Abstract
This protocol describes methods for increasing and evaluating the efficiency of genome editing based on CRISPR/Cas9, TALENs or ZFNs. First, Indel Detection by Amplicon Analysis (IDAA) determines the size and frequency of insertions and deletions elicited by nucleases in cells, tissues or embryos through analysis of fluorophore-labelled PCR amplicons covering the nuclease target site by capillary electrophoresis in a sequenator. Second, FACS enrichment of cells expressing nucleases linked to fluorescent proteins can be used to maximize knockout or knockin editing efficiencies or balance editing efficiency and toxic/off-target effects. The two methods can be combined to form a pipeline for cell line editing, which facilitates the testing of new nuclease reagents and the generation of edited cell pools or clonal cell lines, reducing the number of clones that need to be generated and increasing the ease with which they are screened. The pipeline shortens the timeline, but most prominently reduces the workload of cell line editing.
Keywords: genome editing method, CRISPR/Cas9, TALEN, ZFN, FACS, indel, IDAA, Peak Scanner
EDITORIAL SUMMARY:
This protocol provides a pipeline with which to increase and evaluate the efficiency of genome editing by ZFNs, TALENs or CRISPR-Cas9. The pipeline comprises enrichment of nuclease expressing cells by FACS followed by Indel Detection by Amplicon Analysis (IDAA). (max 250 characters)
INTRODUCTION
Programmable, sequence-specific nucleases that enable precise genome editing are becoming widely exploited in basic life science, medicine and biotechnology because of their capabilities to engineer cells and organisms in virtually any desired way1. At the core of this technology lies the ability of programmable nucleases to create a DNA double-strand break (DSB) at a user-specified genomic site. These breaks are resolved via the cellular DNA damage repair pathways, which are exploited to achieve the desired genome editing result. The non-homologous end joining (NHEJ) and microhomology-mediated end joining (MMEJ) pathways often give rise to small insertions and/or deletions, collectively called indels, at the cut site. If nucleases are targeted to a coding sequence, these repair pathways can thereby be incited to generate frameshift mutations and functional gene knockouts2. Alternatively, DSBs can be resolved by the homology-directed repair (HDR) pathway, which uses homologous DNA sequences as templates for precise repair. HDR can be exploited to mutate the genome in a user-defined manner through co-delivery of nucleases with an exogenous homologous DNA template (called a donor) that contains the desired mutation. The donor can be a plasmid containing the mutation flanked by homology arms of 200–800 base pairs (bp)3, 4. Alternatively, short, homologous single-stranded oligodeoxynucleotides (ssODNs) can be used as effective donors for smaller mutations via repair mechanisms that are unclear5–8.
Three major programmable nuclease systems exist: zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs) and clustered regulatory interspaced short palindromic repeats (CRISPR)-Cas9. ZFNs and TALENs are fusions of the FokI nuclease to programmable DNA-binding protein moieties, which direct the chimeric protein to the targeted genomic site. CRISPR/Cas9 is a two-component system in which the Cas9 nuclease is directed by a user-specified short guide RNA (gRNA) molecule to the targeted genomic site. The three systems have comparable genome editing efficacy, but differ in ease of construction, targetable sites, specificity, indel mutation signature, and intellectual property issues, among other characteristics1, 9, 10. Several reviews and protocols exist on the basic mechanism of action and how to construct or obtain ZFNs11–14, TALENs15, 16 or CRISPR/Cas917–21.
Regardless of the nuclease system used, it is essential to have methods that can detect and characterize mutations in targeted cells and that can ensure expression of nucleases in a sufficiently high fraction of the cells and at a sufficiently high level in individual cells in order to obtain a useful degree of editing. Sometimes, very high nuclease levels cause toxic or off-target effects, so the methods should ideally be able to tune nucleases to optimal levels with regard to desired and undesired effects. However, in contrast to the rapidly increasing number of improvements reported for the nuclease tools, a more limited number of studies have addressed ways of improving mutation detection and targeting enrichment.
This protocol is designed to facilitate these key issues and describes methods that work equally well with all the three major nuclease systems. First, we describe a simple, PCR-based method for detection and characterization of indel mutations in cells, tissues or embryos, which is fast, precise, informative, sensitive and robust, yet inexpensive22. Second, we describe a simple, FACS-based method to enrich for cell populations with defined levels of nuclease expression from a transfected cell pool, either high levels for maximal knockout or knockin editing frequencies or intermediate levels for balancing editing efficiency and undesired effects23. Finally, we describe how the two methods can be combined to create a pipeline for efficient genome editing of cell lines. This pipeline can shorten the timeline of an editing project, but more significantly, it can greatly reduce the workload. By exploiting the two methods throughout an editing project, the pipeline facilitates the initial screening of newly generated nuclease reagents as well as the subsequent generation of edited cell pools or clonal cell lines. For the latter, the pipeline reduces the number of clones that need to be generated and increases the ease with which they are screened. We recently used the pipeline to generate many different CHO cell lines with combinatorial knockout of multiple glycogenes24.
Indel Detection by Amplicon Analysis (IDAA)
DNA fragment sizes are easily and precisely determined using fragment analysis instruments based on capillary electrophoresis, such as standard DNA sequenators. By combining established fragment analysis methods with a PCR amplicon labelling scheme, we devised protocols for sensitive, accurate and fast detection of nuclease-induced indels in genome edited samples22. We have termed the method Indel Detection by Amplicon Analysis (IDAA) and the principle is outlined in Fig. 1. In stage 1, fluorophore-labelled amplicons that cover the nuclease-edited genomic target site are generated. This is accomplished through a tri-primer genomic PCR using a pair of locus-specific forward and reverse primers (Fwd, Rev) along with a third 6-carboxyfluorescein (6-FAM)-labelled primer containing the same sequence as an extension of the Fwd primer (FamFwd). Depending on the repair outcome of the nuclease-induced DNA break, wild-type amplicons, shorter amplicons containing deletions or longer amplicons encompassing insertions will be generated and 6-FAM labelled. In stage 2, the amplicons are size-discriminated by a DNA sequenator or other fragment analytical instrument. Associated software generates easily interpretable graphic depiction of the amplicons as peaks, revealing indel sizes with single-base resolution. Furthermore, the relative frequency of the differently sized amplicons, i.e. indel events, can easily be estimated from software-determined areas of the peaks. A very similar PCR-based indel detection method was reported by Carrington et al., evaluating gRNA activities in zebrafish embryos25. This method only differs from IDAA by the design of the tri-primer setup and PCR conditions.
Figure 1 |. Principle of IDAA.

IDAA detects indel mutations elicited by nucleases at their genomic target site (indicated by yellow lightning). In stage 1 of IDAA, the nuclease target site is amplified by genomic PCR using three primers: a locus-specific pair of forward (Fwd) and reverse (Rev) primers and a universal primer (FamFwd) with same sequence as an extension of the Fwd primer and labelled with 6-FAM (green asterisk), which renders the amplicons fluorescent. Amplicons derived from alleles with a deletion or an insertion will be shorter or longer, respectively, than amplicons derived from the wild-type allele, which typically are designed to be 200–450 bp in size. In stage 2, the amplicons are analyzed in a fragment analyzer, such as a commonplace Sanger sequenator. This reveals the size of the amplicons, and thereby the indels contained in them in bp (x-axis) as well as their frequency in relative fluorescence units (RFU) (y-axis). An IDAA profile of indels at the GALNT10 locus in a HEK293 cell pool FACS-edited using CRISPR/Cas9 as outlined in Fig. 3 is shown. The size and frequency of selected indels contained in the amplicons are indicated. IDAA was performed in an ABI 3130 instrument.
IDAA has been tested on indels elicited by CRISPR/Cas9, TALENs and ZFNs in a variety of applications reported previously22, 24(and here) and demonstrates the following features:
Sensitive.
IDAA can detect indels that occur with frequencies down to ~0.1% and can detect the smallest possible indels (±1 bp)22(and see Fig. 1; Supplementary Fig. 1). A few dozen cells can be used as template for the IDAA PCR and only sub-nanogram amounts of the PCR product need to be analyzed due to the sensitivity of fragment analyzers (Supplementary Fig. 2; Supplementary Fig. 3).
Precise.
IDAA defines the size of the smallest indels of 1 bp as well as the largest, and relatively rare, indels of >100 bp with 1 bp resolution (Fig. 1).
Multi-indel resolution power.
All of the predominant indels in a sample are detected (Fig. 1).
Quantitative.
The frequency of the various indels present in a sample can be estimated (Fig. 1).
Robust and reproducible.
IDAA can establish the “indel signature” of a given nuclease due to the reproducibility of the assay and the pre-defined nature of cellular indel repair at a given cut site; thus IDAA profiles from independent experiments are almost identical and replicate determinations are therefore not needed (Fig. 2; Supplementary Fig. 4). IDAA works essentially every time with any nuclease target site that can be amplified. The indel signature observed during testing of a new nuclease reagent is predictive of the indel outcome that will be obtained in any later editing with same nuclease in the same cell type.
Figure 2 |. IDAA reveals the indel signature of a given gRNA.
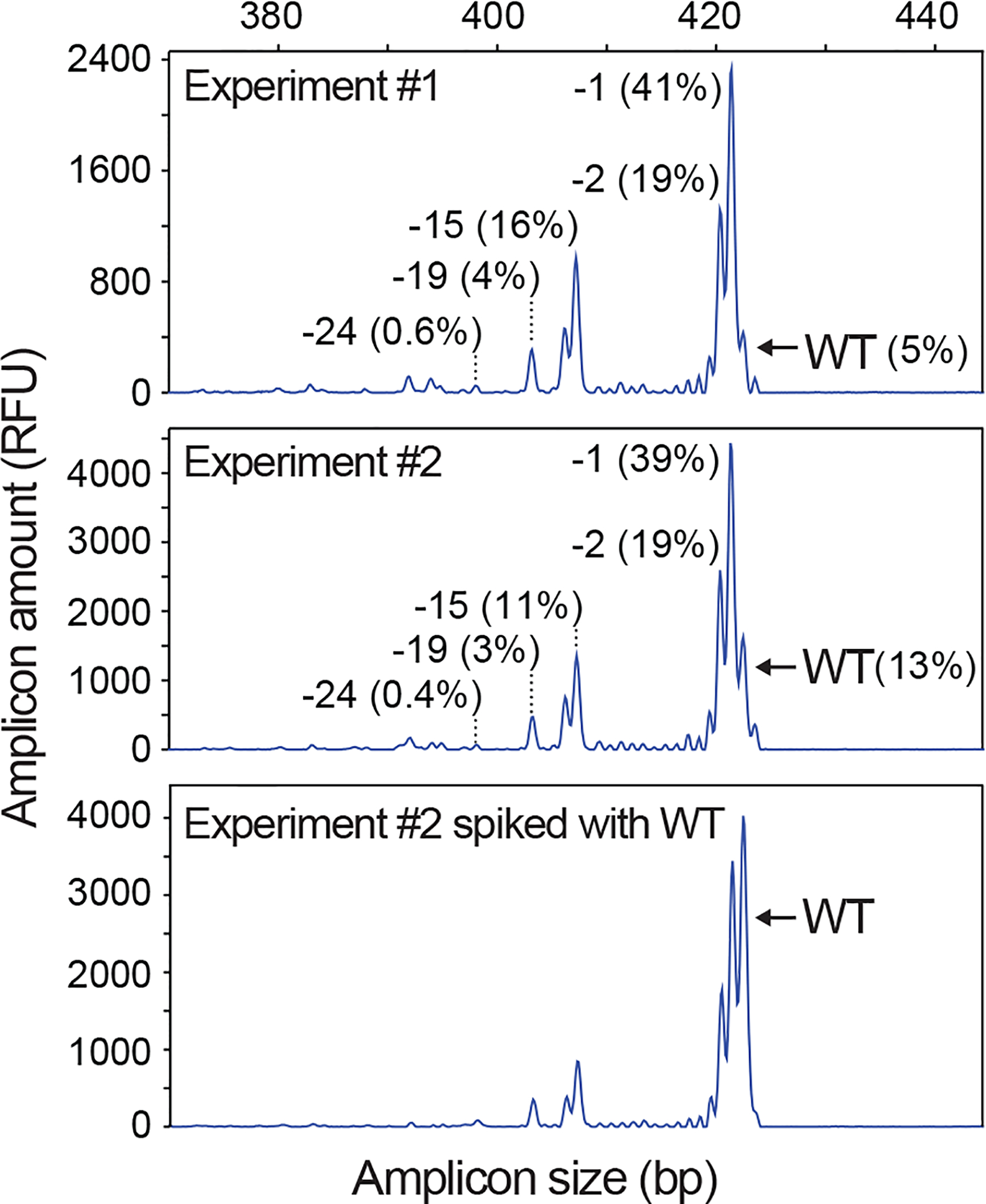
IDAA profiles for two independent experiments performed on different days and targeting the Trp53 locus using the same gRNA in mouse Neuro2A cell pools via FACS-editing as outlined in Fig. 3. Note that the two profiles are almost identical due to the reproducibility of IDAA and the pre-defined nature of indel repair at a given cut site. “Experiment #2 spiked with WT” indicates that the IDAA PCR from the edited sample was mixed 4:1 with an IDAA PCR from an untreated sample, which is a simple means to highlight the wt peak. The size and frequency of selected indels are indicated. IDAA was performed in an ABI 3130 instrument.
Simple and easy.
Crude cell lysates are used as template for the IDAA PCR, which can be loaded directly onto the sequenator; i.e. IDAA does not require purification or quantitation of the genomic template DNA, nor of the PCR products to be analyzed. For a given analysis, only a locus-specific primer pair needs to be designed; the 6-FAM primer remains constant, and universal PCR conditions have been optimized. Hands-on time is therefore minimal.
Fast and high throughput.
All IDAA steps can be performed in 96-well plates and newer sequenators can analyze 2 plates at a time in an automated fashion in less than 4 h. Together with the simplicity of the assay, genotyping of ~200 samples can therefore be completed within 8 h from cell lysis to results.
Cost-effective.
Given access to a standard capillary-based sequenator or other fragment analyzer, IDAA is inexpensive, since it requires only standard PCR reagents. If a fragment analyzer is not available, several DNA service providers can perform the analysis as conveniently and cost-effectively as standard Sanger sequencing service (see Experimental design).
In summary, IDAA can reveal the complete spectrum and frequencies of predominant indels at a nuclease target site in a sample, establishing whether they cause frameshifts and functional gene knockout. Together with extraordinary robustness, simplicity and minimal hands-on time, this makes IDAA a powerful method for analyzing genome editing events that can be used to screen newly generated nucleases for efficiency and indel signature, determine if an edited cell pool is suitable for experimentation or cloning, predict the fraction of clonal derivatives of the pool that will harbor a desired indel(s), and screen the clones, providing indel information on all alleles of the targeted gene.
FACS-based genome editing
Low, high or heterogeneous expression of nuclease in a transfected cell population can pose significant barriers to genome editing. We therefore devised methods for isolation of cell populations with a homogeneous level of nuclease that may be low, medium or high as desired, through FACS of cells transfected with a nuclease linked to a fluorescent protein (FP)23. Specifically, the method is based on constructs that express nuclease and FP in one coding sequence, but separated by a 2A peptide sequence (Fig. 3a). This results in 1:1 co-expression of nuclease and FP as separate entities and faithful marking of nuclease-transfected cells (Fig. 3b). In stage 1 of the method, cells are transfected with the nuclease-2A-FP construct and cultured for 2–3 days (Fig. 3c). In stage 2, the cells are subjected to FACS and those that express the nuclease marker at a desired level are isolated.
Figure 3 |. Principle of FACS-based genome editing.
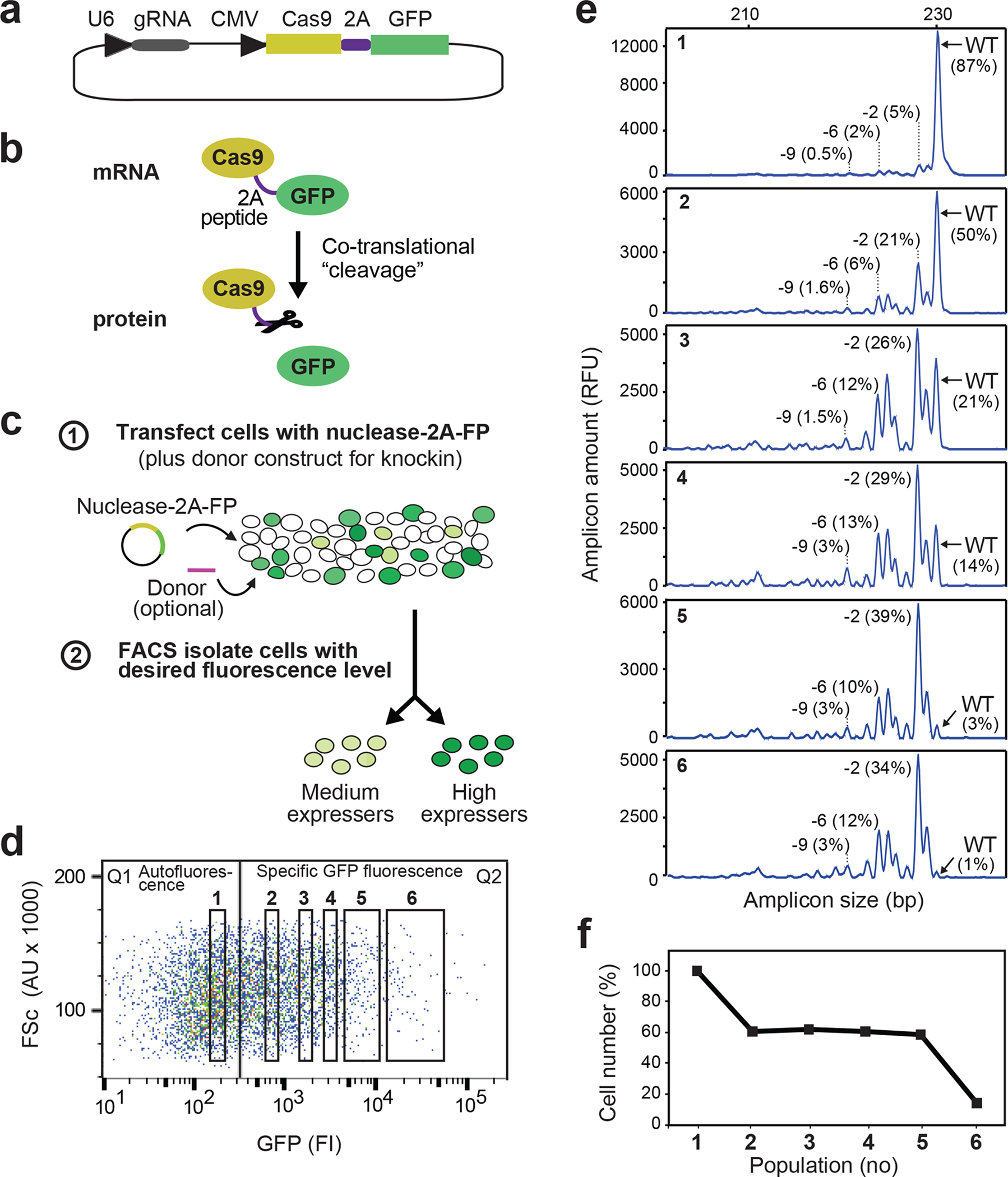
(a) Example of nuclease-2A-fluorescent protein construct for FACS isolation of nuclease-transfected cells. This construct expresses Cas9–2A-GFP and gRNA from CMV and U6 promoters, respectively. (b) 2A peptide undergoes ribosomal codon skipping during translation, resulting in 1:1 co-expression of nuclease and fluorescent protein as separate entities. (c) Schematic of FACS-based genome editing. In stage 1, cells are transfected with nuclease-2A-FP construct. In stage 2, cell populations with a desired level of fluorescence are isolated by FACS 2–3 days later. (d) FACS profile of K562 cells 2 days after transfection with Cas9–2A-GFP construct expressing gRNA targeting the KRAS locus. The GFP expression level of individual cells (represented by dots) is measured as arbitrary fluorescence intensity (FI) units (x-axis) and cell-size as arbitrary forward scatter (FSc) units (y-axis). Cell population 1 displays GFP fluorescence intensities similar to non-transfected cells (autofluorescence level), whereas populations 2–6 display specific GFP levels ranging from low to high (varying ~500-fold). (e) 20,000 cells were isolated from each of populations 1–6 in (d), cultured for 6 days, after which IDAA was used to determine indels at the KRAS locus in an aliquot of the cells. Note that editing levels increase with increasing GFP fluorescence level at the time of FACS and that almost complete editing was obtained at submaximal fluorescence levels (population 5). Though exhibiting background fluorescence, population 1 expresses some nuclease as evidenced by a low level of editing. The size and frequency of selected indels are indicated. IDAA was performed in an ABI 3500 instrument. (f) Prior to IDAA, the total number of live cells in each population analyzed in (e) was determined by trypan blue exclusion and expressed in % of population 1. Note that cell number decreases with increasing GFP fluorescence level at the time of FACS, but with a distinct dose-response relationship than the on-target editing. Note that KRAS is a non-essential gene in K562 cells22, meaning that the decrease in cell number is due to non-defined toxic or off-target effects associated with high transfection levels. Population 5 has an optimal fluorescence level with respect to editing efficiency and number of live cells, as revealed by comparing (e) and (f). Note that dose-response relationships will vary with nuclease construct, cell type and other parameters.
The method essentially sorts for “transfectability” of the cells. Thus, cells that have taken up high levels of nuclease-2A-FP will normally also have taken up high amounts of any co-transfected donor, in particular small oligo donors that are transfected more efficiently than the larger nuclease-2A-FP constructs23. The method can therefore be used for both knockout and knockin editing.
We have explored this nuclease enrichment method with CRISPR/Cas9 and ZFNs targeting many endogenous loci in an array of knockout and knockin applications23, 24. The principle was also explored by Certo et al. studying meganuclease I-SceI editing of reporter genes26, and its use has been reported for CRISPR/Cas9 by Mandal et al.27 and for TALENs by Ding et al.28. A conceptually similar, but technically different, approach has been developed29–31, which FACS-enriches for co-transfected fluorescent “surrogate reporter” genes for the nuclease used (discussed further below). We have applied the nuclease-2A-FP FACS-based method22–24 to increase or otherwise optimize nuclease-based genome editing in the following variations:
Defined low-to-high genome editing frequencies.
Transfection levels often vary >100-fold within a cell population, as exemplified with a Cas9–2A-GFP construct in Fig. 3d. By FACS for cell populations with defined and increasing levels of the FP nuclease marker, increasing levels of genome editing can be achieved (compare populations 1–6 in Fig. 3d and e). Frequently, it is possible to obtain essentially complete editing at the higher nuclease levels (populations 5 and 6 in Fig. 3d).
Isolation of rare nuclease transfectants.
Even if transfection efficiencies are very low (e.g. 0.5%), the FACS procedure enables efficient isolation of the rare nuclease transfected cells and useful levels of editing23.
Building-up genome modification levels in cell pools.
In some cases, the above procedures may not yield a sufficient level of editing, e.g. in connection with difficult knockin modifications, unfavourable features of the targeted locus, repair deficiencies in the given cell type, or the inability to generate sufficiently active nuclease reagents. In such cases, the cells may be subjected to repeated rounds of nuclease delivery and FACS to gradually build up editing levels in the cell population until satisfactory23.
Multiplex genome editing.
Nucleases targeting distinct genes may be linked to distinct FPs and co-sorted for multiplex editing. This strategy is particularly useful for ZFNs and TALENs that otherwise are less amenable to multiplex editing than CRISPR/Cas923.
Balancing editing efficiency and undesired effects.
If the nuclease or other of the transfected reagents have undesirable effects at high levels, the method can be used to select nuclease expression levels, where the adverse effects are minimal but the editing level remains useful (for example population 5 in Fig. 3d). Undesirable effects may include nuclease cutting at off-target sites or cutting-unrelated toxicity associated with the transfected reagents.
Convenient single-cell cloning of nuclease-expressing cells.
Many FACS instruments can sort single cells into 96-well plates, allowing convenient and accurate cloning of nuclease-expressing cells without the technical problems associated with cloning by limiting dilution.
A pipeline for cell line editing combining IDAA and FACS
The IDAA and FACS methods can be combined to form a highly efficient pipeline for generation of knockout cell pools or cell lines, which we have exploited recently24, and is outlined in Fig. 4. The pipeline is equally useful for knockin editing: in this application, a donor is co-transfected along with the nuclease and IDAA is replaced by genotyping by restriction fragment length polymorphism (RFLP)3, where indicated in Fig. 4.
Figure 4 |. Timeline and overview of a pipeline for cell line editing based on IDAA and FACS.
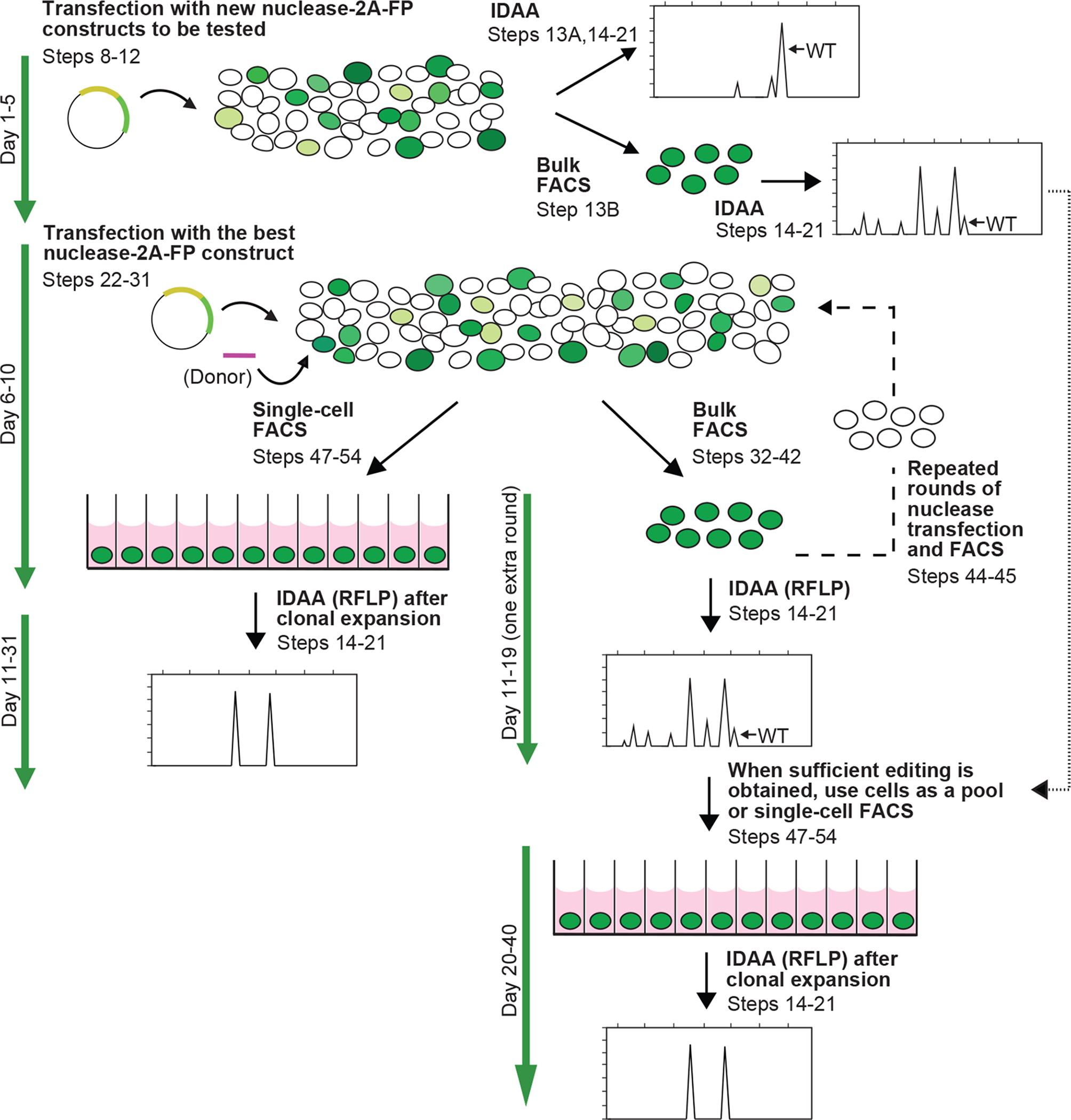
First, newly generated nuclease-2A-FP constructs are screened by transient transfection (Steps 8–12) and IDAA directly on the cell population (Steps 13A, 14–21) or after FACS enrichment of nuclease expressing cells (Steps 13B, 14–21). The best nuclease-2A-FP construct is next used to transfect cells (Steps 22–31) followed by FACS for a desired nuclease level and either single-cell sorting for clonal expansion (Steps 47–54) or bulk sorting (Steps 32–42), followed by IDAA (Steps 14–21) to determine the indel mutation level. If inadequate, the bulk sorted cells may be subjected to further rounds of transfection and FACS (dashed line, Steps 44–45) until a desired level of editing has been reached, where after the cells can be used for experiments as a pool or single-cell sorted for clone derivation (Steps 47–54). Clones may also be derived from edited cell populations obtained when screening nuclease reagents (dotted line). Expanded clones are genotyped by IDAA. For knockin editing, donor construct is incorporated as indicated and IDAA is replaced by RFLP assay, except in the nuclease construct testing step.
In the first stage of the pipeline, a number of newly generated nuclease-2A-FP constructs are screened by transfection into cells of interest and evaluated by IDAA after 2–3 days to identify those that elicit the highest level of frameshifting indels. IDAA may be performed directly on the transfected cell population (Fig. 4, Steps 13A, 14–21), or on cells FACS-enriched for nuclease expression, if low transfection efficiency would otherwise prohibit a clear result (Fig. 4, Steps 13B, 14–21). The best nuclease-2A-FP construct is then used to transfect the cells of interest. After 2–3 days, the cells are subjected to FACS and cells with the desired level of nuclease expression are either single-cell sorted into 96-well plates for expansion to clonal cell lines (Fig. 4, Steps 47–54) or bulk-sorted to generate a pool of edited cells (Fig. 4, Steps 32–42). In either case, an aliquot of the FACS-isolated cells are analyzed by IDAA to estimate the degree of editing and predict the fraction of expanded clones that will contain the desired indel(s). If the level of editing is too low, the cell pool can be subjected to further rounds of nuclease-2A-FP transfection and FACS (Fig. 4, Steps 44–45, dashed line). In this scenario, IDAA after each round is used to monitor if the degree of modification is sufficiently high to allow experimentation with the cellular pool and/or single-cell sorting for generation of clonal cell lines. For low-frequency modifications, such enrichment of edited cells can drastically reduce the number of clones that need to be generated to obtain a cell line with the desired mutation. Note that edited cell populations obtained when screening nuclease constructs may also be single-cell sorted for generation of clonal cell lines, either after FACS or even without application of FACS, if the level of mutation is above 10–30%, as revealed by IDAA (Fig. 4, dotted line). Finally, the genotype of expanded clones is screened by IDAA.
Wider applications of IDAA and FACS-based genome editing
Besides cell line engineering, IDAA can be exploited for indel surveillance in any genome editing application in tissues or embryos. We have used the method in zebrafish genome editing as an easy means to determine the nature and degree of indel mosaicism and hence, probability of germline transmission of loss-of-function mutations from F0 fish, as well as to screen the downstream F1 fish for the mutations (Supplementary Fig. 5). IDAA is also a very useful method for assessment of mutagenesis at specific nuclease off-target sites22 that are predicted based on sequence homology and typically are cut at low frequencies. The prevailing method for off-target analysis at specific sites has been the easy and economical, but rather insensitive and unreliable, enzymatic mismatch cleavage (EMC) assay, whereas next-generation sequencing has been the gold standard due to high sensitivity, but its drawbacks are cost and inconveniency (see further discussion below). IDAA combines the advantages of these two methods by being easy and cost-effective with a detection sensitivity down to ~0.1% (Supplementary Fig. 1), which is comparable to that of next-generation sequencing for detection of off-target cleavage32.
FACS-based genome editing may be used for therapeutic editing of human primary cells ex vivo, taking advantage of the method’s ability to isolate potentially rare nuclease transfectants and among these, select for cells with an optimal nuclease level with respect to the desired editing and off-target mutagenesis.
Limitations of IDAA and FACS-based genome editing
The main limitation of IDAA is lacking sequence information on the nuclease induced mutation, precluding its use for knockin mutagenesis analysis. FACS-based genome editing is limited to cells that can survive the FACS procedure, which fortunately include most of the commonly used cell types, even though optimization of FACS conditions may sometimes be required.
Comparison with other methods
Indel detection and nuclease-enrichment can be achieved by other methods, some of which may be employed at certain steps of the pipeline, but with limitations as discussed below.
The prevailing indel detection method in the field has been EMC of PCR amplicons spanning the nuclease target site12, 33–35 using plant-derived Cel-I endonuclease (a.k.a. Surveyor)36–40 or bacteriophage-derived T7 endonuclease I (T7EI)39, 41. In these assays, the amplicons are denatured and re-annealed, resulting in duplex hybrids with mismatches when non-identical fragments, due to differing indels or unmodified sequences, combine. The hybrids are subsequently digested by the mismatch endonucleases followed by agarose gel electrophoretic separation of the cleaved products. When analyzing a pool of edited cells, the fraction of digested amplicons is roughly proportional to the frequency of indel mutations in the sample. EMC assays are as inexpensive as IDAA, but inferior in all other respects: EMC assays provide no information on indel sizes; cannot reveal whether more than one allele has been mutated in a cell clone and cannot detect clones homozygous for the same indel; will significantly underestimate the editing level in cell pools with high levels of 1 or 2 predominant indels22, a frequent CRISPR/Cas9 indel outcome; may fail to detect certain small mutations and thereby underestimate the mutation rate22, 35; lack robustness and often require replicate determinations; are less amenable to high throughput, requiring more hands-on time than IDAA; and are less sensitive, having a detection limit of 1–2%.
Next-generation sequencing of PCR amplicons spanning the nuclease target site and data analysis using the CRISPR Genome Analyzer or similar software has detection sensitivities down to ~0.1%, i.e. in the range of IDAA, and can also detect knockin mutations, since the sequence of the mutation site is obtained32, 42. Next-generation sequencing in a multiplex format is an alternative to IDAA if a large number of clones need to be genotyped, in particular for knockin modification. It is, however, neither a convenient nor cost-effective alternative at other steps of the pipeline.
Tracking of Indels by DEcomposition (TIDE) is based on two parallel Sanger sequencing reactions of PCR amplicons spanning the nuclease target site from an edited and a wild-type sample, respectively. Subsequent TIDE software analysis of the data reveals the size and frequency of indels in the edited sample43. TIDE is the best alternative to IDAA in the pipeline, providing comparable indel resolution, quantitation ability and flexibility. Like IDAA, TIDE does not provide any sequence information on the indels (except for revealing the nucleotide identity of 1 bp insertions). TIDE software, however, does not allow analysis of TALEN- and ZFN-elicited indels, cannot reveal indels larger than 50 bp, sensitivity is limited to 1–2%, and the analysis cannot be performed on such small amounts of amplicons that can readily be used for IDAA (see Supplementary Fig. 2). TIDE is also limited by a requirement for high-quality sequence chromatograms, without which the software will not work. TIDE therefore requires more hands-on time in sample preparation; amplicons must be purified and quantitated prior to sequencing. Furthermore, sequencing from both ends of the amplicons is recommended to confirm the results. Difficulties in obtaining high-quality Sanger sequencing is one reason why TIDE may work less robustly with some sites.
Sanger sequencing of cloned PCR amplicons of the target site can define the sequence of the indels. This analysis is usually performed on the final selected clone(s) produced by the pipeline.
Finally, a method has been developed (GEF-dPCR) for sensitive and quantitative monitoring of already-defined indel patterns elicited by nucleases based on digital PCR44, which is beyond the scope of the pipeline.
FACS-based editing can be performed by an alternative efficient method, which marks cells with active nuclease by co-transfection with a “surrogate reporter” plasmid that expresses GFP upon indel mutagenesis at a target site of the nuclease placed upstream of GFP29–31. This approach requires cloning of the target site of each nuclease construct used into the reporter and does not allow editing based solely on RNA reagents, as the reporter is a plasmid. The method has not been reported for knockin editing and potentially the homologous reporter may compete with donor for the genomic DNA ends during HDR or be ligated into the genomic cut site45, 46. Nuclease-transfected cells have also been marked indirectly by co-transfection of nuclease construct with a separate vector expressing GFP47. This indirect marking of nuclease transfection is less efficient than 2A-based or surrogate reporter methods, possibly because the GFP constructs are smaller, and thereby transfected more efficiently than the nuclease constructs, resulting in less faithful marking of nuclease-transfected cells23, 29, 31.
Enrichment of nuclease-transfected cells can alternatively be achieved by drug selection or use of antibody-linked beads if the nuclease or surrogate reporter construct carries a drug resistance gene18, 48 or a cell surface antigenic marker30, 31. These constitute valid approaches for cell types not amenable to FACS and the protocol pipeline can easily be adapted to include them. These alternative approaches, however, do not allow for selection of cells with defined low, medium or high levels of nuclease expression. The methods therefore cannot be used to push for extra high knockout or knockin frequencies by selecting top-level transfected cells, nor can they be used to balance on-target editing with undesired effects or to avoid nuclease top-expressers, all of which require the ability to isolate populations with well-defined nuclease levels.
Experimental design
Nuclease-2A-FP expression systems.
A large number of 2A-FP vector systems are now available for CRISPR/Cas9, TALENs and ZFNs. Examples are shown in Fig. 5, and Supplementary Table 1 lists all current vectors to our knowledge. “All-in-one” Cas9–2A-FP vectors co-express Cas9 and a single gRNA or several gRNAs for multiplex editing (Fig. 5a). Alternatively, Cas9 and gRNA can be expressed from separate vectors that are linked to distinct FPs, followed by FACS for both colors (Fig. 5b). In this separate construct format, a gRNA, or more gRNAs for multiplex editing, may alternatively be expressed from vectors or PCR amplicons not carrying FP (Fig. 5b), which also works well, presumably because the smaller gRNA constructs are transfected more efficiently than the larger Cas9–2A-FP construct sorted for. One advantage of the separate construct format is that the components can be transfected in different ratios, if needed for further optimization of editing efficiency. The multiplex editing format can be converted into a Cas9 nickase pair editing format, if using Cas9 harboring the D10A mutation and gRNAs targeting the same locus49–52. Nickase pair editing may also be achieved by two all-in-one Cas9 nickase constructs carrying the gRNAs of the pair and linked to distinct FPs followed by FACS for both colors23 (Fig. 5c). For TALENs and ZFNs, the two monomers of the pair are expressed from separate plasmids and linked to GFP and RFP, respectively, followed by dual-color FACS (Fig. 5d). The predominant ZFN vector format (Sigma-Aldrich), however, allows the monomers to be combined into a GFP-coupled all-in-one construct in one subcloning step22, 23 (Fig. 5e).
Figure 5 |. Nuclease-2A-FP expression systems.
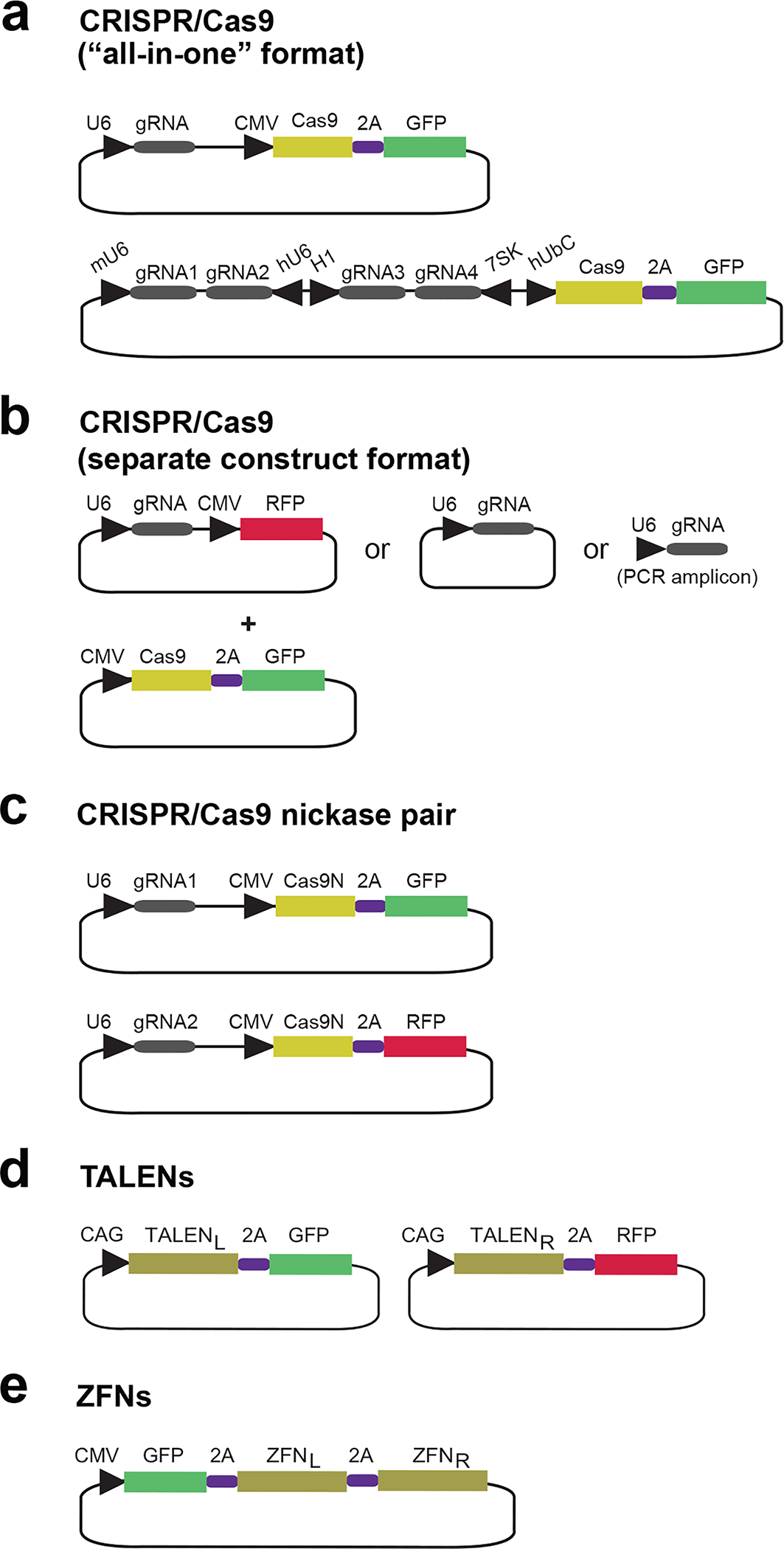
Schematic of some currently available nuclease-2A-FP vectors described in the text and Supplementary Table 1. Promotors are indicated by an arrowhead. L and R denote left and right monomers, respectively, of TALEN and ZFN pairs.
Nuclease-2A-FP delivery.
Delivery of nuclease-2A-FP constructs via transfection23, 27, nucleofection23, 27 or transduction and as plasmid23, 27 or mRNA23 (Cas9–2A-FP mRNA is commercially available: https://www.systembio.com/crispr-cas9-systems/mrna-grna) have all proven successful. Delivery of Cas9 protein along with gRNA as a ribonucleoprotein complex is another efficient delivery form53–56 that should be readily compatible with FACS-based editing, although to our knowledge, delivery of Cas9-FP as protein has not yet been reported. In general, the delivery mode may not be critical, since FACS enrichment can typically compensate for suboptimal transfection levels. Some applications may have special delivery issues: for instance, viral nuclease delivery is not compatible with the ssODN donor format. In other cases, a certain nuclease delivery format, e.g. as modified mRNA57, may be preferred for enhanced cell viability.
IDAA.
IDAA is performed in instruments capable of performing capillary electrophoresis-based fragment analysis, which in practice means the commonplace DNA sequenators for Sanger sequencing: Genetic Analyzer 310, 3100, 3130, 3500 or 3730 instruments (Applied BiosystemsR/Thermo Fisher Scientific), the GenomeLab GeXP Genetic Analysis System (SCIEX/Beckman Coulter) or in a specialized Fragment Analyzer™ (Advanced Analytical Technologies), albeit the latter only gives 5 bp resolution.
IDAA can be provided as a service (often called fragment length analysis) by GeneWiz Inc. (http://www.genewiz.com/index.aspx), Eurofins Genomics (http://www.eurofinsgenomics.eu/fla) or other DNA analysis service providers. For example, the crude IDAA PCR (Step 15) or the ready-to-load IDAA PCR mixed with Hi-Di formamide and size standard (Step 17) can be sent for fragment length analysis at Eurofins Genomics via the standard sequencing drop-box pickup service. In the former case, tick the box in the order form indicating that GSLIZ500 should be used as the size standard. IDAA profiles are then returned as PDF files, as well as FSA files that are compatible with Peak Scanner Software (discussed below), within a time frame and cost roughly comparable to standard DNA sequencing service.
To cover as many users as possible, this protocol employs the Genetic Analyzer 3130 instrument, which is by far the most widely distributed DNA sequenator/fragment analyzer and will serve the vast majority of IDAA needs, as well as the 3500 instrument, which is also becoming commonplace and has somewhat higher sensitivity.
IDAA; PCR.
The locus-specific IDAA Fwd and Rev primers should amplify the genomic cut site of the nuclease and are designed according to common principles for genomic PCR58–60 using free online software, such as Primer3 (http://bioinfo.ut.ee/primer3/) or Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). Sequences and specifications of IDAA Fwd, Rev and FamFwd primers are provided in Table 2 and Supplementary Table 2. Optimal amplicon size depends on capillary length of the fragment analyzer and will typically be 200–450 bp, but up to 900 bp amplicons have successfully been analyzed. Otherwise no restrictions apply, but note that deletions extending beyond the primer binding site will not be amplified/detected (although deletions >100 bp are rare). We recommend testing new IDAA primers in the tri-primer setup (Step 14) on a control sample (cells not transfected with nuclease) with a cell concentration not extremely different than the samples to be genotyped. The present touchdown PCR protocol generally ensures specific amplification, but when genotyping edited samples, such a control sample should also be run in parallel and should ideally produce a single (wild-type) amplicon peak in the subsequent IDAA analysis. Rare, non-specific amplification can often be resolved (see Troubleshooting). If not, however, nonspecific amplicons can usually be ignored; they are revealed by the IDAA analysis of the control sample, and unless they have exactly the same size as an indel amplicon, their presence in the profile of the edited sample will usually not interfere with proper analysis, nor interpretation of the results obtained. The robustness of the IDAA PCR nearly always allows the use of crude lysates of cells or tissues as template, as opposed to purified genomic DNA, and few cells are needed. We find that 1 μl of QuickExtract™ (Epicentre) lysate containing down to 10–20 cells μl−1 yields sufficient amounts of amplicons with various IDAA primers and cell types. To detect 0.1% frequency indel events (the lower limit of IDAA) in a cell pool or tissue, however, a theoretical minimum of at least 500 diploid cells (1000 template chromosomes) is required for the IDAA PCR. If cells are not a limiting factor, we aim for QuickExtract lysates containing 2000–5000 cells μl−1 and up to 20,000 cells μl−1 is compatible with the IDAA PCR.
Table 2 |.
IDAA PCR primers
| Primer | Sequence (5´−3´) | Purpose |
|---|---|---|
| Fwd | AGCTGACCGGCAGCAAAATTGn19–24 | Forward primer with locus specific sequence (n19–24) to amplify the nuclease target site and with an extension (in italics) having the same sequence as FamFwd primer. |
| Rev | n19–24 | Reverse primer with locus specific sequence (n19–24) to amplify the nuclease target site. |
| FamFwd | 6-FAM-AGCTGACCGGCAGCAAAATTG | Constant primer with same sequence as the extension of the Fwd primer and labelled with 6-FAM to render the amplicon fluorescent. |
IDAA; Analytical run.
Importantly, indel frequencies are analyzed within each IDAA profile (as opposed to comparisons between profiles). Therefore, it is not necessary to run equal amounts of IDAA amplicons among samples to be compared. For 3130 and 3500 instruments, the optimal quantitative ranges are 150–4000 RFU and 175–10,000 RFU, with saturation at 8000 RFU and 30,000 RFU, respectively. For the majority of nuclease-edited samples (containing ≤10 predominant amplicons), loading 0.5 μl and 0.05 μl, respectively, of a saturated IDAA PCR onto these instruments will keep most signals, except for the very low, within the dynamic range, when using standard sample injection parameters such as 48 seconds at 10 kV. Supplementary Fig. 2 shows IDAA profiles after loading various amounts of an IDAA PCR in a 3500 instrument. Note that a useful profile can be obtained with amplicon amounts barely visible, or even invisible, in agarose gel electrophoresis, which has practical advantages: occasionally, certain IDAA primers may yield low amounts of PCR product for various reasons, which will be noted in the agarose gel electrophoresis routinely performed after the IDAA PCR (Step 16). In case of a faint or invisible band, run 1–2 μl of the IDAA PCR in the fragment analyzer, which will often produce a useful IDAA profile.
For quantitation of very low-abundant amplicons relative to one major amplicon, only small amounts of the IDAA PCR should be loaded to avoid signal saturation of the major amplicon (perhaps 0.1 μl and 0.02 μl for 3130 and 3500 instruments, respectively). With proper loading, specific indel signals can easily be discriminated from background signals down to ~0.1%, even though this is outside the dynamic range and therefore less quantitative (Supplementary Fig. 1; Supplementary Fig. 3). Above the quantitative range (i.e. when overloading), saturated (overloaded) peaks may give rise to a minor peak artefact appearing 22–24 bp ahead of the true signal (see Supplementary Fig. 3).
The signal intensity of size standard peaks should ideally be 30–100% of the intensity of sample peaks, which cannot be achieved for all peaks in complex samples. Loading 0.3 μl GS500LIZ (the typical size standard in IDAA, when analyzing <500 bp amplicons), along with 0.3 μl saturated IDAA PCR will work in the majority of IDAA runs. The Applied Biosystems Genetic Analyzer manual can advise on further optimization for special needs.
IDAA; Data analysis.
Data are analyzed by software associated with the fragment analyzer used, for ABI instruments typically GeneMapper® or Peak Scanner™ 2, or with general software, such as GeneMarker® that works with many fragment analyzers. Information regarding how to obtain free Peak Scanner and GeneMarker® (Demo) software, as well as some of their features, are described in Supplementary Note. A step-by-step quick-guide to Peak Scanner™ 2 is provided in Supplementary Manual and an exhaustive manual can be found at http://www3.appliedbiosystems.com/sup/URLRedirect/index.htm?xDoD=4382253. The basics of Peak Scanner and GeneMarker® (Demo) take <20 min to learn from import of data files to mouse clicks on peaks to show amplicon sizes and peak areas, from which the relative frequencies of the indels can be calculated. Since PCR efficiency is a function of amplicon size, determinations of indel amplicon frequency relative to the wild-type amplicon become less accurate with increasing indel size, but this bias is insignificant for indels <10 bp and very modest for indels up to 30 bp in size.
FACS-based editing.
FACS of nuclease-2A-FP transfected cells can be used to isolate cells with a specific nuclease level as singlets for clonal derivation or pools of cells (bulk sorting). A step-by-step guide to the FACS procedure is provided in Box 1 and Fig. 6.
Box 1 |. Setting FACS parameters for nuclease-2A-FP genome editing.
FACS parameters for isolation of FP-positive, live, single cells are set up prior to each sort based on analysis of mock-transfected (control plasmid without FP) and nuclease-2A-FP-transfected cells, as illustrated with K562 cells in Fig. 6. In the plots shown, each dot, also called an “event”, represents a cell or a particle, the latter most often being cell debris. Use of a large nozzle orifice size of 100 μm will provide optimal survival for most cell types. Perform the following analyses, preferably based on ~10,000 events:
Determining autofluorescence level of the cell type used by analysis of mock-transfected cells.
Define the live cell population (Fig. 6a). Analyze the cells for forward scatter (FSc), a measure of cell size, and side scatter (SSc), a measure of granularity. Select the live cells, which in a healthy cell line population typically are distributed as illustrated here, showing one bulk of cells (P1) with low FSc and SSc.
Define cell singlets (Fig. 6b). Among the live cells defined in Fig. 6a, the single cells are next selected by analysis for FSc-width signal (pulse width). The plot represents a typical, well-dissociated cell population with the bulk of cells being singlets (P2).
Determine autofluorescence level (Fig. 6c). Analyze the live, singlets (P2) for fluorescence using a filter appropriate for the FP used, GFP in this example. As the mock-transfected cells do not express FP, any fluorescence can be attributed to autofluorescence (background level). The voltage of the FP fluorescence detection system is now adjusted such that the GFP fluorescence signal of the population spans about one decade. A cursor can now be placed to define cells showing autofluorescence levels (cells in Q1) for the subsequent analysis in Fig. 6d. The fluorescence is measured as arbitrary fluorescence intensity (FI) units.
Defining the nuclease-2A-FP expressing cell population.
Next, analyze the nuclease-2A-FP transfected cells for FSc/SSc and FSc-width/FSc-height as described for Fig. 6a and Fig. 6b to define the live (P1) and the live, single cells (P2) (dot plots not shown).
Select populations of nuclease-2A-FP transfected cells with defined FP levels (Fig. 6d). Analyze the live, single-cell population (P2) for fluorescence of the FP used, in this example GFP, using the same voltage setting for the fluorescence detection system as for Fig. 6c. Place cursor at the position defined in Fig. 6c to group background fluorescent cells (cells in Q1) and select sub-populations (also called “gates”) from Q2 with desired FP levels, e.g. A-D.
Calculate cell number in the selected populations (Fig. 6e). The number of available cells with a fluorescence level corresponding to for example “A” can now be calculated based on data output obtained at this stage: First, divide the number of cells in “A” with the number of cells in “P1”: 327/7884 = 0.041. Then, multiply this fraction with the number of live cells that was brought for FACS (determined in Step 32 of Procedure), say for instance 106 cells: 0.041 × 106 = 41,000 cells.
Notes: in Fig. 6e “#Events” signifies live cells, except for “All events” that also include dead cells and cell debris (all dots in Fig. 6a). In the present example, 10,000 events were analyzed.
Selecting cells co-expressing two, different nuclease-2A-FPs (Fig. 6f). Perform analyses as described for Fig. 6a–d, except that the mock-transfected (not shown) as well as the nuclease-2A-FP transfected cells are analyzed with filters for both FPs used, GFP and RFP in this example. The dual FP expressing cells are in Q2.
Figure 6 |. Setting FACS parameters for nuclease-2A-FP genome editing.
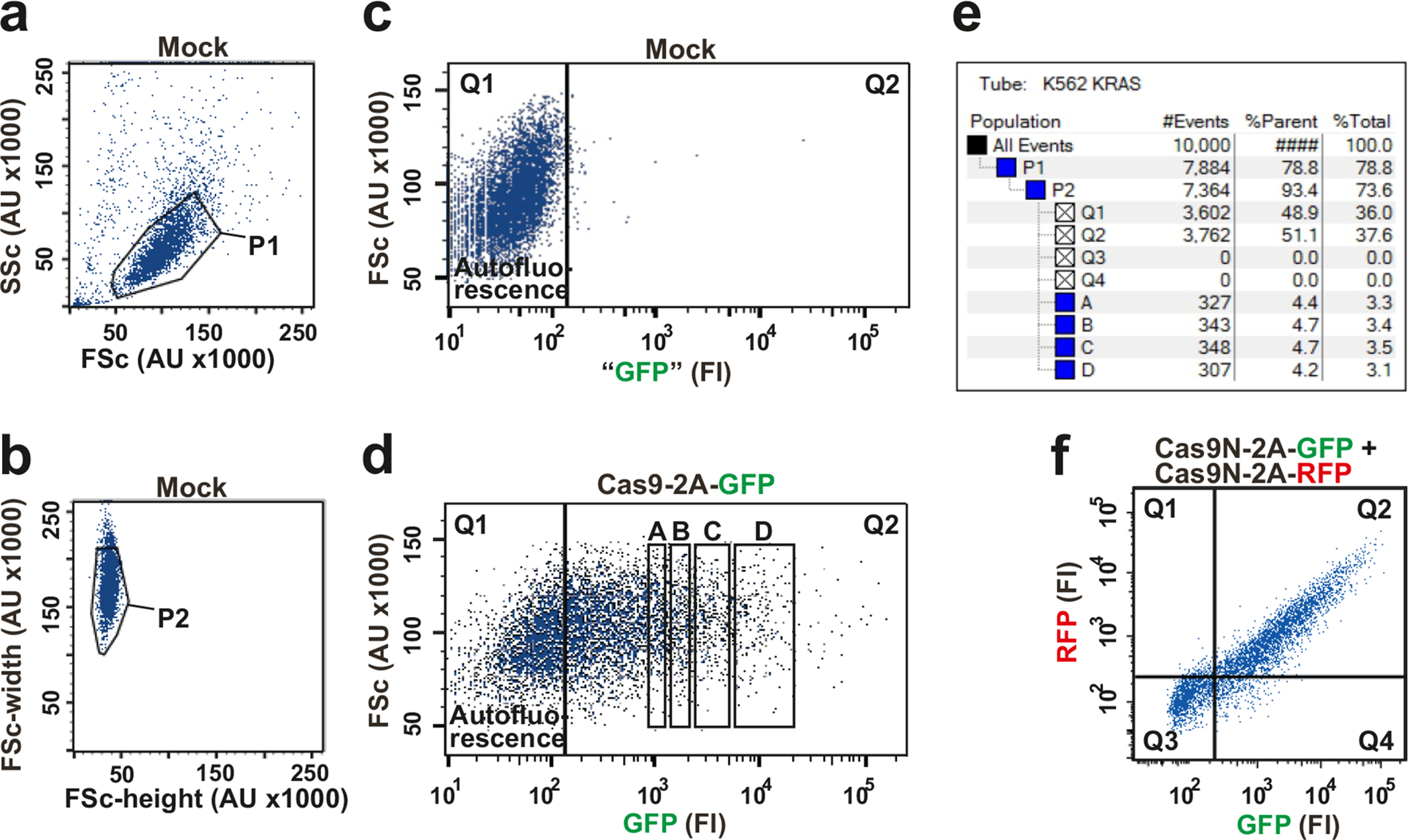
K562 cells were transfected with mock construct or with the indicated Cas9–2A-FP constructs and analyzed in a FACS Aria II instrument. See Box 1 for details on the analysis and how to set up the FACS parameters. (a) Defining the live cell population (P1) in mock-transfected cells by analysis for forward scatter (FSc) and side scatter (SSc). (b) Defining the live single-cell population (P2) in mock-transfected cells by analysis for FSc-width. (c) Determining the autofluorescence level of mock-transfected cells. (d) Selecting populations of nuclease-2A-GFP transfected cells with defined GFP levels (labelled A-D) by gating cells for specific GFP fluorescence intensity (FI) levels. (e) Calculating cell number in the selected populations A-D based on data output obtained from panels (a)-(d). (f) Selecting cells co-expressing two, different nuclease-2A-FP constructs (cells in Q2).
FACS-based editing; Bulk sorting.
To reduce FACS time, the density of the sorted cells should be as high as possible, i.e. 10−15×106 cells ml−1 in e.g. 0.5–1 ml (the volume depending on how many cells that need to be isolated – see calculations in Box 1). Very self-adherent cells may require a lower density to allow a single-cell suspension. Isolation of e.g. 20,000 cells takes from 2–15 min depending on cell density in the gate.
FACS-based editing; Single-cell sorting.
Single-cell sorting requires that the cell type can expand from individual cells. Due to loss of cell suspension associated with single-cell sorting, the volume should be ≥1 ml containing ≥1×106 cells, which will suffice for >20 96-well plates. One dish takes from ~2½−5 min to plate, depending on cell density in the gate.
FACS-based editing; Nuclease level to sort for.
FACS offers the possibility to edit using an optimal nuclease level with respect to a productive balance between on-target editing and a particular undesired effect associated with transfection level. Both phenomena will increase with increasing fluorescence levels, but typically with distinct dose-response relationships, as they are the results of independent processes. Furthermore, the dose-response relationships for both phenomena will vary with cell type, specific nuclease construct, format and delivery method. Cell death is one major undesired effect. The dose-response relationship for cell death can be complex (see the bi-phasic example in Fig. 3f), because it has several components, including chemical/physical transfection stress, double-stranded DNA stress, FP toxicity and cutting at off-target sites. Off-target sites are sequences resembling the target site and generally, they are cut with low frequencies. Increased nuclease levels, however, may overcome the mismatch energy barrier and lead to increased off-target cutting23, 50, 61. This may cause cell death via the DNA damage response pathway if excessive or, potentially more problematic, undesired mutagenesis in surviving cells. We consider it likely that off-target cutting contributes prominently to the excessive cell death sometimes observed at top nuclease levels, implying that the surviving cells may contain substantial off-target mutagenesis.
To optimize efficiency and safety, it is therefore highly recommended to determine the fluorescence level that gives satisfactory on-target editing and acceptable cell death with the construct and cell type in use. This can be rapidly achieved by bulk isolating a defined number of cells from a range of distinct fluorescence levels, culturing for 6 days, then assessing the level of on-target editing by IDAA as well as the number of live cells in each of the populations, as described in Steps 32–43 and exemplified in Fig. 3.
Similarly, the dose-relationship for indel mutagenesis at specific, undesirable off-target sites relative to the target site may be determined by IDAA, and often it is possible to tune nucleases to a level where off-target mutagenesis is minimal, but on-target mutagenesis remains high (Fig. 7). Depending on the FACS instrument, 2–6 cell populations with different fluorescence levels can be isolated simultaneously for the above-mentioned optimizations. Once determined, the optimal fluorescence level can be used for downstream editing.
Figure 7 |. FACS balancing of on-target and off-target mutagenesis.
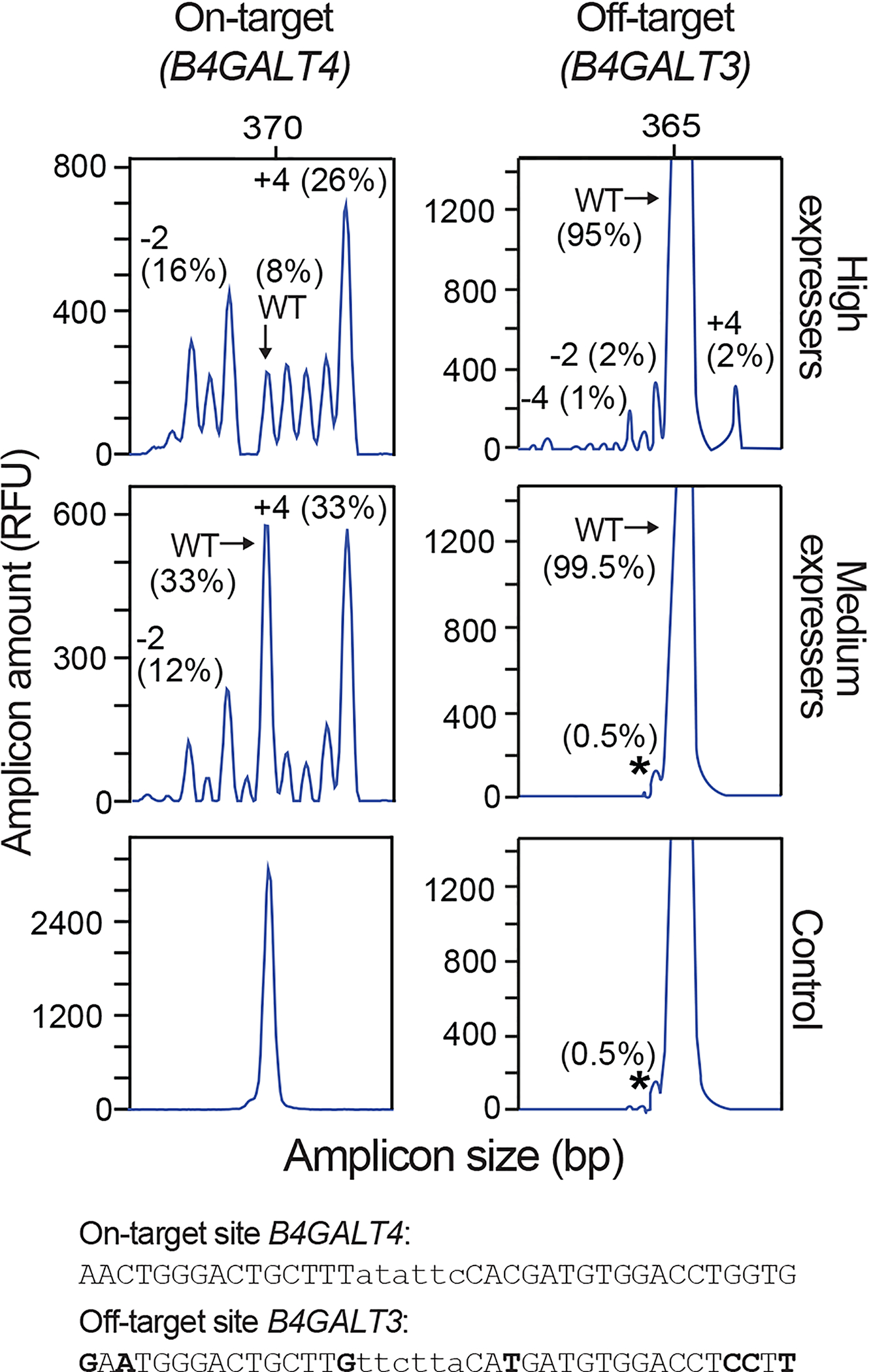
Fifty thousand CHO cells transfected with GFP-2A-ZFNLeft and RFP-2A-ZFNRight targeting B4GALT4 were bulk isolated for high or medium GFP and RFP double-fluorescence levels and analyzed by IDAA for indels at the on-target site in B4GALT4 and an off-target site in B4GALT3. Note that off-target mutagenesis occurs at the high nuclease level but not at the medium nuclease level, where on-target mutagenesis remains high. The size and frequency of selected indels are indicated. The “shoulder” indicated by an asterisk is not an indel, since it is also present in the control sample; it is likely due to incomplete 3´ A nucleotide addition to the IDAA amplicon (see Table 3, Step 21). The sequences of the on- and off-target sites are shown, with nuclease-binding sequence in uppercase and mismatches in boldface. IDAA was performed in an ABI 3130 instrument.
FACS-based editing; Nuclease considerations.
It is recommended to design nucleases with target sites that differ from potential off-target sites by at least 2–3 mismatches, as this will significantly decrease unwanted mutagenesis in FACS-based editing. It may, however, not be eliminated, since off-target sites occasionally can have more than 2–3 mismatches32, 62, 63. For CRISPR/Cas9, it may be considered to use one of the high-fidelity versions of the conventional system, such as CRISPR/Cas9 nickase pairs49–52, 62, truncated gRNAs (tru-gRNAs)32, 64 and gg-gRNAs52, 65 that can readily be incorporated in a FACS-based editing scheme. Other high-fidelity systems are dCas9/FokI fusions66, 67, Cas9 mutants with improved specificity68, 69 and Cas9 orthologs with more restrictive PAM requirements70, for which FP-based vectors, however, need to be generated. Delivery of CRISPR/Cas9 constructs as RNA is another means to reduce off-target mutagenesis. For a discussion of the various strategies for improving CRISPR/Cas9 specificity, see21, 71, 72. TALENs62 or obligate heterodimeric ZFNs73 are generally quite specific nucleases. We observed very limited cutting at select off-target sites when sorting for high levels of obligate heterodimeric ZFNs having off-target sites with more than 3 mismatches or CRISPR/Cas9 nickase pairs23.
FACS-based editing; When to perform FACS.
FACS can be performed 1–3 days after transient nuclease-2A-FP delivery or longer if the construct is genomically integrated after viral delivery. It is worth noting that the fluorescence signal is not a direct read-out for nuclease/gRNA level, because the FP and nuclease/gRNA have different degradation kinetics (we find that FPs are typically more long-lived). This implies that the same time period between nuclease-2A-FP delivery and FACS used for determining the optimal fluorescence level must also be used in the actual editing experiment.
FACS-based editing; Increasing cell viability.
Various factors associated with FACS-based editing may decrease cell viability, including the FACS process itself, cell type characteristics, and the selection for high transfection levels. Several parameters can be optimized to improve cell survival and it is recommended to consult the literature on how to FACS the cell type of interest. Single-cell sorting is most challenging for cell survival, but often the problem can be solved by simply seeding many 96-well plates (e.g. 5–10) to ensure expansion of a sufficient number of clones to obtain one or more with the desired modification. Parameters to consider for improved cell survival include: First and foremost, exploit the ability of FACS-based genome editing to determine the optimal nuclease level regarding editing efficiency and cell survival and use that level for single-cell sorting; Sort cells into conditioned medium (Step 47) and/or onto coated plates; Deliver nuclease constructs as in vitro transcribed mRNA (if the nuclease construct allows) and donor as ssODN, which is less toxic than double-stranded DNA for many cell types; Use the larger nozzle orifice size of the FACS apparatus to reduce mechanical sorting stress; Use optimal FACS temperatures for the cell type of interest (e.g. 4 ˚C, room temperature, 37 ˚C).
Screening of new nuclease reagents using FACS and IDAA.
For rapid generation and testing of new gRNA constructs, we have modified the strategy based on PCR amplicons as gRNA expression cassettes18 to include: gRNA PCR primers that are shorter and less expensive; GFP-linked Cas9; and IDAA. The protocol is robust and has been used to test ~250 new gRNA designs. The generation of the gRNA constructs, which we term QuickChange (QC) gRNA amplicons and the screening procedure are detailed in Fig. 8, Supplementary Fig. 6 and Steps 1–21. In brief, gRNA sequences are designed using a preferred algorithm (see options in Step 1) and incorporated in a tri-primer PCR with a U6 promoter template to generate the QCgRNA amplicons containing the U6 promoter, gRNA design and scaffolding tracrRNA elements (Fig. 8a). The unpurified QCgRNA amplicons (i.e. crude PCRs) are individually co-transfected with Cas9–2A-GFP into the cells of interest, followed by evaluation of their indel-inducing activities by IDAA 2–3 days later either without (Fig. 8b) or with prior FACS for nuclease enrichment (Fig. 8c). Note that FACS enrichment does not grossly change the IDAA profile, i.e. the indel mutagenesis pattern, but lower-frequency indels become clearly recognizable. The TEMPase DNA polymerase used may inadvertently introduce mutations in ~15% of the QCgRNAs, though only a smaller fraction will affect function. If desired however, mutation rates can be reduced to <1% by using high-fidelity DNA polymerases. FACS and IDAA are equally useful for screening new TALEN and ZFN constructs.
Figure 8 |. Screening of gRNA designs using gRNA amplicon expression cassettes, FACS and IDAA.
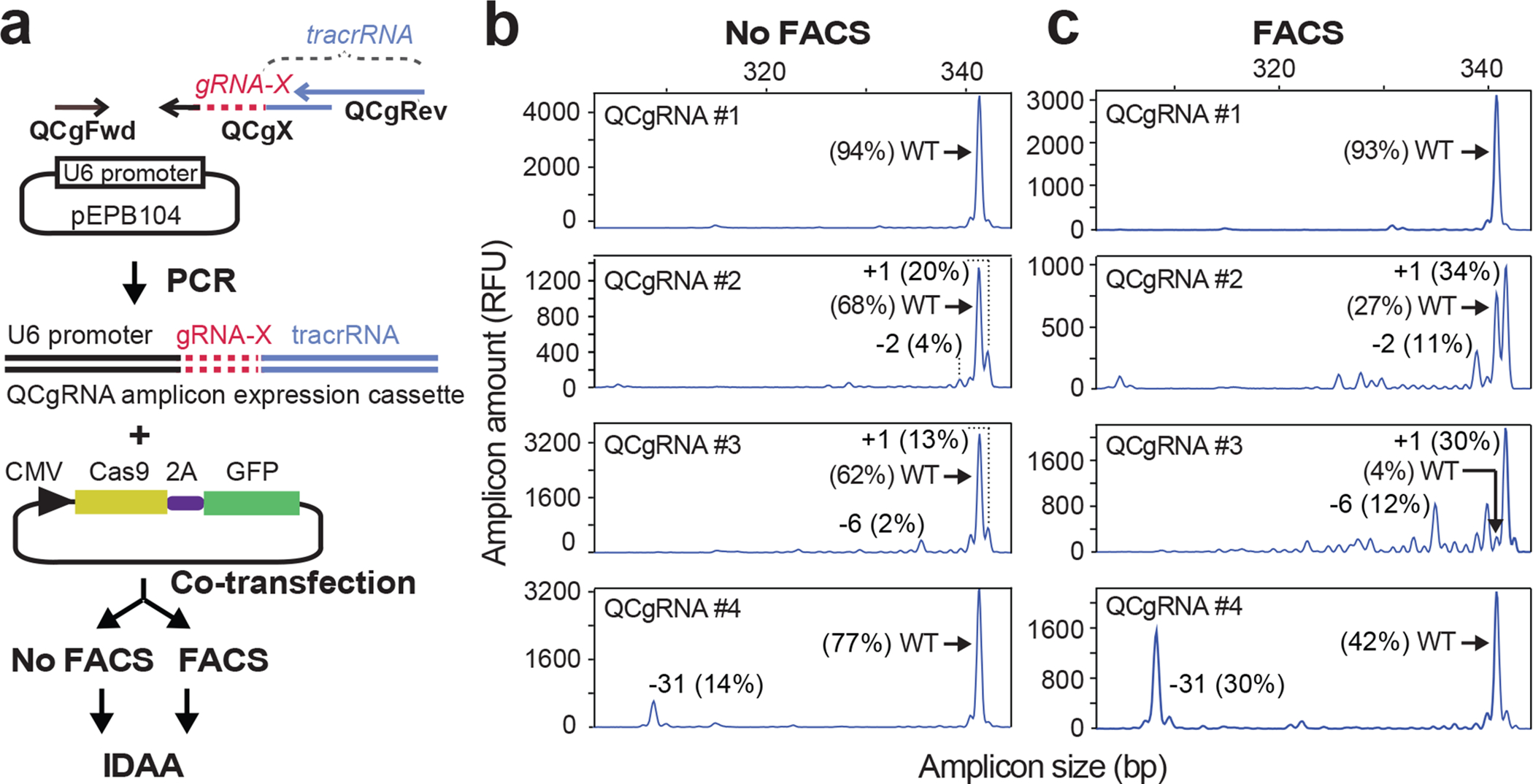
(a) QuickChange (QC) gRNA amplicon expression cassettes are generated through a tri-primer PCR using a QCgX primer encoding the gRNA design (gRNA-X), universal QCgFwd and QCgRev primers and a U6 promoter template. The QCgRNA amplicon and a Cas9–2A-GFP construct are co-transfected into cells of interest, followed by IDAA on unsorted cells or cells enriched for nuclease by FACS 2 days post-transfection. The example shows IDAA for 4 different QCgRNA amplicons targeting GALNT3 and performed on an aliquot of a HEK293 cell pool, (b) without or (c) with FACS for the top 60% most highly GFP-fluorescent cells. Note that QCgRNAs #2, #3 and #4 are functional, as they are able to elicit a high fraction of indels, whereas #1 is not. The size and frequency of selected indels are indicated. IDAA was performed in an ABI 3130 instrument.
The predictive value of an IDAA screening profile requires that IDAA is reproducible and that the same indels are elicited in the subsequent editing experiments, i.e. that indel repair of a specific nuclease-induced DSB has specific, favoured outcomes. Regarding the former requirement, IDAA is amply reproducible and gives essentially the same result every time (Fig. 2; Supplementary Fig. 4). Regarding the latter, the literature contains only scattered information. We have found, however, from screening hundreds of nucleases by IDAA that the indel pattern for a given nuclease conforms to a specific signature that is almost identical from experiment to experiment (Fig. 2; Supplementary Fig. 4). In different cell types, the indel pattern of a specific nuclease may sometimes also be similar (Supplementary Fig. 4), but since the generality of this observation is still uncertain, we recommend screening new nuclease reagents in the cell line to be edited. Though not specific for the present protocol, it is recommended to sequence the intended nuclease target sites in the cells to be edited, since single nucleotide polymorphisms (SNPs) can greatly decrease editing efficiencies.
Donor and knockin editing considerations.
High amounts of nuclease lingering in the cells after FACS may cut the donor after its genomic integration and cause indel mutation along with the knockin mutation. Unwanted indel mutagenesis can be largely prevented by disrupting the nuclease binding site in the donor through incorporation of silent mutations (which can be designed to also serve as diagnostic RFLP sites). We typically introduce 1–2 mutations per monomeric binding site of CRISPR/Cas9 nickase, TALEN or ZFN pairs and 2–3 for conventional CRISPR/Cas9, preferably at the PAM site and the PAM proximal 6–12 bp, where mismatches are least tolerated. Thus, donors should be designed after the best nuclease reagent has been identified, such that the position of silent mutations and homology arms can be optimized accordingly.
Plasmid donors may be co-sorted for by inclusion of an FP expression cassette in the donor construct that is distinct from the FP in the nuclease construct and FACS for both colors26. FP-encompassing donor constructs are likely to be particularly advantageous when knocking in larger DNA sequences74, 75. Alternatively, we find that ssODN-mediated knockin of larger DNA segments without FPs76 can be combined with FACS for co-transfected nuclease-2A-FP constructs to enable efficient knockin of large DNA sequences. For knockin of small mutations, ssODNs may be preferred donors as they are present in the transfection reaction in huge numbers due to their small size, are therefore taken up in great excess of the nuclease-2A-FP construct and are thus not rate-limiting when sorting for the nuclease23. Various additional aspects of ssODN donor design are described elsewhere5–8. Finally, constructs have been created which co-express Cas9, FP and viral proteins that mediate ubiquitination and degradation of DNA ligase IV, a critical NHEJ component (pCas9–2A-FP-2A-Ad4E1B55K/Ad4E4orf6)77. This allows simultaneous sorting for nuclease expression and suppression of NHEJ, leading to greatly elevated knockin of co-transfected plasmid donor constructs.
MATERIALS
REAGENTS
pEPB104 (Addgene plasmid ID: 68369, see Supplementary Data for sequence) or similar plasmid containing the U6 promotor, but no sgRNA elements.
CAS9PBKS (Addgene plasmid ID: 68371, see Supplementary Data for sequence) or similar Cas9–2A-GFP plasmid.
pCas9–2A-GFP/gRNA all-in-one construct for KRAS (Sigma-Aldrich; this construct as well as the similar construct for VEGFA were previously described23).
Plasmids encoding ZFNs in the CompoZr ZFN format (Sigma-Aldrich).
Genomic target sequences for construction of nucleases are listed in Supplementary Table 3.
Primer sequences for construction of QCgRNA amplicons are listed in Table 1 and ordered desalted in 10 nM synthesis scale from TAG Copenhagen A/S (www.TAGC.com) or from another oligo supplier.
5’- 6-FAM-labelled FamFwd, Fwd and Rev IDAA primers are listed in Table 2 and Supplementary Table 2 and can be ordered in a kit-format from TAG/C (www.TAGC.com) or custom-synthesized from other oligo supplier. ▲CRITICAL The 6-FAM fluorophore is light-sensitive and should be stored in the dark at −20 °C.
TEMPase Hot Start DNA Polymerase with 10× Ammonium Buffer and MgCl2 (Ampliqon, cat. no. A221106)
dNTP 100 mM: 25 mM ATP (cat.no. A521102), 25 mM TTP (cat.no. A521402), 25 mM CTP (cat.no. A521202), 25 mM GTP (cat.no. A521302) (Ampliqon)
UltraPure DNase/RNase-free distilled water (Life Technologies, cat. no. 10977–023)
TAE buffer, 10× (Sigma-Aldrich cat. no. 11666690001)
UltraPure agarose (Thermo Fisher Scientific, cat.no. 16500–500)
MassRuler Low Range DNA Ladder, (Thermo Fisher Scientific, cat. no. SM0383)
TrackIt CyanOrange loading buffer (Life Technologies, cat. no. 10482–028)
SYBR Safe DNA Gel Stain (Thermo Fisher Scientific, cat.no. S33102)
-
HEK293T cells (ATCC, cat. no. CRL-11268)
! CAUTION The cell lines used in your research should be regularly checked to ensure they are authentic and are not infected with mycoplasma.
-
K562 cells (ATCC, cat. no. CCL-243)
! CAUTION The cell lines used in your research should be regularly checked to ensure they are authentic and are not infected with mycoplasma.
DMEM, high glucose, GlutaMAX™ Supplement, pyruvate (Thermo Fisher Scientific, cat. no. 31966–047)
Fetal bovine serum (VWR, ch30160.03)
Penicillin-streptomycin (Life Technologies, cat. no. 15140–130)
Trypsin-EDTA (Life Technologies, cat. no. 25200–056)
Lipofectamine 3000 transfection reagent (Thermo Fisher Scientific, cat. no. L3000–015)
Amaxa Nucleofector kit V (Lonza, cat. no. VCA1003)
PBS cell culture grade Ca2+ and Mg2+ free
QuickExtract DNA extraction solution (Epicentre, cat. no. QE09050)
Hi-Di Formamide (Thermo Fisher Scientific, cat. no. 4311320). Aliquot and store at −20 °C for up to 3 months. Only freeze/thaw twice.
GS500LIZ size standard (Thermo Fisher Scientific, cat. no. 4322682)
MilliQ water
1 M Tris (pH 8.0) (Sigma-Aldrich, cat. no. T3038)
Table 1 |.
Primer sequences for QCgRNA amplicon expression cassettes
| Primer | Sequence (5´−3´) | Purpose |
|---|---|---|
| QCgFwd | CTCGATATCGAATTCGAGGGCCTATTTCCCATGATTCC | Constant forward primer annealing to U6 promotor. |
| QCgRev | CGAATTAACGGTACCAAAAAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTC | Constant reverse primer with tracr elements and overlapping with primer QCgX. |
| QCgX | TTTAACTTGCTATTTCTAGCTCTAAAACn17–20(c)GGTGTTTCGTCCTTTCCACAAGAT | Variable reverse primer containing tracr elements and gRNA design (n17–20) as reverse complement of target. Anneals to U6 promoter and overlaps with primer QCgRev. The (c) is only included, when the gRNA design does not contain a G as the first (5´) nucleotide, and is appended to increase gRNA transcription. |
EQUIPMENT
UV Spectrophotometer (e.g. NanoDrop 2000c, Thermo Fisher Scientific)
FACS machine (e.g. FACS Aria II, BD Biosciences)
Centrifuge for 15-ml and 50-ml Falcon tubes (e.g. Thermo Fisher Scientific Heraeus multifuge 3S+)
Falcon tubes, polypropylene, 15 ml (BD Falcon, cat. no. 352097)
Falcon tubes, polypropylene, 50 ml (BD Falcon, cat. no. 352070)
Sterile p10; p100 and p1000 pipette tips with filters (Gilson DF10ST cat. no F171103; DF100ST cat. no. F171403 and DF1000ST cat. no. F171703)
Standard microcentrifuge tubes, 1.5 ml (Eppendorf, cat. no. 0030 125.150)
Tissue culture plate, 6 wells (Corning Falcon, cat. no. 353934)
Tissue culture plate, 24 wells (Corning Falcon, cat. no. 353226)
Tissue culture plate, 96 wells flat bottom (Corning Falcon, cat. no. 353227)
Tissue culture dish, 100 mm (Corning Falcon, cat. no. 353003)
Nunc EasYFlask 225 cm2 (T225 flask), filter cap, 70-ml working volume (Thermo Scientific, cat. no. 159934)
Nunc EasYFlask 75 cm2 (T75 flask), filter cap, 25-ml working volume (Thermo Scientific, cat. no. 156499)
MicroAmp Optical Adhesive Film (Thermo Fisher Scientific, cat.no. 4311971)
50 μm cup Filcons (BD Biosciences, cat. no. 340629)
5-ml round-bottom Falcon tubes (BD Biosciences, cat. no. 352054)
PCR plates, 96 well (Sarstedt, cat. no. 72.1978.202)
Axygen 8-Strip PCR tubes (Fischer Scientific, cat. no. 14-222-250)
Thermocycler with programmable temperature stepping functionality, 96 well (e.g. Veriti Thermocycler cat. no. 4452300 or Eppendorf vapo-protect, cat. no. E950040015)
Benchtop microcentrifuge (e.g. Eppendorf, Centrifuge 5417R)
Eppendorf Thermomixer® R, dry block heating and cooling shaker (T3317)
Gel electrophoresis system (PowerPac basic power supply, Bio-Rad, cat. no. 164–5050), and Sub-Cell GT System gel tray (Bio-Rad, cat. no. 170–4401)
Novex XCell SureLock mini-cell (Life Technologies, cat. no. EI0001)
Digital DNA gel imaging system (e.g GelDoc EZ, Bio-Rad, cat. no. 170–8270), and blue sample tray (Bio-Rad, cat. no. 170–8273)
Blue-light transilluminator and orange filter goggles (SafeImager 2.0; Invitrogen, cat. no. G6600)
Gel quantification software (Bio-Rad, ImageLab or open-source ImageJ from the National Institutes of Health (NIH), USA, available at http://rsbweb.nih.gov/ij/)
Nucleofector 2b (Lonza)
Fluorescence microscope for GFP
REAGENT SETUP
10 mM Tris (pH 8.0).
Mix 10 ml 1 M Tris (pH 8.0) with 90 ml ddH2O.
Primer preparation
Resuspend the QCgFwd and QCgRev primers to a final concentration of 25 μM and QCgX primers to 2.5 μM in 10 mM Tris (pH 8.0).
Resuspend the IDAA Fwd primer to a final concentration of 2.5 μM and the IDAA Rev and FamFwd primers to 25 μM in 10 mM Tris (pH 8.0).
▲CRITICAL The 6-FAM fluorophore is light-sensitive and should be stored in the dark at −20 °C.
TAE electrophoresis solution
Dilute 10× TAE buffer in distilled water to a 1× working solution, and store at room temperature for up to 6 months.
1.5% agarose gels
Dissolve 1.5 g agarose in 100 ml 1× TAE buffer, boil for at least 10 minutes, and store at 65 °C for up to one week. Add DNA stain just prior to use.
PROCEDURE
Generation of QCgRNA amplicon expression cassettes ● TIMING 10 h hands-on time (excluding oligo ordering)
▲CRITICAL Nucleases of choice for a given target site can be obtained in a 2A-FP vector system using the information listed in Supplementary Table 1, in which case proceed directly to Step 9. Alternatively, you may choose to establish your own CRISPR/Cas9 reagents, in which case it is often an efficient route to start by generating and testing gRNA designs in the form of QCgRNA amplicons, by following Steps 1–8.
1 | Obtain candidate gRNA designs. Input target genomic DNA sequence into one of the online gRNA design tools: Cas-Designer, http://www.rgenome.net/cas-designer/78; ZiFiT, http://zifit.partners.org/ZiFiT/79, E-Crisp, http://www.e-crisp.org80; http://crispr.mit.edu18; WGE, http://www.sanger.ac.uk/htgt/wge/81; CRISPRscan, http://www.crisprscan.org82; CROP-IT, http://cheetah.bioch.virginia.edu/AdliLab/CROP-IT/homepage.html83; https://www.dna20.com/eCommerce/cas9/input (for a review and discussion of design tools, see84). Thereafter, evaluate and select suitable target sites. We recommend testing 3 designs for standard knockout and 4–6 designs for the more challenging knockin or nickase pair editing applications.
2 | Preparation of U6 PCR template. As template, we recommend pEPB104 (Addgene #68369, see Supplementary Data for sequence) or similar construct containing only the U6 promotor. Dilute the template with ddH2O to a concentration of 50 pg μl−1.
▲ CRITICAL STEP The absence of any gRNA and tracr elements in pEPB104 avoids their potential interference with same elements in the primers during the PCR. Furthermore, the PCRs can be used directly for transfection without the need of purification to remove the template.
3 | QCgRNA amplicon PCR. Set up one reaction for each QCgX primer design, as described below, using the oligos specified in Table 1 (see also Supplementary Fig. 6a). Include a control reaction containing only QCgFwd and QCgX (at 0.25 μM final concentration in this reaction) alongside the tri-primer reaction to enable validation of full-length product generation by parallel agarose gel analysis of the sample and control.
| Component | Amount (μl) | Final concentration |
|---|---|---|
| Ammonium PCR buffer, 10× | 2.5 | 1× |
| dNTP, 100 mM (25 mM each) | 0.25 | 1 mM |
| QCgFwd primer, 25μM | 0.25 | 0.25 μM |
| QCgRev primer, 25μM | 0.25 | 0.25 μM |
| QCgX primer, 2.5μM | 0.25 | 0.025 μM |
| TEMPase Hot Start DNA polymerase | 0.25 | 1.25 U |
| DNA template from Step 2, 50 pg μl−1 | 1.0 | 50 pg |
| ddH2O | 20.25 | |
| Total | 25 |
▲CRITICAL STEP Observe the molar primer ratios for successful full-length product generation. We also find that >100 pg amounts of template may lead to a significant decrease in amplicon yields.
4 | Perform PCR using the following touchdown cycling conditions:
| Cycle number | Denature | Anneal | Extend |
|---|---|---|---|
| 1 | 95 °C, 15 min | ||
| 2–13 | 95 °C, 30 s | *64 °C, 30 s | 72 °C, 30 s |
| 14–38 | 95 °C, 30 s | 52 °C, 30 s | 72 °C, 30 s |
| 39 | 72 °C, 20 min |
Decrease annealing temperature by 1 °C in each subsequent cycle
▲CRITICAL STEP To reduce the small risk of PCR-introduced mutations, the PCR may optionally be performed with Phusion polymerase in Phusion HF PCR buffer with the other PCR reagents as described above, but with the following cycling conditions: Initial denaturation for 2 min at 98 °C, then 12 cycles of denaturation for 10 s at 98 °C, a combined annealing/extension step for 30 s at 72 °C, then 25 cycles of denaturation for 10 s at 98 °C/annealing for 15 s at 57 °C/extension for 30 s at 72 °C, then a final extension step for 10 min at 72 °C.
5 | Run 3 μl of the PCR and 15 μl, 10 μl and 5 μl MassRuler Low Range DNA ladder on a 10 cm-long, 2% (wt/vol) agarose gel such that the PCR products have migrated ~5 cm (~1½ h at 5 V/cm distance between electrodes), using CyanOrange loading dye and SYBR Safe DNA gel stain. Visualize DNA by UV irradiation and check that the full-length QCgRNA amplicon has been generated by comparison to the control reaction containing only QCgFwd and QCgX primers (see Supplementary Fig. 6b).
? TROUBLESHOOTING
6 | Determine the QCgRNA amplicon concentration in the PCRs based on agarose gel band intensity comparison with the 400 bp DNA ladder band (representing 60 ng, 40 ng and 20 ng with loading suggested in Step 5) or by using ImageJ software (http://rsbweb.nih.gov/ij/).
■ PAUSE POINT The PCRs can be stored at −20 °C for several months.
Functional testing of new nuclease constructs: HEK293T culture and transfection ● TIMING 3 d
▲CRITICAL Newly generated/obtained nucleases are most optimally tested in the cell type of interest. Below, we describe testing the QCgRNA amplicon designs generated in Steps 1–4 in adherent HEK293T cells transfected using Lipofectamine 3000 as an example. However, the nuclease constructs may be delivered by any transfection method useful for HEK293T cells or alternatively, by nucleofection; delivery of nuclease constructs via nucleofection is exemplified with K562 cells in Steps 24–30. In all cases, nuclease testing may be performed without or with application of FACS (see considerations in Step 13).
7 | HEK293T culture. Culture cells in Dulbecco’s Modified Eagle Medium supplemented with 10% (vol/vol) fetal bovine serum, and 1% (vol/vol) penicillin-streptomycin (optional) at 37 ˚C and 5% CO2, and maintain as per the supplier’s instructions (ATCC). Note that FACS is performed under semi-sterile conditions and therefore, we routinely use penicillin-streptomycin in the culture medium.
8 | Preparation of cells for transfection. Seed 1×105 cells per well in 24-well plates in a volume of 0.5 ml 16–24 hours before transfection using Lipofectamine 3000 transfection according to the manufactureŕs instructions. Plan one transfection per gRNA to be tested and one mock transfection (control).
▲CRITICAL STEP Observe the above seeding density for optimal transfection efficiency.
9 | On the day of transfection, add a PCR volume containing 50 ng of QCgRNA amplicon from Step 6 and 0.5 μg Cas9–2A-FP plasmid to 25 μl serum- and penicillin/streptomycin-free culture medium in Eppendorf tubes (one tube per transfection). Then add 1 μl P3000 Reagent to each tube and mix well. Include a mock transfection using irrelevant, neutral plasmid without FP. As Cas9–2A-FP plasmid, we recommend CAS9PBKS (Addgene #68371) or similar that contains Cas9–2A-GFP, which apart from allowing FACS, also enables fluorescence microscopic assessment of transfection efficiency in Step 12.
▲CRITICAL STEP This step allows for testing of higher amounts of QCgRNA amplicons, which may sometimes increase editing efficiency.
10 | Prepare a master mix containing 1.5 μl Lipofectamine 3000 Reagent and 25 μl serum- and penicillin/streptomycin-free culture medium per transfection. Vortex briefly, then aliquot 26.5 μl of this mix into each Eppendorf tube prepared in Step 9. Incubate for 10–15 min at room temperature.
11 | Add the DNA-lipid mix from Step 10 to the cells from Step 8 and place them in the cell incubator at 37 ˚C and 5% CO2.
12 | After 2 days, estimate transfection efficiency by GFP fluorescence microscopy (typically 40–60% for above experiment).
Functional testing of new nuclease constructs: preparation of cell lysates for IDAA
13 | Determine the efficiency and indel signature of each gRNA design by IDAA directly on cells (option A) or after FACS for nuclease-expressing cells (option B), if transfection efficiencies are <20%.
(A) Preparation of cell lysates for IDAA by direct lysis ● TIMING 1 h
Aspirate medium from QCgRNA/Cas9–2A-GFP and mock-transfected cells and add 150 μl QuickExtract solution (yielding a lysate of ~3,000 cells μl−1) Transfer lysates to Eppendorf or PCR tubes and incubate in a heating block or a thermocycler for 20 min at 65 °C followed by 10 min at 98 °C.
■ PAUSE POINT Lysates can be stored at −20 °C for several months.
(B) Preparation of cell lysates for IDAA by FACS for nuclease-expressing cells ● TIMING 2 h
-
Preparation of cells for FACS. Aspirate the medium from QCgRNA/Cas9–2A-GFP and mock-transfected cells, wash 2× with PBS (carefully as HEK293T cells are loosely attached to the substratum) and incubate cells for 10 min in 250 μl 0.25% (wt/vol) trypsin-EDTA in the incubator. Triturate cells with a p500 pipette tip and check under the microscope for dissociation into single cells. Transfer cells into 5 ml of culture medium in a 15-ml Falcon tube and centrifuge at 200g for 5 min at room temperature. Aspirate the supernatant and resuspend in 0.5 ml culture medium.
▲ CRITICAL STEP It is important to obtain a single-cell suspension.
-
Pass the cell suspension through a 50-μm cup Filcon placed in a 15-ml Falcon tube to remove cell aggregates. Collect the dead-volume (~250 μl) at the lower side of the cup Filcon with a p500 pipette tip and pool with the pass-through.
▲ CRITICAL STEP Check under the microscope that cells are dissociated to single cells. Triturate with a p500 pipette tip, if necessary.
-
Place cells on ice until FACS, preferably within 1 h.
▲CRITICAL The optimal temperature to keep cells until FACS vary with cell type.
Subject cells to bulk FACS, as described in Steps 35–38, except that all the top ~80% most GFP-fluorescent cells are isolated. Note the number of isolated cells. If desired, a portion of the sorted cells may be saved for later single-cell cloning, in case high modification levels are observed in Steps 16–26.
Centrifuge the FACS-isolated cells at 200g for 5 min at room temperature and carefully aspirate the supernatant.
-
Add 30 μl QuickExtract solution to each vial of FACS-isolated cells. Note that the volume of QuickExtract solution may be adjusted according to the number of FACS-isolated cells (Step 15B iv): aim to obtain a solution containing 1000–5000 cells μl−1. Transfer lysates to Eppendorf or PCR tubes and incubate in a heating block or thermocycler for 20 min at 65 °C followed by 10 min at 98 °C.
■ PAUSE POINT Lysates can be stored at −20 °C for several months.
Functional testing of new nuclease constructs: Indel characterization by IDAA ● TIMING PCR/capillary electrophoresis 6 h; hands-on time 3 h for ≤96 samples)
14 | Setting up the IDAA PCR. Set up the following IDAA PCR reaction for each sample to be analyzed, as detailed below and using the oligos specified in Table 2 (see examples of full IDAA primer sequences in Supplementary Table 2). Include a PCR for a non-edited (wt) control sample.
| Component | Amount (μl) | Final concentration |
|---|---|---|
| Ammonium PCR buffer, 10× | 1.25 | 1× |
| dNTP, 100 mM (25 mM each) | 0.125 | 1 mM |
| IDAA Fwd primer, 2.5 μM | 0.125 | 0.025 μM |
| IDAA Rev primer, 25 μM | 0.125 | 0.25 μM |
| FamFwd primer, 25 μM | 0.125 | 0.25 μM |
| TEMPase Hot Start DNA polymerase | 0.12 | 0.6 U |
| MgCl2, 25 mM | 0.7 | 2.5 mM* |
| QuickExtract cell lysate from Step 13 | 1 | 20–20,000 cells** |
| ddH2O | 8.93 | |
| Total | 12.5 |
The PCR buffer also contains MgCl2 to give the final concentration.
Most IDAA PCRs work within this range, but see “Experimental design” for discussion. Alternatively, 1–100 ng purified genomic DNA may be used as template.
▲CRITICAL STEP Observe the 10:1:10 molar ratio of the FamFwd:Fwd:Rev primers for optimal amplicon labelling and yields.
15 | Perform PCR using the following touchdown cycling conditions:
| Cycle number | Denature | Anneal | Extend |
|---|---|---|---|
| 1 | 95 °C, 15 min | ||
| 2–16 | 95 °C, 30 s | *72 °C, 30 s | 72 °C, 30 s |
| 17–40 | 95 °C, 30 s | 58 °C, 30 s | 72 °C, 30 s |
| 41 | 72 °C, 20 min |
Decrease annealing temperature by 1 °C in each subsequent cycle.
■ PAUSE POINT PCR product may be stored in the dark at 4 °C for up to 2 weeks or at −20 °C for up to 6 months.
16 | Run 3 μl of the PCRs on a 10 cm-long 1.5% (wt/vol) agarose gel for ~45 min at 5 V/cm distance between electrodes, using CyanOrange loading dye and SYBR Safe DNA gel stain. Visualize DNA by UV irradiation. Ideally, there should be only one clear band and no smear (See example in Supplementary Fig. 2).
? TROUBLESHOOTING
17 | Prepare the IDAA PCRs for analytical run. Create a master mix of the size standard and formamide as detailed below and aliquot 10.3 μl per well in a 96-well PCR plate. Add 0.5 μl of each IDAA PCR from Step 15 to a separate well. The mixing of sample and master mix can also be performed in the reverse order. Seal the plate with MicroAmp Optical Adhesive Film.
| Component | Amount (μl) |
|---|---|
| Hi-Di Formamide | 10 |
| GS500LIZ size standard | 0.3 |
▲ CRITICAL STEP The optimal amount of IDAA PCR to analyze may vary from 0.05–1 μl, depending on the dynamic range of the fragment analyzer used, indel profile complexity and goal of analysis – see “Experimental design” for discussion and Supplementary Fig. 2 for profiles obtained with loading of a range of PCR amounts. If analyzing other than 200–450 bp IDAA PCR amplicons, choose the size standard accordingly.
18 | Denature the PCR products for 5 min at 98 °C in a heating block or a thermocycler.
19 | Spin the plate briefly to ensure that there are no air bubbles at the bottom of the wells.
20 | Load plate onto ABI Genetic analyzer 310, 3100, 3130 or 3730 sequenator and run fragment analysis according to manufacturer’s instructions.
▲CRITICAL STEP The 6-FAM fluorophore is unstable in formamide and IDAA PCRs should therefore be analyzed within a couple of days after preparation for the analytical run.
21 | Analyze data using Peak Scanner (see Supplementary Manual) or other software as appropriate (see Supplementary Note).
? TROUBLESHOOTING
22 | Directly use the best QCgRNA amplicon for the actual genome editing application. Alternatively, introduce the best gRNA design in a 2A-FP expression vector more suitable for the intended task (see 2A-FP vector list in Supplementary Table 1) or subclone the entire QCgRNA expression cassette to pEPB104 using EcoRI/KpnI restriction endonuclease sites present in both constructs (see Supplementary Figure 6).
FACS-based genome editing: delivery of nuclease-2A-FP construct into cells (optional: include donor construct)
● TIMING 1 h hands-on; 3 d incubation
▲CRITICAL The preferred nuclease-2A-FP construct format and delivery mode will vary with the cell type and genome editing application in question. To illustrate a typical example, we describe nucleofection of human leukemic K562 suspension cells with a pCas9–2A-GFP/gRNA all-in-one construct.
23 | K562 culture. Culture cells in Iscovés Modified Dulbecco’s Medium supplemented with 10% (vol/vol) fetal bovine serum and 1% (vol/vol) penicillin-streptomycin (optional) at 37 ˚C and 5% CO2, and maintain as per the supplier’s instructions (ATCC).
▲CRITICAL STEP We recommend use of penicillin-streptomycin in the culture medium, since FACS is normally performed under semi-sterile conditions.
24 | Preparation of cells for nucleofection. Two days before nucleofection, seed out cells at ~1×105 cells ml−1 (e.g. as 2 ml per well in a 6-well plate) so they will be growing exponentially at 3−5×105 cells ml−1 on the day of nucleofection. Plan ahead to have sufficient cells for the desired number of nucleofections (see Step 26).
25 | Medium preparation before nucleofection. On the day of nucleofection, add 2 ml culture medium to each well of 6-well plates (one well per nucleofection) and place them in the cell incubator at 37 ˚C and 5% CO2.
26 | Centrifuge 1.5×106 cells prepared in Step 24 per nucleofection at 200g for 5 min at room temperature in 15-ml Falcon tubes or similar. Remove supernatant completely.
▲CRITICAL STEP It may be necessary to perform several nucleofections per construct, if e.g. the goal is to isolate a sparse high-expresser cell population (to guide calculations on cell number, see Box 1). Prepare also a mock nucleofection with irrelevant, neutral plasmid containing no FP or gRNA for the FACS mock control (see Box 1) and for negative control cells in downstream analyses, if needed.
27 | Resuspend each cell pellet carefully in 100 μl room temperature Nucleofector Solution V. Do not leave the cells for longer than 15 min.
28 | Add 2 μg pCas9–2A-GFP/gRNA construct (optional: and 1–5 μg ssODN donor) to the 100 μl cell suspension and mix gently by pipetting or tapping.
29 | Transfer each cell/DNA suspension to a cuvette provided with the nucleofection kit, insert into Amaxa Nucleofector 2b Device and apply nucleofection program T-016.
30 | After nucleofection, immediately add ~500 μl of the pre-equilibrated culture medium to each cuvette, then gently transfer the cells from each cuvette into separate wells in the prepared 6-well plate (Step 25) and place it in the cell incubator at 37 ˚C and 5% CO2.
▲ CRITICAL STEP Cells are fragile at this stage and should be pipetted gently.
31 | After 2 days, GFP fluorescence microscopy can give an estimate of transfection efficiency, typically 20–40% for the above experiment. Note that the Lonza pmaxGFP transfection control plasmid supplied with the nucleofection kits generally gives much higher fluorescence/transfection levels and therefore is not useful as an additional control for transfectability of the cells.
FACS-based genome editing: Bulk sorting to establish the dose-response relationship for fluorescence level versus editing efficiency and cell survival ● TIMING 4 h hands-on; 1 week expansion
32 | Preparation of cells for FACS. At day 2–3 after nucleofection, determine the number of live, e.g. trypan blue-excluding, cells in the cultures. If more than one nucleofection per construct was performed, pool cells transfected with the same construct prior to cell counting. Then pellet the cells by centrifugation at 200g for 5 min at room temperature. Aspirate supernatant and resuspend cells in normal culture medium to obtain a suspension of ~10×106 cells ml−1. Triturate 15 times with a p500 pipette tip to obtain a single-cell suspension.
▲CRITICAL STEP Optimal cell density will vary depending on cell type: high densities (up 10−15×106 cells ml−1) reduce FACS time, but may increase cell aggregation. Check under the microscope that cells are dissociated to single cells. Phenol red in the medium gives some green background fluorescence. With the high levels of specific GFP fluorescence normally obtained, we have not found it worthwhile to shift to phenol red-free culture medium at this step, but in unusual cases of low GFP expression it may be considered.
33 | Pass the cell suspension through a 50-μm cup Filcon placed in a 15-ml Falcon tube to remove cell aggregates. Collect the dead-volume (~250 μl) at the lower side of the cup Filcon with a p500 pipette tip and pool with the pass-through.
▲CRITICAL STEP Check under the microscope that cells are dissociated to single cells.
34 | Place cells on ice until FACS, preferably within 1 h.
▲CRITICAL STEP The optimal temperature to keep cells until FACS varies with cell type.
35 | Preparation of tubes to collect sorted cells. Prepare 5-ml round-bottom Falcon tubes containing ~1 ml of normal culture medium.
36 | Set up the parameters for FACS based on the mock and nuclease-2A-GFP transfected cells, as described in Box 1.
▲CRITICAL STEP Optimal FACS parameters like nozzle size and temperature vary with cell type.
37 | Set gates to isolate 4 cell populations with non-overlapping fluorescence levels, e.g. corresponding to populations A-D in Fig. 6d. that include >30,000 cells. The number of cells present in each gate is calculated as described in Box 1.
▲CRITICAL STEP For more detailed characterization of the dose-response relationship for fluorescence level versus editing level/cell number, more distinct populations may be isolated. Optimal fluorescence levels sorted for will vary with cell type and nuclease construct.
38 | FACS isolate 20,000 cells from each gate.
39 | Centrifuge the FACS-isolated cells at 200g for 5 min at room temperature. Remove supernatant carefully, leaving ~50 μl to avoid disturbing the cell pellet. Add 400 μl culture medium, transfer each cell population to distinct wells in a 24-well plate and place it in the cell incubator at 37 ˚C and 5% CO2.
40 | At day ~4 after FACS, transfer the cells to a 12-well plate. Add an additional 500 μl culture medium to each well.
41 | At day 6 after FACS, determine the number of live cells, e.g. by counting trypan blue-excluding cells, in each of the 4 sorted cell populations.
▲CRITICAL STEP The appropriate day to evaluate cell survival and editing levels may vary with cell type and nuclease construct.
42 | Transfer 50,000 cells from each population to an Eppendorf or PCR tube, while continuing to expand the remainder of the populations. Centrifuge the 50,000 cells at 1,500g for 5 min at room temperature. Remove the supernatant and add 20 μl QuickExtract solution. Incubate lysates in a heating block or thermocycler for 10 min at 65 °C followed by 10 min at 98 °C.
■ PAUSE POINT Lysates can be stored at −20 °C for several months.
43 | Determine the level of genome editing in the 4 cell populations by subjecting the QuickExtract lysates to IDAA (Steps 14–21) (or to RFLP assay if donor was included in the experiment).
44 | Select the most optimal of the 4 FACS isolated cell populations based on the observed levels of genome editing and cell number. Expand this population using normal culture procedures and use for experimentation, for further editing (Step 45) or for clonal derivation (Step 47).
▲CRITICAL STEP Additional parameters may guide the selection of the best cell population, including mutagenesis at select off-target sites, general cell condition, etc.
45 | If the level of genome editing is not sufficient in the most optimal cell population, then expand cells and re-nucleofect with the nuclease-2A-FP construct (Steps 24–30), FACS isolate the cell population with the optimal nuclease levels, as identified in Steps 41–43, and determine the editing level by IDAA (or RFLP). Continue these cycles as needed (for an example, see Fig. 9).
Figure 9 |. Building up genome modification levels in cell pools by repeated nuclease delivery and FACS.
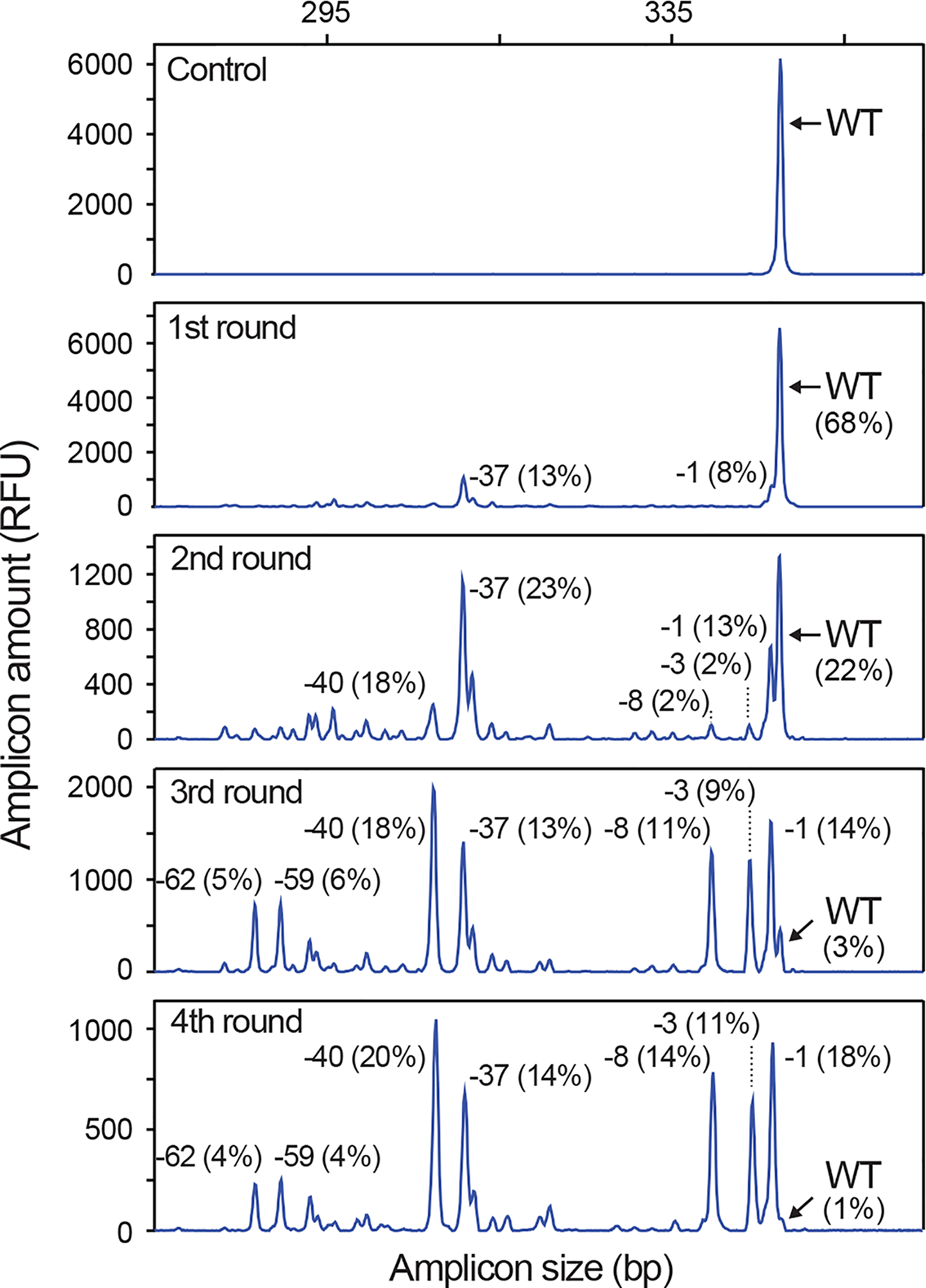
The VEGFA locus was targeted by transfection of 1.5×106 K565 cells with an all-in-one Cas9–2A-GFP construct, illustrating a challenging editing task, since the locus is quadroallelic in these cells and the particular gRNA used is not very active. Two days post-transfection, the top 10% most highly GFP fluorescent cells were isolated by FACS and cultured for 6 days, where after the procedure was repeated. After each round, IDAA was used to monitor the degree of indel editing at the VEGFA locus on an aliquot of the cell pools. Note that editing levels gradually increase and after 4 rounds, a cell pool with almost complete editing of the VEGFA locus has been achieved. The size and frequency of selected indels are indicated. IDAA was performed in an ABI 3130 instrument.
▲CRITICAL STEP To be able to extrapolate the optimal nuclease level identified in Steps 41–43 to the subsequent rounds of nuclease delivery and FACS, it is essential to use the same period of time between nucleofection and FACS as well as all FACS parameters (nozzle size, FACS temperature and voltage setting on FACS instrument – see Box 1), as was used in the first round. Thus, mock-transfected cells are not needed to determine the voltage setting for the subsequent rounds of FACS.
FACS-based genome editing: Generation of clonal cell lines ● TIMING 8 h hands-on; 2–3 weeks expansion
CRITICAL: The edited cell populations generated through QCgRNA amplicon screening (Step 13B iv) or FACS optimization of nuclease levels (Step 43) can be cloned by re-FACS based on FSc/SSc and exclusion of doublets (i.e. cell population P2 described in Box 1) and single-cell seeding into 96-well plates. Alternatively, cells may be cloned during the FP FACS analysis by sorting single cells with a desired FP fluorescence level into 96-well plates, as described below.
46 | Deliver nuclease-2A-FP and mock construct to adherent cells or suspension culture cells, as described in Steps 8–11 and 24–30, respectively, and culture for 2–3 days before FACS.
47 | Preparation of conditioned medium for single-cell cloning. Two days prior to FACS, incubate ~50% confluent adherent cells or suspension cells at ~2×105 cells ml−1 in fresh, normal culture medium. Calculate 5 ml per 96-well plate to be seeded and 20% extra for handling loss. On the day of FACS, collect the conditioned medium (suspension culture cells are removed by centrifugation at 200g for 5 min at room temperature) and pass it through a 0.22-μm filter mounted on a 50-ml syringe. Mix 1:1 with normal culture medium. Conditioned media are best when freshly prepared.
▲CRITICAL STEP The number of plates needed depends on the editing level in the cells (determined in preceding steps) and the number of clones that grow up, the latter depending on cell survival at the chosen FACS condition (estimated in Step 41), capability of cells for clonal expansion and other factors, as discussed in “Experimental design”. From these variables, the needed number of plates can be estimated. We recommend using penicillin-streptomycin in the single-cell cloning medium, since FACS is normally performed under semi-sterile conditions.
48 | Prepare 96-well plates for single-cell sorting. On the day of FACS, prepare the required number of 96-well plates (round-bottomed for suspension cells; flat-bottomed for adherent cells) by adding 100 μl of conditioned medium to each well using a multi-channel pipette.
49 | Prepare the adherent cells or the suspension culture cells for FACS as described in Steps 13B i-iii and 32–34, respectively, except that cell density of the FACS suspension can be in the lower end, e.g. ≥1×106 cells ml−1 and preferably in ≥1 ml.
50 | Set up the parameters for FACS based on the mock and nuclease-2A-GFP transfected cells, as described in Box 1.
▲CRITICAL STEP If FACS is to be performed based on previous FACS optimization of nuclease levels (Steps 41–43), all FACS parameters must be identical; use same interval between transfection and FACS, nozzle size, FACS temperature and voltage setting for the fluorescence detection system. Thus, mock- transfected cells are not needed to determine the voltage setting in this scenario.
51 | Set a gate for the desired fluorescence level to be sorted for and sort cells singly into the prepared 96-well plates.
52 | Place the plates in the cell incubator at 37 ˚C and 5% CO2 and allow cells to expand for 2–4 weeks (expansion time depends on cell type): add 100 μl normal culture medium 5 d after sorting, then change 100 μl of the medium every 5–8 d as appropriate for the cell type. For suspension cells, medium removal must be done carefully to avoid losing cells.
53 | When the cells are 30–50% confluent, prepare replica 96-well plates for passaging containing 100 μl normal culture medium per well (smaller colonies are readily seen when holding the plate up against the light and inspecting the plate from the underside). Triturate clones vigorously by pipetting up and down until clones are dissociated (check under the microscope; very adherent cells may need trypsinization for detachment). Transfer half of the resuspended cells per well to a well in the replicate plate for further expansion.
54 | Transfer the rest of the cells to 96-well PCR plates. Spin down cells at 1,500g for 5 min at room temperature. Carefully remove the supernatant in order to not disturb the cell pellet. Add 10 μl QuickExtract solution and incubate in a thermocycler for 20 min at 65 °C, followed by 10 min at 98 °C. Genotype the clones by IDAA, as described in Steps 14–21 (for an example, see Fig. 10) or by RFLP assay if knockin was the task.
Figure 10 |. Genotype screening of cell clones by IDAA.
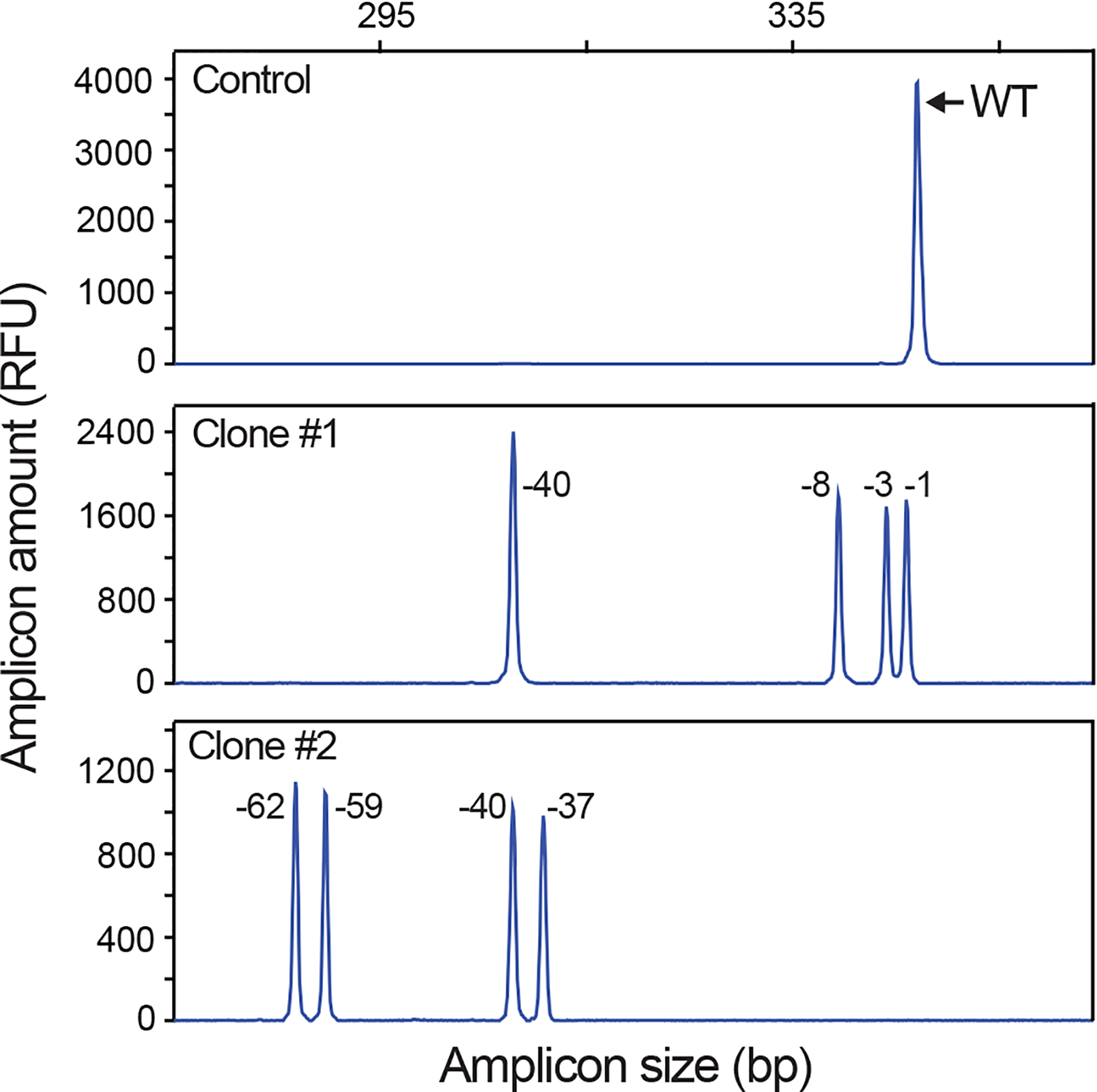
Two representative IDAA profiles at the VEGFA locus in K562 clones derived by single-cell sorting into 96-well plates at 4th round of nuclease delivery and FACS per Fig. 9. Note that IDAA provides indel information on all 4 copies of the VEGFA gene present in K562 cells and that clones with frameshift-causing indels at all copies can readily be identified, e.g. clone #2. The sizes of the indels are indicated. IDAA was performed in an ABI 3130 instrument.
■ PAUSE POINT Lysates can be stored at −20 °C for prolonged periods of time.
? TROUBLESHOOTING
Troubleshooting advice can be found in Table 3.
Table 3 |.
Troubleshooting table.
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 5 | Poor or no amplification. | Ill defined | Due to low complexity, the QCgRNA PCR is robust and has so far never failed to generate full-length product. Yields, however, may vary slightly, possibly due to QCgX primer differences. If yields are very low, purify and concentrate the QCgRNA amplicons prior to transfection. |
| 16 | Multiple bands or smear. | Non-specific priming; non-optimal template concentration. | Include 5% DMSO in the PCR; slightly increase the annealing temperature and/or titrate MgCl2 (test 0.1–2.5 mM final concentration in the PCR); titrate template concentration; in rare cases, re-design primers. |
| No or very little PCR product | Amplified region is particularly GC-rich. | Add 5% DMSO and/or 0.2 M betaine to the PCR; increase the concentration MgCl2in the PCR and/or lower the annealing temperature; Exchange TEMPase DNA polymerase with KOD DNA polymerase; in rare cases, re-design primers so as to amplify a less GC-rich region around the target site. | |
| 21 | Signal intensity is too high/saturated. | Concentration of IDAA PCR sample is too high; | Decrease sample concentration by diluting PCR products further in Step 17 before addition of the formamide/size standard mix; |
| No signal or too low signal intensity. | Too little sample injected. | Increase sample concentration by diluting sample less in Step 15. | |
| Degraded or incorrectly stored formamide. | Use fresh formamide. | ||
| Air bubble at the bottom of sample well. | Centrifuge the sample plate briefly before inserting it into the fragment analyzer. | ||
| Two peaks in control (i.e. wt) samples: a prominent wt peak and a minor −1 bp peak. | Incomplete 3´ A nucleotide addition to the IDAA amplicon by the polymerase. | Increase the final extension step in cycle 41 to 60 min (Step 20). | |
| In the rare cases, when the problem persists, add a single G nucleotide over-hang85 or a GTTTCTT “PIG tail”86 to the 5´ end of the IDAA Rev primer, which generally will promote complete 3´ A nucleotide addition. |
●TIMING
Steps 1–6, generation of QCgRNA amplicon expression cassettes: 10 h hands-on time (excluding oligo ordering)
Steps 7–12, functional testing of new nuclease constructs: HEK293T culture and transfection: 3 d
Step 13A, preparation of cell lysates for IDAA by direct lysis: 1 h
Step 13B, preparation of cell lysates for IDAA by FACS for nuclease-expressing cells: 2 h
Steps 14–22, functional testing of new nuclease constructs: Indel characterization by IDAA: primer ordering 2 d; PCR/capillary electrophoresis 6 h; hands-on time 3 h
Steps 23–31, FACS-based genome editing: delivery of nuclease-2A-FP construct into cells: 1 h hands-on; 3 d incubation
Steps 32–45, FACS-based genome editing: Bulk sorting to establish the dose-response relationship for fluorescence level versus editing efficiency and cell survival: 4 h hands-on; 1 week expansion
Steps 46–54, FACS-based genome editing: Generation of clonal cell lines: 8 h hands-on; 2–3 weeks expansion
ANTICIPATED RESULTS
This protocol (Fig. 4) can increase (Fig. 3) and monitor (Fig. 1) indel mutation frequencies throughout a cell line genome editing project. By bulk sorting for high-level nuclease expressing cells, it is often possible to obtain a pool of almost completely edited cells in the first round of nuclease delivery and FACS (Fig. 3). This may not be possible in other cases, where one or more of a variety of factors may preclude high-efficiency editing, regardless of high nuclease levels. In such cases, repeated rounds of nuclease delivery and FACS on the same cell population may be used to obtain a cell pool with almost complete editing, even when multiple copies of the targeted gene is present in the cells (Fig. 9). Experimentation with such pools of edited cells can be a powerful means to avoid the confounding effects of inherent clonal variation (i.e. clonal effects not associated with the editing) on the results. However, if clones are desirable, the protocol can also be used for FACS derivation of clones of edited cell lines (Fig. 10), and the nuclease-enrichment step greatly reduces the number of clones that need to be generated and screened. For clone screening, IDAA can determine whether an indel mutation has occurred and define indel size on all copies of the targeted gene (Fig. 10). In variant applications of the protocol, FACS for low or intermediate levels of nuclease can be used to ameliorate cell death (Fig. 3) or reduce off-target mutagenesis (Fig. 7) associated with editing.
Supplementary Material
Supplementary Table 1 | Nuclease-2A-FP expression systems.
Supplementary Table 2 | IDAA primers.
Supplementary Table 3 | Nuclease target sites.
Supplementary Note | Peak Scanner™ 2 Software and GeneMarker® (Demo) Software.
Supplementary Data | Sequences of pEPB104 and CAS9PBKS.
Supplementary Manual | Peak Scanner™ 2 Software step-by-step guide.
Supplementary Figure 1 | Sensitivity and quantitative performance of IDAA performed in an ABI 3500 instrument is comparable to Next-Generation Sequencing (MiSeq).
Supplementary Figure 2 | IDAA can generate useful indel profiles from minute amounts of IDAA PCR amplicons.
Supplementary Figure 3 | Upper and lower detection levels for IDAA amplicons in ABI 3130 and 3500 instruments.
Supplementary Figure 4 | IDAA reveals the indel signature of a given gRNA, TALEN or ZFN.
Supplementary Figure 5 | Using IDAA to estimate probability of germline transmission of indels from F0 mosaic fish and to identify F1 mutant fish in Danio rerio genome editing.
Supplementary Figure 6 | QCgRNA amplicon expression cassettes.
ACKNOWLEDGMENTS
We thank Anna Fossum (BRIC), Claire Katrine Beier Holden, Camilla Andersen, Sanae Narimatsu (Copenhagen Center for Glycomics) and Angela Dürr (the Danish Stem Cell Center) for excellent technical assistance. We thank Shondra Miller, Washington University, for critical reading of the manuscript. This work was supported by the Danish Cancer Society (R124-A7632–15-S2 to M.F.), the Danish Council for Independent Research (0602–02368B to M.F.), the University of Copenhagen Excellence Programme for Interdisciplinary Research - CDO2016 (to M.F., E.P.B., H.H.W.), the Novo Nordisk Foundation (to M.F.), the Lundbeck Foundation (R165-2013-15743 to F.N.), the National Institutes of Health (R21CA184656 to S.H.H.) and the Danish National Research Foundation (DNRF107 to E.P.B., H. H. W., Y.N., Z.Y., H.C.).
Footnotes
COMPETING FINANCIAL INTERESTS
E.P.B. declares competing financial interests (see the HTML version of this article for details).E.P.B. possesses the rights to a patent application describing the IDAA methodology.
TWEET: Increasing and evaluating genome editing efficiency by FACS enrichment & Indel Detection by Amplicon Analysis (IDAA)
References
- 1.Carroll D Genome Engineering with Targetable Nucleases. Annual Review of Biochemistry 83, 409–439 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Bae S, Kweon J, Kim HS & Kim J-S Microhomology-based choice of Cas9 nuclease target sites. Nat Meth 11, 705–706 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Urnov FD et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 435, 646–651 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Moehle EA et al. Targeted gene addition into a specified location in the human genome using designed zinc finger nucleases. Proceedings of the National Academy of Sciences 104, 3055–3060 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen F et al. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat Meth 8, 753–755 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang L et al. Optimization of scarless human stem cell genome editing. Nucleic Acids Research 41, 9049–9061 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richardson CD, Ray GJ, DeWitt MA, Curie GL & Corn JE Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotech 34, 339–344 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Renaud J-B et al. Improved Genome Editing Efficiency and Flexibility Using Modified Oligonucleotides with TALEN and CRISPR-Cas9 Nucleases. Cell Reports 14, 2263–2272 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Kim H & Kim J-S A guide to genome engineering with programmable nucleases. Nat Rev Genet 15, 321–334 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Mussolino C, Mlambo T & Cathomen T Proven and novel strategies for efficient editing of the human genome. Current Opinion in Pharmacology 24, 105–112 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Maeder ML, Thibodeau-Beganny S, Sander JD, Voytas DF & Joung JK Oligomerized pool engineering (OPEN): an ‘open-source’ protocol for making customized zinc-finger arrays. Nat. Protocols 4, 1471–1501 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim HJ, Lee HJ, Kim H, Cho SW & Kim J-S Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Research 19, 1279–1288 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sander JD, Maeder ML & Joung JK Engineering Designer Nucleases with Customized Cleavage Specificities, in Current Protocols in Molecular Biology (John Wiley & Sons, Inc., 2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Urnov FD, Rebar EJ, Holmes MC, Zhang HS & Gregory PD Genome editing with engineered zinc finger nucleases. Nat Rev Genet 11, 636–646 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Sanjana NE et al. A transcription activator-like effector toolbox for genome engineering. Nat. Protocols 7, 171–192 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reyon D et al. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotech 30, 460–465 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sander JD & Joung JK CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotech 32, 347–355 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ran FA et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protocols 8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu Patrick D., Lander Eric S. & Zhang F Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell 157, 1262–1278 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wright Addison V., Nuñez James K. & Doudna Jennifer A. Biology and Applications of CRISPR Systems: Harnessing Nature’s Toolbox for Genome Engineering. Cell 164, 29–44 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Tsai SQ & Joung JK Defining and improving the genome-wide specificities of CRISPR-Cas9 nucleases. Nat Rev Genet 17, 300–312 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Z et al. Fast and sensitive detection of indels induced by precise gene targeting. Nucleic Acids Research 43, e59 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duda K et al. High-efficiency genome editing via 2A-coupled co-expression of fluorescent proteins and zinc finger nucleases or CRISPR/Cas9 nickase pairs. Nucleic Acids Research 42, e84 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Z et al. Engineered CHO cells for production of diverse, homogeneous glycoproteins. Nat Biotech 33, 842–844 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Carrington B, Varshney GK, Burgess SM & Sood R CRISPR-STAT: an easy and reliable PCR-based method to evaluate target-specific sgRNA activity. Nucleic Acids Research 43, e157 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Certo MT et al. Tracking genome engineering outcome at individual DNA breakpoints. Nat Meth 8, 671–676 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mandal Pankaj K. et al. Efficient Ablation of Genes in Human Hematopoietic Stem and Effector Cells using CRISPR/Cas9. Cell Stem Cell 15, 643–652 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ding Q et al. A TALEN Genome-Editing System for Generating Human Stem Cell-Based Disease Models. Cell Stem Cell 12, 238–251 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim H et al. Surrogate reporters for enrichment of cells with nuclease-induced mutations. Nat Meth 8, 941–943 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Kim H et al. Magnetic Separation and Antibiotics Selection Enable Enrichment of Cells with ZFN/TALEN-Induced Mutations. PLoS ONE 8, e56476 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramakrishna S et al. Surrogate reporter-based enrichment of cells containing RNA-guided Cas9 nuclease-induced mutations. Nat Commun 5 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Tsai SQ et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotech 33, 187–197 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller JC et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotech 25, 778–785 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Yeung AT, Hattangadi D, Blakesley L & Nicolas E Enzymatic mutation detection technologies. BioTechniques 38, 749–758 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Vouillot L, Thélie A & Pollet N Comparison of T7E1 and Surveyor Mismatch Cleavage Assays to Detect Mutations Triggered by Engineered Nucleases. G3: Genes|Genomes|Genetics 5, 407–415 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oleykowski CA, Bronson Mullins CR, Godwin AK & Yeung AT Mutation detection using a novel plant endonuclease. Nucleic Acids Research 26, 4597–4602 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Till BJ, Burtner C, Comai L & Henikoff S Mismatch cleavage by single‐strand specific nucleases. Nucleic Acids Research 32, 2632–2641 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cross MJ, Waters DLE, Lee LS & Henry RJ Endonucleolytic mutation analysis by internal labeling (EMAIL). ELECTROPHORESIS 29, 1291–1301 (2008). [DOI] [PubMed] [Google Scholar]
- 39.Tsuji T & Niida Y Development of a simple and highly sensitive mutation screening system by enzyme mismatch cleavage with optimized conditions for standard laboratories. ELECTROPHORESIS 29, 1473–1483 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Qiu P et al. Mutation detection using Surveyor™ nuclease. BioTechniques 36, 702–707 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Mashal RD, Koontz J & Sklar J Detection of mutations by cleavage of DNA heteroduplexes with bacteriophage resolvases. Nature Genetics 9, 177–183 (1995). [DOI] [PubMed] [Google Scholar]
- 42.Güell M, Yang L & Church GM Genome editing assessment using CRISPR Genome Analyzer (CRISPR-GA). Bioinformatics 30, 2968–2970 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brinkman EK, Chen T, Amendola M & van Steensel B Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Research 42, e168 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mock U, Hauber I & Fehse B Digital PCR to assess gene-editing frequencies (GEF-dPCR) mediated by designer nucleases. Nat. Protocols 11, 598–615 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Orlando SJ et al. Zinc-finger nuclease-driven targeted integration into mammalian genomes using donors with limited chromosomal homology. Nucleic Acids Research 38, e152 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maresca M, Lin VG, Guo N & Yang Y Obligate Ligation-Gated Recombination (ObLiGaRe): Custom-designed nuclease-mediated targeted integration through nonhomologous end joining. Genome Research 23, 539–546 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carlson DF et al. Efficient TALEN-mediated gene knockout in livestock. Proceedings of the National Academy of Sciences 109, 17382–17387 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frank S, Skryabin BV & Greber B A modified TALEN-based system for robust generation of knock-out human pluripotent stem cell lines and disease models. BMC Genomics 14, 1–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ran FA et al. Double Nicking by RNA-Guided CRISPR Cas9 for Enhanced Genome Editing Specificity. Cell 154, 1380–1389 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mali P et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotech 31, 833–838 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shen B et al. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat Meth 11, 399–402 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Cho SW et al. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Research 24, 132–141 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim S, Kim D, Cho SW, Kim J & Kim J-S Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Research 24, 1012–1019 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zuris JA et al. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat Biotech 33, 73–80 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liang X et al. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. Journal of Biotechnology 208, 44–53 (2015). [DOI] [PubMed] [Google Scholar]
- 56.Yu X et al. Improved delivery of Cas9 protein/gRNA complexes using lipofectamine CRISPRMAX. Biotechnology Letters 38, 919–929 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hendel A et al. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat Biotech 33, 985–989 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dieffenbach CW, Lowe TM & Dveksler GS General concepts for PCR primer design. Genome Research 3, S30–S37 (1993). [DOI] [PubMed] [Google Scholar]
- 59.Kibbe WA OligoCalc: an online oligonucleotide properties calculator. Nucleic Acids Research 35, W43–W46 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Korbie DJ & Mattick JS Touchdown PCR for increased specificity and sensitivity in PCR amplification. Nat. Protocols 3, 1452–1456 (2008). [DOI] [PubMed] [Google Scholar]
- 61.Hsu PD et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotech 31, 827–832 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Frock RL et al. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat Biotech 33, 179–186 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang X et al. Unbiased detection of off-target cleavage by CRISPR-Cas9 and TALENs using integrase-defective lentiviral vectors. Nat Biotech 33, 175–178 (2015). [DOI] [PubMed] [Google Scholar]
- 64.Fu Y, Sander JD, Reyon D, Cascio VM & Joung JK Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotech 32, 279–284 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim D et al. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Meth 12, 237–243 (2015). [DOI] [PubMed] [Google Scholar]
- 66.Tsai SQ et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotech 32, 569–576 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guilinger JP, Thompson DB & Liu DR Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotech 32, 577–582 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kleinstiver BP et al. High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Slaymaker IM et al. Rationally engineered Cas9 nucleases with improved specificity. Science 351, 84–88 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Muller M et al. Streptococcus thermophilus CRISPR-Cas9 Systems Enable Specific Editing of the Human Genome. Mol Ther 24, 636–644 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bolukbasi MF, Gupta A & Wolfe SA Creating and evaluating accurate CRISPR-Cas9 scalpels for genomic surgery. Nat Meth 13, 41–50 (2016). [DOI] [PubMed] [Google Scholar]
- 72.Koo T, Lee J & Kim J-S Measuring and Reducing Off-Target Activities of Programmable Nucleases Including CRISPR-Cas9. Molecules and Cells 38, 475–481 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Doyon Y et al. Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat Meth 8, 74–79 (2011). [DOI] [PubMed] [Google Scholar]
- 74.Nakade S et al. Microhomology-mediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9. Nat Commun 5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sakuma T, Nakade S, Sakane Y, Suzuki K-IT & Yamamoto T MMEJ-assisted gene knock-in using TALENs and CRISPR-Cas9 with the PITCh systems. Nat. Protocols 11, 118–133 (2016). [DOI] [PubMed] [Google Scholar]
- 76.Yoshimi K et al. ssODN-mediated knock-in with CRISPR-Cas for large genomic regions in zygotes. Nat Commun 7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chu VT et al. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotech 33, 543–548 (2015). [DOI] [PubMed] [Google Scholar]
- 78.Park J, Bae S & Kim J-S Cas-Designer: a web-based tool for choice of CRISPR-Cas9 target sites. Bioinformatics 31, 4014–4016 (2015). [DOI] [PubMed] [Google Scholar]
- 79.Sander JD et al. ZiFiT (Zinc Finger Targeter): an updated zinc finger engineering tool. Nucleic Acids Research 38, W462–W468 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Heigwer F, Kerr G & Boutros M E-CRISP: fast CRISPR target site identification. Nat Meth 11, 122–123 (2014). [DOI] [PubMed] [Google Scholar]
- 81.Hodgkins A et al. WGE: a CRISPR database for genome engineering. Bioinformatics 31, 3078–3080 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Moreno-Mateos MA et al. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat Meth 12, 982–988 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Singh R, Kuscu C, Quinlan A, Qi Y & Adli M Cas9-chromatin binding information enables more accurate CRISPR off-target prediction. Nucleic Acids Research 43, e118 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lee CM, Cradick TJ, Fine EJ & Bao G Nuclease Target Site Selection for Maximizing On-target Activity and Minimizing Off-target Effects in Genome Editing. Mol Ther 24, 475–487 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Magnuson VL et al. Substrate nucleotide-determined non-templated addition of adenine by Taq DNA polymerase: implications for PCR-based genotyping and cloning. BioTechniques 21, 700–709 (1996). [DOI] [PubMed] [Google Scholar]
- 86.Brownstein MJ, Carpten JD & Smith JR Modulation of non-templated nucleotide addition by Taq DNA polymerase: primer modifications that facilitate genotyping. BioTechniques 20, 1004–1006 (1996). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1 | Nuclease-2A-FP expression systems.
Supplementary Table 2 | IDAA primers.
Supplementary Table 3 | Nuclease target sites.
Supplementary Note | Peak Scanner™ 2 Software and GeneMarker® (Demo) Software.
Supplementary Data | Sequences of pEPB104 and CAS9PBKS.
Supplementary Manual | Peak Scanner™ 2 Software step-by-step guide.
Supplementary Figure 1 | Sensitivity and quantitative performance of IDAA performed in an ABI 3500 instrument is comparable to Next-Generation Sequencing (MiSeq).
Supplementary Figure 2 | IDAA can generate useful indel profiles from minute amounts of IDAA PCR amplicons.
Supplementary Figure 3 | Upper and lower detection levels for IDAA amplicons in ABI 3130 and 3500 instruments.
Supplementary Figure 4 | IDAA reveals the indel signature of a given gRNA, TALEN or ZFN.
Supplementary Figure 5 | Using IDAA to estimate probability of germline transmission of indels from F0 mosaic fish and to identify F1 mutant fish in Danio rerio genome editing.
Supplementary Figure 6 | QCgRNA amplicon expression cassettes.


