defects. Key defects included in this review include oral clefts, congenital heart defects (CHDs), biliary atresia, pyloric stenosis, hypospadias, craniosynostosis, and clubfoot. Additionally, other issues related to GWAS are considered, including the assessment of polygenic risk scores and issues related to genetic ancestry, as well as utilizing genome-wide single nucleotide polymorphism array data to evaluate gene–environment interactions and Mendelian randomization.
Results:
For some birth defects, including oral clefts and CHDs, several novel susceptibility loci have been identified and replicated through GWAS, including 8q24 for oral clefts, DGKK for hypospadias, and 4p16 for CHDs. Relatively common birth defects for which there are currently no published GWAS include neural tube defects, anotia/microtia, anophthalmia/microphthalmia, gastroschisis, and omphalocele.
Conclusions:
Overall, GWAS have been successful in identifying several novel susceptibility genes and genomic regions for structural birth defects. These findings have provided new insights into the etiologies of these phenotypes. However, GWAS have been underutilized for understanding the genetic etiologies of several birth defects.
Keywords: birth defects, epidemiology, genetics, GWAS
1 |. INTRODUCTION
Approximately one in 33 babies born in the United States each year is affected by a birth defect (Centers for Disease Control and Prevention (CDC), 2008); however, individual types of birth defects are relatively rare. In the United States, birth defects are the leading cause of infant death (Kochanek, Murphy, Xu, & Arias, 2017), contribute substantially to morbidity and disability, and in 2013, the estimated annual cost of birth defect-associated hospitalizations was $22.9 billion (Arth et al., 2017). Substantial stress and disruption of family life accompany this economic burden. Despite their public health significance, the causes of most birth defects remain unknown. There are several lines of evidence indicating that inherited genetic risk factors play an important role in the etiologies of these conditions. Existing evidence from human studies includes increased concordance among monozygotic twins compared to dizygotic twins, among full siblings compared to half siblings, and among first-degree relative compared to second- and third-degree relatives (reviewed in [Webber et al., 2015]). However, candidate gene studies (i.e., where genes are selected based on current understanding of the disease) have largely produced equivocal findings related to genetic susceptibility for structural birth defects (Hobbs et al., 2014; Lupo et al., 2017; Webber et al., 2015).
Advances in technology that permit affordable and reliable genotyping of millions of single nucleotide polymorphisms (SNPs) have provided the opportunity to expand beyond candidate gene association studies to genome-wide studies (GWAS) that do not require prior hypotheses regarding underlying disease biology. Although the first GWAS, for age-related macular degeneration, was not published until 2005, the NHGRI-EBI GWAS Catalog (https://www.ebi.ac.uk/gwas/) currently includes data from over 5,600 GWAS (Buniello et al., 2019). In GWAS, hundreds of thou-sands or millions of genetic variants are tested for association. Because of the large number of tests, the commonly accepted threshold for statistical significance in a GWAS of common variants (i.e., minor allele frequency > 5%) is p < 5 × 10−8 (Fadista, Manning, Florez, & Groop, 2016), and thus very large study populations are required to provide adequate statistical power. Additionally, a tiered approach is typically used in GWAS, where a subset of SNPs from the first stage (i.e., discovery set) is moved to a second stage (i.e., replication set) for confirmation. This process limits the potential for false positives. Meta-analyses performed across studies are commonly used to confirm or refute previously reported associations and can identify novel candidate loci (de Bakker et al., 2008; Willer, Li, & Abecasis, 2010). Meta-analyses also provide opportunities to identify genes with multiple significant SNPs or regions that might not be identified in a single GWAS.
In comparison to other diseases, there have been relatively few GWAS of structural birth defects (Agopian et al., 2014; Birnbaum et al., 2009; Cordell et al., 2013; Mitchell et al., 2015; van der Zanden et al., 2011). This is largely due to the difficulty of assembling the large study populations needed for GWAS (i.e., >1,000 affected individuals), especially as structural birth defects tend to be individually rare. Nonetheless, GWAS have been completed for some of the most common structural birth defects, and these studies have provided new insights regarding the genetic contribution to disease etiology. The purpose of this review is to outline the discoveries made in GWAS of selected structural birth defects and to propose strategies for future genomic studies of these conditions.
2 |. STUDY DESIGNS
The majority of GWAS have used a case–control study design. In this approach, the frequency of variants in putative disease genes is compared between cases (i.e., individuals with the condition) and controls (i.e., individuals without the condition). This study design is particularly useful when studying rare out-comes, such as structural birth defects, and is often used in genetic association studies of other conditions. For instance, the case–control study design was used to identify genes associated with neuroblastoma (a relatively rare pediatric malignancy), which has led to improved therapeutic options for these children (Bosse & Maris, 2016). However, genetic association studies using the case–control design are vulnerable to a type of confounding referred to as population stratification bias, where a false association between a genotype and disease, or the masking of a true genotypic effect, is induced by the existence of subgroups within a population (e.g., different racial or ethnic groups) that have different genotype frequencies and frequencies of disease (Campbell et al., 2005).
Another study design that emerged in the 1990s has proven to be very useful in genetic association studies of structural birth defects: the case-parent trio design (or child-parent trio). Studies of birth defects that employed this design (e.g., [Mitchell, 2008]) were initiated shortly after the advent of this method. The trio is composed of the affected child and his or her biological parents. This design is particularly useful for studies of birth defects and conditions with early disease onset, since parents of children with these conditions are generally available. Several methods for analyzing the data generated in a case-parent trio study have been developed, including the transmission disequilibrium test (Schaid & Sommer, 1993; Spielman, McGinnis, & Ewens, 1993) and approaches using log-linear models (Weinberg, Wilcox, & Lie, 1998). The child–parent trio design has the advantage (as compared to case–control studies) of being immune to population stratification bias when assessing the effects of the inherited genotype. Furthermore, this design can be used to assess maternal genetic effects (i.e., the effect of the maternal genotype on the phenotype of offspring) without incurring additional genotyping expenses (i.e., in the case–control design, evaluation of the maternal and case genotype would require genotyping cases and controls as well as the mothers of these individuals).
3 |. GWAS OF SELECTED BIRTH DEFECTS
3.1 |. Oral clefts
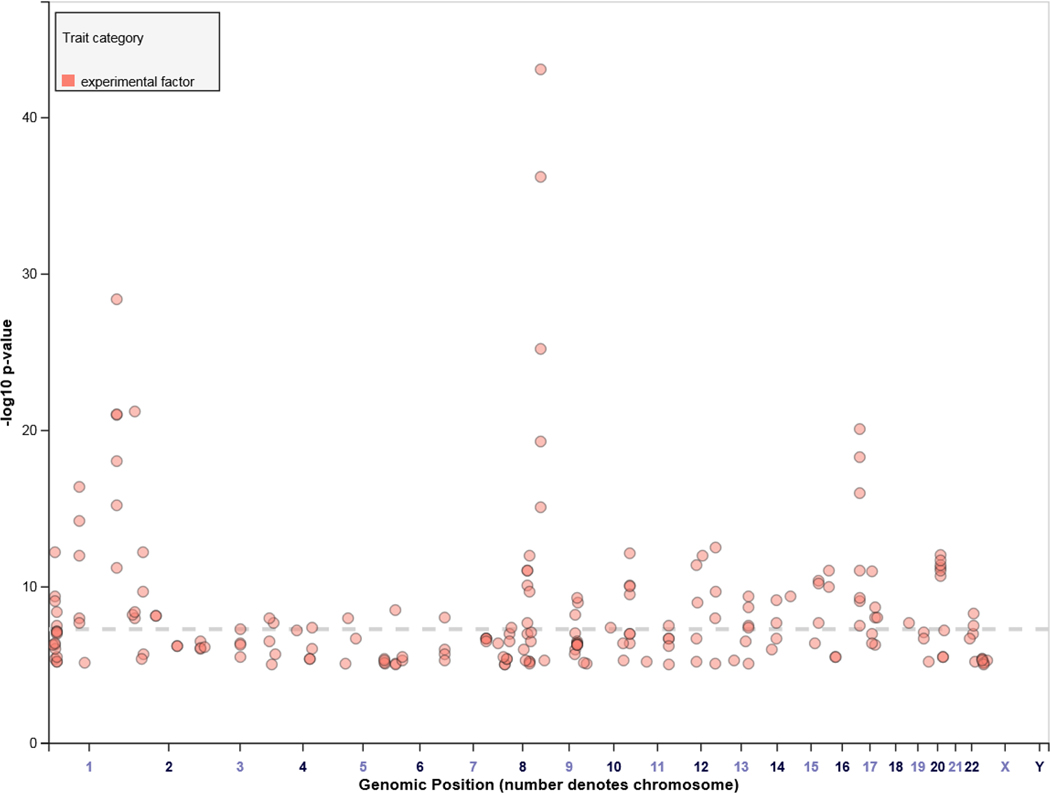
Oral clefts represent a group of structural birth defects where there is a gap or break in normal features of the mouth, most commonly the roof of the mouth (the palate) or the upper lip or both. The most common anatomical forms of orofacial clefts include cleft lip (CL), cleft palate (CP), and cleft lip and palate (CL/P). The overall prevalence of oral clefts is one in 1,000, and because it is one of the most common groups of birth defects, there have been a large number of genetic association studies to map genes underlying susceptibility (Beaty, Marazita, & Leslie, 2016). Not surprisingly, several novel genes and genomic regions underlying the occurrence of oral clefts have been identified through these efforts, including 8q24, IRF6, and NOG. A list of these genes and regions is presented in Table 1. Additionally, oral cleft-associated loci reported in the NHGRI-EBI GWAS Catalog (https://www.ebi.ac.uk/gwas/) are displayed in Figure 1. Of these loci (Figure 1), four (IRF6, 8q24, 17q22, and 10q25.3) appear to account for 20–25% of the estimated heritability to CL/P, a much larger proportion of the estimated heritability attributable to markers identified by GWAS than seen for many other complex disorders (Beaty et al., 2016).
TABLE 1.
A sample of genes and genomic regions identified in GWAS of oral clefts (adapted from Beaty et al., 2016)
| Locus | Gene | Phenotype | p-valuea |
| 1p36.13 | PAX7 | CL/P | 6 × 10−13 |
| 1p36 | GRHL3 | CP | 4 × 10−9 |
| 1q32.2 | IRF6 | CL/P | 9 × 10−22 |
| 2p21 | THADA | CL/P | 9 × 10−8 |
| 2p24 | FAM49A | CL/P | 6 × 10−22 |
| 3p11 | EPHA3 | CL/P | 4 × 10−8 |
| 3q12 | COL8A1/FILIPIL | CL/P | 4 × 10−7 |
| 8q24 | Gene Desert | CL/P | 8 × 10−44 |
| 10q25.3 | VAX1 | CL/P | 7 × 10−13 |
| 13q31.2 | SPRY2 | CLP | 8 × 10−6 |
| 15q22 | TPM1 | CL/P | 4 × 10−7 |
| 15q24 | ARID3B | CL/P | 2 × 10−8 |
| 16p13 | CREBBP | CL/P | 9 × 10−12 |
| 17p13.1 | NTN1 | CL/P | 8 × 10−21 |
| 17q22 | NOG | CL/P | 9 × 10−9 |
| 20q12 | MAFB | CL/P | 9 × 10−13 |
Abbreviations: CL/P, cleft lip with or without cleft palate; CLP, cleft lip and palate; CP, cleft palate.
Lowest p-value for representative SNP in gene or genomic region as reported in the GWAS Catalog (https://www.ebi.ac.uk/gwas/).
FIGURE 1.
Loci identified in genome-wide association studies (GWAS) of oral clefts (obtained from the NHGRI-EBI GWAS Catalog (https://www.ebi.ac.uk/gwas/)
GWAS have confirmed the genetic contribution to the etiology of oral clefts. However, it has also demonstrated that these defects can result from variation in multiple genes. Therefore, compared to other birth defects where less is known in terms of genetic etiologies, the next challenge will be to develop strategies for characterizing the function of these genes in relation to oral cleft development and to trans-late this information into prevention strategies.
3.2 |. Congenital heart defects
With a birth prevalence of ~1%, congenital heart defects (CHDs) are the most common group of birth defects. Consequently, it is not surprising that CHDs are one of the few birth defects for which there have been several GWAS (Table 2). These studies have identified variants in several genes and regions of the genome with genome-wide significant (p < 5×10−8) or suggestive (commonly defined as 5 × 10−8 < p < 1 × 10−5 [Aminkeng et al., 2015; Kraja et al., 2017; Sung et al., 2018]) evidence of association with CHDs as a broad group, narrower subtypes of CHDs (e.g., septal or conotruncal defects), and even individual CHD phenotypes (e.g., tetralogy of Fallot). The top hits reported from these studies do not overlap, which—given the stringent threshold for declaring significance—is not uncommon for GWAS. Several of the implicated variants have, however, showed evidence of association in replication samples evaluated as part of the original GWAS (Cordell et al., 2013; Cordell, Bentham, et al., 2013; Lin et al., 2015), and a few have been independently replicated. In particular, the association of atrial septal defects (ASDs) and a locus at chromosome 4p16 (Cordell, Bentham, et al., 2013), has been independently replicated in two studies of ASDs in Chinese populations (Zhao et al., 2014; Zhao, Li, et al., 2015).
TABLE 2.
Summary of findings from GWAS of congenital heart defects
| Reference | Phenotype | Population | Design | No. of cases | Chromosome/Region | Genes | SNP | p-value | OR (95% CI) |
|---|---|---|---|---|---|---|---|---|---|
| Cordell, Bentham, et al. (2013) | TOF | Europe, Australia, and Canada | CC | 1,633 | 12q24 | 1.6 Mb, evolutionarily conserved, haplotype including 15 genes; strongest candidate, PTPN11 | rs11065987 | 7.7 × 10−11 | 1.3 (1.1–1.4) |
| 13q32 | GPC5 (intron) | rs7982677 | 3.0 × 10−11 | 1.3 (1.2–1.5) | |||||
| Cordell, Topf, et al. (2013) | osASD | Europe, Australia, and Canada | CC | 791 | 4p16 | Intergenic, between STX18 and MSX1 | rs870142 | 2.6 × 10−10 | 1.5 (NR) |
| Hu et al. (2013) | ASD and/or VSD | Han Chinese | CC | 3,097 | 1p12 | Intergenic, between SPAG17 and TBX15 | rs2474937 | 8.4 × 10−10 | 1.4 (1.3–1.6) |
| 4q31.1 | MAML3 | rs1531070 | 5.0 × 10−12 | 1.4 (1.3–1.5) | |||||
| Lin et al. (2015) | CHD | Han Chinese | CC | 6,053 | 4q31.22 | upstream of EDNRA | rs1400558 | 1.6 × 10−9 | 1.2 (1.1–1.2) |
| 9p24.2 | close to SMARCA2 | rs7863990 | 3.7 × 10−14 | 1.3 (1.2–1.4) | |||||
| 12q24.13 | upstream of TBX3 and TBX5 | rs2433752 | 1.0 × 10−10 | 0.8 (0.8–0.9) | |||||
| 20q12 | PTPRT | rs490514 | 1.2 × 10−13 | 1.2 (1.1–1.2) | |||||
| Agopian et al. (2014) | CTD | United States | CPT | 1,108 | 22q13.1 | KCNJ4 (intron) | rs2267386 | 1.4 × 10−6 | 3.1 (1.9–5.1) |
| 20p12.3 | Intergenic, between BMP2 and FERMT1 | rs6140038 | 1.0 × 10−6 | 5.2 (2.5–11.0) | |||||
| Mitchell et al. (2015) | LSL | United States | CPT | 601 | 16q24.2 | Intergenic, between FOXL1 and c16orf95 | rs8061121 | 4.0 × 10−9 | 2.6 (1.9–3.7) |
| 3p14.2 | SYNPR (intron) | rs1975649 | 3.4 × 10−7 | 1.6 (1.4–2.0) | |||||
| Agopian et al. (2017) | CTD | United States | MA (CPT + CC) | 1,431 | 5p12 | LOC648987 (noncoding) | rs6886261 | 1.7 × 10−7 | 1.8 (1.4–2.2) |
| LSL | MA (CPT) | 509 | 2q11.2 | MGAT4A (intron) | rs11894932 | 1.5 × 10−7 | 0.4 (0.3–0.6) | ||
| 6p24.3 | Intergenic, between OFCC1 and HULC | rs72820264 | 2.1 × 10−8 | 2.0 (1.6–2.6) | |||||
| CTD + LSL | MA (CPT + CC) | 1,940 | 2q22.1 | LRP1B (intron) | rs11895588 | 7.3 × 10−7 | 0.5 (0.4–0.7) | ||
| 6p24.3 | Intergenic, between OFCC1 and HULC | rs56409046 | 2.7 × 10−7 | 1.3 (1.2–1.5) | |||||
| 14q21.1 | Intergenic, between SLC35B3 and OFCC1 | rs1176869 | 9.2 × 10−7 | 1.3 (1.2–1.5) | |||||
| Hanchard et al. (2016) | LSL | United States, Austria | CC | 778 | 20q11.22 | 200 kb region, including MYH7B, miR-499A, miR-499B, GSS, and TRPC4AP | rs6088703 | 3.0 × 10−9 | 1.6 (NR) |
| Bjornsson et al. (2018) | CoA | Iceland | CC | 120 | 14q11 | MYH6 | p.Arg721Trp | 5.0 × 10−22 | 44.2 (20.5–95.5) |
Abbreviations: ASD, atrial septal defect; CHD, congenital heart defect (broad definition); coA, coarctation of the aorta; CTD, conotruncal heart defect; LSL, left-sided lesion, osASD, ostium secundum atrial septal defect; TOF, tetralogy of Fallot; VSD, ventricular septal defect; CC, case–control study; CPT, case-parent trio study; MA, meta-analysis.
Attempts to replicate the associations uncovered by GWAS have generally been conducted using data from cases with the same CHD phenotypes that were included in the original GWAS. However, there have been attempts to determine whether associations detected in one phenotypic group (e.g., ASDs) are also observed in other CHD subtypes. These studies indicate that some associations are phenotype-specific, whereas others apply to a range of different phenotypes. For example, the association reported in the 4p16 region appears to be specific to ASDs (Cordell, Bentham, et al., 2013; Zhao et al., 2014), several of the associations reported by Lin and colleagues seem to apply to a broad range of CHD phenotypes (Lin et al., 2015), and the rare variant identified as being strongly associated with coarctation of the aorta is associated with other CHDs (e.g., bicuspid aortic valve, p = 7.3 × 10−8), as well as other cardiovascular disease phenotypes (e.g., atrial fibrillation, p = 1.1 × 10−14; Bjornsson et al., 2018).
In addition to the traditional GWAS of inherited variants (summarized above and in Table 2), several additional GWAS of CHDs have been conducted. These include a study of inherited compound heterozygous genotypes (Jiang et al., 2018), and a study of inherited genotypes and neurodevelopmental outcomes following cardiac surgery in infancy (Kim et al., 2012). In addition, three GWAS have focused on the maternal genotype (Agopian et al., 2014; Agopian et al., 2017; Mitchell et al., 2015). There have also been two GWAS conducted in syndromic populations: Down syndrome (Ramachandran et al., 2015) and the 22q11 deletion syndrome (Guo et al., 2017). Each of these studies focused on the inherited genotype and identified genes and genomic regions with at least suggestive evidence of association. For example, a genome-wide significant association between tetralogy of Fallot and an intronic variant in GPR98 (p = 3.0 × 10−8) was identified in the study of individuals with the 22q11 deletion syndrome. However, given the relatively unique approaches used in each of these studies, there has been little to no internal or external replication of these findings.
In summary, GWAS have provided evidence that common genetic variants are associated with CHDs and identified new candidate CHD genes and genomic regions. These studies provide support for a genetic model of CHDs that includes genes that influence specific CHD phenotypes (e.g., ASD) as well as genes that are associated with a broader spectrum of CHD phenotypes. In addition, GWAS conducted in syndromic populations indicate that common genetic variants may also contribute to variability in CHD phenotypes observed across affected individuals. As has been observed for other complex traits (e.g., autism), additional insights regarding the genetic basis of CHDs are expected to be gained by further analyses of the existing GWAS datasets (e.g., meta-analyses, analyses of specific CHD phenotypes) and through the evaluation of new GWAS samples.
3.3 |. Other defects
3.3.1 |. Biliary atresia
Biliary atresia is a birth defect characterized by inflammation and obliteration of the extrahepatic and intrahepatic bile ducts (Lee, Lewis, Schoen, Brand, & Ricketts, 2001; Sanchez-Valle et al., 2017; Sundaram, Mack, Feldman, & Sokol, 2017). While this condition is relatively rare with an estimated birth prevalence of 0.7–0.9 per 10,000 births (Caton, Druschel, & McNutt, 2004; Yoon, Bresee, Olney, James, & Khoury, 1997), biliary atresia is the most common cause of extrahepatic obstructive jaundice in the newborn and the most frequent indication for liver transplantation in children (Sundaram et al., 2017; Yoon et al., 1997). Four independent GWAS of biliary atresia among relatively small cohorts of patients (35–499 patients) have identified four novel biliary atresia susceptibility loci (Table 3): (a) an intergenic locus on 10q24.2 between ADD3 and XPNPEP1 (Garcia-Barcelo et al., 2010); (b) a deletion in 2q37.3 that included AGXT and GPC1 (Cui et al., 2013); (c) ARF6 (Ningappa et al., 2015); and (d) EFEMP1 (Chen et al., 2018).
TABLE 3.
Summary of findings from GWAS of noncardiac defects
| Reference | Population | Design | No. of cases | Chromosome/Region | Genes | SNP | p-value | OR (95% CI) |
|---|---|---|---|---|---|---|---|---|
| Biliary atresia | ||||||||
| Garcia-Barcelo et al. (2010) | Han Chinese | MA (CC) | 324 | 10q25.1 | Between XPNPEP1 and ADD3 | rs17095355 | 6.9 × 10−9 | 1.8 (1.4–2.3) |
| Cui et al. (2013) | CHOP patients | CC | 61 | 2q27.3 | GPC1 | CNV | 4.4 × 10−10 | NR |
| Ningappa et al. (2015) | Caucasian | CC | 63 | 14q21.3 | 3′ flanking enhancer region for ARF6 | rs3126184 | 5.9 × 10−7 | 2.7 (NR) |
| rs10140366 | 5.6 × 10−7 | 2.7 (NR) | ||||||
| Chen et al. (2018) | European-American | MA (CC) | 499 | 2p16.1 | EFEMP1 | rs10865291 | 3.7 × 10−8 | NR |
| rs6761893 | 3.4 × 10−8 | NR | ||||||
| rs727878 | 4.3 × 10−8 | NR | ||||||
| Pyloric stenosis | ||||||||
| Feenstra et al. (2012) | Denmark | MA (CC) | 1,700 | 3p25.1 | 150 kb upstream of MBNL1 | rs11712066 | 1.5 × 10−17 | 1.6 (1.4–1.8) |
| 1,693 | 3p25.2 | Intergenic,1.3 Mb downstream of MBNL1 | rs573872 | 4.3 × 10−12 | 1.4 (1.3–1.6) | |||
| 1,691 | 5q35.2 | 64 kb downstream of NKX2-5 | rs29784 | 1.1 × 10−15 | 1.4 (1.3–1.6) | |||
| Feenstra et al. (2013) | Denmark, Sweden, United States | MA (CC) | 2,656 | 11q23.3 | 301 bases downstream of APOA1 | rs12721025 | 1.9 × 10−10 | 1.6 (1.4–1.8) |
| Fadista et al. (2019) | Denmark, Sweden, United States | MA (CC) | 3,179 | 2p21 | EML4 | rs6736913 | 3.0 × 10−15 | 2.3 (2.1–2.5) |
| 3,178 | 9q22.32 | 1 kb downstream of BARX1 | rs1933683 | 3.1 × 10−9 | 1.3 (1.2–1.4) | |||
| Hypospadias | ||||||||
| van der Zanden et al. (2010) | Netherlands, Sweden | CC | 835 | X | DGKK | rs1934179 | 2.5 × 10−11 | 2.5 (1.9–3.2) |
| DGKK | rs7063116 | 2.9 × 10−9 | 2.3 (1.7–3.0) | |||||
| Geller et al. (2014) | European | CC | 2,978 | 2 | PKDCC (1.6 kb) | rs988958 | 1.6 × 10−10 | 1.3 (NR) |
| Craniosynostosis, sagittal | ||||||||
| Justice et al. (2012) | European, non-Hispanic White | MA (CPT + CC) | 302 | 20p12.3 | 345 kb downstream of BMP2 | rs1884302 | 1.1 × 10−39 | 4.4 (3.5–5.5) |
| 302 | 7p14.3 | 167 kb region of BBS9 | rs10262453 | 5.6 × 10−20 | 0.2 (0.2–0.3) |
Abbreviations: CC, case–control study; CPT, case-parent trio study; MA, meta-analysis.
3.3.2 |. Pyloric stenosis
Pyloric stenosis is characterized by a narrowing of the pyloris, a sphincter muscle connecting the stomach and the duodenum (Ranells, Carver, & Kirby, 2011). It regulates the movement of food into the small intestine. The incidence of pyloric stenosis varies between two and five per 1,000 live births and there is a four- to five-fold higher risk in males than females (Peters, Oomen, Bakx, & Benninga, 2014). Three GWAS have been conducted using surgery-confirmed cases and controls from Denmark in the discovery phase (Fadista et al., 2019; Feenstra et al., 2012; Feenstra et al., 2013). Replication samples were drawn solely from the same population as the discovery phase in the earliest study (Feenstra et al., 2012) while the 2013 study (Feenstra et al., 2013) also included replication samples from the United States (mostly non-Hispanic white) and Sweden. In the most recent study, Fadista et al. (2019)) conducted a genome-wide meta-analysis, combining their previous GWAS cohort with an additional 427 surgery-confirmed cases and 2,031 controls, all of Danish descent, in their discovery phase. Genome-wide significant results were replicated in populations of European descent followed by confirmation in a Hispanic population. The meta-analysis confirmed the genome-wide significant SNPs identified in the two earlier GWAS (SNPs located close to MBNL1 and NKX2-5 (Feenstra et al., 2012), and APOA1 (Feenstra et al., 2013)) and reported two novel loci that included EML4, MTA3, and BARX1 (Table 3).
MBNL1 is a member of the muscleblind protein family that is involved in regulation of alternative splicing and NKX2–5 is crucial for the formation of pyloric sphincter muscle tissue (Ho et al., 2004; Self, Geng, & Oliver, 2009). The three genome-wide significant SNPs located close to MBNL1 and NKX2–5 collectively explain 1.8% of the variance in liability to pyloric stenosis (Feenstra et al., 2012). Due to the excess risk in males, heterogeneity of effects between the sexes was assessed for the three SNPs with no evidence of a difference between males and females. However, a variant on chromosome 19p13.2 had a strong effect in males with no effect in females, warranting further investigation. Apolipoprotein A-1 is the major protein component of HDL cholesterol in plasma (Davidson & Thompson, 2007) and pyloric stenosis is a clinical feature in patients with Smith–Lemli–Opitz syndrome, which is characterized by low cholesterol levels. The relationship between lipid levels and pyloric stenosis warrants further study. EML4, MTA3, and BARX1 are expressed in the fetal and adult stomach (Kim, Woo, Kanellopoulou, & Shivdasani, 2011; The Human Protein Atlas, 2019), establishing them as strong candidate genes for pyloric stenosis.
3.3.3 |. Hypospadias
Hypospadias is one of the most common genitourinary malformations and occurs when the urethral opening develops ventrally at varying degrees of severity rather than at the distal end of the glans penis (Carmichael, Shaw, & Lammer, 2012). The first GWAS of hypospadias identified two loci in or near DGKK on the X chromosome that were associated with risk (Table 3) (van der Zanden et al., 2011). Notably, these associations were relatively strong (odds ratios [ORs] >2.0). Additionally, there is emerging evidence that these variants have subtype specificity. Specifically, genetic variation in DGKK appears to be limited to mild forms of hypospadias as compared to moderate or severe phenotypes (Richard et al., 2019). This supports hypotheses related to etiologic heterogeneity of hypospadias by classifications of severity.
A subsequent GWAS by Geller et al. (2014)) not only confirmed the role of DGKK (represented by another locus, rs4554617) on hypospadias risk, but also identified 21 additional SNPs (17 of which reached genome-wide level significance) associated with hypospadias. When considering these 22 SNPs together, they jointly explain 9.4% of the variance in liability to hypospadias.
3.3.4 |. Craniosynostosis
Craniosynostosis (CS), the premature fusion of one or several sutures of the skull, is a common birth defect that affects ~1 in 2,250 births (Boulet, Rasmussen, & Honein, 2008; Lajeunie, Le Merrer, Bonaiti-Pellie, Marchac, & Renier, 1995). The defect presents as nonsyndromic (i.e., without unrelated, major birth defects or developmental delay) or as a component of more than 100 genetic syndromes (Boulet et al., 2008; Kimonis, Gold, Hoffman, Panchal, & Boyadjiev, 2007). These syndromes account for only 15% of all cases, leaving the etiology undetermined for most individuals with CS. Major sutures involved in CS include sagittal (40–58%), coronal (20–29%), metopic (4–10%), and lambdoid (2–4%; Kimonis et al., 2007).
To date, the only GWAS completed for CS included cases with sagittal nonsyndromic CS and their parents (Justice et al., 2012). An international consortium identified candidate loci on chromosome 7, within BBS9, and on chromosome 20, near BMP2, and successfully replicated these findings in an independent, population-based case–control sample of newborn residual blood spots. Notably, the loci had opposite effects on risk; the locus on chromosome 7 showed a strong negative association (OR = 0.2; 95% CI: 0.2–0.3) and the locus on chromosome 20 showed a strong positive association (OR = 4.4; 95% CI: 3.5–5.5) with CS risk. In addition, genotyping of the common variant located near BMP2 in CS patients with rare heterozygous SMAD6 mutations has provided the first evidence for a 2-locus disease mechanism (Timberlake et al., 2016). SMAD6 is an inhibitor of BMP-induced osteoblast differentiation and this 2-locus model was estimated to account for approximately 3.5% of all CS cases.
3.3.5 |. Clubfoot
Clubfoot is a birth defect of the lower limb with a birth prevalence of ~1 in 1,000. An important role for genetic factors in clubfoot etiology is supported by high concordance rates in identical twins compared to fraternal twins (33% vs. 3%), and an increased risk to first-degree relatives compared to the general population. We identified one published GWAS of isolated clubfoot, which included 766 cases (discovery + replication) of European ancestry (Zhang et al., 2014). In this assessment, no SNP reached genome-wide level significance. The strongest evidence for genetic association was found with an intergenic SNP on chromosome 12q24.31 between NCOR2 and ZNF664 (rs7969148, combined OR = 0.6, p = 1.9 × 10−7).
Overall, GWAS have demonstrated that common genes are involved in the etiologies of many birth defects and have identified previously unrecognized functional components of the human genome. In fact, these findings may lead to new biological insights and prevention strategies for these conditions. Further analyses of existing GWAS datasets (e.g., meta-analyses), as well as GWAS in new datasets, will continue to mitigate knowledge gaps in birth defects research. Due to the demonstrated genetic and clinical heterogeneity of these birth defects, analyses of specific phenotypes (e.g., individual types of orofacial clefts, CHDs, CS, or hypospadias; isolated defects vs. multiple defects) would be informative.
4 |. THE VALUE OF GWAS IN THE AGE OF SEQUENCING
There is a debate about the utility of GWAS for identifying the role of inherited genetic variation on disease susceptibility utilizing SNP array data in the age of sequencing (Tam et al., 2019). Whole-exome and whole-genome sequencing (WES and WGS, respectively) studies have moved to the forefront of genomic research in recent years. This is largely due to advances in technology that have led to reductions in the cost and time required to sequence DNA. However, there are still important advantages to GWAS in relation to WES/WGS. In terms of WES, the focus is strictly on genetic variants that alter protein sequences, which only constitute 1% of the human genome. While this could be important in clinical settings for highly penetrant pathogenic variants (Yang et al., 2013), these account only for a small proportion of cases. In fact, most replicated SNPs from GWAS are in noncoding regions of the genome and would, thus, likely have been excluded from analyses of WES data. While interpreting findings from noncoding regions is challenging, it has led to new insights into the underlying biology of birth defects (e.g., 8q and oral clefts).
While WGS provides more complete coverage of the genome (like GWAS), there are still advantages to performing GWAS utilizing SNP array data. First, the costs of SNP arrays are lower compared to WGS (Tam et al., 2019). In fact, WGS remains the most costly of these three options (i.e., SNP arrays, WES, and WGS). Therefore, WGS in large sample sizes is often cost prohibitive. Second, the technology underlying SNP arrays is highly accurate and more mature compared to WGS. Third, the analytic pipelines for GWAS of SNP arrays are well-established and require less computational complexity. While some of these factors may be mitigated in the future (e.g., cost), there is still a clear rationale for GWAS using SNP arrays compared to WGS (Tam et al., 2019). Further, as illustrated in the study of coarctation of the aorta (Bjornsson et al., 2018), sequencing data can be used to impute rare variants into array-based data, thereby allowing for the evaluation of both common and rare inherited variants without the need to sequence all study participants.
5 |. OTHER CONSIDERATIONS AND FUTURE DIRECTIONS
5.1 |. Defects in need of GWAS
As previously noted, while GWAS have been successful in identifying genetic susceptibility loci associated with other complex traits (Visscher et al., 2017), there have been relatively few GWAS of birth defects (Agopian et al., 2014; Birnbaum et al., 2009; Cordell, Bentham, et al., 2013; Mitchell et al., 2015; van der Zanden et al., 2011). Those studies that have been conducted are limited by small sample sizes and single ancestry populations, leaving us with much to gain from this approach for future studies of these conditions.
Most birth defects have unknown causes (Nelson & Holmes, 1989); however, there is strong evidence that genetic factors contribute to their etiologies. To date, much of what is known about the genetics of birth defects includes effects of high-risk alleles that cause rare multiple malformation syndromes (Belmont, Mohapatra, Towbin, & Ware, 2004; de Munnik et al., 2015; Lewin, Glass, & Power, 2004; Maslen, 2004; Mori & Bruneau, 2004; Yates, Turner, Firth, Berg, & Pilz, 2017). Such alleles occur at very low frequency in the general population and explain relatively little of the population burden of birth defects. Common modest-risk alleles may explain a much greater proportion of overall cases (Reich & Lander, 2001). However, aside from structural birth defects outlined above, to date, most structural birth defects have not been included as part of GWAS. The main barrier to the GWAS approach for relatively rare individual birth defects is the need for large numbers of specimens. Collaborations between researchers with access to specimens and environmental data, such as the National Birth Defects Prevention Study (NBDPS) (Reefhuis et al., 2015; Yoon et al., 2001), will provide opportunities to discover novel gene-birth defect associations, and environmental factors with which they may interact. Such discoveries will improve the accuracy of risk assessment information, provide information about the biological mechanisms underlying birth defects, and identify potential therapeutic targets. Notably, GWAS is likely to be more useful for birth defects that are relatively common and for which there is evidence of multifactorial etiologies (Agopian, Eastcott, & Mitchell, 2012; Jenkins et al., 2019). A short list of birth defects with no GWAS to date, and for which this approach would be beneficial, include but are not limited to:
congenital anomalies of the nervous system, including spina bifida, encephalocele, anencephaly, and hydrocephalus;
congenital anomalies of the eye, including anophthalmia/microphthalmia and anterior chamber segment defects;
congenital anomalies of the ear, including anotia/microtia; (d) gastroschisis; and (e) omphalocele. Rare birth defects, and birth defects where de novo mutations are likely to play a role, would likely be better candidates for WES/WGS studies (Veltman & Brunner, 2012).
5.2 |. Race/ethnicity in GWAS
Over 80% of subjects included in all published GWAS have been of European ancestry (Bustamante, Burchard, & De la Vega, 2011; Rosenberg et al., 2010). In part, this is to limit the impact of population stratification bias. However, this exclusive focus on a few selected ancestry groups raises a number of critical questions. For example, are findings from studies dominated by those of European ancestry transferable to other populations (Ioannidis, 2009; Ntzani, Liberopoulos, Manolio, & Ioannidis, 2012)? Can disease biology be different among populations and thus characterized by distinct risk factors (Torgerson et al., 2011)? What is the contribution of ancestry-related genetic variation to ethnic differences in birth prevalence? These issues are of particular relevance to structural birth defects, where the prevalence varies substantially by race/ethnicity. While some GWAS of structural birth defects have not been limited to those of European ancestry (Tables 1–3), it is incumbent on genetic epidemiologists to conduct GWAS of structural birth defects among multi-ethnic populations.
5.3 |. Gene–environment interactions
Evidence of gene–environment interactions in birth defect etiologies has been observed for many years (e.g., [Christensen et al., 1999; Etheredge et al., 2012; Lacasana et al., 2012; Padula et al., 2018; Shaw et al., 2005; Wu et al., 2010]). Assessing these interactions is critical to uncovering genetic and/or environmental contributions that might otherwise be undetectable (i.e., genetic variants related to the birth defect might only be expressed in the subgroup of the population that is exposed to a specific environmental factor).
Although many gene–environment interaction studies of birth defects have included small study populations and modest numbers of variants, the approach has been expanded recently using GWAS data to assess common exposures among pregnant women (e.g., maternal alcohol consumption, maternal active and passive smoking, and multivitamin supplement use) (Beaty et al., 2011; Haaland et al., 2018; Wu et al., 2014). In these studies, both increased and reduced orofacial cleft risks were observed between the exposures and genetic variants identified in the GWAS (i.e., association with the genetic variant differed as a function of the environmental exposure). The transition from assessment of candidate genes to genome wide investigations should continue to increase in the foreseeable future due to decreasing costs of GWAS.
Expanded approaches that include analyses to assess gene–environment interactions using GWAS data are referred to as gene–environment wide interaction studies (GEWIS; Khoury & Wacholder, 2009) or genome-wide environmental interactions (GWEI; Aschard et al., 2012). Methods that improve limitations inherent in these early designs have been developed and include approaches that can account for the complex correlations between individuals in admixed populations (Chen et al., 2019), assess the impact of exposure misclassification (Boonstra et al., 2016), evaluate interactions using case subjects only (vs. the traditional method using case and control subjects) (Cornelis et al., 2012; Helbig et al., 2012), and assess interactions using exposed subjects only (Zhao, Fan, et al., 2015).
Of note, these approaches are limited by the exposure data available and novel approaches are needed to measure and estimate exposure. However, it is important to conduct these studies or risk missing a key component to understanding the biological mechanisms causing birth defects, improving the accuracy of risk assessment, and identifying potential targets for prevention.
5.4 |. Polygenic risk scores
Polygenic risk scores (PRS) are quantitative measures of risk summed across multiple risk alleles identified through GWAS. More specifically, the goal of PRS is to utilize aggregated genetic information, often obtained from GWAS, to better estimate the likelihood of a specific outcome (Gibson, 2019; Sugrue & Desikan, 2019). PRS have been generated for several conditions, including coronary artery disease, atrial fibrillation, Crohn’s disease, and Type 2 diabetes, in each case identifying a threshold above which a small percentage of the population has disease risk at least threefold higher than the general population (Khera et al., 2018). In one study of breast cancer risk, a PRS combined with conventional risk factors was able to identify 16% of the population who could benefit from earlier screening and 32% who could delay screening (Maas et al., 2016). While there is promise for the use of PRS in identifying at-risk populations, these tools have not become part of routine clinical care or prevention strategies (Sugrue & Desikan, 2019). However, there have been no large-scale studies to generate PRS for birth defects, much less evaluate the clinical utility of these models.
5.5 |. Gene-level GWAS
The genotype data (e.g., array and imputed) and summary statistics (e.g., association p-values, estimates of relative risk) generated as part of SNP-level (i.e., variant by variant) GWAS can be used for genome-wide studies conducted at the level of the gene. Gene-level analyses are therefore extremely cost-effective and also have the advantage of a reduced multiple correction burden relative to SNP-level GWAS (i.e., correction for approximately 20,000 genes vs. millions of SNPs). Thus, gene-level GWAS provide a useful complement to SNP-level GWAS, providing the opportunity for additional gene discovery (Tam et al., 2019).
Although statistical methods for gene-level GWAS are not as well established as the methods for SNP-level analyses, several approaches have been described and can be implemented using publically available programs (e.g., (de Leeuw, Mooij, Heskes, & Posthuma, 2015); (Wang et al., 2017)). Nonetheless, despite the availability of both data and methods, there has been only one published gene-based GWAS for birth defects (Sewda et al., 2019). This study identified eight candidate genes for conotruncal heart defects and provided additional evidence that genes involved in chromatin-modification and in ribonucleic acid splicing are associated with CHDs.
5.6 |. Mendelian randomization
In Mendelian randomization, investigators use genetic variants to determine whether an observational association between a nongenetic risk factor and an outcome is consistent with a causal effect (Davies, Holmes, & Davey Smith, 2018; Ross et al., 2015). The underlying assumption of Mendelian randomization relies on the natural, random assortment of genetic variants. More specifically, individuals are naturally “assigned” at birth a genetic variant that is associated with certain traits (e.g., elevated body mass index [BMI]). When determining the role of BMI on the risk of a disease, it is often difficult to disentangle the confounding effects of other variables. However, genetic variants associated with BMI are not likely to be associated with the confounders in question. Therefore, by leveraging information from GWAS of BMI or other cardiometabolic traits, investigators can evaluate the impact of these factors on the risk of a given disease. As GWAS of several traits and conditions (e.g., smoking, alcohol intake, infection) continue to grow, this information can be used to more fully characterize associations between nongenetic factors and structural birth defects.
6 |. RESEARCH PRIORITIES AND CONCLUSIONS AND FUTURE DIRECTIONS
Overall, GWAS have been successful in identifying novel susceptibility loci for common structural birth defects. These findings have provided new insights into the etiologies of these phenotypes. In spite of these successes, GWAS have been under-utilized and as a result, understanding of the genetic contribution to birth defects etiologies lags behind that of other conditions. As GWAS continue to evolve to include rare and coding variants, variants optimized for multi-ethnic populations, as well as improved capability to interrogate copy number variants (CNVs), the application of this approach to all structural birth defects will continue to improve. Future assessments could expand on these findings to better ascertain the mechanisms (e.g., evaluation of phenotypic heterogeneity) underlying these associations, lever-age existing GWAS data for additional studies (e.g., gene-level analyses, Mendelian randomization), expand GWAS to individuals of non-European ancestry, and follow up with functional studies on genes that have been identified.
Acknowledgments
Funding information
Eunice Kennedy Shriver National Institute of Child Health and Human Development, Grant/Award Numbers: P01HD070454, R01HD093660, The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention. No replication was needed since this is a review of previously published analyses.
REFERENCES
- Agopian AJ, Eastcott LM, & Mitchell LE. (2012). Age of onset and effect size in genome-wide association studies. Birth Defects Research. Part A, Clinical and Molecular Teratology, 94(11), 908–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agopian AJ, Goldmuntz E, Hakonarson H, Sewda A, Taylor D, Mitchell LE, & Pediatric Cardiac Genomics C. (2017). Genome-wide association studies and meta-analyses for congenital heart defects. Circulation. Cardiovascular Genetics, 10(3), e001449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agopian AJ, Mitchell LE, Glessner J, Bhalla AD, Sewda A, Hakonarson H, & Goldmuntz E. (2014). Genome-wide association study of maternal and inherited loci for conotruncal heart defects. PLoS One, 9(5), e96057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aminkeng F, Bhavsar AP, Visscher H, Rassekh SR, Li Y, Lee JW, … The Canadian Pharmacogenomics Network for Drug Safety Consortium. (2015). A coding variant in RARG confers susceptibility to anthracycline-induced cardiotoxicity in childhood cancer. Nature Genetics, 47(9), 1079–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arth AC, Tinker SC, Simeone RM, Ailes EC, Cragan JD, & Grosse SD. (2017). Inpatient hospitalization costs associated with birth defects among persons of all ages—United States, 2013. MMWR. Morbidity and Mortality Weekly Report, 66(2), 41–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschard H, Lutz S, Maus B, Duell EJ, Fingerlin TE, Chatterjee N, … Van Steen K. (2012). Challenges and opportunities in genome-wide environmental interaction (GWEI) studies. Human Genetics, 131(10), 1591–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaty TH, Marazita ML, & Leslie EJ. (2016). Genetic factors influencing risk to orofacial clefts: today’s challenges and tomorrow’s opportunities. F1000Res, 5, 2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaty TH, Ruczinski I, Murray JC, Marazita ML, Munger RG, Hetmanski JB, … Scott AF. (2011). Evidence for gene-environment interaction in a genome wide study of nonsyndromic cleft palate. Genetic Epidemiology, 35(6), 469–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont JW, Mohapatra B, Towbin JA, & Ware SM. (2004). Molecular genetics of heterotaxy syndromes. Current Opinion in Cardiology, 19(3), 216–220. [DOI] [PubMed] [Google Scholar]
- Birnbaum S, Ludwig KU, Reutter H, Herms S, Steffens M, Rubini M, … Mangold E. (2009). Key susceptibility locus for nonsyndromic cleft lip with or without cleft palate on chromosome 8q24. Nature Genetics, 41(4), 473–477. [DOI] [PubMed] [Google Scholar]
- Bjornsson T, Thorolfsdottir RB, Sveinbjornsson G, Sulem P, Norddahl GL, Helgadottir A, … Stefansson K. (2018). A rare missense mutation in MYH6 associates with nonsyndromic coarctation of the aorta. European Heart Journal, 39(34), 3243–3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonstra PS, Mukherjee B, Gruber SB, Ahn J, Schmit SL, & Chatterjee N. (2016). Tests for Gene-Environment Interactions and Joint Effects With Exposure Misclassification. American Journal of Epidemiology, 183(3), 237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosse KR, & Maris JM. (2016). Advances in the translational genomics of neuroblastoma: From improving risk stratification and revealing novel biology to identifying actionable genomic alterations. Cancer, 122(1), 20–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulet SL, Rasmussen SA, & Honein MA. (2008). A population-based study of craniosynostosis in metropolitan Atlanta, 1989–2003. American Journal of Medical Genetics. Part A, 146A (8), 984–991. [DOI] [PubMed] [Google Scholar]
- Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, … Parkinson H. (2019). The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Research, 47(D1), D1005–D1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante CD, Burchard EG, & De la Vega FM. (2011). Genomics for the world. Nature, 475(7355), 163–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell CD, Ogburn EL, Lunetta KL, Lyon HN, Freedman ML, Groop LC, … Hirschhorn JN. (2005). Demonstrating stratification in a European American population. Nature Genetics, 37(8), 868–872. [DOI] [PubMed] [Google Scholar]
- Carmichael SL, Shaw GM, & Lammer EJ. (2012). Environmental and genetic contributors to hypospadias: a review of the epidemiologic evidence. Birth Defects Research. Part A, Clinical and Molecular Teratology, 94(7), 499–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caton AR, Druschel CM, & McNutt LA. (2004). The epidemiology of extrahepatic biliary atresia in New York State, 1983–98. Paediatric and Perinatal Epidemiology, 18(2), 97–105. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC). (2008). Update on overall prevalence of major birth defects—Atlanta, Georgia, 1978–2005. MMWR. Morbidity and Mortality Weekly Report, 57(1), 1–5. [PubMed] [Google Scholar]
- Chen Y, Adrianto I, Ianuzzi MC, Garman L, Montgomery CG, Rybicki BA, … Li J. (2019). Extended methods for gene-environment-wide interaction scans in studies of admixed individuals with varying degrees of relationships. Genetic Epidemiology, 43(4), 414–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Gilbert MA, Grochowski CM, McEldrew D, Llewellyn J, Waisbourd-Zinman O, … Devoto M. (2018). A genome-wide association study identifies a susceptibility locus for biliary atresia on 2p16.1 within the gene EFEMP1. PLoS Genetics, 14(8), e1007532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen B, Arbour L, Tran P, Leclerc D, Sabbaghian N, Platt R, … Rozen R. (1999). Genetic polymorphisms in methylenetetrahydrofolate reductase and methionine synthase, folate levels in red blood cells, and risk of neural tube defects. American Journal of Medical Genetics, 84(2), 151–157. [DOI] [PubMed] [Google Scholar]
- Cordell HJ, Bentham J, Topf A, Zelenika D, Heath S, Mamasoula C, … Keavney BD. (2013). Genome-wide association study of multiple congenital heart disease phenotypes identifies a susceptibility locus for atrial septal defect at chromosome 4p16. Nature Genetics, 45(7), 822–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordell HJ, Topf A, Mamasoula C, Postma AV, Bentham J, Zelenika D, … Goodship JA. (2013). Genome-wide association study identifies loci on 12q24 and 13q32 associated with tetralogy of Fallot. Human Molecular Genetics, 22(7), 1473–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis MC, Tchetgen EJ, Liang L, Qi L, Chatterjee N, Hu FB, & Kraft P. (2012). Gene-environment interactions in genome-wide association studies: a comparative study of tests applied to empirical studies of type 2 diabetes. American Journal of Epidemiology, 175(3), 191–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui S, Leyva-Vega M, Tsai EA, EauClaire SF, Glessner JT, Hakonarson H, … Matthews RP. (2013). Evidence from human and zebrafish that GPC1 is a biliary atresia susceptibility gene. Gastroenterology, 144(5), 1107–1115 e1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson WS, & Thompson TB. (2007). The structure of apolipoprotein A-I in high density lipoproteins. The Journal of Biological Chemistry, 282(31), 22249–22253. [DOI] [PubMed] [Google Scholar]
- Davies NM, Holmes MV, & Davey Smith G. (2018). Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ, 362, k601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bakker PI, Ferreira MA, Jia X, Neale BM, Raychaudhuri S, & Voight BF. (2008). Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Human Molecular Genetics, 17(R2), R122–R128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leeuw CA, Mooij JM, Heskes T, & Posthuma D. (2015). MAGMA: generalized gene-set analysis of GWAS data. PLoS Computational Biology, 11(4), e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Munnik SA, Hoefsloot EH, Roukema J, Schoots J, Knoers NV, Brunner HG, … Bongers EM. (2015). Meier-Gorlin syndrome. Orphanet Journal of Rare Diseases, 10, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etheredge AJ, Finnell RH, Carmichael SL, Lammer EJ, Zhu H, Mitchell LE, & Shaw GM. (2012). Maternal and infant gene-folate interactions and the risk of neural tube defects. American Journal of Medical Genetics. Part A, 158a(10), 2439–2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadista J, Manning AK, Florez JC, & Groop L. (2016). The (in) famous GWAS P-value threshold revisited and updated for low-frequency variants. European Journal of Human Genetics, 24(8), 1202–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadista J, Skotte L, Geller F, Bybjerg-Grauholm J, Gortz S, Romitti PA, … Feenstra B. (2019). Genome-wide meta-analysis identifies BARX1 and EML4-MTA3 as new loci associated with infantile hypertrophic pyloric stenosis. Human Molecular Genetics, 28(2), 332–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feenstra B, Geller F, Carstensen L, Romitti PA, Korberg IB, Bedell B, … Melbye M. (2013). Plasma lipids, genetic variants near APOA1, and the risk of infantile hypertrophic pyloric stenosis. JAMA, 310(7), 714–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feenstra B, Geller F, Krogh C, Hollegaard MV, Gortz S, Boyd HA, … Melbye M. (2012). Common variants near MBNL1 and NKX2–5 are associated with infantile hypertrophic pyloric stenosis. Nature Genetics, 44(3), 334–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Barcelo MM, Yeung MY, Miao XP, Tang CS, Cheng G, So MT, … Tam PK. (2010). Genome-wide association study identifies a susceptibility locus for biliary atresia on 10q24.2. Human Molecular Genetics, 19(14), 2917–2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geller F, Feenstra B, Carstensen L, Pers TH, van Rooij IA, Korberg IB, … Melbye M. (2014). Genome-wide association analyses identify variants in developmental genes associated with hypospadias. Nature Genetics, 46(9), 957–963. [DOI] [PubMed] [Google Scholar]
- Gibson G. (2019). On the utilization of polygenic risk scores for therapeutic targeting. PLoS Genetics, 15(4), e1008060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T, Repetto GM, DM MDMG, Chung JH, Nomaru H, Campbell CL, … Behavior C. (2017). Genome-Wide Association Study to Find Modifiers for Tetralogy of Fallot in the 22q11.2 Deletion Syndrome Identifies Variants in the GPR98 Locus on 5q14.3. Circulation. Cardiovascular Genetics, 10(5). 10.1161/CIRCGENETICS.116.001690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanchard NA, Swaminathan S, Bucasas K, Furthner D, Fernbach S, Azamian MS, … McBride KL. (2016). A genome-wide association study of congenital cardiovascular left-sided lesions shows association with a locus on chromosome 20. Human Molecular Genetics, 25(11), 2331–2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaland OA, Lie RT, Romanowska J, Gjerdevik M, Gjessing HK, & Jugessur A. (2018). A Genome-Wide Search for Gene-Environment Effects in Isolated Cleft Lip with or without Cleft Palate Triads Points to an Interaction between Maternal Periconceptional Vitamin Use and Variants in ESRRG. Frontiers in Genetics, 9, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helbig KL, Nothnagel M, Hampe J, Balschun T, Nikolaus S, Schreiber S, … Nothlings U. (2012). A case-only study of gene-environment interaction between genetic susceptibility variants in NOD2 and cigarette smoking in Crohn’s disease aetiology. BMC Medical Genetics, 13, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho TH, Charlet BN, Poulos MG, Singh G, Swanson MS, & Cooper TA. (2004). Muscleblind proteins regulate alternative splicing. The EMBO Journal, 23(15), 3103–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs CA, Chowdhury S, Cleves MA, Erickson S, MacLeod SL, Shaw GM, … Tycko B. (2014). Genetic epidemiology and nonsyndromic structural birth defects: from candidate genes to epigenetics. JAMA Pediatrics, 168(4), 371–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Shi Y, Mo X, Xu J, Zhao B, Lin Y, … Shen H. (2013). A genome-wide association study identifies two risk loci for congenital heart malformations in Han Chinese populations. Nat Genet, 45(7), 818–821. [DOI] [PubMed] [Google Scholar]
- Ioannidis JP. (2009). Population-wide generalizability of genome-wide discovered associations. Journal of the National Cancer Institute, 101(19), 1297–1299. [DOI] [PubMed] [Google Scholar]
- Jenkins MM, Almli LM, Pangilinan F, Chong JX, Blue EE, Shapira SK, … National Birth Defects Prevention Study. (2019). Exome sequencing of family trios from the National Birth Defects Prevention Study: Tapping into a rich resource of genetic and environmental data. Birth Defects Research. 10.1002/bdr2.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang T, Huang M, Jiang T, Gu Y, Wang Y, Wu Y, … Hu Z. (2018). Genome-wide compound heterozygosity analysis highlighted 4 novel susceptibility loci for congenital heart disease in Chinese population. Clinical Genetics, 94(3–4), 296–302. [DOI] [PubMed] [Google Scholar]
- Justice CM, Yagnik G, Kim Y, Peter I, Jabs EW, Erazo M, … Boyadjiev SA. (2012). A genome-wide association study identifies susceptibility loci for nonsyndromic sagittal craniosynostosis near BMP2 and within BBS9. Nature Genetics, 44(12), 1360–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, … Kathiresan S. (2018). Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nature Genetics, 50(9), 1219–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury MJ, & Wacholder S. (2009). Invited commentary: from genome-wide association studies to gene-environment-wide interaction studies—Challenges and opportunities. American Journal of Epidemiology, 169(2), 227–230 discussion 234–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BM, Woo J, Kanellopoulou C, & Shivdasani RA. (2011). Regulation of mouse stomach development and Barx1 expression by specific microRNAs. Development, 138(6), 1081–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DS, Stanaway IB, Rajagopalan R, Bernbaum JC, Solot CB, Burnham N, … Jarvik GP. (2012). Results of genome-wide analyses on neurodevelopmental phenotypes at four-year follow-up following cardiac surgery in infancy. PLoS One, 7(9), e45936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimonis V, Gold JA, Hoffman TL, Panchal J, & Boyadjiev SA. (2007). Genetics of craniosynostosis. Seminars in Pediatric Neurology, 14(3), 150–161. [DOI] [PubMed] [Google Scholar]
- Kochanek KD, Murphy S, Xu J, & Arias E. (2017). Mortality in the United States, 2016 NCHS Data Brief, no 293 (pp. 1–8). Hyattsville, MD: National Center for Health Statistics. [PubMed] [Google Scholar]
- Kraja AT, Cook JP, Warren HR, Surendran P, Liu C, Evangelou E, … Charge Exome Bp CHDEEBPGDTDCTUKBC-MTCBPWG. (2017). New blood pressure-associated loci identified in meta-analyses of 475 000 individuals. Circulation. Cardiovascular Genetics, 10(5). 10.1161/CIRCGENETICS.117.001778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacasana M, Blanco-Munoz J, Borja-Aburto VH, Aguilar-Garduno C, Rodriguez-Barranco M, Sierra-Ramirez JA, … Garcia-Cavazos R. (2012). Effect on risk of anencephaly of gene-nutrient interactions between methylenetetrahydrofolate reductase C677T polymorphism and maternal folate, vitamin B12 and homocysteine profile. Public Health Nutrition, 15(8), 1419–1428. [DOI] [PubMed] [Google Scholar]
- Lajeunie E, Le Merrer M, Bonaiti-Pellie C, Marchac D, & Renier D. (1995). Genetic study of nonsyndromic coronal craniosynostosis. American Journal of Medical Genetics, 55(4), 500–504. [DOI] [PubMed] [Google Scholar]
- Lee H, Lewis J, Schoen BT, Brand T, & Ricketts RR. (2001). Kasai portoenterostomy: differences related to race. Journal of Pediatric Surgery, 36(8), 1196–1198. [DOI] [PubMed] [Google Scholar]
- Lewin MB, Glass IA, & Power P. (2004). Genotype-phenotype correlation in congenital heart disease. Current Opinion in Cardiology, 19(3), 221–227. [DOI] [PubMed] [Google Scholar]
- Lin Y, Guo X, Zhao B, Liu J, Da M, Wen Y, … Hu Z. (2015). Association analysis identifies new risk loci for congenital heart disease in Chinese populations. Nature Communications, 6, 8082. [DOI] [PubMed] [Google Scholar]
- Lupo PJ, Agopian AJ, Castillo H, Castillo J, Clayton GH, Dosa NP, … Mitchell LE. (2017). Genetic epidemiology of neural tube defects. Journal of Pediatric Rehabilitation Medicine, 10(3–4), 189–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas P, Barrdahl M, Joshi AD, Auer PL, Gaudet MM, Milne RL, … Chatterjee N. (2016). Breast cancer risk from modifiable and nonmodifiable risk factors among white women in the United States. JAMA Oncology, 2(10), 1295–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslen CL. (2004). Molecular genetics of atrioventricular septal defects. Current Opinion in Cardiology, 19(3), 205–210. [DOI] [PubMed] [Google Scholar]
- Mitchell LE. (2008). Spina Bifida research resource: Study design and participant characteristics. Birth Defects Research. Part A, Clinical and Molecular Teratology, 82(10), 684–691. [DOI] [PubMed] [Google Scholar]
- Mitchell LE, Agopian AJ, Bhalla A, Glessner JT, Kim CE, Swartz MD, … Goldmuntz E. (2015). Genome-wide association study of maternal and inherited effects on left-sided cardiac malformations. Human Molecular Genetics, 24(1), 265–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori AD, & Bruneau BG. (2004). TBX5 mutations and congenital heart disease: Holt-Oram syndrome revealed. Current Opinion in Cardiology, 19(3), 211–215. [DOI] [PubMed] [Google Scholar]
- Nelson K, & Holmes LB. (1989). Malformations due to presumed spontaneous mutations in newborn infants. The New England Journal of Medicine, 320(1), 19–23. [DOI] [PubMed] [Google Scholar]
- Ningappa M, So J, Glessner J, Ashokkumar C, Ranganathan S, Min J, … Sindhi R. (2015). The Role of ARF6 in Biliary Atresia. PLoS One, 10(9), e0138381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntzani EE, Liberopoulos G, Manolio TA, & Ioannidis JP. (2012). Consistency of genome-wide associations across major ancestral groups. Human Genetics, 131(7), 1057–1071. [DOI] [PubMed] [Google Scholar]
- Padula AM, Yang W, Schultz K, Lurmann F, Hammond SK, & Shaw GM. (2018). Genetic variation in biotransformation enzymes, air pollution exposures, and risk of spina bifida. American Journal of Medical Genetics. Part A, 176(5), 1055–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters B, Oomen MW, Bakx R, & Benninga MA. (2014). Advances in infantile hypertrophic pyloric stenosis. Expert Review of Gastroenterology & Hepatology, 8(5), 533–541. [DOI] [PubMed] [Google Scholar]
- Ramachandran D, Zeng Z, Locke AE, Mulle JG, Bean LJ, Rosser TC, … Zwick ME. (2015). Genome-wide association study of down syndrome-associated atrioventricular septal defects. G3 (Bethesda), 5(10), 1961–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranells JD, Carver JD, & Kirby RS. (2011). Infantile hypertrophic pyloric stenosis: epidemiology, genetics, and clinical update. Advances in Pediatrics, 58(1), 195–206. [DOI] [PubMed] [Google Scholar]
- Reefhuis J, Gilboa SM, Anderka M, Browne ML, Feldkamp ML, Hobbs CA, … Honein MA. (2015). The National Birth Defects Prevention Study: A review of the methods. Birth Defects Research. Part A, Clinical and Molecular Teratology, 103(8), 656–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich DE, & Lander ES. (2001). On the allelic spectrum of human disease. Trends in Genetics: TIG, 17(9), 502–510. [DOI] [PubMed] [Google Scholar]
- Richard MA, Sok P, Canon S, Brown AL, Peckham-Gregory EC, Nembhard WN, … Lupo PJ. (2019). The role of genetic variation in DGKK on moderate and severe hypospadias. Birth Defects Res, 111, 932–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA, Huang L, Jewett EM, Szpiech ZA, Jankovic I, & Boehnke M. (2010). Genome-wide association studies in diverse populations. Nature Reviews. Genetics, 11(5), 356–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross S, Gerstein HC, Eikelboom J, Anand SS, Yusuf S, & Pare G. (2015). Mendelian randomization analysis supports the causal role of dysglycaemia and diabetes in the risk of coronary artery disease. European Heart Journal, 36(23), 1454–1462. [DOI] [PubMed] [Google Scholar]
- Sanchez-Valle A, Kassira N, Varela VC, Radu SC, Paidas C, & Kirby RS. (2017). Biliary Atresia: Epidemiology, Genetics, Clinical Update, and Public Health Perspective. Advances in Pediatrics, 64(1), 285–305. [DOI] [PubMed] [Google Scholar]
- Schaid DJ, & Sommer SS. (1993). Genotype relative risks: methods for design and analysis of candidate-gene association studies. American Journal of Human Genetics, 53(5), 1114–1126. [PMC free article] [PubMed] [Google Scholar]
- Self M, Geng X, & Oliver G. (2009). Six2 activity is required for the formation of the mammalian pyloric sphincter. Developmental Biology, 334(2), 409–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewda A, Agopian AJ, Goldmuntz E, Hakonarson H, Morrow BE, Taylor D, … Pediatric Cardiac Genomics C. (2019). Gene-based genome-wide association studies and meta-analyses of conotruncal heart defects. PLoS One, 14(7), e0219926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw GM, Iovannisci DM, Yang W, Finnell RH, Carmichael SL, Cheng S, & Lammer EJ. (2005). Risks of human conotruncal heart defects associated with 32 single nucleotide polymorphisms of selected cardiovascular disease-related genes. American Journal of Medical Genetics. Part A, 138(1), 21–26. [DOI] [PubMed] [Google Scholar]
- Spielman RS, McGinnis RE, & Ewens WJ. (1993). Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM). American Journal of Human Genetics, 52(3), 506–516. [PMC free article] [PubMed] [Google Scholar]
- Sugrue LP, & Desikan RS. (2019). What Are Polygenic Scores and Why Are They Important? JAMA, 321(18), 1820–1821. [DOI] [PubMed] [Google Scholar]
- Sundaram SS, Mack CL, Feldman AG, & Sokol RJ. (2017). Biliary atresia: Indications and timing of liver transplantation and optimization of pretransplant care. Liver Transplantation, 23(1), 96–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung YJ, Winkler TW, de Las Fuentes L, Bentley AR, Brown MR, Kraja AT, … Chasman DI. (2018). A Large-Scale Multi-ancestry Genome-wide Study Accounting for Smoking Behavior Identifies Multiple Significant Loci for Blood Pressure. American Journal of Human Genetics, 102(3), 375–400. 10.1016/j.ajhg.2018.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam V, Patel N, Turcotte M, Bosse Y, Pare G, & Meyre D. (2019). Benefits and limitations of genome-wide association studies. Nature Reviews. Genetics, 20(8), 467–484. [DOI] [PubMed] [Google Scholar]
- The Human Protein Atlas. 2019. EML4. [Google Scholar]
- Timberlake AT, Choi J, Zaidi S, Lu Q, Nelson-Williams C, Brooks ED, … Lifton RP. (2016). Two locus inheritance of nonsyndromic midline craniosynostosis via rare SMAD6 and common BMP2 alleles. eLife, 5, e20125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torgerson DG, Ampleford EJ, Chiu GY, Gauderman WJ, Gignoux CR, Graves PE, … Nicolae DL. (2011). Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nature Genetics, 43(9), 887–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Zanden LF, van Rooij IA, Feitz WF, Knight J, Donders AR, Renkema KY, … Knoers NV. (2011). Common variants in DGKK are strongly associated with risk of hypospadias. Nature Genetics, 43(1), 48–50. [DOI] [PubMed] [Google Scholar]
- Veltman JA, & Brunner HG. (2012). De novo mutations in human genetic disease. Nature Reviews. Genetics, 13(8), 565–575. [DOI] [PubMed] [Google Scholar]
- Visscher PM, Wray NR, Zhang Q, Sklar P, McCarthy MI, Brown MA, & Yang J. (2017). 10 Years of GWAS Discovery: Biology, Function, and Translation. American Journal of Human Genetics, 101(1), 5–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Huang J, Liu Y, Ma L, Potash JB, & Han S. (2017). COMBAT: A Combined Association Test for Genes Using Summary Statistics. Genetics, 207(3), 883–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber DM, MacLeod SL, Bamshad MJ, Shaw GM, Finnell RH, Shete SS, … Hobbs C. (2015). Developments in our understanding of the genetic basis of birth defects. Birth Defects Research. Part A, Clinical and Molecular Teratology, 103(8), 680–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg CR, Wilcox AJ, & Lie RT. (1998). A log-linear approach to case-parent-triad data: assessing effects of disease genes that act either directly or through maternal effects and that may be subject to parental imprinting. American Journal of Human Genetics, 62(4), 969–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Li Y, & Abecasis GR. (2010). METAL: fast and efficient meta-analysis of genome-wide association scans. Bioinformatics, 26(17), 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Liang KY, Hetmanski JB, Ruczinski I, Fallin MD, Ingersoll RG, … Beaty TH. (2010). Evidence of gene-environment interaction for the IRF6 gene and maternal multivitamin supplementation in controlling the risk of cleft lip with/without cleft palate. Human Genetics, 128(4), 401–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Schwender H, Ruczinski I, Murray JC, Marazita ML, Munger RG, … Beaty TH. (2014). Evidence of gene-environment interaction for two genes on chromosome 4 and environmental tobacco smoke in controlling the risk of nonsyndromic cleft palate. PLoS One, 9(2), e88088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Muzny DM, Reid JG, Bainbridge MN, Willis A, Ward PA, … Eng CM. (2013). Clinical whole-exome sequencing for the diagnosis of mendelian disorders. The New England Journal of Medicine, 369(16), 1502–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates TM, Turner CL, Firth HV, Berg J, & Pilz DT. (2017). Baraitser-Winter cerebrofrontofacial syndrome. Clinical Genetics, 92(1), 3–9. [DOI] [PubMed] [Google Scholar]
- Yoon PW, Bresee JS, Olney RS, James LM, & Khoury MJ. (1997). Epidemiology of biliary atresia: a population-based study. Pediatrics, 99(3), 376–382. [DOI] [PubMed] [Google Scholar]
- Yoon PW, Rasmussen SA, Lynberg MC, Moore CA, Anderka M, Carmichael SL, … Edmonds LD. (2001). The National Birth Defects Prevention Study. Public Health Reports (Washington, DC : 1974), 116(Suppl 1), 32–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang TX, Haller G, Lin P, Alvarado DM, Hecht JT, Blanton SH, … Gurnett CA. (2014). Genome-wide association study identifies new disease loci for isolated clubfoot. Journal of Medical Genetics, 51(5), 334–339. [DOI] [PubMed] [Google Scholar]
- Zhao B, Lin Y, Xu J, Ni B, Da M, Ding C, … Ma H. (2014). Replication of the 4p16 susceptibility locus in congenital heart disease in Han Chinese populations. PLoS One, 9(9), e107411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Li B, Dian K, Ying B, Lu X, Hu X, … Qin L. (2015). Association between the European GWAS-identified susceptibility locus at chromosome 4p16 and the risk of atrial septal defect: a case-control study in Southwest China and a meta-analysis. PLoS One, 10(4), e0123959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao LP, Fan W, Goodman G, Radich J, & Martin P. (2015). Deciphering genome environment wide interactions using exposed subjects only. Genetic Epidemiology, 39(5), 334–346. [DOI] [PMC free article] [PubMed] [Google Scholar]