Abstract
Aeromonads are ubiquitous aquatic bacteria that cause opportunistic infections in humans, but their pathogenesis remains poorly understood. A pathogenomic approach was undertaken to provide insights into the emergence and evolution of pathogenic traits in aeromonads. The genomes of 64 Aeromonas strains representative of the whole genus were analyzed to study the distribution, phylogeny, and synteny of the flanking sequences of 13 virulence-associated genes. The reconstructed evolutionary histories varied markedly depending on the gene analyzed and ranged from vertical evolution, which followed the core genome evolution (alt and colAh), to complex evolution, involving gene loss by insertion sequence-driven gene disruption, horizontal gene transfer, and paraphyly with some virulence genes associated with a phylogroup (aer, ser, and type 3 secretion system components) or no phylogroup (type 3 secretion system effectors, Ast, ExoA, and RtxA toxins). The general pathogenomic overview of aeromonads showed great complexity with diverse evolution modes and gene organization and uneven distribution of virulence genes in the genus; the results provided insights into aeromonad pathoadaptation or the ability of members of this group to emerge as pathogens. Finally, these findings suggest that aeromonad virulence-associated genes should be examined at the population level and that studies performed on type or model strains at the species level cannot be generalized to the whole species.
Keywords: Aeromonas, opportunistic pathogens, genomes, pathogenomics, evolution, pathoadaptation
Introduction
Aeromonads are ubiquitous Gram-negative bacilli primarily found in freshwater environments. The population structure of the genus Aeromonas has several characteristics that favor an evolutionary mode of species complexes that are heterogeneous groups of closely related but genetically distinct strains. These characteristics include a high rate of horizontal genetic transfer (HGT), a large genome harboring several ribosomal operons and a large pangenome encoding, in these bacteria, various metabolic capabilities supporting their adaptation to environmental changes and numerous virulence factors (Seshadri et al. 2006; Reith et al. 2008; Georgiades and Raoult 2011; Talagrand-Reboul et al. 2017). Moreover, aeromonads in the same aquatic habitat may be physically related, which allows sympatric speciation. In addition, this genus is a reservoir from which some species or subspecies may have emerged by allopatric speciation and specialized and/or adapted to particular niches, such as the specialized fish pathogen Aeromonas salmonicida subsp. salmonicida (Reith et al. 2008).
Aeromonas are emerging opportunistic pathogens often with an environment-to-human transmission route, resulting in a broad range of infections in humans (Janda and Abbott 2010; Khajanchi et al. 2010; Parker and Shaw 2011). Among the 30 validated species in the genus Aeromonas, 4 are most often associated with human diseases: Aeromonas dhakensis, A. hydrophila, A. veronii, and A. caviae (Janda and Abbott 2010; Chen et al. 2014; Wu et al. 2015). Additionally, aeromonads are able to colonize a wide range of animals, and some species, namely, A. salmonicida, A. hydrophila, A. veronii, A. bestiarum, and A. piscicola, are especially pathogenic to fish, causing septicemia and ulcerative and hemorrhagic diseases (Kozińska 2007; Janda and Abbott 2010; Beaz-Hidalgo and Figueras 2013).
As a characteristic of many opportunistic pathogens, the pathogenesis of Aeromonas infections is complex, multifactorial, and only partially elucidated to date. Aeromonads can express a wide repertoire of virulence factors involved in biofilm formation, cell adherence, invasion, and cytotoxicity, including polar and lateral flagella (Rabaan et al. 2001; Gavín et al. 2003), adhesins (Kirov et al. 1999), lipopolysaccharides (Canals et al. 2007), iron-binding systems (Byers et al. 1991; Massad et al. 1991), numerous extracellular toxins and enzymes (Braun et al. 2002) exported by different types of secretion systems (e.g., type 2 secretion system and type 3 secretion system [T3SS]) (Burr et al. 2002; Sha et al. 2005), and quorum-sensing systems (Swift et al. 1997, 1999; Kozlova et al. 2008; Khajanchi et al. 2012) that are critical for colonization (infection) and disease.
Whatever their provenance, Aeromonas spp. isolates harbor similar assemblages of virulence-associated genes (Chacón et al. 2003; Aguilera-Arreola et al. 2005), which has led to the presumption that exaptation occurred in environmental aeromonads (Cabello and Godfrey 2018), which facilitated host colonization, followed by pathoadaptation, which then resulted in adverse outcomes for the host. The elucidation of Aeromonas pathogenesis has been hampered by a number of impediments, such as an ambiguous correlation between virulence phenotypes and genetic content and low performance of tools for the detection of virulence-associated genes (Talagrand-Reboul et al. 2018). Conversely, current whole-genome sequencing approaches provide high-quality sequences to accurately study virulence genes and their genetic microenvironment.
In this context, we support that Aeromonas is an opportunistic bacterial pathogen characterized by various ecological niches, which can be considered “nurseries” suitable for genomic exchanges and rearrangements concerning genes involved in adaptation and virulence. This genomic flexibility, which is associated with the lifestyle of Aeromonas, may allow lineages to emerge that harbor, among others, capabilities of colonization and adhesion, in escaping innate immunity and in the production and secretion of toxins and exoenzymes. We hypothesized that the evolutionary process of virulence-associated genes will provide insight into pathoadaptation, and several questions were raised: Does the evolution of virulence genes follow the overall evolution of the genus? Are there any HGT of genes encoding virulence factors? Is the genetic microenvironment of these virulence genes conserved among strains? To provide the genomic basis of pathoadaptation in the genus Aeromonas, we studied a panel of virulence-associated genes from a large collection of genomes that reflect the current known diversity in the genus. We estimated the correlation between the genome-based phylogeny and the virulence-associated gene repertoire and used this information to address the questions raised and to form new hypotheses about the gain, maintenance, or loss of virulence factors throughout the genus Aeromonas.
Materials and Methods
Strains and Genomes
A total of 64 Aeromonas genomes were included in this study and are described in supplementary table S1, Supplementary Material online. They covered the 30 validated species and represented the type strain of every species or a reference strain in the case of A. rivipollensis, for which the genome of the type strain is not yet sequenced (supplementary table S1, Supplementary Material online). For the species with clinical relevance, we included several strains: A. hydrophila (n = 7), A. dhakensis (n = 6), A. veronii (n = 7), A. caviae (n = 7), A. salmonicida (n = 7), A. rivipollensis (n = 3), and A. media (n = 3). Whole genome sequencing was performed on four strains (this study), and 40 genomes were sequenced previously (Colston et al. 2014; Mosser et al. 2015; Talagrand-Reboul et al. 2018). The remaining 20 genomes were obtained from the public genome repository of NCBI (supplementary table S1, Supplementary Material online). Genome sequencing was performed at the Microbial Analysis, Resources and Services facility at the University of Connecticut (Storrs, USA) using an Illumina MiSeq benchtop sequencer after preparing libraries from the genomic DNA using a Nextera XT DNA sample preparation kit (Illumina, San Diego, CA). Paired-end reads were trimmed and assembled into scaffolded contigs using a de novo assembler of CLC Genomics Workbench version 6.1.5 (CLC-bio, Aarhus, Denmark) to obtain “improved high-quality draft genomes” (Chain et al. 2009). In all instances in which an isolate was reclassified as a different species (e.g., A. hydrophila subsp. anaerogenes CECT 4221, which was reclassified to the species A. caviae; Miñana-Galbis et al. 2013), we used the validated taxon to avoid any confusion. The draft genomes included are “high-quality draft genomes” (supplementary table S1, Supplementary Material online) based on sequencing and assembly metrics (e.g., average genome coverage and number of scaffolds) and verification of automated annotation (the presence of 15 housekeeping genes: atpD, dnaJ, dnaK, dnaX, gltA, groL, gyrA, gyrB, metG, radA, recA, rpoB, rpoD, tsf, and zipA).
Genome Analysis
Complete and draft genomes were annotated using the RAST server to identify RNAs and protein-coding genes (Overbeek et al. 2014). Based on the quality metrics, all the genomes included in this study were sufficient for the assessment of virulence-associated gene content and the comparison between strains (supplementary table S1, Supplementary Material online) (Chain et al. 2009). The genomes were screened for genes encoding virulence factors acting by various mechanisms (toxins, enzymes, secretion system components, and flagellin) and well characterized in Aeromonas spp. (table 1) by using reference protein sequences and either translated sequences of the validated subset of UniProt (SwissProt) or annotated genes of the previously sequenced Aeromonas spp. in the TrEMBL database. Sequence comparisons with reference protein sequences were performed with SEED viewer, which uses bidirectional protein–protein BLAST (BlastP) sequence comparison of translated open reading frames. Proteins with amino acid sequence similarities ≥65% and E-values ≤10−10 were considered homologs (Altschul and Lipman 1990). All the results of BlastP analysis were manually verified. Sequences homologous to virulence-associated genes were checked to identify coding sequences (CDSs) that harbor an open reading frame without nonsense mutations. The HMMSCAN program (HMMER website, EMBL-EBI, Potter et al. 2018) was used to evaluate the putative impact of nucleotide polymorphisms between homologs on protein functions. Amino acids in aerolysin that are critical for oligomerization (H in 155) and heptamerization (K in 374 and E in 390) were examined in the protein homologs (Degiacomi et al. 2013). The neighboring CDS order of the considered loci, or “flanking gene organization,” was qualitatively compared among the different strains on the basis of RAST annotation when genes were not at the end of a contig or interrupted by contig gaps. The analyzed region on each side of the genes included 1) the right and left directly flanking coding DNA sequences, 2) an extended region up to three CDS positions if these configurations were conserved in the majority of strains, and 3) a more extended region that would be gathered with one flanking gene found in another close position, up to seven CDSs. To detect putative HGT, we manually compared the relative branching order of the 13 virulence gene phylogenies within each phylogroup and within each species when several strains had been included. Mobile genetic elements (MGEs) were searched by using ISsaga2 (Mobile Genetic Elements team- CNRS, UMR5100, Toulouse, France) to detect insertion sequences (ISs) and ICEfinder (Microbial Bioinformatics Group, Shanghai, China) to detect integrative and mobilizable elements (IMEs) and integrative and conjugative elements (ICEs). We manually verified whether virulence genes were located in the predicted IME/ICE regions.
Table 1.
Aeromonad Virulence Factors Studied
| Virulence Factor | Reference Sequences |
Virulence-Associated Gene | Genomic Location | Source | ||
|---|---|---|---|---|---|---|
| Accession No. | Strain | Length (Amino Acids) | ||||
| Aerolysin AerA (syn: Cytolytic enterotoxin Act) |
|
493493 | aer (syn: act) | Chromosome | Howard et al. (1987), Chakraborty et al. (1987), Chopra et al. (1993) | |
| Thermolabile cytotonic enterotoxin | Q44061E | A. dhakensis SSU | 368 | alt | Chromosome | Chopra et al. (1996) |
| Thermostable cytotonic enterotoxin | Q8VRN3E | A. dhakensis SSU | 636 | ast | Chromosome | Sha et al. (2002) |
| Extracellular collagenase | J7FWV3E | A. piscicola AH-3 | 915 | colAh | Chromosome | Duarte et al. (2015) |
| Toxin RtxA (repeat-in-toxin A) | A0KHZ7E | A. hydrophila subsp. hydrophila ATCC 7966T | 4,685 | rtxA | Chromosome | Suarez et al. (2012) |
| Exotoxin A | A0A0W0AX19E | A. salmonicida Y577 | 639 | exoA | Unknown (chromosome in P. aeruginosa PA01) | Tsaur and Clowes (1989), Ponnusamy et al. (2016), Vincent et al. (2016) |
| SST3 needle protein AscF | Q6WG33E | A. veronii 283c | 85 | ascF | Chromosome or plasmid (chromosome in A. hydrophila ANNIH1 and ALO6-06/ plasmid in A. salmonicida subsp. salmonicida A449) | Chacon et al. (2004), Reith et al. (2008); Tekedar et al. (2015),Hughes et al. (2016) |
| SST3 component AscG | Q6WG32E | A. veronii 283c | 116 | ascG | ||
| SST3 Inner membrane channel protein AscV | A4SUH2E | A. salmonicida subsp. salmonicida A449 | 721 | ascV | ||
| ADP-ribosyltransferase toxin AexT | Q93Q17S | A. salmonicida subsp. salmonicida A449 | 475 | aexT | Chromosome | Stuber et al. (2003), Dacanay et al. (2006), Reith et al. (2008), Silver and Graf (2009) |
| ADP-ribosyltransferase toxin AexU | D5LUP3E | A. veronii bv. sobria AeG1 | 512 | aexU | Chromosome | Sha et al. (2007), Silver and Graf (2009), Abolghait et al. (2011) |
| Lateral flagellin A | Q93TL9E | A. caviae Sch3 | 281 | lafA | Chromosome | Kirov et al. (2002), Canals et al. (2006) |
| Serine protease Ahe2 | A4SNU7E | A. salmonicida subsp. salmonicida A449 | 625 | ser (syn.: ahe2) | Chromosome | Reith et al. (2008) |
Note.—S/E, accession numbers correspond to protein sequences in SwissProt or TrEMBL databases.
Table 2.
Summary of Targeted Virulence Gene Analysis Results in Aeromonad Genomes
| Virulence-Associated Genes | RAST Annotation of Coding DNA Sequence (Length in Amino acids, AA) | % of Presence (%) | Present | Absent | Flanking Sequences |
|
|---|---|---|---|---|---|---|
| Upstream | Downstream | |||||
| aer/act | Hemolysin (481–505 AA) | 52 | Most phylogroups | Caviae and Media phylogroups | Hypothetical protein | Mostly hydroxymethyl pyrimidine phosphate synthase ThiC |
| alt | Putative lipase (791–819 AA) | 97 | Most phylogroups | A. fluvialis and A. lacus | Hypothetical protein | GlpG except in the Schubertii phylogroup |
| ast | Predicted exported alpha-N-acetylgalactosaminidase (628–637 AA) | 19 | Hydrophila, Salmonicida, and Veronii phylogroups | Most phylogroups | Putative pyridoxine 5′-phosphate synthase (or hypothetical protein or a putative aspartate amino transferase) | Putative tagatose 1,6 bi-phosphate aldolase |
| colAh | Microbial collagenase secreted (910–920 AA) | 89 | Most phylogroups | Variable | Putative O-succinyl acid-CoA ligase (or hypothetical protein) | SanA protein |
| rtxA | RtxA toxin (4439–4849 AA) | 14 | Hydrophila phylogroup | Most phylogroups | RTX toxin activating lysine acyltransferase | Variable |
| exoA | Putative exotoxin A precursor (639–640 AA) | 14 | < id="625" data-dummy="list" list-type="suimple">
|
Most phylogroups | Putative cyanate transporter protein CynX | Ribosomal large subunit pseudouridine synthase F |
| ascF | Cytoplasmic protein AscF (82–89 AA) | 33 | Most phylogroups | Caviae, Media, and Molluscorum phylogroups | AscE | AscG |
| ascG | AscG (114–118 AA) | AscF | AscH | |||
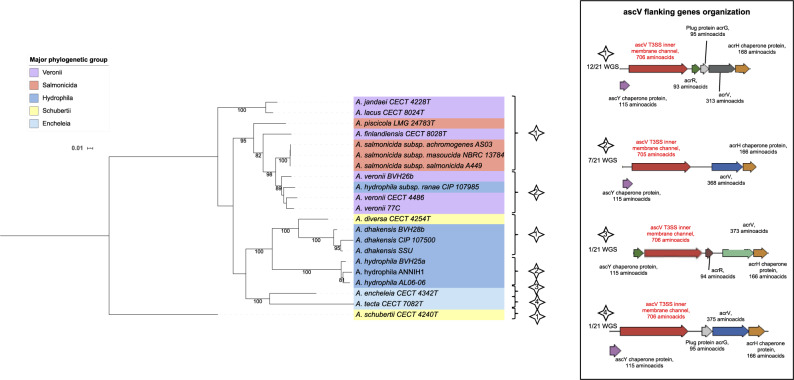
| ascV | AscV T3SS inner membrane channel (705–706 AA) | AscY chaperone protein | Variable | |||
| aexT | ADP-ribosyltransferase (451–476 AA) | 13 (38% of T3SS+ genomes) | A. veronii and A. salmonicida | Most phylogroups | Type 3 secretion chaperone protein | AexU in A. veronii and EAL domain |
| aexU | ADP-ribosyltransferase (446–514 AA) | 20 (62% of T3SS+ genomes) | Encheleia, Salmonicida, Hydrophila, and Veronii phylogroups | Schubertii, Molluscorum, Media, and Caviae phylogroups | Type 3 secretion chaperone protein (or AexT) | EAL domain or phenylalanine tRNA synthetase beta subunit |
| lafA | LafA flagellin protein (280–297 AA) | 50 | Most phylogroups | Variable | Variable | Putative LafB flagellar hook-associated protein |
| ser/ahe2 | Putative extracellular protease (624–634 AA) | 67 | Most phylogroups | < id="692" data-dummy="list" list-type="suimple">
|
Variable | Hypothetical protein |
Genome-Based Phylogenetic Relationships
All genome assemblies used in this study were adequate to reconstruct a single-nucleotide polymorphism (SNP)-based phylogenomic tree using the k-mer method. The aeromonad phylogeny was inferred using the kSNP 3.1 software package in which SNPs were based on k-mer analysis (Gardner et al. 2015). The maximum-likelihood (ML) tree was reconstructed on the basis of 27,856 SNPs identified in whole-genome sequences (WGS) for at least 75% of all strains (k-mer = 19). The aligned SNP sequences were used for decomposition analyses with the neighbor-net algorithm available in SplitsTree 4.0 software (Huson and Bryant 2006).
Phylogenetic Analysis of Virulence-Associated Genes
Phylogeny was inferred from the sequences of 13 virulence-associated genes (aer/act, alt, ast, colAh, rtxA, exoA, ascF, ascG, ascV, aexT, aexU, lafA, and ser). In the case of T3SS genes (ascF, ascG, and ascV), the three genes were concatenated. Nucleotide sequences were aligned using the Clustal ω2 program in the Seaview 4 package (Gouy et al. 2010). ML phylogenetic trees were reconstructed for each gene or concatenated genes using the best-fit model of evolution determined by the Akaike criterion (http://iqtree.cibiv.univie.ac.at/). ML bootstrap supports were calculated after 100 reiterations. To study the phylogenetic inference of the exoA gene, we added the nucleotide sequence of the eta gene (GenBank locus PA1148) coding for exotoxin A (SwissProt PA11439) produced by Pseudomonas aeruginosa PA01.
Statistics
All qualitative variables were compared using a χ2 test, and all quantitative variables were compared using Student’s t-test, wherein a P value ≤0.05 was considered significant. All these computations were performed using R project software (http://www.r-project.org).
Results
Phylogenomic Relationships in Aeromonads
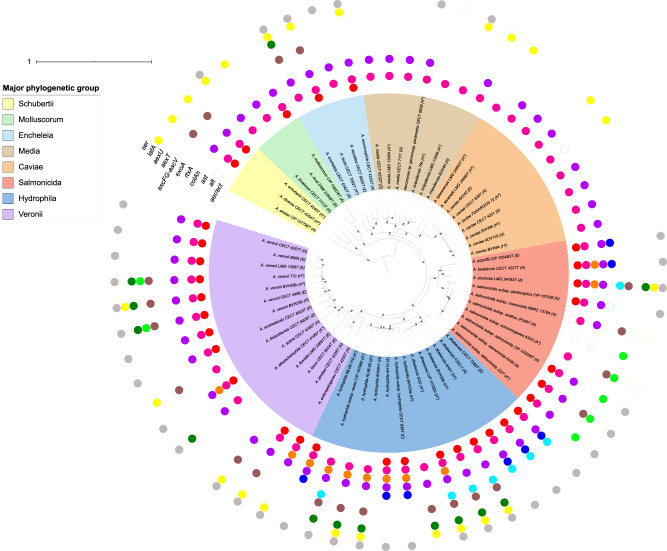
A phylogenetic ML tree based on SNPs present in 75% of the strains provided information on the relative phylogenetic placement of the studied strains (27,856 SNPs; fig. 1). This SNP-based phylogenomic approach led to the reconstruction of a robust tree that delineated the eight major phylogenetic groups (bootstraps = 100) previously clustered by core-based phylogenomics or multilocus phylogeny based on 15 housekeeping genes (Colston et al. 2014), hereafter called phylogroups Schubertii, Hydrophila, Veronii, Caviae, Media, Encheleia, Salmonicida, and Molluscorum. Every taxonomical species represented by several strains was distinctly separated from the others in the phylogenomic ML tree (bootstraps = 100).
Fig. 1.
—ML phylogeny based on 27,856 SNPs and the virulence-associated gene repertoire. The tree shows the phylogeny of 64 Aeromonas strains, including the 30 validated species represented by their type strain or a reference strain (A. rivipollensis). The scale bar is expressed as changes per total number of SNPs. The numbers at the nodes are support values estimated with 100 bootstrap replicates. Only bootstrap values ≥70 were indicated. Eight well-supported clades named “Major phylogenetic groups” are shown by colored ranges on strain labels: group Schubertii, group Molluscorum, group Encheleia, group Media, group Caviae, group Salmonicida, group Hydrophila, and group Veronii. The isolation source is indicated in parentheses after the strain number: environmental (E), animal (A), or human (H). An asterisk denotes whether pathogenic phenotypes have been described for the strain. The external colored circles corresponded to genes that encoded virulence factors detected after genome BLAST analysis, including from the inside to the outside: aer/act for a toxin with two denominations, “Aerolysin” or “cytolytic enterotoxin Act”; alt for a thermolabile cytotonic enterotoxin Alt; ast for a thermostable cytotonic enterotoxin Ast; colAh for an extracellular collagenase ColAh; rtxA for a repeat-in-the-toxin A; exoA for an exotoxin A, ascF, ascG, and ascV for T3SS components; aexT for an ADP-ribosylating transferase and T3SS-effector AexT; aexU for an ADP-ribosylating transferase and T3SS-effector AexU; lafA for a lateral flagellin A LafA; and ser for an extracellular serine protease Ahe2/AspA. Open circles indicate interrupted genes.
The phylogenetic network generated by the neighbor-net analysis (supplementary fig. A1, Supplementary Material online) was overall congruent with the phylogenomic ML tree. In addition, it showed interconnections between phylogroups, species within phylogroups and strains within species. Such reticulations suggest recombination events (e.g., horizontal gene transfers) that may have taken place during evolution. Recent events obviously occurred among the A. salmonicida lineages, with the exception of the mesophilic strain (supplementary fig. A1, Supplementary Material online).
Repertoire and Distribution of Virulence Genes in Aeromonads
The characteristics and annotations of the targeted virulence genes as well as the flanking regions are summarized in table 2. The results of genome screening for the presence of 13 virulence-associated genes are shown for every genome in the phylogenomic ML tree (fig. 1). Virulence-associated genes were distributed unevenly among the phylogroups, from “no gene” in A. fluvialis LMG 24681T to “nine genes out of ten” in A. dhakensis SSU and A. piscicola LMG 24783T. The strains belonging to the phylogroups Hydrophila and Salmonicida contained significantly more virulence-associated genes (means of 6.8 and 5.9 genes detected from the test panel, respectively) than those from the phylogroups Media and Caviae (means of 2.2 and 2.6 genes detected, respectively; P value <0.001).
Each virulence gene fell into one of three categories based on its distribution among genomes (fig. 1 and table 2). The genes of the first group (alt, colAh, and lafA) were well represented in the whole tree except in some scattered genomes. The genes of the second group (aer/act and ser) were widely found among the tree, except in some specific phylogroups, for example, the Caviae and Media groups. In addition, a significant association between the presence/absence of the aer and ser genes was noted with 29 aer+/ser+ and 17 aer−/ser− strains out of the 64 genomes (P value <0.001). The genes of the third group (ast, rtxA, exoA, ascF, ascG, ascV, aexT, and aexU) were more specifically associated with particular genomes or phylogroups. For instance, the distributions of the genes rtxA and exoA were restricted to the phylogroup Hydrophila (A. dhakensis and A. hydrophila) and to the mesophilic species in the phylogroup Salmonicida, that is, A. salmonicida subsp. pectinolytica (fig. 1). None of the virulence-associated genes were present in any of the genomes.
The HMMSCAN program detected a peptide signal in each CDS homologous to the aerolysin, collagenase, Ast, exotoxin A, and serine protease genes. In addition, all aerolysin homolog genes were predicted to encode APT and aerolysin domains. The amino acid positions important for oligomerization (H in 155) and heptamerization (K in 374 and E in 390) were conserved with the exception of a change of unknown consequence with N instead of K at position 374 in the aerolysin gene of A. tecta CECT 7082T. The N-terminus of a bacterial virulence factor lipase and a peptidase M9 domain were detected in all the alt and colAh genes, respectively. The RtxA CDSs were predicted to have 28–30 RtxA repeats, 2–3 coiled coils, an actin-linking domain (except A. bestiarum and A. popoffii strains), a yersinia-like virulence antigen (except A. hydrophila ML09-119), a serine aminopeptidase S33a domain or an alpha/beta hydrolase family domain, a peptidase C80 domain with 2 predicted active sites, and a membrane localization domain (except A. hydrophila ML09-119). All the exotoxin A genes harbored exoA-binding, exoA-targeting, and exoA-catalytic domains with one predicted active site. The T3SS structural genes were predicted to carry their appropriate functional domains. The ADP-ribosylating transferase toxin genes aexT and aexU shared one YopE domain when all the aexT genes also harbored one ADP-ribosyltransferase exoenzyme domain with two predicted active sites. All the flagellar lafA homologs had bacterial flagellin N-terminal and C-terminal helical regions. A peptidase S8 family protein with three predicted active sites and a proprotein convertase P-domain were detected in every serine protease gene. Overall, this predictive analysis supports the presumption that the homologs studied herein may be able to generate functional proteins after the appropriate transcription, translation, and posttranslation modifications.
In most species for which multiple genomes were assessed, there was no obvious relationship between the isolate origin and the virulence gene content. For instance, A. hydrophila and A. dhakensis recovered from the environment and animals (CECT 839 and ML09-119) or humans (CECT 7289 and AAK1) exhibited identical profiles that included the aerolysin gene. Similar observations were made in the species A. veronii, A. media, and A. caviae (fig. 1). An exception was A. dhakensis, for which only three strains recovered from human disease cases harbored the genes for the T3SS apparatus.
In addition to the 340 virulence gene homologs detected without nonsense mutations, RAST identified 19 ISs that interrupted several virulence-associated genes except ascF, ascG, ascV, and aexU, which were not interrupted (fig. 1). Eight of the ISs were located at the ends of contigs; therefore, their annotations remain putative (data not shown). Twelve of the 19 detected ISs (63%) were recovered from A. salmonicida genomes, representing 11% of the studied genomes; this result is consistent with the systematic search of MGEs performed in genomes (supplementary table S1, Supplementary Material online) because the highest numbers of putative ISs were found in the species A. popoffii, A. allosacharophila, and A. salmonicida, with 31, 41, and from 17 to 432 ISs detected, respectively. More generally, ISs were predicted in all the genomes analyzed (min. of 1, max. of 432, median of 15 ISs). IMEs and/or ICEs were predicted in 47% of the strains and belonged to all eight major phylogroups. From one to two IMEs were detected in 38% of the strains, of which no directed repeats were identified for 29%. We observed a genomic colocalization between MGEs and virulence genes only for alt in A. rivipollensis 76c and for colAh in A. salmonicida subsp. smithia.
Phylogeny of Virulence-Associated Genes
The ML phylogenetic trees of the ten targeted virulence genes or gene combinations were reconstructed and compared with the structure of the phylogenomic tree. The content and synteny of the regions flanking the virulence genes were also considered to reconstruct evolution hypotheses of virulence genes in the genus Aeromonas.
Core Genome-like Evolution of the alt, colAh, and ser Virulence Genes
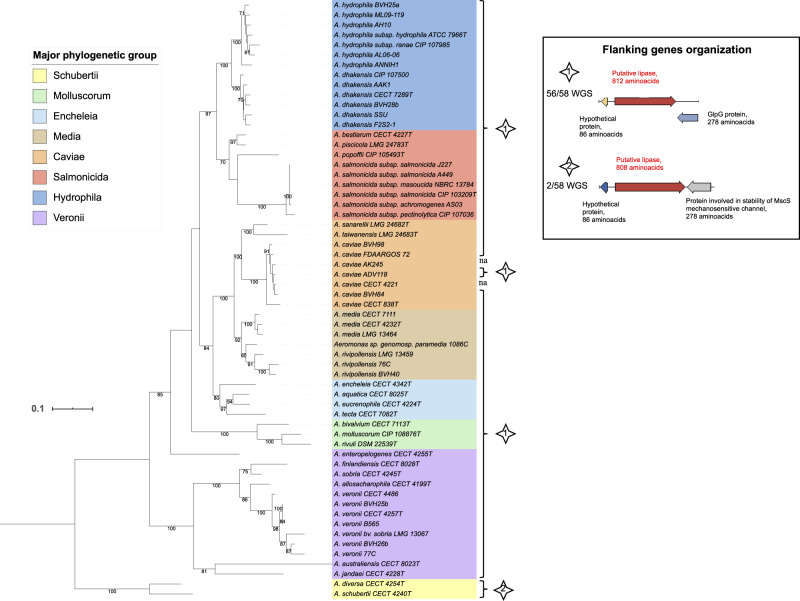
The gene alt was detected in all studied genomes except A. fluvialis LMG 24681T and A. lacus CECT 8024T (i.e., 97% of the genomes). The alt ML tree shown in figure 2 is emblematic of congruence between virulence-associated gene phylogeny and phylogenomics. Indeed, alt gene sequences from the eight phylogroups clustered in eight clades (bootstrap ≥70), in which the sequences from the species A. hydrophila, A. dhakensis, A. rivipollensis, A. media, A. veronii, and A. salmonicida were well separated (bootstrap ≥91). The region flanking the alt gene encoded the same putative GlpG protein in all genomes except in the Schubertii genomes that encoded a putative protein involved in the stability of the MscS mechanosensitive channel in place of GlpG.
Fig. 2.
—ML tree based on alt gene sequences (2,526 nt) reconstructed using the TIM3 model plus gamma distribution and invariant sites as a substitution model with 60 complete nucleotide sequences. The two interrupted sequences (A. simiae and A. salmonicida subsp. smithia homologs of alt) are not represented in this tree. The horizontal lines represent genetic distance, with the scale bar indicating the number of substitutions per nucleotide position. The numbers at the nodes are support values estimated with 100 bootstrap replicates. Only bootstrap values ≥70 are indicated. The major phylogenetic group of each strain is indicated by colored ranges on strain labels. The type of genetic organization of the flanking genes shown in the inserted box is indicated for each strain with a numbered star. Abbreviations: na, not applicable; WGS, whole-genome sequences.
Similar to alt, colAh was widely distributed in all but seven genomes (i.e., 89% of the genomes) and scattered among phylogroups. The colAh phylogeny (supplementary fig. A2, Supplementary Material online) was also congruent with the phylogenomics with the sole exception of A. bivalvium CECT 7113T, whose colAh sequence clustered with the group Salmonicida. In addition, the species A. caviae, A. hydrophila, A. dhakensis, and A. salmonicida were robustly delineated by the colAh phylogeny (bootstrap ≥96). The two colAh-flanking regions were clade specific with a putative O-succinyl acid-CoA ligase in the phylogroups Media, Encheleia, Caviae, and Molluscorum; a hypothetical protein in the groups Hydrophila and Salmonicida; or one or the other one in the groups Veronii and Schubertii.
The gene ser was present in only 67% of the genomes but clearly demarcated (bootstraps = 100) five groups, Veronii, Hydrophila, Salmonicida, Media, and Molluscorum, within which the sequences from the species A. veronii, A. hydrophila, A. dhakensis, A. rivipollensis, and A. salmonicida were well separated (bootstrap >70) (supplementary fig. A3, Supplementary Material online). The ser gene was absent from the Caviae and Schubertii groups and only detected sporadically in the Media phylogroup, where ser was present in A. rivipollensis (3/3), and in the strain Aeromonas sp. genomospecies paramedia 1086C but was absent from the three A. media sensu stricto strains (fig. 1). Eight different ser-flanking regions displayed an overall but partial superposition with phylogroups. In addition to this vertical evolution backbone, some signals of recombination were detected. The group Encheleia was paraphyletic (bootstraps = 98) in ser phylogeny with two different ser-flanking regions in the two Encheleia clades. In addition, A. bivalvium CECT 7113T grouped with A. rivipollensis in ser phylogeny. These findings suggest that ser genes display a core genome-like evolution (ancestral and vertical inheritance) with some significant MGE transfer events between bacteria or “horizontal gene transfers” (HGT).
In conclusion, the prevalence of alt and colAh genes within the genus, the congruence of virulence gene-based trees and phylogenomics, and the conservation of genes and flanking regions according to clades all suggest a common ancestral inheritance of alt and colAh genes and that their evolution was directly linked to that of the core genome and to speciation in the genus. The backbone of ser phylogeny also displayed a core genome-like evolution, but the absence of complete aerolysin-Ser systems in several phylotypes, species, or strains suggests the loss of ser, collectively or independently. Recombination events or HGT were not detected in alt phylogeny, were scarce in colAh, and were more apparent in ser phylogeny.
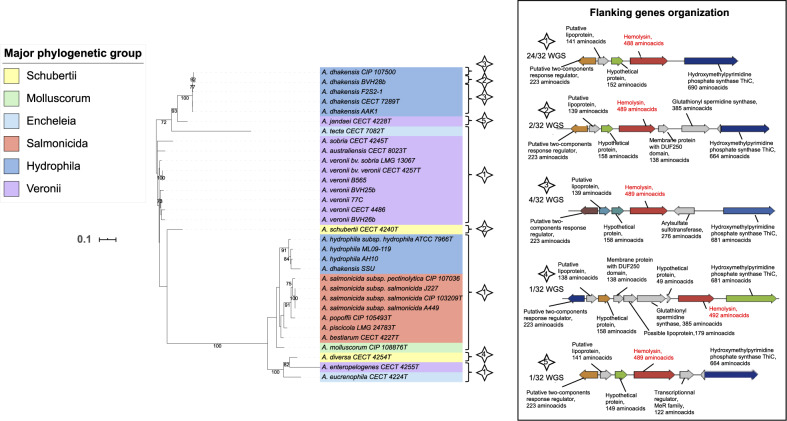
Paraphyly of Aerolysin in Aeromonads
The aer/act gene was present in 52% of the evaluated genomes. The ML phylogeny for aer/act delineated two main clades (bootstrap = 100), where each clade contains members from phylogroups Schubertii, Hydrophila, Veronii, and Encheleia but not from phylogroups Caviae and Media (fig. 3). The distribution within the two clades was dependent on species rather than phylogroup. For instance, the aer/act sequences from A. veronii and A. dhakensis (group Hydrophila) strains (except the strain SSU) belonged in one clade, whereas all the sequences from A. hydrophila (group Hydrophila) and A. salmonicida were in the other clade. The aerolysin genes in aeromonads were split into two clades without phylogenomic congruence. For instance, the homologs in A. dhakensis SSU and A. dhakensis BVH28b shared only 66% sequence identity (78% similarity) with proteins with lengths of 504 and 488 amino acids, respectively. However, the two types of aerolysin genes were clearly homologous and shared the same microenvironment (fig. 3). The most likely hypothesis for aerolysin evolution is that two Aeromonas ancestors each independently acquired the aer/act variants and that they were maintained in the different phylogroups. The near species-specific distribution of the two types of aer/act genes is possibly further evidence for their putative involvement in the speciation process that occurred later in aeromonads.
Fig. 3.
—ML tree based on aer/act gene sequences (1,527 nt) reconstructed using the TIM model plus gamma distribution as a substitution model from the 32 complete nucleotide sequences. One interrupted sequence (A. salmonicida subsp. masoucida homolog of aer/act) is not represented in this tree. The horizontal lines represent genetic distance, with the scale bar indicating the number of substitutions per nucleotide position. The numbers at the nodes are support values estimated with 100 bootstrap replicates. Only bootstrap values ≥70 are indicated. The major phylogenetic group of each strain is indicated by colored ranges on strain labels. The type of genetic organization of the flanking genes shown in the inserted box is indicated for each strain with a numbered star. Abbreviation: WGS, whole-genome sequences.
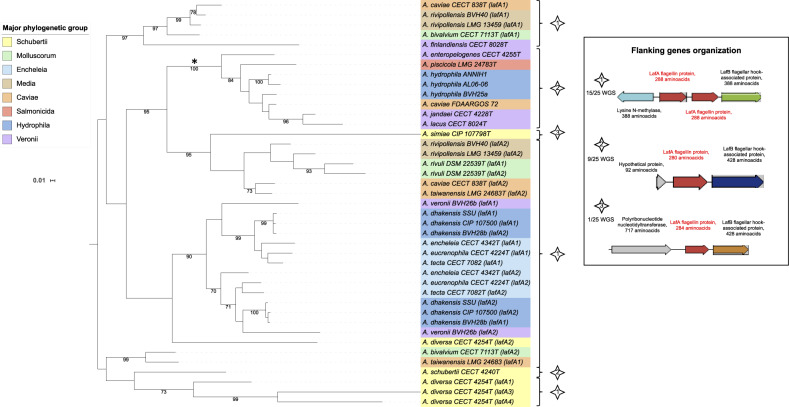
Complex Evolution of Lateral Flagellin among Aeromonads
Multiple copies of the lafA gene, which encodes lateral flagellin, were found in several genomes: 2 different copies in 13 genomes and 4 different copies in the genome of A. diversa CECT 4254T. Within the Hydrophila phylogroup, A. hydrophila strains (n = 3) had a unique copy, whereas the three strains of A. dhakensis harbored two different copies. The lafA ML tree was structured in four clades (bootstrap ≥ 90) and several weakly supported lineages (fig. 4). The lafA genes from the genomes that contained only one copy were mostly grouped in one clade (asterisk in fig. 4) except for the A. finlandiensis, A. simiae, and A. schubertii strains. Duplicated lafA genes formed another clade, which further separated into two subclades (bootstrap = 70) that encompass all strains of the phylogroup Encheleia and of the species A. dhakensis. In all cases, a CDS encoding a putative LafB flagellar hook-associated protein was located downstream (fig. 4). The prevalence of lafA within the genus Aeromonas (all the major phylogroups), the locus organization, and its phylogenetic inference suggest a common ancestral inheritance and that duplication occurred before mutations, insertions, or loss events during the evolutionary history of the genus. Despite ancestral inheritance, the high level of incongruence between the lafA phylogeny and phylogenomics suggests frequent HGT events in aeromonads (fig. 4 and table 2).
Fig. 4.
—ML tree based on lafA gene sequences (906 nt) reconstructed using the TIM model plus gamma distribution and invariant sites as a substitution model from 41 complete lafA nucleotide sequences. Each different copy found in a genome (or a strain) was arbitrarily numbered (lafAx). The same lafA numbering but in two different species corresponded to different lafA sequences. The clade gathering most of the genomes with a monocopy of lafA is indicated by an asterisk (*). The numbers at the nodes are support values estimated with 100 bootstrap replicates. Only bootstrap values ≥70 are indicated. The major phylogenetic group of each strain is indicated by colored ranges on strain labels. The type of genetic organization of the flanking genes shown in the inserted box is indicated for each strain with a numbered star. Abbreviation: WGS, whole-genome sequences.
HGT Drive the Evolution of TSS3, TSS3 Effectors, and Toxin-Encoding Genes in Aeromonads
The ascF, ascG, and ascV genes, which encode the structural components of the T3SS, were never detected separately. Therefore, they have been concatenated for further analysis (ascFGV). They were detected in 21 studied genomes (i.e., 33% of the genomes) covering all the major phylogenetic groups, except the groups Caviae, Media, and Molluscorum (fig. 1). The ML tree of the concatenated sequences showed four major clades (bootstraps ≥ 95; fig. 5), corresponding to the Salmonicida/Veronii, Hydrophila, Encheleia, and Veronii groups. The strain A. schubertii represented an external branch, and the strains A. hydrophila subsp. ranae and A. diversa belonged to the Salmonicida/Veronii and Hydrophila ascFGV clades, respectively. From a functional point of view, the presence of ascFGV was associated with at least one T3SS effector gene (P value <0.001). The T3SS effectors aexT and aexU were absent from six T3SS-encoding genomes (29%). Conversely, in four of the aexU- and/or aexT-positive genomes (21%), no T3SS component-encoding genes were detected.
Fig. 5.
—ML tree based on concatenated sequences of the T3SS coding gene sequences ascF, ascG, and ascV (2,739 nt) reconstructed using a GTR model plus gamma distribution as a substitution model from 21 complete nucleotide sequences. The numbers at the nodes are support values estimated with 100 bootstrap replicates. Only bootstrap values ≥70 are indicated. The major phylogenetic group of each strain is indicated by colored ranges on strain labels. The type of genetic organization of the flanking genes shown in the inserted box is indicated for each strain with a numbered star. Abbreviation: WGS, whole-genome sequences.
Concerning T3SS effectors, the gene aexT was present in five A. salmonicida and two A. veronii strains and was found in the same locus as the gene aexU only in the A. veronii strains. The aexU gene was detected from the genomes of 13 strains belonging to the species A. hydrophila, A. dhakensis, A. veronii, A. allosacharophila, A. piscicola, and A. encheleia (fig. 1). The aexT ML tree showed two clades that aggregate A. veronii strains and A. salmonicida strains and that harbor a specific genetic locus organization (numbered 1 and 2 in supplementary fig. A4, Supplementary Material online). Four organizations of aexU-flanking genes were revealed for the locus (supplementary fig. A5, Supplementary Material online). Among the aexU-positive genomes, the major clade contained strains from the Hydrophila phylogroup, a second clade contained strains from the Veronii phylogroup, and a third contained A. encheleia and A. piscicola strains.
Finally, T3SS component and effector phylogenies showed major discrepancies between them and phylogenomics, which suggested HGT events. The frequency, distribution and location of genes encoding T3SS components and effectors do not support the case for unique ancestral inheritance. The T3SS-related genes were more than likely acquired by HGT events that involved only some ancestors among phylogroups. The nucleotide Blast analysis demonstrated the relatedness of aeromonad genetic sequences to Pseudomonas sp. strains for genes encoding T3SS components and aexT and to Chromobacterium sp. strains for aexU (data not shown).
For other toxins, the gene ast was detected in 19% of strains, exclusively in the Hydrophila, Salmonicida, and Veronii phylogroups (supplementary fig. A6, Supplementary Material online). The ML tree of ast sequences showed one major clade that corresponded to the group Hydrophila. The other ast-positive genomes were more scattered and found close in the Salmonicida group (A. piscicola and A. bestiarum) or separated from each other, as in the Veronii group (A. sobria and A. enteropelogenes). This distribution suggested HGT acquisition of ast in a Hydrophila ancestor and in some isolated lineages. The nucleotide Blast analysis demonstrated the similarity of aeromonad genetic sequences with Enterobacteriaceae strains for ast (data not shown).
CDSs homologous to the toxin RtxA were found in 14% of aeromonads, mainly in the Hydrophila phylogroup. This CDS was associated with a region upstream of an RTX toxin that activates lysine acyltransferase. The genetic clustering and the locus organizations of the rtxA gene were different among the two taxonomic species of the Hydrophila phylogroup, which argues for an acquisition by two lateral transfer events, perhaps from vibrios given the relatedness of rtx sequences seen in Blast analysis (data not shown), one by a common ancestor of the members of A. hydrophila and another to the members of A. dhakensis (supplementary fig. A7, Supplementary Material online).
Nine aeromonad genomes (i.e., 14%) contained a gene coding for a putative exotoxin A that was very closely related to one found in P. aeruginosa (70% identity from 89% of the total sequence length in more than 100 different strains). This CDS was found almost exclusively in the strains within the Hydrophila phylogroup, with a conserved gene sequence and similar genetic organization (supplementary fig. A8, Supplementary Material online, and table 2).
Finally, contrary to the thermolabile cytotonic enterotoxin (Alt) gene that nearly met the threshold for inclusion in the core genome (97%), the genes encoding ADP-ribosylating toxins (AexT, AexU), RtxA toxin, exotoxin A, and thermostable cytotonic enterotoxin (Ast) were quite rare within the studied genomes (11–20%). When present, a low level of divergence was observed between strains from the same phylogroup (supplementary figs. A4–A8, Supplementary Material online). These features for aexT, aexU, exoA, and ast support the presumption of horizontal acquisition via MGEs and with a higher exchange compatibility among closely related strains.
Discussion
The traditional approach to characterize bacterial pathogenesis is mutagenesis/complementation and assessment of phenotypes in model systems. Although they are generally an efficient approach for specialist pathogens, these methods can lead to equivocal results for opportunistic pathogens such as Aeromonas (e.g., Gavín et al. 2002; Sha et al. 2002; Vilches et al. 2008; Sierra et al. 2010; Ponnusamy et al. 2016). Furthermore, these methods have largely failed to attribute virulence-associated genes in Aeromonas in accordance with molecular Koch’s postulates (Falkow 2004). Population studies that have compared the pathogenicity of environmental strains with that of clinical/animal strains using standard molecular biology techniques have delivered disappointing results in part because the high genetic diversity of the so-called virulence-associated genes in these bacteria has rendered polymerase chain reaction-based tools unreliable (Talagrand-Reboul et al. 2018). To better understand the adaptation mechanisms that drive the genome dynamics in these environmental opportunistic bacteria, in this study, we compared the phylogenomics of several virulence genes against the genetic background of the genus Aeromonas. We selected 11 previously described virulence factors with characterized phenotypes for this analysis, that is, T3SS and effector proteins, flagellar protein, collagenase, serine protease, cytotoxic, and cytotonic enterotoxins (Bernheimer et al. 1975; Chopra et al. 1996; Abrami et al. 1998; Braun et al. 2002; Gavín et al. 2002; Sha et al. 2002, 2007; Vilches et al. 2004; Suarez et al. 2012; Duarte et al. 2015; Ponnusamy et al. 2016). We tested the hypothesis that the evolutionary process of virulence-associated genes is informative of pathoadaptation. The method used to study the synteny of genes takes slight rearrangements into consideration to formulate evolution hypotheses. Our analysis revealed a great complexity with diverse evolution modes and gene organization and an uneven distribution of virulence genes in the genus, with evidence of HGT among strains in some cases. Overall, this study provided insights into aeromonad pathoadaptation or the ability of some members of this group to emerge as pathogens.
Pathogenicity versus Pathogenomics
The 30 species within the genus Aeromonas are not equivalent in their clinical importance. Human infections are most often caused by four taxonomic species that represent 96% of the aeromonads found in clinical samples: A. caviae (30%), A. dhakensis (26%), A. veronii (22%), and A. hydrophila (18%) (Figueras and Beaz-Hidalgo 2015). The species A. schubertii, A. enteropelogenes, A. jandaei, A. allosaccharophila, A. encheleia, A. sanarellii, and A. taiwanensis are rarely involved in human aeromonosis (Lai et al. 2007; Alperi et al. 2010; Roger et al. 2012; Latif-Eugenín et al. 2016; Sinclair et al. 2016). The pathogenicity profiles available for the species A. media, A. rivipollensis, A. eucrenophila, A. encheleia, and A. tecta are uncertain (Demarta et al. 2008; Roger et al. 2012; Talagrand-Reboul et al. 2017), whereas other species (e.g., A. salmonicida, A. popoffii, and A. bestiarum) have rarely been isolated from clinical samples (Hua et al. 2004; Yang et al. 2008; Sinclair et al. 2016). From our analysis, virulence-associated gene distribution could help to explain the pathogenic potential of A. hydrophila and A. dhakensis because these strains presumably contained most of the target virulence genes, but this hypothesis suffers from a possible bias related to the nature of the gene panel.
A recent study examined the pathogenicity and extensive virulence arsenal of A. dhakensis (Chen et al. 2016), and the data correlated well with the pathogenomic profile described in our work. Additional genome analyses suggest a greater virulence potential in A. hydrophila in comparison to A. veronii and A. caviae (Ghatak et al. 2016). Certain diseases/conditions have been associated with a particular aeromonad; however, our understanding of this relationship is limited. For instance, A. caviae and A. veronii are highly prevalent in enteritis and bacteremia, whereas A. hydrophila and A. veronii are prevalent in wound infections (Lamy et al. 2009). These observations suggest different capabilities in invasion and tissue damage between phylogroups. In our study, fewer virulence genes were identified in A. veronii and A. caviae strains compared with A. hydrophila. Most notably, aerolysin and T3SS components or effectors were absent from all A. caviae strains. Interestingly, the observed virulence profile of A. caviae was similar to that of A. media, but A. caviae exhibited higher infectious success (Lamy et al. 2009; Figueras and Beaz-Hidalgo 2015). We must also consider that A. caviae or A. veronii strains may possess unknown virulence determinants not included in our study and that the selected strains may not be representative enough of the virulence gene prevalence in these species. An unbiased pangenome survey could strengthen targeted studies and lead to a more robust correlation between pathogenomics and pathogenesis in aeromonads, as demonstrated for P. aeruginosa with the aim of searching for new drugs and vaccines (Mosquera-Rendón et al. 2016) or recently for the identification and characterization of new putative Aeromonas spp. T3SS effectors (Rangel et al. 2019). One limitation of this study, as in any genomic survey without phenotypic and/or proteomic investigation, is that virulence factor gene detection does not ensure appropriate protein expression or efficient secretion, where applicable. Despite these limitations, our work provides novel results that contribute to our understanding of virulence in aeromonads.
Virulence by Exaptation
From our analysis, neither the isolation source nor the degree of reported pathogenicity influenced the results of the virulence screen. The presence of virulence genes in environmental aeromonads is not rare and was also observed by Vázquez-Rosas-Landa et al. (2017) using a similar genomic approach. In that study, the authors reported that the natural selection of these so-called “virulence genes” in aquatic bacteria reflects the integral nature of these factors to the lifestyle within that particular habitat. For instance, secretion systems or motility mechanisms may enhance fitness by facilitating nonpathogenic interactions with other bacteria and eukaryotes (Silver et al. 2007). Thus, adaptive factors that are primarily of ecological importance within one niche are thought to become virulence factors by exaptation, enabling aeromonads to emerge as pathogens and cause disease in incidental hosts.
The analysis of Alt and ColAh may illustrate the hypothesis of exaptation-based virulence in aeromonads. ColAh is a peptidase that belongs to the gluzincin subfamily of the M9 family recently described in A. piscicola that shares low similarity with other known bacterial collagenases. The enzyme exhibits a cytopathic effect on Vero cells (Duarte et al. 2015), but its overall role in aeromonad pathogenicity is mostly unknown. ColAh may play a role in host invasion similar to other bacterial collagenases. The cytotonic enterotoxin Alt belongs to the enterotoxic arsenal of aeromonads. However, Alt contributed to A. hydrophila-induced gastroenteritis in a mouse model to a lesser extent than the toxin Act (Sha et al. 2002). The presence of the alt gene in the Aeromonas “soft-core genome” (≥95% of genomes) (Kaas et al. 2012), including exclusively environmental nonpathogenic species (e.g., A. rivuli), raises the question about its true involvement in pathogenicity. In the literature, the prevalence of alt is relatively low for a gene that we can consider as belonging to the soft-core genome (e.g., 53% of 129 strains, Aravena-Román et al. 2014). One reason for this is that polymerase chain reaction assays can have poor sensitivity at the genus level because of the high polymorphism that results in a biased alt prevalence (Talagrand-Reboul et al. 2018). From our analysis, the genes alt and colAh were highly conserved in the genus Aeromonas because they were found in the large majority of the genomes (i.e., 97% and 89%, respectively), and their phylogenies were congruent with the global genetic background of the whole genus. We assume that these genes may have been acquired by a common ancestor and then followed the general genetic evolution of these bacteria. With their basic functions as lipases and proteases, Alt and ColAh are likely involved in general aeromonad metabolism within the customary environment. In a secondary host environment, these enzymes can be utilized in other biological processes, such as those involved in virulence. These secondary functions represent potential roles in exaptation (Adiba et al. 2010; Cabello and Godfrey 2018) for these two enzymes.
Aer Is Likely a True Virulence Factor
Aerolysin (Aer) is a pore-forming toxin secreted by Aeromonas (Bernheimer and Avigad 1974; Bücker et al. 2011). Aer/Act is considered the major enterotoxin that contributes to aeromonad pathogenicity (Sha et al. 2002), and as such, its presence/absence in a genome should be related to the pathogenic behavior of the strain. Our observations are consistent with this hypothesis. First, act/aer-negative species exhibit little to no virulence. Aeromonas media displays a rather low virulence profile in humans and animals (Talagrand-Reboul et al. 2017). To date, A. fluvialis has not been reported as virulent in any host, and A. simiae has only been described in monkey feces but not associated with any pathology (Harf-Monteil et al. 2004). Second, among the recognized pathogenic aeromonads, the Aer/Act-negative species A. caviae is less cytotoxic than the Aer/Act-positive species A. veronii (Chen et al. 2015). Finally, for almost all studied isolates of A. dhakensis, a species very closely related to A. hydrophila that is one of the most virulent among aeromonads (Chen et al. 2016), a specific act/aer gene was found. The theory that Aer/Act produced by A. dhakensis may contribute to its high virulence potential requires further investigation.
We observed a significant genetic association between the aer/act and ser genes that is probably related to their function (Iacovache et al. 2016). The aer+/ser− pattern suggests that either proteases other than Ser could be involved in the activation of the pore-forming aerolysin or Aer/Act is secreted but not matured in the transmembrane complexes of these strains. Despite their association, the phylogenies of aer/act and ser are complex and distinct. The phylogeny reconstructed from aer/act leads us to assume that two different Aeromonas ancestors acquired variants of the aerolysin genes, and this gene may be associated with the speciation process. In contrast, it seems likely that the ser gene was acquired by a common ancestor of aeromonads and then transmitted by vertical inheritance. It could have been positively coselected in the aer/act-positive Aeromonas clades, but the pleiotropic role of proteases may explain its presence in several aer/act-negative genomes.
LafA, an Evolution Mode toward Multiple Copies
Lateral flagellin is the major component of lateral flagella that are involved in swarming motility and biofilm formation in numerous bacteria. Mesophilic Aeromonas strains display a polar flagellum but can express multiple lateral flagella (Kirov et al. 2002). Mutagenesis and complementation experiments confirmed that the lateral flagella of Aeromonas play a role in adherence and biofilm formation (Gavín et al. 2002). Despite numerous orthologs, the lateral flagellar system in Aeromonas does not share either structural or regulatory genes with the polar flagellar system (Wilhelms et al. 2013). In this work, we hypothesized that after acquisition from a common ancestor, duplication likely drove the evolution of the lafA gene in the genus Aeromonas and that copies of the gene were also involved in HGT events. This confirms the major role of the multiple lafA copies in the genetic evolution of bacterial flagella and agrees with previous studies of Proteobacteria in that phylogenetic analysis and organization of lateral flagellar genes highly suggest that this system originated both from the duplication and horizontal transfer of polar flagella system genes (Liu and Ochman 2007). From a functional view, the four different copies detected in the A. diversa genome are possibly involved in the peculiar swarming ability of the species (Miñana-Galbis et al. 2010), although the swarming capacity has not been studied with the same approach in other members of the genus.
T3SS, an Example of Pathoadaptative Evolution
T3SSs enable the injection of effectors into eukaryotic cells. They are widely distributed in Gram-negative bacteria. The T3SS found in aeromonads is described as a homolog of those reported in P. aeruginosa and Yersinia spp., suggesting its potential role in Aeromonas pathogenesis with the ADP-ribosylating toxins AexT and AexU as translocons, as supported by mutagenesis data (Burr et al. 2002; Vilches et al. 2004; Sierra et al. 2010). The aexU null mutant was attenuated in a mouse model (Sierra et al. 2010). In A. hydrophila, the aexT mutant showed a slight reduction in virulence, whereas mutants without a functional T3SS apparatus displayed significantly reduced virulence in the same assays (Vilches et al. 2008). In addition to causing cellular damage, the T3SS and related effectors in A. salmonicida impair the transcription of immune mediators in rainbow trout (Origgi et al. 2017).
From our phylogenetic analysis and study of the distribution/organization of the loci, we hypothesize that aeromonad T3SS-related genes (T3SS and effectors) were acquired by HGT within phylogroups, which is consistent with another work that investigated the distribution and genetic evolution of 21 Aeromonas T3SS likely effector families from 105 strains covering the whole genus (Rangel et al. 2019) and observed in other genera, for example, Vibrio (Okada et al. 2010) and Pseudomonas (Dillon et al. 2019). The T3SS effectors AexT and AexU likely correspond to two different allelic forms of ADP-ribosyltransferase, similar to the HopZ gene family, which encodes T3SS effectors in the opportunistic plant pathogen Pseudomonas syringae. Briefly, T3SS effectors corresponded to three allelic forms of HopZ, HopZ1, HopZ2, and HopZ3, which display various specific targets or substrates. HopZ proteins are structurally and functionally heterogeneous due to 1) the acquisition of HopZ2 and HopZ3 from members of the genera Xanthomonas and Erwinia by HGT, respectively and 2) the pathoadaptive evolution of the ancestral form HopZ1. The evolution of hopZ1 follows the evolution of the genes from the P. syringae core genome (Ma et al. 2006). Interestingly, previous works on the genetics of T3SS effectors of P. syringae have shown that pathoadaptation is not inconsistent with genomic plasticity or the acquisition of virulence genes by lateral transfer, but T3SS effectors can be affected by mutations, which can then modify the functions of the effectors (Ma et al. 2006; Dillon et al. 2019).
In this work, the examination of the aexU-flanking genes by RAST annotation showed a possible association between aexT and aexU in A. veronii strains, as previously reported (Silver and Graf 2009). We observed that the presence of aexU in addition to aexT was an original feature of A. veronii strains among the genus Aeromonas, but this association was not present in all the strains of the species. Our results were highly consistent with those reported in the recent study of Rangel et al. (2019) on aeromonad T3SS effectors.
Finally, a functional study demonstrated the regulatory crosstalk between the T3SS and lateral flagellum systems (Zhao and Shaw 2016), but no obvious evolutionary link between lafA and the T3SS genes was observed therein (figs. 4 and 5).
In summary, genes encoding the T3SS structural components and the effectors AexT and AexU in the genus Aeromonas have probably followed a pathoadaptive evolution likely guided by their environment and/or their host. The acquisition of the genes and their subsequent evolution may have been driven by interactions with eukaryotic organisms within their native aquatic environments (e.g., amoeba, nematodes, or leech) and by inadvertently encountered circumstances where the T3SS and their effectors act as virulence factors (Yu et al. 2004; Sha et al. 2007). This hypothesis requires further study to discern the selective pressures leading to maintenance, expression, and induction of the T3SS machinery.
HGT as a Major Driver for Toxin Acquisition
Two mechanisms of HGT have been described in Aeromonas bacteria, that is, transformation and conjugative transfer (Piotrowska and Popowska 2015). The interconnected network generated by the neighbor-net analysis, the high number of ISs detected, and the presence of putative ICEs and IMEs are consistent with the probable frequency of these HGT events among aeromonads. Conversely, the analysis of IMEs and ICEs did not show any significant impact of particular conjugative events on the presence of the studied virulence factor genes. Despite the absence of direct imputation of well-identified transfers, the comparative analysis results of the genus phylogeny and the studied genes are still compatible with HGT. These transfers may have occurred, independently of conjugal events, by DNA transformation mechanisms, which have been experimentally demonstrated in Aeromonas, and facilitated exchanges were more frequent between closely related strains (Huddleston et al. 2013).
Cumulative data on the three non-T3SS-related toxins that we evaluated (ast, rtxA, and exoA) showed that they were likely acquired by HGT from other environmental/aquatic bacteria, such as Pseudomonadaceae, Enterobacteriaceae, or vibrios. These toxins are particularly associated with the Hydrophila phylogroup, A. hydrophila species for the gene ast, A. dhakensis species for exoA, and both species for rtxA. The exoA gene detected in A. hydrophila genomes codes for a homolog of P. aeruginosa exotoxin A, a major virulence factor for this bacterium. Mutagenesis experiments showed that exoA is associated with host tissue destruction, which allows invasive deep infections, such as necrotizing fasciitis, in a murine model (Ponnusamy et al. 2016). We found that this gene was particularly frequent in the highly virulent species A. dhakensis (6/6) (Chen et al. 2016). We hypothesize that these three toxins, for which a high degree of virulence in animal models has been demonstrated, contribute to a particular pathogenicity of members of the phylogroup Hydrophila. Moreover, these toxins may exhibit their virulence properties when produced alone and/or in conjunction or sequentially with other toxins, potentially resulting in aggregate adverse effects.
Aeromonads and Pathoadaptation
The substantial diversity of evolutionary modes for virulence-associated genes can give rise to a complex evolutionary network and likely sets the foundation for different assemblages of virulence factors in aeromonads. Most of the virulence-associated genes are chromosomal, which implies that either their functions are fundamentally essential or beneficial or that they have been fixed after HGT as determinants involved in niche adaptation. The dynamics of adaptive evolution through changes in gene sequence, regulation, expression, loss, or acquisition correspond to the pathoadaption phenomenon presumably enhance the fitness of a microbe in its new host niche (Pallen and Wren 2007).
In Aeromonas, the products of the alt, colAh, and, to some extent, ser genes are thought to serve functions essential to aeromonad biology and physiology because these genes follow vertical evolution patterns parallel to the evolution patterns of genes of the core genome. These hydrolytic enzymes are probably involved in general cell metabolism, but by exaptation (Adiba et al. 2010; Cabello and Godfrey 2018), they may also be involved in interactions with the host and virulence. HGT is another mode of evolution detected herein for more specialized virulence-associated genes, such as toxins RtxA, ExoA, Ast, AexT, and AexU. Some of these virulence-associated genes were specific to a species or phylogroup. However, none of them was the sole virulence factor in a pathogenic phenotype. Some of these genes were also probably lost by several strains, for example, aerolysin in A. hydrophila. Another signature of gene loss in aeromonads is the interruption of several virulence-associated genes by ISs. The accumulation of pseudogenes and insertion elements for A. salmonicida strains corresponds to genomic markers of the shift to a new niche (Reith et al. 2008; Aujoulat et al. 2012; Vincent et al. 2016). Indeed, the psychrophilic subspecies of A. salmonicida are considered specialized pathogens with a narrow host range limited to fish (Vincent et al. 2016). In addition, recent works have highlighted the degree of diversity of virulence traits among mesophilic A. salmonicida strains (Vincent et al. 2019), and these results may indicate that further studies of virulence gene evolution are needed to increase the understanding of pathoadaptation in A. salmonicida lineages.
Conclusion
In conclusion, virulence-associated gene content by itself does not fully explain the pathogenic behavior of Aeromonas taxonomic species either in vitro or in clinical contexts. However, the high complexity of Aeromonas virulence genes in terms of uneven distribution, diversity of organization, and variable evolutionary modes revealed in this study likely explains why Aeromonas pathogenicity is so difficult to assess in terms of attribution. The pathogenomics overview in this study postulates that Aeromonas virulence-associated genes should be studied at the population level and that studies performed on type or model strains in a species should not be generalized to the whole species. In addition, correlations between pathogenomics and epidemiological data should also be considered. In a recent study, the analysis of WGS for 101 A. salmonicida subsp. salmonicida strains has revealed four major lineages of this fish pathogen that emerged in Denmark, and the genomic variations of these strains were associated with virulence factors carried on and disseminated by plasmids (Bartkova et al. 2017). We emphasize that pathoadaptation resulting in diverse phenotypic variations is a presumptive key element to consider in the challenging effort to determine virulence profiles and discrete pathotypes in the Aeromonas genus.
Finally, the present study provides a novel point of view of evolutionary processes concerning virulence genes in a model of environmental opportunistic pathogen bacteria. We anticipate that the dichotomies of vertical/lateral inheritance, prevalence in the genus/lineages, and single/multiple copies of the virulence genes will be established in future genomic analyses to characterize newly described virulence factors. Our pathogenomic analysis allows a better understanding of the dynamics of the emergence and evolution of pathogenic traits in aeromonads. The varied patterns of evolution suggested by our study, exaptation processes, fixation in the chromosome of virulence factors acquired by HGT, virulence-associated genes that evolve according to core genome phylogeny, and loss of virulence genes in specific niches are converging characteristics consistent with the role of niche adaptation, including pathoadaptation, in aeromonad speciation. This vision of aeromonads evolution meets current evolution theories such as the extended evolutionary synthesis that is influenced by various disciplines such as ecology. This synthesis suggests that organisms do not evolve to fit into preexisting environments but coconstruct and coevolve with their environments, thereby considered as ecological niches (Laland et al. 2014). For a pathogen, changing the structure of its ecosystem involves coevolution with the host and the construction of a niche that allows contact with this host.
Supplementary Material
Acknowledgments
This work was supported by the Association des Biologistes de l’Ouest (ABO) and the Association pour la recherche et le développement en microbiologie et pharmacie (ADEREMPHA) to E.T.-R. Part of this research was supported by U.S. Department of Agriculture (USDA) / Agricultural Research Service (ARS) agreement 58-1930-4-002 and USDA/ARS CRIS Project 1930-32000-005-00 to J.G.
Data deposition: New bacterial WGS released from this project have been deposited at NCBI Genome under the accession numbers JAAALX000000000, JAAALW000000000, JAAALV000000000, and JAAALU000000000.
Literature Cited
- Abolghait SKet al. . 2011. Recombinant AexU effector protein of Aeromonas veronii bv. sobria disrupts the actin cytoskeleton by downregulation of Rac1 and induces direct cytotoxicity to β4-integrin expressing cell lines. Microb Pathog. 51(6):454–465. [DOI] [PubMed] [Google Scholar]
- Abrami L, et al. 1998. The pore-forming toxin proaerolysin is activated by furin. J Biol Chem. 273(49):32656–32661. [DOI] [PubMed] [Google Scholar]
- Adiba S, Nizak C, van Baalen M, Denamur E, Depaulis F.. 2010. From grazing resistance to pathogenesis: the coincidental evolution of virulence factors. PLoS One 5(8):e11882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera-Arreola MG, Hernández-Rodríguez C, Zúñiga G, Figueras MJ, Castro-Escarpulli G.. 2005. Aeromonas hydrophila clinical and environmental ecotypes as revealed by genetic diversity and virulence genes. FEMS Microbiol Lett. 242(2):231–240. [DOI] [PubMed] [Google Scholar]
- Alperi A, et al. 2010. Aeromonas taiwanensis sp. nov. and Aeromonas sanarellii sp. nov., clinical species from Taiwan. Int J Syst Evol Microbiol. 60(9):2048–2055. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Lipman DJ.. 1990. Protein database searches for multiple alignments. Proc Natl Acad Sci U S A. 87(14):5509–5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravena-Román M, Inglis TJJ, Riley TV, Chang BJ.. 2014. Distribution of 13 virulence genes among clinical and environmental Aeromonas spp. in Western Australia. Eur J Clin Microbiol Infect Dis. 33(11):1889–1895. [DOI] [PubMed] [Google Scholar]
- Aujoulat F, et al. 2012. From environment to man: genome evolution and adaptation of human opportunistic bacterial pathogens. Genes (Basel) 3(2):191–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkova S, Leekitcharoenphon P, Aarestrup FM, Dalsgaard I.. 2017. Epidemiology of Danish Aeromonas salmonicida subsp. salmonicida in fish farms using whole genome sequencing. Front Microbiol. 8:2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaz-Hidalgo R, Figueras MJ.. 2013. Aeromonas spp. whole genomes and virulence factors implicated in fish disease. J Fish Dis. 36(4):371–388. [DOI] [PubMed] [Google Scholar]
- Bernheimer AW, Avigad LS.. 1974. Partial characterization of aerolysin, a lytic exotoxin from Aeromonas hydrophila. Infect Immun. 9(6):1016–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernheimer AW, Avigad LS, Avigad G.. 1975. Interactions between aerolysin, erythrocytes, and erythrocyte membranes. Infect Immun. 11(6):1312–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun M, et al. 2002. Characterization of an ADP-ribosyltransferase toxin (AexT) from Aeromonas salmonicida subsp. salmonicida. J Bacteriol. 184(7):1851–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bücker R, et al. 2011. Aerolysin from Aeromonas hydrophila perturbs tight junction integrity and cell lesion repair in intestinal epithelial HT-29/B6 cells. J Infect Dis. 204(8):1283–1292. [DOI] [PubMed] [Google Scholar]
- Burr SE, Stuber K, Wahli T, Frey J.. 2002. Evidence for a type III secretion system in Aeromonas salmonicida subsp. salmonicida. J Bacteriol. 184(21):5966–5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers BR, Massad G, Barghouthi S, Arceneaux JE.. 1991. Iron acquisition and virulence in the motile aeromonads: siderophore-dependent and -independent systems. Experientia 47(5):416–418. [PubMed] [Google Scholar]
- Cabello FC, Godfrey HP.. 2018. Aquaculture, exaptation, and the origin of mcr-positive colistin resistance. Antimicrob Agents Chemother. 62:pii: e01903–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canals Ret al. . 2006. Analysis of the lateral flagellar gene system of Aeromonas hydrophila AH-3. J Bacteriol. 188(3):852–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canals R, et al. 2007. Role of Gne and GalE in the virulence of Aeromonas hydrophila serotype O34. J Bacteriol. 189(2):540–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacon MR, Soler L, Groisman EA, Guarro J, Figueras MJ.. 2004. Type III Secretion System Genes in Clinical Aeromonas Isolates. J Clin Microbiol. 42(3):1285–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacón MR, Figueras MJ, Castro-Escarpulli G, Soler L, Guarro J.. 2003. Distribution of virulence genes in clinical and environmental isolates of Aeromonas spp. Antonie Van Leeuwenhoek 84(4):269–278. [DOI] [PubMed] [Google Scholar]
- Chain PSG, et al. 2009. Genomics. Genome project standards in a new era of sequencing. Science 326(5950):236–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty T, Huhle B, Hof H, Bergbauer H, Goebel W.. 1987. Marker exchange mutagenesis of the aerolysin determinant in Aeromonas hydrophila demonstrates the role of aerolysin in A. hydrophila-associated systemic infections. Infect Immun. 55(9):2274–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P-L, Lamy B, Ko W-C.. 2016. Aeromonas dhakensis, an increasingly recognized human pathogen. Front Microbiol. 7:793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P-L, et al. 2014. A comparative study of clinical Aeromonas dhakensis and Aeromonas hydrophila isolates in southern Taiwan: A. dhakensis is more predominant and virulent. Clin Microbiol Infect. 20(7):O428–O434. [DOI] [PubMed] [Google Scholar]
- Chen P-L, et al. 2015. Aeromonas stool isolates from individuals with or without diarrhea in southern Taiwan: predominance of Aeromonas veronii. J Microbiol Immunol Infect. 48(6):618–624. [DOI] [PubMed] [Google Scholar]
- Chopra AK, Houston CW, Peterson JW, Jin G-F.. 1993. Cloning, expression, and sequence analysis of a cytolytic enterotoxin gene from Aeromonas hydrophila. Can J Microbiol. 39(5):513–523. [DOI] [PubMed] [Google Scholar]
- Chopra AK, Peterson JW, Xu XJ, Coppenhaver DH, Houston CW.. 1996. Molecular and biochemical characterization of a heat-labile cytotonic enterotoxin from Aeromonas hydrophila. Microb Pathog. 21(5):357–377. [DOI] [PubMed] [Google Scholar]
- Colston SM, et al. 2014. Bioinformatic genome comparisons for taxonomic and phylogenetic assignments using Aeromonas as a test case. MBio 5(6):e02136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dacanay Aet al. . 2006. Contribution of the type III secretion system (TTSS) to virulence of Aeromonas salmonicida subsp. salmonicida. Microbiology(Reading, Engl.). 152(6):1847–1856. [DOI] [PubMed] [Google Scholar]
- Degiacomi MT, et al. 2013. Molecular assembly of the aerolysin pore reveals a swirling membrane-insertion mechanism. Nat Chem Biol. 9(10):623–629. [DOI] [PubMed] [Google Scholar]
- Demarta A, et al. 2008. Aeromonas tecta sp. nov., isolated from clinical and environmental sources. Syst Appl Microbiol. 31(4):278–286. [DOI] [PubMed] [Google Scholar]
- Dillon MM, et al. 2019. Molecular evolution of Pseudomonas syringae type III secreted effector proteins. Front Plant Sci. 10:418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte AS, et al. 2015. Aeromonas piscicola AH-3 expresses an extracellular collagenase with cytotoxic properties. Lett Appl Microbiol. 60(3):288–297. [DOI] [PubMed] [Google Scholar]
- Falkow S. 2004. Molecular Koch’s postulates applied to bacterial pathogenicity—a personal recollection 15 years later. Nat Rev Microbiol. 2(1):67–72. [DOI] [PubMed] [Google Scholar]
- Figueras MJ, Beaz-Hidalgo R.. 2015. Aeromonas infections in humans In: Graf J, editor. Aeromonas. Norfolk (United Kingdom: ): Caister Academic Press; p. 65–68. [Google Scholar]
- Gardner SN, Slezak T, Hall BG.. 2015. kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 31(17):2877–2878. [DOI] [PubMed] [Google Scholar]
- Gavín R, et al. 2002. Lateral flagella of Aeromonas species are essential for epithelial cell adherence and biofilm formation. Mol Microbiol. 43(2):383–397. [DOI] [PubMed] [Google Scholar]
- Gavín R, et al. 2003. Lateral flagella are required for increased cell adherence, invasion and biofilm formation by Aeromonas spp. FEMS Microbiol Lett. 224(1):77–83. [DOI] [PubMed] [Google Scholar]
- Georgiades K, Raoult D.. 2011. Defining pathogenic bacterial species in the genomic era. Front Microbiol. 1:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghatak S, et al. 2016. Pan-genome analysis of Aeromonas hydrophila, Aeromonas veronii and Aeromonas caviae indicates phylogenomic diversity and greater pathogenic potential for Aeromonas hydrophila. Antonie Van Leeuwenhoek 109(7):945–956. [DOI] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O.. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 27(2):221–224. [DOI] [PubMed] [Google Scholar]
- Harf-Monteil C, et al. 2004. Aeromonas simiae sp. nov., isolated from monkey faeces. Int J Syst Evol Microbiol. 54(2):481–485. [DOI] [PubMed] [Google Scholar]
- Howard SP, Garland WJ, Green MJ, Buckley JT.. 1987. Nucleotide sequence of the gene for the hole-forming toxin aerolysin of Aeromonas hydrophila. J Bacteriol. 169(6):2869–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua HT, Bollet C, Tercian S, Drancourt M, Raoult D.. 2004. Aeromonas popoffii urinary tract infection. J Clin Microbiol. 42(11):5427–5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huddleston JR, Brokaw JM, Zak JC, Jeter RM.. 2013. Natural transformation as a mechanism of horizontal gene transfer among environmental Aeromonas species. Syst Appl Microbiol. 36(4):224–234. [DOI] [PubMed] [Google Scholar]
- Hughes HYet al. . 2016. Detection and Whole-Genome Sequencing of Carbapenemase-Producing Aeromonas hydrophila Isolates from Routine Perirectal Surveillance Culture. J Clin Microbiol. 54(4):1167–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Bryant D.. 2006. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 23(2):254–267. [DOI] [PubMed] [Google Scholar]
- Iacovache I, et al. 2016. Cryo-EM structure of aerolysin variants reveals a novel protein fold and the pore-formation process. Nat Commun. 7(1):12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda JM, Abbott SL.. 2010. The genus Aeromonas: taxonomy, pathogenicity, and infection. Clin Microbiol Rev. 23(1):35–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaas RS, Friis C, Ussery DW, Aarestrup FM.. 2012. Estimating variation within the genes and inferring the phylogeny of 186 sequenced diverse Escherichia coli genomes. BMC Genomics. 13:577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khajanchi BK, Kozlova EV, Sha J, Popov VL, Chopra AK.. 2012. The two-component QseBC signalling system regulates in vitro and in vivo virulence of Aeromonas hydrophila. Microbiology 158(1):259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khajanchi BK, et al. 2010. Distribution of virulence factors and molecular fingerprinting of Aeromonas species isolates from water and clinical samples: suggestive evidence of water-to-human transmission. Appl Environ Microbiol. 76(7):2313–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov SM, O’Donovan LA, Sanderson K.. 1999. Functional characterization of type IV pili expressed on diarrhea-associated isolates of Aeromonas species. Infect Immun. 67(10):5447–5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov SM, et al. 2002. Lateral flagella and swarming motility in Aeromonas species. J Bacteriol. 184(2):547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozińska A. 2007. Dominant pathogenic species of mesophilic aeromonads isolated from diseased and healthy fish cultured in Poland. J Fish Dis. 30(5):293–301. [DOI] [PubMed] [Google Scholar]
- Kozlova EV, et al. 2008. Mutation in the S-ribosylhomocysteinase (luxS) gene involved in quorum sensing affects biofilm formation and virulence in a clinical isolate of Aeromonas hydrophila. Microb Pathog. 45(5–6):343–354. [DOI] [PubMed] [Google Scholar]
- Lai C-C, Ding L-W, Hsueh P-R.. 2007. Wound infection and septic shock due to Aeromonas trota in a patient with liver cirrhosis. Clin Infect Dis. 44(11):1523–1524. [DOI] [PubMed] [Google Scholar]
- Laland K, et al. 2014. Does evolutionary theory need a rethink? Nature 514(7521):161–164. [DOI] [PubMed] [Google Scholar]
- Lamy B, Kodjo AcolBVH Study GroupLaurent F.. 2009. Prospective nationwide study of Aeromonas infections in France. J Clin Microbiol. 47(4):1234–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latif-Eugenín F, Beaz-Hidalgo R, Figueras MJ.. 2016. First record of the rare species Aeromonas schubertii from mussels: phenotypic and genetic reevaluation of the species and a review of the literature. Arch Microbiol. 198(4):333–345. [DOI] [PubMed] [Google Scholar]
- Liu R, Ochman H.. 2007. Origins of flagellar gene operons and secondary flagellar systems. J Bacteriol. 189(19):7098–7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Dong FFT, Stavrinides J, Guttman DS.. 2006. Type III effector diversification via both pathoadaptation and horizontal transfer in response to a coevolutionary arms race. PLoS Genet. 2(12):e209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massad G, Arceneaux JE, Byers BR.. 1991. Acquisition of iron from host sources by mesophilic Aeromonas species. J Gen Microbiol. 137(2):237–241. [DOI] [PubMed] [Google Scholar]
- Miñana-Galbis D, Farfán M, Gaspar Lorén J, Carmen Fusté M.. 2010. Proposal to assign Aeromonas diversa sp. nov. as a novel species designation for Aeromonas group 501. Syst Appl Microbiol. 33(1):15–19. [DOI] [PubMed] [Google Scholar]
- Miñana-Galbis D, et al. 2013. Reclassification of Aeromonas hydrophila subspecies anaerogenes. Syst Appl Microbiol. 36(5):306–308. [DOI] [PubMed] [Google Scholar]
- Mosquera-Rendón J, et al. 2016. Pangenome-wide and molecular evolution analyses of the Pseudomonas aeruginosa species. BMC Genomics. 17(1):45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosser T, et al. 2015. Exposure to pairs of Aeromonas strains enhances virulence in the Caenorhabditis elegans infection model. Front Microbiol. 6:1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada N, et al. 2010. Presence of genes for type III secretion system 2 in Vibrio mimicus strains. BMC Microbiol. 10(1):302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Origgi FC, et al. 2017. Aeromonas salmonicida type III secretion system-effectors-mediated immune suppression in rainbow trout (Oncorhynchus mykiss). Fish Shellfish Immunol. 60:334–345. [DOI] [PubMed] [Google Scholar]
- Overbeek R, et al. 2014. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 42(D1):D206–D214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallen MJ, Wren BW.. 2007. Bacterial pathogenomics. Nature 449(7164):835–842. [DOI] [PubMed] [Google Scholar]
- Parker JL, Shaw JG.. 2011. Aeromonas spp. clinical microbiology and disease. J Infect. 62(2):109–118. [DOI] [PubMed] [Google Scholar]
- Piotrowska M, Popowska M.. 2015. Insight into the mobilome of Aeromonas strains. Front Microbiol. 6:494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnusamy D, et al. 2016. Cross-talk among flesh-eating Aeromonas hydrophila strains in mixed infection leading to necrotizing fasciitis. Proc Natl Acad Sci U S A. 113(3):722–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter SC, et al. 2018. HMMER web server: 2018 update. Nucleic Acids Res. 46(W1):W200–W204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabaan AA, Gryllos I, Tomás JM, Shaw JG.. 2001. Motility and the polar flagellum are required for Aeromonas caviae adherence to HEp-2 cells. Infect Immun. 69(7):4257–4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangel LT, et al. 2019. Identification and characterization of putative Aeromonas spp. T3SS effectors. PLoS One 14(6):e0214035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reith ME, et al. 2008. The genome of Aeromonas salmonicida subsp. salmonicida A449: insights into the evolution of a fish pathogen. BMC Genomics. 9(1):427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger F, et al. 2012. Multilocus genetics to reconstruct aeromonad evolution. BMC Microbiol. 12(1):62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshadri R, et al. 2006. Genome sequence of Aeromonas hydrophila ATCC 7966T: jack of all trades. J Bacteriol. 188(23):8272–8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha J, Kozlova EV, Chopra AK.. 2002. Role of various enterotoxins in Aeromonas hydrophila-induced gastroenteritis: generation of enterotoxin gene-deficient mutants and evaluation of their enterotoxic activity. Infect Immun. 70(4):1924–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha J, et al. 2005. The type III secretion system and cytotoxic enterotoxin alter the virulence of Aeromonas hydrophila. Infect Immun. 73(10):6446–6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha J, et al. 2007. Further characterization of a type III secretion system (T3SS) and of a new effector protein from a clinical isolate of Aeromonas hydrophila—part I. Microb Pathog. 43(4):127–146. [DOI] [PubMed] [Google Scholar]
- Sierra JC, et al. 2010. Unraveling the mechanism of action of a new type III secretion system effector AexU from Aeromonas hydrophila. Microb Pathog. 49(3):122–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver AC, Graf J.. 2009. Prevalence of genes encoding the type three secretion system and the effectors AexT and AexU in the Aeromonas veronii group. DNA Cell Biol. 28(8):383–388. [DOI] [PubMed] [Google Scholar]
- Silver AC, et al. 2007. Interaction between innate immune cells and a bacterial type III secretion system in mutualistic and pathogenic associations. Proc Natl Acad Sci U S A. 104(22):9481–9486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair HA, et al. 2016. Genotypic and phenotypic identification of Aeromonas species and CphA-mediated carbapenem resistance in Queensland, Australia. Diagn Microbiol Infect Dis. 85(1):98–101. [DOI] [PubMed] [Google Scholar]
- Stuber K, Burr SE, Braun M, Wahli T, Frey J.. 2003. Type III Secretion Genes in Aeromonas salmonicida subsp. salmonicida Are Located on a Large Thermolabile Virulence Plasmid. J Clin Microbiol. 41(8):3854–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez G, et al. 2012. Actin cross-linking domain of Aeromonas hydrophila repeat in toxin A (RtxA) induces host cell rounding and apoptosis. Gene 506(2):369–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift S, et al. 1997. Quorum sensing in Aeromonas hydrophila and Aeromonas salmonicida: identification of the LuxRI homologs AhyRI and AsaRI and their cognate N-acylhomoserine lactone signal molecules. J Bacteriol. 179(17):5271–5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift S, et al. 1999. Quorum sensing-dependent regulation and blockade of exoprotease production in Aeromonas hydrophila. Infect Immun. 67(10):5192–5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talagrand-Reboul E, et al. 2017. Delineation of taxonomic species within complex of species: Aeromonas media and related species as a test case. Front Microbiol. 8:621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talagrand-Reboul E, et al. 2018. Genome-driven evaluation and redesign of PCR tools for improving the detection of virulence-associated genes in aeromonads. PLoS One 13(8):e0201428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekedar HC, et al. 2015. Complete genome sequence of fish pathogen Aeromonas hydrophila AL06-06. Genome Announc. 3(2):e00368–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsaur ML, Clowes RC.. 1989. Localization of the control region for expression of exotoxin A in Pseudomonas aeruginosa. J Bacteriol. 171(5):2599–2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez-Rosas-Landa M, Ponce-Soto GY, Eguiarte LE, Souza V.. 2017. Comparative genomics of free-living Gammaproteobacteria: pathogenesis-related genes or interaction-related genes? Pathog Dis. 75(5). [DOI] [PubMed] [Google Scholar]
- Vilches S, et al. 2004. Complete type III secretion system of a mesophilic Aeromonas hydrophila strain. Appl Environ Microbiol. 70(11):6914–6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilches S, et al. 2008. Aeromonas hydrophila AH-3 AexT is an ADP-ribosylating toxin secreted through the type III secretion system. Microb Pathog. 44(1):1–12. [DOI] [PubMed] [Google Scholar]
- Vincent AT, et al. 2016. Increasing genomic diversity and evidence of constrained lifestyle evolution due to insertion sequences in Aeromonas salmonicida. BMC Genomics. 17(1):44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent AT, et al. 2019. Investigation of the virulence and genomics of Aeromonas salmonicida strains isolated from human patients. Infect Genet Evol. 68:1–9. [DOI] [PubMed] [Google Scholar]
- Wilhelms M, Gonzalez V, Tomás JM, Merino S.. 2013. Aeromonas hydrophila lateral flagellar gene transcriptional hierarchy. J Bacteriol. 195(7):1436–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C-J, et al. 2015. Clinical implications of species identification in monomicrobial Aeromonas bacteremia. PLoS One 10(2):e0117821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, et al. 2008. Aeromonas salmonicida peritonitis after eating fish in a patient undergoing CAPD. Perit Dial Int. 28(3):316–317. [PubMed] [Google Scholar]
- Yu HB, et al. 2004. A type III secretion system is required for Aeromonas hydrophila AH-1 pathogenesis. Infect Immun. 72(3):1248–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y-H, Shaw JG.. 2016. Cross-talk between the Aeromonas hydrophila type III secretion system and lateral flagella system. Front Microbiol. 7:1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.