Abstract
The present-day world is severely suffering from the recently emerged SARS-CoV-2. The lack of prescribed drugs for the deadly virus has stressed the likely need to identify novel inhibitors to alleviate and stop the pandemic. In the present high throughput virtual screening study, we used in silico techniques like receptor-ligand docking, Molecular dynamic (MD), and ADME properties to screen natural compounds. It has been documented that many natural compounds display antiviral activities, including anti–SARS-CoV effect. The present study deals with compounds of Natural Product Activity and Species Source (NPASS) database with known biological activity that probably impedes the activity of six essential enzymes of the virus. Promising drug-like compounds were identified, demonstrating better docking score and binding energy for each druggable targets. After an extensive screening analysis, three novel multi-target natural compounds were predicted to subdue the activity of three/more major drug targets simultaneously. Concerning the utility of natural compounds in the formulation of many therapies, we propose these compounds as excellent lead candidates for the development of therapeutic drugs against SARS-CoV-2.
Keywords: COVID-19, Natural compounds, Muti-target, Molecular docking, Molecular dynamics
1. Introduction
Coronaviruses (CoV) have a wide range of hosts, including several species of animals and birds [1]. They belong to a group of enveloped viruses that have positive-sense RNA as genetic material [2]. In 1965, Tyrrell and Bynoe isolated the first Human coronaviruses (HCoV) from the nasal washings of a male child suffering from symptoms of a common cold [3]. HCoV came into the spotlight during the outbreak of Severe Acute Respiratory Syndrome (SARS) in China during the fall of 2002 [[4], [5], [6]]. The SARS-CoV appeared to have originated from the bat and civet reservoir [7,8], claimed 8096 probable cases and 774 deaths in >30 countries [9,10]. Ten years after of SARS episode, in 2012, there was another outbreak of SARS-like disease caused by a different verity of HCoV, officially named as Middle-East Respiratory Syndrome Coronavirus (MERS-CoV) [11]. In 2012, MERS-CoV claimed 2279 laboratory-confirmed cases in 27 different countries [12]. Genome sequencing analysis has revealed MERS-CoV was also of bat origin [13,14]. In Dec 2019, complicated cases of pneumonia were reported in Wuhan, China [15]. World Health Organization (WHO) announced the name of the causative virus as 2019 novel coronavirus (2019-nCoV) on January 12, 2020. Later, the Coronavirus Study Group (CSG) of the International Committee on Taxonomy of Viruses named 2019-nCoV as Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) [16]. As of March 24, 2020, SARS-CoV-2 has cumulatively infected over 381,653 people worldwide and killed over 16,558 individuals since it is first detected. Recent evidence reveals that SARS-CoV-2 was a chimeric virus of a bat coronavirus and an unknown coronavirus [17]. The current epidemic and faster transmission to almost 190 countries and territories are making the situation alarming worldwide. The threat of the currently emerging SARS-CoV-2 necessitates therapeutic strategies targeting proteins involved in the replication and transcription processes of the viral RNA genome.
Since the SARS outbreak of 2002, a tremendous amount of basic and clinical research on coronaviruses has been performed, which led to the identification of many potential drug targets [[18], [19], [20]]. Out of the proteases involved in polyprotein proteolysis, 3C-like protease (3CLpro) (EC 3.4.22.69) has been well studied [21]. Antiviral activities of a series of 3CLpro inhibitors have been reported for Feline Coronaviruses and Feline Caliciviruses [22]. Furthermore, recently, the X-ray crystal structure of 3CLpro of SARS-CoV-2 and its complex with an α-ketoamide inhibitor have been reported, one of the α-ketoamide inhibitors has shown pronounced inhibition of SARS-CoV replication in human Calu-3 lung cells [23].
Coronavirus helicase catalyses the processive separation of double-stranded DNA and RNA in a 5′-to-3′ direction [24,25]. Moreover, helicase (EC 3.6.4.12) inhibitors such as bananin and its derivatives have shown promising anti-cornonaviridae activity [26]. The non-structural protein 15 have endoribonuclease (EC 3.1.26.3) activity that cleaves preferentially at uridine residues [27,28]. A non-structural protein (nsp) 14 has also been shown to have exoribonuclease (ExoN) (EC 3.1.13.1) activity and involved in CoV replicative machinery. The active site mutations of the protein revealed severe defects in CoV RNA synthesis [29]. In the SARS-CoV replication study, it has been shown that AdoMet dependent methyltransferase inhibitors inhibit SARS-CoV replication [30] by targeting N-methyltransferases (EC 2.1.1.35) of the virus [31]. Due to strictly virus-specific activity, the RNA-dependent RNA polymerase (RdRp) (EC 2.7.7.6) has been considered as a promising wide-spectrum drug target for antiviral drug development. Several studies discovered that inhibitors against RdRp could effectively intervene in the coronavirus lifecycle [32,33]. RNA viruses are unique pathogens in terms of exceptionally genetically variable. They accumulate mutations in their genome by employing viral reverse transcriptase that lacks a proofreading mechanism. This unique feature of RNA viruses makes it challenging to design active therapeutic agents. Most of the antiviral drugs in use are designed to specifically target single viral enzyme, which is essential for viral replication or invasion [34,35]. However, the high rate of mutations in the viral drug targets has been accounted for the reduced susceptibility of currently available antiviral drugs [36]. A combination of drugs that have different molecular targets can be a better choice; however, some times, the combination therapy is not safe due to unwanted drug interactions [37]. Besides, drugs designed for multiple protein targets are extensively used for the treatment of both infectious and inherited diseases [[38], [39], [40], [41], [42]]. For the immediate drug requirement for SARS-CoV-2, biologically active drug-like molecules that target multiple viral enzymes of the viral replication cycle are highly wanted.
Natural compounds of plant-based origin have been studied as an exciting class of pharmacologically active molecules, some of them have an ancient history of antiviral activity [[43], [44], [45], [46], [47], [48], [49], [50]]. Extensive studies have been performed over the past decades to identify anti-CoV agents using natural products [51]. The saikosaponins (A, B2, C, and D), for example, exerts antiviral activity by interfering viral attachment and penetration, against HCoV-22E9 [52]. Besides, several natural products such as Lycoris radiata, Artemisia annua, Pyrrosia lingua, and Lindera aggregate have been reported to display significant anti–SARS-CoV properties [53]. Moreover, inhibitors from natural origin have been identified against the SARS-CoV enzymes, such as helicase and 3CLpro and viral RdRp [[54], [55], [56], [57]]. NPASS database is freely accessible (http://bidd2.nus.edu.sg/NPASS/) that provides the literature-reported experimentally-determined activity (e.g., IC50, Ki, EC50, GI50, and MIC) values of the natural products against macromolecule or cell targets along with the taxonomy of the species sources of 35,032 unique natural products [58]. In the heart of the current Corona Virus Disease 2019 (COVID-19) outbreak, these NPASS compounds may be used for capable therapy as they can remarkably reduce the time taken to design a therapeutic regimen.
Structure-based drug design by virtual screening and molecular docking studies has become a valuable primary step in the identification of novel lead molecules for the treatment of diseases [59,60], and proven to be a very efficient tool for antiviral [[61], [62], [63], [64]] and antibacterial [65,66] and antiprotozoal [67,68] drug discovery. Therefore, a virtual screening experiment was conducted to determine the interaction of natural ligands of the NPASS database within the binding pocket of putative drug targets of the virus that was calculated in terms of docking scores and MM-GBSA values. Our high throughput virtual screening revealed 21 natural compounds having higher docking scores and MM-GBSA values for six potent therapeutic targets of SARS-CoV-2 over the known chemical inhibitors. Remarkably, we suggested three natural compounds that able to bind the catalytic site of three/more crucial viral enzymes with a relatively high affinity, which ultimately can be used for the development of instant drugs for the emerging COVID-19.
2. Material and methods
2.1. Selection of different drug targets of SARS-CoV-2 and its sequence similarity with SARS coronavirus
For developing the structure of SARS-CoV-2 functional enzymes, the amino acid sequences of SARS-CoV-2 (accession NC_045512.1) were downloaded from the NCBI database (https://www.ncbi.nlm.nih.gov/) in the FASTA format. All of the six proteins namely helicase (accession YP_009725308.1), endoribonuclease (accession YP_009725310.1), exoribonuclease (accession YP_009725309.1), RNA dependent RNA polymerase (RdRp) (accession YP_009725307.1), methyltransferase (accession YP_009725311.1) and 3C-like proteinase (accession YP_009725301.1) belong to the replication complex of the deadly virus SARS-CoV-2. The amino acid sequences obtained from NCBI were aligned with SARS coronavirus using the BLASTp server (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins) [69].
2.2. Homology modeling of the selected drug targets, refinement, and validation of structure
Since the crystal structures of the selected drug targets were not available on the protein data bank (PDB), the 3D structures were modeled using SWISS-MODEL (https://swissmodel.expasy.org/). For this purpose, the amino acid sequences of each target were submitted in the SWISS-MODEL server to develop the tertiary structure [70]. Here, we had selected the template similar to the query sequence coverage and identity of the sequence-based upon their Global Model Quality Estimate (GMQE) [71] and Quaternary Structure Quality Estimate (QSQE) scores. The homology modeling technique, which we use to predict the tertiary structure of the protein, is the widely used method. However, accurate estimation of the three-dimensional position of individual atoms in a protein sequence is tough even with the best-matched template and target sequence alignment [[72], [73], [74], [75], [76]]. The homology model generally deviates from their native structure concerning their atomic coordinates [77]. Therefore, the refinement of the homology model is a very crucial step to identify the accurate near-native structure [78]. It is known that the geometrical/predicted structure of the target sequence affects the function of the protein, which also includes pharmacophore drug designing.
Here, we have used the 3D-refine server (http://sysbio.rnet.missouri.edu/3Drefine/) for the refinement of the modeled structures of each target protein of SARS-CoV-2. This refinement server works on the two-step protocol, which reliably brings the predicted homology model closer to its native structure [[79], [80], [81], [82], [83], [84]]. Where the first step is the optimization of the hydrogen bond network, and second is the minimization of atomic-level energy of optimized homology models using the knowledge-based force fields [85]. This server requires a homology model in PDB format as an input query. The server provides the five refined models as an output with best on the top in PDB format. There are several parameters for the selection of best-refined models, which include the Molprobity score, which is the measure of the local quality of the structure [86], the lower score shows the excellent model. Another given parameters as output scores are GDT-TS score, GDT-HA score, and RMSD, which tells about the positioning of Calpha atoms. GDT-TS and GDT-HA score of the refined model, which ranges from 0 to 1, where the lower score shows a good quality model [87]. The .pdb files of refined structures of respective target proteins were uploaded on the RAMPAGE server (http://mordred.bioc.cam.ac.uk/~rapper/rampage.php), which had generated the Ramachandran plot. This plot is generated to obtain the value of the total count of amino acids present in the allowed, disallowed, and favorable regions [88].
2.3. Preparation of protein and ligands using Schrödinger suite
Refined homology models of the COVID-19 drug targets (proteins) were used for the further downstream steps. These protein structures were defined as a receptor and opened via protein preparation wizard in maestro v11.9 [89]. The standard refined model of protein does not fit for the docking with ligand molecules vis-a-vis calculation of the binding affinity. Therefore, models were preprocessed and optimized by filling the missing side chain using prepwizard module of the Schrodinger suite 2019-1. Heavy water molecules were removed with less than 3H-bonds followed by energy minimization, which convert the heavy atoms to RMSD 0.30 Å.
Natural compounds from the NPASS database [58] were used as ligands to target the COVID-19 replication complex proteins. The compounds having druggable properties were identified (3963 natural compounds), and 2D structures in .sdf format were downloaded from the NPASS database. For using these compounds as ligands for docking purposes, there is a need to remove the salt, addition of hydrogen atoms, and deprotonation. LigPrep v4.7 module of the Schrodinger suite was used to prepare the ligands in the three-dimensional structure. Charge neutralization to attain the biological relevant pH (pH 7), desalting was done using Epik v4.7, which is based upon Hammett and Taft methodologies. Torsional bond of ligands was released, and a maximum one stereoisomer per ligand was generated, which resulted in the final 4570 ligands upon LigPrep of natural compounds from the NPASS database.
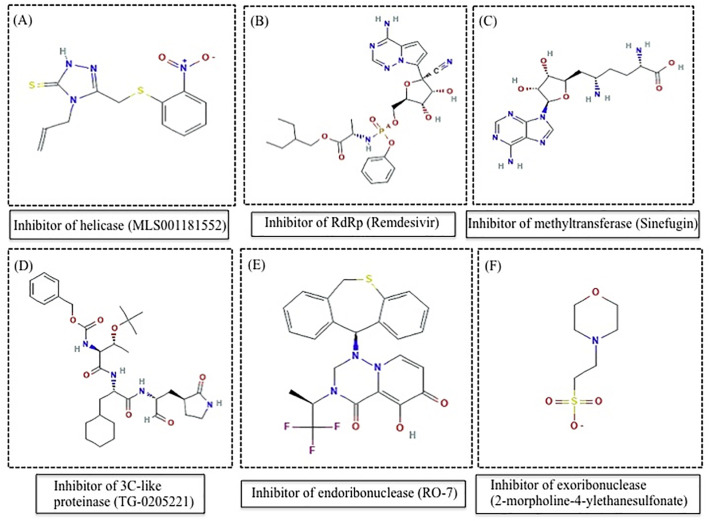
In this study, one reference/control ligand for each target protein was included. Those ligands were chosen based upon the previous studies. Out of six, four inhibitors namely MLS001181552 (inhibitor of helicase) [90], remdesivir (inhibitor ofRdRp) [91], sinefungin (inhibitor of methyltransferase) & TG-0205221 (3C-like proteinase) [92] were well-known for SARS coronavirus protein targets while rest two namely RO-7 (inhibitor of endoribonuclease) [93] and 2-morpholine-4-ylethanesulfonate (inhibitor of exonuclease) [94] inhibitors were known for Influenza virus protein and member of DEddH family of exonucleases (Fig. 1 ).
Fig. 1.
Showing the chemical formula of reference/control ligand for each target protein.
2.4. Generation of grid
The generation of the grid is a very crucial step for the binding of a ligand to the receptor. Here, a 3-dimensional boundary for the ligand binding was generated in the receptor using Glide, version 8.2 [95] of Maestro, Schrodinger. The receptor grid for each target protein was generated by indicating the active sites amino acid residues, which were searched from previously reported studies appropriate to each target. Amino acid residues involved as the active site with respect to the target, along with their references, are available in Supplementary Table 1. The size of the receptor grid was set at default, i.e., 20 Å. Active site amino acids are crucial because they may affect the entry of the ligand, followed by binding to the target protein.
2.5. Molecular interaction/docking studies of six target proteins of COVID-19 against natural compounds
After grid generation, the natural compounds/ligands were docked to the target protein/receptor using the protocol of Glide grid version v8.2 [95]. The software internally generates different conformations, which passes across several filters viz. euler angles, grid-based force field evaluation, and Monte Carlo energy minimization. Lastly, the evaluation of conformers takes place based upon the docking score, and one of the best conformations per ligand is generated as an output. The docking was performed in three steps, which include high throughput virtual screening (HTVS), standard precision (SP), and extra precision (XP). The first two steps of docking, HTVS, and SP utilize the self-same scoring function, whereas XP reduces the intermediate conformations and thoroughness of the torsional refinement and sampling [96]. A total of 4570 natural compounds were docked against each target protein with their respective positive control inhibitor (mentioned earlier). Subsequently, top scorers were set forth for SP docking, and the output of SP docking was put forward in XP docking with positive control. The docking score is a binding energy/affinity given in the kcal/mol. All the statistical data of top scorers for each target protein wrt control obtained is presented in result tables.
2.6. Absorption, Distribution, Metabolism, Excretion (ADME) properties evaluation/calculation
It has been shown that the natural compounds do not follow Lipinski's rule as these compounds tend to keep low hydrophobicity as well as the potential of donating the intermolecular H-bond [97]. We have used QikPropv5.9 of the Schrodinger suite, which analyzes the druggable properties of the ligands. This module takes ligprep file as an input and predicts the ADME properties in the form of several physicochemical properties and principal descriptor values, which were found in the acceptable range.
2.7. Generation of molecular mechanics, the generalized born solvent accessibility (MM-GBSA) score and ROC plot analysis
Another application of Schrodinger suite software viz., Prime, was used to estimate the binding energy and represented as MM-GBSA score of the selected docked complex of the natural compound and target protein obtained from extra precision (XP) step of docking. This post-docking application provides the correct ranking of ligands binding based upon the MM-GBSA energy score [98].
Further, there are several other metrics, which are currently in use to evaluate the performance of virtual screening docking protocol, for instance, enrichment factor (EF), area under the ROC curve, and Boltzmann-enhanced discrimination of receiver operating characteristic (BEDROC) etc. [99]. In this study, receiver operating characteristic (ROC) plots were also generated to assess the performance of the SP and XP docking methodology, which differentiates the active compounds with false positives means decoys [100]. These statistical curves represented the probability and were plotted to correctly classify the ligands into actives and decoys that finally attested our docking performance. It gives the value between 0 and 1, where 0 represents the worst performance of docking, and 1 represents the best performance [99,101]. To plot these curves, sensitivity (on Y-axis) was plotted against 1-specificity (on X-axis). Here, sensitivity signifies the true positive, whereas 1-specificity signifies the decoys/false positive. All-natural compounds/ligands performed best on SP and XP docking were subjected to the docking protocol validation. Separate ROC curves to validate for both XP and SP docking protocols were generated. In addition to this, we have also calculated the Enrichment factor (an enrichment calculator from Schrodinger suite was used), Area Under Accumulation (AUC), and BEDROC values were also predicted.
2.8. Molecular dynamics
For comprehending the properties of the structure and their microscopic interaction, molecular dynamics (MD) simulation was performed. For this, systems were built for best three multi-target natural compounds with one of their potent targets viz. helicase and NPC270578, exoribonuclease, and NPC214620 & methyltransferase and NPC52382 complex using Desmond v5.6, system builder panel (Schrodinger 2019-1). The optimized potentials were used for the liquid simulations at the OPLS3e force field, and these solvated systems were opened in the molecular dynamics panel [102]. Firstly, the solvated system builder was used to selecting a single point charge as a water model, and ligand-protein complexes were neutralized by adding sodium or chloride ions. The NPC214620-exonuclease complex model system consisted of 80,840 atoms and 24,147 water molecules and was neutralized by adding 7 Cl− ions. NPC52382-methyltransferase model system contained 34,012 atoms and 9781water molecules and was neutralized by adding 2 Cl− ions. Whereas, the NPC270578-helicase complex model system consisted of 79,549 atoms and 23,354 water molecules and was neutralized by adding 9Cl− ions. System equilibration was done at a constant temperature, i.e., 300.0 K and standard atmospheric pressure, i.e., 1 bar. Upon the equilibration step, the total time for each simulation was appointed ten nanoseconds with trajectories analysis at regular intervals for the protein-ligand complex. Ultimately, the RMSD was analyzed to identify the stability of the target protein in its natural motion.
3. Results
3.1. Developing the structure of SARS-CoV-2 functional enzymes
The amino acid lengths of the selected protein targets for SARS-CoV-2 were found to be 601 for helicase, 346 for endoribonuclease, 527 for exoribonuclease, 932 for RNA-dependent RNA polymerase, 298 for methyltransferase and 306 for 3C-like proteinase. It was observed that these target proteins from SARS coronavirus were also carrying almost similar amino acid length, i.e., 603, 346, 527, 955, 344, and 308, respectively. In this piece of study, we wanted to investigate the identity, similarity, and percentage of conserved sequences between COVID-19 and SARS coronavirus.
3.2. Homology modeling of the selected drug targets, refinement, and validation of structure
Models for each enzyme such as helicase, endoribonuclease, exoribonuclease, RNA dependent RNA polymerase, methyltransferase, and 3-C like proteinase were generated, using template 6jyt.2.A, 2h85.1.A, 5nfy.1.A, 6nur.1.A, 2xyr.1.A and 2z9j.1, respectively. The model quality of the generated 3D models was also estimated by the QMEAN score [103] function of SWISS-MODEL, which relies on modeling error quantification and expected model accuracy. The GMQE and QSQE scores generally lie between 0 and 1, where higher the score, the higher the reliability of the modeled structure means expected accuracy of interchain interactions for a given template, whereas, in case of QMEAN, a score lower than 4.0 gives reliability [104]. After the model generation and refinement, each enzyme's structure was validated through the generation of the Ramachandran plot (Table 1 ). Ramachandran plot confirmed that all target models after refinement would be used for molecular docking with ligand except 3C-like proteinase. In case of 3C-like proteinase the model obatined from Swiss-modeling was used for molecular docking. The structure generation confirms the amino acid mapping in the allowed region, which shows the stable nature of existing proteins.
Table 1.
Details of scores obtained for target proteins via RAMPAGE and Swiss-model servers.
| Proteins | Template used for homology modeling | Sequence identity (%) by Blast (protein seq. with template seq.) | Ramachandran score (% of residue) | GMQE score | RMSD (Å) | Ramachandran score after refinement (% of residue) |
|---|---|---|---|---|---|---|
| Helicase | 6jyt.2. A | 97.95 | 84.34 | 0.98 | 0.304 | 88.6 |
| Endoribonuclease | 2h85.1. A | 88.0 | 97.96 | 0.98 | 0.249 | 98.5 |
| Exoribonuclease | 5nfy.1. A | 94.07 | 89.44 | 0.98 | 0.258 | 93.1 |
| RNA-dependent RNA polymerase | 6nur.1. A | 96.24 | 97.5 | 0.83 | 0.228 | 98.1 |
| Methyltransferase | 2xyr.1. A | 92.57 | 96.18 | 0.97 | 0.252 | 96.2 |
| 3C-like proteinase | 2z9j.1.A | 96.0 | 96.51 | 0.99 | 0.269 | 96.4 |
3.3. Computational Virtual Screening of Natural Ligands against potential therapeutic targets of SARS-CoV-2
To screen effective inhibitors for SARS-CoV-2, we used compounds of NPASS database against six potential therapeutic targets of the virus, such as helicase, endoribonuclease, exoribonuclease, RNA-dependent RNA polymerase, N-methyltransferase, and 3C-like protease in our molecular docking experiment. HTVS, SP, and XP step-wise screening protocol were followed to find potent compounds having higher docking scores and binding energy as follows.
3.4. Molecular docking with helicase
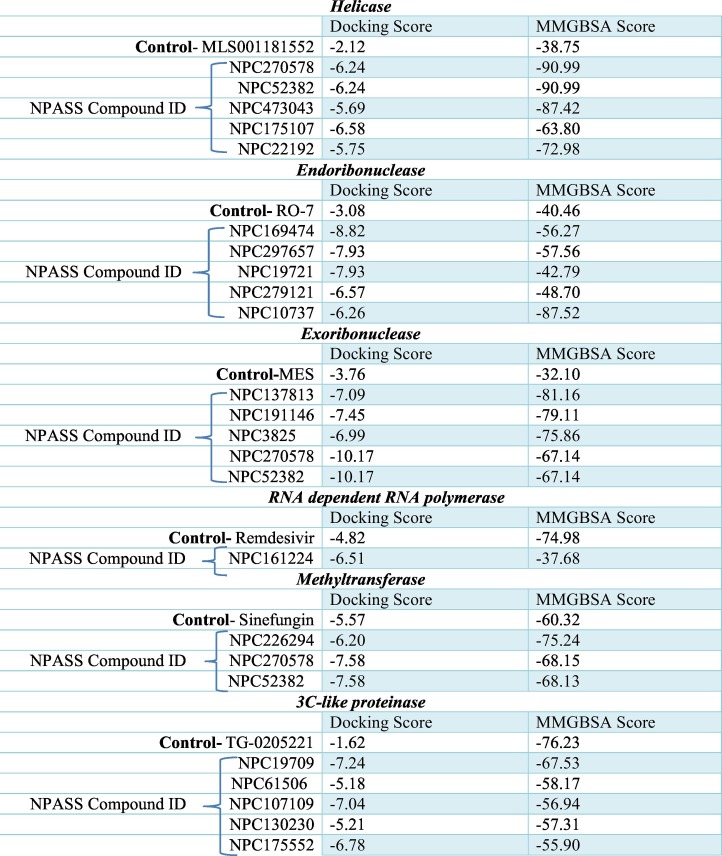
In a SARS-CoV replicon assay, it has been shown that the specific inhibition of SARS-CoV helicase by SSYA10-001 blocked viral replication [90]. Therefore, we used this compound as a control to find out more effective inhibitors for COVID-19 during our high throughput virtual screening experiment of natural compounds. Interestingly, we found five compounds such as NPC270578, NPC52382, NPC473043, NPC175107, and NPC22192 displayed docking score and MM-GBSA value 2.5 and 1.5 times greater than the control, respectively (Table 2 ). Notably, NPC52382 has been shown to have biological activity against the malaria parasite in the NPASS database.
Table 2.
Docking and MMGBSA score of the top targeted compounds.
3.5. Molecular docking with endoribonuclease
The antiviral property of endoribonuclease inhibitors has been validated for the influenza virus in multiple in vitro assay [93]. The essential function of endonuclease in the initiation of viral transcription supports its potential as a promising target for the development of antiviral agents. We find five compounds such as NPC169474, NPC297657, NPC19721, NPC279121, and NPC10737, which have more than two-fold docking scores and better MM-GBSA value than the known viral endoribonuclease inhibitor (Table 2). Subsequent biological activity analysis of these compounds in the NPASS database reveals that NPC279121 has antiviral activity against Hepatitis C virus by inhibition of RdRp [105], Influenza A virus by inhibition of neuraminidase [106], HIV-1 by inhibition of integrase [107] and Simian virus 40 by unknown mechanism whereas NPC10737 has antiviral property (IC50 = 34 μg/ml) against HIV1 through suppression of transcription from 5′-long terminal repeat including activation via NF-kB [108].
3.6. Molecular docking with exoribonuclease
Inhibitors for exonuclease of the Lassa fever virus have been identified [94]. Our molecular docking studies identified five compounds, such as NPC137813, NPC191146, NPC3825, NPC270578, and NPC52382, bearing 1.8 to 2.7fold higher docking score and 2 to 2.5fold higher MM-GBSA value as compared to the general lead exonuclease inhibitor MES (2-(N-morpholino) ethanesulfonic acid) (Table 2). Surprisingly, we did not get any hints of the antiviral studies of these compounds in the NPASS database.
3.7. Molecular docking with RdRP
It is essential to find out novel compounds against COVID-19 based on broad-spectrum antiviral compounds. Several studies revealed the potent antiviral activities of remdesivir through the inhibition of RdRp [[109], [110], [111]]. Further, remdesivir has also shown antiviral activities against SARS-CoV and MERS-CoV [111]. To study whether the NPASS database has some compounds active against RdRP of SARS-CoV-2, we conducted molecular docking studies. We found a compound, NPC161224, having a 1.35 fold higher docking score but with a lower MM-GBSA value than remdesivir (Table 2). According to the NPASS database, this compound has also shown RdRP inhibition property against HIV1.
3.8. Molecular Docking with 3C-like proteinase (3CLpro)
3CLpro is an attractive target for antiviral therapeutics because of its essential role in the processing of CoV polyprotein [112,113]. Numerous 3CLpro inhibitors have been shown to block CoV replication in cell culture [114,115]. TG-0205221, a known inhibitor of 3CLpro of SARS coronavirus, could significantly reduce the viral titer of SARS CoV in cell culture [92]. The binding mode information of TG-0205221 is employed here towards the discovery of new 3CLpro inhibitors from the NPASS database. From our intensive docking studies, we report five molecules such as NPC19709, NPC61506, NPC107109, NPC130230, and NPC175552 having three-fold higher docking score than TG-0205221. However, these molecules have a relatively lower MM-GBSA value than the control (Table 2). Notable, the antiviral activity (IC50 = 14.48 μg/ml) against Influenza A virus of NPC130230 by inhibition of neuraminidase [116] has been described in the NPASS database.
3.9. Molecular docking with methyltransferase
Methylation of the viral mRNA cap structure at the N7 position of the guanine is indispensable for the synthesis of viral proteins [117]. N7- methyltransferase has been shown as a potential antiviral target for SARS-CoV in a yeast-based screening assay [118]. In our docking experiment, three natural compounds such as NPC226294, NPC270578, and NPC52382 has shown better docking scores and MM-GBSA value than, Sinefungin, the known inhibitor of N7- methyltransferase (Table 2). Besides, we get one compound NPC70622 that has antiviral activities such as IC50 = 300 μg/ml against the Hepatitis B virus by inhibition of HBV-DNA production [119] and EC50 = 0.2 μM against Hepatitis C virus by inhibition of RdRp [105] as described in the NPASS database.
3.10. Identification of potential multi-target inhibitors of SARS-CoV-2
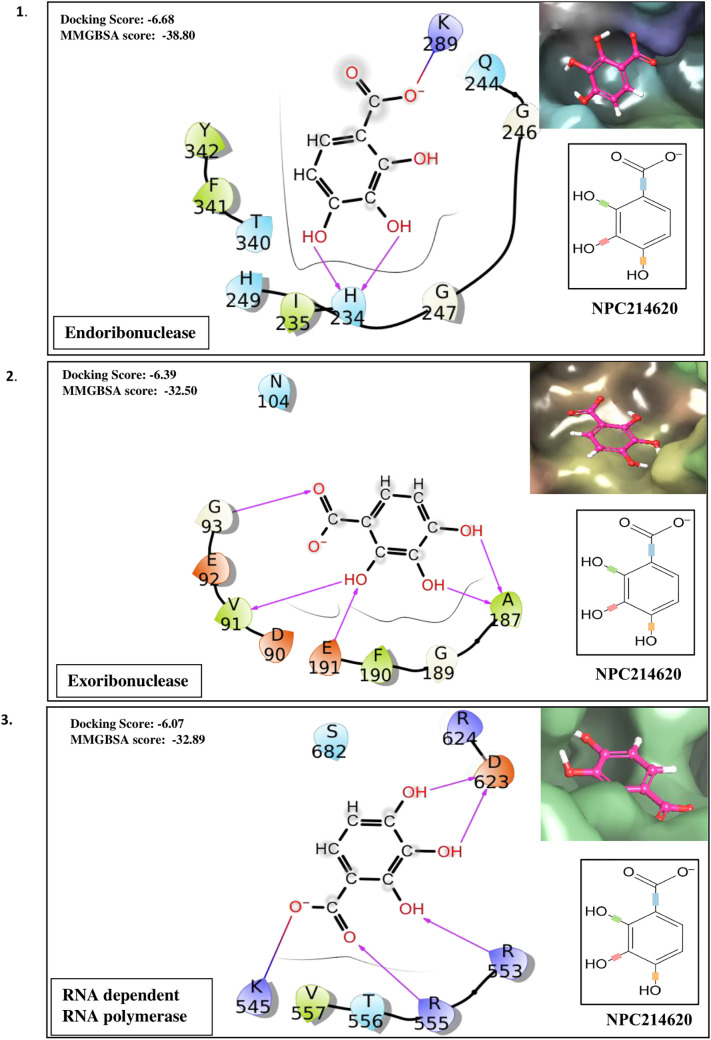
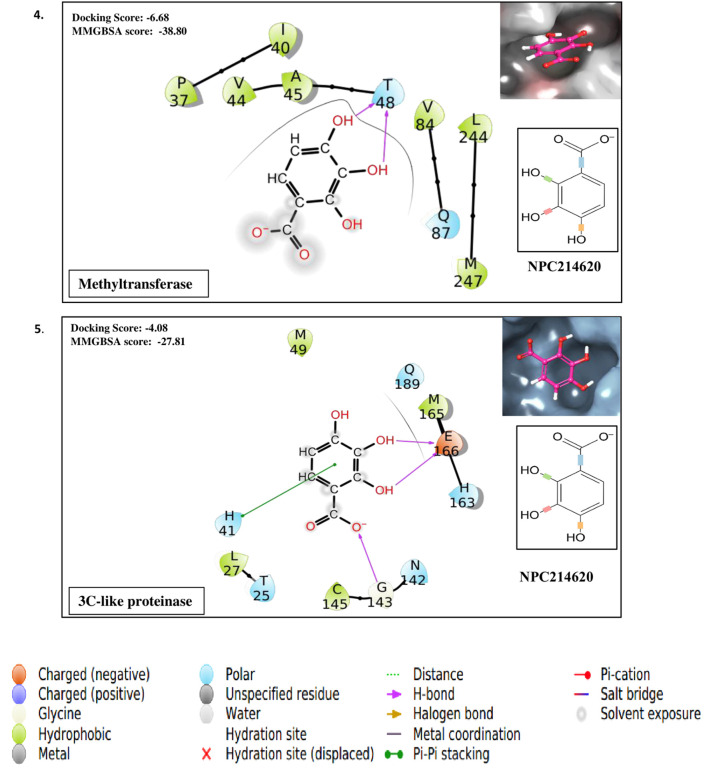
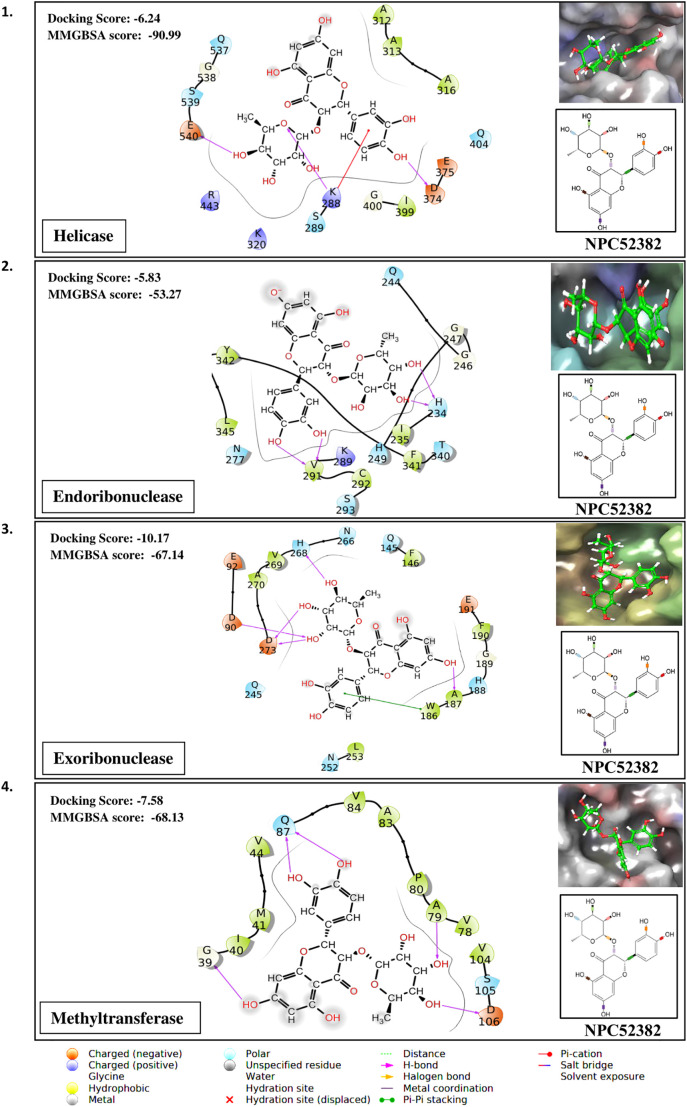
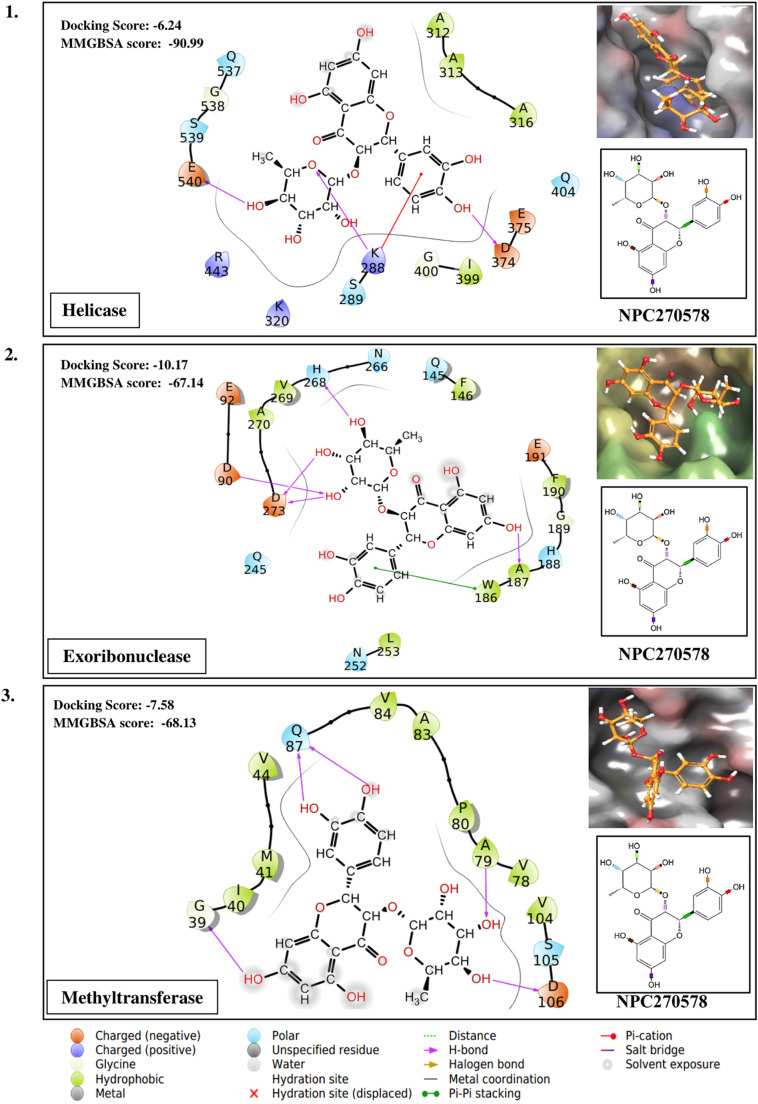
Taking the docking scores and MM-GBSA values displayed by natural compounds for three/more targets into consideration, we propose three compounds as multi-target inhibitors against SARS-CoV-2. Compound NPC214620 secures better docking scores than respective control molecules against five different drug targets. The docking scores and MM-GBSA values of the molecule in the binding pocket of respective targets are shown in Fig. 2A. As compared to controls, the second compound NPC52382 showed higher docking scores and MM-GBSA values for four therapeutic targets, as shown in Fig. 2B. The third multi-target compound that exhibits higher docking scores and MM-GBSA values towards helicase, endoribonuclease, and N-methyltransferase is NPC270578, shown in Fig. 2C.
Fig. 2.
(A). Showing the contacts between multi-target NPASS compound, NPC214620 with (1) Endoribonuclease (2) Exoribonuclease (3) RNA dependent RNA polymerase (RdRp) (4) Methyltransferase and (5) 3C-like proteinase. The ligand 2D representation in the side panel is showing the rotatable bonds of the ligand in different colors. (B). Showing the contacts between multi-target NPASS compound, NPC52382 with (1) Helicase (2) Endoribonuclease (3) Exoribonuclease (4) Methyltransferase. The ligand 2D representation in the side panel is showing the rotatable bonds of the ligand in different colors. (C). Showing the contacts between multi-target NPASS compound, NPC270578 with (1) Helicase (2) Exoribonuclease (3) Methyltransferase. The ligand 2D representation in the side panel is showing the rotatable bonds of the ligand in different colors.
3.11. QikProp analysis of ADME and principal descriptor values
Lipinski's Rule of Five is not a strict criterion for natural compounds [120]. Therefore, some crucial ADME properties for the lead compounds were evaluated using Schrӧdinger QikProp module. Various basic physiochemical properties such as PlogPo/w (Predicted octanol/water partition coefficient), PlogS (Predicted aqueous solubility), PlogHERG (Predicted IC50 value for the blockage of HERG K+ channels), PPCaco (Predicted apparent Caco-2 cell permeability for the gut blood barrier), PlogBB (predicted brain/blood partition coefficient), PPMDCK (Predicted apparent MDCK cell permeability in nm/s), and PlogKhsa (prediction of binding to human serum albumin) of these compounds were predicted. The values of those compounds found in the acceptable range were shown in Table 3 . The principal descriptor values including MW (molecular weight of molecule), SASA(Total solvent accessible surface area), FOSA (Hydrophobic component of the SASA), FISA (Hydrophilic component of the SASA), PISA (π (carbon and attached hydrogen) component of the SASA), PHOA (Percent human oral absorption), DonorHB (Estimated number of hydrogen bonds that would be donated by the solute to water molecules in an aqueous solution), and Accept HB (Estimated number of hydrogen bonds that would be accepted by the solute from water molecules in an aqueous solution) were evaluated for the drug-like behavior of the lead compounds. The values of the compounds fallen in the suitable range were displayed in Table 4 .
Table 3.
ADME Properties of the selected NPASS compound.
| NPASS compound ID | QPlogPo/w (−2.5 to 6.5) | QPlogS (−6.5 to 0.5) | QPlogHERG (above −5.0) | QPPCaco (<25%-poor, >500-great) | QPlogBB (−3 to 1.2) | QPPMDCK (<25%-poor, >500-great) | QPlogKp (−8.0 to −0.1) | QPlogKhsa (−1.5 to 1.5) |
|---|---|---|---|---|---|---|---|---|
| NPC214620 | −1.19 | −0.09 | −2.24 | 180.67 | −0.96 | 77.83 | −4.61 | −0.91 |
| NPC52382 | −0.78 | −1.97 | −3.55 | 384.25 | −0.91 | 175.94 | −3.97 | −0.88 |
| NPC270578 | −0.78 | −1.97 | −3.55 | 384.25 | −0.91 | 175.94 | −3.97 | −0.88 |
| NPC473043 | −1.02 | −1.92 | −3.24 | 113.29 | −1.32 | 46.99 | −5.01 | −0.78 |
| NPC175107 | −2.75 | −1.41 | −3.34 | 65.68 | −1.56 | 26.07 | −5.47 | −1.29 |
| NPC22192 | 1.97 | −5.89 | −5.45 | 246.55 | −1.52 | 108.91 | −4.45 | 0.10 |
| NPC169474 | 0.52 | −3.52 | −4.92 | 870.98 | −0.68 | 426.10 | −3.48 | −0.56 |
| NPC297657 | 0.38 | −2.61 | −3.92 | 131.17 | −1.50 | 55.06 | −4.79 | −0.43 |
| NPC19721 | 0.02 | −1.53 | −3.38 | 54.84 | −1.76 | 21.45 | −5.43 | −0.61 |
| NPC279121 | 0.55 | −1.89 | −2.97 | 100.45 | −1.31 | 41.27 | −5.11 | −0.33 |
| NPC10737 | 0.42 | −3.37 | −4.68 | 516.13 | −0.95 | 242.04 | −3.82 | 77.97 |
| NPC137813 | −1.52 | −2.57 | −5.05 | 713.22 | −0.74 | 352.70 | −3.72 | −1.48 |
| NPC191146 | 3.08 | −4.94 | −4.32 | 1406.61 | −0.43 | 715.33 | −2.98 | 0.36 |
| NPC3825 | 0.98 | −3.29 | −4.12 | 316.75 | −1.05 | 142.79 | −4.23 | −0.32 |
| NPC161224 | −8.84 | 2.00 | −4.83 | 5.58 | −1.58 | 4.22 | −8.63 | −3.37 |
| NPC226294 | −1.15 | −1.88 | −3.67 | 178.04 | −1.31 | 76.60 | −4.53 | −0.95 |
| NPC19709 | −0.891 | −1.973 | −3.159 | 259.635 | −0.931 | 115.172 | −4.401 | −0.766 |
| NPC61506 | 1.18 | −2.972 | −4.431 | 85.894 | −1.889 | 34.841 | −5.047 | −0.226 |
| NPC107109 | 0.436 | −0.906 | −2.725 | 607.8894 | −0.55 | 288.864 | −3.683 | −0.769 |
| NPC130230 | 0.50 | −1.06 | −2.60 | 154.93 | −1.13 | 65.92 | −4.65 | −0.58 |
| NPC175552 | 0.08 | −1.76 | −3.51 | 203.24 | −1.22 | 88.39 | −4.42 | −0.60 |
Table 4.
Principal drug characteristic predictions for NPASS compounds.
| NPASS compound ID | MW (130–725) | SASA (300−1000) | FOSA (0–750) | FISA (7–330) | PISA (0–450) | Volume (500–2000) | PHOA (<25%-poor, >80%-high) |
|---|---|---|---|---|---|---|---|
| NPC214620 | 170.12 | 273.73 | 90.35 | 183.38 | 0.00 | 398.07 | 60.39 |
| NPC52382 | 450.40 | 492.93 | 344.11 | 148.82 | 0.00 | 842.51 | 42.72 |
| NPC270578 | 405.40 | 492.93 | 344.11 | 148.82 | 0.00 | 842.51 | 42.72 |
| NPC473043 | 448.38 | 468.84 | 264.08 | 204.76 | 0.00 | 807.40 | 31.81 |
| NPC175107 | 492.39 | 505.02 | 275.29 | 229.29 | 0.00 | 893.00 | 17.48 |
| NPC22192 | 440.48 | 710.51 | 541.37 | 169.15 | 0.00 | 1175.41 | 68.36 |
| NPC169474 | 320.34 | 544.04 | 432.70 | 111.35 | 0.00 | 838.95 | 69.64 |
| NPC297657 | 326.35 | 486.40 | 288.36 | 198.05 | 0.00 | 793.81 | 67.08 |
| NPC19721 | 320.26 | 413.49 | 175.51 | 237.99 | 0.00 | 659.32 | 45.03 |
| NPC279121 | 286.24 | 284.77 | 174.51 | 210.26 | 0.00 | 617.49 | 65.98 |
| NPC10737 | 358.39 | 573.79 | 438.48 | 135.31 | 0.00 | 934.07 | 77.97 |
| NPC137813 | 458.46 | 633.15 | 513.79 | 119.36 | 0.00 | 1037.56 | 56.35 |
| NPC191146 | 396.44 | 589.20 | 499.81 | 89.39 | 0.00 | 1013.24 | 88.35 |
| NPC3825 | 354.36 | 525.85 | 368.18 | 157.67 | 0.00 | 872.99 | 64.50 |
| NPC161224 | 491.18 | 529.71 | 191.73 | 279.05 | 0.00 | 884.39 | 0.00 |
| NPC226294 | 448.38 | 497.96 | 313.91 | 184.05 | 0.00 | 844.39 | 34.60 |
| NPC19709 | 432.383 | 461.385 | 294.609 | 166.776 | 0.00 | 794.934 | 51.982 |
| NPC61506 | 304.256 | 475.401 | 257.966 | 217.435 | 0.00 | 724.859 | 55.51 |
| NPC107109 | 300.267 | 375.147 | 247.332 | 127.815 | 0.00 | 609.057 | 79.324 |
| NPC130230 | 286.24 | 372.91 | 182.49 | 190.42 | 0.00 | 611.19 | 69.09 |
| NPC175552 | 290.27 | 415.65 | 237.66 | 177.99 | 0.00 | 655.12 | 55.75 |
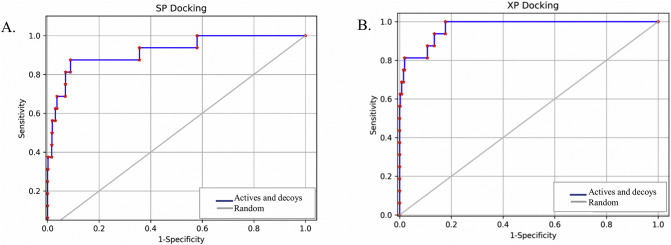
3.12. ROC curve plot and enrichment study
In machine learning and data mining research, Receiver Operating Characteristic (ROC) curves are usually used to study the performance of scoring function [121]. The ROC curve value detects predictive ability, which varies in the range 0.5 for random prediction to 1.0 for perfect prediction [122,123]. For analyzing the performance of our scoring functions, SP and XP data were evaluated by (ROC) curve, and the ROC curve was generated for the virtual screening experiment by employing the pose viewer (pv.- maegz) file of docking output (16 actives and 1000 decoys) of SP and XP results. SP and XP docking reveal ROC values 0.92 and 0.97, respectively (Fig. 3 ), indicating a high predictive performance of the used methods. In the later stage, the overall performance of SP and XP docking methods was measured by the values of the area under the curve(AUC). The discrimination of active and inactive compounds are measured as 100% with an AUC value equal to 1 and random with a value of 0.5 [121]. The value of the AUC was calculated as 0.91 and 0.96 for SP and XP docking, respectively. The BEDROC values at α = 160.9 for SP and XP docking were calculated as 0.658 and 0.866, respectively, indicating the better performance of XP against SP docking, as shown in Table 5 .
Fig. 3.
SP and XP docking validation and representation of both actives and decoys compounds by plotting the ROC curve.
Table 5.
SP and XP docking validation on the basis of their active counts and ROC value.
| Docking mode | Percentage of results | 1% | 2% | 5% | 10% | 20% | Enrichment | Matric value | |
|---|---|---|---|---|---|---|---|---|---|
| SP docking | Active counts | 6 | 6 | 11 | 13 | 14 | ROC | 0.92 | |
| % of Actives | 37.5 | 37.5 | 68.8 | 81.2 | 87.5 | Area under accumulation | 0.91 | ||
| BEDROC | 160.9 | 0.658 | |||||||
| 20.0 | 0.646 | ||||||||
| 8.0 | 0.752 | ||||||||
| XP docking | Active counts | 9 | 11 | 13 | 13 | 16 | ROC | 0.97 | |
| % of Actives | 56.2 | 68.8 | 81.2 | 81.2 | 100 | Area under accumulation | 0.96 | ||
| BEDROC | 160.9 | 0.866 | |||||||
| 20.0 | 0.804 | ||||||||
| 8.0 | 0.863 | ||||||||
3.13. MD simulation of multi-target inhibitors of SARS-CoV-2
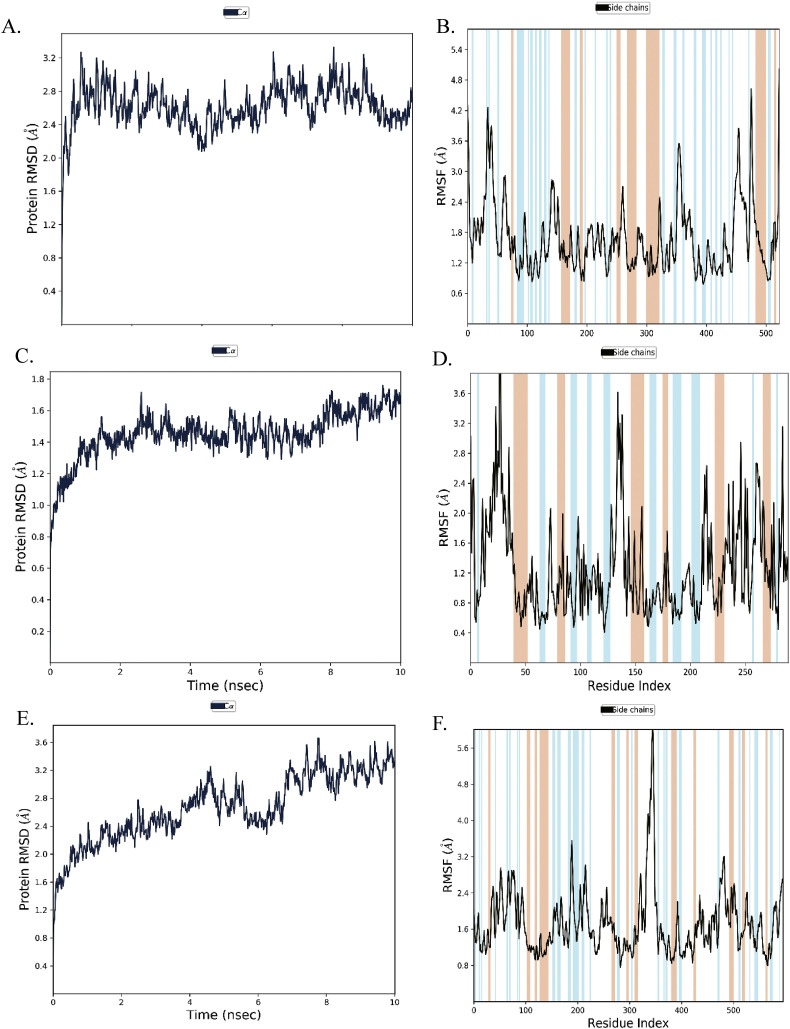
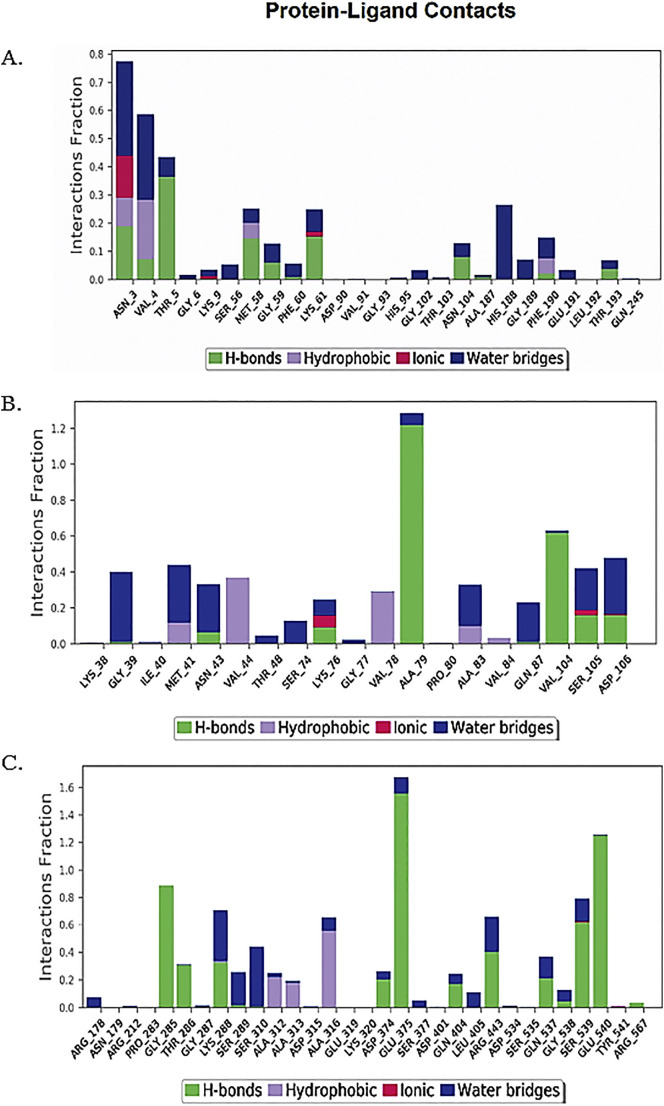
To understand the conformational changes of the protein and ligand during the course on interactions, we have performed ten nanoseconds of MD simulation using Desmond v 5.7 for each multi-target inhibitor with one of their potent targets. Root mean square deviation (RMSD), Root mean square fluctuations (RMSF) have been analyzed throughout the simulation trajectory. The RMSD computed for NPC214620 and exonuclease complex most of the time found below 3 Å, indicating minimum conformational changes (Fig. 4A) of the complex during the simulation. The RMSD of NPC52382 protein-ligand complex presented below 1.8 Å remained within the acceptable range (0–3 Å) (Fig. 4C) indicates little induction of conformational changes of the protein during the simulation. The RMSD of NPC270578 protein-ligand complex above 3 Å (Fig. 4E) suggests the protein went substantial conformational changes during ligand interaction. To monitor the dynamic behavior of amino acid residues during protein-ligand interaction, we have analyzed the RMSF fluctuations of the simulation studies. Along with the typical N- and C-terminal fluctuations, the internal fluctuation detected for the three complexes were observed in the unstructured regions of the protein (Fig. 4B, D, and F). Next, the graphical analysis of the total number of protein-ligand interactions such as hydrogen bond, ionic interaction, hydrophobic interaction, and water bridges during the simulation for the three compounds with their respective target proteins are presented in Fig. 5 .
Fig. 4.
Molecular dynamics simulation. (A) and (B) showing the RMSD and RMSF plot of Exoribonuclease and NPC214620 complex (C) and (D) representing the RMSD and RMSF of Methyltransferase and NPC52382 complex whereas (E) and (F) showing the RMSD and RMSF of Helicase and NPC270578 complex, respectively. The blue and red lines in the background of the RMSF plot showing the beta and alpha helices, respectively.
Fig. 5.
Graphical representation of protein-ligand contacts. (A) Showing the interaction between the helicase and NPC270578 (B), showing the interaction between Exoribonuclease and NPC214620, whereas (C) showing the interaction between Methyltransferase and NPC52382 compound.
4. Discussion
The highly contagious nature of SARS-CoV-2 enables the virus to spread 190 countries and territories of the globe within a short period of two and a half months. In response to the global pandemic situation, it is urgent and reasonable to employ sophisticated in silico approaches for facilitating the drug discovery for SARS-CoV-2. The highly error-prone replication [124] cycle and exceptionally shorter generation times have become a pressing issue for designing new effectual compounds for the highly evolving RNA virus. In a computational study, it has been shown that herbal leads displayed better binding potential towards probable drug targets of the Ebola virus than the known chemical analogs [125]. In the current effort, we have searched for effective inhibitors against six potent therapeutic targets of SARS-CoV-2 throughout the virtual screening on the NPASS database. Amongst 35,032 screened compounds, twenty-one compounds showed maximum docking scores compared to their respective known inhibitors. The detailed information regarding the natural source and biological activity is described in Supplementary Table 2. Notably, the antiviral properties of compounds NPC279121, NPC10737, NPC161224, NPC130230, and NPC70622 have been documented for many viruses, including Hepatitis B virus, Hepatitis C virus, Simian virus 40, Influenza A virus and HIV1 in the NPASS database. The compounds NPC214620, NPC52382, and NPC270578 are targeting five, four, and three-drug targets, respectively. These molecules are the significant marks in the multi-target-based drug design for the rapidly spreading virus. NPC214620 (PubChem CID: 11874) is 2,3,4-trihydroxybenzoic acid, identified in the course of biomarker analysis of flavonoid intake during metabolic profiling of human urine [126]. Trihydroxybenzene motif has been applied to design anti-inflammatory drugs targeting selectin molecules [127]. NPC52382 (PubChem CID: 442437) is known as neoastilbin and has 14 natural sources, including fungi, sea slug, and plants (NPASS database). The compound has biological activities with five proteins (mitochondrial Glutaminase kidney isoform, Protein disulfide-isomerase, Transcriptional regulator ERG, DNA dC->dU-editing enzyme APOBEC-3G, and Peptidyl-prolyl cis-trans NIMA-interacting1) and with two organisms (malaria parasite and human being) as per the NPASS database. According to the NPASS database, NPC270578 (PubChem CID: 119258) is known as astilbin and can be obtained from 93 natural sources and have biological activity against Xanthine dehydrogenase. Neoastilbin and astilbin are enantiomers and display the same docking score and MMGBSA value for three protein targets, including helicase, exoribonuclease, and methyltransferase as shown in Fig. 2B and C. However, unlike astilbin, neoastilbin shows better docking score and MMGBSA value than the known inhibitor (RO-7) for endoribonuclease of SARS-CoV-19 (Fig. 2C). Similar observation was indicated for the two thiocarbazate enantiomers of cathepsin L. Molecular docking studies revealed the docking score of S-enantioner (9.03 kcal/mol) more than the R-enantiomer (7.02 kcal/mol), the difference was also validated in their biological activity; S-enantiomer, with an IC50 of 56 nM and R-enantiomer, with an IC50 of 33 μM [128]. The importance of enantiomerism in the interaction of compounds with their binding sites, evaluated by docking scores, was also mentioned in other works [129,130]. On the basis of the earlier reports, it can be accepted that endoribonuclease has shown a better binding affinity for one enantiomer than the other in the present molecular docking study.
Reports from in silico work have aided in ranking the molecule/compound, thus plummeting the chances related to the poor selection of lead molecules. Therefore, such studies are worthwhile for research groups working on wet-lab experiments intending to identify novel antiviral lead compounds. Recent advances in computational approaches are playing a substantial role in the screening of drug and their design. Here, we will discuss a story of successful drug identified via structure-based virtual screening of compounds, i.e., a researcher's team successfully discovered neuraminidase inhibitor against influenza virus [131]. This is a potent, highly selective, novel, and orally active compound against influenza A and influenza B named BCX-1812, RWJ-270201 (cyclopentane peramivir). Further, the group conducted the in vitro studies and experiments on mice, rats, and ferrets and found higher potency than known drug named zanamivir and oseltamivir [131].
After human clinical trials, the drug, premavir found safe and effective [132], and the team got US patent [133]. This cyclopentane drug premavir, was approved by US-FDA to be administered as an intravenous under an Emergency Use Authorization during H1N1 pandemic in 2009 [134,135] and later on IV peramivir was approved by FDA under the brand name Rapivab TM [136]. Several other major successful drugs are existing in the market, which was discovered by in silico structure-based work includes drug (retroviral protease inhibitor) against HIV-1 [137]. Potential drugs, namely ratirexed [138], an inhibitor of thymidylate synthetase against HIV-1, and amprenavir target the protease of HIV-1 [137,139]. Other popular examples are drugs; namely, Isoniazid targets the InhA enzyme works against tuberculosis, which is also a major global health problem discovered by virtual screening [140] and Norfloxacin targets topoisomerase II & IV enzyme and treats the Urinary Tract infection [141]. All these successful drugs pave us the way to work upon the virtual screening based strategy to combat the ongoing pandemic. By considering this work now, researchers will be able to plan the experimental work on lead compounds identified in this study. However, the only concern is that there are several failure stories also reported in the documents; for instance, an antidepressant (RPX00023) was declared to be an agonist of 5-HT1A receptor while it was found to be an inhibitor [142]. Therefore, in vitro activity experiments with individual protein targets followed by SARS-CoV-2 replication assays of the compound identified from this study may support the development of novel and highly potent therapeutics for the treatment of rapidly spreading COVID-19.
Authors statement
The authors appreciate and extend thanks for the kind consideration and thorough review of this manuscript that provided us the opportunity to revise and improve it. Keeping the reviewers useful and validly raised concerns and questions in mind we revised and improved the manuscript. We are extremely thankful to the editor for the selection of five reviewers and humbly thankful to all reviewers for giving your valuable suggestion which will not only be useful for the improvement of this manuscript but also useful in the fight against minacious COVID-19 present situation. We have addressed the questions raised or corrections suggested by reviewers point by point.
Acknowledgments
Acknowledgments
DP is thankful to UGC for providing a start-up grant.
Declaration of competing interest
The authors declare no conflicts of interest.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijbiomac.2020.05.184.
Appendix A. Supplementary data
Supplementary tables
References
- 1.Vabret A., Dina J., Brison E., Brouard J., Freymuth F. Human coronaviruses. Pathologie-biologie. 2009;57(2):149–160. doi: 10.1016/j.patbio.2008.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gorbalenya A.E., Enjuanes L., Ziebuhr J., Snijder E.J. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006;117(1):17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tyrrell D., Bynoe M. Cultivation of a novel type of common-cold virus in organ cultures. Br. Med. J. 1965;1(5448):1467. doi: 10.1136/bmj.1.5448.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drosten C., Günther S., Preiser W., Van Der Werf S., Brodt H.-R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 5.Fouchier R.A., Kuiken T., Schutten M., Van Amerongen G., Van Doornum G.J., Van Den Hoogen B.G., Peiris M., Lim W., Stöhr K., Osterhaus A.D. Koch's postulates fulfilled for SARS virus. Nature. 2003;423(6937):240. doi: 10.1038/423240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 7.Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H. 2191341. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310(5748):676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 8.Guan Y., Zheng B., He Y., Liu X., Zhuang Z., Cheung C., Luo S., Li P., Zhang L., Guan Y. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302(5643):276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 9.Braden C.R., Dowell S.F., Jernigan D.B., Hughes J.M. Progress in global surveillance and response capacity 10 years after severe acute respiratory syndrome. Emerg. Infect. Dis. 2013;19(6):864. doi: 10.3201/eid1906.130192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cherry J.D. The chronology of the 2002–2003 SARS mini pandemic. Paediatr. Respir. Rev. 2004;5(4):262–269. doi: 10.1016/j.prrv.2004.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coleman C.M., Frieman M.B. Emergence of the Middle East respiratory syndrome coronavirus. PLoS Pathog. 2013;9(9) doi: 10.1371/journal.ppat.1003595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.W.H. Organization . WHO; 2019. Countries Agree Next Steps to Combat Global Health Threat by MERS-CoV. [Google Scholar]
- 13.Annan A., Baldwin H.J., Corman V.M., Klose S.M., Owusu M., Nkrumah E.E., Badu E.K., Anti P., Agbenyega O., Meyer B. Human betacoronavirus 2c EMC/2012–related viruses in bats, Ghana and Europe. Emerg. Infect. Dis. 2013;19(3):456. doi: 10.3201/eid1903.121503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.W. Cape, Close Relative of Human Middle East Respiratory Syndrome Coronavirus in Bat, South Africa. [DOI] [PMC free article] [PubMed]
- 15.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorbalenya A.E. The species severe acute respiratory syndrome related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5(4):536–544. doi: 10.1038/s41564-020-0695-z. PMC7095448, In this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020:1–4. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prajapat M., Sarma P., Shekhar N., Avti P., Sinha S., Kaur H., Kumar S., Bhattacharyya A., Kumar H., Bansal S., Medhi B. Drug targets for corona virus: a systematic review. Indian Journal of Pharmacology. 2020;52(1):56–65. doi: 10.4103/ijp.IJP_115_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang C.K., Lo S.C., Wang Y.S., Hou M.H. Recent insights into the development of therapeutics against coronavirus diseases by targeting N protein. Drug Discov. Today. 2016;21(4):562–572. doi: 10.1016/j.drudis.2015.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pillaiyar T., Manickam M., Namasivayam V., Hayashi Y., Jung S.H. An overview of severe acute respiratory syndrome-coronavirus (SARS-CoV) 3CL protease inhibitors: peptidomimetics and small molecule chemotherapy. J. Med. Chem. 2016;59(14):6595–6628. doi: 10.1021/acs.jmedchem.5b01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu Y., Lu X., Denison M.R. Identification and characterization of a serine-like proteinase of the murine coronavirus MHV-A59. J. Virol. 1995;69(6):3554–3559. doi: 10.1128/jvi.69.6.3554-3559.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim Y., Shivanna V., Narayanan S., Prior A.M., Weerasekara S., Hua D.H., Kankanamalage A.C.G., Groutas W.C., Chang K.-O. Broad-spectrum inhibitors against 3C-like proteases of feline coronaviruses and feline caliciviruses. J. Virol. 2015;89(9):4942–4950. doi: 10.1128/JVI.03688-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368(6489):409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adedeji A.O., Marchand B., te Velthuis A.J., Snijder E.J., Weiss S., Eoff R.L., Singh K., Sarafianos S.G. Mechanism of nucleic acid unwinding by SARS-CoV helicase. PLoS One. 2012;7(5) doi: 10.1371/journal.pone.0036521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adedeji A.O., Lazarus H. Biochemical characterization of Middle East respiratory syndrome coronavirus helicase. mSphere. 2016;1(5) doi: 10.1128/mSphere.00235-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanner J.A., Zheng B.-J., Zhou J., Watt R.M., Jiang J.-Q., Wong K.-L., Lin Y.-P., Lu L.-Y., He M.-L., Kung H.-F. The adamantane-derived bananins are potent inhibitors of the helicase activities and replication of SARS coronavirus. Chem. Biol. 2005;12(3):303–311. doi: 10.1016/j.chembiol.2005.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhardwaj K., Guarino L., Kao C.C. The severe acute respiratory syndrome coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor. J. Virol. 2004;78(22):12218–12224. doi: 10.1128/JVI.78.22.12218-12224.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ivanov K.A., Hertzig T., Rozanov M., Bayer S., Thiel V., Gorbalenya A.E., Ziebuhr J. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc. Natl. Acad. Sci. 2004;101(34):12694–12699. doi: 10.1073/pnas.0403127101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Minskaia E., Hertzig T., Gorbalenya A.E., Campanacci V., Cambillau C., Canard B., Ziebuhr J. Discovery of an RNA virus 3′→ 5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. 2006;103(13):5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He R., Adonov A., Traykova-Adonova M., Cao J., Cutts T., Grudesky E., Deschambaul Y., Berry J., Drebot M., Li X. Potent and selective inhibition of SARS coronavirus replication by aurintricarboxylic acid. Biochem. Biophys. Res. Commun. 2004;320(4):1199–1203. doi: 10.1016/j.bbrc.2004.06.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bouvet M., Debarnot C., Imbert I., Selisko B., Snijder E.J., Canard B., Decroly E. In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLoS Pathog. 2010;6(4) doi: 10.1371/journal.ppat.1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Agostini M.L., Andres E.L., Sims A.C., Graham R.L., Sheahan T.P., Lu X., Smith E.C., Case J.B., Feng J.Y., Jordan R. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. MBio. 2018;9(2) doi: 10.1128/mBio.00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Te Velthuis A.J., van den Worm S.H., Sims A.C., Baric R.S., Snijder E.J., van Hemert M.J. Zn2+ inhibits coronavirus and arterivirus RNA polymerase activity in vitro and zinc ionophores block the replication of these viruses in cell culture. PLoS Pathog. 2010;6(11) doi: 10.1371/journal.ppat.1001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dinesh D.C., Tamilarasan S., Rajaram K., Boura E. Antiviral drug targets of single-stranded RNA viruses causing chronic human diseases. Curr. Drug Targets. 2020;21(2):105–124. doi: 10.2174/1389450119666190920153247. [DOI] [PubMed] [Google Scholar]
- 35.Alhammad Y.M.O., Fehr A.R. The viral macrodomain counters host antiviral ADP-ribosylation. Viruses. 2020;12(4) doi: 10.3390/v12040384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pillay D., Zambon M. Antiviral drug resistance. Bmj. 1998;317(7159):660–662. doi: 10.1136/bmj.317.7159.660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gleason L.J., Luque A.E., Shah K. Polypharmacy in the HIV-infected older adult population. Clin. Interv. Aging. 2013;8:749. doi: 10.2147/CIA.S37738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Csermely P., Agoston V., Pongor S. The efficiency of multi-target drugs: the network approach might help drug design. Trends Pharmacol. Sci. 2005;26(4):178–182. doi: 10.1016/j.tips.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 39.Filgueira de Azevedo W., Jr., Canduri F., Simoes de Oliveira J., Basso L.A., Palma M.S., Pereira J.H., Santos D.S. Molecular model of shikimate kinase from Mycobacterium tuberculosis. Biochem. Biophys. Res. Commun. 2002;295(1):142–148. doi: 10.1016/S0006-291X(02)00632-0. [DOI] [PubMed] [Google Scholar]
- 40.de Azevedo W.F., Jr., Dias R. Experimental approaches to evaluate the thermodynamics of protein-drug interactions. Curr. Drug Targets. 2008;9(12):1071–1076. doi: 10.2174/138945008786949441. [DOI] [PubMed] [Google Scholar]
- 41.Canduri F., Fadel V., Basso L.A., Palma M.S., Santos D.S., de Azevedo W.F., Jr. New catalytic mechanism for human purine nucleoside phosphorylase. Biochem. Biophys. Res. Commun. 2005;327(3):646–649. doi: 10.1016/j.bbrc.2004.12.052. [DOI] [PubMed] [Google Scholar]
- 42.de Azevedo J., Walter F., Dias R. Experimental approaches to evaluate the thermodynamics of protein-drug interactions. Curr. Drug Targets. 2008;9(12):1071–1076. doi: 10.2174/138945008786949441. [DOI] [PubMed] [Google Scholar]
- 43.Kitazato K., Wang Y., Kobayashi N. Viral infectious disease and natural products with antiviral activity. Drug Discov. Ther. 2007;1(1):14–22. [PubMed] [Google Scholar]
- 44.Ak T., Gülçin İ. Antioxidant and radical scavenging properties of curcumin. Chem. Biol. Interact. 2008;174(1):27–37. doi: 10.1016/j.cbi.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 45.Gülçin İ. Antioxidant activity of eugenol: a structure–activity relationship study. J. Med. Food. 2011;14(9):975–985. doi: 10.1089/jmf.2010.0197. [DOI] [PubMed] [Google Scholar]
- 46.Gülcin I. Antioxidant activity of food constituents: an overview. Arch. Toxicol. 2012;86(3):345–391. doi: 10.1007/s00204-011-0774-2. [DOI] [PubMed] [Google Scholar]
- 47.Gulcin İ. Antioxidants and antioxidant methods: an updated overview. Arch. Toxicol. 2020:1–65. doi: 10.1007/s00204-020-02689-3. [DOI] [PubMed] [Google Scholar]
- 48.Sarwar M.S., Zhang H.J., Tsang S.W. Perspectives of plant natural products in inhibition of cancer invasion and metastasis by regulating multiple signaling pathways. Curr. Med. Chem. 2018;25(38):5057–5087. doi: 10.2174/0929867324666170918123413. [DOI] [PubMed] [Google Scholar]
- 49.Luo S., Lenon G.B., Gill H., Yuen H., Yang A.W.H., Hung A., Nguyen L.T. Do the natural chemical compounds interact with the same targets of current pharmacotherapy for weight management?-a review. Curr. Drug Targets. 2019;20(4):399–411. doi: 10.2174/1389450119666180830125958. [DOI] [PubMed] [Google Scholar]
- 50.Panda S.S., Jhanji N. Natural products as potential anti-Alzheimer agents. Curr. Med. Chem. 2019;26 doi: 10.2174/0929867326666190618113613. [DOI] [PubMed] [Google Scholar]
- 51.Lin L.-T., Hsu W.-C., Lin C.-C. Antiviral natural products and herbal medicines. J. Tradit. Complement. Med. 2014;4(1):24–35. doi: 10.4103/2225-4110.124335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheng P.W., Ng L.T., Chiang L.C., Lin C.C. Antiviral effects of saikosaponins on human coronavirus 229E in vitro. Clin. Exp. Pharmacol. Physiol. 2006;33(7):612–616. doi: 10.1111/j.1440-1681.2006.04415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li S.-y., Chen C., Zhang H.-q., Guo H.-y., Wang H., Wang L., Zhang X., Hua S.-n., Yu J., Xiao P.-g. Identification of natural compounds with antiviral activities against SARS-associated coronavirus. Antivir. Res. 2005;67(1):18–23. doi: 10.1016/j.antiviral.2005.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin C.-W., Tsai F.-J., Tsai C.-H., Lai C.-C., Wan L., Ho T.-Y., Hsieh C.-C., Chao P.-D.L. Anti-SARS coronavirus 3C-like protease effects of Isatis indigotica root and plant-derived phenolic compounds. Antivir. Res. 2005;68(1):36–42. doi: 10.1016/j.antiviral.2005.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ryu Y.B., Jeong H.J., Kim J.H., Kim Y.M., Park J.-Y., Kim D., Naguyen T.T.H., Park S.-J., Chang J.S., Park K.H. Biflavonoids from Torreya nucifera displaying SARS-CoV 3CLpro inhibition. Bioorg. Med. Chem. 2010;18(22):7940–7947. doi: 10.1016/j.bmc.2010.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu M.-S., Lee J., Lee J.M., Kim Y., Chin Y.-W., Jee J.-G., Keum Y.-S., Jeong Y.-J. Identification of myricetin and scutellarein as novel chemical inhibitors of the SARS coronavirus helicase, nsP13. Bioorg. Med. Chem. Lett. 2012;22(12):4049–4054. doi: 10.1016/j.bmcl.2012.04.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lau K.-M., Lee K.-M., Koon C.-M., Cheung C.S.-F., Lau C.-P., Ho H.-M., Lee M.Y.-H., Au S.W.-N., Cheng C.H.-K., Bik-San Lau C. Immunomodulatory and anti-SARS activities of Houttuynia cordata. J. Ethnopharmacol. 2008;118(1):79–85. doi: 10.1016/j.jep.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zeng X., Zhang P., He W., Qin C., Chen S., Tao L., Wang Y., Tan Y., Gao D., Wang B. NPASS: natural product activity and species source database for natural product research, discovery and tool development. Nucleic Acids Res. 2018;46(D1):D1217–D1222. doi: 10.1093/nar/gkx1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kuntz I.D. Structure-based strategies for drug design and discovery. Science. 1992;257(5073):1078–1082. doi: 10.1126/science.257.5073.1078. [DOI] [PubMed] [Google Scholar]
- 60.Lin X., Li X., Lin X. A review on applications of computational methods in drug screening and design. Molecules. 2020;25(6):1375. doi: 10.3390/molecules25061375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Murgueitio M.S., Bermudez M., Mortier J., Wolber G. In silico virtual screening approaches for anti-viral drug discovery. Drug Discov. Today Technol. 2012;9(3):e219–e225. doi: 10.1016/j.ddtec.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Santos F., Nunes D.A.d.F., Lima W.G., Davyt D., Santos L.L., Taranto A.G., Maria Siqueira Ferreira J. Identification of Zika virus NS2B-NS3 protease inhibitors by structure-based virtual screening and drug repurposing approaches. J. Chem. Inf. Model. 2019 doi: 10.1021/acs.jcim.9b00933. [DOI] [PubMed] [Google Scholar]
- 63.Zheng X., Liang C., Wang L., Wang B., Liu Y., Feng S., Wu J.Z., Gao L., Feng L., Chen L. Discovery of benzoazepinequinoline (BAQ) derivatives as novel, potent, orally bioavailable respiratory syncytial virus fusion inhibitors. J. Med. Chem. 2018;61(22):10228–10241. doi: 10.1021/acs.jmedchem.8b01394. [DOI] [PubMed] [Google Scholar]
- 64.Ma L., Zhang Z., Liu Z., Pan Q., Wang J., Li X., Guo F., Liang C., Hu L., Zhou J. Identification of small molecule compounds targeting the interaction of HIV-1 Vif and human APOBEC3G by virtual screening and biological evaluation. Sci. Rep. 2018;8(1):1–12. doi: 10.1038/s41598-018-26318-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nirwan S., Chahal V., Kakkar R. Structure-based virtual screening, free energy of binding and molecular dynamics simulations to propose novel inhibitors of Mtb-MurB oxidoreductase enzyme. J. Biomol. Struct. Dyn. 2020:1–16. doi: 10.1080/07391102.2020.1712258. [DOI] [PubMed] [Google Scholar]
- 66.Tatum N.J., Liebeschuetz J.W., Cole J.C., Frita R., Herledan A., Baulard A.R., Willand N., Pohl E. New active leads for tuberculosis booster drugs by structure-based drug discovery. Organic & Biomolecular Chemistry. 2017;15(48):10245–10255. doi: 10.1039/c7ob00910k. [DOI] [PubMed] [Google Scholar]
- 67.Pereira C.A., Sayé M., Reigada C., Silber A.M., Labadie G.R., Miranda M.R., Valera-Vera E. Computational approaches for drug discovery against trypanosomatid-caused diseases. Parasitology. 2020:1–23. doi: 10.1017/S0031182020000207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Melo-Filho C.C., Braga R.C., Muratov E.N., Franco C.H., Moraes C.B., Freitas-Junior L.H., Andrade C.H. Discovery of new potent hits against intracellular Trypanosoma cruzi by QSAR-based virtual screening. Eur. J. Med. Chem. 2019;163:649–659. doi: 10.1016/j.ejmech.2018.11.062. [DOI] [PubMed] [Google Scholar]
- 69.Johnson M., Zaretskaya I., Raytselis Y., Merezhuk Y., McGinnis S., Madden T.L. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008;36(suppl_2):W5–W9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1):W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cardoso J.M., Fonseca L., Egas C., Abrantes I. Cysteine proteases secreted by the pinewood nematode, Bursaphelenchus xylophilus: in silico analysis. Comput. Biol. Chem. 2018;77:291–296. doi: 10.1016/j.compbiolchem.2018.10.011. [DOI] [PubMed] [Google Scholar]
- 72.McNitt D.H., De Water L.V., Marasco D., Berisio R., Lukomski S. Streptococcal collagen-like protein 1 binds wound fibronectin: implications in pathogen targeting. Curr. Med. Chem. 2019;26(11):1933–1945. doi: 10.2174/0929867325666180831165704. [DOI] [PubMed] [Google Scholar]
- 73.Bitencourt-Ferreira G., de Azevedo W.F. Docking Screens for Drug Discovery. Springer; 2019. Homology modeling of protein targets with MODELLER; pp. 231–249. [DOI] [PubMed] [Google Scholar]
- 74.Silveira N.J. da, Arcuri H.A., Bonalumi C.E., de Souza F.P., Mello I.M., Rahal P., Pinho J.R., de Azevedo W.F. Molecular models of NS3 protease variants of the Hepatitis C virus. BMC Struct. Biol. 2005;5(1):1. doi: 10.1186/1472-6807-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Canduri F., de Azevedo J., Walter F. Protein crystallography in drug discovery. Curr. Drug Targets. 2008;9(12):1048–1053. doi: 10.2174/138945008786949423. [DOI] [PubMed] [Google Scholar]
- 76.Pereira J.H., Canduri F., de Oliveira J.S., Silveira N.J. da, Basso L.A., Palma M.S., de Azevedo W.F., Jr., Santos D.S. Structural bioinformatics study of EPSP synthase from Mycobacterium tuberculosis. Biochem. Biophys. Res. Commun. 2003;312(3):608–614. doi: 10.1016/j.bbrc.2003.10.175. [DOI] [PubMed] [Google Scholar]
- 77.Cheng J. A multi-template combination algorithm for protein comparative modeling. BMC Struct. Biol. 2008;8(1):18. doi: 10.1186/1472-6807-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.van Gelder C.W., Leusen F.J., Leunissen J.A., Noordik J.H. A molecular dynamics approach for the generation of complete protein structures from limited coordinate data. Proteins: Structure, Function, and Bioinformatics. 1994;18(2):174–185. doi: 10.1002/prot.340180209. [DOI] [PubMed] [Google Scholar]
- 79.Heo L., Feig M. What makes it difficult to refine protein models further via molecular dynamics simulations? Proteins: Structure, Function, and Bioinformatics. 2018;86:177–188. doi: 10.1002/prot.25393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bhattacharya D., Cheng J. i3Drefine software for protein 3D structure refinement and its assessment in CASP10. PLoS One. 2013;8(7) doi: 10.1371/journal.pone.0069648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Modi V., Dunbrack R.L., Jr. Assessment of refinement of template-based models in CASP11. Proteins: Structure, Function, and Bioinformatics. 2016;84:260–281. doi: 10.1002/prot.25048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang Y. Protein structure prediction: when is it useful? Curr. Opin. Struct. Biol. 2009;19(2):145–155. doi: 10.1016/j.sbi.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Heo L., Feig M. Experimental accuracy in protein structure refinement via molecular dynamics simulations. Proc. Natl. Acad. Sci. 2018;115(52):13276–13281. doi: 10.1073/pnas.1811364115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Park H., DiMaio F., Baker D. CASP 11 refinement experiments with ROSETTA. Proteins: Structure, Function, and Bioinformatics. 2016;84:314–322. doi: 10.1002/prot.24862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mass T., Drake J.L., Haramaty L., Kim J.D., Zelzion E., Bhattacharya D., Falkowski P.G. Cloning and characterization of four novel coral acid-rich proteins that precipitate carbonates in vitro. Curr. Biol. 2013;23(12):1126–1131. doi: 10.1016/j.cub.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 86.Chen V.B., Arendall W.B., Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66(1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bhattacharya A., Cui Y. SomamiR 2.0: a database of cancer somatic mutations altering microRNA–ceRNA interactions. Nucleic Acids Res. 2016;44(D1):D1005–D1010. doi: 10.1093/nar/gkv1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lovell S.C., Davis I.W., Arendall W.B., III, De Bakker P.I., Word J.M., Prisant M.G., Richardson J.S., Richardson D.C. Structure validation by Cα geometry: ϕ, ψ and Cβ deviation. Proteins: Structure, Function, and Bioinformatics. 2003;50(3):437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 89.ödinger S. 2019. Release, 1: Schrödinger Suite 2019-1 Protein Preparation Wizard, Epik, Schrödinger, LLC, New York, NY. [Google Scholar]
- 90.Adedeji A.O., Singh K., Calcaterra N.E., DeDiego M.L., Enjuanes L., Weiss S., Sarafianos S.G. Severe acute respiratory syndrome coronavirus replication inhibitor that interferes with the nucleic acid unwinding of the viral helicase. Antimicrob. Agents Chemother. 2012;56(9):4718–4728. doi: 10.1128/AAC.00957-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gordon C.J., Tchesnokov E.P., Feng J.Y., Porter D.P., Gotte M. The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J. Biol. Chem. 2020;295(15):4773–4779. doi: 10.1074/jbc.AC120.013056. (jbc. AC120. 013056) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yang S., Chen S.-J., Hsu M.-F., Wu J.-D., Tseng C.-T.K., Liu Y.-F., Chen H.-C., Kuo C.-W., Wu C.-S., Chang L.-W. Synthesis, crystal structure, structure–activity relationships, and antiviral activity of a potent SARS coronavirus 3CL protease inhibitor. J. Med. Chem. 2006;49(16):4971–4980. doi: 10.1021/jm0603926. [DOI] [PubMed] [Google Scholar]
- 93.Jones J.C., Marathe B.M., Lerner C., Kreis L., Gasser R., Pascua P.N.Q., Najera I., Govorkova E.A. A novel endonuclease inhibitor exhibits broad-spectrum anti-influenza virus activity in vitro. Antimicrob. Agents Chemother. 2016;60(9):5504–5514. doi: 10.1128/AAC.00888-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Huang K.-W., Hsu K.-C., Chu L.-Y., Yang J.-M., Yuan H.S., Hsiao Y.-Y. Identification of inhibitors for the DEDDh family of exonucleases and a unique inhibition mechanism by crystal structure analysis of CRN-4 bound with 2-morpholin-4-ylethanesulfonate (MES) J. Med. Chem. 2016;59(17):8019–8029. doi: 10.1021/acs.jmedchem.6b00794. [DOI] [PubMed] [Google Scholar]
- 95.Friesner R.A., Banks J.L., Murphy R.B., Halgren T.A., Klicic J.J., Mainz D.T., Repasky M.P., Knoll E.H., Shelley M., Perry J.K. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004;47(7):1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 96.Pandey R.K., Sharma D., Bhatt T.K., Sundar S., Prajapati V.K. Developing imidazole analogues as potential inhibitor for Leishmania donovani trypanothione reductase: virtual screening, molecular docking, dynamics and ADMET approach. J. Biomol. Struct. Dyn. 2015;33(12):2541–2553. doi: 10.1080/07391102.2015.1085904. [DOI] [PubMed] [Google Scholar]
- 97.Ganesan A. The impact of natural products upon modern drug discovery. Curr. Opin. Chem. Biol. 2008;12(3):306–317. doi: 10.1016/j.cbpa.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 98.Lyne P.D., Lamb M.L., Saeh J.C. Accurate prediction of the relative potencies of members of a series of kinase inhibitors using molecular docking and MM-GBSA scoring. J. Med. Chem. 2006;49(16):4805–4808. doi: 10.1021/jm060522a. [DOI] [PubMed] [Google Scholar]
- 99.Truchon J.-F., Bayly C.I. Evaluating virtual screening methods: good and bad metrics for the “early recognition” problem. J. Chem. Inf. Model. 2007;47(2):488–508. doi: 10.1021/ci600426e. [DOI] [PubMed] [Google Scholar]
- 100.Fawcett T. ROC graphs with instance-varying costs. Pattern Recogn. Lett. 2006;27(8):882–891. [Google Scholar]
- 101.Lu S.-H., Wu J.W., Liu H.-L., Zhao J.-H., Liu K.-T., Chuang C.-K., Lin H.-Y., Tsai W.-B., Ho Y. The discovery of potential acetylcholinesterase inhibitors: a combination of pharmacophore modeling, virtual screening, and molecular docking studies. J. Biomed. Sci. 2011;18(1):8. doi: 10.1186/1423-0127-18-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Harder E., Damm W., Maple J., Wu C., Reboul M., Xiang J.Y., Wang L., Lupyan D., Dahlgren M.K., Knight J.L. OPLS3: a force field providing broad coverage of drug-like small molecules and proteins. J. Chem. Theory Comput. 2016;12(1):281–296. doi: 10.1021/acs.jctc.5b00864. [DOI] [PubMed] [Google Scholar]
- 103.Benkert P., Künzli M., Schwede T. QMEAN server for protein model quality estimation. Nucleic Acids Res. 2009;37(suppl_2):W510–W514. doi: 10.1093/nar/gkp322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Patel B., Singh V., Patel D. volume I. Springer; 2019. Structural Bioinformatics, Essentials of Bioinformatics; pp. 169–199. [Google Scholar]
- 105.Liu M.-M., Zhou L., He P.-L., Zhang Y.-N., Zhou J.-Y., Shen Q., Chen X.-W., Zuo J.-P., Li W., Ye D.-Y. Discovery of flavonoid derivatives as anti-HCV agents via pharmacophore search combining molecular docking strategy. Eur. J. Med. Chem. 2012;52:33–43. doi: 10.1016/j.ejmech.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 106.Liu A.-L., Wang H.-D., Lee S.M., Wang Y.-T., Du G.-H. Structure–activity relationship of flavonoids as influenza virus neuraminidase inhibitors and their in vitro anti-viral activities. Bioorg. Med. Chem. 2008;16(15):7141–7147. doi: 10.1016/j.bmc.2008.06.049. [DOI] [PubMed] [Google Scholar]
- 107.Li B.-W., Zhang F.-H., Serrao E., Chen H., Sanchez T.W., Yang L.-M., Neamati N., Zheng Y.-T., Wang H., Long Y.-Q. Design and discovery of flavonoid-based HIV-1 integrase inhibitors targeting both the active site and the interaction with LEDGF/p75. Bioorg. Med. Chem. 2014;22(12):3146–3158. doi: 10.1016/j.bmc.2014.04.016. [DOI] [PubMed] [Google Scholar]
- 108.Mitsuhashi S., Kishimoto T., Uraki Y., Okamoto T., Ubukata M. Low molecular weight lignin suppresses activation of NF-κB and HIV-1 promoter. Bioorg. Med. Chem. 2008;16(5):2645–2650. doi: 10.1016/j.bmc.2007.11.041. [DOI] [PubMed] [Google Scholar]
- 109.Siegel D., Hui H.C., Doerffler E., Clarke M.O., Chun K., Zhang L., Neville S., Carra E., Lew W., Ross B. ACS Publications; 2017. Discovery and Synthesis of a Phosphoramidate Prodrug of a Pyrrolo [2, 1-f][triazin-4-amino] Adenine C-Nucleoside (GS-5734) for the Treatment of Ebola and Emerging Viruses. [DOI] [PubMed] [Google Scholar]
- 110.Lo M.K., Jordan R., Arvey A., Sudhamsu J., Shrivastava-Ranjan P., Hotard A.L., Flint M., McMullan L.K., Siegel D., Clarke M.O. GS-5734 and its parent nucleoside analog inhibit Filo-, Pneumo-, and Paramyxoviruses. Sci. Rep. 2017;7 doi: 10.1038/srep43395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Warren T.K., Jordan R., Lo M.K., Ray A.S., Mackman R.L., Soloveva V., Siegel D., Perron M., Bannister R., Hui H.C. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531(7594):381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.-H. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. science. 2003;300(5624):1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 113.Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y. The genome sequence of the SARS-associated coronavirus. Science. 2003;300(5624):1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 114.Jain R.P., Pettersson H.I., Zhang J., Aull K.D., Fortin P.D., Huitema C., Eltis L.D., Parrish J.C., James M.N., Wishart D.S. Synthesis and evaluation of keto-glutamine analogues as potent inhibitors of severe acute respiratory syndrome 3CLpro. J. Med. Chem. 2004;47(25):6113–6116. doi: 10.1021/jm0494873. [DOI] [PubMed] [Google Scholar]
- 115.Ghosh A.K., Xi K., Ratia K., Santarsiero B.D., Fu W., Harcourt B.H., Rota P.A., Baker S.C., Johnson M.E., Mesecar A.D. Design and synthesis of peptidomimetic severe acute respiratory syndrome chymotrypsin-like protease inhibitors. J. Med. Chem. 2005;48(22):6767–6771. doi: 10.1021/jm050548m. [DOI] [PubMed] [Google Scholar]
- 116.Mercader A.G., Pomilio A.B. QSAR study of flavonoids and biflavonoids as influenza H1N1 virus neuraminidase inhibitors. Eur. J. Med. Chem. 2010;45(5):1724–1730. doi: 10.1016/j.ejmech.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 117.Case J.B., Ashbrook A.W., Dermody T.S., Denison M.R. Mutagenesis of S-adenosyl-l-methionine-binding residues in coronavirus nsp14 N7-methyltransferase demonstrates differing requirements for genome translation and resistance to innate immunity. J. Virol. 2016;90(16):7248–7256. doi: 10.1128/JVI.00542-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sun Y., Wang Z., Tao J., Wang Y., Wu A., Yang Z., Wang K., Shi L., Chen Y., Guo D. Yeast-based assays for the high-throughput screening of inhibitors of coronavirus RNA cap guanine-N7-methyltransferase. Antivir. Res. 2014;104:156–164. doi: 10.1016/j.antiviral.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Li Z., Li L.-j., Sun Y., Li J. Identification of natural compounds with anti-hepatitis B virus activity from Rheum palmatum L. ethanol extract. Chemotherapy. 2007;53(5):320–326. doi: 10.1159/000107690. [DOI] [PubMed] [Google Scholar]
- 120.Lipinski C.A. Chris Lipinski discusses life and chemistry after the Rule of Five. Drug Discov. Today. 2003;8(1):12–16. doi: 10.1016/s1359-6446(02)02556-4. [DOI] [PubMed] [Google Scholar]
- 121.Fawcett T. An introduction to ROC analysis. Pattern Recogn. Lett. 2006;27(8):861–874. [Google Scholar]
- 122.Vyas V.K., Ghate M., Goel A. Pharmacophore modeling, virtual screening, docking and in silico ADMET analysis of protein kinase B (PKB β) inhibitors. J. Mol. Graph. Model. 2013;42:17–25. doi: 10.1016/j.jmgm.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 123.Nair S.B., Teli M.K., Pradeep H., Rajanikant G.K. Computational identification of novel histone deacetylase inhibitors by docking based QSAR. Comput. Biol. Med. 2012;42(6):697–705. doi: 10.1016/j.compbiomed.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 124.Holmes E.C. The evolutionary genetics of emerging viruses. Annu. Rev. Ecol. Evol. Syst. 2009;40:353–372. [Google Scholar]
- 125.Setlur A.S., Naik S.Y., Skariyachan S. Herbal lead as ideal bioactive compounds against probable drug targets of Ebola virus in comparison with known chemical analogue: a computational drug discovery perspective. Interdisciplinary Sciences: Computational Life Sciences. 2017;9(2):254–277. doi: 10.1007/s12539-016-0149-8. [DOI] [PubMed] [Google Scholar]
- 126.Loke W.M., Jenner A.M., Proudfoot J.M., McKinley A.J., Hodgson J.M., Halliwell B., Croft K.D. A metabolite profiling approach to identify biomarkers of flavonoid intake in humans. J. Nutr. 2009;139(12):2309–2314. doi: 10.3945/jn.109.113613. [DOI] [PubMed] [Google Scholar]
- 127.Kranich R., Busemann A.S., Bock D., Schroeter-Maas S., Beyer D., Heinemann B., Meyer M., Schierhorn K., Zahlten R., Wolff G. Rational design of novel, potent small molecule pan-selectin antagonists. J. Med. Chem. 2007;50(6):1101–1115. doi: 10.1021/jm060536g. [DOI] [PubMed] [Google Scholar]
- 128.Beavers M.P., Myers M.C., Shah P.P., Purvis J.E., Diamond S.L., Cooperman B.S., Huryn D.M., Smith Iii A.B. Molecular docking of cathepsin L inhibitors in the binding site of papain. J. Chem. Inf. Model. 2008;48(7):1464–1472. doi: 10.1021/ci800085c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chen Z., Ma Y., He M., Ren H., Zhou S., Lai D., Wang Z., Jiang L. Semi-rational directed evolution of monoamine oxidase for kinetic resolution of rac-mexiletine. Appl. Biochem. Biotechnol. 2015;176(8):2267–2278. doi: 10.1007/s12010-015-1716-x. [DOI] [PubMed] [Google Scholar]
- 130.Eryanni-Levin S., Khatib S., Levy-Rosenzvig R., Tamir S., Szuchman-Sapir A. 5, 6-δ-DHTL, a stable metabolite of arachidonic acid, is a potential substrate for paraoxonase 1. Biochimica et Biophysica Acta (BBA)-Molecular and Cell Biology of Lipids. 2015;1851(9):1118–1122. doi: 10.1016/j.bbalip.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 131.Babu Y.S., Chand P., Bantia S., Kotian P., Dehghani A., El-Kattan Y., Lin T.-H., Hutchison T.L., Elliott A.J., Parker C.D. BCX-1812 (RWJ-270201): discovery of a novel, highly potent, orally active, and selective influenza neuraminidase inhibitor through structure-based drug design. J. Med. Chem. 2000;43(19):3482–3486. doi: 10.1021/jm0002679. [DOI] [PubMed] [Google Scholar]
- 132.Smee D.F., Sidwell R.W. Peramivir (BCX-1812, RWJ-270201): potential new therapy for influenza. Expert Opin. Investig. Drugs. 2002;11(6):859–869. doi: 10.1517/13543784.11.6.859. [DOI] [PubMed] [Google Scholar]
- 133.S.B. Yarlagadda, P. Chand, S. Bantia, S. Arnold, J.M. Kilpatrick, Antiviral treatments, Google Patents, 2019.
- 134.Alame M.M., Massaad E., Zaraket H. Peramivir: a novel intravenous neuraminidase inhibitor for treatment of acute influenza infections. Front. Microbiol. 2016;7:450. doi: 10.3389/fmicb.2016.00450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Birnkrant D., Cox E. The emergency use authorization of peramivir for treatment of 2009 H1N1 influenza. N. Engl. J. Med. 2009;361(23):2204–2207. doi: 10.1056/NEJMp0910479. [DOI] [PubMed] [Google Scholar]
- 136.Food and Drug Administration . 2015. RAPIVABTM (Peramivir Injection), for Intravenous Use Initial U.S. Approval 2014: Highlights of Prescribing Information. [Google Scholar]
- 137.Wlodawer A., Vondrasek J. Inhibitors of HIV-1 protease: a major success of structure-assisted drug design. Annu. Rev. Biophys. Biomol. Struct. 1998;27(1):249–284. doi: 10.1146/annurev.biophys.27.1.249. [DOI] [PubMed] [Google Scholar]
- 138.Anderson A.C. The process of structure-based drug design. Chem. Biol. 2003;10(9):787–797. doi: 10.1016/j.chembiol.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 139.Clark D.E. What has computer-aided molecular design ever done for drug discovery? Expert Opin. Drug Discovery. 2006;1(2):103–110. doi: 10.1517/17460441.1.2.103. [DOI] [PubMed] [Google Scholar]
- 140.Marrakchi H., Lanéelle G., Quémard A.k. InhA, a target of the antituberculous drug isoniazid, is involved in a mycobacterial fatty acid elongation system, FAS-II. Microbiology. 2000;146(2):289–296. doi: 10.1099/00221287-146-2-289. [DOI] [PubMed] [Google Scholar]
- 141.Rutenber E.E., Stroud R.M. Binding of the anticancer drug ZD1694 to E. coli thymidylate synthase: assessing specificity and affinity. Structure. 1996;4(11):1317–1324. doi: 10.1016/s0969-2126(96)00139-6. [DOI] [PubMed] [Google Scholar]
- 142.de Paulis T. Drug evaluation: PRX-00023, a selective 5-HT1A receptor agonist for depression. Current Opinion in Investigational Drugs (London, England: 2000) 2007;8(1):78. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables