Abstract
Coprophilous fungi are saprotrophic organisms that show great diversity, mainly on herbivore dung. The physico-chemical characteristics of this peculiar substrate combined with the high level of fungal adaptation to different environmental conditions offer the perfect setting for discovering new taxa. This study focused on the species diversity of penicillium-like fungi isolated mainly from herbivore dung collected at different Spanish locations. From 130 samples, a total of 104 isolates were obtained, and 48 species were identified. Preliminary identifications were based on morphology and partial β-tubulin (tub2) gene sequences. Putative new taxa were characterized by a multi-gene sequencing analysis testing the tub2, the internal transcribed spacer rDNA (ITS), calmodulin (cmdA), and RNA polymerase II second largest subunit (rpb2) genes, and a detailed phenotypic study. Using this polyphasic approach and following the genealogical concordance phylogenetic species recognition (GCPSR) method, we propose the new genera Penicillago (for Penicillium nodositatum) and Pseudopenicillium (for Penicillium megasporum and P. giganteum) in the family Aspergillaceae, and 11 new species, including seven Penicillium, three Talaromyces and one Pseudopenicillium. A lectotype and epitype are designed for Penicillium nodositatum. Our results show that the species diversity of penicillium-like fungi on herbivore dung has not been widely studied and that this substrate seems to be a good reservoir of interesting Eurotialean fungi.
Keywords: Aspergillaceae, coprophilous fungi, Eurotiales, phylogeny, Trichocomaceae
INTRODUCTION
Herbivore dung is a complex substrate that has a high amount of readily available carbohydrates with high nitrogen content, water-soluble vitamins, growth factors and mineral ions (Bell 1983). Due to the nutritional properties, together with its physical structure, pH, and the varying moisture content, it is an excellent substrate for a great diversity of fungi to grow (Bell 1983, Richardson 2001, Herrera et al. 2011).
Ascomycetes accounts for the largest number of species found on dung from herbivorous animals; genera such as Ascobolus, Lasiobolus, Podospora or Thelebolus practically only occur on this substrate (Webster 1970, Bell 1983, Richardson 2001). On the contrary, other ascomycetes like Eurotialean penicillium-like fungi are not considered predominantly coprophilous (Krug et al. 2004), but they can be commonly found on this substrate when its soluble materials are exhausted, and its relative humidity has reduced to below 85 % (Kuthubutheen & Webster 1986). However, species diversity of this latter group has been poorly studied on herbivore dung. Although Seifert et al. (1983) provided a list of near 20 Penicillium species isolated from animal faeces, their identification was based exclusively on phenotypic features.
Traditionally, based on similarities in ecology, morphology and extrolite production, most of the Penicillium species considered coprophilous (i.e. P. brevistipitatum, P. clavigerum, P. concentricum, P. coprobium, P. coprophilum, P. formosanum, P. glandicola and P. vulpinum) were classified in series Claviformia (Seifert & Samson 1985, Frisvad & Samson 2004, Wang & Zhuang 2005). Currently, the new concept of the genus, which is based on molecular taxonomy, coprophilous penicillia belongs in section Robsamsonia (Houbraken et al. 2016), which also includes other species previously assigned to series Urticicolae (Frisvad & Samson 2004). Furthermore, other species of Penicillium previously found on dung currently belong to the genus Talaromyces (Houbraken et al. 2016). Coprophilous species reported in this latter genus are T. atroroseus, T. helicus, T. flavus, T. muroii and T. trachyspermus, and two others T. dupontii and T. emersonii, which have been transferred to respectively, Thermomyces and Rasamsonia (Masunga et al. 2006, Frisvad et al. 2013, Yilmaz et al. 2014).
The taxonomy of penicillium-like fungi has changed dramatically due to phylogenetic data. Houbraken & Samson (2011) demonstrated that Penicillium is phylogenetically more-closely related to Aspergillus (family Aspergillaceae) than to Talaromyces (family Trichochomaceae). In the last decade, the species of these genera have been delineated on the basis of a polyphasic approach that has mainly included the evaluation of morphological and multi-locus sequence analyses testing with the internal transcribed spacer region rDNA (ITS), and the β-tubulin (tub2), calmodulin (cmdA), and/or the DNA-dependent RNA polymerase II largest subunit (rpb2) genes (Peterson 2008, Houbraken et al. 2014a, Samson et al. 2014, Visagie et al. 2014b, Yilmaz et al. 2014). In addition, tub2 was recommended as an identification marker in Penicillium (Visagie et al. 2014b) and Talaromyces (Yilmaz et al. 2014). The use of this already generalised approach has proven to be able to discriminate very close species and to discover and characterise new taxa (Visagie et al. 2014b, Guevara-Suarez et al. 2017, Barbosa et al. 2018).
Given the scarcity of data on species isolated from dung and identified by modern techniques, we consider this the perfect setting for discovering new taxa. Therefore, the present work focuses on the study of the species diversity of penicillium-like fungi from herbivore dung samples collected across Spain. Identification was made using both morphology and by comparing multi-locus sequences with a database of ex-type and verified reference strains. For multisequence analyses, the Genealogical Phylogenetic Species Recognition (GCPSR) method was applied to data in support of discovering putative new species (Taylor et al. 2000).
MATERIALS AND METHODS
Sampling and fungal isolation
Dung samples were collected in 2016 and 2017 from Spanish regions that have different climates and biodiversity, such as Andalusia, the Balearic and Canary Islands, Cantabria, Castilla-Leon, Catalonia, Extremadura and Galicia. The dung collected was mostly from herbivorous animals, such as rabbits, sheep, deer, goats, cattle and horses. However, some samples from other animals, such as wild pigs or foxes, and even soil mixed with dung was also studied. The samples were placed in individual paper or plastic bags and processed no later than 3 d after collection. Individual samples were divided into two parts: one processed using moist chambers (Richardson 2001) and the other by a modified Waksman (1922) dilution series method. For moist chambers, pieces of the sample were placed on moist filter paper with sterile distilled water in individual Petri dishes and incubated at room temperature (22–25 °C) for up to 30 d. For the dilution series, approximately one gram of dung or associated soil was diluted 1:10 (w/v) in sterile water and shaken for approximately 10 min. Aliquots of the suspensions were pipetted into Petri dishes and mixed with 20 mL of melted cooled agar medium. Culture media used for isolation were potato dextrose agar (PDA; Pronadisa, Madrid, Spain), potato carrot agar (PCA; 20 g potatoes, 20 g carrot, 20 g agar, 1000 mL distilled water), both supplemented with chloramphenicol (200 mg/L), and dichloran rose-bengal chloramphenicol agar (DRBC; 5 g peptone, 10 g glucose, 1 g KH2PO4, 0.5 g MgSO4, 25 mg rose-bengal, 2 mg dichloran, 200 mg chloramphenicol, 20 g agar, 1 000 mL distilled water). All media were supplemented with dieldrin in dimethyl-ketone (1 %). Petri dishes were incubated at room temperature for up to 30 d. The moist chambers and Petri dishes were examined at regular intervals with the aid of a stereo-microscope, and conidia from sporulating colonies were transferred to PDA supplemented with chloramphenicol.
Isolates identified morphologically as penicillium-like were preserved and deposited in the culture collection of the Faculty of Medicine, Reus (FMR). Cultures of interesting species as well as type material and cultures of the new species were deposited at the Westerdijk Fungal Biodiversity Institute (CBS, Utrecht, the Netherlands). Nomenclatural novelties and descriptions were deposited in MycoBank (Crous et al. 2004).
Molecular identification and phylogenetic analysis
Isolates were cultured on PDA or malt extract agar (MEA; Difco, Detroit, USA) for 7–14 d at 25 °C. DNA was extracted using the FastDNA® kit protocol (MP Biomedicals, Solon, OH) and for the lysis step done with a FastPrep® FP120 cell disrupter (Thermo Savant, Holbrook, NY).
Preliminary species identifications were carried out BLASTing tub2 DNA sequences using GenBank. In the case of putative new species, the ITS region, including the 5.8S rRNA gene, and fragments of cmdA and/or rpb2 genes were also amplified and sequenced. The primer pairs used were: ITS5/ITS4 for ITS (White et al. 1990), Bt2a/Bt2b for tub2 (Glass & Donaldson 1995), CMD5/CMD6 for cmdA (Hong et al. 2006), and RPB2-5F/RPB2-7Cr for rpb2 (Liu et al. 1999). The amplification protocol and PCR conditions were performed using methods and primers previously described (Peterson 2008, Houbraken & Samson 2011, Samson et al. 2014, Visagie et al. 2014b, Yilmaz et al. 2014). The amplified products were purified and sequenced at Macrogen Corp. Europe (Amsterdam, the Netherlands) with a 3730xl DNA analyzer (Applied Biosystems, Foster City, CA). Consensus sequences were obtained using SeqMan v. 7.0.0 (DNASTAR, Madison, WI). Newly generated sequences and their GenBank/EMBL accession numbers are summarized in Table 1.
Table 1.
Isolates of Penicillium, Talaromyces and related genera included in the study and their GenBank/EMBL accession numbers.
| Genus/Species1 | Section | Collection numbers2 | Substrate and Origin | GenBank/EMBL accession numbers | |||
|---|---|---|---|---|---|---|---|
| ITS | tub2 | cmdA | rpb2 | ||||
| P. arabicum | Exilicaulis | FMR 15298 | Dung, Castile and Leon | – | LT898226 | – | – |
| FMR 15095 | Dung, Catalonia | – | LT898225 | – | – | ||
| P. atramentosum | Paradoxa | FMR 15309 | Dung, Castile and Leon | – | LT898224 | – | – |
| FMR 15046 | Dung, Catalonia | – | LT898221 | – | – | ||
| FMR 15092 | Dung, Catalonia | – | LT898222 | – | – | ||
| FMR 15102 | Dung, Catalonia | – | LT898223 | – | – | ||
| P. balearicum | Paradoxa | FMR 15191T = CBS 143044 | Dung, Balearic Islands | LT899762 | LT898227 | LT899758 | LT899760 |
| FMR 15196 | Dung, Balearic Islands | LT899763 | LT898228 | LT899759 | LT899761 | ||
| P. beceitense | Ramosa | FMR 15038T = CBS 142989 | Dung, Catalonia | LT899780 | LT898229 | LT899764 | LT899798 |
| P. biforme | Fasciculata | FMR 15312 | Dung, Castile and Leon | – | LT898230 | – | – |
| FMR 15313 | Dung, Castile and Leon | – | LT898231 | – | – | ||
| P. brasilianum | Lanata-Divaricata | FMR 15483 | Dung, Galicia | – | LT898232 | – | – |
| P. brevicompactum | Brevicompacta | FMR 15105 | Dung, Catalonia | – | LT898233 | – | – |
| P. brevistipitatum | Robsamsonia | FMR 15103 | Dung, Catalonia | – | LT898234 | – | – |
| P. burguense | Exilicaulis | FMR 15493 | Dung, Galicia | – | LT898235 | – | – |
| P. canariense | Stolkia | FMR 15838 | Dung, Canary Islands | – | LT898236 | – | – |
| P. canescens | Canescentia | FMR 15028 | Dung, Catalonia | – | LT898237 | – | – |
| P. caprifimosum | Turbata | FMR 15041T = CBS 142990 | Dung, Catalonia | LT899781 | LT898238 | LT899765 | LT899799 |
| P. chrysogenum | Chrysogena | FMR 15100 | Dung, Catalonia | – | LT898244 | – | – |
| P. cinereoatrum | Exilicaulis | FMR 15033 | Dung, Catalonia | – | LT898284 | – | – |
| P. citrinum | Citrina | FMR 15646 | Dung, Castile and Leon | – | LT898242 | – | – |
| FMR 15647 | Dung, Castile and Leon | – | LT898243 | – | – | ||
| FMR 15094 | Dung, Catalonia | – | LT898239 | – | – | ||
| FMR 15486 | Dung, Galicia | – | LT898240 | – | – | ||
| FMR 15520 | Dung, Galicia | – | LT898241 | – | – | ||
| P. concentricum | Robsamsonia | FMR 15195 | Dung, Balearic Islands | – | LT898245 | – | – |
| FMR 15840 | Dung, Castile and Leon | – | LT898246 | – | – | ||
| P. coprobium | Robsamsonia | FMR 15201 | Dung, Balearic Islands | – | LT898247 | – | – |
| FMR 15311 | Dung, Castile and Leon | – | LT898248 | – | – | ||
| P. coprophilum | Robsamsonia | FMR 15187 | Dung, Balearic Islands | – | LT898249 | – | – |
| P. cremeogriseum | Lanata-Divaricata | FMR 15487 | Dung, Galicia | – | LT898250 | – | – |
| FMR 15488 | Dung, Galicia | – | LT898251 | – | – | ||
| P. crustosum | Fasciculata | FMR 15185 | Dung, Balearic Islands | – | LT898259 | – | – |
| FMR 15186 | Dung, Balearic Islands | – | LT898260 | – | – | ||
| FMR 15189 | Dung, Balearic Islands | – | LT898261 | – | – | ||
| FMR 15194 | Dung, Balearic Islands | – | LT898262 | – | – | ||
| FMR 15197 | Dung, Balearic Islands | – | LT898263 | – | – | ||
| FMR 15200 | Dung, Balearic Islands | – | LT898264 | – | – | ||
| FMR 15213 | Dung, Balearic Islands | – | LT898265 | – | – | ||
| FMR 15034 | Dung, Catalonia | – | LT898252 | – | – | ||
| FMR 15036 | Dung, Catalonia | – | LT898253 | – | – | ||
| FMR 15037 | Dung, Catalonia | – | LT898254 | – | – | ||
| FMR 15042 | Dung, Catalonia | – | LT898255 | – | – | ||
| FMR 15043 | Dung, Catalonia | – | LT898256 | – | – | ||
| FMR 15045 | Dung, Catalonia | – | LT898257 | – | – | ||
| FMR 15098 | Dung, Catalonia | – | LT898258 | – | – | ||
| FMR 15494 | Dung, Galicia | – | LT898266 | – | – | ||
| P. cvjetkovicii | Cinnamopurpurea | FMR 15310 | Dung, Castile and Leon | – | LT898267 | – | – |
| P. expansum | Penicillium | FMR 15097 | Dung, Catalonia | – | LT898268 | – | – |
| FMR 15484 | Dung, Galicia | – | LT898269 | – | – | ||
| P. fimosum | Paradoxa | FMR 15104T = CBS 142991 | Dung, Catalonia | LT970836 | LT898273 | LT970837 | |
| P. flavigenum | Chrysogena | FMR 15096 | Dung, Catalonia | – | LT898270 | – | – |
| P. frequentans | Aspergilloides | FMR 15193 | Dung, Balearic Islands | – | LT898271 | – | – |
| FMR 15212 | Dung, Balearic Islands | – | LT898272 | – | – | ||
| P. glabrum | Aspergilloides | FMR 15184 | Dung, Balearic Islands | – | LT898274 | – | – |
| FMR 15190 | Dung, Balearic Islands | – | LT898275 | – | – | ||
| FMR 15209 | Dung, Balearic Islands | – | LT898276 | – | – | ||
| P. griseofulvum | Robsamsonia | FMR 15203 | Dung, Balearic Islands | – | LT898280 | – | – |
| FMR 15204 | Dung, Balearic Islands | – | LT898281 | – | – | ||
| FMR 15207 | Dung, Balearic Islands | – | LT898282 | – | – | ||
| FMR 15314 | Dung, Castile and Leon | – | LT898283 | – | – | ||
| FMR 15029 | Dung, Catalonia | – | LT898277 | – | – | ||
| FMR 15030 | Dung, Catalonia | – | LT898278 | – | – | ||
| FMR 15093 | Dung, Catalonia | – | LT898279 | – | – | ||
| P. ibericum | Paradoxa | FMR 15040T = CBS 142992 | Dung, Catalonia | LT899782 | LT898285 | LT899766 | LT899800 |
| FMR 15107 | Soil, Galicia | LT899783 | LT898286 | LT899767 | LT899801 | ||
| P. lilacinoechinulatum | Sclerotiora | FMR 15492 | Dung, Galicia | – | LT898287 | – | – |
| P. magnielliptisporum | Paradoxa | FMR 15044 | Dung, Catalonia | – | LT898288 | – | – |
| P. mediterraneum | Roquefortorum | FMR 15188T = CBS 142754 | Dung, Balearic Islands | LT899784 | LT898291 | LT899768 | LT899802 |
| FMR 15031 = CBS 142755 | Dung, Catalonia | LT899785 | LT898289 | LT899769 | LT899803 | ||
| FMR 15032 | Dung, Catalonia | LT899786 | LT898290 | LT899770 | LT899804 | ||
| P. momoii | Exilicaulis | FMR 15208 | Dung, Balearic Islands | – | LT898292 | – | – |
| P. murcianum | Canescentia | FMR 15304 | Dung, Andalusia | – | LT898293 | – | – |
| FMR 15305 | Dung, Andalusia | – | LT898294 | – | – | ||
| FMR 15308 | Dung, Andalusia | – | LT898295 | – | – | ||
| FMR 15491 | Dung, Galicia | – | LT898296 | – | – | ||
| FMR 15845 | Dung, Galicia | – | LT898297 | – | – | ||
| P. polonicum | Fasciculata | FMR 15099 | Dung, Catalonia | – | LT898298 | – | – |
| P. radiolubatum | Canescentia | FMR 15485 | Dung, Canary Islands | – | LT898299 | – | – |
| P. roseoviride | Aspergilloides | FMR 15645 | Dung, Castile and Leon | – | LT898300 | – | – |
| P. rubefaciens | Exilicaulis | FMR 15202 | Dung, Balearic Islands | – | LT898301 | – | – |
| FMR 15297 | Dung, Castile and Leon | – | LT898302 | – | – | ||
| P. rudallense | Aspergilloides | FMR 15843 | Dung, Canary Islands | – | LT898303 | – | – |
| P. sizovae | Citrina | FMR 15300 | Dung, Castile and Leon | – | LT898304 | – | – |
| FMR 15521 | Dung, Galicia | – | LT898305 | – | – | ||
| P. synnematicola | Robsamsonia | FMR 15192T = CBS 142669 | Dung, Balearic Islands | LT898167 | LT898172 | LT898137 | LT898142 |
| FMR 15210 | Dung, Balearic Islands | LT898168 | LT898173 | LT898138 | LT898143 | ||
| FMR 15211 | Dung, Balearic Islands | LT898169 | LT898174 | LT898139 | LT898144 | ||
| FMR 16481 = CBS 143045 | Soil, Catalonia | LT898170 | LT898175 | LT898140 | LT898145 | ||
| FMR 16491 = CBS 143046 | Dung, Catalonia | LT898171 | LT898176 | LT898141 | LT898146 | ||
| Penicillium sp. | Exilicaulis | FMR 15841 | Dung, Castile and Leon | LT898311 | – | – | |
| Pgo. nodositata | - | FMR 15296 = CBS 142988 | Dung, Balearic Islands | LT899787 | LT898312 | LT899771 | LT899805 |
| - | FMR 16442 | Dung, Extremadura | LT899788 | LT898313 | LT899772 | LT899806 | |
| Pse. cervifimosum | - | FMR 15299T = CBS 142670 | Dung, Castile and Leon | LT899789 | LT898315 | – | LT899807 |
| Pse. giganteum | - | FMR 14718 | Soil, unknown | LT899790 | LT898314 | – | LT899808 |
| T. angelicus | Talaromyces | FMR 15489 | Dung, Galicia | LT899791 | LT898316 | LT899773 | LT899809 |
| FMR 15490 | Dung, Galicia | LT899792 | LT898317 | LT899774 | LT899810 | ||
| T. catalonicus | Trachyspermi | FMR 16441T = CBS 143039 | Dung, Catalonia | LT899793 | LT898318 | LT899775 | LT899811 |
| T. coprophilus | Talaromyces | FMR 15199T = CBS 142756 | Dung, Balearic Islands | LT899794 | LT898319 | LT899776 | LT899812 |
| T. muroii | Talaromyces | FMR 15496 | Dung, Galicia | – | LT898321 | – | – |
| T. pseudofuniculosus | Talaromyces | FMR 15307T = CBS 143041 | Dung, Andalusia | LT899796 | LT898323 | LT899778 | LT899814 |
| FMR 15035 | Dung, Catalonia | LT899797 | LT898322 | LT899779 | LT899815 | ||
| T. ruber | Talaromyces | FMR 15839 | Dung, Castile and Leon | – | LT898324 | – | – |
| T. sayulitensis | Talaromyces | FMR 15842 | Dung, Canary Islands | – | LT898325 | – | – |
1P. = Penicillium; Pgo. = Penicillago; Pse. = Pseudopenicillium; T. = Talaromyces; new taxa proposed in this study are in bold;
T = ex-type strain.
2CBS: Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands; FMR: Facultat de Medicina i Ciencies de la Salut, Reus, Spain.
Sequences were retrieved from GenBank taking into account the last update of the database of the International Commission of Penicillium and Aspergillus (http://www.aspergilluspenicillium.org), which includes all the accepted species for Penicillium and Talaromyces (Samson et al. 2014, Visagie et al. 2014b, Yilmaz et al. 2014). Single and concatenated phylogenetic analyses were performed to delineate putative new species and the phylogenies corresponding to each section of those genera were properly reconstructed.
Data sets for each locus were aligned individually using ClustalW (Thompson et al. 1994), in MEGA v. 6.0 software (Tamura et al. 2013), refined with MUSCLE (Edgar 2004) under the same platform, and manually adjusted when needed. Larger alignments including different sections of each genus was performed using the MAFFT tool in the EMBL-EBI Web Services portal (https://www.ebi.ac.uk/Tools/msa/mafft/) and manually adjusted in MEGA v. 6.0. Phylogenetic reconstructions by maximum likelihood (ML) and Bayesian inference (BI) were carried out using MEGA v. 6.0 and MrBayes v. 3.1.2 (Huelsenbeck & Ronquist 2001, Ronquist & Huelsenbeck 2003, Tamura et al. 2013), respectively. For ML analyses, the trees were inferred using Nearest-Neighbour-Interchange as a heuristic method and gaps were treated as partial deletion with a 95 % site coverage cut-off. Phylogenetic support for internal branches was assessed by 1 000 ML bootstrapped pseudoreplicates and bootstrap support (bs) ≥ 70 % was considered significant. The BI analyses were performed using five million Markov chain Monte Carlo (MCMC) generations, with two runs (one cold and three heated chains) and samples were stored every 1 000 generations. The 50 % majority-rule consensus tree and posterior probability values (pp) were calculated after discarding the first 25 % of the samples. A pp value ≥ 0.95 was considered significant. The best substitution models for each data partition were estimated using jModelTest v. 2.1.3 according to the Akaike criterion (Darriba et al. 2012, Guindon & Gascuel 2003). The length, number of variable and phylogenetic informative sites, and substitution models for each data partition are summarized in Table 2. The resulting trees were plotted using FigTree v. 1.3.1 (http://tree.bio.ed.ac.uk/software/figtree/) and edited in Adobe Illustrator CS3. The alignments and trees were deposited in TreeBASE (www.treebase.org) under the submission number 21345.
Table 2.
Overview and details used for phylogenetic analyses in sections of Penicillium and Talaromyces, and related taxa with taxonomic novelties proposed in the study.
| Penicillium | Talaromyces | Miscellaneous Aspergillaceae | ||||||
|---|---|---|---|---|---|---|---|---|
| Turbata and Paradoxa | Ramosa | Robsamsonia | Roquefortorum | Talaromyces | Trachyspermi | |||
| ITS dataset | Length (bp) | 504 | 485 | 428 | 506 | 360 | 474 | 440 |
| Pvar | 29 | 47 | 207 | 24 | 72 | 119 | 124 | |
| Pi | 18 | 16 | 18 | 13 | 45 | 86 | 104 | |
| Model* | K80+G | GTR+I | K80+G | TPM2uf+I | TrN+I+G | TIM2+I+G | TrN+I+G | |
| tub2 dataset | Length (bp) | 412 | 387 | 352 | 409 | 370 | 398 | 401 |
| Pvar | 151 | 112 | 122 | 68 | 178 | 158 | 223 | |
| Pi | 100 | 64 | 67 | 29 | 140 | 104 | 187 | |
| Model* | TIM2ef+G | TIM2ef+G | GTR+G | TIM1ef | HKY+I+G | TPM3uf+G | HKY+I+G | |
| CmdA dataset | Length (bp) | 492 | 544 | 486 | 501 | 475 | 482 | – |
| Pvar | 195 | 223 | 164 | 50 | 250 | 247 | – | |
| Pi | 88 | 92 | 96 | 8 | 205 | 176 | – | |
| Model* | TIM2+G | TIM2+G | GTR+I | TPM1+G | HKY+I+G | TPM1+I+G | – | |
| rpb2 dataset | Length (bp) | 915 | – | 804 | 915 | – | – | 956 |
| Pvar | 239 | – | 243 | 106 | – | – | 463 | |
| Pi | 159 | – | 127 | 55 | – | – | 375 | |
| Model* | TrN+I+G | – | GTR+I | TIM3ef+I | – | – | TrN+I+G | |
| Concatenated dataset | Length (bp) | 2323 | 1416 | 2068 | 2331 | 1205 | 1354 | 1797 |
| Pvar | 614 | 382 | 574 | 248 | 503 | 524 | 810 | |
| Pi | 365 | 172 | 303 | 105 | 395 | 366 | 666 | |
Pvar = variable sites; Pi = phylogenetic informative sites;
* = substitution model for Bayesian inference
Phenotypic characterisation
Phenotypic characterization was carried out using standard growth conditions suggested by Visagie et al. (2014b) and Yilmaz et al. (2014). Briefly, isolates were cultured onto MEA 2 % (Samson et al. 2010), oatmeal agar (OA; Samson et al. 2010), Czapek yeast autolysate agar (CYA; Pitt 1979), yeast extract sucrose agar (YES; Frisvad 1981), creatine sucrose agar (CREA; Frisvad 1981) and dichloran 18 % glycerol agar (DG18; Hocking & Pitt 1980), incubated at 25 °C for 7 d in darkness. Colony growth rates were also measured after 7 d at 30 and 37 °C on MEA, CYA, YES and OA. Colours used for descriptions refer to Kornerup & Wanscher (1978). Microscopic features were examined on colonies grown on MEA after 1–2 wk, mounted on slides with Shear’s solution or 60 % lactic acid, and excess conidia removed with 70 % ethanol. Microscopic features were captured with a Zeiss Axio-Imager M1 light microscope using Nomarski differential interference contrast and phase-contrast optics (Zeiss, Oberkochen, Germany) with a DeltaPix Infinity x digital camera and DeltaPix InSight v. 5.3.11 software. Photoplates were assembled from separate photographs using PhotoShop CS3.1.
RESULTS
During the study, 130 dung samples were processed and 104 isolates of penicillium-like fungi were recovered, including three from soil (Table 1). Preliminary identification based on morphology and tub2 sequences showed that most of the isolates belonged to Penicillium (n = 91; 87.5 %) and a few to Talaromyces (n = 9; 8.65 %), while four of them (3.85 %) could not be assigned to any genus despite exhibiting a penicillium-like morphology. To maximize the quality of the analyses, three separate tub2 alignments were done for the different genera studied. We also carried out additional analyses of single-gene ITS, cmdA and rpb2 and combined datasets from the different genes, corresponding to the genera or sections where putative new species were included (Table 2). The topology of the trees was similar for both methods, with ML trees used to represent the results. Bootstrap and BI posterior probability values are indicated on relevant branches. Final identification of the isolates, resulting from the phylogenetic and phenotypic characterization, is shown in Table 1.
Penicillium phylogeny
The phylogenetic tree based on the tub2 locus with the 91 isolates of Penicillium is shown in Fig. 1. The aligned dataset was 404 bp long, with 257 variable sites and 237 phylogenetic informative. The best substitution model for ML was K2+G, and for BI it was GTR+G+I. In general, the topology of the phylogenetic tree showed well-delimitated sections.
Fig. 1.
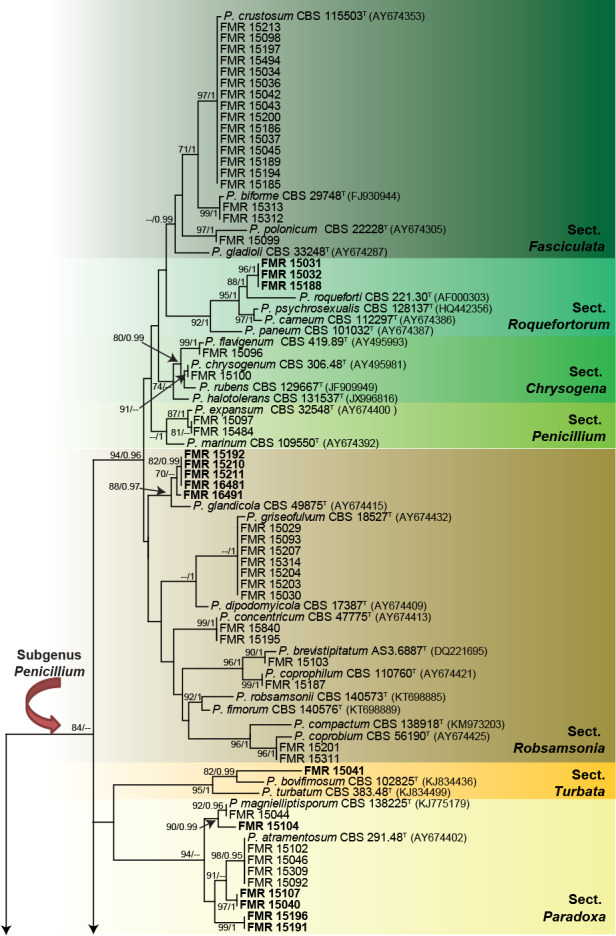
ML tree of Penicillium inferred from tub2 including the sections recovered from dung in this work. Branch lengths are proportional to phylogenetic distance. Some of the larger branches were condensed, with the proportions showed above the parallel diagonal lines. Bootstrap support values/Bayesian posterior probability scores above 69 %/0.94 are indicated on the nodes. Isolates corresponding to new species are shown in bold. Between parentheses, GenBank accession numbers of tub2 sequences. The tree is rooted to Talaromyces flavus CBS 310.38 and Talaromyces duclauxii CBS 322.48. T = ex-type strain.
The analysis distributed the Penicillium isolates in at least 38 species, belonging to 17 sections. The two major clades coincided with the two subgenera currently accepted in Penicillium, i.e. Aspergilloides and Penicillium (Houbraken & Samson 2011). Subgenus Penicillium (84 % bs/-- pp) is represented by the sections that according to the numbers of isolates obtained are: Fasciculata and Robsamsonia with 18 isolates (19.78 %) each, followed by Paradoxa (n = 10; 11 %), Canescentia (n = 7; 7.69 %), Roquefortorum (n = 3; 3.29 %), Chrysogena (n = 2; 2.19 %), Penicillium (n = 2; 2.19 %), and Brevicompacta, Ramosa, and Turbata each with one isolate (1.09 %). Subgenus Aspergilloides (72 % bs/0.99 pp) is represented by the sections Exilicaulis (n = 8; 8.79 %), Aspergilloides (n = 7; 7.69 %), Citrina (n = 7; 7.69 %), Lanata-Divaricata (n = 3; 3.29 %), and Sclerotiora and Stolkia each with one isolate (1.09 %).
Isolates of section Fasciculata (--% bs/0.99 pp) were identified as P. crustosum (n = 15), P. biforme (n = 2), and P. polonicum (n = 1), the former being the most prevalent species (14.42 %) found in the study. In fact, P. crustosum is a relatively common species commonly isolated from nuts, meat, cheese, feeds, vegetables, and pomaceous and stone fruits (Sonjak et al. 2005).
In section Roquefortorum (92 % bs/1 pp) three isolates (FMR 15031, FMR 15032, FMR 15188) were clustered in the same terminal clade. This is a small section of closely related species (i.e. P. carneum, P. paneum, P. psychrosexualis and P. roqueforti) commonly associated with the cheese industry (Houbraken et al. 2010, 2016). Our isolates were related to P. roqueforti, but formed an independent and distant branch that could represent an undescribed species. In order to evaluate possible intra- and inter-specific variability within the species and confirm the phylogenetic position of our putative new species, we performed an additional analysis with tub2 gene (see Fig. S1 in supplemental material) with more sequences of the species available in GenBank. This demonstrated that P. roqueforti is divided in two clades with an intra-specific variability of around 0.08 %, with our isolates forming a separate branch from the P. roqueforti complex. The concatenated analysis using ITS, tub2, cmdA and rpb2 (Fig. 2) supported the novelty of these three isolates, being therefore proposed as the new species P. mediterraneum. Although, P. mediterraneum and P. roqueforti have identical ITS barcodes, both species have unique tub2, cmdA and rpb2 sequences.
Fig. 2.
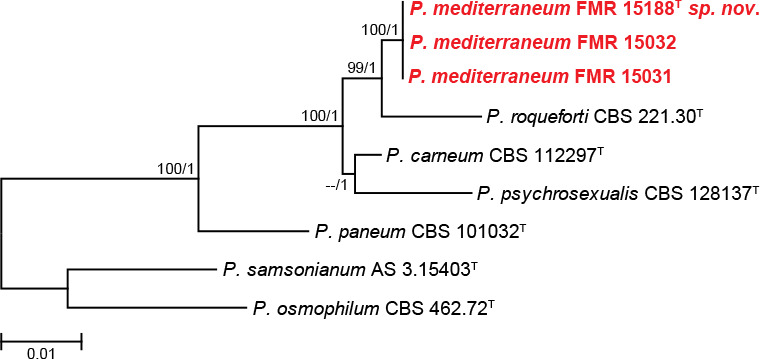
ML tree of Penicillium section Roquefortorum inferred from the combined ITS, tub2, cmdA, and rpb2 loci. Branch lengths are proportional to phylogenetic distance. Bootstrap support values/Bayesian posterior probability scores above 70 %/0.95 are indicated on the nodes. The tree is rooted to P. samsonianum AS 3.15403 and P. osmophilum CBS 462.72. The name in red is the new species described in this study. T = ex-type strain.
In section Chrysogena (74 % bs/--pp), which includes well-known species usually isolated from soil and also from indoor environments (Houbraken et al. 2012), two dung isolates were identified as P. chrysogenum and P. flavigenum, respectively. Whereas the two isolates of the section Penicillium (--% bs/1 pp), a group that currently mainly includes plant pathogenic penicillia (Houbraken et al. 2016), were both identified as P. expansum.
Isolates of section Robsamsonia (88 % bs/0.97 pp) were identified as P. brevistipitatum (n = 1), P. concentricum (n = 2), P. coprobium (n = 2), P. coprophilum (n = 1), and P. griseofulvum (n = 7); however, five isolates (FMR 15192, FMR 15210, FMR 15211, FMR 16481 and FMR 16491) could not be assigned to any known species. The preliminary tub2 analysis, but also the concatenate phylogeny (Fig. 3) including the currently accepted species in the section, showed that the five isolates grouped together in a supported and undescribed lineage closely related to the ex-type strain of P. glandicola. An additional tub2 analysis (see Fig. S2 in supplemental material) with more sequences of P. glandicola showed that our isolates and P. glandicola were 97.4 % similar. The genetic differences and morphological peculiarities observed in this group of dung isolates justify the description of a new species, namely P. synnematicola.
Fig. 3.
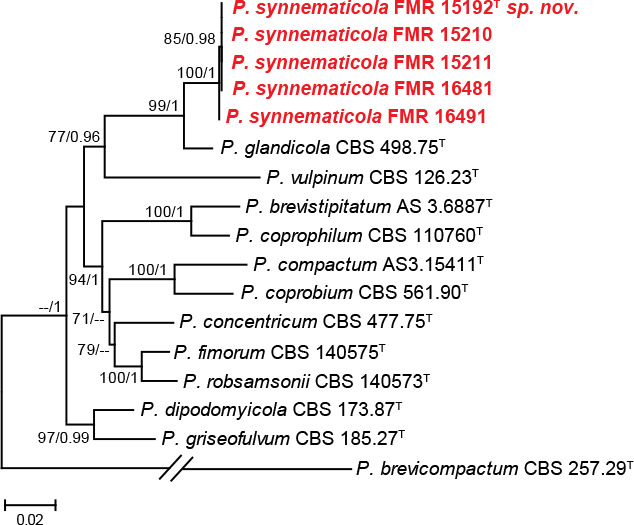
ML tree of Penicillium section Robsamsonia inferred from the combined ITS, tub2, cmdA, and rpb2 loci. Branch lengths are proportional to phylogenetic distance. Bootstrap support values/Bayesian posterior probability scores above 70 %/0.95 are indicated on the nodes. The tree is rooted to P. brevicompactum CBS 257.29. The name in red is the new species described in this study. T = ex-type strain.
The only isolate (FMR 15041) of section Turbata (95 % bs/1 pp) did not fit with any species of the section, although it was closely related to P. bovifimosum. In Paradoxa (94 % bs/-- pp) 10 isolates were included; four were identified as P. atramentosum, one as P. magnielliptisporum, and the rest (FMR 15040, FMR 15104, FMR 15107, FMR 15191 and FMR 15196) were allocated in three single branches which represent three putative new species for the genus. The concatenated phylogeny of sections Paradoxa and Turbata with the six unidentified isolates is shown in Fig. 4. In the former section, the isolates FMR 15107 and FMR 15040 formed a terminal clade separated from other one formed by FMR 15191 and 15196. The lineages of these two clades were in turn distant from the two terminal branches where FMR 15104 and the ex-type strain of P. atramentosum were located, respectively. Genetic differences of the three undescribed lineages with respect to P. atramentosum (97.07 % and 98.23 % similar with tub2 and concatenate dataset for FMR 15107 and FMR 15040, respectively; 96.2 % and 96.2 % with tub2 and concatenate dataset for FMR 15191 and FMR 15196; 94.68 % and 95 % with tub2 and concatenate dataset for FMR 15104) justifies them as distinct taxa. The three species are proposed below as P. ibericum, P. balearicum and P. fimosum. An additional phylogenetic analysis with tub2 including more sequences of the most closely related species confirms our proposal (see Fig. S3 in supplemental material). In section Turbata, P. bovifimosum and FMR 15041 were located in the same clade, but the latter formed a long terminal branch that proved to be a distinct species, described below as P. caprifimosum.
Fig. 4.
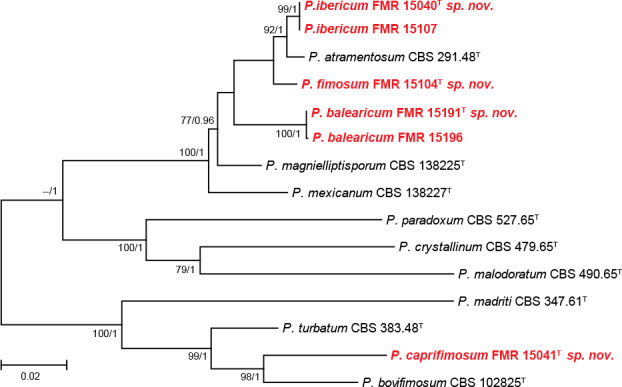
ML tree of Penicillium sections Paradoxa and Turbata inferred from the combined ITS, tub2 and cmdA loci. Branch lengths are proportional to phylogenetic distance. Bootstrap support values/Bayesian posterior probability scores above 70 %/0.95 are indicated on the nodes. The names in red are the new species described in this study. T = ex-type strain
Section Ramosa (97 % bs/0.98 pp) only included the unidentified isolate FMR 15038. Currently, the section comprises 13 species (Visagie et al. 2014a, 2016a, Rong et al. 2016), although two of them, P. lanosum and P. kojigenum, were considered conspecific by Samson & Pitt (2000). The concatenate analysis performed with ITS, tub2, and cmdA (Fig. 5), rpb2 was not included due to the lack of sequences for all species in the section, showed that FMR 15038 was placed in an independent branch between two clades, one with P. kojigenum and P. lanosum, and the other with P. jamesonlandense and P. swiecickii. The similarity among our isolate and its phylogenetic sisters was 98.3 %, proving that our isolate is an undescribed species, which is proposed below as P. beceitense.
Fig. 5.
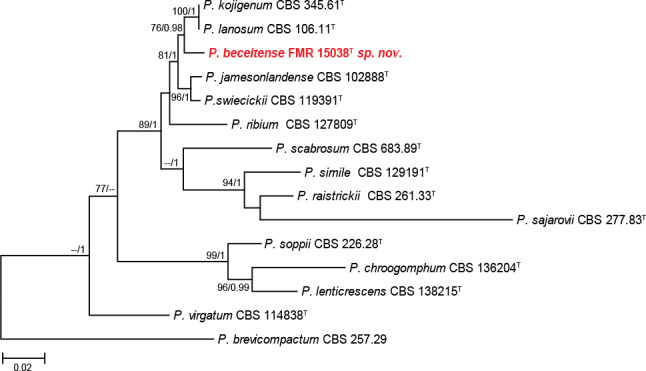
ML tree of Penicillium section Ramosa inferred from the combined ITS, tub2 and cmdA loci. Branch lengths are proportional to phylogenetic distance. Bootstrap support values/Bayesian posterior probability scores above 70 %/0.95 are indicated on the nodes. The tree is rooted to P. brevicompactum CBS 257.29. The name in red is the new species described in this study. T = ex-type strain.
The only isolate of section Brevicompacta (99 % bs/1 pp) was identified as P. brevicompactum, a species commonly inhabiting in soil and decaying vegetation, but also described from food, cereals, textiles, clinical specimens and faeces of snake (Pitt 1979, Guevara-Suarez et al. 2016). The seven isolates classified in section Canescentia (98 % bs/1 pp) were identified as P. canescens (n = 1), P. murcianum (n = 3) and P. radiatolobatum (n = 3). Despite tub2 being unable to discriminate between P. radiatolobatum and P. murcianum, these two species can be recognized with the cmdA and rpb2 barcodes.
Section Exilicaulis (99 % bs/1 pp) included eight of our isolates. It is noteworthy that the ML general tree based only on tub2 presented some doubtful results in the species identification of some isolates, especially those belonging to the P. restrictum-clade. However, when we carried out a restricted alignment with only members of the section and reconstructing the ML tree, most of them were satisfactory identified (see Fig. S4 in supplemental material). These were P. arabicum (n = 2), P. burgense (n = 1), P. cinereoatrum (n = 1), P. momoii (n = 1), and P. rubefaciens (n = 2). The only isolate that could not be identified was FMR 15841, precisely located in the P. restrictumclade. The most recent review of the section by Visagie et al. (2016b) indicates that such a clade needs further revision due to the high variability between strains currently belonging to the clade. Thus, the identification of FMR 15841 remains uncertain until its taxonomy is resolved.
Section Lanata-Divaricata (92 % bs/1 pp) included three dung isolates. One of them was identified as P. brasilianum, a widespread species commonly found in soil (Pitt 1979) and recently also reported from human clinical specimens (Guevara-Suarez et al. 2016). The other two isolates were identified as P. cremeogriseum, a species previously found in forest soil in Ukraine (Houbraken & Samson 2011, Visagie et al. 2016a). To our knowledge, this is the first report of P. brasilianum and P. cremeogriseum associated with herbivore dung.
The only species identified in our study belonging to section Stolkia (99 % bs/1 pp) was P. canariense. This species was described from soil in Canary Islands (Peterson & Sigler 2002), the same geographical origin as our isolate. This is the second isolate of the species found so far.
Isolates of section Citrina (98 % bs/1 pp) were identified as P. citrinum (n = 5) and P. sizovae (n = 2). Although we have not found previous records of these species from dung, they have a worldwide distribution, being isolated from soil, foodstuff, and many other types of substrates (Houbraken et al. 2011). This section comprises nearly 40 species, but only those identified here were included in the analysis to simplify the tub2 phylogenetic tree (Fig. 1).
In sections Sclerotiora (77 % bs/-- pp) and Cinnamopurpurea (96 % bs/-- pp) P. lilacinoechinulatum (n = 1) and P cvjetkovicii (n = 1) were respectively identified. Although the former species was previously considered synonym of P. bilaiae, they are currently recognised as distinct penicillia (Visagie et al. 2013). P. lilacinoechinulatum is an uncommon species which type was collected from Japanese soil (Abe 1956). Penicillium cvjetkovicii is a recently described from indoor air samples in the USA (Peterson et al. 2015).
Aspergilloides (99 % bs/1 pp) was the last section in Penicillium that included dung isolates. The species identified were P. glabrum (n = 3), P. frequentans (n = 2), P. roseoviride (n = 1), and P. rudallense (n = 1). Although the former two are currently accepted as distinct species, they were considered synonyms for a long time (Houbraken et al. 2014b). Penicillium frequentans together with P. spinulosum and P. glabrum are generally the most common species in the section, being isolated from a wide range of substrates, including soil, food, bark, and indoor environments (Houbraken et al. 2014b).
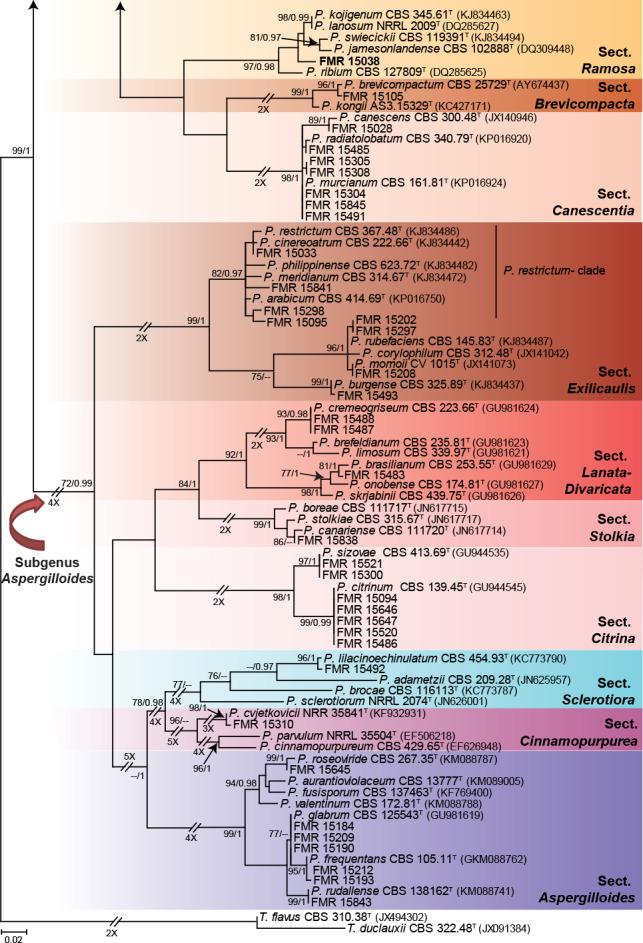
Talaromyces phylogeny
The phylogenetic tree based on the tub2 locus (Fig. 6) shows the relationships of the nine Talaromyces isolates included in the study. The aligned dataset was 388 bp long, from which 217 were variable sites and 170 phylogenetic informative. The ML substitution model was K2+G+I, while for BI it was HKY+G+I. Two main clades were formed, representing sections Talaromyces (98 % bs/1 pp) and Trachyspermi (90 % bs/1 pp). Only three sect. Talaromyces species were identified, namely T. muroii (FMR 15496), T. ruber (FMR 15839) and T. sayulitensis (FMR 15842); the rest (FMR 15489, FMR15490, FMR 15199, FMR 15035 and FMR 15307) could not be identified with this single marker, just like the only isolate (FMR 16441) included in section Trachyspermi.
Fig. 6.
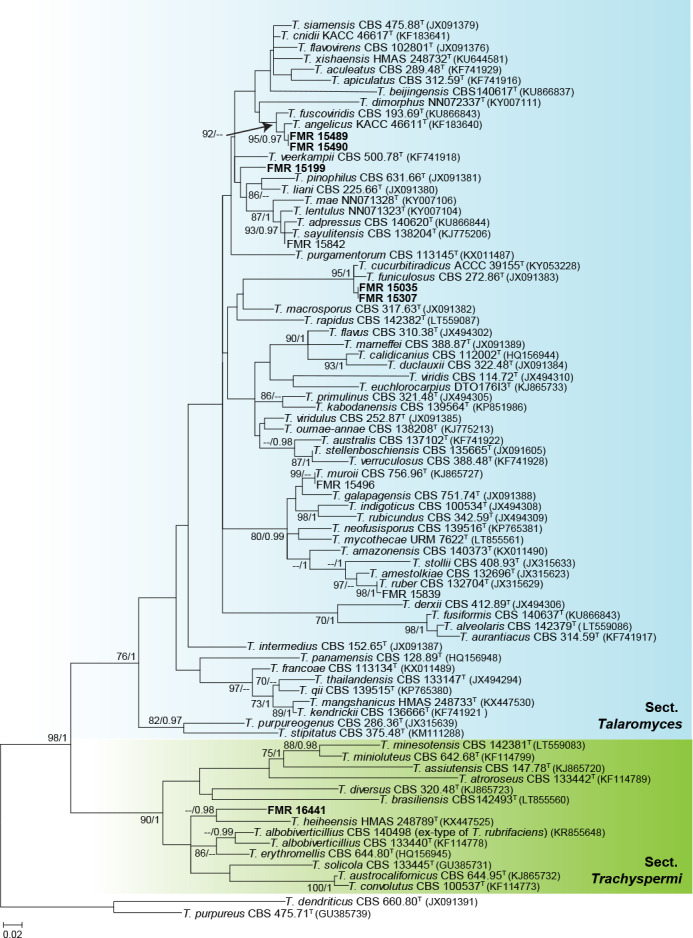
ML tree of Talaromyces inferred from tub2 including the sections recovered from dung in this work (Talaromyces and Trachyspermi). Branch lengths are proportional to phylogenetic distance. Some of the larger branches were condensed, with the proportions showed above the parallel diagonal lines. Bootstrap support values/Bayesian posterior probability scores above 69 %/0.95 are indicated on the nodes. Isolates corresponding to new species are shown in bold. Between parentheses, GenBank accession numbers of tub2 sequences. The tree is rooted to T. dendriticus CBS 660.80 and T. purpureus CBS 475.71 (Section Purpurei). T = ex-type strain.
To resolve the taxonomy of these unidentified isolates, we calculated a phylogeny for each section using sequences of the ITS, tub2 and cmdA genes from the accepted species in both sections, except for T. amyrossmaniae in section Trachyspermi. This is a recently described species that morphologically differs from our isolates and species of Trachyspermi by producing short determinate synnemata on both the natural substrate and in vitro (Rajeshkumar et al. 2019). Although rpb2 was sequenced for all our isolates (Table 1), this marker was not incorporated in the concatenated analysis due to the lack of sequences for all ex-type strains. The combined phylogeny of the members of section Talaromyces is shown in Fig. 7. This analysis showed that the unidentified isolates FMR 15489 and FMR 15490 exhibited both a similarity of 99.42 % with respect to the ex-type strain of T. angelicus, a monotypic species described by Sang et al. (2013). Although most of their morphological features matched with those of the protologue of T. angelicus, we observed some phenotypic variation with respect to the description of the species by Yilmaz et al. (2014); i.e, production of diffusible red pigment in one of our isolates (FMR 15490), the colonies of our isolates were deep turquoise and pastel red rather than yellow to white on MEA, they grew faster at 37 °C (on CYA 34–37 mm diam in 7 d vs 25–27 mm in Yilmaz et al. 2014), and exhibited stipes up to 100 μm long (up to 120 μm in Yilmaz et al. 2014). On the other hand, while FMR 15199 was located in an independent branch clearly separated from the other species of the section; FMR 15035 and FMR 15307 clustered together and were closely related to T. curcubitiradicus and T. funiculosus, but with enough genetic difference to be considered a distinct species (see also Fig. S5 in supplemental material). These genetic differences and the particular phenotypic features observed in the three FMR isolates allow us to propose the following new species, T. coprophilus (FMR 15199) and T. pseudofuniculosus (FMR 15035 and FMR 15307). Both can be easily identified with the tub2 marker.
Fig. 7.
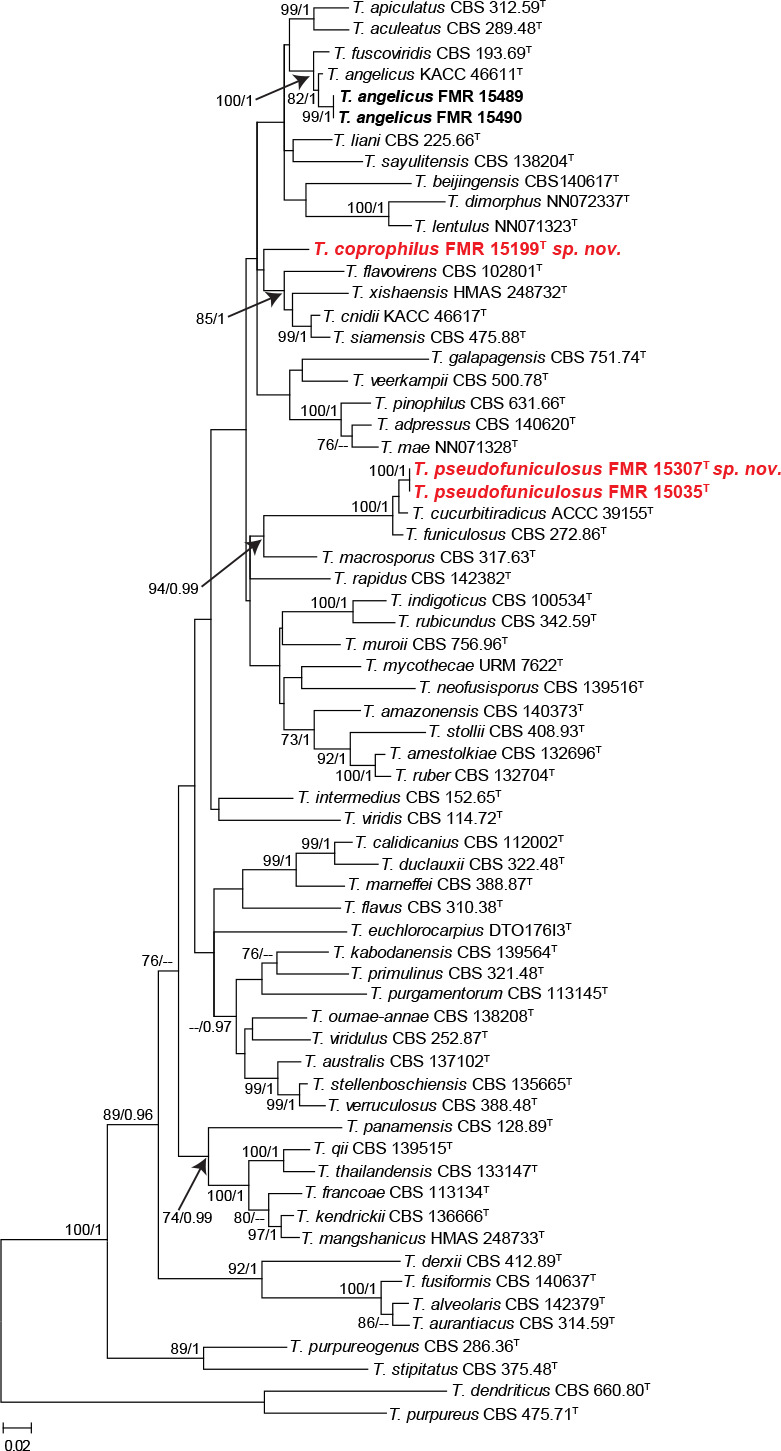
ML tree of Talaromyces section Talaromyces inferred from the combined ITS, tub2 and cmdA loci. Branch lengths are proportional to phylogenetic distance. Bootstrap support values/Bayesian posterior probability scores above 70 %/0.95 are indicated on the nodes. The tree is rooted to T. dendriticus CBS 660.80 and T. purpureus CBS 475.71 (Section Purpurei). The names in red are the new species described in this study. T = ex-type strain.
The combined phylogenetic analysis of the section Trachyspermi (Fig. 8) showed that the isolate FMR 16441 was located in a single branch within the same clade as T. albobiverticillius, T. erythromellis, T. heiheensis, and T. solicola (88 % bs/0.99 pp). Considering the phylogenetic position of our isolate and the morphological differences observed, the new species T. catalonicus is proposed.
Fig. 8.
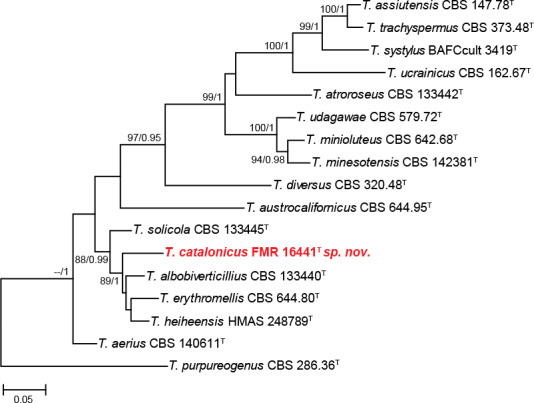
ML tree of selected of Talaromyces section Trachyspermi inferred from the combined ITS, tub2 and cmdA loci. Branch lengths are proportional to phylogenetic distance. Bootstrap support values/Bayesian posterior probability scores above 70 %/0.94 are indicated on the nodes. The tree is rooted to T. purpureogenus CBS 286.36 (Section Talaromyces). The name in red is the new species described in this study. T = ex-type strain.
Phylogeny of miscellaneous Aspergillaceae
According to the tub2 phylogeny, isolates FMR 15296, FMR 16442, FMR 14718 and FMR 15299 were related to some penicillia currently excluded from the genus Penicillium (i.e. P. giganteum, P. megasporum and P. nodositatum), but belonging to the family Aspergillaceae (Fig. 9). Previous molecular data on these fungi suggested that they could represent distinct genera in the family (Peterson et al. 2010, Visagie et al. 2013). The aligned dataset of our analysis was 393 bp long, from which 242 were variable sites and 216 phylogenetic informative. The best substitution model for ML was K2 +G+I and for BI it was GTR +G+I. While FMR 15296 and FMR 16442 were located in a well-supported clade along with the ex-type strain of P. nodositatum (CBS 333.90), the other two (FMR 14718 and FMR 15299) were in a fully supported distant clade together with the ex-type strains of P. giganteum (NRRL 3553) and P. megasporum (NRRL 2232). The phylogeny inferred with sequences of the ITS, tub2 and rpb2 genes (Fig. 10), including the isolates under study and members of different genera of Aspergillaceae, confirms the proposal of two novel genera, which are described below as Penicillago (Pgo.), typified by Penicillium nodositatum, and Pseudopenicillium (Pse.) with Penicillium giganteum and Penicillium megasporum, this latter species here selected as the type. The former genus is phylogenetically more related to Penicillium, whereas the latter to the genus Hamigera.
Fig. 9.
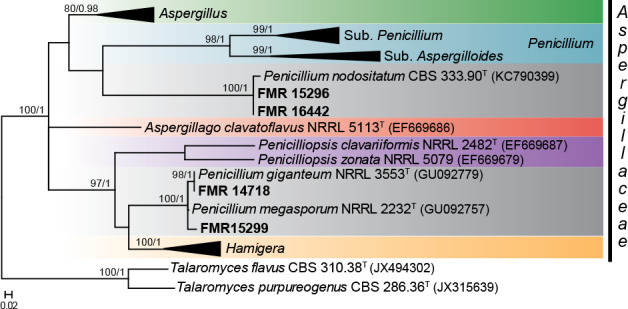
ML tree of selected members of Aspergillaceae family inferred from tub2. Branch lengths are proportional to phylogenetic distance. Some clades were collapsed to facilitate layout and illustrate the phylogenetic relationships between the shown genera. Bootstrap support values/Bayesian posterior probability scores above 70 %/0.95 are indicated on the nodes. Isolates corresponding to new or interesting taxa are shown in bold. Between parentheses, GenBank accession numbers of tub2 sequences. The tree is rooted to T. flavus CBS 310.38 and T. purpureogenus CBS 286.36. T = ex-type strain.
Fig. 10.
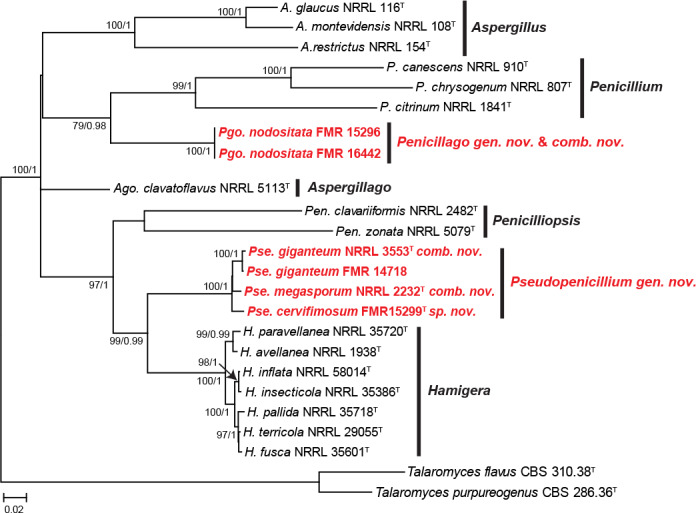
ML tree of members of Aspergillaceae family inferred from the combined ITS, tub2, and rpb2 loci. Branch lengths are proportional to phylogenetic distance. Bootstrap support values/Bayesian posterior probability scores above 70 %/0.95 are indicated on the nodes. The tree is rooted to Talaromyces flavus CBS 310.38 and Talaromyces purpureogenus CBS 286.36. The names in red are the new taxa proposed in this study. T = ex-type strain.
The analysis of ITS and tub2 showed that the ex-type strain of P. nodositatum (CBS 333.90) and the isolates FMR 15296 and FMR 16442 have practically identical sequences. However, CBS 333.90 was not included in the concatenated phylogeny with the three markers (Fig. 10) since its rpb2 sequence was not available for comparison. Despite this, we observed some cultural difference between our isolates respect to the protologue of P. nodositatum (Valla et al. 1989), microscopically they fit with the features of the species. Differences include better growth for FMR 15296 and FMR 16442 on CYA and MEA at 25 °C (40–44 mm and 34–36 mm diam in 7 d, respectively, vs 13–19 mm and 16–30 mm in CBS 333.90) and showed the ability to grow at 37 °C. Considering the phylogenetic position of our isolates and their morphological similarity with P. nodositatum, they were the base to propose the new combination Pgo. nodositata. Other fungus excluded from Penicillium s. str. and closely related to Pgo. nodositata was P. kabunicum (Visagie et al. 2013). Since only the ITS barcode was available for the ex-type strain of the latter species (CBS 575.90) in the GenBank database (MH862240), we constructed an ITS phylogenetic tree with different genera in Aspergillaceae showing that P. kabunicum clustered with Pgo. nodositata (see Fig. S6 in supplemental material).
The isolate FMR 14718 matched morphologically and molecularly with P. giganteum, whereas FMR 15299 was placed in a new lineage into the Pseudopenicillium clade (Figs 9, 10). The morphological differences observed, such as having smaller conidia (up to 6 μm) than those of the other two species in the genus (conidia up to 12 μm in Pse. giganteum, and up to 10 μm in Pse. megasporum), and the genetic differences obtained (98 % similar to Pse. giganteum, and 98.16 % similar to Pse. megasporum) allow us to propose the new species Pse. cervifimosum.
Taxonomy
Penicillium balearicum Guevara-Suarez, Cano & Gené, sp. nov. MycoBank MB822061. Fig. 11.
Fig. 11.
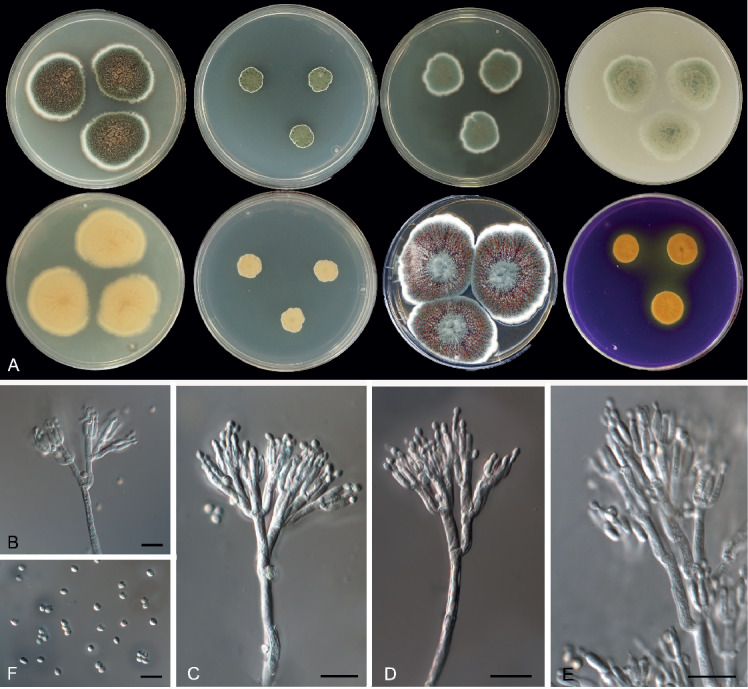
Morphological characters of Penicillium balearicum (FMR 15191T). A. Colonies from left to right (top row) CYA, MEA, DG18 and OA; (bottom row) CYA reverse, MEA reverse, YES, and CREA. B–E. Conidiophores. F. Conidia. Scale bars = 10 μm.
Etymology: Name refers to the geographical area (the Balearic Islands) where the species was found.
In: Section Paradoxa.
Typus: Spain, Balearic Islands, Mallorca, Pollença, from herbivore dung, Feb. 2016, J. Guarro, M. Guevara-Suarez & J.P.Z. Siqueira (holotype CBS H-23215; culture ex-type FMR 15191 = CBS 143044; ITS barcode LT899762, alternative markers: tub2 LT898227, cmdA LT899758, rpb2 LT899760).
Additional material examined: Spain, Balearic Islands, Mallorca, Pollença, from herbivore dung, Feb. 2016, J. Guarro, M. Guevara-Suarez & J.P.Z. Siqueira (FMR 15196).
Colony diameter in 7 d (mm): On CYA: 25 °C 25–36, 30 °C 5–6, 37 °C no growth; on MEA: 25 °C 14–17, 30 °C 4–6; on YES: 25 °C 36–45, 30 °C 4–6; on OA: 25 °C 26–30, 30 °C 2–6; on DG18: 25 °C 14–27, on CREA: 25 °C 14–20.
Colony characters at 25 °C in 7 d: On CYA, colonies velvety, flat, mycelium white, sporulation dense, conidial masses dark dull green (28E3), margin dentate; reverse pale orange (5A3); exudate present, with pale droplets; soluble pigment absent. On MEA, colonies velvety, elevated, sporulation dense, conidial masses dark dull green (28E3), margin lobate; reverse light orange (5A4); exudate and soluble pigment absent. On DG18, colonies flat and velvety, mycelium white, sporulation dense, conidial masses dull green (27D4); reverse greenish white (27B2). On YES, colonies cerebriform, raised at the centre, radially sulcate, mycelium white, sporulation dense at the centre, with conidial masses greenish white (27C2), margin lobate; reverse bright orange-red (8A8); exudate present, small brown (7E6-8) droplets; soluble pigment absent. On OA, colonies fasciculate, mycelium white, sporulation dense, with conidial masses greenish grey (28B2); exudate and soluble pigment absent. On CREA, acid production moderately strong.
Micromorphology: On MEA, conidiophores typically terverticillate, minor proportion biverticillate and quaterverticillate; stipes smooth, 250–500 × 2–2.5 μm; metula 3–4 per branch, divergent, 10–14 × 2–3 μm; phialides 3–4 per metula, ampulliform, 7–10 × 2–2.5 μm; conidia globose to subglobose, 2.5–3 × 2–3 μm, smooth-walled, brownish yellow.
Distinguishing characters: Penicillium balearicum is resolved in the P. atramentosum-clade containing six species, P. atramentosum, P. balearicum, P. fimosum, P. ibericum (the latter three described here), P. magnielliptisporum and P. mexicanum (Visagie et al. 2014a). Penicillium atramentosum, P. balearicum, P. magnielliptisporum and P. mexicanum have smooth-walled stipes and conidia, and the conidia of the latter two are larger (5 × 4 μm in P. magnielliptisporum, 4 × 3.5 μm in P. mexicanum); while P. atramentosum can be differentiated by its production of red soluble pigment on CYA and by the lack of growth at 30 °C (Pitt 1979, Frisvad & Samson 2004). Penicillium balearicum differs from P. fimosum and P. ibericum mainly by having longer stipes (up to 500 μm long), by the production of abundant brown exudate droplets on YES, and by the moderately strong acid production on CREA.
Penicillium beceitense Guevara-Suarez, Gené & Guarro, sp. nov. MycoBank MB822063. Fig. 12.
Fig. 12.
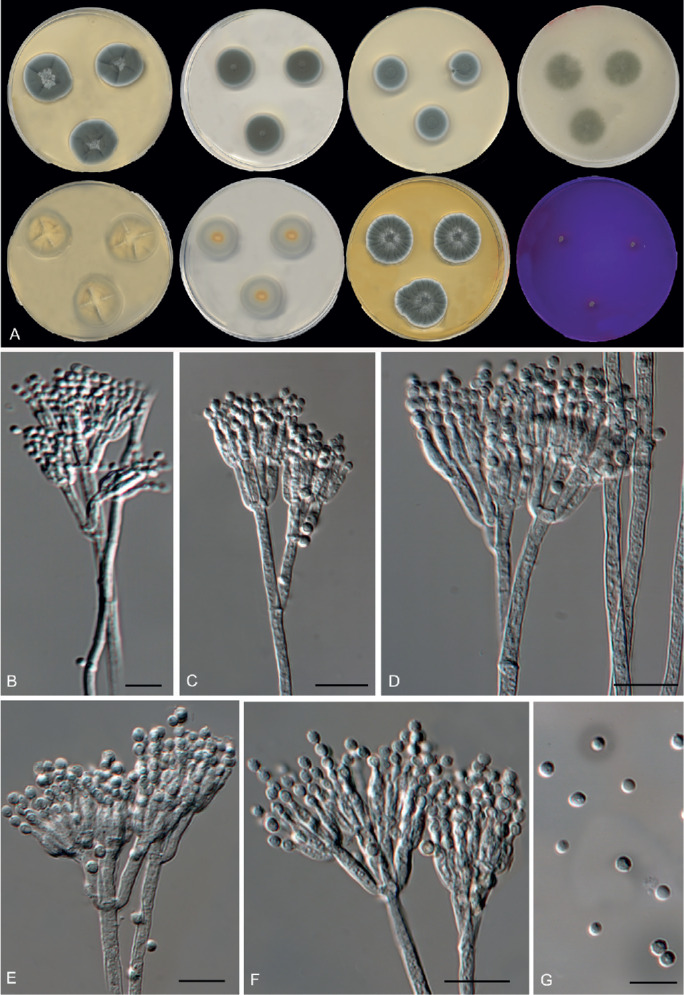
Morphological characters of Penicillium beceitense (FMR 15038T). A. Colonies from left to right (top row) CYA, MEA, DG18 and OA; (bottom row) CYA reverse, MEA reverse, YES, and CREA. B–F. Conidiophores. G. Conidia. Scale bars = 10 μm.
Etymology: Name refers to Beceite, the vilage where the fungus was found.
In: Section Ramosa.
Typus: Spain, Aragon, Beceite, from herbivore dung, Feb. 2016, J. Guarro, M. Guevara-Suarez, E. Rosas (holotype CBS H-23183; cultures ex-type FMR 15038 = CBS 142989; ITS barcode LT899780, alternative markers: tub2 LT898229, cmdA LT899764, rpb2 LT899798).
Colony diameter in 7 d (mm): On CYA: 25 °C 25–27, 30 °C no growth, 37 °C no growth; on MEA: 25 °C 23–25; on YES: 25 °C 26–28; on OA: 25 °C 26–30; on DG18: 25 °C 19–20, on CREA: 25 °C 3–5.
Colony characters at 25 °C in 7 d: On CYA, colonies sunken at the centre, slightly radially sulcate, velvety, mycelium white, sporulation dense, with conidial masses dull green (25E4), margin entire; reverse greyish green (28C5); exudate and soluble pigment absent. On MEA, colonies flat, velvety, mycelium white, sporulation dense, with conidial masses conidial masses dull green (25E4), margin entire; reverse orange (5A6) to greenish grey (30C2); exudate and soluble pigment absent. On DG18, colonies flat, velvety, mycelium white, sporulation dense, conidial masses dull green (26E4), margin entire; reverse greyish green (30B5). On YES, colonies radially sulcate, velvety, mycelium white, sporulation dense, with conidial masses dark green (27F14), margin lobate; reverse greyish yellow (4B5); exudate and soluble pigment absent. On OA, colonies flat, velvety, sporulation dense, conidial masses dark dull green (28D4), margin entire; reverse greenish grey (30C2); exudate and soluble pigment absent. On CREA, acid production absent.
Micromorphology: On MEA, conidiophores predominantly terverticillate, minor proportion biverticillate; stipes 190–250 × 3.5–4 μm, smooth-walled; branches 30–40 μm; metula 3–5 per branch, divergent, cylindrical, 10–12(–15) × 2.5–3 μm; phialides 4–6 per metula, ampulliform, 8–10 × 2.5–3 μm; conidia globose, 2.5–3 × 2.5–3 μm, smooth to finely roughened, greenish.
Distinguishing characters: Penicillium beceitense is closely related to P. lanosum and P. kojigenum, two species that are indistinguishable according to our phylogeny of the section Ramosa (Fig. 5). To date, there is no updated description for either P. lanosum or P. kojigenum. The protologue of P. kojigenum described conidia that are rough-walled, measuring 2.2–2.6 μm (Smith 1961). The absence of growth at 30 °C is the relevant feature to distinguish P. beceitense from other species of the section, such as P. chroogomphum, P. jamesonlandense, P. lanosum, P. ribium, and P. soppii (Rong et al. 2016).
Penicillium caprifimosum Guevara-Suarez, Dania García & Cano, sp. nov. MycoBank MB822064. Fig. 13.
Fig. 13.
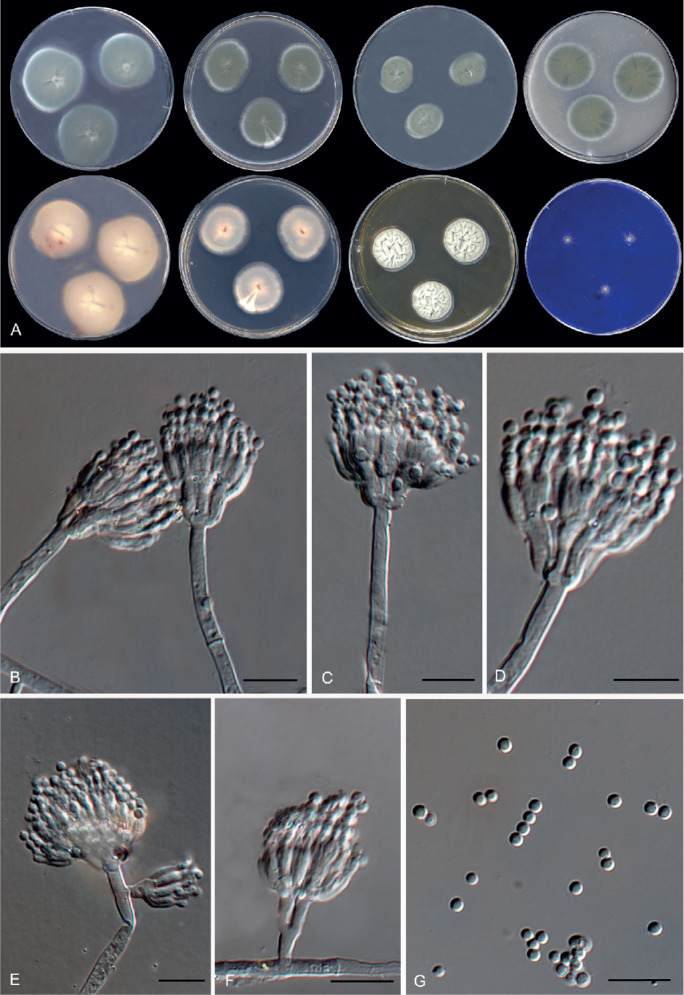
Morphological characters of Penicillium caprifimosum (FMR 15041T). A. Colonies from left to right (top row) CYA, MEA, DG18 and OA; (bottom row) CYA reverse, MEA reverse, YES, and CREA. B–F. Conidiophores. G. Conidia. Scale bars = 10 μm.
Etymology: From the Latin capra = “goat”, and fimosum = “dungd-welling”, describing the substrate from where the species was isolated.
In: Section Turbata.
Typus: Spain, Catalonia, Els Ports Natural Park, from goat dung, Feb. 2016, J. Guarro, M. Guevara-Suarez & E. Rosas (holotype CBS H-23184; culture ex-type FMR 15041 = CBS 142990; ITS barcode: LT899781, alternative markers: tub2 LT898238, cmdA LT899765, rpb2 LT899799).
Colony diameter in 7 d (mm): On CYA: 25 °C 30–35, 30 °C 35–38, 37 °C 18–20; on MEA: 25 °C 24–26, 30 °C 19–21, 37 °C 8–10; on YES: 25 °C 25–24, 30 °C 30–31; on OA: 25 °C 23–25, 30 °C 28–30, 37 °C 9–11; on DG18: 25 °C 16–21, on CREA: 25 °C 3–6.
Colony characters at 25 °C in 7 d: On CYA, colonies flat, velvety, mycelium white, sporulation dense, with conidial masses dull green (28E5), margin entire; reverse purplish grey (14B2); exudate absent; soluble pigment pinkish (12A2). On MEA, colonies flat, velvety, sporulation dense, conidial masses dark dull green (28E3), margin lobate; reverse pale yellow (4A3) to greenish grey (26B3); exudate and soluble pigment absent. On DG18, colonies velvety, flat, mycelium white, sporulation dense, conidial masses dull green (27D4); reverse greyish green (27B5). On YES, colonies velvety, raised at the centre, cerebriform, mycelium greenish white (27A2), sporulation sparse, margin lobate; reverse light brown (7D5); exudate and soluble pigment absent. On OA, colonies granulose and lat, radially sulcate towards the periphery, mycelium white, sporulation dense, conidial masses dull green (28E2), margin entire; exudate and soluble pigment absent. On CREA, acid not produced.
Micromorphology: On MEA, conidiophores biverticillate, sometimes with subterminal branches; stipes smooth-walled, 100–250 × 3–3.5 μm; metula 3–4 per branch, appressed, 10–12 × 2–3 μm; phialides 3–4 per metula, ampulliform, 8–10 × 2–2.5 μm; conidia globose to subglobose, 2.5–3 × 2–2.5 μm, smooth-walled, brownish yellow.
Distinguishing characters: Penicillium caprifimosum can be differentiated from its closest relative, P. bovifimosum described from dry cow manure (Tuthill & Frisvad 2002), by its better growth at 25 and 37 °C on MEA and CYA. The maximum colony diameter reported for P. bovifimosum is 16–21 mm on MEA and 25–29 mm on CYA in 7 d at 25 °C and 6 mm on CYA at 37 °C (Tuthill & Frisvad 2002). In addition, this species produces cleistothecia in culture, which are absent in P. caprifimosum.
Penicillium fimosum Guevara-Suarez, Guarro & Dania García, sp. nov. MycoBank MB822069. Fig. 14.
Fig. 14.
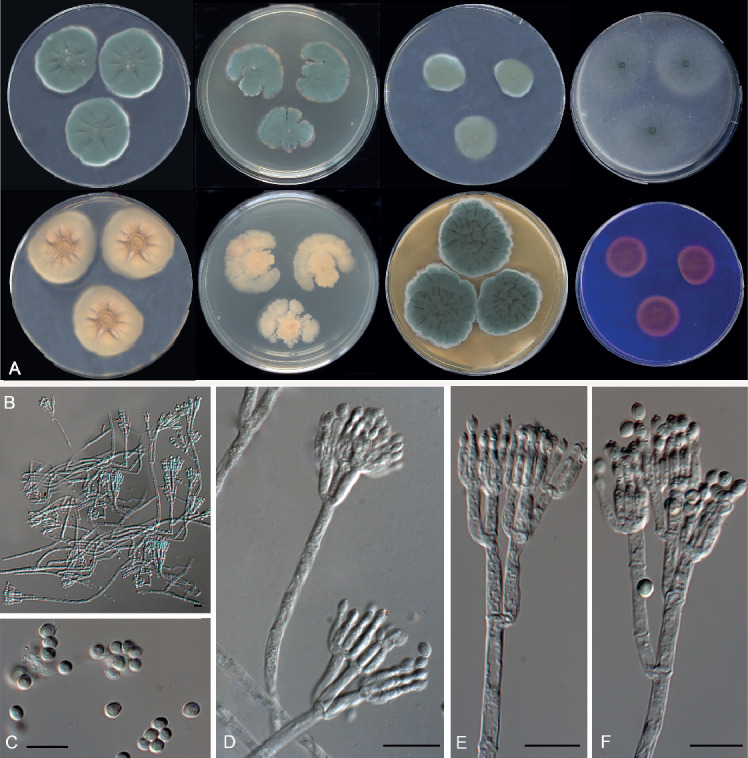
Morphological characters of Penicillium fimosum (FMR 15104T). A. Colonies from left to right (top row) CYA, MEA, DG18 and OA; (bottom row) CYA reverse, MEA reverse, YES, and CREA. B, D–F. Conidiophores. C. Conidia. Scale bars = 10 μm.
Etymology: Name refers to the substrate from where the species was isolated.
In: Section Paradoxa.
Typus: Spain, Catalonia, Pratdip, from hervibore dung, Feb. 2016, D. García (holotype CBS H-23185; culture ex-type FMR 15104 = CBS 142991; ITS barcode: LT970836, alternative markers: tub2 LT898273, cmdA LT970837).
Colony diameter in 7 d (mm): On CYA: 25 °C 31–34, 30 °C 3–4, 37 °C no growth; on MEA: 25 °C 25–30, 30 °C no growth; on YES: 25 °C 50–55, 30 °C 5–7; on OA: 25 °C 25–27, 30 °C no growth; on DG18: 25 °C 16–20, on CREA: 25 °C 20–23.
Colony characters at 25 °C in 7 d: On CYA, colonies velvety, slightly radially sulcate at the centre, mycelium white, sporulation dense, conidial masses dark dull green (26E4), margin slightly lobate and fimbriate; reverse greyish orange (6B4) at the centre, greyish green (28C3) towards the periphery; exudate and soluble pigment absent. On MEA, colonies flat, velvety, sporulation dense, conidial masses dark dull green (28E3), irregular, margin slightly crenate; reverse light orange (5A4) to dull green (28D4); exudate and soluble pigment absent. On DG18, colonies velvety, flat, mycelium white, sporulation dense, conidial masses dull green (27D4); reverse greenish white (27B2). On YES, colonies irregularly sulcate, slightly raised at the centre, mycelium white, sporulation dense, with conidial masses dull green (26E5), margin slightly crenate; reverse bright orange-red (8A8) at the centre; exudate and soluble pigment absent. On OA, colonies slightly granulose, mycelium white, sporulation dense, conidial masses greyish green (28E7); exudate and soluble pigment absent. On CREA, moderate acid production.
Micromorphology: On MEA, conidiophores predominantly biverticillate, minor proportion terverticillate and some irregularly branched; stipes 70–130 × 2.5–3(–4) μm, smooth; metula 3–4 per branch, rather divergent, 8–14 × 2.5–3(–4) μm; phialides 3–4 per metula, ampulliform, (6–)8–10 × 2.5–3 μm; conidia globose to subglobose, 2.5–3 × 2.5–3(–4) μm, smooth, brownish yellow.
Distinguishing characters: Penicillium fimosum is closely related to P. atramentosum and P. ibericum (described below). Penicillium fimosum and P. atramentosum differ from P. ibericum by the absence or limited growth at 30 °C. Penicilium atramentosum can be distinguished by the production of reddish brown exudate, soluble pigment and colony reverse on CYA, features absent in P. fimosum. Moreover, P. fimosum has shorter stipes (70–130 μm) than P. atramentosum (300–500 μm) (Pitt 1979).
Penicillium ibericum Guevara-Suarez, Cano & Dania García, sp. nov. MycoBank MB822070. Fig. 15.
Fig. 15.
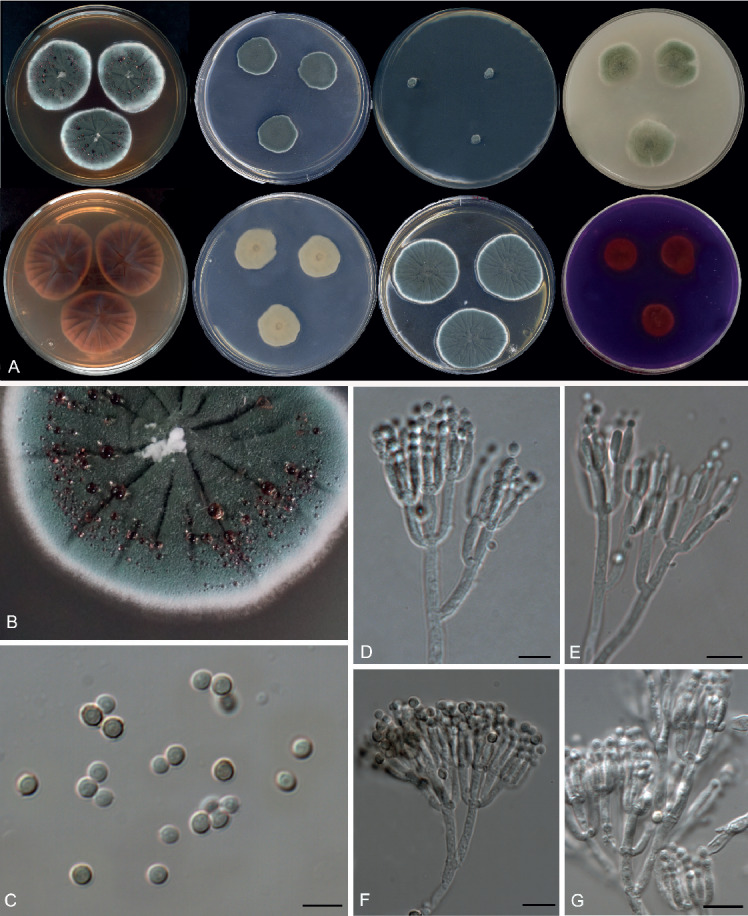
Morphological characters of Penicillium ibericum (FMR 15040T). A. Colonies from left to right (top row) CYA, MEA, DG18 and OA; (bottom row) CYA reverse, MEA reverse, YES, and CREA. B. Colony texture on CYA at 25 °C after 1 wk incubation. C. Conidia. D–G. Conidiophores. Scale bars = 10 μm.
Etymology: Name refers to the occurrence of the species on the Iberian Peninsula.
In: Section Paradoxa.
Typus: Spain, Catalonia, Els Ports Natural Park, from herbivore dung, Feb. 2016, J. Guarro, M. Guevara-Suarez & E. Rosas (holotype CBS H-23186; culture ex-type FMR 15040 = CBS 142992; ITS barcode: LT899782, alternative markers: tub2 LT898285, cmdA LT899766, rpb2 LT899800).
Additional material examined: Spain, Galicia, Las Dunas de Corrubedo Natural Park, from soil, Feb. 2016, D. García (FMR 15107).
Colony diameter in 7 d (mm): On CYA: 25 °C 31–35, 30 °C 18–21, 37 °C no growth; on MEA: 25 °C 18–20, 30 °C 8–9; on YES: 25 °C 33–35, 30 °C 18–20; on OA: 25 °C 21–23, 30 °C no growth; on DG18: 25 °C 5–6; on CREA: 25 °C 21–22.
Colony characters at 25 °C in 7 d: On CYA, colonies flat, velvety, slightly radially sulcate, mycelium white, sporulation dense, conidial masses dark dull green (26E4), margin slightly lobate; reverse reddish brown (9E5) to reddish (9A2); exudate present, with pale red (10A3) droplets; soluble pigment reddish brown. On MEA, colonies, velvety and flat, sporulation dense, conidial masses dark dull green (28E3); reverse light orange (5A4), margin slightly irregular; exudate and soluble pigment absent. On DG18, colonies sunken in the middle, velvety, mycelium white, sporulation moderate, conidial masses dull green (27D4); reverse greenish white (27B2). On YES, velvety, sulcate, mycelium white, sporulation dense, conidial masses dull green (26E5), margin crenate; reverse orange-grey (5B3); exudate and soluble pigment absent. On OA, colonies irregular, granulose, mycelium white, sporulation dense, conidial masses greyish green (28E7); exudate and soluble pigment absent. On CREA, moderate acid production.
Micromorphology: On MEA, conidiophores predominantly terverticillate, minor proportion quarterverticillate; stipes 70–150 × 2–3(–4) μm, smooth; metula 2–4 per branch, divergent, 8–12 × 2–2.5 μm; phialides 3–4 per metula, ampulliform, 8–10 × 2.5–3 μm; conidia globose, 2.5–3 × 2.5–3 μm, smooth to finely roughened, subhyaline to pale green.
Distinguishing characters: Penicillium ibericum is closely related to P. atramentosum and P. fimosum. It can be distinguished by having shorter stipes (70–150 μm) than P. atramentosum (300–500 μm) (Pitt 1979), and by its more restricted growth on YES (33–35 mm diam) and DG18 (5–6 mm) compared to P. fimosum (YES 50–55 mm, DG18 16–20 mm).
Penicillium mediterraneum Guevara-Suarez, Gené & Cano, sp. nov. MycoBank MB822071. Fig. 16.
Fig. 16.
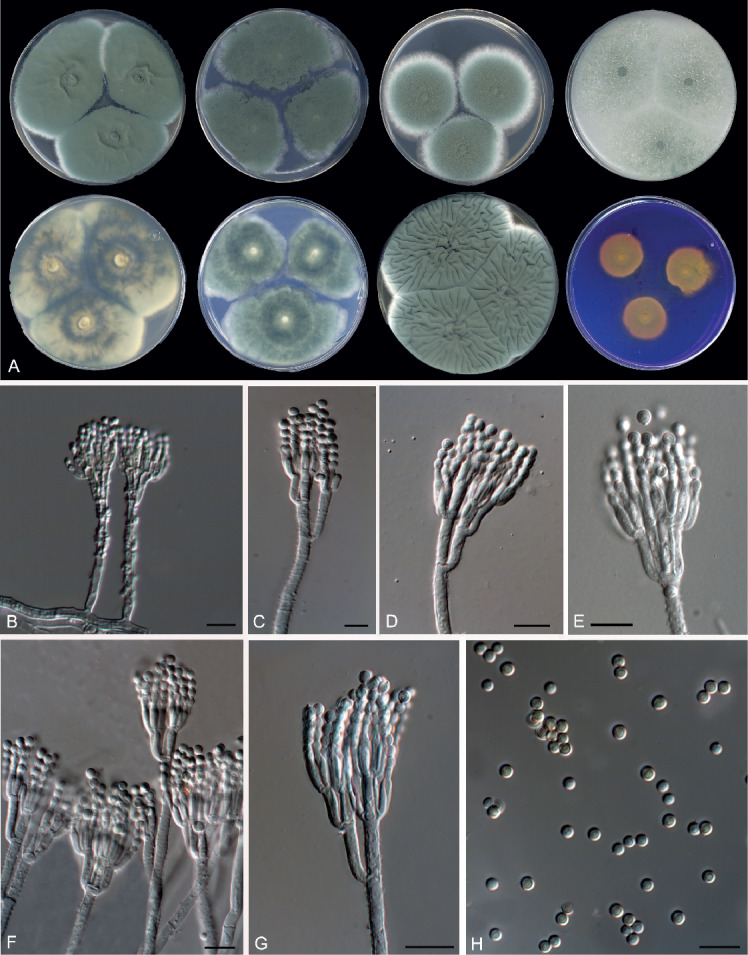
Morphological characters of Penicillium mediterraneum (FMR 15188T). A. Colonies from left to right (top row) CYA, MEA, DG18 and OA; (bottom row) CYA reverse, MEA reverse, YES, and CREA. B–G. Conidiophores. H. Conidia. Scale bars = 10 μm.
Etymology: Name refers to the geographical area where the species was found.
In: Section Roquefortorum.
Typus: Spain, Balearic Islands, Mallorca, Puigpuñent, from herbivore dung, Feb. 2016, J. Guarro, M. Guevara-Suarez & J.P.Z. Siqueira (holotype CBS H-23143; culture ex-type FMR 15188 = CBS 142754; ITS barcode: LT899784, alternative markers: tub2 LT898291, cmdA LT899768, rpb2 LT899802).
Additional materials examined: Spain, Catalonia, Els Ports Natural Park, herbivore dung, Feb. 2016, J. Guarro, M. Guevara-Suarez & E. Rosas (FMR 15031 = CBS 142755); Catalonia, Els Ports Natural Park, herbivore dung, Feb. 2016, J. Guarro, M. Guevara-Suarez & E. Rosas (FMR 15032).
Colony diameter in 7 d (mm): On CYA: 25 °C 45–60, 30 °C 24–26, 37 °C no growth; on MEA: 25 °C 45–60, 30 °C 23–26; on YES: 25 °C >60, 30 °C 35–40; on OA: 25 °C 31–38, 30 °C 8–15; on DG18: 25 °C 43–45; on CREA: 25 °C 22–25.
Colony characters at 25 °C in 7 d: On CYA, colonies flat, velvety, mycelium white, sporulation dense, conidial masses dull green (25E4), margin entire; reverse greyish green (28C5); exudate and soluble pigment absent. On MEA, colonies flat, velvety, mycelium white, margin crenate, sporulation dense, with conidial masses conidial masses dull green (25E4), margin irregular, slightly lobulate, reverse greyish green (28E5); exudate and soluble pigment absent. On DG18, colonies flat, velvety, mycelium white, sporulation dense, conidial masses dull green (26E4), margin entire; reverse greyish green (30C6). On YES, colonies velvety, raised at the centre, irregularly sulcate, mycelium white, sporulation dense, conidial masses dull green (26E4), margin entire and fimbriate; reverse greyish green (28C6); exudate and soluble pigment absent. On OA, colony cottony, mycelium white, sporulation dense, conidial masses dark dull green (28D4), margin entire; exudate and soluble pigment absent. On CREA, moderate acid production.
Micromorphology: On MEA, conidiophores bi- and terverticillate; stipes rough-walled, 50–100 × 2.5–3 μm; metula 2–3 per branch, appressed, cylindrical, 10–14 × 2–3 μm; phialides 2–4 per metula, ampulliform, 10–13 × 2–3 μm; conidia globose, 2.5–4(–5) × 2–4 μm, smooth-walled, greenish.
Distinguishing characters: All the species in section Roquefortorum show fast growth on CYA and MEA and produce rough-walled conidiophores. Penicillium roqueforti is the closest-related species to P. mediterraneum, and it can be distinguished by a more restricted growth on CYA at 30 °C (5–15 mm diam) and by its blackish green colony reverse on YES (Frisvad & Samson 2004, Houbraken et al. 2010).
Penicillium synnematicola Guevara-Suarez, Dania García & Guarro, sp. nov. MycoBank MB822072. Fig. 17.
Fig. 17.
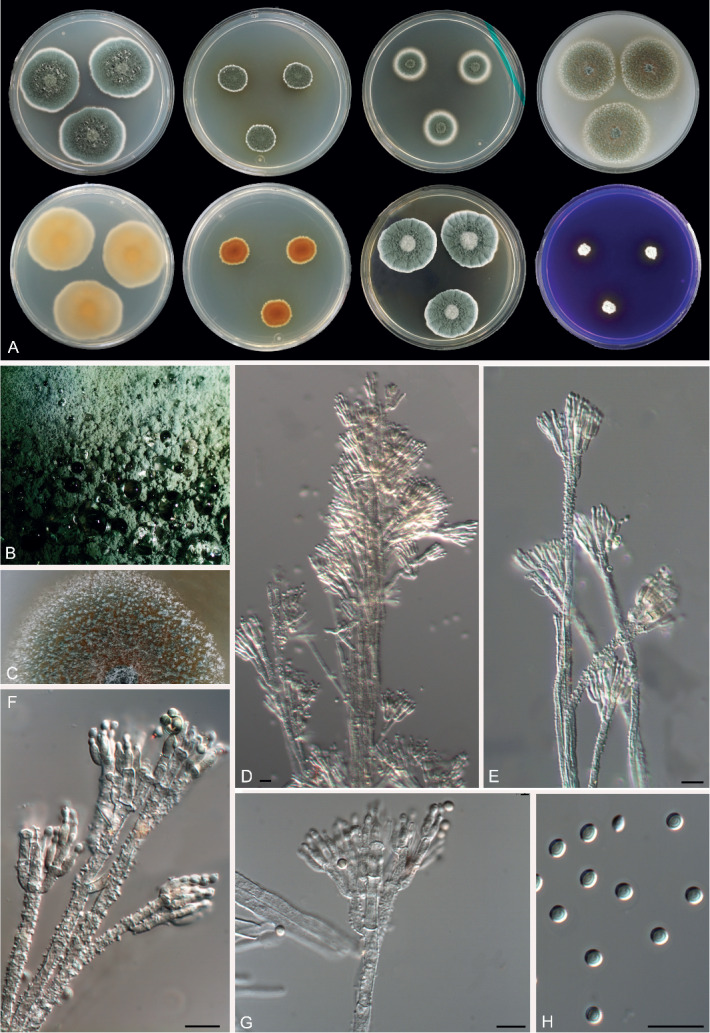
Morphological characters of Penicillium synnematicola (FMR 15192T). A. Colonies from left to right (top row) CYA, MEA, DG18 and OA; (bottom row) CYA reverse, MEA reverse, YES, and CREA. B. Colony texture on CYA at 25 °C after 1 wk incubation. C. Colony texture on OA at 25 °C after 1 wk incubation. D–G. Conidiophores. H. Conidia. Scale bars = 10 μm.
Etymology: Name refers to the formation of synnemata.
In: Section Robsamsonia.
Typus: Spain, Balearic Islands, Mallorca, Camí Vell d’Orient, from goat dung, Feb. 2016, J. Guarro, M. Guevara-Suarez & J.P.Z. Siqueira (holotype CBS H-23132, culture ex-type FMR 15192 = CBS 142669; ITS barcode: LT898167, alternative markers: tub2 LT898172, cmdA LT898137, rpb2 LT898142).
Additional materials examined: Spain, Mallorca, Escorca, from goat dung, Feb. 2016, J. Guarro, M. Guevara-Suarez & J.P.Z. Siqueira (FMR 15210); Mallorca, Camí Vell d’Orient, from goat dung, Feb. 2016, J. Guarro, M. Guevara-Suarez & J.P.Z. Siqueira (FMR 15211); Catalonia, Barcelona, Montseny Natural Park, from herbivore dung, Apr. 2017, J. Gené, M. Guevara-Suarez & I. Iturrieta-González (FMR 16481 = CBS 143045); Catalonia, Poblet, from herbivore dung, Mar. 2017, J. Guarro, Guevara-Suarez & I. Iturrieta-González (FMR 16491 = CBS 143046).
Colony diameter in 7 d (mm): On CYA: 25 °C 33–37, 30 °C 9–10, 37 °C no growth; on MEA: 25 °C 11–13, 30 °C 5–7; on YES: 25 °C 30–34, 30 °C 9–17; on OA: 25 °C 31–38, 30 °C 8–15; on DG18: 25 °C 17–19; on CREA: 25 °C 6–9.
Colony characters at 25 °C in 7 d: On CYA, colonies granular, raised at the centre, mycelium white, sporulation dense, conidial masses dark dull green (25E4), margin dentate; reverse light orange (5A5); exudate present, consisting of small hyaline droplets; soluble pigment absent. On MEA, colonies fasciculate due to the presence of small feathery synnemata, mycelium white, sporulation dense, with conidial masses dark green (28F5), margin crenate; reverse orange (6B8); exudate absent and soluble pigment deep orange (5A8). On DG18, colonies velvety, mycelium white, sporulation dense, conidial masses dull green (27E4); reverse greyish orange (5B3). On YES, colonies granular, raised at the centre, radially sulcate, mycelium white, sporulation dense, conidial masses dull green (27E4), margin dentate; reverse brownish orange (6C8); exudate and soluble pigment absent. On OA, colony strongly fasciculate, mycelium white, sporulation dense, conidial masses dark dull green (27E4), margin crenate; reverse yellowish white (4A); exudate and soluble pigment absent. On CREA, weak acid production.
Micromorphology: On MEA, synnemata present, up to 1 mm long; conidiophores ter- to quaterverticillate, minor proportion biverticillate; stipes 100–218 × 3–4 μm (up to 1 mm long in synnemata), coarsely roughened; metula 3–4 per branch, rather appressed, cylindrical, smooth to conspicuously roughened, 10–14 × 2–3 μm; phialides 2–4 per metula, ampulliform, 10–13 × 2–3 μm; conidia globose, subglobose to broadly ellipsoidal, 2.5–3 × 2–2.5 μm, smooth-walled, subhyaline to pale green.
Distinguishing characters: Section Robsamsonia includes some species that produce synnemata, e.g. P. coprophilum, P. glandicola and P. vulpinum. Penicillium coprophilum and P. vulpinum can be differentiated from our new species by their smooth-walled stipes (Houbraken et al. 2016). Penicillium synnematicola is closely related to P. glandicola, a species with world-wide distribution isolated from oak trees and forest soil, including dungy soil and mouse dung (Houbraken et al. 2016), and both produce coarsely roughened synnematous conidiophores. However, P. glandicola can be distinguished by having shorter phialides (7.5–10.5 µm) and ellipsoidal and yellow-green conidia. In addition, the colonies of P. glandicola show orange-brown to brown and bright orange-red reverse on CYA and YES, respectively, and tend to have a more restricted growth on both media (17–30 mm on CYA; 19–36 mm on YES) (Houbraken et al. 2016).
Penicillago Guevara-Suarez, Gené & Dania García, gen. nov. MycoBank MB822073.
Etymology: Name refers to the phylogenetic closeness to Penicillium (Fig. 10).
In: Family Aspergillaceae.
Diagnosis: Morphologically differs from Penicillium by its symmetrically biverticillate conidiophores, ampulliform to acerose phialides with a fine long neck, and conidia with conspicuous disjunctors. Conidiophore structure resembles Talaromyces, but phylogenetically distant.
Mycelium superficial and immersed, composed of septate, branched, canary yellow to chrome yellow hyphae. Conidiophores symmetrically branched, penicillium-like, composed of long, septate, hyaline stipes, often terminating in a small vesicle giving rise to a verticil of metulae, occasionally irregularly branched; metulae appressed to divergent, cylindrical to somewhat obpyriform, bearing a compact verticil of conidiogenous cells. Conidiogenous cells phialidic, ampulliform to acerose, with a fine and long neck. Conidia dry, catenate, with conspicuous disjunctors, subglobose to ellipsoid, coarsely echinulate, subhyaline to brown; conidial chains short to moderately long. Sexual morph unknown.
Type species: Penicillago nodositata (Valla) Guevara-Suarez, Gené & Dania García.
Notes: The genus Penicillago is introduced to accommodate Penicillium nodositatum. It is noteworthy that this species has had a confusing taxonomic history. Initially, Valla et al. (1989) classified P. nodositatum in the subgenus Biverticillium from its morphological affinity with members of the series Islandica (Pitt 1979). Later, based on molecular data, it was considered a member of the Penicillium section Sclerotiora, and tentatively placed in synonymy with P. bilaiae by Houbraken & Samson (2011), despite morphological differences in the conidiophore structure (biverticillate in P. nodositatum vs monoverticillate in P. bilaiae). However, Visagie et al. (2013) noticed that in the previous study that synonymy was based on a culture of P. bilaiae (CBS 330.90) mislabeled as P. nodositatum. The origin of the problem was probably introduced by Pitt & Samson (2000) since they erroneously designated on pag. 66 "No. 330.90 (CBS)" as neotype of P. nodositatum. In fact, the International Commission of Penicillium and Aspergillus (http://www.aspergilluspenicillium.org/) included the living ex-type strain of the herbarium specimen of P. nodositatum as CBS 333.90, this therefore being the correct ex-type strain for the species. Newly sequence data of that strain showed that P. nodositaum was closely related to P. kabunicum, and neither of them belonged in Penicillium s. str. (Houbraken & Samson 2011, Visagie et al. 2013). We observed that the only sequence available for P. kabunicum (CBS 574.90, ITS sequence GenBank MH862240) showed a 99.64 % of similarity with that of P. nodositatum (CBS 333.90, ITS sequence GenBank NR_103703) (Fig. S6 in supplemental material), suggesting they could be conspecific. However, the protologue of P. kabunicum (Baghdadi 1968) disagree with that of P. nodosotatum (Valla et al. 1989) in features such as the conidial morphology (globose, smooth-walled and hyaline in P. kabunicum vs ellipsoid to subglobose, echinulate and brown in P. nodosotatum). Therefore, further comparative studies will be needed to determine whether both species should be treated as synonym.
The possibility of examining two Spanish isolates that fit morphologically with P. nodositatum and their DNA sequences coincide with those of the ex-type strain of this species, has allowed us to delineate a new genus of penicillium-like fungi in the family Aspergillaceae (Figs 9, 10). Because herbarium type material for P. nodositatum is lost according to Visagie et al. (2013), the illustrations included in the protologue of the species (Valla et al. 1989) are selected here as lectotype, and the metabolically inactive culture CBS 333.90 as epitype deposited in the Westerdijk Fungal Biodiversity Institute.
Penicillago nodositata (Valla) Guevara-Suarez, Gené & Dania García, comb. nov. MycoBank MB822074. Fig. 18.
Fig. 18.
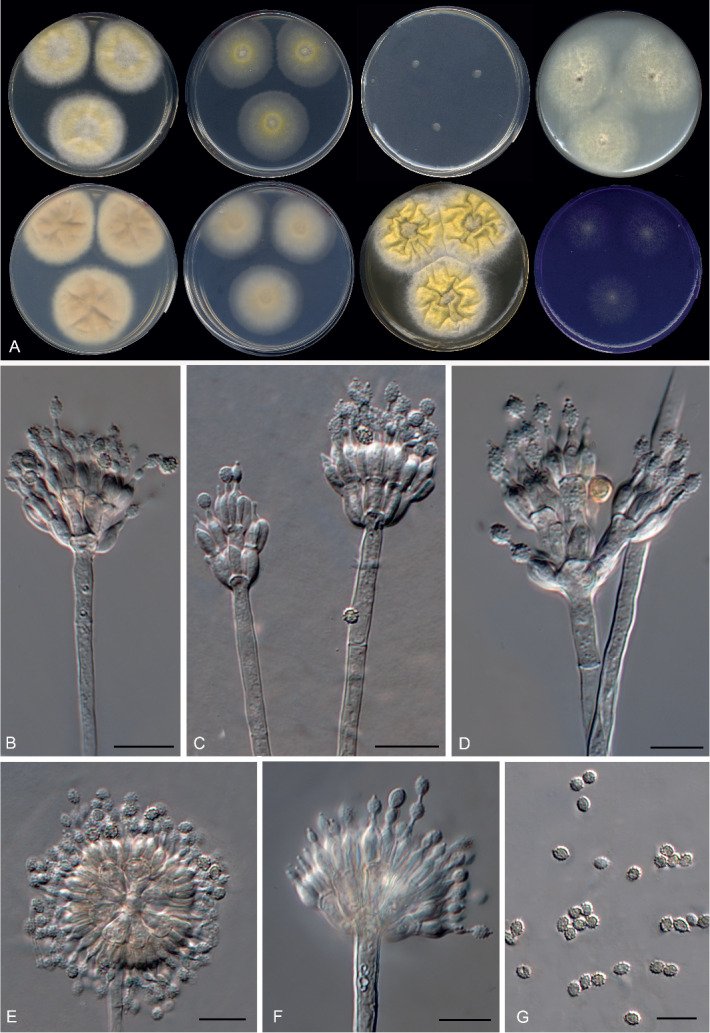
Morphological characters of Penicillago nodositata (FMR 15296). A. Colonies from left to right (top row) CYA, MEA, DG18 and OA; (bottom row) CYA reverse, MEA reverse, YES, and CREA. B–F. Conidiophores. G. Conidia. Scale bars = 10 μm.
Basionym: Penicillium nodositatum Valla, Pl. Soil 114: 142. 1989.
Typus: Lectotype designed here: figs 1–4 in Valla et al. (1989), Penicillium nodositatum Valla, a new species inducing myconodules on Alnus roots, Plant & Soil 114: 142–146, MBT388228; Epitype designed here: metabolically inactive culture CBS 333.90, France, Isère, Col d’Ornon, from myconodules in roots of Alnus incana, 1986, G. Valla, MBT388229; culture exepitype CBS 333.90.
Additional materials examined: Spain, Balearic Islands, Mallorca, from wild pig dung, Feb. 2016, J. Guarro, M. Guevara-Suarez & J.P.Z. Siqueira (FMR 15296 = CBS 142988; ITS barcode: LT899787, alternative markers: tub2 LT898312, cmdA LT899771, rpb2 LT899805); Extremadura, Badajoz, Granja de Torrehermosa, from herbivore dung, Dec. 2016, J. Cano-Lira (FMR 16442).
Colony diameter in 7 d (mm): On CYA: 25 °C 40–44, 30 °C 34–36, 37 °C 7–8; on MEA: 25 °C 34–36, 30 °C 24–27, 37 °C 0–3; on YES: 25 °C 50–55, 30 °C 45–55, 37 °C 11–10; on OA: 25 °C 24–26, 30 °C 22–24, 37 °C 4–6; on DG18: 25 °C 2–3; on CREA: 25 °C 22–30.
Colony characters at 25 °C in 7 d: On CYA, colonies cottony, slightly sulcate, mycelium light yellow (3A5) to white, sporulation sparse after 7d, conidial masses yellowish brown (5E8) after 14 d, margin fimbriate; reverse greyish orange (5B4) at the centre, white to pale yellow towards the periphery; exudate and soluble pigment absent. On MEA, colonies slightly cottony, flat, mycelium light yellow (3A5) to white, sporulation dense after the 14 d, with conidial masses dark green (28F5), margin slightly fimbriate; reverse light yellow (4A4); exudate and soluble pigment absent. On DG18, colonies restricted, flat, mycelium white, sporulation absent, margin entire; reverse light yellow (4A4); exudate and soluble pigment absent. On YES, colonies velvety, raised at the centre, irregularly sulcate, mycelium yellow (3A7) to white, sporulation sparse, margin fimbriate; reverse greyish orange (5B5); exudate and soluble pigment absent. On OA, colony slightly cottony, mycelium white, sporulation sparse, margin entire; reverse yellowish white (4A2); exudate and soluble pigment absent. On CREA, acid production absent.
Micromorphology: On MEA, conidiophores symmetrically biverticillate, sometimes irregularly branched; stipes smooth, hyaline, often with a small vesicle at the apex, 3.5–5.5 μm wide, 200–300 × 2.5–3 μm; metulae divergent, cylindrical to somewhat obpyriform, 9–10(–15) × 3–4(–5) μm; phialides four to seven per metula, ampulliform or tending towards acerose, often tapering abruptly to a long, slender neck, 6–8(–9) × 2.5–3.5 μm; conidia in short to moderately long disorder chains, subglobose to broadly ellipsoidal, echinulate, 2.5–4 × 3–4(–5) μm, pale brown.
Pseudopenicillium Guevara-Suarez, Cano & Guarro, gen. nov. MycoBank MB822076.
Etymology: Pseudo- meaning “false”-, name refers to the morphological similarity but not belonging to Penicillium.
In: Family Aspergillaceae.
Diagnosis: Phylogenetic closeness to Hamigera, but differs in the lack of sexual morph and by its brown globose conidia with conspicuous disjunctors. Morphologically differs from Penicillium and Penicillago by its short and often irregularly branched conidiophores producing large conidia in short chains.
Mycelium superficial and immersed, composed of septate, branched, hyaline to subhyaline hyphae. Conidiophores undifferentiated, reduced to conidiogenous cells arising directly from the main hyphae, or differentiated composed of short to moderately long, aseptate or few septate, hyaline to brown stipes, terminating in an irregular penicilli; penicilli varying from monoverticillate to partly biverticillate, with few metulae bearing each a verticil of conidiogenous cells; metulae terminal or subterminal, generally divergent, cylindrical or apically inflated. Conidiogenous cells phialidic, ampulliform, with swollen base and tapering abruptly to a slender neck of variable length. Conidia dry, catenate, with conspicuous disjunctors, globose to subglobose, with spinulose walls, brown, dark in masses; conidial chains short. Sexual morph unknown.
Type species: Pseudopenicillium megasporum (P.A. Orpurt & D.I. Fennell) Guevara-Suarez, Cano & Gené
Notes: The genus Pseudopenicillium is proposed to accommodate a new species and two species previously classified in the genus Penicillium, P. giganteum and P. megasporum, the latter being selected as the type since it was the first described. Penicillium giganteum and P. megasporum were recovered from soil samples in India and the UK, respectively. Despite their different geographical origin, both were considered conspecific and classified in series Megasporus (Pitt 1979). Penicillium asperosporum, also included in this series, was later shown to be a synonym of P. montanense in section Aspergilloides (Houbraken et al. 2014b). Multi-gene phylogenies (Peterson et al. 2010, Houbraken & Samson 2011) demonstrated that P. megasporum and P. giganteum were distinct species, closely related to the genus Hamigera. Our phylogeny confirms this relationship and shows that Hamigera and the afore-mentioned Penicillium species comprise two sister clades with enough genetic differences to be considered distinct genera (Figs 9, 10).
Pseudopenicillium cervifimosum Guevara-Suarez, Dania García & Gené, sp. nov. MycoBank MB822079. Fig. 19.
Fig. 19.
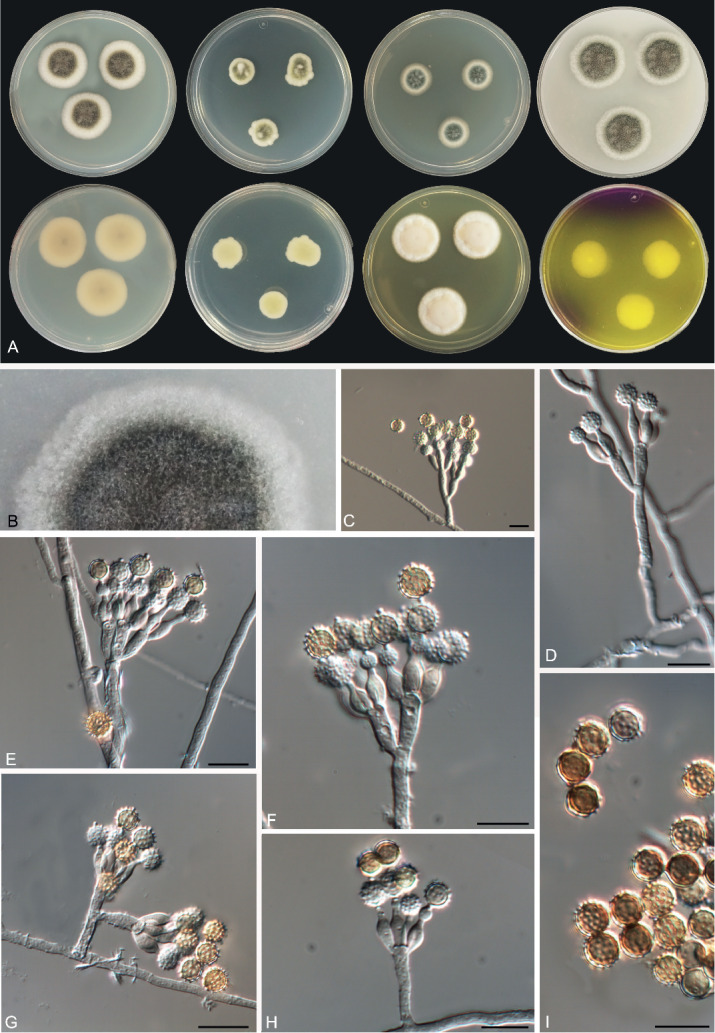
Morphological characters of Pseudopenicillium cervifimosum (FMR 15299T). A. Colonies from left to right (top row) CYA, MEA, DG18 and OA reverse; (bottom row) CYA reverse, MEA reverse, YES, and CREA. B. Colony texture on OA at 25 °C after 1 wk incubation. C–H. Conidiophores. I. Conidia. Scale bars = 10 μm.
Etymology: From the Latin cervus = “deer”, and fimosum = “dung-dwelling”, describing the substrate from where the species was isolated.
Typus: Spain, Castile and Leon, Palencia, Monte el Viejo, Deer Reserve Park, from deer dung, Mar. 2016, J. Guarro & M. Guevara-Suarez (holotype CBS H-23133; culture ex-type FMR 15299 = CBS 142670; ITS barcode: LT899789, alternative markers: tub2 LT898315, rpb2 LT899807).
Colony diameter in 7 d (mm): On CYA: 25 °C 26–30, 30 °C 30–32, 37 °C 6–9; on MEA: 25 °C 13–17, 30 °C 22–24, 37 °C 3–5; on YES: 25 °C 24–26, 30 °C 35–38, 37 °C 23–25; on OA: 24–26, 30 °C –34, 36 °C 14–16; on DG18: 25 °C 16–17; on CREA: 25 °C 18–20.
Colony characters at 25 °C in 7 d: On CYA, colonies flat, velvety, mycelium white, sporulation dense, central area with conidial masses dark green (29F3), margin entire; reverse pale yellow (4A3); exudate and soluble pigment absent. On MEA, colonies growing moderately, velvety, slightly umbonate, mycelium white, sporulation dense, with conidial masses olive-yellow (3C8) to olive (3F8), margin lobate; exudate and soluble pigment absent; reverse greyish yellow (2B4). On YES, colonies dome-shaped, velvety, mycelium white, sporulation absent, margin entire; reverse pale yellow (4A3); exudate and soluble pigment absent. DG18, colonies velvety, slightly sulcate, mycelium white, sporulation dense, with conidial masses dull green (28E3), margin entire; reverse yellowish white (4A3). On OA, colonies cottony, mycelium white, sporulation dense, with conidial masses dark green (29F3), margin fimbriate; exudate and soluble pigment absent. On CREA, strong acid production.
Micromorphology: On MEA, conidiophores short, arising as lateral branches from hyphae, biverticillate, sometimes monoverticillate; stipes 10–35 × 2.5–3 μm, cylindrical, without vesicle, smooth to finely verruculose, hyaline; metulae divergent, cylindrical, 9.5–10 × 2.5–3 μm; phialides in verticils of 2–4, ampulliform with swollen base, tapering abruptly to a slender neck, (6–)7.5–9 × 2.5–3 μm; conidia globose, 5–5.5 × 6 μm, spinulose, thick-walled, pale brown to brown.
Distinguishing characters: Pseudopenicillium cervifimosum is closely related to Pse. giganteum and Pse. megasporum. This group of species have a similar conidiogenous apparatus and produce globose spinulose conidia. However, they can be distinguished by their conidial sizes (i.e., up to 6 μm in Pse. cervifimosum, up to 10 μm in Pse. megasporum and up to 12 μm in Pse. giganteum). In addition, Pse. megasporum has a less abundant growth on CYA at 25 °C (15–25 mm) and shows vesiculate stipes (Pitt 1979, Peterson et al. 2010).
Pseudopenicillium giganteum (R.Y. Roy & G.N. Singh) Guevara-Suarez, Gené & Cano, comb. nov. MycoBank MB822077. Basionym: Penicillium giganteum R.Y. Roy & G.N. Singh, Trans. Br. Mycol. Soc. 51: 805. 1968.
Typus: India, West Bengal, Varanasi, Banaras Hindu University, Botanical Garden, from soil, Nov. 1967, G.N. Singh (holotype IMI 132774; cultures ex-type NRRL 3553 = ATCC 48996 = CBS 144.69 = FRR 535 = IMI 132774).
Descriptions and illustrations: See Roy & Singh (1968) and Peterson et al. (2010).
Additional material examined: Spain, unknown geographic region, from soil (FMR 14718).
Notes: The new isolate is the second specimen known for this taxon. In general, its morphological features are consistent with those of the species as described by Roy & Singh (1968).
Pseudopenicillium megasporum (P.A. Orpurt & D.I. Fennell) Guevara-Suarez, Cano & Gené, comb. nov. MycoBank MB822078. Basionym: Penicillium megasporum P.A. Orpurt & D.I. Fennell, Mycologia 47: 233. 1955.
Typus: UK, England, Suffolk, Lakenheath Warren, from heath soil, J.H. Warcup (holotype IMI 216904; cultures ex-type NRRL 2232 = ATCC 12322 = CBS 256.55 = FRR 2232 = MUCL 38804 = QM 6879 = WE 2232 = IMI 216904).
Descriptions and illustrations: See Orpurt & Fennell (1955), Pitt (1979) and Peterson et al. (2010).
Talaromyces catalonicus Guevara-Suarez, Gené & Guarro, sp. nov. MycoBank MB822080. Fig. 20.
Fig. 20.
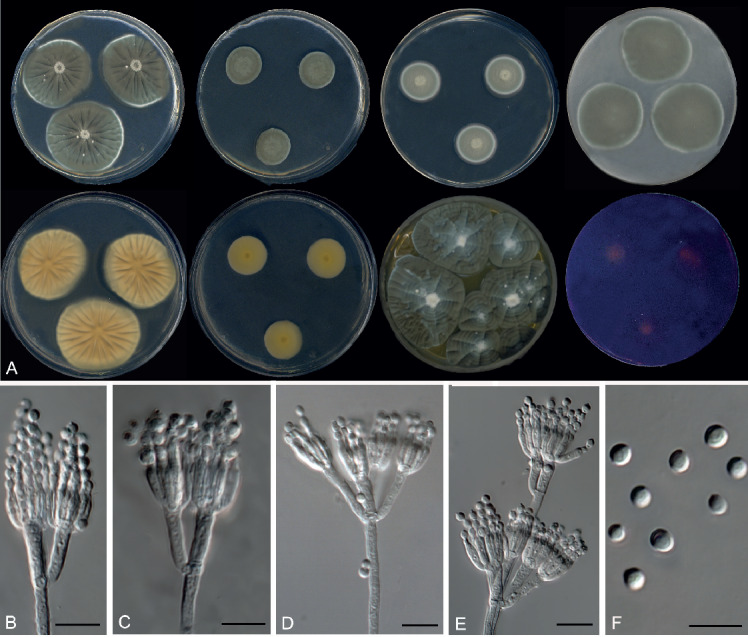
Morphological characters of Talaromyces catalonicus (FMR 16441T). A. Colonies from left to right (top row) CYA, MEA, DG18 and OA reverse; (bottom row) CYA reverse, MEA reverse, YES, and CREA. B–E. Conidiophores. F. Conidia. Scales bars = 10 μm.
Etymology: Name refers to the region (Catalonia) from where the fungus was isolated.
In: Section Trachyspermi.
Typus: Spain, Catalonia, Poblet, from herbivore dung, Feb. 2017, J. Guarro, M. Guevara-Suarez & I. Iturrieta-González (holotype CBS H-23212; culture ex-type FMR 16441 = CBS 143039; ITS barcode: LT899793, alternative identification markers: tub2 LT898318, cmdA LT899775, rpb2 LT899811).
Colony diameter in 7 d (mm): On CYA: 25 °C 35–40, 30 °C 38–40, 37 °C 15–18; on MEA: 25 °C 17–19, 30 °C 19–20, 37 °C 16–18; on YES: 25 °C 45–50, 30 °C 50–55, 37 °C 25–30; on OA: 25 °C 40–45, 30 °C 40–42, 37 °C 12–14; on DG18: 25 °C 20–22, on CREA: 25 °C 17–19.
Colony characters at 25 °C in 7 d: On CYA, colonies velvety, radially sulcate, mycelium white, sporulation dense, conidial masses dull green (27E4), margin lobate; reverse greyish orange (5B4); exudate absent, soluble pigment only at 30 and 37 °C, light yellow (3A5) to light orange (5A6). On MEA, colonies velvety, flat, sporulation dense, with conidial masses greyish green (27E5), margin entire; reverse yellow (2B8); exudate and soluble pigment absent. On DG18, colonies flat, slightly cotton at the centre, velvety in the periphery, mycelium white, sporulation dense, conidial masses dull green (26E5), margin entire; reverse greyish green (28B4). On YES, colonies raised at the centre, irregularly sulcate, mycelium white, sporulation dense, with conidial masses dark green (30F3), margin entire; reverse orange-yellow (4B8); exudate and soluble pigment absent. On OA, colonies velvety, flat, mycelium white, sporulation dense, conidial masses dark green (27F3), margin entire; reverse greenish grey (29C2); exudate and soluble pigment absent. On CREA, weak acid production.
Micromorphology: On MEA, conidiophores biverticillate, sometimes irregularly branched, stipes (40–)100–130 × 2–2.5 μm, smooth-walled, hyaline; metulae divergent, 10–11 × 2–2.5 μm; phialides 4–6 per metula, ampulliform to acerose, (7–)8–10 × 2–2.5 μm; conidia globose to subglobose, 2–2.5 × 1.8–2 μm, smooth-walled, brownish yellow. Ascomata not observed.
Distinguishing characters: The most closely related species to T. catalonicus are T. albobiverticillius, T. erythromellis, T. heiheensis, and T. solicola (Fig. 8). Talaromyces catalonicus is characterised by its ability to grow at 37 °C; in contrast, the above-mentioned species do not grow or grow very restricted at this temperature (Samson et al. 2011, Yilmaz et al. 2014).
Talaromyces coprophilus Guevara-Suarez, Cano & Dania García, sp. nov. MycoBank MB822088. Fig. 21.
Fig. 21.
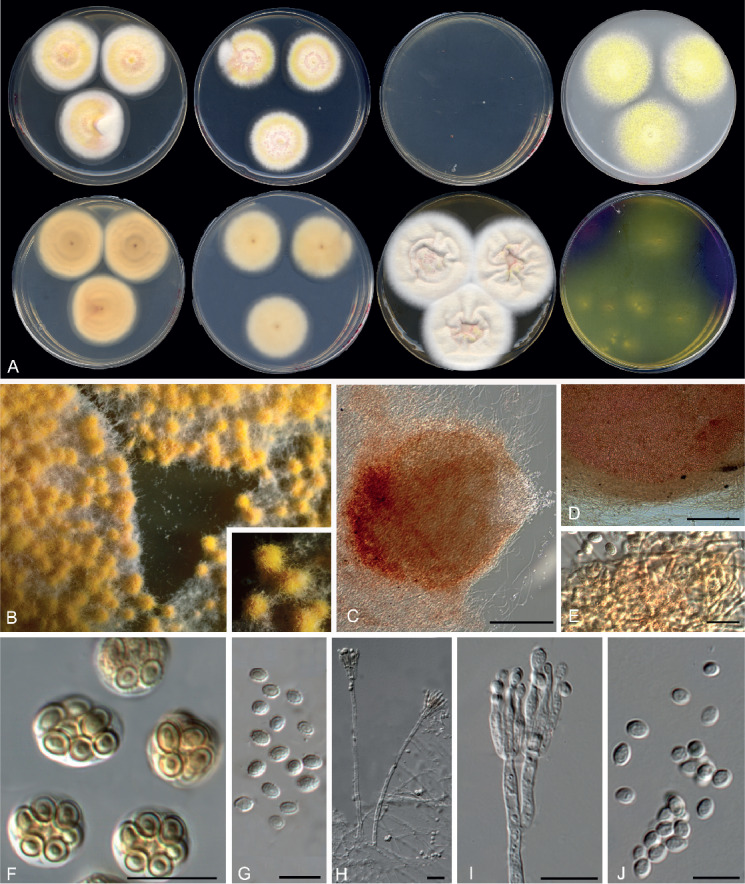
Morphological characters of Talaromyces coprophilus (FMR 15199T). A. Colonies from left to right (top row) CYA, MEA, DG18 and OA reverse; (bottom row) CYA reverse, MEA reverse, YES, and CREA. B. Detail of the colony texture with ascomata on MEA after 2 wk incubation. C. Ascoma. D, E. Part of an ascoma and peridial hyphae. F. Asci. G. Ascospores. H, I. Conidiophores. J. Conidia. Scales bars C, D = 100 μm, E–J = 10 μm.
Etymology: Name refers to the source of isolation of the fungus.
In: Section Talaromyces.
Typus: Spain, Balearic Islands, Mallorca, Escorca, from herbivore dung, Feb. 2016, J. Guarro, M. Guevara-Suarez & J.P.Z. Siqueira (holotype CBS H-23144; culture ex-type FMR 15199 = CBS 142756; ITS barcode: LT899794, alternative identification markers: tub2 LT898319, cmdA LT899776, rpb2 LT899812).
Colony diameter in 7 d (mm): On CYA: 25 °C 38–40, 30 °C 40–45, 37 °C 30–35; on MEA: 25 °C 33–35, 30 °C 30–35, 37 °C 21–24; on YES: 25 °C 45–50, 30 °C 50–55, 37 °C 38–42; on OA: 25 °C 35–40, 30 °C 40–45, 37 °C 25–26; on DG18: 25 °C no growth, on CREA: 25 °C 8–10.
Colony characters at 25 °C in 7 d: On CYA, colonies slightly raised at the centre, velvety, mycelium light yellow (4A5) to white, sporulation absent to sparse, with margin entire and fimbriate; reverse light orange (5A4); soluble pigment and exudate absent. On MEA, colonies slightly raised at centre, velvety, flat, mycelium light yellow (4A5) to white, conidial sporulation absent to sparse, young ascomata visible, margin fimbriate; reverse light orange (5A4); soluble pigment and exudate absent. On YES, colonies raised at centre, velvety, mycelium pinkish (7A3) to white, sporulation absent, with margin entire; reverse brownish orange (7C6); exudate absent and soluble pigment brownish red (10C7). On OA, colonies granular, flat, with young bright orange ascomata visible, conidial sporulation sparse, margin fimbriate; reverse greenish grey (28C3); exudate and soluble pigment absent. On CREA, strong acid production.
Micromorphology: On MEA, conidiophores mono-to biverticillate, stipes 40–70 × 2–2.5 μm, smooth, hyaline; metulae rather appressed, 10–12 × 2.5–3 μm; phialides 3–4 per metula, acerose, 9–12(–14) × 2–2.5 μm; conidia ellipsoidal, 2–3 × 2.5–3 μm, smooth-walled, brownish yellow. Ascomata after 1–2 wk of incubation on OA and MEA at 25 °C, bright orange to orange-red, globose, 200–310 μm diam, with peridial hyphae branched and verruculose; asci globose to subglobose, 6–7 × 7–9(–10) μm; ascospores ellipsoidal, 3.5–4 × 3–4 μm, spiny, thick-walled, golden yellow.
Distinguishing characters: Talaromyces coprophilus is characterised by the production of orange-red ascomata with spiny ellipsoidal ascospores, the lack of growth on DG18, and a good growth on other culture media tested. Phylogenetically, it forms an independent and distant branch in an unsupported clade where the ex-type strains of T. cnidii, T. flavovirens, T. siamensis, and T. xishaensis are found (Fig. 7). Morphologically, T. coprophilus resembles T. flavovirens in the production of a sexual morph; however, the latter differs in slower growth on CYA at 25 °C (19–20 mm after 7d), and larger ascospores (4–7 × 3–4 μm) (Yilmaz et al. 2014).
Talaromyces pseudofuniculosus Guevara-Suarez, Dania García & Gené, sp. nov. MycoBank MB822090. Fig. 22.
Fig. 22.
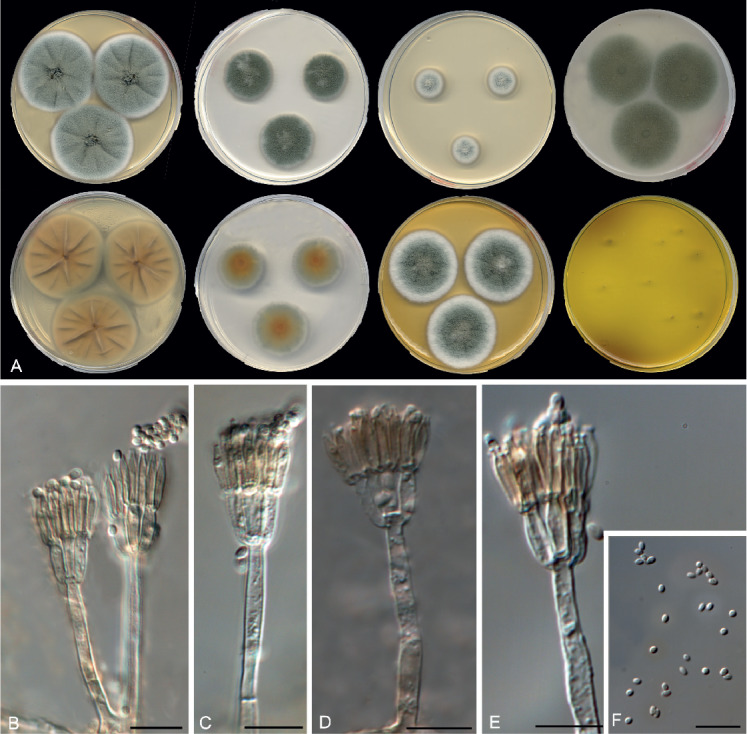
Morphological characters of Talaromyces pseudofuniculosus (FMR 15307T). A. Colonies from left to right (top row) CYA, MEA, DG18 and OA reverse; (bottom row) CYA reverse, MEA reverse, YES, and CREA. B–E. Conidiophores. F. Conidia. Scale bars = 10 μm.
Etymology: –pseudo, meaning “false”-, name refers to its phylogenetic relationship with T. funiculosus.
In: Section Talaromyces.
Typus: Spain, Andalusia, La Rocina, Doñana National Park, from herbivore dung, Mar. 2016, D. García (holotype CBS H-23214; culture ex-type FMR 15307 = CBS 143041; ITS barcode: LT899796, alternative markers: tub2 LT898323, cmdA LT899778, rpb2 LT899814).
Additional material examined: Spain, Catalonia, Els Ports Natural Park, from herbivore dung, Feb. 2016, J. Guarro, M. Guevara-Suarez & E. Rosas (FMR 15035).
Colony diameter in 7 d (mm): On CYA: 25 °C 36–43, 30 °C 45–40, 37 °C 31–35; on MEA: 25 °C 24–29, 30 °C 43–45, 37 °C 23–26; on YES: 25 °C 34–35, 30 °C 40–44, 37 °C 32–34; on OA: 25 °C 33–38, 30 °C 42–45, 37 °C 27–30; on DG18: 25 °C 12–15, on CREA: 25 °C 2–4.
Colony characters at 25 °C in 7 d: On CYA, colonies slightly raised at centre, radially sulcate, cottony, mycelium white, sporulation dense, conidial masses greyish green (29D5), with margin entire; reverse dark orange (5D8); soluble pigments and exudate absent. On MEA, colonies velvety to cottony, flat, sporulation dense, conidial masses greyish green (28D5), margin entire to slightly crenate; reverse deep yellow (5A4) to greyish green (30C5); exudate and soluble pigment absent. On YES, colonies slightly raised at the centre, sulcate, cottony, mycelium white, sporulation dense in the centre, conidial masses greyish green (27D4), margin entire; reverse olive yellow (3C7); exudate and soluble pigments absent. On DG18, colonies flat and velvety, mycelium white; moderate sporulation, conidial masses dull green (27D4); reverse greenish white (27B2). On OA, colonies slightly granular, sporulation dense, conidial masses greenish grey (28B2), margin entre; reverse greyish green (30C5); exudate and soluble pigment absent. On CREA, acid production strong.
Micromorphology: On MEA, conidiophores biverticillate; stipes 40–70 × 2.5–3 μm, smooth-walled, brownish yellow; metulae 3–4 per branch, appressed, 8–10 × 2–3 μm; phialides 3–4 per metula, acerose, 8–10(–11) × 2–3 μm; conidia mostly ellipsoidal, 2–3 × 1.5–2(–2.5) μm, smooth-walled. Ascomata not observed.
Distinguishing characters: Talaromyces cucurbitiradicus, recently described by Su & Niu (2018), and T. funiculosus are the closest related species to T. pseudofuniculosus. The former can be distinguished from the other two by the production of chlamydospores and absence of growth at 37 °C. Talaromyces funiculosus differs from the new species by its divergent metulae (appressed in T. pseodofuniculosus) and by the pigmentation of the conidiophores, being olivaceous in T. funiculosus and brownish yellow in T. pseudofuniculosus. Moreover, T. pseudofuniculosus has a restricted growth on CREA (2–4 mm diam; 20–30 mm diam in T. funiculosus) and does not produce funiculose colonies, a typical feature of T. funiculosus mainly on MEA and OA (Yilmaz et al. 2014).
DISCUSSION
In recent years, the phylogeny of the genera Penicillium, Talaromyces and other members of family Trichocomaceae sensu lato was studied in order to clarify their taxonomic relationships. Subsequently, many species were re-classified and new taxonomic approaches proposed for species delineation and identification. Considering that several of these fungi have been discovered from specific and poorly studied substrates using modern taxonomic techniques (Houbraken et al. 2014b, Barbosa et al. 2018), we explored the species diversity of penicillium-like fungi from herbivore dung collected in different Spanish regions (Andalusia, Balearic and Canary Islands, Castile and Leon, Catalonia and Galicia) using the gene markers recommended for their study (Visagie et al. 2014b, Yilmaz et al. 2014).
Our results agree with other studies in which a large set of isolates of penicillium-like fungi could be identified with the tub2 sequence analysis (Guevara-Suarez et al. 2016, Visagie et al. 2014a, Chen et al. 2016, Barbosa et al. 2018). It is a good marker, easy to amplify and sequence, and useful for the classification at the sectional and species level, and even for detection of putative undescribed species. However, in addition to the ITS and tub2 barcodes, the characterization of penicillia requires multi-gene analyses along with other markers such as fragments of cmdA and/or rpb2 loci. Based on this molecular approach, we were able to find a remarkable diversity of penicillium-like fungi that have not been described on herbivore dung so far. Among the 104 isolates recovered, we identified a total of 48 species, 38 Penicillium including seven new taxa (i.e. P. balearicum, P. beceitense, P. caprifimosum, P. fimosum, P. ibericum, P. mediterraneum, and P. synnematicola), seven Talaromyces including three new taxa (T. catalonicus, T. coprophilus, and T. pseudofuniculosus), and three species in the newly prosposed genera Penicilago (Pgo. nodositata) and Pseudopenicillium (Pse. giganteum, and the new species Pse. cervifimosum). In addition, of note is that the highest number of species identified (34 out of 48) and novel taxa described (9 out of 11) were recovered from samples collected in the Mediterranean area, namely Catalonia and the Balearic island of Mallorca (Table 1). This is not surprising if we consider that this area is recognized as one of the most species rich regions in Southern Europe (Cuttellot et al. 2008, Rundel et al. 2018).
Among Penicillium isolates, those of the sections Fasciculata and Robsamsonia were the most frequently isolated, accounting nearly 20 % of the isolates in each section. It is of note, however, that in Fasciculata most isolates identified were of P. crustosum, while in Robsamsonia, as expected, we found the most species diversity among the sections included here. This latter section currently comprises 11 species (Houbraken et al. 2016) and we identified five of these taxa, in addition to the new P. synnematicola. This species would seem to be a common coprophilous fungus in the Mediterranean area, as its isolates were recovered exclusively from samples collected in different locations from Mallorca and Catalonia. As mentioned before, Houbraken et al. (2016) restricted the majority of coprophilous Penicillium species in section Robsamsonia. However, our results clearly show that the coprophilous penicillia are distributed across different and phylogenetically distant sections of the genus (Fig. 1). In sections such as Aspergilloides, Exilicatus and Paradoxa, we even found a similar diversity to the section Robsamsonia. Of particular note is Paradoxa with the highest number of new penicillia proposed in this study (P. ibericum, P. fimosum and P. balearicum). However, we also identified P. atramentosum and P. magnielliptisporum as first records on dung. The former species is commonly associated to cheese and P. magnielliptisporum was described from house dust (Visagie et sal. 2014a). The only species in the section reported previously from animal faeces was P. paradoxum, an aspergillus-like Penicillium collected in different countries such as Denmark, India, New Zealand, the Netherlands and UK (Houbraken et al. 2014b, Rajeshkumar et al. 2016). Paradoxa is a very small section (nine species, including our new proposal) in comparison to sections Aspergilloides and Exillicaulis, which have 50 species each, but without any species reported from dung (Houbraken et al. 2014d, Visagie et al. 2016).
The most Talaromyces isolates found in our survey were from section Talaromyces. This is the largest section in the genus, with nearly 60 species, 22 of them described in the last four years from different substrates, including human clinical specimens (Visagie et al. 2015, Crous et al. 2016, Cheng et al. 2016, Wang et al. 2016a, b, 2017, Yilmaz et al. 2016, Guevara-Suarez et al. 2017, Barbosa et al. 2018, Jiang et al. 2018). We added two more new taxa in the section (T. coprophilus and T. pseudofuniculosus), but also identified several interesting species firstly reported from dung, such as T. angelicus, T. muroii, T. ruber, and T. sayulitensis. Unlike T. ruber, which is a worldwide species isolated from soil, air and fuel (Yilmaz et al. 2012), the other three are considered uncommon fungi previously found in the roots of Angelica gigas in Korea (Sang et al. 2013), in soil in Taiwan (Chen et al. 2016), and in house dust in Mexico (Visagie et al. 2014a), respectively. The other new Talaromyces species described is T. catalonicus in section Trachyspermi. This is a much smaller section than Talaromyces, with 19 species including our new one (Yaguchi et al. 1996, Yilmaz et al. 2014, Chen et al. 2016, Luo et al. 2016, Wang et al. 2016b, Guevara-Suarez et al. 2017, Barbosa et al. 2018, Rajeshkumar et al. 2019). The most recently described are T. aerius from indoor air in China (Chen et al. 2016), T. amyrossmaniae from decaying fruits in India (Rajeshkumar et al. 2019), T. brasiliensis from honey and nest of bee in Brazil (Barbosa et al. 2018), T. heiheensi from rotten wood in China (Wang et al. 2017), T. minnesotensis from human specimens in the USA (Guevara-Suarez et al. 2017), and T. rubrifaciens from air outlets of public buildings also in China (Luo et al. 2016), although this latter was considered a synonym of T. albobiverticillius by Chen et al. (2016). To date, however, there are no reports of species in either of these two sections isolated from dung samples.
Penicilago nodositata seems to be a common fungus usually associated with the roots of Alnus in Europe (Valla 1989, Sequerra et al. 1997); it is not surprising therefore to recover this fungus from animal dung, in our case from a wild pig, which feeds on a wide variety of plant material including roots, shoots and fruit. Conversely, Pse. giganteum is a rare species only known from its type collection, a garden soil sample from India (Roy & Singh 1968). In addition to the latter species, we also recovered from herbivore dung other uncommon species of Penicillium and Talaromyces, which are monotypic or have very few isolates available for investigation. These are P. canariense (section Stolkia), P. cremeogriseum (section Lanata-Divaricata), P. momoii (section Exilicaulis), and P. roseoviride (section Aspergilloides), including the three Talaromyces species mentioned before from section Talaromyces (i.e. T. angelicus, T. muroii and T. sayulitensis). Therefore, our study highlights that herbivore dung is a good substrate for recovering interesting species of penicillium-like fungi, although their ecological role on that substrate remains unclear and would merit further investigation.
ACKNOWLEDGEMENT
This study was supported by the Spanish Ministerio de Economía y Competitividad, grant CGL2017-88094-P.
Supplementary Material: http://fuse-journal.org/
Fig. S1.
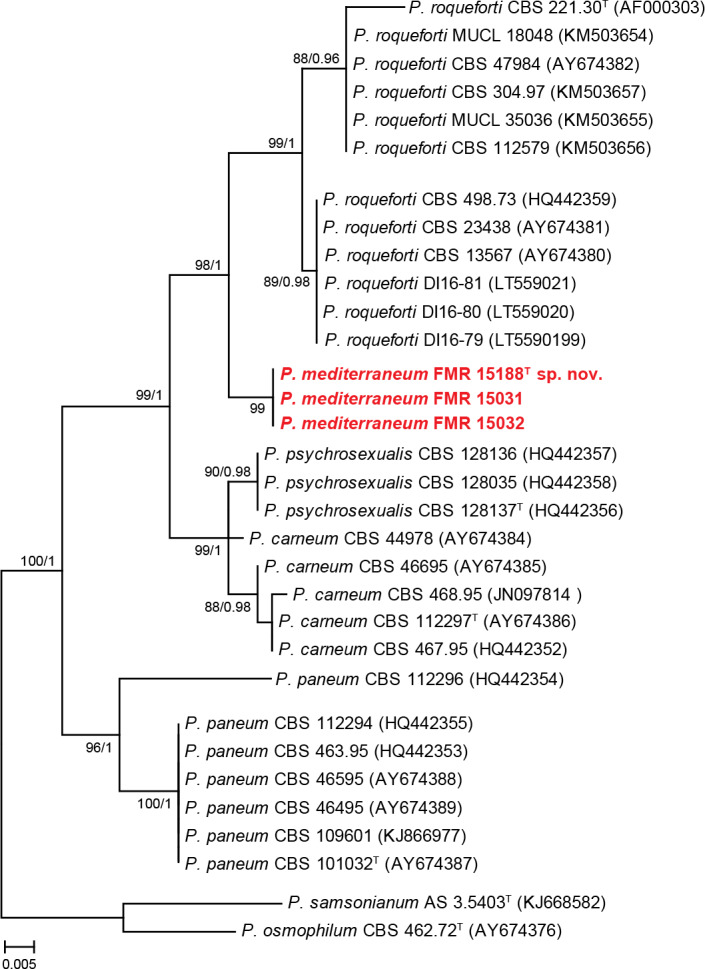
ML tree of selected Penicillium section Roquefortorum species, inferred from tub2. Branch lengths are proportional to phylogenetic distance. Bootstrap support values/Bayesian posterior probability scores above 70 %/0.95 are indicated on the nodes. The tree is rooted to P. sansonianum AS 3.5403 and P. osmophilum CBS 462.72. The name in red is the new species described in this study. T = ex-type strain.
Fig. S2.

ML tree of selected Penicillium section Robsamsonia species, inferred from tub2. Branch lengths are proportional to phylogenetic distance. Bootstrap support values/Bayesian posterior probability scores above 70 %/0.95 are indicated on the nodes. The tree is rooted to P. brevicompactum CBS 257.29. The name in red is the new species described in this study. T = ex-type strain.
Fig. S3.
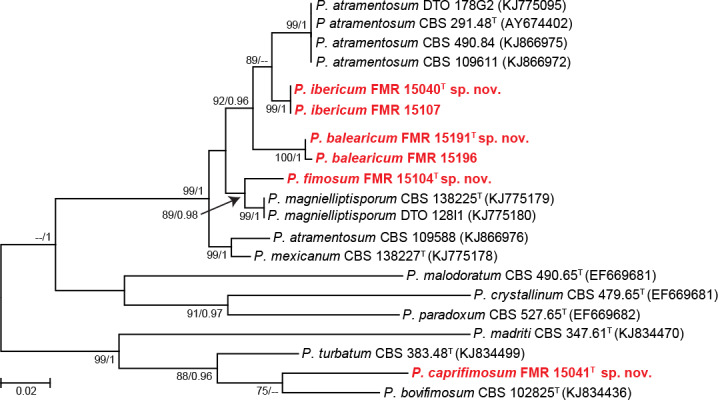
ML tree of selected Penicillium section Paradoxa species, inferred from tub2. Branch lengths are proportional to phylogenetic distance. Bootstrap support values/Bayesian posterior probability scores above 70 %/0.95 are indicated on the nodes. The tree is rooted to species of section Tubata. The names in red are the new species described in this study. T = ex-type strain.
Fig. S4.
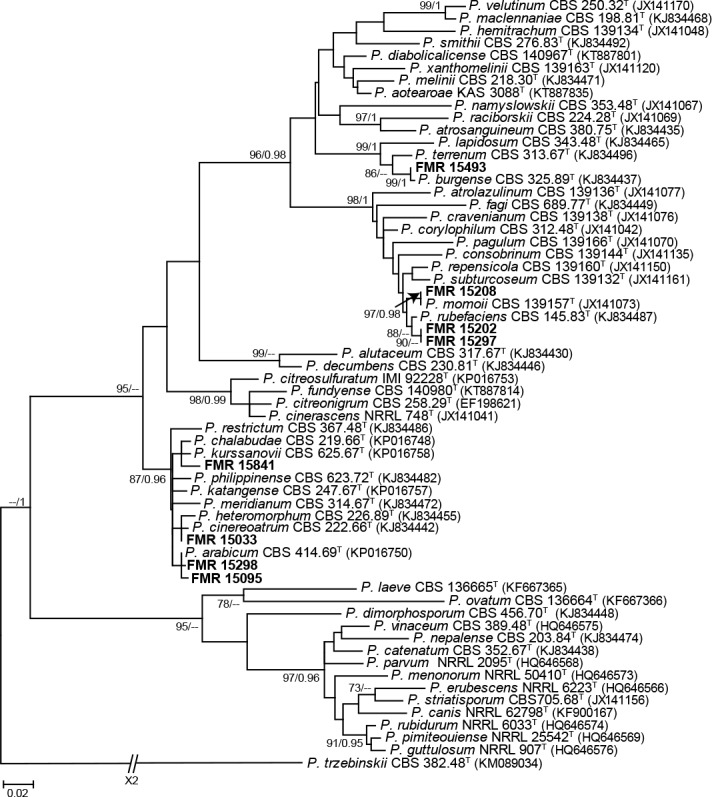
ML tree of selected Penicillium section Exilicaulis species, inferred from tub2, including the isolates belonging to this section recovered in this work. Branch lengths are proportional to phylogenetic distance. Bootstrap support values/Bayesian posterior probability scores above 70 %/0.95 are indicated on the nodes. The tree is rooted to P. trzebinskii CBS 382.48. T = ex-type strain.
Fig. S5.
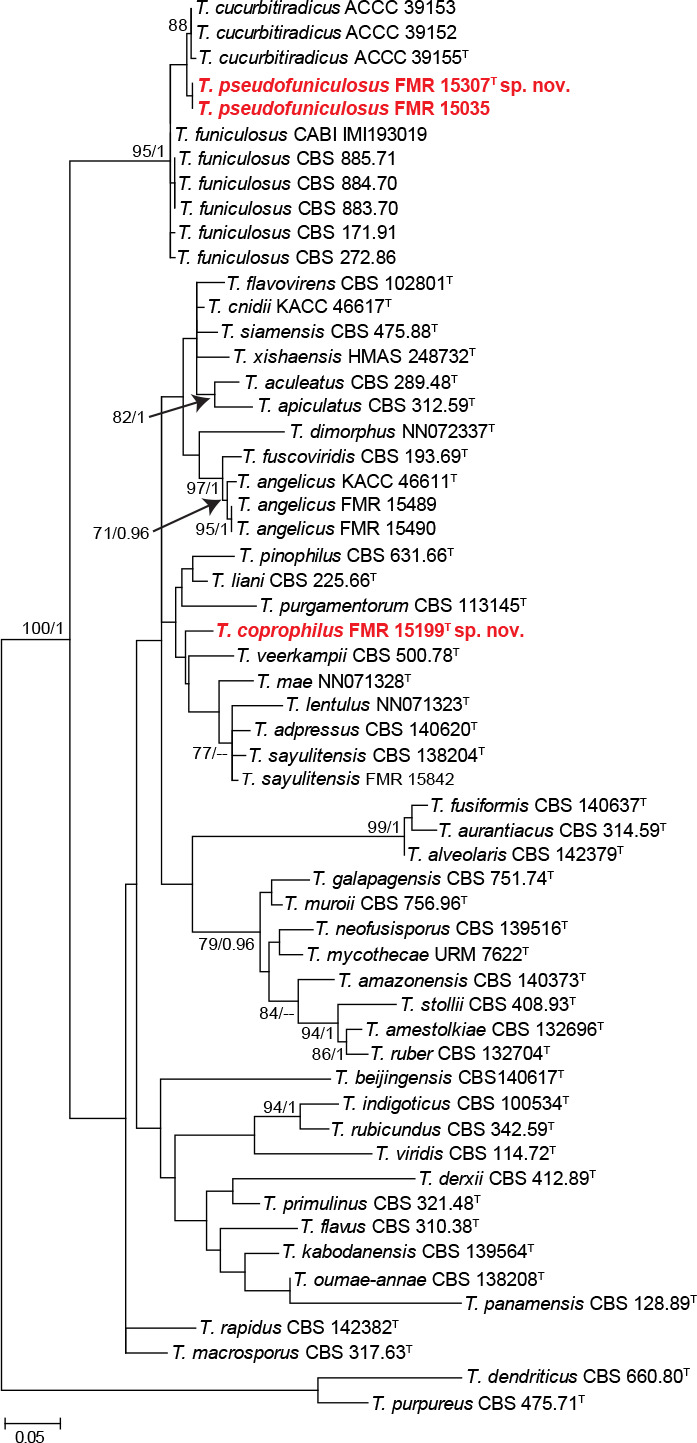
ML tree of selected Talaromyces section Talaromyces species, inferred from tub2, including the isolates belonging to this section recovered in this work. Branch lengths are proportional to phylogenetic distance. Bootstrap support values/Bayesian posterior probability scores above 70 %/0.95 are indicated on the nodes. The tree is rooted to T. dendriticus CBS 660.80 and T. purpureus CBS 475.71. The names in red are the new species described in this study. T = ex-type strain.
Fig. S6.
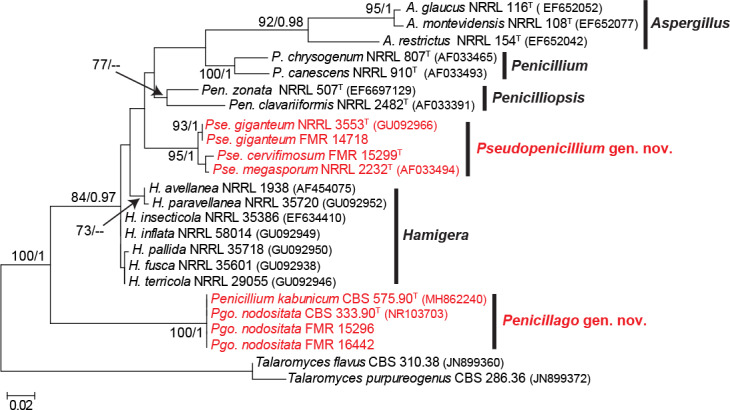
ML tree of selected members of Aspergillaceae family inferred from ITS, including the ex-type strain of Penicillium kabuniccum (CBS 575.90). Branch lengths are proportional to phylogenetic distance. Bootstrap support values/Bayesian posterior probability scores above 70 %/0.95 are indicated on the nodes. Isolates corresponding to novel taxa are shown in red. Between parentheses, GenBank accession numbers of ITS sequences. The tree is rooted to T. flavus CBS 310.38 and T. purpureogenus CBS 286.36. T = ex-type strain.
REFERENCES
- Abe S. (1956). Studies on the classification of the penicillia. Journal of General and Applied Microbiology 2: 1–344. [Google Scholar]
- Baghdadi VCh. (1968). De speciebus novis Penicilli Fr. et Aspergilli Fr. e terris Syriae isolatis notula. Novosti Sistematiki Vysshikh i nizshikh Rastenii 5: 96–114. [Google Scholar]
- Barbosa RN, Bezerra JD, Souza-Motta CM, et al. (2018). New Penicillium and Talaromyces species from honey, pollen and nests of stingless bees. Antonie van Leeuwenhoek Journal of Microbiology 111: 1883–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell A. (1983). Dung fungi: an illustrated guide to coprophilous fungi in New Zealand. Victoria University Press, New Zealand. [Google Scholar]
- Chen AJ, Sun BD, Houbraken J, et al. (2016). New Talaromyces species from indoor environments in China. Studies in Mycology 84: 119–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Gams W, Stalpers JA, et al. (2004). MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology 50: 19–22. [Google Scholar]
- Crous PW, Wingfield MJ, Burgess TI, et al. (2016). Fungal Planet description sheets: 469—557. Persoonia 37: 252–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuttelod A, García N, Abdul Malak D, et al. (2008). The Mediterranean: a biodiversity hotspot under threat. In: The 2008 Review of The IUCN Red List of Threatened Species (Vié JC, Hilton-Taylor C, Stuart SN, eds). IUCN Gland, Switzerland: 1–13. [Google Scholar]
- Darriba D, Taboada GL, Doallo R, et al. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9: 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisvad JC. (1981). Physiological criteria and mycotoxin production as aids in identification of common asymmetric penicillia. Applied and Environmental Microbiology 41: 568–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisvad JC, Samson RA. (2004). Polyphasic taxonomy of Penicillium subgenus Penicillium. A guide to identification of the food and air-borne terverticillate Penicillia and their mycotoxins. Studies in Mycology 49: 1–173. [Google Scholar]
- Frisvad JC, Yilmaz N, Thrane U, et al. (2013). Talaromyces atroroseus, a new species efficiently producing industrially relevant red pigments. PLoS ONE 8: e84102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass NL, Donaldson GC. (1995). Development of premier sets designed for use with the PCR to amplify conserved genes from filamentous Ascomycetes. Applied and Environmental Microbiology 61: 1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guevara-Suarez M, Sutton DA, Cano-Lira JF, et al. (2016). Identification and antifungal susceptibility of penicillium-like fungi from clinical samples in the United States. Journal of Clinical Microbiology 54: 2155–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guevara‐Suarez M, Sutton DA, Gené J, et al. (2017). Four new species of Talaromyces from clinical sources. Mycoses 60: 651–662. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. (2003). A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Systematic Biology 52: 696–704. [DOI] [PubMed] [Google Scholar]
- Herrera J, Poudel R, Khidir HH. (2011). Molecular characterization of coprophilous fungal communities reveals sequences related to root-associated fungal endophytes. Microbial Ecology 61: 239–244. [DOI] [PubMed] [Google Scholar]
- Hocking AD, Pitt JI. (1980). Dichloran-glycerol medium for enumeration of xerophilic fungi from low-moisture foods. Applied and Environmental Microbiology 39: 488–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SB, Cho HS, Shin HD. (2006). Novel Neosartorya species isolated from soil in Korea. International Journal of Systematic and Evolutionary Microbiology 56: 477–486. [DOI] [PubMed] [Google Scholar]
- Houbraken J, de Vries RP, Samson RA. (2014a). Chapter four – Modern taxonomy of biotechnologically important Aspergillus and Penicillium species. Advances in Applied Microbiology 86: 199–249. [DOI] [PubMed] [Google Scholar]
- Houbraken J, Frisvad JC, Samson RA. (2010). Sex in Penicillium series Roqueforti. IMA Fungus 1: 171–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J, Frisvad JC, Samson RA. (2011). Taxonomy of Penicillium section Citrina. Studies in Mycology 70: 53–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J, Frisvad JC, Seifert KA, et al. (2012). New penicillinproducing Penicillium species and an overview of section Chrysogena. Persoonia 29: 78–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J, Samson RA. (2011). Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Studies in Mycology 70: 1–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J, Visagie CM, Meijer M, et al. (2014b). A taxonomic and phylogenetic revision of Penicillium section Aspergilloides. Studies in Mycology 78: 373–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J, Wang L, Lee HB, et al. (2016). New sections in Penicillium containing novel species producing patulin, pyripyropens or other bioactive compounds. Persoonia 36: 299–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. (2001). MRBAYES: bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- Jiang IZ, Yu ZD, Ruan YM, Wang L. (2018). Three new species of Talaromyces sect. Talaromyces discovered from soil in China. Scientific Reports 8: 4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornerup A, Wanscher JH. (1978). Methuen handbook of colour, 3rd edition Eyre Methuen Ltd Press, London. [Google Scholar]
- Krug JC, Benny GL, Keller HW. (2004). Coprophilous fungi. In: Biodiversity of fungi: inventory and monitoring methods (Mueller G, Bills M, Foster G, eds). Elsevier Academic Press, USA: 467–499. [Google Scholar]
- Kuthubutheen AJ, Webster J. (1986). Water availability and the coprophilous fungus succession. Transactions of the British Mycological Society 86: 63–76. [Google Scholar]
- Liu YJ, Whelen S, Hall BD. (1999). Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Molecular Biology and Evolution 16: 1799–1808. [DOI] [PubMed] [Google Scholar]
- Luo Y, Lu X, Bi W, et al. (2016). Talaromyces rubrifaciens, a new species discovered from heating, ventilation and air conditioning systems in China. Mycologia 108: 773–779. [DOI] [PubMed] [Google Scholar]
- Masunga GS, Andresen Ø, Taylor JE, et al. (2006). Elephant dung decomposition and coprophilous fungi in two habitats of semi-arid Botswana. Mycological Research 110: 1214–1226. [DOI] [PubMed] [Google Scholar]
- Orpurt PA, Fennell DI. (1955). A new species of Penicillium from soil. Mycologia 47: 233–237. [Google Scholar]
- Peterson SW. (2008). Phylogenetic analysis of Aspergillus species using DNA sequences from four loci. Mycologia 100: 205–226. [DOI] [PubMed] [Google Scholar]
- Peterson SW, Jurjević Ž, Bills GF, et al. (2010). Genus Hamigera, six new species and multilocus DNA sequence based phylogeny. Mycologia 102: 847–864. [DOI] [PubMed] [Google Scholar]
- Peterson SW, Jurjević Ž, Frisvad JC. (2015). Expanding the species and chemical diversity of Penicillium section Cinnamopurpurea. PLoS ONE 10: e0121987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson SW, Sigler L. (2002). Four new Penicillium species having Thysanophora-like melanized conidiophores. Mycological Research 106: 1109–1118. [Google Scholar]
- Pitt JI. (1979). The genus Penicillium and its teleomorphic states Eupenicillium and Talaromyces. Academic Press, UK. [Google Scholar]
- Pitt JI, Samson RA. (2000). Types of Aspergillus and Penicillium and their teleomorphs in current use. In: Integration of modern taxonomic methods for Penicillium and Aspergillus classification (Samson RA, Pitt JI, eds), Harwood Academic Publishers Press, The Netherlands: 51–72. [Google Scholar]
- Rajeshkumar KC, Marathe SD, Lad SS, et al. (2016). Rediscovery of Penicillium paradoxum (Ascomycete: Aspergillaceae) from Maharashtra, India. Journal of Threatened Taxa 8: 8919–8922. [Google Scholar]
- Rajeshkumar KC, Yilmaz N, Marathe SD, et al. (2019) Morphology and multigene phylogeny of Talaromyces amyrossmaniae, a new synnematous species belonging to the section Trachyspermi from India. MycoKeys 45: 41–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson MJ. (2001). Diversity and occurrence of coprophilous fungi. Mycological Research 105: 387–402. [Google Scholar]
- Rong C, Ma Y, Wang S, et al. (2016). Penicillium chroogomphum, a new species in Penicillium section Ramosa isolated from fruiting bodies of Chroogomphus rutilus in China. Mycoscience 57: 79–84. [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Roy RY, Singh GN. (1968). Penicillium giganteum sp. nov. from soil. Transactions of the British Mycological Society 51: 805–806. [Google Scholar]
- Rundel PW, Arroyo MTK, Cowling RM, et al. (2018) Fire and plant diversification in Mediterranean-climate regions. Frontiers in Plant Science 9: 851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson RA, Houbraken J, Thrane U, et al. (2010). Food and indoor fungi. CBS Laboratory Manual Series 2. CBS-KNAW Fungal Biodiversity Centre, The Netherlands. [Google Scholar]
- Samson RA, Pitt JI. (2000). Integration of modern taxonomic methods for Penicillium and Aspergillus classification. Harwood Academic Publishers Press, The Netherlands. [Google Scholar]
- Samson RA, Visagie CM, Houbraken J, et al. (2014). Phylogeny, identification and nomenclature of the genus Aspergillus. Studies in Mycology 78: 141–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson RA, Yilmaz N, Houbraken J, et al. (2011) Phylogeny and nomenclature of the genus Talaromyces and taxa accommodated in Penicillium subgenus Biverticillium. Studies in Mycology 70: 159–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang H, An TJ, Kim CS, et al. (2013). Two novel Talaromyces species isolated from medicinal crops in Korea. Journal of Microbiology 51: 704–708. [DOI] [PubMed] [Google Scholar]
- Seifert KA, Kendrick B, Murace G. (1983). A key to hyphomycetes on dung. University of Waterloo, Biology Series 27: 1–61. [Google Scholar]
- Seifert KA, Samson RA. (1985). The genus Coremium and the synnematous penicillia. In: Advances in Penicillium and Aspergillus systematics (Samson RA, Pitt JI, eds). Plenum Press, USA: 143–154. [Google Scholar]
- Sequerra J, Marmeisse R, Valla G, et al. (1997) Taxonomic position and intraspecific variability of the nodule forming Penicillium nodositatum inferred from RFLP analysis of the ribosomal intergenic spacer and Random Amplified Polymorphic DNA. Mycological Research 101: 466–472. [Google Scholar]
- Smith G. (1961). Some new and interesting species of micro-fungi. II. Transactions of the British Mycological Society 44: 42–50. [Google Scholar]
- Sonjak S, Frisvad JC, Gunde‐Cimerman N. (2005). Comparison of secondary metabolite production by Penicillium crustosum strains, isolated from Arctic and other various ecological niches. FEMS Microbiology Ecology 53: 51–60. [DOI] [PubMed] [Google Scholar]
- Su L, Niu YC. (2018) Multi-locus phylogenetic analysis of Talaromyces species isolated from cucurbit plants in China and description of two new species, T. cucurbitiradicus and T. endophyticus. Mycologia 110: 375–386. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, et al. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JW, Jacobson DJ, Kroken S, et al. (2000). Phylogenetic species recognition and species concepts in fungi. Fungal Genetics and Biology 31: 21–32. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuthill DE, Frisvad JC. (2002). Eupenicillium bovifimosum, a new species from dry cow manure in Wyoming. Mycologia 94: 240–246. [PubMed] [Google Scholar]
- Valla G, Capellano A, Hugueney R, et al. (1989). Penicillium nodositatum Valla, a new species inducing myconodules on Alnus roots. Plant and Soil 114: 142–146. [Google Scholar]
- Visagie CM, Hirooka Y, Tanney JB, et al. (2014a). Aspergillus, Penicillium and Talaromyces isolated from house dust samples collected around the world. Studies in Mycology 78: 63–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie CM, Houbraken J, Frisvad JC, et al. (2014b). Identification and nomenclature of the genus Penicillium. Studies in Mycology 78: 343–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie CM, Houbraken J, Rodriques C, et al. (2013). Five new Penicillium species in section Sclerotiora: a tribute to the Dutch Royal family. Persoonia 31: 42–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie CM, Renaud JB, Burgess KMN, et al. (2016a). Fifteen new species of Penicillium. Persoonia 36: 247–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie CM, Seifert KA, Houbraken J, et al. (2016b). A phylogenetic revision of Penicillium sect. Exilicaulis, including nine new species from fynbos in South Africa. IMA Fungus 7: 75–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie CM, Yilmaz N, Frisvad JC, et al. (2015). Five new Talaromyces species with ampulliform-like phialides and globose rough walled conidia resembling T. verruculosus. Mycoscience 56: 486–502. [Google Scholar]
- Waksman SA. (1922). A method for counting the number of fungi in the soil. Journal of Bacteriology 7: 339–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhuang WY. (2005). Penicillium brevistipitatum, a new species isolated from Jilin Province, China. Mycotaxon 93: 233–240. [Google Scholar]
- Wang QM, Zhang YH, Wang B, et al. (2016a). Talaromyces neofusisporus and T. qii, two new species of section Talaromyces isolated from plant leaves in Tibet, China. Scientific Reports 6: 18622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XC, Chen K, Xia YW, et al. (2016b) A new species of Talaromyces (Trichocomaceae) from the Xisha Islands, Hainan, China. Phytotaxa 267: 187–200. [Google Scholar]
- Wang XC, Chen K, Qin WT, et al. (2017). Talaromyces heiheensis and T. mangshanicus, two new species from China. Mycological Progress 16: 73–81. [Google Scholar]
- Webster J. (1970). Presidential address: coprophilous fungi. Transactions of the British Mycological Society 54: 161–180. [Google Scholar]
- White TJ, Bruns T, Lee S, et al. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR protocols: a guide to methods and applications (Innis MA, Gelfand DH, Sninsky JJ, White TJ, eds). Academic Press, USA: 315–322. [Google Scholar]
- Yaguchi T, Someya A, Udagawa SI. (1996). A reappraisal of intrageneric classification of Talaromyces based on the ubiquinone systems. Mycoscience 37: 55–60. [Google Scholar]
- Yilmaz N, Houbraken J, Hoekstra ES, et al. (2012). Delimitation and characterization of Talaromyces purpurogenus and related species. Persoonia 29: 39–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz N, López-Quintero CA, Vasco-Palacios AM, et al. (2016). Four novel Talaromyces species isolated from leaf litter from Colombian Amazon rain forests. Mycological Progress 15: 1041–1056. [Google Scholar]
- Yilmaz N, Visagie CM, Houbraken J, et al. (2014). Polyphasic taxonomy of the genus Talaromyces. Studies in Mycology 78: 175–341. [DOI] [PMC free article] [PubMed] [Google Scholar]


