Abstract
Penicillium species were commonly isolated during a fungal survey of bat hibernacula in New Brunswick and Quebec, Canada. Strains were isolated from arthropods, bats, rodents (i.e. the deer mouse Peromyscus maniculatus), their dung, and cave walls. Hundreds of fungal strains were recovered, of which Penicillium represented a major component of the community. Penicillium strains were grouped by colony characters on Blakeslee’s malt extract agar. DNA sequencing of the secondary identification marker, beta-tubulin, was done for representative strains from each group. In some cases, ITS and calmodulin were sequenced to confirm identifications. In total, 13 species were identified, while eight strains consistently resolved into a unique clade with P. discolor, P. echinulatum and P. solitum as its closest relatives. Penicillium speluncae is described using macroand micromorphological characters, multigene phylogenies (including ITS, beta-tubulin, calmodulin and RNA polymerase II second largest subunit) and extrolite profiles. Major extrolites produced by the new species include cyclopenins, viridicatins, chaetoglobosins, and a microheterogenous series of cyclic and linear tetrapeptides.
Keywords: Thysanophora, sect. Fasciculata, Genealogical Concordance Phylogenetic Species Recognition (GCPSR) concept, new taxon, Pseudogymnoascus destructans (Pd), secondary metabolites
INTRODUCTION
The study of fungi associated with bats and their habitats has become important after the spread of White-nose Syndrome (WNS) caused by Pseudogymnoascus destructans (Pd), resulted in an ongoing rapid decline of bat populations in North America. Much effort has focused on populations of Pd from positive caves. White-nose Syndrome is named for characteristic white growth caused by P. destructans, which was previously known as Geomyces destructans (Gargas et al. 2009, Minnis & Lindner 2013). Characterization of fungal populations and identification of other fungal species may reveal possible antagonists to Pd (Micalizzi et al. 2017).
White-nose Syndrome was first reported in New York in 2006 (Blehert et al. 2009), while the first report from Canada was from Ontario in 2010. In both cases, it led to mass mortality of the hibernating bat populations (McAlpine et al. 2012). The disease only occurs while bats hibernate. Pseudogymnoascus destructans cannot grow at temperatures above ± 20 °C (Gargas et al. 2009), and it is thought that the cool caves and mines inhabited by bats during hibernation serve as environmental reservoirs of Pd (Lorch et al. 2013, Reynolds et al. 2015). The presence of Pd in bat populations was confirmed in many countries in Europe and Asia but no significant mortality was observed, despite the fact that some European bats have been found with clinical WNS (Wibbelt et al. 2010, Puechmaille et al. 2011). Why bats remain healthy in these areas is unclear.
The study of fungal diversity is important to determine the true impact of a potential invasive species such as Pd on fungal community structure among bats and hibernacula (Johnson et al. 2013). Understudied environments such as caves are rich sources of undescribed microbial species. Many new fungi have recently been described from underground environments as more studies are conducted, although it is still unknown whether obligate troglobiotic fungi exist (Zhang et al. 2017). Previous studies commonly reported the isolation of Cladosporium, Fusarium, Mortierella, and Penicillium species from bat wings, caves and mines (Johnson et al. 2013, Vanderwolf et al. 2013a, b). Penicillium is one of the most common genera isolated from caves on multiple substrates, particularly sediment and air, although no new Penicillium species have been described from caves apart from P. cavernicola, which has also been found outside of caves on dairy products (Frisvad & Samson 2004, Vanderwolf et al. 2013a, b), and P. gravinicasei recently described from a cave in Italy from ripening Apulian cave cheeses (Anelli et al. 2018). Vanderwolf et al. (2016) studied the fungi associated with over-wintering arthropods in Pd positive hibernacula in Canada. They isolated 87 fungal taxa from four arthropod genera. In the current study, we report Penicillium isolated from these arthropods, but also include strains isolated from various other substrates associated with caves and/or bats. The aims of this study were (1) to determine the Penicillium species diversity in bat caves and hibernacula in New Brunswick and Quebec, and (2) formally describe the new species that was isolated during the survey.
MATERIALS AND METHODS
Strains, sampling and isolations
Strains were isolated from arthropods, bats, rodents, rodent dung, and walls of bat hibernacula in New Brunswick (Berryton Cave, Dallings Cave, Dorchester Mine, Glebe Mine, Markhamville Mine, White Cave) and Quebec (Grotte à la Patate), Canada (Vanderwolf et al. 2013b, 2016, 2017). Fungi were also isolated from a dead big brown bat that was found in a parking garage in Fredericton, New Brunswick. Isolation media included dextrose-peptone yeast extract agar (DPYA), sabouraud agar (SD) or malt extract agar (MEA), with plates incubated at 7 °C. Representative strains for each species found were submitted to the Canadian Collection of Fungal Cultures (DAOMC) and the holotype specimen of the new species deposited in the Canadian National Mycological Herbarium (DAOM). Strains isolated during this study are summarized in Table 1.
Table 1.
Species isolated from Canadian bat caves.
| Species | Section | Strain | Date collected | Isolation medium | Province | Location | Cave name | Substrate | ITS | BenA | CaM | RPB2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P. biolowiezense | Brevicompacta | KAS 7465, W7430A | 31-Mar~2015 | MEA | New Brunswick | Dorchester | Dorchester Mine | Cave wall | n/a | MG490896 | n/a | n/a |
| DAOMC 252097, KAS 7466, W72102 | 07-Jul-2014 | DPYA | Quebec | Anticosti Island | Grotte à la Patate | Cave wall | n/a | MG490897 | n/a | n/a | ||
| DAOMC 252098, KAS 7476, W29304 | 30-Apr-2015 | MEA | New Brunswick | Moncton | Berryton Cave | Cave wall | n/a | MG490903 | n/a | n/a | ||
| KAS 7480, W24103 | 30-Apr-2015 | DPYA | New Brunswick | Moncton | Berryton Cave | Cave wall | n/a | MG490906 | n/a | n/a | ||
| KAS 7511, S11101 | 18-Mar-2013 | DPYA | New Brunswick | Dorchester | Dorchester Mine | Spider (Meta ovalis) | n/a | MG490924 | n/a | n/a | ||
| KAS 7517, M50103 | ll-Apr-2014 | DPYA | New Brunswick | Sussex | Glebe Mine | Gnat (Exechiopsis sp) | n/a | MG490929 | n/a | n/a | ||
| KAS 7522, H27101 | ll-Apr-2014 | DPYA | New Brunswick | Sussex | Glebe Mine | Harvestman (Nelima elegans) | n/a | MG490933 | n/a | n/a | ||
| KAS 7523, H26208 | ll-Apr-2014 | SD | New Brunswick | Sussex | Glebe Mine | Harvestman (Nelima elegans) | n/a | MG490934 | n/a | n/a | ||
| DAOMC 252099, KAS 7525, H09108 | 18-Mar-2013 | DPYA | New Brunswick | Dorchester | Dorchester Mine | Harvestman (Nelima elegans) | n/a | MG490936 | n/a | n/a | ||
| KAS 7542, 742102 | 16-Apr-2014 | DPYA | New Brunswick | Fredericton | Fredericton parking garage | Bat (Eptesicus fuscus) | n/a | MG490949 | n/a | n/a | ||
| P. brevistipitotum | Robsamsonia | DAOMC 252100, KAS 7514, P06101 | 14-Mar-2014 | DPYA | New Brunswick | Dorchester | Dorchester Mine | Rodent (Peromyscus maniculotus) | MG490876 | MG490926 | MG490966 | n/a |
| DAOMC 252101, KAS 7520, M26108 | 16-Apr-2013 | DPYA | New Brunswick | Sussex | Dal lings Cave | Moth (Scoliopteryx libatrix) | MG490878 | MG490932 | MG490968 | n/a | ||
| DAOMC 252102, KAS 7531, D3303 | 25-Mar-2014 | DPYA | New Brunswick | Dorchester | Dorchester Mine | Rodent dung (Peromyscus maniculotus) | MG490879 | MG490938 | MG490969 | n/a | ||
| DAOMC 252103, KAS 7534, D2203 | 21-Mar-2014 | DPYA | New Brunswick | Dorchester | Dorchester Mine | Rodent dung (Peromyscus maniculotus) | MG490881 | MG490941 | MG490971 | n/a | ||
| DAOMC 252104, KAS 7538, D1007A | 21-Mar-2014 | DPYA | New Brunswick | Dorchester | Dorchester Mine | Rodent dung (Peromyscus maniculotus) | MG490882 | MG490945 | MG490972 | n/a | ||
| DAOMC 252105, KAS 7539, D1007 | 21-Mar-2014 | DPYA | New Brunswick | Dorchester | Dorchester Mine | Rodent dung (Peromyscus maniculotus) | MG490883 | MG490946 | MG490973 | n/a | ||
| P. chrysogenum | Chrysogena | DAOMC 252106, KAS 7505, W05100 | 21-Apr-2015 | DPYA | New Brunswick | Hillsborough | White Cave | Cave wall | n/a | MG490919 | n/a | n/a |
| DAOMC 252107, KAS 7540, 742110 | 16-Apr-2014 | DPYA | New Brunswick | Fredericton | Fredericton parking garage | Bat (Eptesicus fuscus) | n/a | MG490947 | n/a | n/a | ||
| P. concentricum | Robsamsonia | KAS 7459, W98105 | 16-Apr-2015 | DPYA | New Brunswick | Sussex | Glebe Mine | Cave wall | n/a | MG490890 | n/a | n/a |
| DAOMC 252108, KAS 7467, W72101 | 07-Jul-2014 | DPYA | Quebec | Anticosti Island | Grotte à la Patate | Cave wall | n/a | MG490898 | n/a | n/a | ||
| KAS 7470, W59104 | 07-Jul-2014 | DPYA | Quebec | Anticosti Island | Grotte à la Patate | Cave wall | n/a | MG490900 | n/a | n/a | ||
| KAS 7471, W59104 | 07-Jul-2014 | DPYA | Quebec | Anticosti Island | Grotte à la Patate | Cave wall | n/a | MG490901 | n/a | n/a | ||
| KAS 7478, W29203 | 30-Apr-2015 | SD | New Brunswick | Moncton | Berryton Cave | Cave wall | n/a | MG490904 | n/a | n/a | ||
| DAOMC 252109, KAS 7479, W24103A | 30-Apr-2015 | DPYA | New Brunswick | Moncton | Berryton Cave | Cave wall | n/a | MG490905 | n/a | n/a | ||
| DAOMC 252110, KAS 7483, W22102A | 30-Apr-2015 | DPYA | New Brunswick | Moncton | Berryton Cave | Cave wall | n/a | MG490908 | n/a | n/a | ||
| KAS 7486, W20408 | 30-Apr-2015 | DPYA | New Brunswick | Moncton | Berryton Cave | Cave wall | n/a | MG490910 | n/a | n/a | ||
| KAS 7513, P06102 | 14-Mar-2014 | DPYA | New Brunswick | Dorchester | Dorchester Mine | Rodent (Peromyscus maniculotus) | n/a | MG490925 | n/a | n/a | ||
| KAS 7515, P05201 | 14-Mar-2014 | SD | New Brunswick | Dorchester | Dorchester Mine | Rodent (Peromyscus maniculotus) | n/a | MG490927 | n/a | n/a | ||
| KAS 7532, D3301 | 25-Mar-2014 | DPYA | New Brunswick | Dorchester | Dorchester Mine | Rodent dung (Peromyscus maniculotus) | n/a | MG490939 | n/a | n/a | ||
| KAS 7535, D2111 | 21-Mar-2014 | DPYA | New Brunswick | Dorchester | Dorchester Mine | Rodent dung (Peromyscus maniculotus) | n/a | MG490942 | n/a | n/a | ||
| KAS 7536, D1204 | 21-Mar-2014 | DPYA | New Brunswick | Dorchester | Dorchester Mine | Rodent dung (Peromyscus maniculotus) | n/a | MG490943 | n/a | n/a | ||
| KAS 7537, D1106 | 21-Mar-2014 | DPYA | New Brunswick | Dorchester | Dorchester Mine | Rodent dung (Peromyscus maniculotus) | n/a | MG490944 | n/a | n/a | ||
| P. consobrinum | Exilicoulis | DAOMC 252111, KAS 7464, W76401 | 31-Mar-2015 | DPYA | New Brunswick | Dorchester | Dorchester Mine | Cave wall | MG490873 | MG490895 | MG490963 | n/a |
| DAOMC 252112, KAS 7491, W19102 | 30-Apr-2015 | DPYA | New Brunswick | Moncton | Berryton Cave | Cave wall | MG490874 | MG490913 | MG490964 | n/a | ||
| P. corylophilum | Exilicoulis | DAOMC 252113, KAS 7481, W24100 | 30-Apr-2015 | DPYA | New Brunswick | Moncton | Berryton Cave | Cave wall | n/a | MG490907 | n/a | n/a |
| DAOMC 252114, KAS 7484, W22102 | 30-Apr-2015 | DPYA | New Brunswick | Moncton | Berryton Cave | Cave wall | n/a | MG490909 | n/a | n/a | ||
| KAS 7489, W20103 | 30-Apr-2015 | DPYA | New Brunswick | Moncton | Berryton Cave | Cave wall | n/a | MG490912 | n/a | n/a | ||
| KAS 7493, W16200B | 30-Apr-2015 | SD | New Brunswick | Moncton | Berryton Cave | Cave wall | n/a | MG490914 | n/a | n/a | ||
| P. exponsum | Penicillium | DAOMC 252115, KAS 7501, W07104 | 21-Apr-2015 | DPYA | New Brunswick | Hillsborough | White Cave | Cave wall | n/a | MG490917 | n/a | n/a |
| KAS 7502, W05406 | 21-Apr-2015 | DPYA | New Brunswick | Hillsborough | White Cave | Cave wall | n/a | MG490918 | n/a | n/a | ||
| KAS 7506, W04407 | 21-Apr-2015 | DPYA | New Brunswick | Hillsborough | White Cave | Cave wall | n/a | MG490920 | n/a | n/a | ||
| KAS 7510, W00200 | 21-Apr-2015 | SD | New Brunswick | Hillsborough | White Cave | Cave wall | n/a | MG490923 | n/a | n/a | ||
| DAOMC 252116, KAS 7519, M26109 | 16-Apr-2013 | DPYA | New Brunswick | Sussex | Dallings Cave | Moth (Scoliopteryx libatrix) | n/a | MG490931 | n/a | n/a | ||
| KAS 7529, H06108 | 18-Mar-2013 | DPYA | New Brunswick | Dorchester | Dorchester Mine | Harvestman (Nelima elegans) | n/a | MG490937 | n/a | n/a | ||
| KAS 7545, 702115 | 04-Apr-2013 | DPYA | New Brunswick | Markhamville | Markhamville Mine | Bat (Perimyotis subflavus) | n/a | MG490951 | n/a | n/a | ||
| KAS 7546, 701115 | 04-Apr-2013 | DPYA | New Brunswick | Markhamville | Markhamville Mine | Bat (Perimyotis subflavus) | n/a | MG490952 | n/a | n/a | ||
| KAS 7547, 701106 | 04-Apr-2013 | DPYA | New Brunswick | Markhamville | Markhamville Mine | Bat (Perimyotis subflavus) | n/a | MG490953 | n/a | n/a | ||
| P. globrum | Aspergilloides | DAOMC 252117, KAS 7475, W54101 | 07-Jul-2014 | DPYA | Quebec | Anticosti Island | Grotte à la Patate | Cave wall | n/a | MG490902 | n/a | n/a |
| DAOMC 252118, KAS 7494, W16200A | 30-Apr-2015 | SD | New Brunswick | Moncton | Berryton Cave | Cave wall | n/a | MG490915 | n/a | n/a | ||
| KAS 7524, H11101 | 18-Mar-2013 | DPYA | New Brunswick | Dorchester | Dorchester Mine | Harvestman (Nelima elegans) | n/a | MG490935 | n/a | n/a | ||
| P. gloucoolbidum | Thysonophoro | DAOMC 252119, KAS 7460, W88411 | 31-Mar-2015 | DPYA | New Brunswick | Dorchester | Dorchester Mine | Cave wall | MG490870 | MG490891 | MG490960 | n/a |
| DAOMC 252120, KAS 7461, W88405 | 31-Mar-2015 | DPYA | New Brunswick | Dorchester | Dorchester Mine | Cave wall | MG490871 | MG490892 | MG490961 | n/a | ||
| KAS 7462, W88405 | 31-Mar-2015 | DPYA | New Brunswick | Dorchester | Dorchester Mine | Cave wall | MG490872 | MG490893 | MG490962 | n/a | ||
| DAOMC 252122, KAS 7508, W04210 | 16-Apr-2015 | SD | New Brunswick | Sussex | Glebe Mine | Cave wall | MG490875 | MG490921 | MG490965 | n/a | ||
| P. rubens | Chrysogeno | DAOMC 252123, KAS 7488, W20104 | 30-Apr-2015 | DPYA | New Brunswick | Moncton | Berryton Cave | Cave wall | n/a | MG490911 | n/a | n/a |
| KAS 7495, W16200 | 30-Apr-2015 | SD | New Brunswick | Moncton | Berryton Cave | Cave wall | n/a | MG490916 | n/a | n/a | ||
| KAS 7509, W02400 | 16-Apr-2015 | DPYA | New Brunswick | Sussex | Glebe Mine | Cave wall | n/a | MG490922 | n/a | n/a | ||
| DAOMC 252124, KAS 7543, 741102 | 16-Apr-2014 | DPYA | New Brunswick | Fredericton | Fredericton parking garage | Bat (Eptesicus fuscus) | n/a | MG490950 | n/a | n/a | ||
| P. spothulotum | Brevicompacta | KAS 7468, W71101 | 07-Jul-2014 | DPYA | Quebec | Anticosti Island | Grotte à la Patate | Cave wall | n/a | MG490899 | n/a | n/a |
| DAOMC 252125, KAS 7541, 742105 | 16-Apr-2014 | DPYA | New Brunswick | Fredericton | Fredericton parking garage | Bat (Eptesicus fuscus) | n/a | MG490948 | n/a | n/a | ||
| P. speluncoe | Fasciculata | DAOMC 251696, KAS 7473, W54119 | 07-Jul-2014 | DPYA | Quebec | Anticosti Island | Grotte à la Patate | Cave wall | MG490864 | MG490884 | MG490954 | MN170736 |
| DAOMC 251697, KAS 7474, W54102 | 07-Jul-2014 | DPYA | Quebec | Anticosti Island | Grotte à la Patate | Cave wall | MG490865 | MG490885 | MG490955 | MN170737 | ||
| DAOMC 251698, KAS 7500, W07302 | 21-Apr-2015 | MEA | New Brunswick | Hillsborough | White Cave | Cave wall | MG490866 | MG490886 | MG490956 | MN170738 | ||
| DAOMC 251699, KAS 7503, W05404 | 21-Apr-2015 | DPYA | New Brunswick | Hillsborough | White Cave | Cave wall | MG490867 | MG490887 | MG490957 | MN170739 | ||
| DAOMC 251700, KAS 7504, W05202 | 16-Apr-2015 | SD | New Brunswick | Sussex | Glebe Mine | Cave wall | MG490868 | MG490888 | MG490958 | MN170740 | ||
| DAOMC 251701T, KAS 7512, P06201 | 14-Mar-2014 | SD | New Brunswick | Dorchester | Dorchester Mine | Rodent (Peromyscus maniculatus) | MG490869 | MG490889 | MG490959 | MN170741 | ||
| DAOMC 252126, KAS 7516, P01202 | 12-Mar-2014 | SD | New Brunswick | Dorchester | Dorchester Mine | Rodent (Peromyscus maniculatus) | MG490877 | MG490928 | MG490967 | MN170742 | ||
| DAOMC 252127, KAS 7533, D3108 | 25-Mar-2014 | DPYA | New Brunswick | Dorchester | Dorchester Mine | Rodent dung (Peromyscus maniculatus) | MG490880 | MG490940 | MG490970 | MN170743 | ||
| P. westlingii | Citrina | DAOMC 252128, KAS 7463, W77200 | 31-Mar-2015 | SD | New Brunswick | Dorchester | Dorchester Mine | Cave wall | n/a | MG490894 | n/a | n/a |
| DAOMC 252129, KAS 7518, M34108 | 14-Mar-2014 | DPYA | New Brunswick | Dorchester | Dorchester Mine | Moth (Scoliopteryx libatrix) | n/a | MG490930 | n/a | n/a |
DAOMC: Culture collection of the National Mycological Collections, Agriculture & Agri-Food Canada, Ottawa, Canada; KAS: Internal working culture collection at DAOMC; Remaining acronyms represent internal isolate numbers from the personal collection of Karen Vanderwolf and Dave Malloch.
DNA extraction, sequencing and phylogenetic analysis
Strains were grown on Blakeslee’s (1915) malt extract agar (MEAbl) for 7 d and DNA extracted using the UltracleanTM Microbial DNA isolation Kit (MoBio Laboratories Inc., Solana Beach, USA). DNA was amplified with a PCR master mix consisting of 0.5 µL dNTPs (2 µM), 0.04 µL for each primer (20 µM), 1 µL 10× Titanium Taq buffer (Clontech, California, USA), 0.1 µL 50× Titanium Taq enzyme (Clontech, California, USA), 0.5 µL template DNA and 7.82 µL sterile purified water. ITS barcodes (Schoch et al. 2012), partial beta-tubulin (BenA), partial calmodulin (CaM) and RNA polymerase II second largest subunit (RPB2) genes were amplified using PCR conditions and primers suggested by Visagie et al. (2014b). PCR products were verified by agarose gel electrophoresis and subsequently sequenced with the BigDye Terminator Cycle Premix Kit (Applied Biosystems, Waltham, USA). Contigs were assembled and edited in Geneious v. 8.1.5 (BioMatters Ltd., Auckland, New Zealand). Newly generated sequences were submitted to GenBank and accession numbers provided in Table 1. Gene sequences of the new species were compared to a reference sequence dataset built around the ex-type sequences published in Visagie et al. (2014b), also including reference sequences from (Samson et al. 2004, Houbraken et al. 2011, 2012, 2014, 2016, Frisvad et al. 2013a, b, Visagie et al. 2014a) where needed (Suppl. Table S1). Additional unpublished sequences related to the new species were included and originate from various past projects. Sequences were aligned in MAFFT v. 7.407 (Katoh & Standley 2013), with the G-INS-i option and manually trimmed and adjusted in Geneious where needed. Datasets were subsequently analysed using Maximum Likelihood (ML) and Bayesian tree inference (BI). For concatenated phylogenies, each gene was treated as a separate partition. ML trees were calculated in IQtree v. 1.6.8 (Nguyen et al. 2015) with the most suitable model for each gene and/or partition calculated using Modelfinder (Kalyaanamoorthy et al. 2017) and bootstrapping done using UFBoot (Minh et al. 2013), both integrated into IQtree. Bayesian inference trees were calculated in MrBayes v. 3.2.6 (Ronquist et al. 2012) with the most suitable model selected by ParitionFinder v. 2.1.1 (Lanfear et al. 2017) using the corrected Akaike information criterion (Akaike 1974). Alignments and command blocks used for analyses were uploaded to TreeBASE (https://treebase.org) with accession 23575. Trees were visualized in Figtree v. 1.4.4 (http://tree.bio.ed.ac.uk/software/figtree) and visually edited in Affinity Designer v. 1.7.1 [Serif (Europe) Ltd, Nottingham, UK].
Table. S1.
Supplementary Table S1. Strains used for phylogenetic analyses.
| Species | Strain | Section | ITS_GB | BenA_GB | CaM_GB | RBP2_GB |
|---|---|---|---|---|---|---|
| P. armarii | CBS 138171 (ex-type) | Aspergilloides | KM189758 | KM089007 | KM089394 | KM089781 |
| P. bussumense | CBS 138160 (ex-type) | Aspergilloides | KM189458 | KM088685 | KM089070 | KM089457 |
| P. frequentans | CBS 105.11 (ex-type) | Aspergilloides | KM189525 | KM088762 | KM089147 | KM089534 |
| P. glabrum | CBS 125543 (ex-type) | Aspergilloides | KM189530 | KM088767 | KM089152 | KM089539 |
| P. glabrum | CBS 129784 | Aspergilloides | KM189736 | KM088985 | KM089372 | KM089759 |
| P. glabrum | CBS 129606 | Aspergilloides | KM189733 | KM088982 | KM089369 | KM089756 |
| P. glabrum | CBS 126336 | Aspergilloides | KM189537 | KM088775 | KM089160 | KM089547 |
| P. glabrum | CBS 127700 | Aspergilloides | KM189540 | KM088778 | KM089163 | KM089550 |
| P. glabrum | CBS 126333 | Aspergilloides | KM189536 | KM088774 | KM089159 | KM089546 |
| P. glabrum | CBS 127704 | Aspergilloides | KM189533 | KM088771 | KM089156 | KM089543 |
| P. glabrum | CBS 328.48 | Aspergilloides | KM189790 | KM089040 | KM089427 | KM089814 |
| P. glabrum | CBS 138165 | Aspergilloides | KM189546 | KM088784 | KM089169 | KM089556 |
| P. glabrum | CBS 138166 | Aspergilloides | KM189611 | KM088855 | KM089242 | KM089629 |
| P. glabrum | CBS 129602 | Aspergilloides | KM189732 | KM088981 | KM089368 | KM089755 |
| P. glabrum | CBS 131040 | Aspergilloides | KM189785 | KM089035 | KM089422 | KM089809 |
| P. glabrum | CBS 115810 | Aspergilloides | KM189477 | KM088712 | KM089097 | KM089484 |
| P. glabrum | CBS 138164 | Aspergilloides | KM189544 | KM088782 | KM089167 | KM089554 |
| P. glabrum | CBS 171.81 | Aspergilloides | KM189468 | KM088700 | KM089085 | KM089472 |
| P. glabrum | CBS 127703 | Aspergilloides | KM189534 | KM088772 | KM089157 | KM089544 |
| P. pulvis | CBS 138432 (ex-type) | Aspergilloides | KM189632 | KM088876 | KM089263 | KM089650 |
| P. purpurascens | CBS 366.48 (ex-type) | Aspergilloides | KM189561 | KM088801 | KM089186 | KM089573 |
| P. rudallense | CBS 130049 (ex-type) | Aspergilloides | KM189744 | KM088993 | KM089380 | KM089767 |
| P. spinulosum | CBS 374.48 (ex-type) | Aspergilloides | KM189448 | KM088672 | KM089057 | KM089444 |
| P. thomii | CBS 225.81 (ex-type) | Aspergilloides | KM189560 | KM088799 | KM089184 | KM089571 |
| P. bialowiezense | CBS 227.28 (ex-type) | Brevicompacta | EU587315 | AY674439 | AY484828 | JN406604 |
| P. bialowiezense | NRRL 32205 | Brevicompacta | AY484904 | DQ645791 | AY484836 | |
| P. bialowiezense | NRRL 32207 | Brevicompacta | AY484905 | DQ645792 | AY484837 | |
| P. brevicompactum | CBS 257.29 (ex-type) | Brevicompacta | AY484912 | AY674437 | AY484813 | JN406594 |
| P. spathulatum | CBS 117192 (ex-type) | Brevicompacta | JX313165 | JX313183 | JX313149 | JN406636 |
| P. spathulatum | CBS 116976 | Brevicompacta | JX313161 | JX313179 | JX313145 | |
| P. spathulatum | CBS 116975 | Brevicompacta | JX313160 | JX313178 | JX313144 | |
| P. spathulatum | CBS 116977 | Brevicompacta | JX313162 | JX313180 | JX313146 | |
| P. spathulatum | CBS 116972 | Brevicompacta | JX313157 | JX313175 | JX313141 | |
| P. canescens | CBS 300.48 (ex-type) | Canescentia | AF033493 | JX140946 | KJ867009 | JN121485 |
| P. charlesii | CBS 304.48 (ex-type) | Charlesia | AF033400 | JX091508 | AY741727 | JN121486 |
| P. allii-sativi | CBS 132074 (ex-type) | Chrysogena | JX997021 | JX996891 | JX996232 | JX996627 |
| P. allii-sativi | DTO 149A9 | Chrysogena | JX997022 | JX996892 | JX996233 | JX996628 |
| P. chrysogenum | CBS 306.48 (ex-type) | Chrysogena | AF033465 | AY495981 | JX996273 | JN121487 |
| P. chrysogenum | CBS 776.95 | Chrysogena | JX997114 | JX996933 | JX996283 | JX996678 |
| P. chrysogenum | CBS 132217 | Chrysogena | JX996997 | JX996871 | JX996211 | JX996606 |
| P. chrysogenum | CBS 111215 | Chrysogena | JX997070 | JX996922 | JX996266 | JX996661 |
| P. chrysogenum | CBS 259.29 | Chrysogena | JX997089 | JX996924 | JX996270 | JX996665 |
| P. chrysogenum | CBS 906.70 | Chrysogena | JX997117 | JX996934 | JX996284 | JX996679 |
| P. chrysogenum | CBS 282.97 | Chrysogena | JX996925 | JX996271 | JX996666 | |
| P. chrysogenum | CBS 109613 | Chrysogena | KJ866978 | KJ866990 | ||
| P. rubens | CBS 129667 (ex-type) | Chrysogena | JX997057 | JF909949 | JX996263 | JX996658 |
| P. rubens | CBS 339.52 | Chrysogena | JX997098 | JX996929 | JX996277 | JX996672 |
| P. rubens | CBS 111216 | Chrysogena | JX997071 | JX996923 | JX996267 | JX996662 |
| P. rubens | CBS 132210 | Chrysogena | JX996984 | JX996859 | JX996198 | JX996593 |
| P. rubens | CBS 319.59 | Chrysogena | JX997097 | JX996928 | JX996276 | JX996671 |
| P. rubens | CBS 349.48 | Chrysogena | JX997100 | JX996930 | JX996278 | JX996673 |
| P. rubens | CBS 401.92 | Chrysogena | JX997103 | JX996931 | JX996280 | JX996675 |
| P. rubens | CBS 478.84 | Chrysogena | JX997109 | JX996932 | JX996282 | JX996677 |
| P. tardochrysogenum | CBS 132200 (ex-type) | Chrysogena | JX997027 | JX996898 | JX996239 | JX996634 |
| P. cinnamopurpureum | CBS 429.65 (ex-type) | Cinnamopurpurea | EF626950 | EF626948 | EF626949 | JN406533 |
| P. citrinum | CBS 139.45 (ex-type) | Citrina | AF033422 | GU944545 | GU944638 | JF417416 |
| P. cosmopolitanum | CBS 126995 (ex-type) | Citrina | JN617691 | JN606733 | JN606472 | |
| P. cosmopolitanum | CBS 122406 | Citrina | JN606754 | JN606481 | ||
| P. westlingii | CBS 231.28 (ex-type) | Citrina | GU944601 | JN606718 | JN606500 | JN606625 |
| P. westlingii | CBS 127037 | Citrina | JN606720 | JN606496 | ||
| P. westlingii | CBS 127003 | Citrina | JN606711 | JN606490 | ||
| P. sacculum | CBS 231.61 (ex-type) | Eladia | KC411707 | KJ834488 | KU896849 | JN121462 |
| P. consobrinum | CBS 139144 (ex-type) | Exilicaulis | JX140888 | JX141135 | JX157453 | KP064619 |
| P. consobrinum | CV 1457 | Exilicaulis | JX141146 | JX157486 | KP064630 | |
| P. corylophilum | CBS 312.48 (ex-type) | Exilicaulis | AF033450 | JX141042 | KP016780 | KP064631 |
| P. corylophilum | CBS 127808 | Exilicaulis | KP016813 | KP016752 | KP016776 | KP064613 |
| P. albocoremium | CBS 472.84 (ex-type) | Fasciculata | AJ004819 | AY674326 | KU896819 | KU904344 |
| P. allii | CBS 131.89 (ex-type) | Fasciculata | AJ005484 | AY674331 | KU896820 | KU904345 |
| P. aurantiogriseum | CBS 249.89 (ex-type) | Fasciculata | AF033476 | AY674296 | KU896822 | JN406573 |
| P. biforme | CBS 297.48 (ex-type) | Fasciculata | KC411731 | FJ930944 | KU896823 | KU904346 |
| P camemberti | CBS 299.48 (ex-type) | Fasciculata | AB479314 | FJ930956 | KU896825 | |
| P. caseifulvum | CBS 101134 (ex-type) | Fasciculata | KJ834504 | AY674372 | KU896826 | KU904347 |
| P. cavernicola | CBS 100540 (ex-type) | Fasciculata | KJ834505 | KJ834439 | KU896827 | KU904348 |
| P. cavernicola | DTO 046I3 = IBT 25514 | Fasciculata | MN149916 | MN149935 | MN149955 | |
| P. cavernicola | DTO 046I8 = IBT 25513 | Fasciculata | MN149920 | MN149939 | MN149959 | |
| P. cellarum | NRRL 66633 (ex-type) | Fasciculata | KM249068 | KM249108 | KM249117 | |
| P. commune | CBS 311.48 (ex-type) | Fasciculata | AY213672 | AY674366 | KU896829 | KU904350 |
| P. crustosum | CBS 115503 (ex-type) | Fasciculata | AF033472 | AY674353 | DQ911132 | |
| P. crustosum | CV 0241 | Fasciculata | JX091403 | JX091536 | JX141576 | |
| P. crustosum | CV 0251 | Fasciculata | JX091404 | JX091530 | JX141577 | |
| P. cyclopium | CBS 144.45 (ex-type) | Fasciculata | JN942742 | AY674310 | KU896832 | JN985388 |
| P. discolor | CBS 474.84 (ex-type) | Fasciculata | AJ004816 | AY674348 | KU896834 | KU904351 |
| P. discolor | DTO 046I4 = IBT 22523 | Fasciculata | MN149917 | MN149936 | MN149956 | |
| P. discolor | DTO 047A2 = IBT 5736 | Fasciculata | MN149922 | MN149941 | MN149961 | |
| P. discolor | DTO 047A3 = IBT 5744 | Fasciculata | MN149923 | MN149942 | MN149962 | |
| P. echinulatum | CBS 101027 | Fasciculata | AY674342 | |||
| P. echinulatum | CBS 317.48 (ex-type) | Fasciculata | AF033473 | AY674341 | DQ911133 | KU904352 |
| P. echinulatum | CBS 337.59 | Fasciculata | KC411742 | AY674340 | ||
| P. echinulatum | DTO 228I4 | Fasciculata | MN149925 | MN149944 | MN149964 | |
| P. freii | CBS 476.84 (ex-type) | Fasciculata | JN942696 | AY674290 | KU896836 | JN985430 |
| P. gladioli | CBS 332.48 (ex-type) | Fasciculata | AF033480 | AY674287 | KU896837 | JN406567 |
| P. hirsutum | CBS 135.41 (ex-type) | Fasciculata | AY373918 | AF003243 | KU896840 | JN406629 |
| P. hordei | CBS 701.68 (ex-type) | Fasciculata | AJ004817 | AY674347 | KU896841 | KU904355 |
| P. melanoconidium | CBS 115506 (ex-type) | Fasciculata | AJ005483 | AY674304 | KU896843 | KU904358 |
| P. neoechinulatum | CBS 169.87 (ex-type) | Fasciculata | JN942722 | AF003237 | KU896844 | JN985406 |
| P. nordicum | ATCC 44219 (ex-type) | Fasciculata | KJ834513 | KJ834476 | KU896845 | KU904359 |
| P. palitans | CBS 107.11 (ex-type) | Fasciculata | KJ834514 | KJ834480 | KU896847 | KU904360 |
| P. palitans | DTO 046I5 | Fasciculata | MN149918 | MN149937 | MN149957 | |
| P. polonicum | CBS 222.28 (ex-type) | Fasciculata | AF033475 | AY674305 | KU896848 | JN406609 |
| P. radicicola | CBS 112430 (ex-type) | Fasciculata | KJ834516 | AY674357 | ||
| P. robsamsonii | CBS 140573 (ex-type) | Fasciculata | KU904339 | KT698885 | KT698894 | KT698904 |
| P. solitum | CBS 146.86 | Fasciculata | AY674356 | |||
| P. solitum | CBS 147.86 | Fasciculata | HQ225713 | AY674355 | ||
| P. solitum | CBS 424.89 (ex-type) | Fasciculata | AY373932 | AY674354 | KU896851 | KU904363 |
| P. solitum | DTO 046I6 = IBT 22216 | Fasciculata | MN149919 | MN149938 | MN149958 | |
| P. solitum | DTO 161H9 | Fasciculata | MN149924 | MN149943 | MN149963 | |
| P. solitum | DTO 234I5 | Fasciculata | MN149926 | MN149945 | MN149965 | |
| P. solitum | DTO 235G1 | Fasciculata | KJ775670 | KJ775163 | MN149946 | MN149966 |
| P. solitum | DTO 247B8 | Fasciculata | MN149927 | MN149947 | MN149967 | |
| P. solitum | DTO 321F7 | Fasciculata | MN149928 | MN149948 | MN149968 | |
| P. solitum | DTO 376D5 | Fasciculata | MN149930 | MN149950 | ||
| P. speluncae | CBS 271.97 | Fasciculata | AY674350 | MN170734 | ||
| P. speluncae | CBS 278.97 | Fasciculata | AY674349 | MN170735 | ||
| P. speluncae | DAOMC 251696 | Fasciculata | MG490864 | MG490884 | MG490954 | MN170736 |
| P. speluncae | DAOMC 251697 | Fasciculata | MG490865 | MG490885 | MG490955 | MN170737 |
| P. speluncae | DAOMC 251698 | Fasciculata | MG490866 | MG490886 | MG490956 | MN170738 |
| P. speluncae | DAOMC 251699 | Fasciculata | MG490867 | MG490887 | MG490957 | MN170739 |
| P. speluncae | DAOMC 251700 | Fasciculata | MG490868 | MG490888 | MG490958 | MN170740 |
| P. speluncae | DAOMC 251701 (ex-type) | Fasciculata | MG490869 | MG490889 | MG490959 | MN170741 |
| P. speluncae | DAOMC 252126 | Fasciculata | MG490877 | MG490928 | MG490967 | MN170742 |
| P. speluncae | DAOMC 252127 | Fasciculata | MG490880 | MG490940 | MG490970 | MN170743 |
| P. speluncae | CBS 551.95 = DTO 037C9 | Fasciculata | MN149912 | MN149931 | MN149951 | |
| P. speluncae | CBS 112559 = DTO 037D2 | Fasciculata | MN149913 | MN149932 | MN149952 | |
| P. speluncae | CBS 112569 = DTO 046G4 | Fasciculata | MN149914 | MN149933 | MN149953 | |
| P. speluncae | CBS 112568 = DTO 046G5 | Fasciculata | MN149915 | MN149934 | MN149954 | |
| P. speluncae | DTO 046I9 = IBT 22369 | Fasciculata | MN149921 | MN149940 | MN149960 | |
| P. speluncae | DTO 332H8 | Fasciculata | MN149929 | MN149949 | MN149973 | |
| P. thymicola | CBS 111225 (ex-type) | Fasciculata | KJ834518 | AY674321 | FJ530990 | KU904364 |
| P. tricolor | CBS 635.93 (ex-type) | Fasciculata | JN942704 | AY674313 | KU896852 | JN985422 |
| P. tulipae | CBS 109555 (ex-type) | Fasciculata | KJ834519 | AY674344 | ||
| P. venetum | IBT 10661 (ex-type) | Fasciculata | AJ005485 | AY674335 | KU896855 | KU904366 |
| P. verrucosum | CBS 603.74 (ex-type) | Fasciculata | AY373938 | AY674323 | DQ911138 | JN121539 |
| P. viridicatum | CBS 390.48 (ex-type) | Fasciculata | AY373939 | AY674295 | KU896856 | KY989209 |
| P. fractum | CBS 124.68 (ex-type) | Fracta | KC411674 | KJ834452 | JN121441 | |
| P. gracilentum | CBS 599.73 (ex-type) | Gracilenta | KC411768 | KJ834453 | JN121537 | |
| P. javanicum | CBS 341.48 (ex-type) | Lanata-Divaricata | GU981613 | GU981657 | KF296387 | JN121498 |
| P. ochrosalmoneum | CBS 489.66 (ex-type) | Ochrosalmonea | EF626961 | EF506212 | EF506237 | JN121524 |
| P. osmophilum | CBS 462.72 (ex-type) | Osmophila | EU427295 | AY674376 | KU896846 | JN121518 |
| P. paradoxum | CBS 527.65 (ex-type) | Paradoxa | EF669707 | EF669683 | EF669692 | EF669670 |
| P. expansum | CBS 325.48 (ex-type) | Penicillium | AY373912 | AY674400 | DQ911134 | JF417427 |
| P. expansum | CV 2860 | Penicillium | FJ230989 | JX091539 | JX141580 | |
| P. expansum | CV 2861 | Penicillium | FJ230990 | JX091540 | JX141581 | |
| P. expansum | CBS 481.84 | Penicillium | AY674399 | |||
| P. expansum | CBS 281.97 | Penicillium | AY674401 | |||
| P. cyaneum | CBS 315.48 (ex-type) | Ramigena | AF033427 | JX091552 | JN406575 | |
| P. soppii | CBS 226.28 (ex-type) | Ramosa | AF033488 | DQ285616 | KJ867002 | JN406606 |
| P. brevistipitatum | AS 3.6887 (ex-type) | Robsamsonia | DQ221696 | DQ221695 | KU896824 | JN406528 |
| P. concentricum | CBS 477.75 (ex-type) | Robsamsonia | KC411763 | AY674413 | DQ911131 | KT900575 |
| P. concentricum | CBS 191.88 | Robsamsonia | AY674412 | |||
| P. robsamsonii | CBS 140573 (ex-type) | Robsamsonia | KU904339 | KT698885 | KT698894 | KT698904 |
| P. roqueforti | CBS 221.30 (ex-type) | Roquefortorum | EU427296 | AF000303 | HQ442332 | JN406611 |
| P. sclerotiorum | CBS 287.36 (ex-type) | Sclerotiora | JN626132 | JN626001 | JN626044 | JN406585 |
| P. stolkiae | CBS 315.67 (ex-type) | Stolkia | AF033444 | JN617717 | AF481135 | JN121488 |
| P. glaucoalbidum | WCN 1129 | Thysanophora | AB175275 | |||
| P. glaucoalbidum | WCN 1128 | Thysanophora | AB175273 | |||
| P. glaucoalbidum | WCN 1043 | Thysanophora | AB175259 | |||
| P. glaucoalbidum | WCN 1246 | Thysanophora | AB175268 | |||
| P. glaucoalbidum | WCN 1016 | Thysanophora | AB175254 | |||
| P. glaucoalbidum | CBS 314.56 | Thysanophora | AB213277 | |||
| P. glaucoalbidum | CBS 348.64 | Thysanophora | AB213275 | |||
| P. glaucoalbidum | WCN 1152 | Thysanophora | AB175272 | |||
| P. glaucoalbidum | WCN 1077 | Thysanophora | AB175262 | |||
| P. glaucoalbidum | NBRC 9011 | Thysanophora | AB213279 | |||
| P. hennebertii | CBS 334.68 (ex-type) | Thysanophora | KJ834507 | KJ834454 | JN121493 | |
| P. taxi | CBS 206.57 (ex-type) | Thysanophora | KJ834517 | KJ834495 | JN121454 | |
| P. lagena | CBS 185.65 (ex-type) | Torulomyces | KF303665 | KF303619 | KF303634 | JN121450 |
| P. turbatum | CBS 383.48 (ex-type) | Turbata | AF034454 | KJ834499 | KU896853 | JN406556 |
| Talaromyces pinophilus (outgroup) | CBS 631.66 (ex-type) | JN899382 | JX091381 | KF741964 | KM023291 |
AS: Internal culture collection at CGMCC, China General Microbiological Culture Collection Centre, Beijing, China; ATCC: American Type Culture Collection, Manassas, VA, USA; CBS: Culture collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, Netherlands; CV: Working collection of Prof Karin Jacobs, from the Dept of Microbiology, Stellenbosch University, Stellenbosch, South Africa; DAOMC: Culture collection of the National Mycological Collections, Agriculture & Agri-Food Canada, Ottawa, Canada; DTO: Internal culture collection of Westerdijk Fungal Biodiversity Institute; IBT: Culture collection of Center for Microbial Biotechnology (CMB) at Department of Systems Biology, Technical University of Denmark; NRBC: Culture collection of the National Institute of Technology and Evaluation, Tokyo, Japan; NRRL: ARS Culture Collection, U.S. Department of Agriculture, Peoria, Illinois, USA; WCN: Working collection of Susumu Iwamoto, Tokyo, Japan.
Morphology
Morphological characters were captured using standardized protocols proposed by Visagie et al. (2014b). Colony characters were captured on Czapek yeast autolysate agar (CYA), MEAbl, yeast extract sucrose agar (YES), oatmeal agar (OA) and creatine sucrose agar (CREA). Strains were inoculated in a three-point pattern on these media in 90 mm Petri dishes. Plates were incubated for 7 d at 25 °C in darkness in perforated plastic bags. Colour names and codes used in descriptions are from Kornerup & Wanscher (1967). Microscopic observations were made using an Olympus SZX12 dissecting microscope and Olympus BX50 compound microscope equipped with Infinity3 and InfinityX cameras driven by Infinity Analyze v. 6.5.1 software (Lumenera Corp., Ottawa, Canada). Colonies were captured with a Sony NEX-5N camera. Plates were prepared in Affinity Photo v. 1.6.6 [Serif (Europe) Ltd, Nottingham, UK]. For aesthetic purposes, micrographs were adjusted using the “inpainting brush tool” without altering areas of scientific significance. Line drawings were prepared in Affinity Photo v. 1.7.1 [Serif (Europe) Ltd, Nottingham, UK] running on an iPad Pro with an Apple Pencil.
Extrolites
For extrolite analyses, all strains were grown in 9 cm polystyrene Petri dishes on CYA (Pitt 1980) and YES (Frisvad 1981, Filtenborg et al. 1990) incubated at 25 °C for 14 d. Six agar plugs from each fungal isolate were excised with a sterilized 7 mm cork-borer and transferred to a 13 mL polypropylene tube. Two mL of ethyl acetate was then added and vortexed for 30 s, followed by sonication at 30 °C for 30 min and vortexed again for 30 s. The supernatants were transferred into new polypropylene tubes and dried on a centrifugal vacuum concentrator at 35 °C. Extracts were then reconstituted in 1 mL of methanol:water (8:2) and filtered into 2 mL amber glass HPLC vials using a 0.45 µm PVDF syringe filter. Extracts were immediately stored at -20 °C until analysis by liquid chromatography mass spectrometry (LC-MS). Extracts were analyzed in both positive and negative polarities using a Q-Exactive Orbitrap coupled to an Agilent 1290 HPLC. The chemical formula of observed extrolites were determined with Xcalibur® software using accurate mass measurements (< 3.0 ppm) and manually verified by isotopic pattern. The chemical formulae were then searched against microbial extrolite databases [AntiBase2013 (Wiley-VCH, Weinheim, Germany)] and KNApSAcK (Afendi et al. 2012) and putative matches were scrutinized by comparing their MS/MS fragmentation with those published in the literature or predicted by CFM-ID (Allen et al. 2014).
RESULTS
Sampling, isolations & identifications
During the survey, 70 Penicillium strains were isolated from six different caves in New Brunswick and one in Quebec, Canada. Eight strains of the new Penicillium species were isolated from walls of the Glebe Mine and White Cave in New Brunswick and Grotte à la Patate in Quebec, and three strains were isolated from a deer mouse (Peromyscus maniculatus) and its dung from the Dorchester Mine in New Brunswick (Table 1). Based on the BenA phylogeny (Fig. 1), and in some cases additional ITS and CaM BLAST searches, the remaining strains were identified as Penicillium bialowiezense (n = 10), P. brevistipitatum (n = 6), P. chrysogenum (n = 2), P. concentricum (n = 14), P. consobrinum (n = 2), P. corylophilum (n = 4), P. expansum (n = 9), P. glabrum (n = 3), P. glaucoalbidum (n = 4), P. rubens (n = 4), P. spathulatum (n = 2), and P. westlingii (n = 2).
Fig. 1.
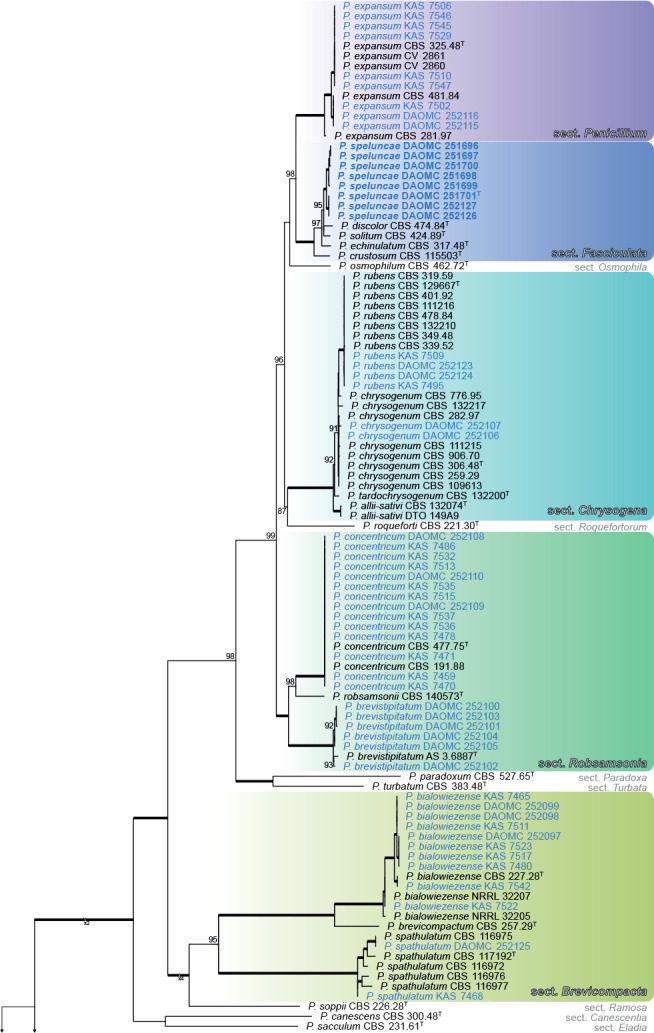
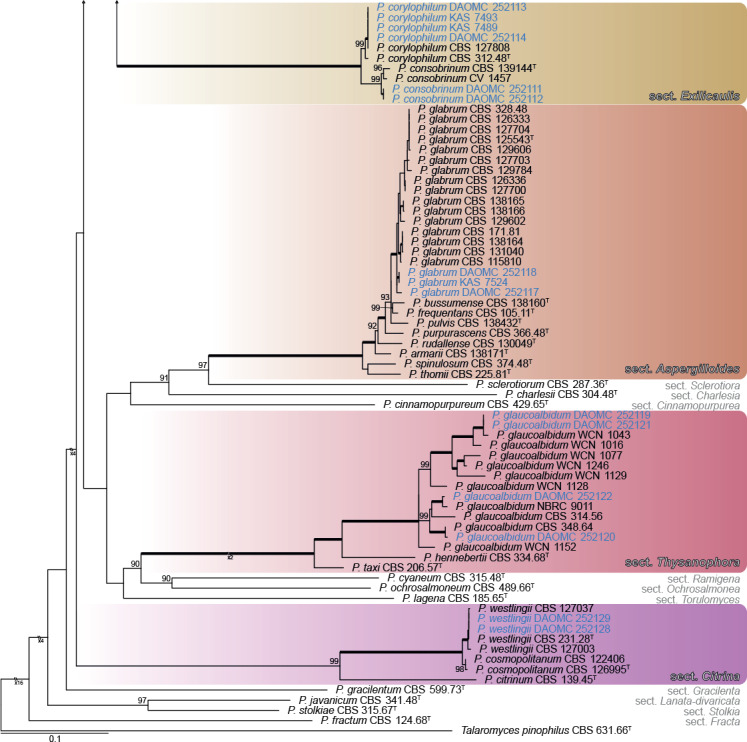
ML tree based on ITS, BenA, CaM & RPB2 showing identities and diversity of Penicillium associated with bats or bat caves. Bootstrap values ≥ 80% are shown above branches while thickened branches indicate 100 % support. Sequences obtained from ex-type cultures are indicated by T. Sequences obtained from strains during this study are indicated by blue text, while the new species, P. speluncae, is in bold blue text. The tree was rooted to Talaromyces pinophilus.
Phylogeny
A multigene phylogeny was used to show identities of strains isolated during this study (Fig. 1). The alignment contained 175 taxa and was 2 375 bp long (BenA 1–453; CaM 454–1019; RPB2 1020–1823; ITS 1824–2375). The most appropriate substitution model for each partition was: BenA TIM2e+I+G4; CaM TNe+I+G4; RPB2 TNe+R3; ITS TIM2+F+I+G4. Generally, BenA sequences from strains isolated during this study matched well with reference sequences in terms of resolving in a particular clade. However, many of the newly generated sequences represented minor deviations from previously known sequences. In some cases, variation was such that calmodulin was sequenced to make a final identification of a species (e.g. P. consobrinum, P. brevistipitatum). One clade was found to represent a new species in section Fasciculata.
To demonstrate the genealogical concordance of the new species in relation to its close relatives, phylogenies of all known species from section Fasciculata were calculated based on BenA, CaM and RPB2 (Fig. 2). To demonstrate the overall phylogenetic relationship, a concatenated dataset, based on ITS, BenA, CaM and RPB2 was calculated. Alignment metadata is summarised in Suppl. Table S2.
Fig. 2.
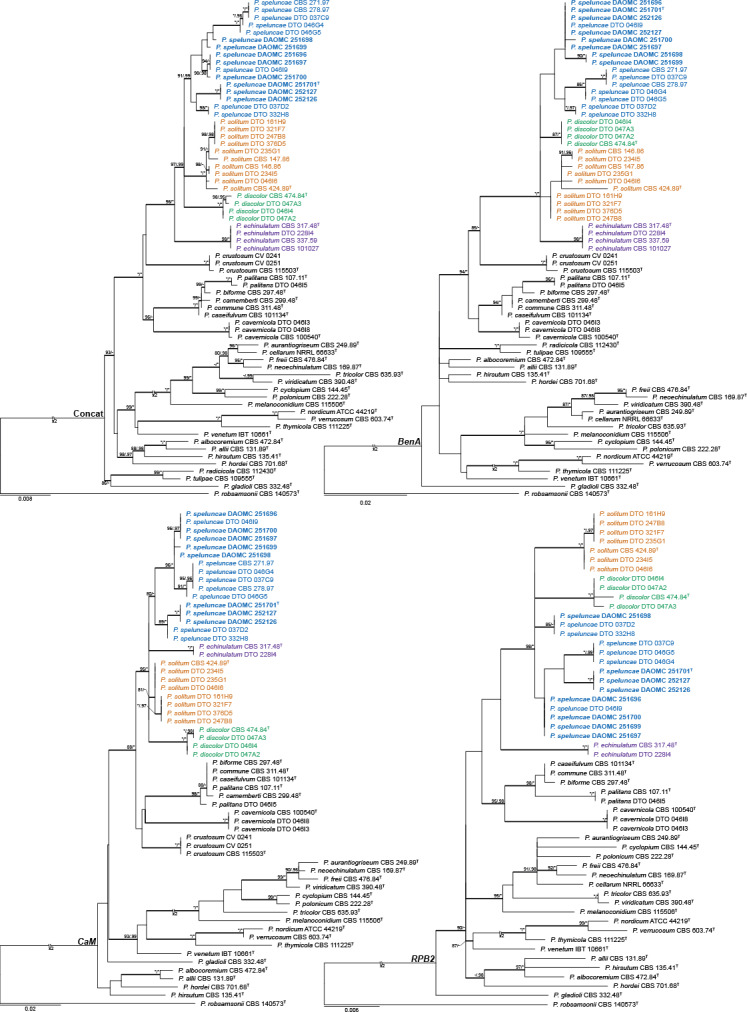
ML trees of Penicillium section Fasciculata, based on concatenated, BenA, CaM and RPB2 alignments, showing the relationship of P. speluncae within the section. PP and BS values ≥ 0.95/80 are shown above thickened branches (* = 1.00/100; - = <0.95/80). Sequences obtained from ex-type cultures are indicated by T. Strains of the new species characterised based on morphology and extrolites is indicated by bold blue text. Trees were rooted to Penicillium robsamsonia.
Table. S2.
Supplementary Table S2. Metadata related to the phylogenetic analysis of sect. Fasciculata.
| Dataset | Nr of taxa | Bp length | Partition scheme |
|---|---|---|---|
| BenA | 67 | 406 | 1st, 2nd & 3rd codon positions (HKY+G) |
| CaM | 60 | 500 | 1st, 2nd & 3rd codon positions (SYM+G) |
| ITS | 44 | 505 | 1st, 2nd & 3rd codon positions (GTR+I) |
| RPB2 | 54 | 937 | 1st codon position (F81), 2nd codon position (HKY+G), 3rd codon position (GTR+I) |
| Concat | 67 | 2348 | 1st, 2nd & 3rd codon positions of ITS (GTR+I), 1st codon position of BenA (SYM+I); 2nd & 3rd codon positions of CaM & BenA (SYM+G); 1st codon position of CaM (K80+I); 1st codon position of RPB2 (F81); 2nd codon position of RPB2 (HKY+G); 3rd codon position of RPB2 (GTR+I) |
As expected, ITS (not shown) lacked sufficient variation to distinguish among species. For example, P. speluncae shared similar ITS sequences with P. cavernicola, P. echinulatum, P. discolor and P. solitum, noting that strains DAOMC 251696 and DAOMC 251697 formed a distinct clade because of an A-T transversion. BenA, CaM and RPB2 distinguished among close relatives much better. The exception was the clade associated with cheese; P. biforme, P. camemberti (1 bp difference), P. caseifulvum, P. commune and P. palitans had identical CaM sequences, while P. camemberti had 1 bp difference from these species. RPB2 sequences for P. caseifulvum and P. commune were identical (albeit with limited sampling). BenA was not helpful to distinguish between P. camemberti and P. commune, supporting the hypothesis that the former is a domesticated form of the latter (Pitt et al. 1986, Polonelli et al. 1987). Much sequence variation was observed within the clade containing P. speluncae, P. discolor, P. echinulatum and P. solitum. This resulted in all phylogenies having poor backbone support in both ML and BI, mainly because of the strains identified as P. speluncae. Nonetheless, all phylogenies resulted in three distinct clades corresponding with P. discolor, P. solitum and P. echinulatum. CBS 271.97 and CBS 278.97 previously considered typical of P. discolor (Frisvad & Samson 2004, Samson et al. 2004) were phylogenetically resolved distinct from the ex-type CBS 474.84T within the broad concept applied to P. speluncae.
Extrolites
As analysed by LC-MS, there were five classes of compounds produced by P. speluncae under the reported growth conditions: cyclopenins, viridicatins, chaetoglobosins, cyclic dipeptides, and tetrapeptides. Cyclopenins and viridicatins are derived from a shared biosynthetic pathway (Simonetti et al. 2016) and are among the most widely distributed extrolites across species in Penicillium subgenus Penicillium (Frisvad et al. 2004). Chaetoglobosins are a large class of metabolites biosynthesised by a polyketide derived macrocycle fused to a modified tryptophan amino acid and are produced by Chaetomium globosum as well as P. discolor (Frisvad et al. 1997), P. expansum (Frisvad & Filtenborg 1989) and P. marinum (Frisvad et al. 2004). One of the major chaetoglobosins produced by these isolates is a newly described natural product, tetrahydrochaetoglobosin (Walsh et al. 2018). In addition to these extrolites, a series of cyclic and linear tetrapeptides, composed of combinations of valine, phenylalanine, leucine/isoleucine, tyrosine and tryptophan were consistently detected across all tested strains (Table 2). These peptides could be putatively characterized by de novo sequencing and are likely similar to the series of linear and cyclic tetra peptides previously identified in cultures of P. chrysogenum, including fungisporin; cyclo(D-Phe-L-Phe-D-Val-L-Val) (Ali et al. 2014).
Table 2.
Extrolites produced by Penicillium speluncae.
| Extrolite name | Formula | m/z [M+H]+ | RT | % strains producing |
|---|---|---|---|---|
| cyclopenin | C17H14N2O3 | 295.1076 | 3.03 | 86 % |
| cyclopenol | C17H14N2O4 | 311.1025 | 2.69 | 86 % |
| cyclopeptine | C17H16N2O2 | 281.1285 | 3.11 | 100 % |
| dehydrocyclopeptine | C17H14N2O2 | 279.1130 | 3.17 | 86 % |
| viridicatin | C15H11NO2 | 238.0865 | 3.36 | 100 % |
| viridicatol | C15H11NO3 | 254.0812 | 2.95 | 100 % |
| chaetoglobosin F | C32H38N2O5 | 531.2852 | 3.54 | 100 % |
| tetrahydrochaetoglobosin A | C32H40N2O5 | 533.3009 | 3.27 | 100 % |
| chaetoglobosin A | C32H38N2O5 | 531.2852 | 3.63 | 100 % |
| chaetoglobosin C | C32H36N2O5 | 529.2698 | 3.78 | 57 % |
| prochaetoglobosin I | C32H38N2O2 | 483.3005 | 4.41 | 100 % |
| cyclo(VP) | C10H16N2O2 | 197.1286 | 2.32 | 100 % |
| cyclo(LP) | C11H18N2O2 | 211.1441 | 2.50 | 100 % |
| cyclo(IP) | C11H18N2O2 | 211.1443 | 2.55 | 100 % |
| cyclo(FP) | C14H16N2O2 | 245.1285 | 2.62 | 100 % |
| fungisporin | C28H36N4O4 | 493.2809 | 3.77 | 100 % |
| cyclo(Phe-Val-Phe-Val) | C28H36N4O4 | 493.2809 | 3.55 | 100 % |
| Val-Phe-Val-Phe | C28H38N4O5 | 511.2919 | 2.87 | 100 % |
| cyclo(Phe-Phe-Val-Ile) | C29H38N4O4 | 507.2360 | 4.00 | 100 % |
| Phe-Val-Ile-Phe | C29H4N4O5 | 525.3074 | 2.96 | 100 % |
| cyclo(Phe-Tyr-Val-Val) | C28H36N4O5 | 509.2761 | 3.42 | 100 % |
| Phe-Val-Val-Tyr | C28H38N4O6 | 527.2866 | 2.68 | 100 % |
| Phe-Ile-Val-Tyr | C29H40N4O6 | 541.3022 | 2.74 | 100 % |
| cyclo(Phe-Trp-Val-Val) | C30H37N5O4 | 532.2914 | 3.68 | 100 % |
| Phe-Val-Val-Trp | C30H39N5O5 | 550.3024 | 2.89 | 100 % |
| cyclo(Tyr-Trp-Val-Val) | C30H37N5O5 | 548.2866 | 3.39 | 100 % |
| Tyr-Val-Val-Trp | C30H39N5O6 | 566.2973 | 2.68 | 100 % |
| cyclo (Trp-Trp-Val-Val) | C32H38N6O4 | 571.3025 | 3.62 | 100 % |
Taxonomy
Penicillium speluncae Visagie & Yilmaz, sp. nov. MycoBank MB828614. Figs 3, 4.
Fig. 3.
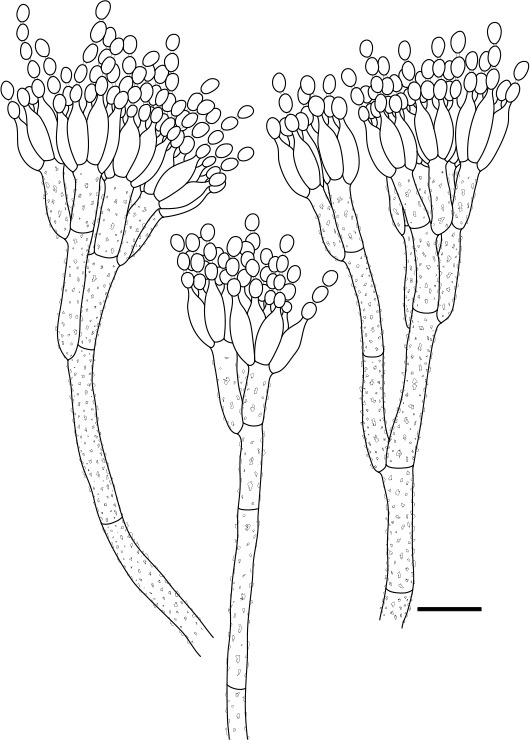
Line drawing ofPenicillium speluncae. Scale bar = 10 µm.
Fig. 4.
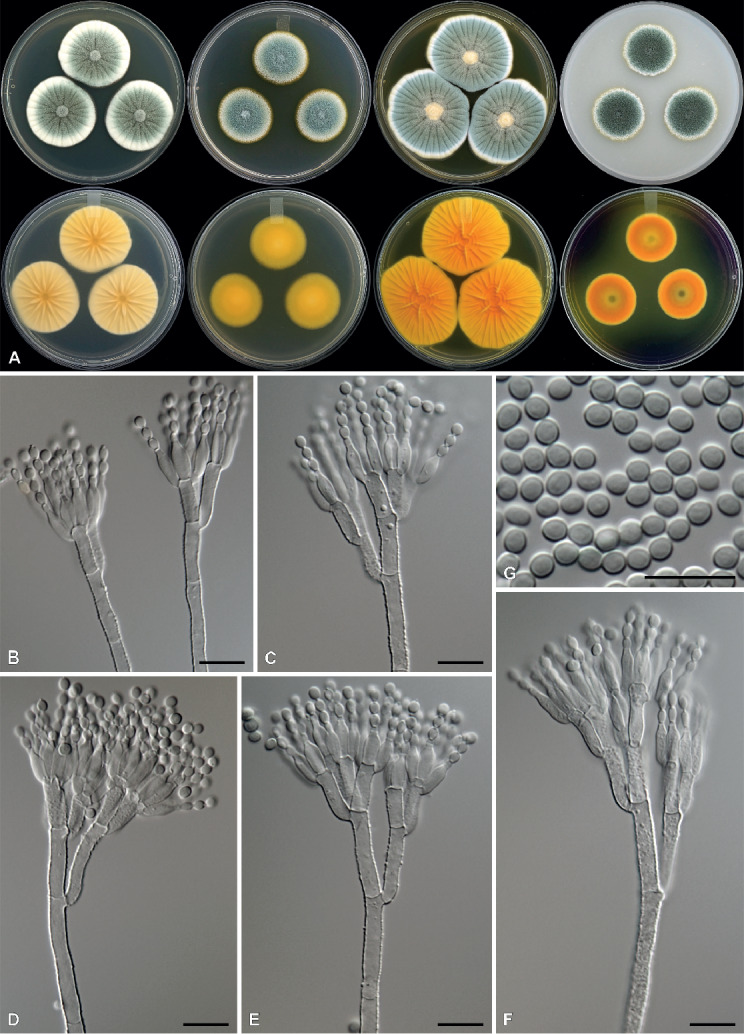
Penicillium speluncae. A. Colonies, from left to right, top row: CYA, MEA, YES, OA; bottom row: reverse on CYA, MEA, YES, CREA. B–F. Conidiophores. G. Conidia. Scale bars = 10 μm.
Etymology: Latin, speluncae, meaning from a cave.
ITS barcode: MG490869. Alternative identification markers: BenA = MG490889, CaM = MG490959, RPB2 = MN170741.
Colony diam, 7 d (at 25 °C; in mm): CYA 30–35; CYA 15 °C (12–) 17–22(–25); CYA 30 °C 12–21(–26); CYA 37°C no growth; CYAS 29–32(–37); MEAbl 25–30; YES 45–47; OA 25–27; CREA 25–26.
Colony characters: CYA 25 °C, 7 d: Colonies moderately deep, sulcate; margins low, narrow to wide, entire; mycelia white; texture velutinous to fasciculate; sporulation moderately dense, conidia en masse greyish green (25E7), dull green (26D3–4); soluble pigments absent; exudates absent; reverse greyish yellow (4B6), greyish orange (5B4), yellowish white (4A2). MEA 25 °C, 7 d: Colonies low, plain; margins low, narrow, entire; mycelia white; texture velutinous to fasciculate; sporulation moderately dense, conidia en masse greyish green (25E5–26E5); soluble pigments forming a yellow halo surrounding colony; exudates absent; reverse greyish yellow (4B6), light yellow (3A5), greyish green (29C6). YES 25 °C, 7 d: Colonies low, sulcate; margins low, wide, entire; mycelia white; texture velutinous to fasciculate; sporulation moderately dense, conidia en masse greyish green (25C5–D5), dull green (26D3); soluble pigments absent; exudates absent; reverse orange yellow to orange (4A7–6A7). OA 25°C, 7 days: Colonies moderately deep, plain; margins low, narrow, entire; mycelia white; texture fasciculate; sporulation dense, conidia en masse greyish to dark green (25E7–F7); soluble pigments forming a yellowish halo surrounding colony; exudates absent. CREA 25 °C, 7 d: Growth strong, acid produced, colony reverse orange.
Micromorphology: Conidiophores terverticillate, minor proportion bi- and quarterverticillate; stipes rough, 180–600 × 3.5–4.5 μm; branches 15–29 μm; metulae (2–)3–4, 10–16 × 3–4.5 μm; phialides ampulliform, 4–6 per metula, 8.5–11 × 3–4 μm (9.9±0.7 × 3.3±0.2); average length metula/phialide 1.3; conidia smooth, broadly ellipsoidal, 3–4 × 2.5–3.5 μm (3.6±0.2 × 3±0.2), average width/length = 0.82, n = 72.
Extrolites: cyclopenins, viridicatins, chaetoglobosins, fungisporin, cyclic and linear tetrapeptides (See Table 2).
Typus: Canada, New Brunswick, Dorchester, Dorchester mine, from a swab of deer mouse fur (live Peromyscus maniculatus), 14 Mar. 2014, K. Vanderwolf (holotype, DAOM 745788 (dried culture); ex-type strain DAOMC 251701 = KAS 7512 = P06201).
Notes: Penicillium speluncae is resolved in a clade with P. discolor, P. echinulatum and P. solitum (Fig. 2). Of these, P. speluncae showed relatively good growth on CYA at 30 °C, compared to poor growth observed for the others. Both P. discolor and P. echinulatum produce roughened globose to subglobose conidia, in contrast to the new species’ smooth, broadly ellipsoidal conidia. Penicillium solitum is morphologically most similar to the new species. Both species have smooth conidia and produce a striking yellow orange reverse on YES. However, P. speluncae produces broadly ellipsoidal conidia (globose to subglobose in P. solitum), grows faster on YES compared to P. solitum (45–47 mm vs 25–39 mm) and has the ability to grow on CYA at 30 °C. Penicillium solitum has several synonyms examined before (Frisvad & Samson 2004), and showed no growth on CYA at 30 °C. Of the extrolites produced in this clade, chaetoglobosins are produced by only P. speluncae and P. discolor, territrems only by P. echinulatum, compactin only by P. solitum, while penitrem and roquefortine are produced by P. crustosum and other distantly related Penicillia. Penicillium speluncae produces cytoglobosin and prochaetoglobosin, which are absent in P. discolor, while palitantin was not detected for the new species (comparisons summarised in Table 3; data from Frisvad et al. 2004).
Table 3.
Distinguishing features of species closely related to Penicillium speluncae.
| Conidia | CYA texture | YES soluble pigment | Yes reverse | Chaetoglobosins | Compactin | Penitrem | Roquefortine | Cytoglobosin | Prochaetoglobosin | Palitantin | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| P. speluncae | Smooth, broadly ellipsoidal | Velutinous to fasciculate | None | Orange yellow to orange | + | - | - | - | + | + | - |
| P. crustosum | Smooth, globose to subglobose | Velutinous to weakly fasciculate, becoming crustose | Pale brown or none | Strongly yellow | - | - | + | + | - | - | - |
| P. discolor | Rough, globose to subglobose | Velutinous to fasciculate | Brilliant red diffusible colour on YES | Orange turning into deep red with age | + | - | - | - | - | - | + |
| P. echinulatum | Rough, globose to subglobose | Velutinous to weakly fasciculate | None | yellow | - | - | - | - | - | - | + |
| P. solitum | Smooth to finely rough, globose to subglobose | Velutinous | None | yellow to orange | - | + | - | - | - | - | + |
Additional materials examined: Canada, New Brunswick, Dorchester, Dorchester copper mine, from rodent fur (Peromyscus maniculatus), 12 Mar. 2014, K. Vanderwolf (culture DAOMC 252126 = KAS 7516 = P01202); Dorchester mine, from rodent dung (Peromyscus maniculatus), 25 Mar. 2014, K. Vanderwolf (culture DAOMC 252127 = KAS 7533 = D3108); Hillsborough, White Cave (gypsum), from cave wall, 21 Apr. 2015, K. Vanderwolf (cultures DAOMC 251698 = KAS 7500 = W07302, DAOMC 251699 = KAS 7503 = W05404); Sussex, Glebe mine (limestone), from cave wall, 16 Apr. 2015, K. Vanderwolf (culture DAOMC 251700 = KAS 7504 = W05202); Quebec, Anticosti Island, Grotte à la Patate (limestone), from cave wall, K. Vanderwolf (cultures DAOMC 251696 = KAS 7473 = W54119, DAOMC 251697 = KAS 7474 = W54102).
DISCUSSION
This study focused on Penicillium species isolated from six Pdpositive bat hibernacula in New Brunswick, Canada and one Pdnegative bat hibernaculum in Quebec, Canada. The isolates were collected from arthropods, bats, rodents and their dung (i.e. the deer mouse Peromyscus maniculatus), cave walls, and one dead bat found in a parking garage. During the survey, hundreds of fungal strains were obtained and Penicillium represented one of the most frequently isolated genera, probably because of the ability of these species to grow at low temperatures (Frisvad & Samson 2004, Vanderwolf et al. 2016). Previous studies had similar results, but the diversity of Penicillium in caves is even greater than previously reported and several of these species have never been reported from caves or mines, including P. bialowiezense, P. brevistipitatum, P. consobrinum, P. rubens, P. spathulatum and P. westlingii (Nováková 2009, 2018, Vanderwolf et al. 2013b, 2016, Anelli et al. 2018). Other species identified were P. chrysogenum, P. concentricum, P. corylophilum, P. expansum, P. glabrum and P. glaucoalbidum. One species could not be identified based on DNA reference sequences and further study showed it to represent a new species, described above as P. speluncae, classified in section Fasciculata.
Phylogenetic analyses of sect. Fasciculata revealed a large degree of genetic variation within P. speluncae. Single gene trees based on BenA, CaM and RPB2 resulted in inconsistent groupings and poor backbone support for this clade meaning that genealogical concordance could not be applied to delimit segregate species. The basal branch encompassing this clade was relatively well supported in the concatenated tree. DAOMC strains were characterized based on morphology and extrolite data, with very few differences noted. For example, colony growth rates varied on CYA at 30 °C and CYAS, but a similar variation was previously observed in P. solitum (Frisvad & Samson 2004). Extrolite data also distinguish among this group of species. Chaetoglobosins are produced only by P. speluncae and P. discolor, while the former produces cytoglobosin and prochaetoglobosin, which are absent in P. discolor. Chaetomium globosum is the best-known producer of chaetoglobosins, including the major chaetoglobosins A, C, and F, also shown here to be produced by P. speluncae. A distinguishing feature between chaetoglobosin production by C. globosum and P. speluncae is a newly described natural product, tetrahydrochaetoglobosin A (Walsh et al. 2019), which was not observed in C. globosum. Our data hint that P. speluncae may be a species complex with so far cryptic species that may be resolved with additional data. Considering the available data, we conservatively propose the name P. speluncae for this clade. In principle, an analogous situation occurred with P. glabrum (sect Aspergilloides). This complex was studied several times morphologically but a satisfactory conclusion was never found (Pitt et al. 1990). Houbraken et al. (2014) provided an extensive phylogenetic analysis and distinguished between P. glabrum and P. frequentans using a concatenated phylogeny of BenA, CaM and RPB2, even though these species had poor backbone support in the single gene trees for BenA and CaM and could not be distinguished.
Even though we adopt a consilient species concept for Penicillium, there is often bias towards DNA sequences for making a species identification or deciding whether strains are new or not. This situation is a direct consequence of the accepted species list and associated ex-type reference sequences published by Visagie et al. (2014b) and resulted in a generally aggressive approach to describing new species or reinstating old names. Many of the resulting taxa are often based on a single strain. This in turn complicates sequence-based identifications because the reference data do not encapsulate infraspecies variation. We thus encourage a more holistic approach to introducing new species, noting that singleton species will always be a part of our science. An example is P. brevistipitatum, which before this study was known only from ex-type sequences. BenA sequences obtained from our strains differed at several nucleotide positions and only after CaM was sequenced could we identify strains as this species. The additional reference sequences generated here will thus aid future identifications of P. brevistipitatum. Several new genotypes were also discovered for P. bialowiezense, P. consobrinum, P. glabrum, P. glaucoalbidum and P. spathulatum (Fig. 1). Several strains were identified as P. glaucoalbidum (≡ Thysanophora glaucoalbida). Although this species is often encountered as an endophyte of conifer needles, the name is not currently accepted because no type material is available (Visagie et al. 2014b). Lectotypification is complicated by the large degree of variation observed in available sequences (Iwamoto et al. 2005); this will be the focus of a future study.
ACKNOWLEDGEMENTS
We acknowledge DNA sequencing support from Lisa James of the Molecular Technologies Laboratory of ORDC, AAFC, Ottawa, and research support from AAFC project J-1848. We also acknowledge Martin Meijer for sequencing support provided at the Westerdijk Fungal Biodiversity Institute. This manuscript was written while the first author worked at ORDC, and was subsequently first submitted while he worked at the Agricultural Research Council, South Africa.
Supplementary Material: http://fuse-journal.org/
REFERENCES
- Afendi FM, Okada T, Yamazaki M, et al. (2012). KNApSAcK Family Databases: Integrated Metabolite–Plant Species Databases for Multifaceted Plant Research. Plant and Cell Physiology 53: e1. [DOI] [PubMed] [Google Scholar]
- Akaike H. (1974). A new look at the statistical model identification. IEEE Transations on Automatic Control 19: 716–723. [Google Scholar]
- Ali H, Ries MI, Lankhorst PP, et al. (2014). A non-canonical NRPS is involved in the synthesis of fungisporin and related hydrophobic cyclic tetrapeptides in Penicillium chrysogenum. PLoS ONE 9: e98212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen F, Pon A, Wilson M, et al. (2014). CFM-ID: A web server for annotation, spectrum prediction and metabolite identification from tandem mass spectra. Nucleic Acids Research 42: W94–W99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anelli P, Peterson SW, Haidukowski M, et al. (2018). Penicillium gravinicasei, a new species isolated from cave cheese in Apulia, Italy. International Journal of Food Microbiology 282: 66–70. [DOI] [PubMed] [Google Scholar]
- Blakeslee AF. (1915). Lindner’s roll tube method of separation cultures. Phytopathology 5: 68–69. [Google Scholar]
- Blehert DS, Hicks AC, Behr M, et al. (2009). Bat White-Nose Syndrome: An Emerging Fungal Pathogen? Science 323: 227. [DOI] [PubMed] [Google Scholar]
- Frisvad JC, Filtenborg O. (1989). Terverticillate Penicillia: Chemotaxonomy and mycotoxin production. Mycologia 81: 837–861. [Google Scholar]
- Frisvad JC, Samson RA, Rassing BR, et al. (1997). Penicillium discolor, a new species from cheese, nuts and vegetables. Antonie van Leeuwenhoek 72: 119–126. [DOI] [PubMed] [Google Scholar]
- Frisvad JC, Samson RA. (2004). Polyphasic taxonomy of Penicillium subgenus Penicillium. A guide to identification of food and air-borne terverticillate Penicillia and their mycotoxins. Studies in Mycology 49: 1–174. [Google Scholar]
- Frisvad JC, Smedsgaard J, Larsen TO, et al. (2004). Mycotoxins, drugs and other extrolites produced by species in Penicillium subgenus Penicillium. Studies in Mycology 49: 201–241. [Google Scholar]
- Frisvad JC, Houbraken J, Popma S, et al. (2013a). Two new Penicillium species Penicillium buchwaldii and Penicillium spathulatum, producing the anticancer compound asperphenamate. FEMS Microbiology Letters 339: 77–92. [DOI] [PubMed] [Google Scholar]
- Frisvad JC, Yilmaz N, Thrane U, et al. (2013b). Talaromyces atroroseus, a new species efficiently producing industrially relevant red pigments. PLoS ONE 8: e84102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargas A, Trest MT, Christensen M, et al. (2009). Geomyces destructans sp. nov. associated with bat white-nose syndrome. Mycotaxon 108: 147–154. [Google Scholar]
- Houbraken J, Frisvad JC, Samson RA. (2011). Taxonomy of Penicillium section Citrina. Studies in Mycology 70: 53–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J, Frisvad JC, Seifert KA, et al. (2012). New penicillinproducing Penicillium species and an overview of section Chrysogena. Persoonia 29: 78–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J, Visagie CM, Meijer M, et al. (2014). A taxonomic and phylogenetic revision of Penicillium section Aspergilloides. Studies in Mycology 78: 373–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J, Wang L, Lee HB, et al. (2016). New sections in Penicillium containing novel species producing patulin, pyripyropens or other bioactive compounds. Persoonia 36: 299–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto S, Tokumasu S, Suyama Y, et al. (2005). Thysanophora penicillioides includes multiple genetically diverged groups that coexist respectively in Abies mariesii forests in Japan. Mycologia 97: 1238–1250. [DOI] [PubMed] [Google Scholar]
- Johnson LJAN, Miller AN, McCleery RA, et al. (2013). Psychrophilic and psychrotolerant fungi on bats and the presence of Geomyces spp. on bat wings prior to the arrival of White Nose Syndrome. Applied and Environmental Microbiology 79: 5465–5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, et al. (2017). ModelFinder: Fast model selection for accurate phylogenetic estimates. Nature Methods 14: 587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornerup A, Wanscher JH. (1967). Methuen Handbook of Colour. 2nd edn Methuen & Co Ltd, London, England. [Google Scholar]
- Lanfear R, Frandsen PB, Wright AM, et al. (2017). PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Molecular Biology and Evolution 34: 772–773. [DOI] [PubMed] [Google Scholar]
- Lorch JM, Muller LK, Russell RE, et al. (2013). Distribution and environmental persistence of the causative agent of white-nose syndrome, Geomyces destructans, in bat hibernacula of the eastern United States. Applied and Environmental Microbiology 79: 1293–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlpine DF, Vanderwolf KJ, Forbes GJ, et al. (2012). Consumption of bats (Myotis spp.) by raccoons (Procyon lotor) during an outbreak of white-nose syndrome in New Brunswick, Canada: Implications for estimates of bat mortality. Canadian Field-Naturalist 125: 257–260. [Google Scholar]
- Micalizzi EW, Mack JN, White GP, et al. (2017). Microbial inhibitors of the fungus Pseudogymnoascus destructans, the causal agent of white-nose syndrome in bats. Plos One 12: e0179770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh BQ, Nguyen MAT, Von Haeseler A. (2013). Ultrafast approximation for phylogenetic bootstrap. Molecular Biology and Evolution 30: 1188–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnis AM, Lindner DL. (2013). Phylogenetic evaluation of Geomyces and allies reveals no close relatives of Pseudogymnoascus destructans, comb. nov., in bat hibernacula of eastern North America. Fungal Biology 117: 638–649. [DOI] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, Von Haeseler A, et al. (2015). IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32: 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nováková A. (2009). Microscopic fungi isolated from the Domica Cave system (Slovak Karst National Park, Slovakia). A review. International Journal of Speleology 38: 71–82. [Google Scholar]
- Nováková A, Hubka V, Valinová Š, et al. (2018). Cultivable microscopic fungi from an underground chemosynthesis-based ecosystem: a preliminary study. Folia Microbiologica 63: 43–55. [DOI] [PubMed] [Google Scholar]
- Pitt JI, Cruickshank RH, Leistner L. (1986). Penicillium commune, P. camembertii, the origin of white cheese moulds, and the production of cyclopiazonic acid. Food Microbiology 3: 363–371. [Google Scholar]
- Pitt JI, Klich MA, Shafer GP, et al. (1990). Differentiation of Penicillium glabrum from Penicillium spinulosum and other closely related species: an integrated taxonomic approach. Systematic and Applied Microbiology 13: 304–309. [Google Scholar]
- Polonelli L, Morace G, Rosa R, et al. (1987). Antigenic characterization of Penicillium camemberti and related common cheese contaminants. Applied and Environmental Microbiology 53: 872–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puechmaille SJ, Wibbelt G, Korn V, et al. (2011). Pan-European distribution of white-nose syndrome fungus (Geomyces destructans) not associated with mass mortality. PLoS ONE 6: e19167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds HT, Ingersoll T, Barton HA. (2015). Modeling the environmental growth of Pseudogymnoascus destructans and its impact on the white-nose syndrome epidemic. Journal of Wildlife Diseases 51: 318–331. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, et al. (2012). MrBayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson RA, Seifert KA, kuijpers AFA, et al. (2004). Phylogenetic analysis of Penicillium subgenus Penicillium using partial B-tubulin sequences. Studies in Mycology 49: 175–200. [Google Scholar]
- Schoch CL, Seifert KA, Huhndorf S, et al. (2012). Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proceedings of the National Academy of Sciences of the USA 109: 6241–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetti SO, Larghi EL, Kaufman TS. (2016). The 3,4-dioxygenated 5-hydroxy-4-aryl-quinolin-2(1H)-one alkaloids. Results of 20 years of research, uncovering a new family of natural products. Natural Product Reports 33: 1425–1446. [DOI] [PubMed] [Google Scholar]
- Vanderwolf KJ, Malloch D, Mcalpine DF, et al. (2013a). A world review of fungi, yeasts, and slime molds in caves. International Journal of Speleology 42: 77–96. [Google Scholar]
- Vanderwolf KJ, McAlpine DF, Malloch D, et al. (2013b). Ectomycota associated with hibernating bats in Eastern Canadian caves prior to the emergence of White-Nose Syndrome. Northeastern Naturalist 20: 115–130. [Google Scholar]
- Vanderwolf KJ, Malloch D, McAlpine DF. (2016). Ectomycota associated with arthropods from bat hibernacula in Eastern Canada, with particular reference to Pseudogymnoasucs destructans. Insects 7: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderwolf KJ, Malloch D, McAlpine DF. (2017). Psychrotolerant microfungi associated with deer mice (Peromyscus maniculatus) in a white-nose syndrome positive bat hibernaculum in eastern Canada. Canadian Field-Naturalist 131: 238–245. [Google Scholar]
- Visagie CM, Hirooka Y, Tanney JB, et al. (2014a). Aspergillus, Penicillium and Talaromyces isolated from house dust samples collected around the world. Studies in Mycology 78: 63–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie CM, Houbraken J, Frisvad JC, et al. (2014b). Identification and nomenclature of the genus Penicillium. Studies in Mycology 78: 343–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh JP, Renaud JB, Hoogstra S, et al. (2019). Diagnostic fragmentation filtering for the discovery of new chaetoglobosins and cytochalasins. Rapid Communications in Mass Spectrometry 33: 133–139. [DOI] [PubMed] [Google Scholar]
- Wibbelt G, Kurth A, Hellmann D, et al. (2010). White-nose syndrome fungus (Geomyces destructans) in bats, Europe. Emerging Infectious Diseases 16: 1237–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZF, Liu F, Zhou X, et al. (2017). Culturable mycobiota from Karst caves in China, with descriptions of 20 new species. Persoonia 39: 1–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


