Abstract
The cytosolic sulfotransferases (SULTs) metabolize a variety of xenobiotic and endogenous substrates. Several SULTs are expressed in the fetus, implying that these enzymes have important functions during human development. We recently reported that while SULT1C4 mRNA is abundant in prenatal human liver specimens, SULT1C4 protein is barely detectable. Two coding transcript variants (TVs) of SULT1C4 are indexed in GenBank, TV1 (full-length) and TV2 (lacking exons 3 and 4). The purpose of this study was to evaluate expression of the individual TVs as a clue for understanding the discordance between mRNA and protein levels. Reverse-transcription polymerase chain reaction was initially performed to identify TVs expressed in intestinal and hepatic cell lines. This analysis generated fragments corresponding to TV1, TV2, and a third variant that lacked exon 3 (E3DEL). Using reverse-transcription quantitative polymerase chain reaction assays designed to quantify TV1, TV2, or E3DEL individually, all three TVs were more highly expressed in prenatal than postnatal specimens. TV2 levels were ∼fivefold greater than TV1, while E3DEL levels were minimal. RNA sequencing (RNA-seq) analysis of another set of liver specimens confirmed that TV1 and TV2 levels were highest in prenatal liver, with TV2 higher than TV1. RNA-seq also detected a noncoding RNA, which was also more abundant in prenatal liver. Transfection of HEK293T cells with plasmids expressing individual Asp-Tyr-Lys-Asp-Asp-Asp-Asp-Lys–tagged SULT1C4 isoforms demonstrated that TV1 produced much more protein than did TV2. These data suggest that the lack of correspondence between SULT1C4 mRNA and protein levels in human liver is likely attributable to the inability of the more abundant TV2 to produce stable protein.
SIGNIFICANCE STATEMENT
Cytosolic sulfotransferases (SULTs) metabolize a variety of xenobiotic and endogenous substrates, and several SULTs are highly expressed in the fetus, implying that they have important functions during human development. SULT1C4 is highly expressed in prenatal liver at the mRNA level but not the protein level. This study provides an explanation for this discordance by demonstrating that the predominant SULT1C4 transcript is a variant that produces relatively little protein.
Introduction
The cytosolic sulfotransferases (SULTs) catalyze the conjugation of a sulfonate moiety to a wide variety of endogenous and xenobiotic substrates, including hormones, neurotransmitters, drugs, and environmental chemicals. These enzymes therefore play important roles in both the regulation of physiologic processes and the disposition of drugs and other xenobiotics. SULTs are classified into six gene families, although only four of these (families 1, 2, 4, and 6) are present in humans, and only the SULT1 and SULT2 family genes are reported to be expressed in human liver (Runge-Morris et al., 2013).
Of the xenobiotic-metabolizing enzymes, the SULTs are particularly likely determinants of xenobiotic-metabolizing capacity in the developing human because several SULTs are prominently, if not predominantly, expressed during prenatal and/or early postnatal life (Barker et al., 1994; Richard et al., 2001; Miki et al., 2002; Stanley et al., 2005; Duanmu et al., 2006; Dubaisi et al., 2019). We recently characterized the developmental expression of SULT1 and SULT2 mRNAs and proteins in human liver using three sets of human liver specimens (Dubaisi et al., 2019). In one set, SULT mRNA levels were quantified by reverse-transcription quantitative polymerase chain reaction (RT-qPCR) analysis in liver specimens from prenatal, infant, and adult donors; in another set, SULT mRNA levels were determined by RNA sequencing (RNA-seq) analysis of specimens from prenatal, infant, and pediatric donors. SULT protein levels were measured by targeted quantitative proteomics in a panel of 193 liver cytosolic fractions. The results demonstrated that SULT1A1 expression was high throughout development; SULT1A3, 1C2, 1C4, and 1E1 expression was highest in prenatal and/or infant liver; and SULT1A2, 1B1, and 2A1 expression was highest in infant and/or adult liver. For most of the SULTs, mRNA and protein showed comparable patterns of expression. SULT1C4 was a clear exception, however, because while RT-qPCR and RNA-seq analyses indicated that SULT1C4 mRNA was abundant in prenatal liver, SULT1C4 protein levels were very low (Dubaisi et al., 2019).
SULT1C4 is a member of a gene subfamily that includes three human members, SULT1C2, 1C3, and 1C4. Several studies have suggested that SULT1C4 has the highest sulfonation capacity of the SULT1C enzymes toward xenobiotics, including a wide range of drugs (e.g., acetaminophen), environmental chemicals (e.g., bisphenol A), and procarcinogens (e.g., hydroxymethyl furans) (Sakakibara et al., 1998; Glatt et al., 2004, 2012; Allali-Hassani et al., 2007; Yasuda et al., 2007; Yamamoto et al., 2015, 2016; Guidry et al., 2017; Rasool et al., 2017). SULT1C4 is also capable of metabolizing estrogenic compounds, such as catechol and methoxy estrogens (Allali-Hassani et al., 2007; Hui et al., 2008). Guidry et al. (2017) recently reported that SULT1C4 has high sulfonation capacity toward dietary flavonoids and environmental estrogens.
The discrepancy that we observed between SULT1C4 mRNA and protein levels in human liver prompted us to seek the underlying mechanism. It is increasingly appreciated that many genes are transcribed and processed into multiple transcript variants (TVs), the sequence and abundancy of which are important determinants of the protein isoforms that are produced and the functionality of these proteins (Her et al., 1998; Gardner-Stephen et al., 2004; Duniec-Dmuchowski et al., 2014; Meloto et al., 2015). Several SULT1C4 TVs are indexed in GenBank, including the full-length mRNA containing seven exons (TV1, NM_006588), a variant mRNA lacking exons 3 and 4 (TV2, NM_001321770), two noncoding RNA variants (TV3, NR_135776 and TV4, NR_135779), and a predicted TV (TVX1, XM_017003807). An additional noncoding RNA (ENST00000494122.1) that consists of two exons and a retained intron is indexed in the Ensembl database. Because the specific SULT1C4 TVs that are expressed could have a major impact on the amount of SULT1C4 protein that is present, we decided to characterize and quantify the SULT1C4 transcripts that are present in human hepatic and intestinal cells and liver specimens.
Materials and Methods
Human Liver Specimens and Cell Lines.
The human liver specimens that were used to measure SULT1C4 TV levels by RT-qPCR or RNA-seq in this study are the same as those described in our recent publication (Dubaisi et al., 2019). HepaRG cells were obtained and cultured as previously described (Dubaisi et al., 2018). Caco-2 colorectal adenocarcinoma cells were obtained from the American Type Culture Collection (Manassas, VA) and maintained in Dulbecco’s Modified Eagle Medium (high glucose) supplemented with 10% heat-inactivated fetal bovine serum, 2 mM glutamine, 1 mM sodium pyruvate, 1× Minimum Essential Medium nonessential amino acids, 100 U/ml penicillin, and 100 μg/ml streptomycin (Thermo Fisher Scientific, Waltham, MA).
5′-Rapid Amplification of cDNA Ends.
Total RNA was isolated from Caco-2 or confluent HepaRG cells using the Purelink RNA Mini Kit (Thermo Fisher Scientific). 5′-rapid amplification of cDNA ends (RACE) was performed using the SMARTer RACE 5′/3′ kit (Takara Bio USA Inc., Mountain View, CA), RACE-ready cDNA prepared from Caco-2 or HepaRG total RNA, and a SULT1C4-specific reverse primer [located within exon 2 at nucleotide (nt) 573] (Supplemental Table 1), according to the manufacturer’s recommendations. PCR conditions were five cycles of 94°C for 30 seconds and 72°C for 2 minutes; five cycles of 94°C for 30 seconds, 70°C for 30 seconds, and 72°C for 2 minutes; and 35 cycles of 94°C for 30 seconds, 65°C for 30 seconds, and 72°C for 2 minutes. PCR reactions were then resolved on a 0.8% agarose gel, and ethidium bromide–stained amplified bands were identified by UV illumination, purified using the QIAquick Gel Extraction Kit (Qiagen, Germantown, MD), and ligated into the pUC19 plasmid. Individual clones (23 clones total from HepaRG and Caco-2 cells) were sequenced by the Wayne State University Applied Genomics Technology Center.
Gene Expression Analysis.
RNA was isolated from human liver specimens as previously described (Dubaisi et al., 2019). RNA (1.5 µg) was reverse transcribed to cDNA using the High Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific), and a primer set predicted to amplify an approximately 1.2-kb fragment from the SULT1C4 TV1 sequence (NM_006588.3; Supplemental Table 1) was used to detect SULT1C4 transcripts in Caco-2 and HepaRG cells by standard reverse-transcription polymerase chain reaction (RT-PCR). PCR was performed using HotStar Taq DNA Polymerase (Qiagen) and the following conditions: initial activation at 95°C for 5 minutes; 35 cycles of 94°C for 30 seconds, 61°C for 45 seconds, and 72°C for 2 minutes; and final extension at 72°C for 7 minutes. The resulting fragments were ligated into the XhoI site of the pGL4.10[luc2] plasmid (Promega Corporation, Madison, WI) using the In-Fusion HD Cloning Plus Kit (Takara Bio USA Inc.) and sequenced, and these plasmids were subsequently used as synthetic standards for RT-qPCR analysis. RT-qPCR was performed using SYBR Green (ThermoFisher Scientific), a common forward primer (50 nM) spanning the exon 1 to 2 junction, and reverse primers (100 nM) designed to span the unique exon-exon junction for each TV, i.e., exons 2 to 3 for TV1, exons 2–5 for TV2, and exons 2–4 for E3DEL. Primers were designed using Oligo 7 (Molecular Biology Insights, Cascade, CO) and visualized in silico against SULT1C4 TV sequences using SnapGene 4.1 (GSL Biotech LLC, Chicago, IL). Primer sequences are shown in Supplemental Table 1. RT-qPCR was performed using a QuantStudio 3 system (Thermo Fisher Scientific). The annealing temperature was 60°C for the TV1 and E3DEL primer pairs and 63°C for the TV2 primer pair. A standard curve of threshold cycle versus attomole plasmid DNA was prepared for each SULT1C4 TV, using the plasmid standards described above. Least squares lines for these standard curves were generated using Prism version 6 (GraphPad Software, La Jolla, CA) and used to calculate RNA content of the TVs in the liver specimens (expressed as attomole SULT1C4 TV/microgram total RNA).
The procedure for RNA-seq analysis was described previously (Dubaisi et al., 2018). SULT1C4 TV information was obtained using StringTie (Pertea et al., 2015).
Expression of SULT1C4 TVs in HEK293T Cells.
5′-Asp-Tyr-Lys-Asp-Asp-Asp-Asp-Lys (DDK)-tagged SULT1C4 TV1 coding sequence was prepared using 100 ng pKK233-2-SULT1C4 (Guidry et al., 2017) as template, Herculase II Fusion DNA Polymerase (Agilent Technologies, Santa Clara, CA), the SULT1C4 primer pair indicated in Supplemental Table 1 (SULT1C4-HindIII-DDK-F and SULT1C4-XhoI-R), and the following PCR conditions: 95°C for 2 minutes; 20 cycles of 95°C for 20 seconds, 64°C for 20 seconds, and 72°C for 30 seconds; and 72°C for 3 minutes. DDK-tagged SULT1C4 TV2 and E3DEL were prepared using the TV2 and E3DEL plasmid standards (described in the section above) as templates, HotStar Taq DNA Polymerase (Qiagen), and the same primer pair used for TV1. The amplified fragments were digested with HindIII and XhoI and ligated into pcDNA3.1 (Thermo Fisher Scientific) using the LigaFast Rapid DNA Ligation System (Promega).
HEK293T cells were obtained from Thermo Fisher Scientific and maintained in Dulbecco’s Modified Eagle Medium supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 µg/ml streptomycin. To express the individual SULT1C4 TVs, 1.5 million HEK293T cells were plated into 100-mm culture dishes and the following day were transfected with a complex containing 5 µg of one of the DDK-tagged SULT1C4 TV expression plasmids or empty pcDNA3.1, 15 µg pBluescript II KS+ (Agilent Technologies), and 50 µl Lipofectamine 2000 in 2 ml of Opti-MEM (Thermo Fisher Scientific) for 72 hours. Total RNA was then prepared from one set of dishes using the Purelink RNA Mini Kit (Thermo Fisher Scientific) and reverse transcribed to cDNA as described above. SULT1C4 TV levels were determined using TaqMan Gene Expression Assay Hs00602560_m1 (Thermo Fisher Scientific), which targets the exon 6 to 7 boundary region of SULT1C4 mRNA and is therefore capable of detecting the three SULT1C4 TVs.
To measure SULT1C4 TV-derived protein levels in the transfected HEK293T cells by Western blot, whole-cell lysates were prepared as previously described (Rondini et al., 2014). Protein concentrations were measured using the BCA Protein Assay Kit (Thermo Fisher Scientific), and lysate samples containing 15 or 60 µg protein were resolved on 12.5% sodium dodecyl sulfate-polyacrylamide gels and transferred to polyvinylidene difluoride membranes. The membranes were incubated for 1 hour with blocking buffer [2.5% nonfat dry milk in Tris-buffered saline with Tween 20 (Sigma-Aldrich)], incubated overnight at 4°C with mouse monoclonal anti-DDK antibody (Clone OTI4C5; Origene, Rockville, MD) diluted 1:2000, and then incubated for 2 hours with horseradish peroxidase–conjugated goat anti-mouse IgG (sc-2005; Santa Cruz Biotechnology) diluted 1:20,000. Chemiluminescent bands were visualized using Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific) and a FluorChem E imager (Protein Simple, San Jose, CA). The blots were then incubated in 60 mM Tris-HCl, 70 mM sodium dodecyl sulfate, and 100 mM β-mercaptoethanol at 37°C to remove antibodies and reprobed with β-actin antibody (clone AC15; Sigma-Aldrich) diluted 1:40,000 followed by horseradish peroxidase–conjugated goat anti-mouse IgG diluted 1:100,000.
To measure SULT1C4 TV-derived protein levels by mass spectrometry, samples containing 100 µg whole-cell lysate protein were resolved on a 4%–12% Bis-Tris gel (Thermo Fisher Scientific) in duplicate. The region between 20 and 43 kDa was excised, and proteins were reduced, alkylated, and digested with trypsin in-gel. Peptides were eluted from the gel pieces and dried. The mass spectrometry analysis and quantification of SULT1C4 was performed as previously described (Dubaisi et al., 2019). Samples were analyzed in triplicate; therefore, results represent the average of three samples.
Statistical Analysis.
Samples were grouped according to age, and scatter plots were prepared for the mRNA data. Statistical comparisons among groups were performed using the Kruskal-Wallis nonparametric analysis of variance and Dunn’s multiple comparison test with Prism version 6 (GraphPad Software).
Results
Identification of SULT1C4 TVs Expressed in Human Intestinal and Hepatic Cell Lines.
SULT1C4 (termed humanSULT1C#2 in the original publication) cDNA was originally cloned from a fetal lung cDNA library (Sakakibara et al., 1998), and Freimuth et al. (2000) subsequently used 5′-RACE to determine the 5′-end of the transcript in fetal lung. We performed 5′-RACE on the Caco-2 and HepaRG cell lines, commonly used cellular models of human intestine and liver, respectively, to determine whether the 5′-end of the SULT1C4 transcript in these cell lines was the same as that previously determined in fetal lung. We and others have found that SULT1C4 is expressed at a high level in Caco-2 cells (Meinl et al., 2008; Bourgine et al., 2012), and we recently reported that SULT1C4 is expressed in confluent HepaRG cells (Dubaisi et al., 2018). Ten 5′-RACE clones from Caco-2 cells and 13 clones from confluent HepaRG cells were sequenced and aligned to human chromosome 2 at the 5′-region of the SULT1C4 gene (SULT1C4 gene address: NC_000003.12:108377954-108388989). The 5′-positions of the 23 5′-RACE clones, as well as of the SULT1C4 reference sequences for TV1 and TV2 (NM_006588 and NM_001321770), are shown in Supplemental Table 2. The 5′-ends of the 10 Caco-2 clones were located 317–400 nt upstream of the translation start site, with six of these clones beginning at nt −380 to −384 relative to the start codon, which was approximately the same position as the 5′-end of the SULT1C4 mRNA reference sequences (nt −384 relative to the start codon). The 5′-ends of 10 of the HepaRG clones were located 345–372 nt upstream of the translation start site, with seven of these clones beginning at nt −353 or −368 relative to the start codon. The 5′-ends of three of the HepaRG clones were further upstream at nt −460, −545, and −690 relative to the start codon. Freimuth et al. (2000) previously reported that the SULT1C4 gene does not contain a TATA box sequence and suggested that transcription was likely initiated through an initiator element. Consistent with this possibility, 11 of the 23 5′-RACE clones had 5′-adenosines that were properly located within initiator element consensus sequences, as defined in a recent report (Vo Ngoc et al., 2017). Overall, our data did not demonstrate major differences in transcription start site between the Caco-2 cells and HepaRG cells or between our findings and those previously reported by Freimuth et al. (2000) and confirmed that the translation initiation codon is located in exon 1.
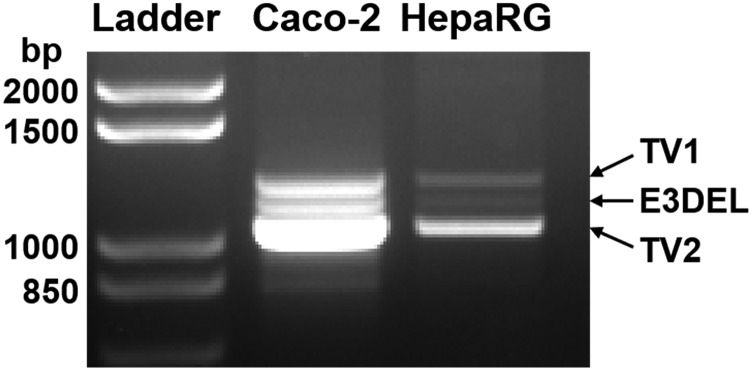
Using this information, primers for RT-PCR were designed that were predicted to amplify an approximately 1.2-kb fragment of the full-length SULT1C4 transcript (TV1), with the forward primer located in exon 1 (118 nt upstream of the translation start site) and the reverse primer in exon 7 (192 nt downstream of the stop codon). Use of these primers resulted in amplification of three fragments from both Caco-2 and HepaRG cells: the expected 1.2-kb fragment, a 1.1-kb fragment, and a 1.0-kb fragment (Fig. 1). These fragments were isolated, cloned, sequenced, and aligned with the reported SULT1C4 reference sequences. The results indicated that the 1.2- and 1.0-kb fragments corresponded to TV1 (NM_006588, full-length) and TV2 (NM_001321770, lacking exons 3 and 4), respectively. The 1.1-kb fragment lacked exon 3; this fragment was therefore termed E3DEL. These data suggested the potential expression of three SULT1C4 mRNA variants in human intestinal and liver cells. Based on the relative band intensities, the data further suggested that TV2, lacking exons 3 and 4, might actually be more abundant than the full-length TV1 in the cell lines.
Fig. 1.
Amplification of three SULT1C4 TVs from Caco-2 and HepaRG cells. RNA was isolated from Caco-2 and HepaRG cells and reverse transcribed to cDNA. PCR was performed using primers predicted to amplify an approximately 1.2-kb fragment from the full-length (TV1) cDNA sequence. PCR products were resolved on a 1% agarose gel, stained with ethidium bromide, and visualized under UV transillumination. The bands corresponding to SULT1C4 TV1, TV2, and E3DEL are indicated.
Developmental Expression of SULT1C4 TVs in Liver.
Using two sets of human liver specimens, we recently reported that SULT1C4 mRNA is expressed at high levels in prenatal human liver and at much lower levels postnatally (Dubaisi et al., 2019). One set of specimens was analyzed by RT-qPCR, using a TaqMan Gene Expression Assay that targets the exon 6 to 7 boundary region. Therefore, this assay would detect all of the SULT1C4 TVs identified in the Caco-2 and HepaRG cells (i.e., TV1, TV2, and E3DEL). We therefore designed primers for RT-qPCR that would amplify TV1, TV2, or E3DEL individually (Fig. 2A) and used these primers to characterize the developmental expression patterns of the three TVs. Plasmids containing the individual SULT1C4 TVs were used to generate standard curves to allow conversion of threshold cycle values to absolute amounts of the SULT1C4 transcripts (Fig. 2B). As shown in Fig. 2C (values provided in Supplemental Table 3), prenatal, infant, and adult liver specimens expressed the three SULT1C4 TVs, and all TVs were present at the highest levels in the prenatal samples. TV2 was the most abundant SULT1C4 transcript at all stages of development. The amount of TV2 in prenatal liver (43.6 amol/μg) was more than fivefold higher than that of TV1 (7.64 amol/μg). The amount of E3DEL transcript was very low in all liver specimens.
Fig. 2.
Developmental expression of SULT1C4 TVs in human liver. (A) Schematic representation of the SULT1C4 TVs that were identified in Caco-2 and HepaRG cells and the primers designed to detect these transcripts individually (primers not drawn to scale). (B) A plasmid standard was prepared for each SULT1C4 TV, and standard curves were prepared to permit quantification of transcript amounts. (C) RNA was isolated from specimens of prenatal (n = 10), infant (n = 7), and adult (n = 7) human liver, and SULT1C4 TV levels were measured using RT-qPCR. For each TV, data were grouped according to developmental stage and are expressed as attomole SULT1C4 transcript/microgram of RNA. Data are shown as scatter plots with the horizontal lines representing the median values. ***Significantly different, P < 0.001. (D) RNA was isolated from prenatal (n = 10) and pediatric (n = 52) human liver specimens and analyzed by RNA-seq. Data are shown as scatter plots with the horizontal lines representing the median values. Groups not sharing a letter are significantly different from each other, P < 0.05.
We also previously evaluated SULT expression in a separate panel of prenatal and pediatric (i.e., infants and children 1–18 years old) human liver specimens by RNA-seq (Dubaisi et al., 2019). We therefore evaluated those data for information about levels of SULT1C4 TVs. The analysis identified three TVs expressed in liver, TV1, TV2, and a noncoding TV (Ensembl Transcript ENST00000494122.1). Again, these TVs were preferentially expressed in the prenatal livers, and TV2 was the most abundant transcript, although the level of TV2 was only ∼twofold greater than TV1 in the prenatal samples (Fig. 2D; Supplemental Table 4). The noncoding TV was present at approximately the same level as TV1 (Supplemental Table 4).
SULT1C4 Protein Expression from Individual TVs
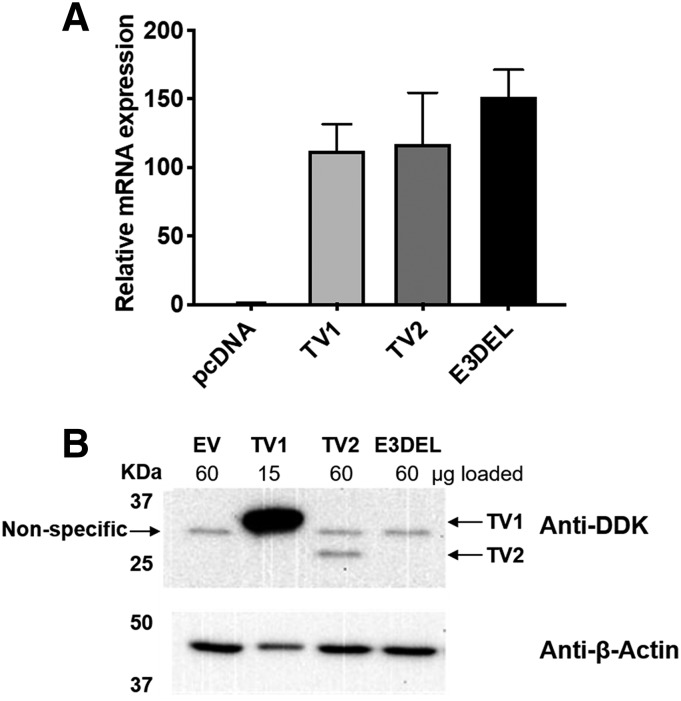
While we previously reported that SULT1C4 mRNA levels were high in prenatal liver, SULT1C4 protein levels were very low (Dubaisi et al., 2019). The SULT1C4 peptide sequence that was used for targeted quantitative proteomics is located in exon 6 of the full-length protein sequence (i.e., isoform derived from TV1, referred to as isoform 1, NP_006579, in the GenPept database). Because deletion of exons 3 and 4 does not change the reading frame, this same peptide sequence would be present in an isoform derived from TV2 (isoform 2, NP_001308699). By contrast, deletion of only exon 3 would alter the reading frame and introduce a premature stop codon, so a protein derived from E3DEL would not be detected by our proteomics assay. To determine the abilities of the TV1, TV2, and E3DEL transcripts to generate proteins in cells, expression plasmids containing DDK-tagged TVs were transfected into HEK293T cells, and TV-derived mRNA and protein levels were determined by RT-qPCR and Western blot, respectively. While all transfected TVs produced comparable levels of mRNA (Fig. 3A), the amount of TV1-derived DDK-immunoreactive protein in the HEK293T cell lysates was much greater than was the amount of protein derived from TV2 (Fig. 3B). No DDK-immunoreactive protein was produced after transfection with E3DEL. As confirmation of these findings, quantitative proteomics analysis revealed that TV1 transfection produced a quantifiable amount of SULT1C4 protein (5.31 fmol/mg), whereas the TV2-derived protein level was below the limit of quantification (Supplemental Table 5).
Fig. 3.
SULT1C4 mRNA and protein levels after transfection of expression plasmids for individual TVs into HEK293T cells. Total RNA (A) and whole-cell lysates (B) were prepared from HEK293T cells that were transfected with DDK-tagged TV1, TV2, or E3DEL expression plasmid (or with empty vector, EV) and analyzed by RT-qPCR and Western blot, respectively. (A) RNA levels are expressed as mean ± range (from two independent experiments) relative to the amount measured in cells transfected with TV1. (B) A representative Western blot image shows DDK-tag and β-actin (endogenous loading control) immunoreactivity. Locations of the TV1- and TV2-derived proteins are indicated, as is the location of a nonspecific band that was present in all groups. Note that one-fourth the amount of TV1 lysate (15 µg total protein) was loaded compared with the other transfectant lysates (60 µg protein). Similar results were obtained in two additional experiments.
Discussion
Xenobiotic-metabolizing enzymes that are expressed during early development are essential determinants of environmental impacts on the developing fetus. Although they generally play a protective role, some xenobiotic-metabolizing enzymes can bioactivate certain compounds to mutagenic species and could therefore enhance the susceptibility of the fetus to cancer development (Banoglu, 2000; Perera et al., 2002; Murray et al., 2007). Many studies have detected SULT mRNA, protein, and activity in various human tissues isolated from prenatal donors (Hines, 2008), and therefore these enzymes presumably metabolize endogenous and xenobiotic compounds during gestation.
SULT1C4 mRNA was previously reported to be abundantly expressed in fetal lung and kidney (Sakakibara et al., 1998). Using human liver specimens and in vitro models of human liver development (i.e., HepaRG cells and primary cultures of fetal hepatocytes), we demonstrated that SULT1C4 mRNA is primarily expressed in prenatal liver or undifferentiated HepaRG cells (Dubaisi et al., 2018, 2019). However, while SULT1C4 mRNA levels were relatively high in prenatal liver specimens, SULT1C4 protein levels were very low (Dubaisi et al., 2019). In the current analysis, we describe three SULT1C4 mRNA variants, TV1, TV2, and E3DEL, that are present in human hepatic and intestinal cells and are coordinately expressed during human liver development, with preferential expression in prenatal liver. TV2 and E3DEL specifically lack exons 3 and 4 or exon 3, respectively, and are therefore produced by the form of alternative splicing known as exon skipping (Zhao, 2019).
Whether an exon is included in a mature mRNA depends on information contained within the primary transcript. For example, the flanking introns contain the canonical splice elements, i.e., 5′ donor splice site, branch site, and 3′ acceptor splice site (Kornblihtt et al., 2013). Constitutive exons, which are always present in the mature mRNA, are well defined because they are demarcated by strong splice sites. By comparison, alternative exons are less well defined, and their proper recognition requires additional information, such as splicing enhancer and silencer elements that are located within the alternative exons themselves or within the flanking introns (Kornblihtt et al., 2013). For example, exonic splicing enhancers function as binding sites for the serine/arginine-rich family of proteins, which promote exon definition by recruiting spliceosomal components and/or by antagonizing splicing silencers (Cartegni et al., 2003). It therefore seems probable that the alternative mRNA variants of SULT1C4 arise from the presence and interactions of cis-acting splicing enhancers/silencers in the SULT1C4 gene. Several computational tools have been developed to analyze gene sequences for features that control splicing, including donor and acceptor splice sites, branch point sequences, and cis-acting splicing enhancer and silencer elements (Brunak et al., 1991; Burge and Karlin, 1997; Cartegni et al., 2003; Desmet et al., 2009). While these analyses might provide clues to the mechanisms controlling splicing of an alternative exon, defining the actual mechanism requires substantial experimental investigation.
Both RT-qPCR and RNA-seq indicated that TV2, lacking exons 3 and 4, is the most abundant transcript in human liver. This deletion does not introduce a frameshift, and TV2 is predicted to encode a protein that has a 75-amino-acid deletion relative to the full-length protein encoded by TV1. Because the active site of SULT1C4 (catalytic histidine residue) is located within exon 3, TV2 cannot encode a catalytically active sulfotransferase enzyme (Allali-Hassani et al., 2007).
Our analysis of the abilities of SULT1C4 TV1, TV2, and E3DEL to generate protein after transfection of expression plasmids into HEK293T cells indicated that TV1 produced much larger amounts of protein than did TV2 and that E3DEL produced no detectable protein, even though all TVs produced comparable amounts of mRNA. The lack of protein in E3DEL-transfected cells can be explained by the fact that deletion of exon 3 causes a frameshift that introduces a stop codon into exon 4 of the transcript. Introduction of this premature stop codon would likely cause nonsense-mediated decay of the mRNA, which might also explain, at least in part, why only small amounts of E3DEL were detected in Caco-2 and HepaRG cells and human liver samples. Our finding in the transiently transfected HEK293T cells that E3DEL-derived mRNA levels were not lower than TV1- or TV2-derived mRNA levels does not rule out this possibility, as nonsense-mediated mRNA decay from transiently transfected constructs was recently reported to be deficient in HEK293 cells (Gerbracht et al., 2017). The lower protein content observed in TV2-transfected cells is likely attributable to instability of the generated protein, which would have a massive loss of 75 amino acids relative to the wild-type protein.
The noncoding RNA that was detected in this study, ENST00000494122.1, is described as containing two exons and one retained intron, and it spans nt NC_000002:108,377,911 – 108,382,922 (i.e., 5012 nt in length) (Ensembl.org website). Alignment of ENST0000494122.1 to the SULT1C4 gene sequence indicates that exon 1 and intron 1 of ENST0000494122.1 are the same as exon 1 and intron 1 of TV1/2, whereas exon 2 of ENST0000494122.1 consists of exon 2, intron 2, exon 3, and ∼70% of intron 3, where it terminates within a 29-nt–long tract of adenosine residues. The biologic function of this noncoding RNA is unknown, including whether it plays any role in controlling the expression and/or alternative splicing of SULT1C4 mRNA.
It is now recognized that the vast majority of human genes are alternatively spliced (Johnson et al., 2003; Kampa et al., 2004; Matlin et al., 2005; Zhao, 2019). It is also recognized that many genes are transcribed into both protein-coding mRNAs and noncoding RNAs. This study establishes SULT1C4 as such a gene. While the gene can be expressed as a transcript (TV1) that encodes a full-length protein with robust sulfotransferase activity (isoform 1), a substantial portion of the mRNA consists of a variant that lacks two internal exons (TV2). The functional significance of TV2 is currently unclear, but this transcript cannot encode a functional sulfotransferase enzyme and does not even appear to generate a stable protein. It remains to be determined whether TV2 can function as a regulatory RNA. The main conclusion of the current study is that it provides a plausible explanation for our previously observed discrepancy between the levels of SULT1C4 mRNA and protein that were measured in human liver specimens: the more abundant TV2 produces relatively little protein.
Acknowledgments
The authors thank the University of Maryland Brain and Tissue Bank, the Central Laboratory for Human Embryology at the University of Washington, the Liver Tissue Cell Distribution System, and Xenotech, LLC for providing the tissue specimens that made this study possible.
Abbreviations
- DDK
Asp-Tyr-Lys-Asp-Asp-Asp-Asp-Lys
- nt
nucleotide
- RACE
rapid amplification of cDNA ends
- RNA-seq
RNA sequencing
- RT-PCR
reverse-transcription polymerase chain reaction
- RT-qPCR
reverse-transcription quantitative polymerase chain reaction
- SULT
cytosolic sulfotransferase
- TV
transcript variant
Authorship Contributions
Participated in research design: Dubaisi, Gaedigk, Vyhlidal, Kocarek, Runge-Morris.
Conducted experiments: Dubaisi, Fang, Caruso, Gaedigk, Vyhlidal
Performed data analysis: Dubaisi, Caruso, Vyhlidal, Kocarek.
Wrote or contributed to the writing of the manuscript: Dubaisi, Caruso, Vyhlidal, Kocarek, Runge-Morris.
Footnotes
This research was supported by the National Institutes of Health National Institute of Environmental Health Sciences [Grant R01 ES022606 (to M.R.-M.) and Center Grant P30 ES020957], NIH National Cancer Institute [Cancer Center Support Grant P30 CA022453], and the NIH Office of the Director [Shared Instrumentation Grant S10 OD 010700].
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Allali-Hassani A, Pan PW, Dombrovski L, Najmanovich R, Tempel W, Dong A, Loppnau P, Martin F, Thornton J, Edwards AM, et al. (2007) Structural and chemical profiling of the human cytosolic sulfotransferases [published correction appears in PLoS Biol (2007) 5:e165]. PLoS Biol 5:e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banoglu E. (2000) Current status of the cytosolic sulfotransferases in the metabolic activation of promutagens and procarcinogens. Curr Drug Metab 1:1–30. [DOI] [PubMed] [Google Scholar]
- Barker EV, Hume R, Hallas A, Coughtrie WH. (1994) Dehydroepiandrosterone sulfotransferase in the developing human fetus: quantitative biochemical and immunological characterization of the hepatic, renal, and adrenal enzymes. Endocrinology 134:982–989. [DOI] [PubMed] [Google Scholar]
- Bourgine J, Billaut-Laden I, Happillon M, Lo-Guidice JM, Maunoury V, Imbenotte M, Broly F. (2012) Gene expression profiling of systems involved in the metabolism and the disposition of xenobiotics: comparison between human intestinal biopsy samples and colon cell lines. Drug Metab Dispos 40:694–705. [DOI] [PubMed] [Google Scholar]
- Brunak S, Engelbrecht J, Knudsen S. (1991) Prediction of human mRNA donor and acceptor sites from the DNA sequence. J Mol Biol 220:49–65. [DOI] [PubMed] [Google Scholar]
- Burge C, Karlin S. (1997) Prediction of complete gene structures in human genomic DNA. J Mol Biol 268:78–94. [DOI] [PubMed] [Google Scholar]
- Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. (2003) ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res 31:3568–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmet FO, Hamroun D, Lalande M, Collod-Béroud G, Claustres M, Béroud C. (2009) Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res 37:e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duanmu Z, Weckle A, Koukouritaki SB, Hines RN, Falany JL, Falany CN, Kocarek TA, Runge-Morris M. (2006) Developmental expression of aryl, estrogen, and hydroxysteroid sulfotransferases in pre- and postnatal human liver. J Pharmacol Exp Ther 316:1310–1317. [DOI] [PubMed] [Google Scholar]
- Dubaisi S, Barrett KG, Fang H, Guzman-Lepe J, Soto-Gutierrez A, Kocarek TA, Runge-Morris M. (2018) Regulation of cytosolic sulfotransferases in models of human hepatocyte development. Drug Metab Dispos 46:1146–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubaisi S, Caruso JA, Gaedigk R, Vyhlidal CA, Smith PC, Hines RN, Kocarek TA, Runge-Morris M. (2019) Developmental expression of the cytosolic sulfotransferases in human liver. Drug Metab Dispos 47:592–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duniec-Dmuchowski Z, Rondini EA, Tibbs ZE, Falany CN, Runge-Morris M, Kocarek TA. (2014) Expression of the orphan cytosolic sulfotransferase SULT1C3 in human intestine: characterization of the transcript variant and implications for function. Drug Metab Dispos 42:352–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freimuth RR, Raftogianis RB, Wood TC, Moon E, Kim UJ, Xu J, Siciliano MJ, Weinshilboum RM. (2000) Human sulfotransferases SULT1C1 and SULT1C2: cDNA characterization, gene cloning, and chromosomal localization. Genomics 65:157–165. [DOI] [PubMed] [Google Scholar]
- Gardner-Stephen D, Heydel JM, Goyal A, Lu Y, Xie W, Lindblom T, Mackenzie P, Radominska-Pandya A. (2004) Human PXR variants and their differential effects on the regulation of human UDP-glucuronosyltransferase gene expression. Drug Metab Dispos 32:340–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbracht JV, Boehm V, Gehring NH. (2017) Plasmid transfection influences the readout of nonsense-mediated mRNA decay reporter assays in human cells. Sci Rep 7:10616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatt H, Pabel U, Meinl W, Frederiksen H, Frandsen H, Muckel E. (2004) Bioactivation of the heterocyclic aromatic amine 2-amino-3-methyl-9H-pyrido [2,3-b]indole (MeAalphaC) in recombinant test systems expressing human xenobiotic-metabolizing enzymes. Carcinogenesis 25:801–807. [DOI] [PubMed] [Google Scholar]
- Glatt H, Schneider H, Murkovic M, Monien BH, Meinl W. (2012) Hydroxymethyl-substituted furans: mutagenicity in Salmonella typhimurium strains engineered for expression of various human and rodent sulphotransferases. Mutagenesis 27:41–48. [DOI] [PubMed] [Google Scholar]
- Guidry AL, Tibbs ZE, Runge-Morris M, Falany CN. (2017) Expression, purification and characterization of human cytosolic sulfotransferase (SULT) 1C4. Horm Mol Biol Clin Investig 29:27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Her C, Wood TC, Eichler EE, Mohrenweiser HW, Ramagli LS, Siciliano MJ, Weinshilboum RM. (1998) Human hydroxysteroid sulfotransferase SULT2B1: two enzymes encoded by a single chromosome 19 gene. Genomics 53:284–295. [DOI] [PubMed] [Google Scholar]
- Hines RN. (2008) The ontogeny of drug metabolism enzymes and implications for adverse drug events. Pharmacol Ther 118:250–267. [DOI] [PubMed] [Google Scholar]
- Hui Y, Yasuda S, Liu MY, Wu YY, Liu MC. (2008) On the sulfation and methylation of catecholestrogens in human mammary epithelial cells and breast cancer cells. Biol Pharm Bull 31:769–773. [DOI] [PubMed] [Google Scholar]
- Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD. (2003) Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science 302:2141–2144. [DOI] [PubMed] [Google Scholar]
- Kampa D, Cheng J, Kapranov P, Yamanaka M, Brubaker S, Cawley S, Drenkow J, Piccolboni A, Bekiranov S, Helt G, et al. (2004) Novel RNAs identified from an in-depth analysis of the transcriptome of human chromosomes 21 and 22. Genome Res 14:331–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt AR, Schor IE, Alló M, Dujardin G, Petrillo E, Muñoz MJ. (2013) Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat Rev Mol Cell Biol 14:153–165. [DOI] [PubMed] [Google Scholar]
- Matlin AJ, Clark F, Smith CW. (2005) Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol 6:386–398. [DOI] [PubMed] [Google Scholar]
- Meinl W, Ebert B, Glatt H, Lampen A. (2008) Sulfotransferase forms expressed in human intestinal Caco-2 and TC7 cells at varying stages of differentiation and role in benzo[a]pyrene metabolism. Drug Metab Dispos 36:276–283. [DOI] [PubMed] [Google Scholar]
- Meloto CB, Segall SK, Smith S, Parisien M, Shabalina SA, Rizzatti-Barbosa CM, Gauthier J, Tsao D, Convertino M, Piltonen MH, et al. (2015) COMT gene locus: new functional variants. Pain 156:2072–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki Y, Nakata T, Suzuki T, Darnel AD, Moriya T, Kaneko C, Hidaka K, Shiotsu Y, Kusaka H, Sasano H. (2002) Systemic distribution of steroid sulfatase and estrogen sulfotransferase in human adult and fetal tissues. J Clin Endocrinol Metab 87:5760–5768. [DOI] [PubMed] [Google Scholar]
- Murray TJ, Maffini MV, Ucci AA, Sonnenschein C, Soto AM. (2007) Induction of mammary gland ductal hyperplasias and carcinoma in situ following fetal bisphenol A exposure. Reprod Toxicol 23:383–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera F, Hemminki K, Jedrychowski W, Whyatt R, Campbell U, Hsu Y, Santella R, Albertini R, O’Neill JP. (2002) In utero DNA damage from environmental pollution is associated with somatic gene mutation in newborns. Cancer Epidemiol Biomarkers Prev 11:1134–1137. [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. (2015) StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33:290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasool MI, Bairam AF, Kurogi K, Liu MC. (2017) On the sulfation of O-desmethyltramadol by human cytosolic sulfotransferases. Pharmacol Rep 69:953–958. [DOI] [PubMed] [Google Scholar]
- Richard K, Hume R, Kaptein E, Stanley EL, Visser TJ, Coughtrie MW. (2001) Sulfation of thyroid hormone and dopamine during human development: ontogeny of phenol sulfotransferases and arylsulfatase in liver, lung, and brain. J Clin Endocrinol Metab 86:2734–2742. [DOI] [PubMed] [Google Scholar]
- Rondini EA, Fang H, Runge-Morris M, Kocarek TA. (2014) Regulation of human cytosolic sulfotransferases 1C2 and 1C3 by nuclear signaling pathways in LS180 colorectal adenocarcinoma cells. Drug Metab Dispos 42:361–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runge-Morris M, Kocarek TA, Falany CN. (2013) Regulation of the cytosolic sulfotransferases by nuclear receptors. Drug Metab Rev 45:15–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara Y, Yanagisawa K, Katafuchi J, Ringer DP, Takami Y, Nakayama T, Suiko M, Liu MC. (1998) Molecular cloning, expression, and characterization of novel human SULT1C sulfotransferases that catalyze the sulfonation of N-hydroxy-2-acetylaminofluorene. J Biol Chem 273:33929–33935. [DOI] [PubMed] [Google Scholar]
- Stanley EL, Hume R, Coughtrie MW. (2005) Expression profiling of human fetal cytosolic sulfotransferases involved in steroid and thyroid hormone metabolism and in detoxification. Mol Cell Endocrinol 240:32–42. [DOI] [PubMed] [Google Scholar]
- Vo Ngoc L, Cassidy CJ, Huang CY, Duttke SH, Kadonaga JT. (2017) The human initiator is a distinct and abundant element that is precisely positioned in focused core promoters. Genes Dev 31:6–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Debrah-Pinamang M, DiModica NJ, Kurogi K, Naqvi AA, Hui Y, Sakakibara Y, Suiko M, Liu MC. (2016) Identification and characterization of the human cytosolic sulfotransferases mediating the sulfation of clioquinol and iodoquinol. Drug Metab Lett 10:200–205. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Liu MY, Kurogi K, Sakakibara Y, Saeki Y, Suiko M, Liu MC. (2015) Sulphation of acetaminophen by the human cytosolic sulfotransferases: a systematic analysis. J Biochem 158:497–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda S, Idell S, Fu J, Carter G, Snow R, Liu MC. (2007) Cigarette smoke toxicants as substrates and inhibitors for human cytosolic SULTs. Toxicol Appl Pharmacol 221:13–20. [DOI] [PubMed] [Google Scholar]
- Zhao S. (2019) Alternative splicing, RNA-seq and drug discovery. Drug Discov Today 24:1258–1267. [DOI] [PubMed] [Google Scholar]