Abstract
Processes to accomplish this useful solution include biphasic equilibrium (chromatographies, extractions), mechanical, bulk property, chemical equilibria, and molecular recognition. Ultimately, the goal of all of these is to physically remove all non-like protein molecules—to the finest detail: all atoms in the full three dimensional structure being identical down the chemical bond and bulk structure chirality. One strategy which has not been effectively pursued is exploiting the higher order subtle electrical properties of the protein-solvent system. The advent of microfluidic systems has enabled the use of very high electric fields and well defined gradients such that extremely high resolution separations of protein mixtures are possible. These advances and recognition of these capabilities has caused a re-evaluation of the underlying theoretical models and they were found to be inadequate. New theoretical descriptions are being considered which align more closely to the total forces present and the subtlety of differences between similar proteins. These are focused on the interfacial area between the protein and hydrating solvent molecules, as opposed to the macroscale assumptions of homogeneous solutions and particles. This critical review examines all data which has been published that place proteins in electric field gradients which induce collection of those proteins, demonstrating a force greater than dispersive effects or countering forces. Evolving theoretical constructs are presented and discussed, and a general estimate of future capabilities using the higher order effects and the high fields and precise gradients of microfluidic systems is discussed.
Keywords: dielectrophoresis, proteins, electrostatics
Graphical abstract
Introduction
There are any number of basic research needs and clinical applications where a pure, isolated and concentrated solution of a specific protein are required. High information content techniques like mass spectrometry, electron microscopy and nuclear magnetic resonance are utterly dependent upon samples with very rigorous requirements. These samples must be within a set concentration range, have limited interfering proteins—often several orders of magnitude difference between the target and other species—and the bulk solution properties must match the technique used. Further, lower information content bioanalytical techniques, like immunoassays or spectroscopies, always function better with a purified sample and, generally, with an increased concentration. To obtain these specific solutions, efforts have been expended on developing separations techniques to match the complex and difficult problem of removing just one protein from the intensely complex samples typically encountered. In these samples, there may be millions of proteins spanning eight orders of concentration range. Techniques include biphasic equilibrium (chromatographies, extractions), mechanical, bulk property (density), chemical equilibria (pH, ion-specific precipitation/solubilization), and molecular recognition (immunocapture, SELEX, imprinted polymers), where these are used in creative multistep benchtop processes.
Electric field based separations have also played a prominent role, where they exploited the first order or monopole effects (total charge state, Coulombic, electrophoresis) to induce differential velocity, drive the targets through porous media or provide a restorative force in altered charge state chemical equilibria (isoelectric focusing). Beyond the forces generated by the charge of the proteins interacting with an electric field are the higher order native and induced moments. The most prominent work has been done on induced polarization or dielectrophoresis which is studied by dielectric spectroscopy [1–5]. In this technique the conductivity and permittivity of concentrated pure solutions are probed. These studies provide fundamental mechanistic information but do not attempt to change the location or concentration of the protein. Directly related, but not coupled with these studies is the generation of a force based on these higher order moments. These forces can be effectively created and exploited on the micro-and nanoscale to selectively move proteins based on their inherent native properties (Figure 1). Further, when precisely coupled with counter forces (flow, electrophoresis), highly efficient separations may be obtained to the level of 1:108.[6] For comparison, the highest resolution mass spectrometers obtain mass/charge resolution on the order of 1:106. These higher order moments reflect all of the specific and subtle changes in the properties of the protein dissolved in its native state in condensed phase. This review comes at a time of re-evaluation of the capabilities of manipulating proteins using these non-linear electric fields, as the evidence is mounting that the classic theory applied to dielectrophoresis is not adequate to describe the protein-solvent-electric field system and new theoretical approaches are being examined which more closely match the data published.
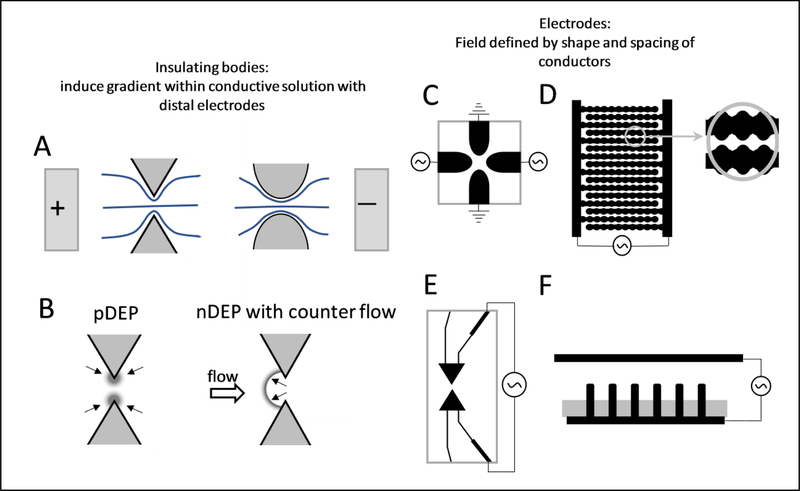
Figure 1.
Schematic representations of various forms of dielectrophoresis experimental platforms used to manipulate proteins. A) Insulator-based dielectrophoresis where the electric field gradient (represented by the relative closeness of the electric field lines (blue)) is induced by constricting the conductive solution using structural elements (of a variety of shapes and spacings, two general classes shown) and the electrodes are at a distance. See Table 1, section ‘iDEP’ for specific realizations. B) Insulator-based system can induce positive (pDEP) and negative (nDEP) dielectrophoresis, where pDEP (left) causes the proteins to be attracted to the higher electric field gradient and nDEP (right) to the lower areas. The nDEP is commonly set to oppose one or more forces (flow, Coulombic). C) Quadrupole electrode-based (eDEP) system, where pDEP induces movement to the electrodes (black) and nDEP towards the center of the structure within the solution.[9] D) Sinusoidally corrugated electrodes used in the original 1994 eDEP work led by Washizu.[7] E) Highly refined, small radius of curvature electrodes used for capture of single molecules in the works of Hölzel.[11] F) Nanometer sized array of electrodes used to capture protein in the work of Laux from the Hölzel laboratory.[55]
It is a well-accepted paradigm, repeated in numerous studies, that small particles and molecules are difficult to influence with dielectrophoretic force with respect to overcoming diffusive effects.[7–14] Accordingly, there have only been only sporadic efforts to observe and quantify the behavior of proteins using dielectrophoretic (DEP) approaches (Table 1).[10, 15] However, experimental evidence showing DEP forces dominating diffusion for proteins such that localized concentrated boluses are formed (termed capture or trapping) reaches back nearly thirty years, to 1994 with the work of Washizu (Figures 1D and 2A).[7] Working on a foundation of successful manipulation of DNA polymers [16], they unequivocally showed trapping of avidin (68 kDa), concanavalin A (52 kDa), chymotripsinogen A (25 kDa), and ribonuclease A (13.7 kDa) using an interdigitated array of sinusoidally corrugated electrodes invoking electrode-based DEP (eDEP)(1 kHz to 1 MHz, 4, 15 and 55 micron gap, up to 7 Vp-p). Very interestingly, in discussing the mechanism which enabled capture, the authors avoid speculating and noted that, “…we do not go further into biophysical discussions here…”. Noting this work and others, it was observed that dielectrophoretic forces appear to create movement at values well below the “…simplistic diffusion [force] argument of Pohl [17].” for macromolecules (nominal 10−7 m radius).[15]
TABLE 1:
List of proteins captured or trapped with DEP forces
| DEP Type and protein | MW (kDa) | †Estimated (V2/m3) | Frequency (MHz) |
cite |
|---|---|---|---|---|
| eDEP | ||||
| ribonuclease A (RNase A) | 13.7 | 1018 | 0.1–1 | [7] |
| chymotripsinogen A | 25 | “ | “ | [7] |
| concanavalin A | 52 | “ | “ | [7] |
| avidin | 68 | “ | “ | [7] |
| “ | “ | 1019 | 1–20 | [18] |
| insulin | 6 | 1018 | 1 | [8] |
| bovine serum albumin (BSA) | 66 | “ | “ | |
| IgM | 900 | “ | “ | |
| BSA | 66 | 1018 | 0.01–30 | [9] |
| R-phycoerythrin | 250 | 1021 | 1 | [11] |
| avidin | 68 | indeterminant | 0.8 | [43] |
| BSA | 66 | 1019 | 1 | [20] |
| streptavidin | 55 | indeterminant | 1 | [22] |
| lectin protein | 120 | 1019 | 0.1 | [36] |
| BSA | 66 | “ | “ | [36] |
| fibrinogen | 340 | “ | “ | [36] |
| R-phycoerythrin | 250 | 1015 | 0.1–0.5 | [25] |
| Horseradish peroxidase | 44 | 1013 | 0.01 | [26] |
| R-phycoerythrin | 250 | 1024 | 1 | [27] |
| BSA | 66 | 1018 | 2.5 | [30] |
| BSA | 66 | 1021 | 0.01 | [55] |
| cardiac troponin I | 24 | 1014 | 0.001–1 | [41] |
| e-green fluor. protein | 36 | 1020 | 0.1 | [32] |
| avidin | 68 | indeterminant | 0.01 | [42] |
| iDEP | ||||
| protein G | 22 | 1021 | DC | [19] |
| immunoglobulin G (IgG) | 150 | “ | “ | [19] |
| yellow fluor. protein | 24 | 1020 | DC | [21] |
| BSA | 66 | 1012 | DC | [35] |
| BSA | 66 | 1018 | DC | [12] |
| A-beta fibrils | Large | 1016 | DC | [47] |
| PEGlyted RNase A | 13.7+ | 1019 | DC | [40] |
| b-galactosidase | 520 | 1018 | DC | [29] |
| IgG | 150 | “ | “ | [29] |
| streptavidin | 55 | 1020 | 1 | [14] |
| streptavidin | 55 | 1021 | 1 | [13] |
| goat anti-human IgG | 150 | “ | “ | [13] |
| Streptavidin | 55 | 1020 | 0.1 | [23] |
| neuropeptide Y | 1.4 | 1021 | 3 | [24] |
| orexin A | 3.5 | “ | “ | [24] |
| strepavidin | 55 | 1021 | 10−5 | [31] |
| phycoerythrin | 240 | 1022 | 10−5 | [31] |
| BSA | 66 | 1018 | 0.1 | [28] |
| prostate specific antigen | 32 | 1019 | 1–6 | [33] |
| Anti-mouse immunoglobulin antibodies |
150 | “ | “ | [33] |
| BSA | 66 | 1023 | 0.001 | [34] |
Bolded value quoted from cite, others are estimated minimum values from characteristic distance (gap for eDEP, constriction ratio iDEP) and the applied electric field.
Figure 2.
Seminal dielectrophoretic collections of proteins. A) Original DEP trapping data for protein (fluorescently labeled avidin) using interdigitated sinusoidally corrugated electrodes (Figure 1D). Proteins (avidin and chymotripsinogen) were shown to be reversibly collected (© 1994 by the Institute of Electrical and Electronics Engineers, reproduced with permission).[7] B) Using a nanopipette tip as a gradient inducing structure, labeled protein G is captured exactly at the tip of the entrance (white spot in sub-image ‘A’ in original figure letter label). The protein is highly localized in the first micron of the pipette tip (‘E’ in original figure letter label) (© Wiley-VCH Verlag GmbH & Co. KGaA. Reproduced with permission).[19] C) Single molecule trapping at sharp nanoelectrodes using dielectrophoretic forces. R-phycoerythrin is held at the strongest gradient near the tip of electrodes spaced 0.5 μm apart (© 2005 by the American Physical Society. Reprinted figure with permission from [11]). D) Fluorescently labeled bovine serum albumin captures in an array of insulating posts at applied global potentials of (letter labeled from original cite) a: 700, b:1000, c: 1600 and d: 900 V/m (© Elsevier 2008, with permission from Elsevier, adapted from [35]).
A variety of works followed which can be grouped in three categories: 1) creating very high gradients and fields, consistent with values estimated by the Clausius-Mossotti factor (CMf) that would be required to overcome diffusion of proteins [9, 11–14, 18–34], 2) applied projects where DEP effects were observed, but the underlying theoretical limitations were not addressed [35–42], and 3) streaming or capture at mild gradients (for large protein structures and aggregates)[8, 43–47].
Pertinent reviews and position papers have been written which contain valuable perspectives of dielectrophoresis in general with varying degrees of attention being paid to proteins.[48–53]
High field/gradient works
Following and noting the work of Washizu, Morgan’s group showed images consistent with positive (pDEP) and negative (nDEP) dielectrophoresis (9 MHz crossover frequency) for avidin using eDEP at quadruple electrodes and a discussion of DEP for proteins followed in chapter form.[18, 54] Somewhat later, Burke’s group was investigating circuit assembly and showed bovine serum albumin (BSA) (a very popular choice, Table 1), could be trapped by positive dielectrophoresis at 30 MHz across five to fifteen micron gaps using quadrupole eDEP (Figure 1C).[9] Alternatively, proteins were captured by ‘pinching’ the electric field between insulators to induce the needed gradient using a glass nanopipette tip in the landmark work of Clarke et al. (Figures 1A, 1B and 2B). [19] They probed Alexa-488 labeled protein G and immunoglobulin G captured near and in a nanopipette tip approximately 50 nm internal diameter, showing a trapped zone of protein approximately one micron in length. They note, “There seems to be no theoretical basis for this observation…and…there…appears to be a significant contribution from the protein…” and speculate that the forces may be due to surrounding solution organization rather than from the protein structure itself. They followed up these proteins with yellow fluorescence protein with similar results, using them to calculate a classic theory-based surface conductivity.[21]
Accurate and precise nanoscale electrode fabrication allowed for large gradients to be induced, such that DEP forces exceeding those calculated by established theory could be induced, generating a factor exceeding 1021 V2/m3, resulting in a calculated force on R-phycoerythrin (RPE) molecule of 0.1 pN (Figures 1E and 2C).[11] Interestingly, in determining the needed force to overcome diffusion, it was estimated that only 3 × 10−16 N/molecule would be needed, showing a graphic indicating a large volume relative to the tip-gap zone about the electrode structures should induce capture (see Figure 6 in cite). A surface conductance and electric double layer structure argument was noted to suggest DEP forces are larger than expected, even though the experimental data suggest fewer molecules were captured than could be expected. Other works showed evidence of immobilization of fluorescently labeled BSA at nanoelectrodes, retention of enzymatic activity for immobilized horseradish peroxidase and quantum dot-labeled streptavidin at carbon nanotubules.[20, 22]
Streptavidin was used to probe a nanoconstriction (50 nm) device, resulting in highly non-linear and dynamic 2D spatially diverse distributions of the protein as a result of the interplay of several forces near the nano gap (Figure 3A).[14] For the DEP related components of these interactions, classic theory was invoked.
Figure 3.
Sub-micron gap insulators, frequency-based differentiation and nanoscale protein collections using dielectrophoresis. A) Collection of streptavidin near nanoconstriction (50 nm) using DC offset and 1 MHz field showing collection of the protein and complex dynamic behaviors near the entrance (© Wiley-VCH Verlag GmbH & Co. KGaA. Reproduced with permission from [14]). B) Paired nanogaps of varying width (100 – 500 nm) between micronscale triangle insulators showing collections of β-galactosidase at varying applied potentials (Reproduced from Ref. [29] with permission from the Royal Society of Chemistry.). C) Frequency-dependent differential collection of separate proteins, enrichment of prostate specific antigen versus anti-mouse immunoglobulin G at 0.8–5 MHz AC-and 1.5 V DC-applied voltage (Reproduced from Ref. [33] with permission from the Royal Society of Chemistry.). D) Bovine serum albumin collected near a nanogap (the diameter of a double stranded DNA molecule, about 2 nm) insulating body constriction at various potentials (© IOP Publishing. Reproduced with permission. All rights reserved).[34] Images shown here are for illustrative purposes: to assess the details of these works, the reader is referred to the original reference material.
A similar structure was used to create ‘molecular dams’, where AC electrokinetic forces were used to concentrate streptavidin, invoking similar physical forces.[13] Others followed with similar structures fabricated via more economic means, probing the system with streptavidin and phycoerythrin.[31] Streptavidin was also used as a probe of positive (pDEP) and negative (nDEP) dielectrophoresis (Figures 1A and 1B) in the presence of varying amounts of electrothermal flow using physiologic buffers. Much smaller molecular targets were tested in a biosensing-oriented study where frequency dependent capture behaviors were observed for neuropeptide Y.[24] β-galactosidase and micellar IgG in nanoconstriction device under DC conditions resulted in an intensely complex, voltage dependent patterning of concentration behaviors (Figure 3B).[29]
Frequency was used at a nanoconstriction to differentiate two proteins, prostate specific antigen (PSA, 150 kDa) and anti-mouse immunoglobin antibody (150 kDa), where pDEP and nDEP were observed (Figure 3C). The physical basis for the differentiation was qualitatively attributed to differences in surface conductance. Significant asymmetries and non-linear concentration distribution near the constriction were not discussed.[33] An array of electrodes fabricated at the nanoscale was able to capture, RPE, IgG antibodies, and BSA (Figure 1F), while showing retention of biological activity for RPE and horseradish peroxidase.[25, 26, 55] Gold nanohole arrays used for plasmonic sensing [28] and, separately, nanotips were exploited to concentrate BSA using DEP forces.[30] Very closely spaced conductors, forming a nanoscale gap, gave enhanced spectroscopic performance in addition to collection of R-phycoerythrin at very high values, 1024 V2/m3[27] and aligned enhanced green fluorescent protein was generated on an interdigitated array of parallel electrodes.[32] In another nanoconstriction study, where the narrow channel was formed via gold coated DNA nanowire negative impression, fluorescein isothiocyanate conjugated BSA was trapped by a nDEP mechanism using DC offset AC DEP (Figure 3D).[34] The fabrication difficulties and the appearance of spatially and temporally dynamic non-linear processes (concentration polarization, electrothermal effects, local electroosmotic effects, etc.) of the high gradient/field systems has hampered their adoption.
Experimental results without note of theoretical limitations
Washizu’s original works from 1994 is included in this category (Figures 1D and 2A), [7] along with the work of Morgan’s group.[18] Multipost insulator based dielectrophoresis was used to trap BSA under constant voltage conditions by the Lapizco-Encinas research group (Figures 1A, 1B and 2D).[35] pHs of 8 and 9 were used with buffers of 25–100 μS/cm conductivity. The mechanism of capture, again, remained elusive and was not quantitatively discussed, although they noted that the trends of the CMf were consistent with the results. Some aggregates were observed, and it was speculated that these larger particles aided in capture. Among others, BSA was also a test probe for a droplet (liquid-gas) DEP system where an asymmetric concentration profile was induced across the fluid by eDEP forces.[36] In this study, the other proteins were lectin protein (120 kDa) and fibrinogen (340 kDa). The concentration changes were modest (~2:1), induced by a 34 Vrms 100 kHz voltage across a 25 micron eDEP gap. No discussion of the mechanism of influence was presented.
A functional exploitation of nDEP and pDEP on an eDEP system was shown by Gong, increasing sensitivity for a biosensor sensitive to PSA (33 kDa), where the mechanisms were not discussed in detail and much of the improved signal most likely resulted from non-selective AC electroosmotic transport.[37] An insulator-based DEP device fabricated to high resolution with ion beam techniques was able to trap BSA.[12] An example with finite value to the current discussion shows DEP of PSA and A Beta 42 for a sensor application [38] and, in a separate study, cardiac troponin I was sensed.(41) A silver nanoparticle dendritic growth electrode scheme was used to generate surfaced enhance Raman spectra from DEP collected avidin.[42]
A more mechanistic study on BSA using a label-free, impedance-based strategy showed at least two dispersions as a function of frequency using eDEP quadrupole electrodes. The authors note that there was a belief that nanoparticles and proteins could not be manipulated by DEP, without further comment.[39]
Streaming and mild field/gradient works
An attractive force was demonstrated in a field flow fractionation system for insulin, BSA and IgM [8], and variances in concentration distribution were noted for avidin on ‘Zipper’ electrodes.[43] Streaming, but not trapping, was observed for immunoglobin G (IgG) on an insulator-based dielectrophoresis device within a study aimed at monitoring the pH of the buffer filling the device and no discussion of mechanism of influence was offered.[44] Two common targets, BSA and IgG were tested in a shaped post array iDEP device by the Ros group and streaming was observed, noting that the effects were stronger than the CMf forces indicate.[45] The Ros group followed up with further studies on IgG where streaming was observed. Their treatment of the theory and literature was thorough and complete, but like other researchers, little foundational support for the observed behaviors could be identified.[46] Mata-Gomez and co-workers subjected PEGylated Ribonuclease A to dielectrophoresis and observed streaming and some capture along with behaviors consistent with concentration polarization.[40]
Fibrils associated with Alzheimer’s disease, along with A-beta monomer, were studied by the Hayes group. The fibrils were well-captured, and the monomers could only be streamed. These behaviors were interpreted using traditional theoretical constructs.[47]
Theory
The theoretical constructs underlying dielectrophoresis for particles larger than approximately one micron have been shown to be quite robust.[53] They have been incredibly valuable and accurate for a wide range of particles.[17, 48, 56–60] An excellent summary and description of this foundational theoretical work can be found in the first section of Pethig’s recent position paper.[10]
These theoretical constructs assumed a homogeneous particle was placed into a homogeneous solution with an infinitesimally thin interface between the two. These constructs modeled the homogeneous materials to extend all the way to the interface, and the boundary conditions set the potential, current density and flux to be continuous across that interface (Figure 4A).[10]
Figure 4.
Representations of theoretical approaches used to describe dielectrophoresis. A) Classic Clausius-Mossotti approach where a homogeneous medium contacts a homogeneous particle and the properties are continuous across the interface (adapted from [10]) (© Wiley-VCH Verlag GmbH & Co. KGaA. Reproduced with permission). This approach typically assigns less than 10−19 N of force for common experimental conditions for proteins. B) Alternative theoretical approach which includes the permanent dipole moment and the discreet properties of the polarized interface in a polar solvent (adapted from [64] © American Institute of Physics. Reproduced with permission of AIP Publishing), giving approximately 10−15 N-range forces. C) A completely different approach based on calculating the solvation free energy in the presence of an electric field gradient specific to individual proteins (personal communication [72], reproduced with permission; manuscript in preparation). For the proteins examined, the forces are in the range of 10−16 N for common experimental conditions.
The constructs predicted that the force generated would be small compared to the Brownian motion, or so-called force of diffusion, under anything but the most extreme experimental conditions.[9, 11] In this context, other forces besides diffusion have been considered and Coulombic force (including electrophoresis and electroosmosis), concentration polarization, and electrohydrodynamic and electrothermal effects [40, 61–63] are significant under some experimental constructs.[15]
The processes within the interfacial region and more subtle aspects of proteins in polar solvents (their inherent flexibility on the molecular scale, for instance) are now being included in theories describing smaller bioparticles in the presence of an external electric field gradient.[10, 64–66] These new approaches are founded on ion solvation asymmetry, spontaneous polarization of the protein-solvent interface, and the dipole moment of the protein. From these works, two major features are now supported by analytical theory: the forces imparted on proteins are much larger than previously thought and the very modest changes to the structure at the molecular scale will definitively alter those forces. These forces are predicted to dominate diffusion force and more effectively compete with other induced forces. The complexity and interplay of the biomolecules with its surrounding buffer have been recognized and discussed.[21, 67]
The new approaches recognize that molecular scale properties and events are not necessarily reflected in traditional bulk property measurements of permittivity and conductivity. Specifically, the spontaneous and asymmetric polarization of the interfacial region requires introduction of an independent parameter (∈int interface dielectric constant, in [64]) that is reflected by the interaction of molecular dipole moment, M0, and the solution permittivity, ∈ (Figure 4B). According to this new construct, the dielectrophoretic force (fDEP) can be described as (from [64], converted to SI units):
| (1) |
where χc is the Lorentz cavity susceptibility, β is the reciprocal of the thermodynamic temperature (1/kBT, kB is the Boltzmann constant and T is the experimental temperature), and E is the electric field. This relationship can be directly compared to the traditional Clausius-Mossotti approach (Figure 4A):
| (2) |
where R is the radius of the particle and CMf* is the classic Clausius-Mossotti factor (in its simplest form, a sphere, is equal to is the complex permittivity of the particle and the medium, respectively) (abbreviated as CMf hereafter). It is quite notable that eqn. 1 does not have a direct dependence on the radius of the particle, although the dipole moment is a function of particle volume. This contrasts markedly with eqn. 2, which predicts a severely reduced force for small values of R. Further, the molecular dipole moment is quadratic and will have a powerful influence on the overall force generated. This derivation arrives at this rather simple result by examining the cross correlation of the dipole moment of the particle and local solvent molecules using a simplification via mean-field theories of dielectrics by introducing the concept of the ‘cavity field’ (see ‘Dipole in Solution’, section IV in [64]).
Another approach which includes interfacial features to calculate dielectrophoretic forces is massive molecular dynamics simulations.[65, 68–71] These are focused on the solvation free energy which also can be used to determine the force on a solvated particle in an electric field gradient (Figure 4C).[72] While a completely different approach, it focuses on adding similar features to the model (complex interfacial phenomena), and it also predicts a much larger dielectrophoretic force than CMf-based theory. No overarching equation can emerge from this approach, as it provides highly detailed localized information that is summed. From these localized interactions, very subtle changes between two proteins may be studied theoretically, while providing an estimated force which can be experimentally determined.
Discussion
It is completely clear in looking at the published data that proteins can be effectively controlled using dielectrophoresis in a variety of formats and conditions. These results show that dielectrophoresis induces effects that far outpace diffusion and countering forces. For particles somewhat larger than proteins (100s of nanometers), Ramos, Morgan, Green and Castellanos thoroughly and quantitatively discussed the various forces for eDEP systems back in 1998.[15] Comparing experimental results with CMf calculations and classic diffusion theory, the opposing forces were understood to be on the same order of magnitude for these somewhat larger particles.
While the data for protein capture by DEP has been sparse, there has been a long period of time to build up data. From these data, the gradients and the corresponding factor shows an extremely large range of values, from 1012 to 1023 V2/m3 (Table 1), the molecular weights include 1 to 500 kDa and the frequency was varied from DC to 10s of megahertz. Both iDEP and eDEP showed strong effects, and pDEP and nDEP were observed. Among these data, some highly preliminary queries may be posed, such as molecular weight (as a proxy for diameter) versus the magnitude of the , where there is no correlation (R2=0.0004). Given the range of experimental systems and specific device and solution details, this lack of correlation is hardly surprising. However, given the vast range for the magnitude of forces (seven orders of magnitude) some relationship may have been expected to be observed. Overall, there is not enough organized and relatable data available to assess trends or validity of any particular theory versus another. The protein BSA (Table 1) was studied in six different works, at DC to 30 MHz, using eDEP and iDEP and demonstrated trapping at values ranging from 1012 to 1024, as example of the diversity of results. As noted, the available conclusion is that proteins respond to dielectrophoretic forces to a greater extent than diffusion or other forces present, and this fact alone is counter to prevailing historical theory.
As there is a realization that native proteins in the condensed phase are a much more accessible target for accurate, precise and selective manipulation at the micro-and nano-scale, several capabilities are enabled. The separatory process takes place over a several-microns or less distance, allowing multiple steps to be performed in a tiny footprint. These processes are independent of each other and may be performed in parallel or in series, or any combination thereof. The actual separatory process occurs over a very short time period, the protein either is trapped or passes through the zone in a matter of seconds or less. This enables the probing (separation and concentration) of relatively fast events (protein binding, equilibria and kinetics, conformational states, etc.). A secondary limiting factor will be overall transport rates, but as devices become more compact and detection elements are made more intimate, these issues will be diminished. The exploitation of the higher order moments and interfacial subtleties of the protein-solvent-electric field for separations provides a very sensitive differentiation, and promises very high fidelity capabilities (supported by single amino acid substitution separations based on the monopole moment and resolution predicted at 1:108 [6]). These higher order moments probe a unique set of properties of the proteins compared to classic separations schemes and are more universal in that internal structures contribute to the forces generated. Taken together, it is reasonable to envision reverse engineered separations on the microscale, where the requirements of the detection element or clinical need are established and the separation system designed around those needs.[73]
Conclusion
Recent efforts to update theoretical constructs for dielectrophoretic forces on proteins in polar solvents has initiated a re-examination of pertinent experimental data. These data were generated in a large variety of formats and buffers, preventing an organized interrogation towards comparing and contrasting the proposed theories. However, an overarching result is that the force on proteins is larger than predicted using the CMf approach. New approaches, which include the permanent dipole and interfacial polarizability and other subtle interfacial features, appear to match the observed experimental forces. These new theories provide for much more nuanced differentiations of similar protein structures compared to previous constructs and an improved capability for correlating those molecular structural differences into changes in the force present.
Acknowledgements
Dmitry Matyushov and Yameng Liu are acknowledged for their insightful and valuable discussions, and Alex Ramirez for proofreading. The referees are also acknowledged for their valuable comments. This work was supported by URI ASU Innovation Hub Collaborative Research Seed Grant and National Institutes of Health grant 5R03AI133397–02.
Abbreviations
- (DEP)
dielectrophoresis
- (iDEP)
insulator-based dielectrophoresis
- (eDEP)
electrode-based dielectrophoresis
- (pDEP)
positive dielectrophoresis
- (nDEP)
negative dielectrophoresis
- (BSA)
bovine serum albumin
- (CMf)
Clausius-Mossotti factor
- ,
dielectrophoretic gradient factor
Biography

Mark A. Hayes is Professor in the School of Molecular Sciences at Arizona State University in Tempe, Arizona (USA) and W.W. Clyde Visiting Chair, College of Engineering, University of Utah in Salt Lake City, Utah (USA). His current work is focused on ultrahigh resolution separation and concentration of biomolecules, bioparticles and cells using microfluidic approaches, commonly employing electric fields.
Footnotes
Conflict of Interest
The author declares a conflict of interest with regards to Charlot Biosciences.
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
References:
- 1.Cametti C, Marchetti S, Gambi CMC, Onori G. Dielectric Relaxation Spectroscopy of Lysozyme Aqueous Solutions: Analysis of the delta-Dispersion and the Contribution of the Hydration Water. Journal of Physical Chemistry B. 2011;115(21):7144–53. [DOI] [PubMed] [Google Scholar]
- 2.Wolf M, Gulich R, Lunkenheimer P, Loidl A. Relaxation dynamics of a protein solution investigated by dielectric spectroscopy. BBA-Proteins Proteomics. 2012;1824(5):723–30. [DOI] [PubMed] [Google Scholar]
- 3.Roy S, Richert R. Dielectric spectroscopy study of myoglobin in glycerol-water mixtures. BBA-Proteins Proteomics. 2014;1844(2):323–9. [DOI] [PubMed] [Google Scholar]
- 4.Laux EM, Ermilova E, Pannwitz D, Gibbons J, Holzel R, Bier FF. Dielectric Spectroscopy of Biomolecules up to 110 GHz. Frequenz. 2018;72(3–4):135–40. [Google Scholar]
- 5.Laux EM, Gibbons J, Ermilova E, Bier FF, Holzel R. Broadband dielectric spectroscopy of bovine serum albumin in the GHz range. European Biophysics Journal with Biophysics Letters. 2017;46:S347–S. [Google Scholar]
- 6.Jones PV, Hayes MA. Development of the Resolution Theory for Gradient insulator-based Dielectrophoresis. Electrophoresis. 2015;36(9–10):1098–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Washizu M, Suzuki S, Kurosawa O, Nishizaka T, Shinohara T. Molecular Dielectrophoresis of Biopolymers. IEEE Transactions on Industry Applications. 1994;30(4):835–43. doi: 10.1109/28.297897 [DOI] [Google Scholar]
- 8.Kawabata T, Washizu M. Dielectrophoretic detection of molecular bindings. IEEE Transactions on Industry Applications. 2001;37(6):1625–33. [Google Scholar]
- 9.Zheng LF, Brody JP, Burke PJ. Electronic manipulation of DNA, proteins, and nanoparticles for potential circuit assembly. Biosensors & Bioelectronics. 2004;20(3):606–19. doi: 10.1016/j.bios.2004.03.029 [DOI] [PubMed] [Google Scholar]
- 10.Pethig R. Limitations of the Clausius-Mossotti function used in dielectrophoresis and electrical impedance studies of biomacromolecules. Electrophoresis. 2019;40(18–19):2575–83. [DOI] [PubMed] [Google Scholar]
- 11.Holzel R, Calander N, Chiragwandi Z, Willander M, Bier FF. Trapping single molecules by dielectrophoresis. Physical Review Letters. 2005;95(12). 10.1103/PhysRevLett.95.128102 [DOI] [PubMed] [Google Scholar]
- 12.Camacho-Alanis F, Gan L, Ros A. Transitioning streaming to trapping in DC insulator-based dielectrophoresis for biomolecules. Sensors and Actuators B-Chemical. 2012;173:668–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liao KT, Chou CF. Nanoscale Molecular Traps and Dams for Ultrafast Protein Enrichment in High-Conductivity Buffers. Journal of the American Chemical Society. 2012;134(21):8742–5. [DOI] [PubMed] [Google Scholar]
- 14.Liao KT, Tsegaye M, Chaurey V, Chou CF, Swami NS. Nano-constriction Device for Rapid Protein Preconcentration in Physiological Media Through a Balance of Electrokinetic Forces. Electrophoresis. 2012;33(13):1958–66. [DOI] [PubMed] [Google Scholar]
- 15.Ramos A, Morgan H, Green NG, Castellanos A. AC electrokinetics: a review of forces in microelectrode structures. Journal of Physics D-Applied Physics. 1998;31(18):2338–53. [Google Scholar]
- 16.Washizu M, Kurosawa O. Electrostatic Manipulation of DNA in Microfabricated Structures. IEEE Transactions on Industry Applications. 1990;26(6):1165–72. [Google Scholar]
- 17.Pohl HA. Dielectrophoresis of the behavior of neutral matter in nonuniform electric fields. Cambridge: Cambridge University Press; 1978. [Google Scholar]
- 18.Bakewell DJG, Hughes MP, Milner JJ, Morgan H. Dielectrophoretic Manipulation of Avidin and DNA. Biomedical Engineering Towards the Year 2000 and Beyond (Cat No98CH36286), Hong Kong. 1998;20 1079–82 vol. 2. [Google Scholar]
- 19.Clarke RW, White SS, Zhou DJ, Ying LM, Klenerman D. Trapping of proteins under physiological conditions in a nanopipette. Angew Chem Int Ed. 2005;44(24):3747–50. doi: 10.1002/anie.200500196 [DOI] [PubMed] [Google Scholar]
- 20.Yamamoto T, Fujii T. Active immobilization of biomolecules on a hybrid three-dimensional nanoelectrode by dielectrophoresis for single-biomolecule study. Nanotechnol. 2007;18:495503–10. [DOI] [PubMed] [Google Scholar]
- 21.Clarke RW, Piper JD, Ying L, Klenerman D. Surface Conductivity of Biological Macromolecules Measured by Nanopipette Dielectrophoresis. Phys Rev Lett. 2007;98(19):198102(1–4). [DOI] [PubMed] [Google Scholar]
- 22.Maruyama H, Nakayama Y. Trapping Protein Molecules at a Carbon Nanotube Tip using Dielectrophoresis. Applied Physics Express. 2008;1(12). [Google Scholar]
- 23.Chaurey V, Rohani A, Su YH, Liao KT, Chou CF, Swami NS. Scaling down constriction-based (electrodeless) dielectrophoresis devices for trapping nanoscale bioparticles in physiological media of high-conductivity. Electrophoresis. 2013;34(7):1097–104. [DOI] [PubMed] [Google Scholar]
- 24.Sanghavi BJ, Varhue W, Chavez JL, Chou CF, Swami NS. Electrokinetic Preconcentration and Detection of Neuropeptides at Patterned Graphene-Modified Electrodes in a Nanochannel. Analytical Chemistry. 2014;86(9):4120–5. [DOI] [PubMed] [Google Scholar]
- 25.Otto S, Kaletta U, Bier FF, Wenger C, Holzel R. Dielectrophoretic immobilisation of antibodies on microelectrode arrays. Lab on a Chip. 2014;14(5):998–1004. [DOI] [PubMed] [Google Scholar]
- 26.Laux EM, Kaletta UC, Bier FF, Wenger C, Holzel R. Functionality of dielectrophoretically immobilized enzyme molecules. Electrophoresis. 2014;35(4):459–66. [DOI] [PubMed] [Google Scholar]
- 27.Lesser-Rojas L, Ebbinghaus P, Vasan G, Chu ML, Erbe A, Chou CF. Low-Copy Number Protein Detection by Electrode Nanogap-Enabled Dielectrophoretic Trapping for Surface-Enhanced Raman Spectroscopy and Electronic Measurements. Nano Letters. 2014;14(5):224250. [DOI] [PubMed] [Google Scholar]
- 28.Barik A, Otto LM, Yoo D, Jose J, Johnson TW, Oh SH. Dielectrophoresis-Enhanced Plasmonic Sensing with Gold Nanohole Arrays. Nano Letters. 2014;14(4):2006–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakano A, Camacho-Alanis F, Ros A. Insulator-based dielectrophoresis with beta-galactosidase in nanostructured devices. Analyst. 2015;140(3):860–8. doi: 10.1039/c4an01503g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schafer C, Kern DP, Fleischer M. Capturing molecules with plasmonic nanotips in microfluidic channels by dielectrophoresis. Lab on a Chip. 2015;15(4):1066–71. [DOI] [PubMed] [Google Scholar]
- 31.Chiou CH, Chien LJ, Kuo JN. Nanoconstriction-based electrodeless dielectrophoresis chip for nanoparticle and protein preconcentration. Applied Physics Express. 2015;8(8):3. [Google Scholar]
- 32.Laux EM, Knigge X, Bier FF, Wenger C, Holzel R. Aligned Immobilization of Proteins Using AC Electric Fields. Small. 2016;12(11):1514–20. [DOI] [PubMed] [Google Scholar]
- 33.Rohani A, Sanghavi BJ, Salahi A, Liao KT, Chou CF, Swami NS. Frequency-selective electrokinetic enrichment of biomolecules in physiological media based on electrical doublelayer polarization. Nanoscale. 2017;9(33):12124–31. doi: 10.1039/c7nr02376f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang P, Liu YX. DC biased low-frequency insulating constriction dielectrophoresis for protein biomolecules concentration. Biofabrication. 2017;9(4):11. doi: 10.1088/17585090/aa82d6 [DOI] [PubMed] [Google Scholar]
- 35.Lapizco-Encinas BH, Ozuna-Chacon S, Rito-Palomares M. Protein manipulation with insulator-based dielectrophoresis and direct current electric fields. Journal of Chromatography A. 2008;1206(1):45–51. 10.1016/j.chroma.2008.05.077 [DOI] [PubMed] [Google Scholar]
- 36.Agastin S, King MR, Jones TB. Rapid enrichment of biomolecules using simultaneous liquid and particulate dielectrophoresis. Lab on a Chip. 2009;9(16):2319–25. [DOI] [PubMed] [Google Scholar]
- 37.Gong JR. Label-Free Attomolar Detection of Proteins Using Integrated Nanoelectronic and Electrokinetic Devices. Small. 2010;6(8):967–73. [DOI] [PubMed] [Google Scholar]
- 38.Kim HJ, Kim J, Yoo YK, Lee JH, Park JH, Hwang KS. Sensitivity improvement of an electrical sensor achieved by control of biomolecules based on the negative dielectrophoretic force. Biosensors & Bioelectronics. 2016;85:977–85. [DOI] [PubMed] [Google Scholar]
- 39.Mohamad AS, Hamzah R, Hoettges KF, Hughes MP. A dielectrophoresis-impedance method for protein detection and analysis. AIP Adv. 2017;7(1):7. [Google Scholar]
- 40.Mata-Gomez MA, Gallo-Villanueva RC, Gonzalez-Valdez J, Martinez-Chapa SO, Rito-Palomares M. Dielectrophoretic behavior of PEGylated RNase A inside a microchannel with diamond-shaped insulating posts. Electrophoresis. 2016;37(3):519–28. [DOI] [PubMed] [Google Scholar]
- 41.Sharma A, Han CH, Jang J. Rapid electrical immunoassay of the cardiac biomarker troponin I through dielectrophoretic concentration using imbedded electrodes. Biosensors & Bioelectronics. 2016;82:78–84. [DOI] [PubMed] [Google Scholar]
- 42.Dies H, Raveendran J, Escobedo C, Docoslis A. In situ assembly of active surface-enhanced Raman scattering substrates via electric field-guided growth of dendritic nanoparticle structures. Nanoscale. 2017;9(23):7847–57. [DOI] [PubMed] [Google Scholar]
- 43.Hubner Y, Hoettges KF, McDonnell MB, Carter MJ, Hughes MP. Applications of dielectrophoretic/ electrohydrodynamic “zipper” electrodes for detection of biological nanoparticles. Int J Nanomed. 2007;2(3):427–31. [PMC free article] [PubMed] [Google Scholar]
- 44.Gencoglu A, Camacho-Alanis F, Nguyen VT, Nakano A, Ros A, Minerick AR. Quantification of pH gradients and implications in insulator-based dielectrophoresis of biomolecules. Electrophoresis. 2011;32(18):2436–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakano A, Chao TC, Camacho-Alanis F, Ros A. Immunoglobulin G and bovine serum albumin streaming dielectrophoresis in a microfluidic device. Electrophoresis. 2011;32(17):2314–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakano A, Camacho-Alanis F, Chao TC, Ros A. Tuning direct current streaming dielectrophoresis of proteins. Biomicrofluidics. 2012;6(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Staton SJR, Jones PV, Ku G, Gilman SD, Kheterpal I, Hayes MA. Manipulation and capture of A beta amyloid fibrils and monomers by DC insulator gradient dielectrophoresis (DCiGDEP). Analyst. 2012;137(14):3227–9. [DOI] [PubMed] [Google Scholar]
- 48.Pethig R. Dielectrophoresis: Status of the theory, technology, and applications (vol 4, 022811, 2010). Biomicrofluidics. 2010;4(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakano A, Ros A. Protein dielectrophoresis: Advances, challenges, and applications. Electrophoresis. 2013;34(7):1085–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Camacho-Alanis F, Ros A. Protein dielectrophoresis and the link to dielectric properties. Bioanalysis. 2015;7(3):353–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Modarres P, Tabrizian M. Alternating current dielectrophoresis of biomacromolecules: The interplay of electrokinetic effects. Sensors and Actuators B-Chemical. 2017;252:391–408. [Google Scholar]
- 52.Laux EM, Bier FF, Holzel R. Electrode-based AC electrokinetics of proteins: A minireview. Bioelectrochemistry. 2018;120:76–82. [DOI] [PubMed] [Google Scholar]
- 53.Kim D, Sonker M, Ros A. Dielectrophoresis: From Molecular to Micrometer-Scale Analytes. Analytical Chemistry. 2019;91(1):277–95. [DOI] [PubMed] [Google Scholar]
- 54.Hughes MP. Nanoelectromechanics in engineering and biology. London: CRC Press; 2003. 322 p. [Google Scholar]
- 55.Laux EM, Knigge X, Bier FF, Wenger C, Holzel R. Dielectrophoretic immobilization of proteins: Quantification by atomic force microscopy. Electrophoresis. 2015;36(17):2094–101. doi: 10.1002/elps.201500108 [DOI] [PubMed] [Google Scholar]
- 56.Pohl HA. The Motion and Precipitation of Suspensoids in Divergent Electric Fields. J Appl Phys. 1951;22:869–71. [Google Scholar]
- 57.Jones TB. Dielectrophoretic Force In Axisymmetrical Fields. Journal of Electrostatics. 1986;18(1):55–62. [Google Scholar]
- 58.Jones TB. Dielectrophoretic Force Calculation. Journal of Electrostatics. 1979;6(1):69–82. [Google Scholar]
- 59.Pethig R. Review-Where Is Dielectrophoresis (DEP) Going? J Electrochem Soc. 2017;164(5):B3049–B55. [Google Scholar]
- 60.Pethig R. Dielectrophoresis : theory, methodology and biological applications. First Edition ed Hoboken, NJ: John Wiley & Sons, LTD; 2017. [Google Scholar]
- 61.Rohani A, Varhue W, Liao KT, Chou CF, Swami NS. Nanoslit design for ion conductivity gradient enhanced dielectrophoresis for ultrafast biomarker enrichment in physiological media. Biomicrofluidics. 2016;10(3):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Slouka Z, Senapati S, Chang HC. Microfluidic Systems with Ion-Selective Membranes In: Cooks RG, Pemberton JE, editors. Annual Review of Analytical Chemistry, Vol 7. Annual Review of Analytical Chemistry. 7 Palo Alto: Annual Reviews; 2014. p. 317–35. [DOI] [PubMed] [Google Scholar]
- 63.Marczak S, Smith E, Senapati S, Chang HC. Selectivity enhancements in gel-based DNA-nanoparticle assays by membrane-induced isotachophoresis: thermodynamics versus kinetics. Electrophoresis. 2017;38(20):2592–602. [DOI] [PubMed] [Google Scholar]
- 64.Matyushov DV. Electrostatic solvation and mobility in uniform and non-uniform electric fields: From simple ions to proteins. Biomicrofluidics. 2019;13:064106. doi: 10.1063/1.5124390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Seyedi S, Matyushov DV. Dipolar susceptibility of protein hydration shells. Chem Phys Lett. 2018;713:210–4. [Google Scholar]
- 66.Matyushov DV. Dipolar response of hydrated proteins. J Chem Phys. 2012;136(8). [DOI] [PubMed] [Google Scholar]
- 67.Holze R. Dielectric and dielectrophoretic properties of DNA. IET Nanobiotechnology. 2009;3(2):28–45. [DOI] [PubMed] [Google Scholar]
- 68.Heyden M. Heterogeneity of water structure and dynamics at the protein-water interface. J Chem Phys. 2019;150(9):10. [DOI] [PubMed] [Google Scholar]
- 69.Heyden M. Disassembling solvation free energies into local contributions-Toward a microscopic understanding of solvation processes. Wiley Interdiscip Rev-Comput Mol Sci. 2019;9(2):16. [Google Scholar]
- 70.Pattni V, Vasilevskaya T, Thiel W, Heyden M. Distinct Protein Hydration Water Species Defined by Spatially Resolved Spectra of Intermolecular Vibrations. Jf Phys Chem B. 2017;121(31):7431–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Persson RAX, Pattni V, Singh A, Kast SM, Heyden M. Signatures of Solvation Thermodynamics in Spectra of Intermolecular Vibrations. J Chem Theory Comput. 2017;13(9):4467–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Waskasi M, Heyden M. Microscopic driving forces of protein dielectrophoresis. personal communication. 2019. [Google Scholar]
- 73.Zhu FY, Nannenga BL, Hayes MA. Electrophoretic exclusion microscale sample preparation for cryo-EM structural determination of proteins. Biomicrofluidics. 2019;13(5):11. [DOI] [PMC free article] [PubMed] [Google Scholar]