Abstract
Prairie dogs (genus Cynomys) are a charismatic symbol of the American West. Their large social aggregations and complex vocalizations have been the subject of scientific and popular interest for decades. A large body of literature has documented their role as keystone species of western North America’s grasslands: They generate habitat for other vertebrates, increase nutrient availability for plants, and act as a food source for mammalian, squamate, and avian predators. An additional keystone role lies in their extreme susceptibility to sylvatic plague (caused by Yersinia pestis), which results in periodic population extinctions, thereby generating spatiotemporal heterogeneity in both biotic communities and ecological processes. Here, we report the first Cynomys genome for a Gunnison’s prairie dog (C. gunnisoni gunnisoni) from Telluride, Colorado (USA). The genome was constructed using a hybrid assembly of PacBio and Illumina reads and assembled with MaSuRCA and PBJelly, which resulted in a scaffold N50 of 824 kb. Total genome size was 2.67 Gb, with 32.46% of the bases occurring in repeat regions. We recovered 94.9% (91% complete) of the single copy orthologs using the mammalian Benchmarking Universal Single-Copy Orthologs database and detected 49,377 gene models (332,141 coding regions). Pairwise Sequentially Markovian Coalescent showed support for long-term stable population size followed by a steady decline beginning near the end of the Pleistocene, as well as a recent population reduction. The genome will aid in studies of mammalian evolution, disease resistance, and the genomic basis of life history traits in ground squirrels.
Keywords: biodiversity genomics, hybrid assembly, repeat evolution, ground squirrels, PSMC
Introduction
Recent years have seen the completion of large scale projects to sequence the genomes of divergent lineages across the tree of life, such as representatives from all neognath avian orders (Jarvis et al. 2014; Zhang et al. 2014), 24 divergent eutherian mammal orders (Lindblad-Toh et al. 2011), diverse squamate species (Tzika et al. 2015), and 159 spider species from diverse lineages (Fernández et al. 2018). Despite these advances, existing genomic resources can be characterized by underrepresentation of the most diverse families and orders. For instance, although they are the most diverse mammalian order—containing 40% of all mammalian species (2,561 out of 6,399 extant species, Burgin et al. 2018)—relatively few rodent genomes have been published (e.g., Kim et al. 2011; Couger et al. 2018; Thybert et al. 2018). For instance, the 84 Rodentia genomes available on GenBank represent <3.3% of the Order’s taxa, in comparison to 15.1% representation of Primates and 18.7% of Carnivora. Rodents are biologically diverse, and some possess medically relevant adaptations (e.g., resistance to cancer and reduced senescence [Buffenstein 2008; Manov et al. 2013]). Among mammals, they provide unparalleled ecological study systems due to the relative ease of catching, housing, and relocating these animals. Rodents vary widely in sociality, longevity, size, and life history traits. In addition, they are thought to be common sources of emerging diseases in humans (Han et al. 2015). Thus, the development of additional genomic resources for rodents would aid in evolutionary, ecological, and epidemiological studies.
Some of the most widely studied wild rodents are North America’s prairie dogs (Sciuridae, genus Cynomys). A charismatic emblem of the American frontier, prairie dogs were historically some of the most abundant animals in western grasslands (Merriam 1902). Their large population sizes, diurnal activity, and loud vocalizations have inspired decades of research on social behavior (Hoogland 1979, 1981, 1998, 1999, 2001, 2013; Haynie et al. 2003; Dobson et al. 1998; Verdolin and Slobodchikoff 2009), call complexity (Grady and Hoogland 1986; Perla and Slobodchikoff 2002; Slobodchikoff et al. 1998; Placer and Slobodchikoff 2004; Slobodchikoff and Placer 2006), and the ecosystem consequences of prairie dog activity (Coppock et al. 1983; Detling and Whicker 1987; Whicker and Detling 1988; Kotliar et al. 1999; Davidson et al. 2012). Prairie dogs are considered “ecosystem engineers” (Van Nimwegen et al. 2008) because their burrows provide shelter for amphibians, burrowing owls, and other species (Ceballos et al. 1999; Augustine and Baker 2013), and their burrow construction aerates the soil, bringing nutrients to the surface where they are available for plants (Coppock et al. 1983; Detling and Whicker 1987; Whicker and Detling 1988). The fate of endangered black-footed ferrets (Mustela nigripes) is inextricably tied to prairie dogs, as prairie dogs comprise >95% of their diet; prairie dogs are also important prey for golden eagles, ferruginous hawks, coyotes, snakes, and other animals (Kotliar et al. 1999; Davidson et al. 2012). As a result, species composition differs on prairie dog colonies, leading to increased beta diversity across the landscape (Bangert and Slobodchikoff 2000; Smith and Lomolino 2004).
In the past two centuries, prairie dogs have declined by 98% as a result of eradication campaigns—due to their public perception as pests (Burnett and McCampbell 1926; Roemer and Forrest 1996; Reading et al. 1999)—and sylvatic plague (caused by the bacterium Yersinia pestis). Plague was introduced to North America from Asia in the early 1900s (Eskey and Haas 1939; Perry and Fetherston 1997; Gage and Kosoy 2005). Plague outbreaks cause 95–99% mortality in prairie dog populations (Cully et al. 1997; Cully and Williams 2001; Sackett et al. 2013); however, there is increasing evidence from natural populations (Cully et al. 1997; Pauli et al. 2006; Sackett et al. 2013) and experimental studies (Rocke et al. 2012, 2015; Busch et al. 2013) that resistance to plague may be evolving in at least two species of prairie dogs (Cynomys ludovicianus and Cynomys gunnisoni). Because the closest relative to have its genome sequenced (Ictidomys tridecemlineatus) diverged from Cynomys 4.67 (95% highest posterior density (HPD) 4.18–6.31) Ma (Upham et al. 2019), a reference genome for prairie dogs would aid in our understanding of the genetic basis of evolved resistance.
In summary, Gunnison’s prairie dogs are an important target for the development of a genome for several reasons: 1) They are ecologically important species in North American grasslands; 2) The species has been the object of intense study on life history, behavior, and the consequences of sociality for decades and thus a genome should be of broad interest; and 3) Elucidating the genomic basis of plague resistance is of both scientific and conservation interest for prairie dogs and associated species.
Materials and Methods
Sample Preparation
Several candidate individuals with low heterozygosity were chosen from available frozen DNA (Sackett et al. 2014) to facilitate genome assembly, and a low-heterozygosity individual (microsatellite Ho = 0.182) with a large amount of tissue was selected from a roadkill animal found near Telluride, CO. Tissue was stored frozen in a dimethyl sulfoxide–ethylenediaminetetraacetic acid buffer until extraction. DNA was extracted primarily from ear tissue using the Qiagen DNeasy Blood & Tissue Kit, using 40 replicate extractions from the roadkill individual to ensure sufficient DNA. Each DNA aliquot was examined for size distribution on an agarose gel and for purity via Nanodrop and Qubit, and 20 μg of the highest-quality replicates were pooled. Libraries were prepared and samples were sequenced to 20× on a PacBio Sequel and 80× on an Illumina HiSeq 4000 (2× 150-bp reads) at Duke University’s Sequencing and Genomic Technologies Shared Resource core facility.
Genome Assembly and Variant Calling
Genomes were constructed by a hybrid assembly of low-coverage PacBio long-read (∼mean 9.5 kb) sequencing for generating scaffolds and high-coverage Illumina short read (150 bp) sequencing for inferring the consensus sequence. We performed a hybrid de novo assembly using MaSurCA (v. 3.2.1, Zimin et al. 2017) and additional scaffolding with SSPACE-LongRead (Boetzer and Pirovano 2014). Gaps were filled using PBJelly (English et al. 2012), and polishing was performed in Pilon (Walker et al. 2014). We used Kraken (Wood et al. 2019) to filter out scaffolds classified as bacteria and remove them from the final assembly (see Supplementary Material online). We used Benchmarking Universal Single-Copy Orthologs (BUSCO v. 3.0.2, Simao et al. 2015) to assess the assembly completeness by comparing it to 4,104 orthologs from 50 species contained in the mammalia_odb9 gene database (Zdobnov et al. 2017). We used Bowtie2 (Langmead and Salzberg 2012) to align the raw reads to the final assembly, and samtools v1.9 (Li et al. 2009) to generate a sorted bam file. Then, we removed polymerase chain reaction duplicates with picard-tools v2.5 (http://broadinstitute.github.io/picard/, last accessed December 28, 2019) and realigned indels and called variants using the GATK v4 (McKenna et al. 2010) following standard pipelines (e.g., DePristo et al. 2011; Cassin-Sackett et al. 2019).
To assemble the mitogenome, we imported the final whole genome assembly into Geneious Prime (Biomatters, 2019.1.3), and then mapped the scaffolds to the C. gunnisoni gunnisoni mitochondrial reference genome, available on GenBank (accession number MG450794, Streich et al. 2019).
Genome Structural Contents
We estimated genome-wide heterozygosity of the Gunnison’s prairie dog using jellyfish v2.3.0 (Marçais and Kingsford 2011) with both the default settings (removing kmers with coverage >1,000×) and with the removal of kmers with coverage >10,000×. Finally, we obtained the genome sequences of four high-quality ground squirrel genomes from GenBank (Marmota flaviventris, estimated 7.59 [95% HPD 6.40–9.33] Myr divergence from Cynomys; M. marmota, 7.59 [95% HPD 6.40 –9.33] My divergence; Urocitellus parryi, 5.66 [95% HPD 4.98–7.34] My divergence; and I. tridecemlineatus, 4.67 [95% HPD 4.18–6.31] My divergence; Upham et al. 2019) and analyzed both repeat content and the relative proportion of CG sites (see Supplementary Material online) in each genome.
Genome Annotation
The genome was annotated using a multipronged approach that included repeat identification, a combination of ab initio and evidence-driven gene prediction using AUGUSTUS (v. 3.3.2; Stanke et al. 2006), and functional gene annotation using Blast2GO (Götz et al. 2008). First, we used RepeatMasker (open-4.0.6, Smit et al. 2013–2015) with the Rodentia database to identify repetitive elements in the genome and soft-mask the assembly. Next, we generated a hints file for AUGUSTUS from two different lines of evidence: 1) alignment of the I. tridecemlineatus transcriptome (Hampton et al. 2011) to our assembly using BLAT (Kent 2002) and 2) conversion of the RepeatMasker .out to GFF (RepeatMasker script rmOutToGFF3.pl) and then GFF to hints (available at http://arthropods.eugenes.org/EvidentialGene/evigene/scripts/gff2hints.pl, last accessed April 19, 2020). AUGUSTUS training was performed during the BUSCO run using the –long flag. To speed up the analysis, we partitioned our assembly into scaffolds using the script partition_EVM_inputs.pl from EVM (Evidence Modeler, Haas et al. 2008). We ran AUGUSTUS in each scaffold individually, allowing genes to be predicted independently on both strands. We concatenated the results using the script join_aug_pred.pl and extracted both the protein and nucleotide sequences of the gene models identified, as well as the individual coding sequences, using the AUGUSTUS script getAnnoFasta.pl. Finally, we used Blast2GO (v5.2.5, Götz et al. 2008) to functionally annotate the genome. To do so, we ran Blast (v2.6.0+, Altschul et al. 1990) on the gene models identified by AUGUSTUS and used the final .xml file as an input to Blast2GO.
We used Blobtools to assess the degree of microbial contamination in the de novo genome assembly. To do so, we subsetted the assembly into multiple fasta files and ran blastn on each. Matches were categorized according to species at the lowest taxonomic level and according to phylum at the highest taxonomic level.
Demographic Inference
All species of prairie dogs are thought to have experienced drastic population declines in the past two centuries as a result of persecution and disease. To infer whether we could detect such changes in historical population size, we estimated the effective population size history using the Pairwise Sequentially Markovian Coalescent implemented in Pairwise Sequentially Markovian Coalescent (PSMC) (Li and Durbin 2011). We generated the input file according to the recommendations of the author (described here https://github.com/lh3/psmc, last accessed December 28, 2019) and ran the analysis using the default settings, performing 100 bootstrap replicates. We scaled the PSMC plots assuming a mean generation time of 2 years and compared two different mutation rates based on estimates from the literature: 1) 2.2 × 10−9 per site per year (Kumar and Subramanian 2002), an estimated genome-wide rate for all mammals (“mammal rate”) and 2) 8.8 × 10−10 per site per year (Nabholz et al. 2008), which is the estimated rate for a single nuclear gene (IRBP) in Cynomys (“Cynomys rate”).
Results and Discussion
Genome Assembly and Variant Calling
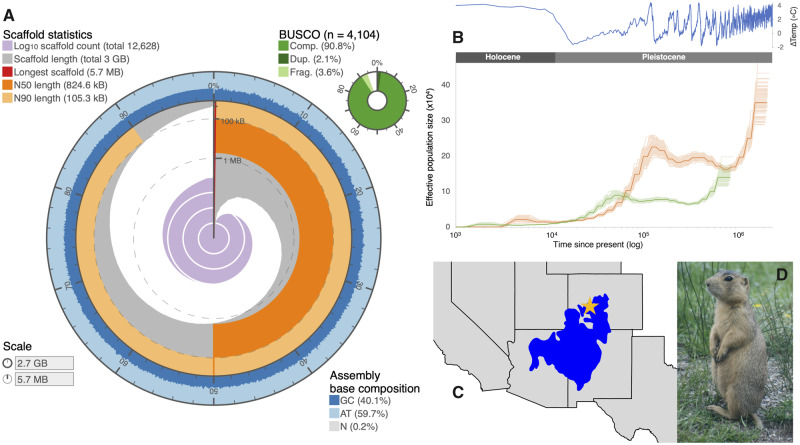
Long-read sequencing resulted in 52.5 GB of data from 14 PacBio SMRT cells, with an average read length of 9 kb. The genome was estimated to be 2.67 Gb in length (supplementary table S1, Supplementary Material online), similar to other rodents, particularly other ground squirrels (e.g., Accessions PRJNA399425, PRJNA516936, and PRJNA477386). The assembly resulted in 15,346 contigs (with a contig N50 of 686,670 bp) and 12,628 scaffolds (with a scaffold N50 of 824,613 bp; supplementary table S1, Supplementary Material online). In comparison with other ground squirrel genomes available on GenBank, this assembly resulted in the second highest scaffold N50 and L50 (after I. tridecemlineatus) and the third fewest number of scaffolds (after M. himalayana and I. tridecemlineatus). Final coverage averaged 66×. We recovered 3,811 (91%) complete and 148 (3.6%) fragmented BUSCOs out of 4,104 mammalian orthologs searched (fig. 1). A single scaffold (∼29 kb) mapped to the reference mitochondrial genome (Streich et al. 2019) with 99.66% similarity. Variant calling produced a set of 2,336,054 single-nucleotide polymorphisms.
Fig. 1.
—(A) Assembly statistic visualization (https://github.com/rjchallis/assembly-stats) showing the genome N50 (dark orange), N90 (light orange), base composition (percentage of GC in dark blue, AT in light blue, and N in light grey), and BUSCO results (top right, in shades of green). (B) PSMC reconstruction of population size estimates over time, estimated using generation time of 2 years (g = 2) and two mutation rates: µ = 2.2 × 10−9 (green; “mammal rate”) and µ = 8.8 × 10−10 (orange; “Cynomys rate”). Shaded lines correspond to 100 bootstrap estimates. The ΔTemp (°C) was calculated using benthic d18O records (Lisiecki and Raymo 2005) and extrapolated using the formula from Epstein et al. (1953). (C) Map depicting the species distribution of C. gunnisoni (blue) in the western United States, with a star denoting the location where the sample was collected (Sackett et al. 2014). (D) Image of C. gunnisoni (LCS).
Genome Structural Contents
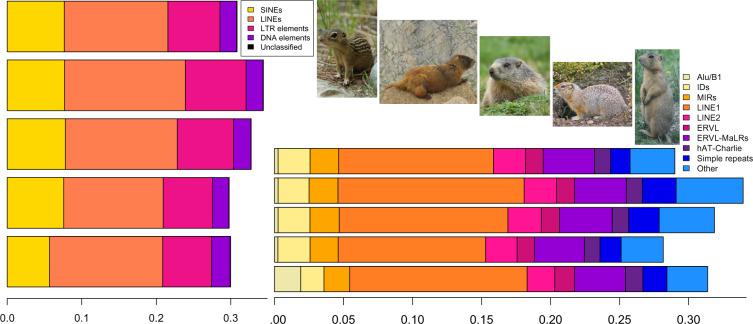
Genome-wide heterozygosity was low, estimated at 0.315% under both kmer settings; this inference is consistent with previously estimated microsatellite heterozygosity (0.18; Sackett et al. 2014). Repeat Masking indicated that 32.47% of the genome consisted of repetitive sequences, primarily LINEs (15.17%), SINEs (5.69%), and LTR elements (6.57%). Repeat content was nearly identical to four other ground squirrel species with divergence times to C. gunnisoni ranging from 9.1–13.4 Myr, both in terms of total repeat content and the proportion of each type of repeat (fig. 2 and supplementary table S5, Supplementary Material online). In all five species, repeat sequences comprised approximately one-third of the genome.
Fig. 2.
—Percent repeat content (repeat classes, left; repeat subclasses, right) in ground squirrel genomes. Top to bottom, and pictured left to right: I. tridecemlineatus, M. flaviventris, M. marmota, Urocitellus parryii, C. gunnisoni. M. flaviventris and C. gunnisoni images, copyright: Loren Cassin-Sackett; others publicly available from Wiki Commons.
Genome Annotation
AUGUSTUS identified 332,141 coding DNA sequences/exons and a total of 49,377 gene models. The number of coding sequences identified for C. gunnisoni was within the range of those found for the other four ground squirrel species, which varied from 324,927 for M. marmota to 463,195 for I. tridecemlineatus. Out of the total number of gene models analyzed, ∼1% (559) returned with Blast hits but without associated Gene Ontology entries. Blast2GO assigned functional labels to ∼82% (40,255), with enzyme codes assigned to 17.32% (8,553) of the sequences (supplementary fig. S2, Supplementary Material online).
Our assessment of contamination in Blobtools indicated that 92.02% of the Illumina reads mapped to the assembly were classified as Chordata, whereas 0.63% of reads mapped to microbial taxa, including bacteria (Proteobacteria, 0.03% and Bacteroidetes, 0.05%), fungi (Ascomycota, 0.10%) and viruses (0.45%; supplementary fig. S3a, Supplementary Material online). The remaining reads either had no blast hits (0.92%) or did not map to the assembly (6.41%). At the lowest taxonomic level, 85.53% of reads mapped to ground squirrels and 5.11% to Hominidae (4.79% human and 0.32% to the genus Pan), likely a function of the completeness of the blast database, which contains more complete human than squirrel sequences. Two microbial taxa present in the assembly were identified to genus: Pseudogymnoascus (0.09%) and Orthohepadnavirus (0.44%) (supplementary fig. S3b, Supplementary Material online). Pseudogymnoascus are a genus of fungi typically found in soil and rotting wood; thus, it is likely that this taxon is a contaminant present on the substrate on which the prairie dog was collected that was isolated along with the specimen. Orthohepadnavirus is a genus of viruses naturally hosted by humans and other mammals.
Demographic Inference
PSMC showed support for long-term stable population size followed by a steady decline beginning during the late Pleistocene and continuing into the present (fig. 1). Using the Cynomys rate, population decline occurred from ∼127 to 13 thousand years ago (ka), and with the mammal rate, populations declined from ∼51 to 9 ka. This time period corresponds approximately to increased glaciation experienced across the planet beginning ∼115 ka (potentially causing population declines). Under the Cynomys rate scenario, population size recovered slightly around 8 ka (a smaller recovery was inferred with the mammal rate at 3 ka), a time marked by the widespread expansion of grasslands across North America, which facilitated grassland specialists (Wisely et al. 2008; Oh et al. 2019) such as prairie dogs. This small increase in effective size may also correspond to divergence (Li and Durbin 2011; Cahill et al. 2016) between subspecies of Gunnison’s prairie dogs. Although the exact magnitude of effective population size inferred by using the genome of a low-heterozygosity individual may not be exact throughout all historical time periods, the patterns (i.e., shape of the curve) of changing population size should be robust to genome-wide heterozygosity levels (Li and Durbin 2011).
The assembly and annotation of the Gunnison’s prairie dog genome will facilitate future study on the genetic basis of social (Wilson-Henjum et al. 2019) and mating behavior (Hoogland et al. 2019), disease resistance (Busch et al. 2011, 2013), divergence and introgression (Sackett et al. 2014), coevolution (Holding et al. 2016), hibernation ecology (Lane et al. 2011, 2012), landscape genetics (Anderson et al. 2015; Kierepka and Latch 2016), phylogeography (Castellanos-Morales et al. 2016), keystone roles (Lindtner et al. 2018), and genomic variation in ground squirrels (Gedeon et al. 2017). A deeper understanding of genomic variation will enable scientists to inform management of threatened and endangered species, for instance, by lending insight into the optimal degree of gene flow among populations in the presence of disease (Sackett et al. 2013), or by identifying populations with “resistance” alleles or high genetic diversity as potential sources for the reintroduction of diversity (Venesky et al. 2012; Strauss et al. 2017).
Supplementary Material
Acknowledgments
We are grateful to Ramona and Kent Gaylord for providing the prairie dog specimen that was used to obtain the genome sequence and to Erin Arnold and Jeanette Calarco for preparing the sample for sequencing. This work was supported by startup funds to L.C.-S. by the University of South Florida and by a crowdfunding campaign through Instrumentl. Computing was conducted by the Smithsonian Institution High Performance Cluster (SI/HPC), Smithsonian Institution. https://doi.org/10.25572/SIHPC
Data deposition: The raw data and genome assembly have been deposited at NCBI under BioProject PRJNA573923. The VCF and GFF files, along with a markdown document detailing each step of the postassembly data processing, have been deposited at FigShare under doi:10.25573/data.c.4806264.
Literature Cited
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Anderson SJ, Kierepka EM, Swihart RK, Latch EK, Rhodes OEJ.. 2015. Assessing the permeability of landscape features to animal movement: using genetic structure to infer functional connectivity. PLoS One 10(2):e0117500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustine DJ, Baker BW.. 2013. Associations of grassland bird communities with black-tailed prairie dogs in the North American Great Plains. Conserv Biol. 27(2):324–334. [DOI] [PubMed] [Google Scholar]
- Bangert R, Slobodchikoff C.. 2000. The Gunnison’s prairie dog structures a high desert grassland landscape as a keystone engineer. J Arid Environ. 46(4):357–369. [Google Scholar]
- Boetzer M, Pirovano W.. 2014. SSPACE-LongRead: scaffolding bacterial draft genomes using long read sequence information. BMC Bioinformatics 15(1):211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffenstein R. 2008. Negligible senescence in the longest living rodent, the naked mole-rat: insights from a successfully aging species. J Comp Physiol B 178(4):439–445. [DOI] [PubMed] [Google Scholar]
- Burgin CJ, Colella JP, Kahn PL, Upham NS.. 2018. How many species of mammals are there? J Mammal. 99(1):1–14. [Google Scholar]
- Burnett WL, McCampbell SC.. 1926. The zuni prairie dog in Montezuma County, Colorado. Fort Collins (CO): Office of State Entomologist, Colorado Agricultural College.
- Busch JD, et al. 2011. Population differences in host immune factors may influence survival of Gunnison’s prairie dogs (Cynomys gunnisoni) during plague outbreaks. J Wildl Dis. 47(4):968–973. [DOI] [PubMed] [Google Scholar]
- Busch JD, et al. 2013. The innate immune response may be important for surviving plague in wild Gunnison’s prairie dogs. J Wildl Dis. 49(4):920–931. [DOI] [PubMed] [Google Scholar]
- Cahill JA, Soares AER, Green RE, Shapiro B.. 2016. Inferring species divergence times using pairwise sequential Markovian coalescent modelling and low-coverage genomic data. Philos Trans R Soc B 371(1699):20150138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassin-Sackett L, Callicrate TE, Fleischer RC.. 2019. Parallel evolution of gene classes, but not genes: evidence from Hawai’ian honeycreeper populations exposed to avian malaria. Mol Ecol. 28(3):568–583. [DOI] [PubMed] [Google Scholar]
- Castellanos-Morales G, Gámez N, Castillo-Gámez RA, Eguiarte LE.. 2016. Peripatric speciation of an endemic species driven by Pleistocene climate change: the case of the Mexican prairie dog (Cynomys mexicanus). Mol Phylogenet Evol. 94:171–181. [DOI] [PubMed] [Google Scholar]
- Ceballos G, Pacheco J, List R.. 1999. Influence of prairie dogs (Cynomys ludovicianus) on habitat heterogeneity and mammalian diversity in Mexico. J Arid Environ. 41(2):161–172. [Google Scholar]
- Coppock ADL, Ellis JE, Detling JK, Dyer MI.. 1983. Plant–herbivore interactions in a North American mixed-grass prairie. I. Effects of black-tailed prairie dogs on intraseasonal aboveground plant biomass and nutrient dynamics and plants species diversity. Oecologia 56(1):1–9. [DOI] [PubMed] [Google Scholar]
- Couger MB, Arevalo L, Campbell P.. 2018. A high quality genome for Mus spicilegus, a close relative of house mice with unique social and ecological adaptations. G3 (Bethesda) 8:2145–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cully JF, Williams ES.. 2001. Interspecific comparisons of sylvatic plague in prairie dogs. J Mammal. 82(4):894–905. [Google Scholar]
- Cully JFJ, Barnes AM, Quan TJ, Maupin G.. 1997. Dynamics of plague in a Gunnison’s prairie dog colony complex New Mexico. J Wildl Dis. 33(4):706–719. [DOI] [PubMed] [Google Scholar]
- Davidson AD, Detling JK, Brown JH.. 2012. Ecological roles and conservation challenges of social, burrowing, herbivorous mammals in the world’s grasslands. Front Ecol Environ. 10(9):477–486. [Google Scholar]
- DePristo MA, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 43(5):491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detling J, Whicker A.. 1987. Control of ecosystem processes by prairie dogs and other grassland herbivores. In: Great Plains Wildlife Damage Control Workshop Proceedings Paper 57. Rapid City, South Dakota: US Forest Service.
- Dobson FS, Chesser RK, Hoogland JL, Sugg DW, Foltz W.. 1998. Breeding groups and gene dynamics in a socially structured population of prairie dogs. J Mammal. 79(3):671–680. [Google Scholar]
- English AC, et al. 2012. Mind the gap: upgrading genomes with Pacific biosciences RS long-read sequencing technology. PLoS One 7(11):e47768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein S, Buchsbaum R, Lowenstam HA, Urey HC.. 1953. Revised carbonate-water isotopic temperature scale. Geol Soc Am Bull. 64(11):1315–1326. [Google Scholar]
- Eskey CR, Haas VH.. 1939. Plague in the western part of the United States: infection in rodents, experimental transmission by fleas, and inoculation tests for infection. Public Health Rep. 54(32):1467–1481.19315725 [Google Scholar]
- Fernández R, et al. 2018. Comparative transcriptomics across the spider tree of life phylogenomics, diversification dynamics, and comparative transcriptomics across the spider tree of life. Curr Biol. 28:1–9. [DOI] [PubMed] [Google Scholar]
- Gage KL, Kosoy MY.. 2005. Natural history of plague: perspectives from more than a century of research. Annu Rev Entomol. 50(1):505–528. [DOI] [PubMed] [Google Scholar]
- Gedeon CI, et al. 2017. The role of landscape history in determining allelic richness of European ground squirrels (Spermophilus citellus) in Central Europe. Hystrix 28(2):231–239. [Google Scholar]
- Götz S, et al. 2008. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36(10):3420–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grady RM, Hoogland JL.. 1986. Why do male black-tailed prairie dogs (Cynomys ludovicianus) give a mating call? Anim Behav. 34:108–112. [Google Scholar]
- Haas BJ, et al. 2008. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 9(1):R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton M, et al. 2011. Deep sequencing the transcriptome reveals seasonal adaptive mechanisms in a hibernating mammal. PLoS One 6(10):e27021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han BA, Schmidt JP, Bowden SE, Drake JM.. 2015. Rodent reservoirs of future zoonotic diseases. Proc Natl Acad Sci U S A. 112(22):7039–7044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynie ML, van Den Bussche RA, Hoogland JL, Gilbert DA.. 2003. Parentage, multiple paternity, and breeding success in Gunnison’s and Utah prairie dogs. J Mammal. 84(4):1244–1253. [Google Scholar]
- Holding ML, Biardi JE, Gibbs HL.. 2016. Coevolution of venom function and venom resistance in a rattlesnake predator and its squirrel prey. Proc R Soc B 283(1829):20152841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogland JL. 1979. Aggression, ectoparasitism, and other possible costs of prairie dog (Sciuridae, Cynomys ssp) coloniality. Behaviour 69(1–2):1–35. [Google Scholar]
- Hoogland JL. 1981. The evolution of coloniality in white-tailed and black-tailed prairie dogs (Sciuridae: Cynomys leucurus and C. ludovicianus). Ecology 62(1):252–272. [Google Scholar]
- Hoogland JL. 1998. Why do female Gunnison’s prairie dogs copulate with more than one male? Anim Behav. 55(2):351–359. [DOI] [PubMed] [Google Scholar]
- Hoogland JL. 1999. Philopatry, dispersal, and social organization of Gunnison’s prairie dogs. J Mammal. 80(1):243–251. [Google Scholar]
- Hoogland JL. 2001. Black-tailed, Gunnison’s, and Utah prairie dogs reproduce slowly. J Mamm Evol. 82(4):917–927. [Google Scholar]
- Hoogland JL. 2013. Prairie dogs disperse when all close kin have disappeared. Science 339(6124):1205–1207. [DOI] [PubMed] [Google Scholar]
- Hoogland JL, Trott R, Keller SR.. 2019. Polyandry and polygyny in a social rodent: an integrative perspective based on social organization, copulations, and genetics. Front Ecol Evol. 7:3. [Google Scholar]
- Jarvis ED, et al. 2014. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 346(6215):1320–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. 2002. BLAT—The BLAST-Like Alignment Tool. Genome Res. 12(4):656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierepka EM, Latch EK.. 2016. High gene flow in the American badger overrides habitat preferences and limits broadscale genetic structure. Mol Ecol. 25(24):6055–6076. [DOI] [PubMed] [Google Scholar]
- Kim EB, et al. 2011. Genome sequencing reveals insights into physiology and longevity of the naked mole rat. Nature 479(7372):223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotliar N, Baker B, Whicker A, Plumb G.. 1999. A critical review of assumptions about the prairie dog as a keystone species. Environ Manage. 24(2):177–192. [DOI] [PubMed] [Google Scholar]
- Kumar S, Subramanian S.. 2002. Mutation rates in mammalian genomes. Proc Natl Acad Sci U S A. 99(2):803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane JEet al. . 2011. A quantitative genetic analysis of hibernation emergence date in a wild population of Columbian ground squirrels. J Evol Biol. 24(9):1949–1959. [DOI] [PubMed] [Google Scholar]
- Lane JE, Kruuk LEB, Charmantier A, Murie JO, Dobson FS.. 2012. Delayed phenology and reduced fitness associated with climate change in a wild hibernator. Nature 489(7417):554–558. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL.. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Het al. . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2011. Inference of human population history from individual whole-genome sequences. Nature 475(7357):493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh K, et al. 2011. A high-resolution map of human evolutionary constraint using 29 mammals. Nature 478(7370):476–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindtner P, Ujházy K, Svitok M, Kubovčík V.. 2018. The European ground squirrel increases diversity and structural complexity of grasslands in the Western Carpathians. Mamm Res. 63(2):223–229. [Google Scholar]
- Lisiecki LE, Raymo ME.. 2005. A Pliocene-Pleistocene stack of 57 globally distributed benthic δ18O records. Paleoceanography 20(1):PA1003. [Google Scholar]
- Manov I, et al. 2013. Pronounced cancer resistance in a subterranean rodent, the blind mole-rat, Spalax: in vivo and in vitro evidence. BMC Biol. 11(1):91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais G, Kingsford C.. 2011. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27(6):764–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20(9):1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merriam CH. 1902. The prairie dog of the great plains. Yearb. United States Dep. Agric. Washington: Government Printing Office. p. 257–270. [Google Scholar]
- Nabholz B, Glémin S, Galtier N.. 2008. Strong variations of mitochondrial mutation rate across mammals—the longevity hypothesis. Mol Biol Evol. 25(1):120–130. [DOI] [PubMed] [Google Scholar]
- Oh KP, Aldridge CL, Forbey JS, Dadabay CY, Oyler-McCance SJ.. 2019. Conservation genomics in the Sagebrush Sea: population divergence, demographic history, and local adaptation in sage-grouse (Centrocercus spp.). Genome Biol Evol. 11(7):2023–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli JN, Buskirk SW, Williams ES, Edwards WH.. 2006. A plague epizootic in the black-tailed prairie dog (Cynomys ludovicianus). J Wildl Dis. 42(1):74–80. [DOI] [PubMed] [Google Scholar]
- Perla BS, Slobodchikoff CN.. 2002. Habitat structure and alarm call dialects in Gunnison’s prairie dog (Cynomys gunnisoni). Behav Ecol. 13(6):844–850. [Google Scholar]
- Perry RD, Fetherston JD.. 1997. Yersinia pestis—etiologic agent of plague. Microbiology 10(1):35–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Placer J, Slobodchikoff CN.. 2004. A method for identifying sounds used in the classification of alarm calls. Behav Processes 67(1):87–98. [DOI] [PubMed] [Google Scholar]
- Reading RP, Miller BJ, Kellert SR.. 1999. Values and attitudes toward prairie dogs. Anthrozoös 12(1):43–52. p [Google Scholar]
- Rocke TE, et al. 2012. Resistance to plague among black-tailed prairie dog populations. Vector-Borne Zoonotic Dis. 12(2):111–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocke TE, et al. 2015. Age at vaccination may influence response to sylvatic plague vaccine (SPV) in Gunnison’s prairie dogs (Cynomys gunnisoni). Ecohealth 12(2):278–287. [DOI] [PubMed] [Google Scholar]
- Roemer DM, Forrest SC.. 1996. Prairie dog poisoning in northern Great Plains: an analysis of programs and policies. Environ Manage. 20(3):349–359. [DOI] [PubMed] [Google Scholar]
- Sackett LC, Collinge SK, Martin AP.. 2013. Do pathogens reduce genetic diversity of their hosts? Variable effects of sylvatic plague in black-tailed prairie dogs. Mol Ecol. 22(9):2441–2455. [DOI] [PubMed] [Google Scholar]
- Sackett LC, et al. 2014. Evidence for two subspecies of Gunnison’s prairie dogs (Cynomys gunnisoni), and the general importance of the subspecies concept. Biol Conserv. 174:1–11. [Google Scholar]
- Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. Genome analysis BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Slobodchikoff CN, Ackers SH, Ert MV.. 1998. Geographic variation in alarm calls of Gunnison’s prairie dogs. J Mammal. 79(4):1265–1272. [Google Scholar]
- Slobodchikoff CN, Placer J.. 2006. Acoustic structures in the alarm calls of Gunnison’s prairie dogs. J Acoust Soc Am. 119(5):3153–3160. [DOI] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P.. 2013. –2015. RepeatMasker Open-4.0. Seattle, Washington: Institute for Systems Biology. Available from: www.repeatmasker.org.
- Smith GA, Lomolino MV.. 2004. Black-tailed prairie dogs and the structure of avian communities on the shortgrass plains. Oecologia 138(4):592–602. [DOI] [PubMed] [Google Scholar]
- Stanke M, et al. 2006. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 34(Web Server):W435–W439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss AT, et al. 2017. Rapid evolution rescues hosts from competition and disease but—despite a dilution effect—increases the density of infected hosts. Proc R Soc B 284(1868):20171970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streich SP, Keepers KG, Griffin KA, Kane NC, Martin AP.. 2019. The complete mitochondrial genome of Gunnison’s prairie dog subspecies (Cynomys gunnisoni gunnisoni) and phylogenetic relationship within the genus Cynomys. Mitochondrial DNA B: Resour. 4(1):397–398. [Google Scholar]
- Thybert D, et al. 2018. Repeat associated mechanisms of genome evolution and function revealed by the Mus caroli and Mus pahari genomes. Genome Res. 28(4):448–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzika AC, Ullate-Agote A, Grbic D, Milinkovitch MC.. 2015. Reptilian transcriptomes v2.0: an extensive resource for Sauropsida genomics and transcriptomics. Genome Biol Evol. 7(6):1827–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upham NS, Esselstyn JA, Jetz W.. 2019. Inferring the mammal tree: species-level sets of phylogenies for questions in ecology, evolution, and conservation. PLoS Biol. 17(12):e3000494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nimwegen RE, Kretzer J, Cully JF Jr. 2008. Ecosystem engineering by a colonial mammal: how prairie dogs structure rodent communities. Ecology 89(12):3298–3305. p [DOI] [PubMed] [Google Scholar]
- Venesky MD, Mendelson JRI, Sears BF, Stiling P, Rohr JR.. 2012. Selecting for tolerance against pathogens and herbivores to enhance success of reintroduction and translocation. Conserv Biol. 26(4):586–592. [DOI] [PubMed] [Google Scholar]
- Verdolin JL, Slobodchikoff CN.. 2009. Resources, not kinship, determine social patterning in the territorial Gunnison’s prairie dog (Cynomys gunnisoni). Ethology 115(1):59–69. [Google Scholar]
- Walker BJ, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9(11):e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whicker AD, Detling JK.. 1988. Ecological consequences of prairie dog disturbances. Bioscience 38(11):778–785. [Google Scholar]
- Wilson-Henjum GE, et al. 2019. Alarm call modification by prairie dogs in the presence of juveniles. J Ethol. 37(2):167–168. [Google Scholar]
- Wisely SM, Statham MJ, Fleischer RC.. 2008. Pleistocene refugia and Holocene expansion of a grassland-dependent species, the black-footed ferret (Mustela nigripes). J Mammal. 89(1):87–96. [Google Scholar]
- Wood DE, Lu J, Langmead B.. 2019. Improved metagenomic analysis with Kraken 2. Genome Biol. 20(1):257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov EMet al. . 2017. OrthoDB v9.1: cataloging evolutionary and functional annotations for animal, fungal, plant, archaeal, bacterial and viral orthologs. Nucleic Acids Res. 45(D1):D744–D749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Get al. . 2014. Comparative genomics reveals insights into avian genome evolution and adaptation. Science. 346(6215):1311–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimin AVet al. . 2017. Hybrid assembly of the large and highly repetitive genome of Aegilops tauschii, a progenitor of bread wheat, with the MaSuRCA mega-reads algorithm. Genome Res. 27(5):787–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.