Abstract
The misfolding avoidance hypothesis postulates that sequence mutations render proteins cytotoxic and therefore the higher the gene expression, the stronger the operation of selection against substitutions. This translates into prediction that relative toxicity of extant proteins is higher for those evolving faster. In the present experiment, we selected pairs of yeast genes which were paralogous but evolving at different rates. We expressed them artificially to high levels. We expected that toxicity would be higher for ones bearing more mutations, especially that overcrowding should rather exacerbate than reverse the already existing differences in misfolding rates. We did find that the applied mode of overexpression caused a considerable decrease in fitness and that the decrease was proportional to the amount of excessive protein. However, it was not higher for proteins which are normally expressed at lower levels (and have less conserved sequence). This result was obtained consistently, regardless whether the rate of growth or ability to compete in common cultures was used as a proxy for fitness. In additional experiments, we applied factors that reduce accuracy of translation or enhance structural instability of proteins. It did not change a consistent pattern of independence between the fitness cost caused by overexpression of a protein and the rate of its sequence evolution.
Keywords: protein misfolding, gene overexpression, maximum growth rate, competitive fitness
Introduction
The rate of molecular evolution—defined as the number of fixed mutations per unit of time per site, over a stretch of DNA—differs between genes by orders of magnitude (Zuckerkandl and Pauling 1965; Koonin and Wolf 2010). It seemed natural to expect that genes which continue to maintain the same and indispensable functions should be among those most conserved as their roles appear fixed. On the contrary, selection would rather guard them against even minor drops in functional efficiency following amino acid substitutions (Kimura and Ohta 1974; Hurst and Smith 1999). However, as subsequent and increasingly abundant data have revealed, the DNA sequence is most conserved not when a gene is important (essential for viability) but abundantly expressed (even if functionally compensable by another gene) (Pál et al. 2001, 2003; Rocha and Danchin 2004; Subramanian and Kumar 2004). The pattern is so clear and universal that it is likely grounded in some elementary constraints at the level of molecules. Specific hypotheses building on this premise have been fully reviewed elsewhere (Pál et al. 2006; Rocha 2006; Drummond and Wilke 2009; Zhang and Yang 2015; Echave et al. 2016; Echave and Wilke 2017). Of two chief explanations, one says that proteins which are needed at highest levels drain highest amounts of resources and therefore are under strongest selection to be functional and thus avoid the cost of resource misuse (Cherry 2010; Gout et al. 2010). The other posits that most abundant proteins are most efficiently conserved by selection because they would turn into highest amounts of toxic polypeptides if destabilized (Drummond et al. 2005; Yang et al. 2010). Both mechanisms appear plausible and may coexist. The present work focuses on the latter because experimental material was chosen in such a way that the compared groups of proteins were equally costly in terms of resource expenditure but potentially different in terms of toxicity.
Former tests of the outlined hypotheses have been typically indirect and comparative. The action of natural selection has not been demonstrated but only inferred from observed correlations between genes’ evolutionary rates and properties of their products. Direct tests would require to measure how differences in DNA sequences translate into differences in fitness under different levels of expression. However, genomes consist of thousands of genes each being responsible for only a small fraction of macromolecules present in a cell. Individual contributions of single genes to the cost of metabolism are therefore small, and any variation in these contributions caused by mutations must be also small. Even small differences in fitness do not escape operation of natural selection in large populations over long time periods (Lanfear et al. 2014) but are far from being detectable experimentally with currently available fitness assays (Blomberg 2011).
One potential remedy would be to overproduce gene variants so abundantly that any difference in fitness cost between them would increase sufficiently to make it detectable. Admittedly, superfluous protein is not just passive burden; it can deregulate the cell in an often unpredictable way. This problem would be circumvented if proteins under comparison were similar to each other in terms of function and structure. Then, not only the “material” but also “functional” costs of overexpression should be similar. On this background, an important difference could be the tendency to get destabilized, and then likely misfolded and aggregated, because this would change overproduced molecules from waste to toxin (Geiler-Samerotte et al. 2011; Farkas et al. 2018). This “toxicity” cost is, at least partly, uniform in its nature for many different proteins because aggregates of any polypeptides tend to interact with hydrophobic regions of other proteins and membranes in a generally similar way (Dobson 2003). Importantly, the cost of toxicity can increase or decrease substantially in response to even small alterations of amino acid sequence (Tomala et al. 2014). It is also unlikely that relative (in)stability of proteins will be reversed under overexpression, that is, those of them which tend to be more unstable when expressed at native levels will most likely remain such when overexpressed. We therefore assume that overexpression could be effective in revealing differences in toxicity due to increased instability of mistranslated chains (Drummond et al. 2005; Drummond and Wilke 2008) as well as due to normal instability of correctly synthesized ones (Yang et al. 2010). In fact, misfolding may be not necessary here. Overexpression could also amplify the negative effects of within-cell misinteractions of natively and stably folded proteins. It has been proposed that more abundant proteins evolve at lower rates because they are selected more intensely against inflicting such interactions (Yang et al. 2012). Therefore, we will use the term “toxicity” in a broad sense, meaning all possible negative consequences of the very presence of a protein in the cell.
Genes that are similar to each other in terms of sequence and function can be found among paralogs. In yeast, there are hundreds of paralogous gene pairs descending from a single whole-genome duplication. Pairs of paralogs often evolve at different rates and the more conserved ones are typically expressed at higher levels. This in itself has been regarded as evidence that more abundant proteins are more strictly policed by natural selection to maintain them more stable and therefore less toxic (Drummond et al. 2005). However, the critical link, lower toxicity of slow-evolving proteins, has been only postulated. In the present experiment, we overexpressed both slow- and fast-evolving paralogs to levels much higher than those seen under normal conditions. To boost potential differences in toxicity, we added compounds known to lower the accuracy of translation or stability of mature proteins. We then compared the fitness effect of overexpression of the slow- and fast-evolving paralogs by measuring their rate of growth and by directly competing them in pairs. We were able to demonstrate that the applied here overexpression was abundant and clearly damaging to fitness. However, the slow-evolving proteins were not less toxic than the fast-evolving ones.
Materials and Methods
Media, Strains, and Plasmids
Standard media, lysogeny broth for bacteria and synthetic complete (SC) for yeast, were used. Thorough the experiment, SC with glucose was used as repressing medium, SC with raffinose served to derepress the GAL1 promoter, and SC with rafinose and galactose was used to induce high expression of the cloned genes (Gelperin et al. 2005). The last medium served for fitness assays and, when needed, could be supplemented with 500 µg/ml azetidine-2-carboxylic acid (AZC), 200 µM paromomycin, or 5% ethanol. Cultures were grown in standard flat-bottom 96-well titration plates containing 150-µl aliquots of media and incubated without agitation at 30 °C (or 37 °C when specified).
We started our work with the MORF collection of single yeast open reading frames, each fused to an inducible promoter PGAL1 and C-terminus fused affinity tag His6-HA-ZZ. The constructs were cloned into a 2-μm plasmid containing the URA3 marker and hosted by a haploid yeast strain Y258, MATa pep4-3 his4-580 ura3-52 leu2-3 (Gelperin et al. 2005) derived from the S288c background (http://dharmacon.gelifesciences.com/resources/faqs/y258-used-yeast-orf-collection-derived-s288c). Of this collection, we selected 788 single strains, or 394 pairs, with cloned paralogous genes. Plasmids were extracted from them and used to transform Escherichia coli DH5α. Plasmids isolated from the resulting bacterial cultures were used to transform the Saccharomyces cerevisiae BY4741 strain, MATa his3 leu2 ura3. Due to discovered errors and omission in the plasmid collection and repeated failures in transformations of individual genes, the final BY4741 collection contained 311 complete pairs of paralogous genes. These strains were stored at −70 °C as 200-µl aliquots with 15% glycerol added and arrayed in titration plates. They were thawed and transferred with 96-pin replicator into fresh media to initiate every individual replicate of every experiment.
Measurements of Maximum Growth Rate
Samples of thawed strains were pinned into 150-µl aliquots of SC with 2% glucose and without uracil. After 24 h of incubation, 3 µl of the resulting cultures were transferred to 147 µl of SC with 2% raffinose and no uracil. After 48 h, 4.5-µl aliquots were transferred to 145.5 µl of SC lacking uracil but with both 2% raffinose and 2% galactose. Cultures initiated in this way were subject to periodic measurements of optical density every 1.5 h (absorbance of λ = 620 nm). Prior to each measurements, titration plates containing microcultures were agitated at 1,000 rpm for 2 min. Inspection of growth curves obtained in this way showed that growth was exponential within the range of OD 0.12–0.40 and therefore only such measurements were used to calculate the maximum growth rate (MGR) as regression of ln(OD) over time. Each strain was assayed twice in every environment. A few single estimates of regression were discarded as potentially erroneous but only when the associated with them squared Pearson’s correlation coefficients exceeded 0.98. Only a few individual estimates of MGR were higher than MGRmax = 0.32 (1/h), the rate of growth unaffected by overexpression (consult fig. 2). They were regarded random effects and not removed from analyses.
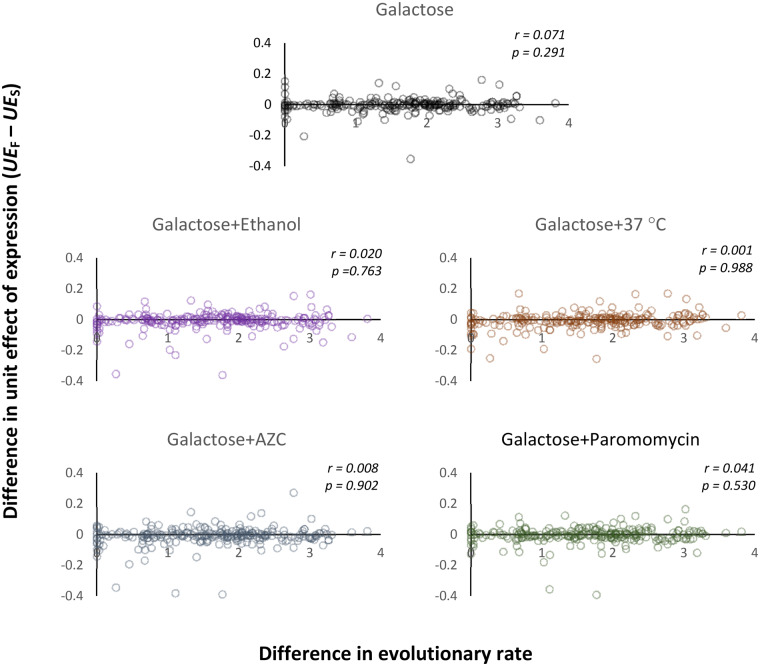
Fig. 2.—
The relation between the difference in the rate of molecular evolution within a pair of paralog genes and the difference in the toxicity of proteins overproduced from them. The within-pair difference in the ER is shown as transformed to ln[1 + 100(ERF − ERS)]. The UE of protein overexpression is calculated as the decrease in the MGR divided by the OL defined as percent of the total cellular protein. r stands for Pearson’s correlation coefficient.
Competition Experiments
Strains were thawed and brought to overnight stationary state cultures in SC without. From these, 311 cultures consisting of two paralogs in about equal numbers were composed. These paired cultures were transferred to fresh SC with raffinose and without uracil (5–145 µl) for 48 h. Samples of the resulting cultures were saved as initial mixes od competitors, other samples, whereas other samples, 5 µl, were transferred to 145 µl of test media (containing raffinose and galactose and supplemented as required). Samples of the resulting cultures were saved as final mixes of competitors. The stored samples of paired competitors (50 µl of each) were then gathered into ten pools and each pool subject to whole DNA extraction and then amplification of ORFs with common primers. Each of the ten groups contained cloned genes of similar length in order to minimize distortion of genes’ relative numbers at the time of polymerase chain reaction. Amplified DNA was fragmented and subject to NGS. Counts of identified gene fragments were used to estimate frequencies of competitors.
Protein Assays
Strains were transferred through media based on glucose, raffinose, and raffinose with galactose in the way described above. In the last medium, cells were harvested after 24 h of incubation, washed with ice cold water, and frozen. To start protein extraction, the cells were beaten with glass beads in 100 μl of lysis buffer (50 mM Tris–HCl, pH 7.5, 0.5% SDS, 0.1 mM ethylenediaminetetraacetic acid, and protease inhibitors) for about 1 h at 4 °C. Afterward, cell remnants were spun down and supernatant was collected. Total protein content was determined using a protocol developed in our earlier work (Tomala and Korona 2013). In short, proteins overexpressed from the MORF plasmids were tagged with the ZZ domain of the protein A. ELISA plates were coated with normal rabbit serum. Each strain lysate was mixed with equal amounts of protein A conjugated with HRP. During the assay, tags of the overproduced proteins and those of HRP-A conjugate competed for the Fc fragments of nonspecific antibodies from rabbit serum. Therefore, obtained for different proteins signal intensities were stronger for those less abundant. To convert those signals into the number of tags, we prepared a calibration curve for a purified tagged protein from the same MORF collection. Resulting values were multiplied by respective molar masses to obtain the mass of overproduced protein. The latter was then divided by the mass of total protein obtained for every tested strain in a BCA assay. This produced the final overexpression level (OL) estimates reported in the Results.
Results
Induced Overexpression of Paralogs
We started with the MORF collection of plasmids each containing a single yeast gene cloned after a promoter inducible by galactose. All plasmids were hosted by the same Y258 yeast strain. This strain has been previously used to estimate the fitness effect of gene overexpression applying methods that were either qualitative (Gelperin et al. 2005; Vavouri et al. 2009) or quantitative in intention but not sufficiently precise in execution and interpretation (Tomala and Korona 2013). In the present research, we planned to get as exact as possible estimates of two clearly defined traits: the MGR of individual strains and their competitive ability in direct confrontations. The Y258 strain proved unsuitable for this purpose. A major problem was poor and nonreproducible growth in media required to prepare and then induce overexpression. We therefore moved the plasmids into a BY strain which grew more robustly and steadily for a large majority of cloned genes. The new collection totaled 311 complete pairs of paralogous yeast genes (supplementary table 1, Supplementary Material online).
We then asked how suitable is this particular set of strains for the planned tests. First, we obtained a measure of relative evolution rate, ER, calculated here as a proportion of substitutions in every Saccharomyces cerevisiae gene aligned with its closest homolog in Kluyveromyces waltii (supplementary table 2, Supplementary Material online). We then asked whether these estimates correlate with estimates of native cellular protein content as listed in a data set integrating results of multiple previous gene expression studies (Wang et al. 2015). The expected negative correlation was manifestly present: Pearson’s r = −0.684; t = −23.272; df = 616, P ≪ 0.0001 (supplementary fig. 1, Supplementary Material online).
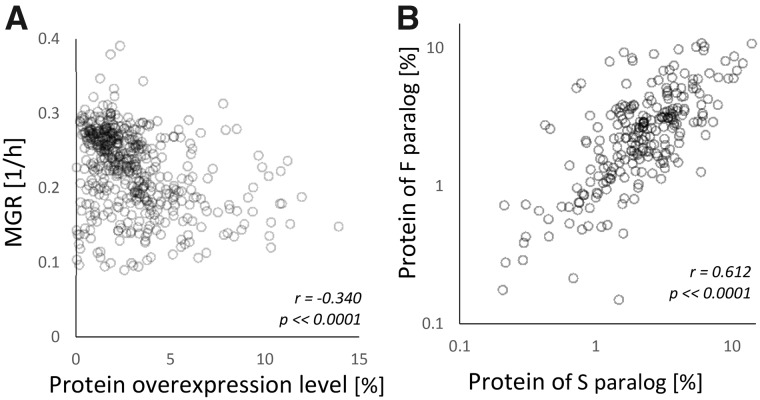
Another assumption of this work was that overexpression could be made abundant to a point where the load of superfluous polypeptides had a negative impact on fitness. We measured the level of overexpression and the MGR of overexpressing strains in our basic overexpression medium (galactose only). Figure 1A shows that overproduced proteins were abundant, up to several percent of the total cellular protein content. There was also a significant negative correlation between the OL and MGR indicating that the very amount of overproduced protein had substantial effects on fitness. Figure 1B demonstrates that the induced expression of the slow- and fast-evolving genes within pairs tended to resemble each other as correlation between them was evident. However, the variation within pairs was nontrivial (fig. 1B). Therefore, the following analyses of the fitness effects of overexpression will take into account both the potential toxicity (predicted from the rate of evolutionary change) and the amount of superfluous protein.
Fig. 1.—
Cellular level of protein overexpression. (A) Pearson’s correlation coefficient between the single-gene OL (% of total cellular protein) and MGR of an overexpressing strain. (B) Pearson’s correlation coefficient between protein level of S(low)- and F(ast)-evolving genes within pairs of paralogs.
Rate of Evolution and Growth Effect of Overexpression
Supplementary figure 2, Supplementary Material online, shows that correlation between replicate MGR estimates was high but only when overexpression was activated, verifying the expectation that fitness was determined by the overexpressing plasmid and not its host. Replicate estimates of the OL were also well correlated (supplementary fig. 3, Supplementary Material online). Strain’s means of both measures are used in the following statistical tests. (Individual MGRs and OLs are listed in supplementary tables 3 and 4, Supplementary Material online.) With these data, we could calculate the unit effect (UE) of protein overexpression for every fast (F) and slow (S) paralog within each pair. This was done by subtracting the measured MGR from MGRmax (unaffected by overexpression) and dividing it by the overproduction level: UE = (MGRmax − MGR)/OL. It was then possible to test the toxicity hypothesis: the larger the difference in the ER (ERF − ERS), the larger the difference in the damaging effect of the compared paralogs (UEF − UES). Figure 2 demonstrates that this prediction was not met: there was no significant association between UEF − UES and ERF − ERS. The result was the same in the plain overexpression medium (galactose only) and in four additional, independently tested, environments: galactose together with two factors depressing accuracy of translation (AZC or paromomycin) and two other ones promoting misfolding of polypeptides (heat stress at 37 °C or addition of ethanol). Thus, environmental conditions which were meant to increase the fitness cost of harboring excessive protein did not change the pattern of independence between toxicity and evolutionary rate.
In an alternative analysis, we did not calculate the UE of overexpression from estimates of the MGR and OL but kept the two latter separate. That is, we asked in a multiple regression analysis whether the between paralog divergence in growth rate (MGRS − MGRF) was explained by (OLS − OLF) or (ERF − ERS). Results for all tested environments are summarized in supplementary table 5, Supplementary Material online. They clearly point to the overproduction level and not the rate of evolution, confirming conclusions derived from former tests.
The fact that the additional media did not change the overall result does not mean that they had no effect. In the basic test environment, an average MGR of all overexpressing strains was 0.2210 ± 0.0063 (1/h) (mean and 99% confidence interval). Growth was always slowed down under stress, that is, after addition of AZC (0.2098 ± 0.0076), ethanol (0.1757 ± 0.0052), and paromomycin (0.1781 ± 0.0060) or shift from 30 to 37 °C (0.1900 ± 0.0069). Importantly, the observed downward shift was largely parallel for individual strains. We saw this when we compared the fitness distance between paralogs, MGRS − MGRF, across environments. Of all possible ten pairwise correlations between five environments, the lowest Pearson’s r was 0.625 and the highest 0.808; of the associated P values, the highest was 10−31. Thus, the impact of additional factors was to decrease the rate of growth in a mostly uniform way and not to introduce significantly new patterns of variation.
Competitive Ability of the Slow- and Fast-Evolving Paralogs under Overexpression
We then tested every pair of paralogs in competition, that is, direct confrontation in a shared batch culture. It tested not only the maximum rate but also other traits, such as time needed to leave the lag phase. The effect of competition was measured by estimating the change in relative abundance of competitors, slow- and fast-evolving paralogs (S and F), in both the basic test environment (galactose) as well as modified ones (galactose plus AZC, ethanol or 37 °C). Counts of NS and NF were obtained by polymerase chain reaction amplification followed by identification of the resulting fragments by NGS and are listed in supplementary table 6, Supplementary Material online.
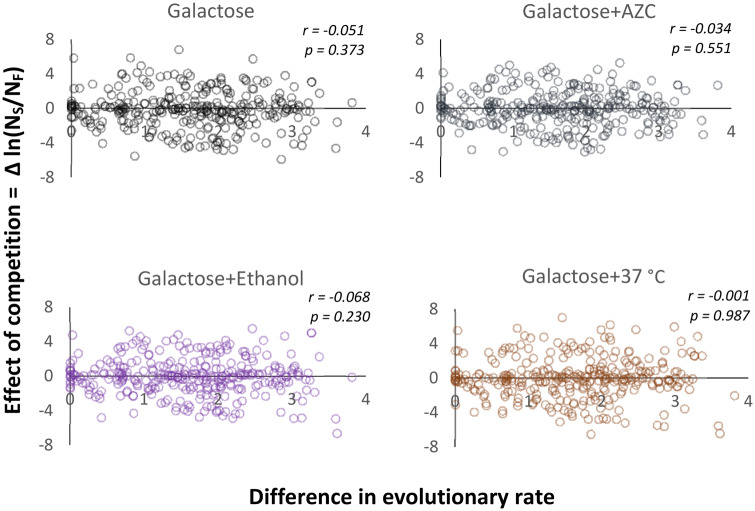
We estimated log-ratios of paralogous pairs, ln(NS/NF), at the beginning of growth in the galactose-based medium and its end, that is, after about 6.6 generations (cell division) of competition. Figure 3 shows that strains with slow-evolving paralogs had no visible competitive advantage over those with fast-evolving ones, that is, Δln(NS/NF) did not increase positively with ERF − ERS in any of the four applied test environments. We then asked whether the result would change if not only differences in the evolutionary distance but also expression level between paralogs were accounted for (multiple regression). Note that, unlike MGR, Δln(NS/NF) is a measure of fitness tied to both compared strains and therefore any adjustments for the unequal OL of superfluous protein cannot be done for individual strains. We applied either the ratio OLS/OLF or difference OLS − OLF as an explaining variable but none of them yielded statistically significant results (supplementary table 7, Supplementary Material online).
Fig. 3.—
The relation between the difference in the rate of molecular evolution within a pair of paralog genes and the result of competition under overexpression. The within-pair difference in the ER is shown as transformed to ln[1 + 100(ERF − ERS)]. The effect of competition is shown as a change in the log-ratio of relative abundance over a period of common growth. r stands for Pearson’s correlation coefficient.
We then compared the effect of competition, Δln(NS/NF), across four test environments and found that individual pairs of paralogs behaved similarly in all of them. Of all possible six pairwise correlations between four environments, the lowest Pearson’s r was 0.685 and the highest 0.888; of the associated P values, the highest was on the order of 10−44. This high uniformity of results highlights three points. First, our estimates of competitive ability came out repeatable, and thus reliable, even though estimation of frequencies of competitors was done through massive amplification and sequencing which are both prone to errors. Second, the impact of overexpression on competitive ability did not correlate with the rate of molecular evolution of the overexpressed protein. The third conclusion is that the impact of chemicals added to destabilize proteins was not higher for those fast evolving.
Finally, we asked whether the two applied here proxies for fitness, growth rate and competitive ability, yielded consistent results. MGRS − MGRF averaged over five test environments correlated positively with Δln(NS/NF) averaged over four test environments: Pearson’s r = 0.248, t = 4.478, P = 1.06E-05. A likely explanation for the relatively modest correlation is that the outcome of competition is only partly determined by the rate of growth while the length of lag is also important. The latter was apparently not tied to the former and, more importantly, did not introduce any systematic difference between the slow- and fast-evolving paralogs.
Discussion
Our first finding was that gene overexpression affected fitness negatively and that this negative effect increased with the amount of overexpressed protein. This general trend was accompanied by substantial variation in individual cases but it has been already reported that the response of fitness to protein overproduction can be remarkably heterogeneous (Keren et al. 2016). Our main question was more specific: Is the burden of overexpression lower for proteins which are under more intense purifying selection? We compared fitness of slow- and fast-evolving genes within pairs of paralogs. We applied abundant overexpression and conditions promoting misfolding. Nevertheless, we saw no indications that the fast-evolving proteins tended to be more harmful than slow-evolving ones.
The hypotheses invoking misfolding toxicity are founded on the observation that a relatively large fraction of mutations destabilize protein structures (Pakula and Sauer 1989; Chakshusmathi et al. 2004). But, how much is this true for the substitutions which actually reside in the coding sequences of existing genes (Wang and Moult 2001)? More specifically, do the destabilizing mutations constitute a sizable portion of the “excess” substitutions present in the fast-evolving genes? Our experiment failed to provide a positive answer. Interestingly, a similar conclusion emerges from another recent study in which entirely different experimental approach was applied. Proteomes of four species were assayed for melting temperatures of individual proteins in vivo (Leuenberger et al. 2017). As it has turned out, highly expressed proteins do not tend to have higher melting temperatures and thus are not less likely to get destabilized. If so, misfolding toxicity is unlikely to constrain protein ER (Plata and Vitkup 2018). In response to this claim, it has been pointed out that melting temperature does not relate very well to energy of (un)folding which is a standard measure of stability in vitro and that the measurements of melting temperature may be not sufficiently accurate when applied simultaneously to all proteins of a living cell (Razban 2019). Nevertheless, we think that the lack of correlation between melting temperature and protein abundance is much telling. First, in vitro measurements of the folding energy of a purified protein can well be superior to in vivo measurements of the melting temperature in terms of precision and conceptual clarity. However, this does not necessarily make them more relevant to assess cytotoxicity, that is, an improper behavior toward other macromolecules in the crowded cell interior. Second, the use of an imperfect measure should decrease any correlation, but not destroy it. The absence of any detectable correlation with a sample size in the thousands and a wide range of protein abundance is a point against the misfolding toxicity hypothesis. We suggest that there is potentially more questionable aspect of the discussed study: It measures protein stability (melting temperature) but not toxicity (negative fitness effect). One might argue that this compromises its relevance because even if fast-evolving proteins do not misfold more frequently they can be more damaging to fitness after misfolding. Our experiments address this variant of the protein toxicity hypothesis as well, and find that it, too, is not empirically supported. In this way, the two experiments complement each other in providing negative evidence for the hypotheses linking the rate of sequence evolution with the toxicity constraint.
The fact that some proteins evolve fast whereas others evolve slow is not an evolutionary enigma of ancient origin which can be studied only by comparison and speculation. The pattern of negative rate-expression correlation is seen not only when evolutionary distant organisms are involved but also when analyses are restricted to individuals of a single species (Marek and Tomala 2018). Thus, it must be constantly recreated by regularly operating mechanism(s) which will be eventually identified. The search promises to be engaging as the arguably most prominent of current hypotheses, those invoking protein toxicity constraint, do no find support in initial experimental tests.
Supplementary Material
Acknowledgments
Funds were provided by grants from National Science Centre of Poland (NCN 2013/11/B/NZ2/00122 and NCN 2017/25/B/NZ2/01036 to R.K. and NCN 2012/07/D/NZ8/03975 to P.S.).
Literature Cited
- Blomberg A. 2011. Measuring growth rate in high-throughput growth phenotyping. Curr Opin Biotechnol. 22(1):94–102. [DOI] [PubMed] [Google Scholar]
- Chakshusmathi G, et al. 2004. Design of temperature-sensitive mutants solely from amino acid sequence. Proc Natl Acad Sci U S A. 101(21):7925–7930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry JL. 2010. Expression level, evolutionary rate, and the cost of expression. Genome Biol Evol. 2(0):757–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson CM. 2003. Protein folding and misfolding. Nature 426(6968):884–890. [DOI] [PubMed] [Google Scholar]
- Drummond DA, Bloom JD, Adami C, Wilke CO, Arnold FH.. 2005. Why highly expressed proteins evolve slowly. Proc Natl Acad Sci U S A. 102(40):14338–14343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO.. 2008. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell 134(2):341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO.. 2009. The evolutionary consequences of erroneous protein synthesis. Nat Rev Genet. 10(10):715–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echave J, Spielman SJ, Wilke CO.. 2016. Causes of evolutionary rate variation among protein sites. Nat Rev Genet. 17(2):109–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echave J, Wilke CO.. 2017. Biophysical models of protein evolution: understanding the patterns of evolutionary sequence divergence. Annu Rev Biophys. 46(1):85–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas Z, et al. 2018. Hsp70-associated chaperones have a critical role in buffering protein production costs. Elife 7:e29845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiler-Samerotte KA, et al. 2011. Misfolded proteins impose a dosage-dependent fitness cost and trigger a cytosolic unfolded protein response in yeast. Proc Natl Acad Sci U S A. 108(2):680–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelperin DM, et al. 2005. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev. 19(23):2816–2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gout J-F, Kahn D, Duret L; Paramecium Post-Genomics Consortium. 2010. The relationship among gene expression, the evolution of gene dosage, and the rate of protein evolution. PLoS Genet. 6(5):e1000944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst LD, Smith NG.. 1999. Do essential genes evolve slowly? Curr Biol. 9(14):747–750. [DOI] [PubMed] [Google Scholar]
- Keren L, et al. 2016. Massively parallel interrogation of the effects of gene expression levels on fitness. Cell 166(5):1282–1294.e1218. [DOI] [PubMed] [Google Scholar]
- Kimura M, Ohta T.. 1974. On some principles governing molecular evolution. Proc Natl Acad Sci U S A. 71(7):2848–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Wolf YI.. 2010. Constraints and plasticity in genome and molecular-phenome evolution. Nat Rev Genet. 11(7):487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear R, Kokko H, Eyre-Walker A.. 2014. Population size and the rate of evolution. Trends Ecol Evol. 29(1):33–41. [DOI] [PubMed] [Google Scholar]
- Leuenberger P, et al. 2017. Cell-wide analysis of protein thermal unfolding reveals determinants of thermostability. Science 355(6327):eaai7825. [DOI] [PubMed] [Google Scholar]
- Marek A, Tomala K.. 2018. The contribution of purifying selection, linkage, and mutation bias to the negative correlation between gene expression and polymorphism density in yeast populations. Genome Biol Evol. 10:2986–2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakula AA, Sauer RT.. 1989. Genetic analysis of protein stability and function. Annu Rev Genet. 23(1):289–310. [DOI] [PubMed] [Google Scholar]
- Pál C, Papp B, Hurst LD.. 2001. Highly expressed genes in yeast evolve slowly. Genetics 158(2):927–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pál C, Papp B, Hurst LD.. 2003. Genomic function (communication arising): rate of evolution and gene dispensability. Nature 421(6922):496–497. [DOI] [PubMed] [Google Scholar]
- Pál C, Papp B, Lercher MJ.. 2006. An integrated view of protein evolution. Nat Rev Genet. 7(5):337–348. [DOI] [PubMed] [Google Scholar]
- Plata G, Vitkup D.. 2018. Protein stability and avoidance of toxic misfolding do not explain the sequence constraints of highly expressed proteins. Mol Biol Evol. 35(3):700–703. [DOI] [PubMed] [Google Scholar]
- Razban RM. 2019. Protein melting temperature cannot fully assess whether protein folding free energy underlies the universal abundance–evolutionary rate correlation seen in proteins. Mol Biol Evol. 36(9):1955–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha EP. 2006. The quest for the universals of protein evolution. Trends Genet. 22(8):412–416. [DOI] [PubMed] [Google Scholar]
- Rocha EP, Danchin A.. 2004. An analysis of determinants of amino acids substitution rates in bacterial proteins. Mol Biol Evol. 21(1):108–116. [DOI] [PubMed] [Google Scholar]
- Subramanian S, Kumar S.. 2004. Gene expression intensity shapes evolutionary rates of the proteins encoded by the vertebrate genome. Genetics 168(1):373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomala K, Korona R.. 2013. Evaluating the fitness cost of protein expression in Saccharomyces cerevisiae. Genome Biol Evol. 5(11):2051–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomala K, Pogoda E, Jakubowska A, Korona R.. 2014. Fitness costs of minimal sequence alterations causing protein instability and toxicity. Mol Biol Evol. 31(3):703–707. [DOI] [PubMed] [Google Scholar]
- Vavouri T, Semple JI, Garcia-Verdugo R, Lehner B.. 2009. Intrinsic protein disorder and interaction promiscuity are widely associated with dosage sensitivity. Cell 138(1):198–208. [DOI] [PubMed] [Google Scholar]
- Wang M, Herrmann CJ, Simonovic M, Szklarczyk D, von Mering C.. 2015. Version 4.0 of PaxDb: protein abundance data, integrated across model organisms, tissues, and cell-lines. Proteomics 15(18):3163–3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Moult J.. 2001. SNPs, protein structure, and disease. Hum Mutat. 17(4):263–270. [DOI] [PubMed] [Google Scholar]
- Yang J-R, Liao B-Y, Zhuang S-M, Zhang J.. 2012. Protein misinteraction avoidance causes highly expressed proteins to evolve slowly. Proc Natl Acad Sci U S A. 109(14):E831–E840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JR, Zhuang SM, Zhang J.. 2010. Impact of translational error-induced and error-free misfolding on the rate of protein evolution. Mol Syst Biol. 6(1):421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Yang J-R.. 2015. Determinants of the rate of protein sequence evolution. Nat Rev Genet. 16(7):409–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerkandl E, Pauling L.. 1965. Evolutionary divergence and convergence in proteins. In: Bryson V, Vogel HJ, editors. Evolving genes and proteins. New York: Academic Press. p. 97–166. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.