Abstract
Endogenous viral elements (EVEs), derived from all major types of viruses, have been discovered in many eukaryotic genomes, representing “fossil records” of past viral infections. The endogenization of nudiviruses has been reported in several insects, leading to the question of whether genomic integration is a common phenomenon for these viruses. In this study, genomic assemblies of insects and other arthropods were analyzed to identify endogenous sequences related to Nudiviridae. A total of 359 nudivirus-like genes were identified in 43 species belonging to different groups; however, none of these genes were detected in the known hosts of nudiviruses. A large proportion of the putative EVEs identified in this study encode intact open reading frames or are transcribed as mRNAs, suggesting that they result from recent endogenization of nudiviruses. Phylogenetic analyses of the identified EVEs and inspections of their flanking regions indicated that integration of nudiviruses has occurred recurrently during the evolution of arthropods. This is the first report of a comprehensive screening for nudivirus-derived EVEs in arthropod genomes. The results of this study demonstrated that a large variety of arthropods, especially hemipteran and hymenopteran insects, have previously been or are still infected by nudiviruses. These findings have greatly extended the host range of Nudiviridae and provide new insights into viral diversity, evolution, and host–virus interactions.
Keywords: Nudiviridae, arthropods, endogenous viral elements, host range, genomic data
Introduction
Nudiviruses (Latin nudi = naked, uncovered) are large, double-stranded (ds) DNA viruses, with rod-shaped and enveloped nucleocapsids. They were previously considered to be “nonoccluded baculoviruses” and have been reported in a wide range of host species, including insects and arthropods (Huger and Krieg 1991; Wang et al. 2007). Although baculoviruses are among the best-studied insect DNA viruses, only a few nudiviruses have been well characterized to date. The identified hosts of previously described nudiviruses include crickets (Gryllus bimaculatus nudivirus, GbNV) (Reinganum et al. 1970), palm rhinoceros beetles (Oryctes rhinoceros nudivirus, OrNV) (Wang et al. 2011), corn earworm moths (Heliothis zea nudivirus, HzNV-1 and HzNV-2) (Burand et al. 2012), fruit flies (Drosophila innubila nudivirus, DiNV; Kallithea virus) (Unckless 2011; Webster et al. 2015), crane flies (Tipula oleracea nudivirus, ToNV) (Bézier et al. 2015), and tiger prawns (Penaeus monodon nudivirus, PmNV) (Yang et al. 2014). Nudivirus-like genes have also been identified in a hemipteran insect, the brown planthopper (Nilaparvata lugens), where the virus was found to be integrated into the host genome and was named Nilaparvata lugens endogenous nudivirus (NlENV) (Cheng et al. 2014).
The endogenization of viral sequences was long thought to be limited to retroviruses. Retroviruses are the only known eukaryotic viruses that integrate into the host genome as an obligate step in their life cycles (Feschotte and Gilbert 2012). Occasionally, virus integration occurs in a host’s germline, resulting in the vertical transmission and fixation of the integrated viral sequence in the host population. These endogenous retroviruses accumulate in vertebrate genomes and contribute to the evolution of host genomes (Johnson 2010).
Although endogenous retroviruses constitute the vast majority of endogenous viral elements (EVEs) that have been recognized so far, EVEs derived from other viruses have also been reported (Crochu et al. 2004; Tang and Lightner 2006). Different types of EVEs represent “fossil records” of past viral infections and can provide insights into the evolution of both the viruses and their hosts (Johnson 2010). Over the past decade, with the increasing availability of whole-genome sequence data, a wide range of nonretroviral EVEs have been identified in the nuclear genomes of various eukaryotic organisms (Horie et al. 2010; Katzourakis and Rj 2010; Liu et al. 2010; Cui and Holmes 2012; Metegnier et al. 2015). The discovery and analysis of these EVEs has revealed new information regarding the histories and origins of modern viruses. These findings have also extended the host ranges of many viruses, in contrast with their known ranges as exogenous viruses.
The endogenization of nudiviruses into the genomes of insects may have occurred several times during evolution. Bracoviruses are thought to be originated from an ancient nudivirus which integrated into the genome of an ancestor wasp (Hymenoptera: Braconidae) ∼100 Ma (Bézier et al. 2009). The endogenized nudivirus was then utilized by the braconid wasps as a virulence gene delivery system, facilitating the protection of wasp larvae in parasitized hosts (Strand and Burke 2012). A similar phenomenon has been reported in the parasitic wasp, Venturia canescens (Hymenoptera: Ichnomeunidae) (Pichon et al. 2015; Drezen et al. 2017), which incorporates virulence proteins into viral liposomes to protect their eggs against host immune responses (Rotheram 1967). In the case of NlENV, a large set of nudivirus genes was detected in the genome of N. lugens, many of which are expressed (Cheng et al. 2014). However, whether the brown planthopper profits from the endogenization of the nudivirus, as has occurred in the parasitoid wasps, remains unclear.
Here, we screened the sequenced genomes of insects and other arthropods from the National Center of Biotechnology Information (NCBI)’s whole-genome shotgun (WGS) database. More than 300 nudivirus-like genes were identified, some of which may have been integrated into host genomes. We also investigated the coding capacity and expression of these sequences and their relationships with existing nudiviruses. The identification of these nudivirus-like genes revealed a novel degree of viral diversity and showed that nudiviruses are (or were at one time) capable of infecting a wide range of arthropod groups.
Materials and Methods
Genome Screening
WGS assemblies of 240 arthropod species (supplementary table S1, Supplementary Material online) were screened in silico, using the TBlastN program (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and a library of the deduced amino acid sequence derived from known viruses belonging to the family Nudiviridae (NCBI: txid1511852, accession numbers: GbNV, YP_001111268.1 to YP_001111365.1; OrNV, YP_002321312.1 to YP_002321450.1; HzNV, AAN04300.1 to AAN04447.1; PmNV, YP_009051839.1 to YP_009051953.1; DiNV, ATZ81483.1 to ATZ81589.1; ToNV, YP_009116648.1 to YP_009116778.1; Esparto virus, AUQ43936.1 to AUQ44022.1; and Tomelloso virus, ATY70176.1 to ATY70268.1). All nonredundant matches for viral peptides with E-values ≤1e-3 were extracted and used to screen the GenBank nonredundant (nr) protein database in a reciprocal BlastX search. Fragments from the host genome assembly were considered to be candidate endogenous viral sequences if they unambiguously matched viral protein sequences. All database searches were completed in July 2019.
Examination of Possible Contamination or Misassembly
To eliminate the possibility of misassembly, we compared the candidate EVEs and their flanking cellular sequences with the NCBI Trace Archive databases ( https://trace.ncbi.nlm.nih.gov/Traces/sra/; last accessed April 20, 2020). The megaBLAST program was used, with a cutoff value of >95% nucleotide identity, and the junctions between EVEs and cellular sequences were carefully examined. The flanking sequences were scanned for adjacent transposable elements (TEs) or repetitive sequences using RepeatMasker (http://www.repeatmasker.org/; last accessed April 20, 2020) based on a combined database of Dfam_3.0 (Wheeler et al. 2013) and RepBase (Bao et al. 2015). In addition, LTR_retriever (Ou and Jiang 2018) was used for identification of long terminal repeat (LTR) retrotransposons, and the results were merged with those of RepeatMasker.
Expression of Nudivirus-Related Sequences
All of the candidate endogenous viral sequences were extracted along with their flanking regions (1 kb on each end of the sequence). Putative viral open reading frames (ORFs) were inferred using the NCBI’s ORF Finder program (https://www.ncbi.nlm.nih.gov/orffinder/; last accessed April 20, 2020), followed by manual editing based on the most closely related exogenous viral sequences in the nr database. To investigate whether these integrated sequences are expressed in the hosts, we searched the Expressed Sequence Tag (EST, http://www.ncbi.nlm.nih.gov/nucest; last accessed April 20, 2020) and transcriptome shotgun assembly (TSA, http://www.ncbi.nlm.nih.gov/genbank/tsa; last accessed April 20, 2020) databases at NCBI for the corresponding mRNA sequences.
Phylogenetic Analysis
Putative peptides encoded by the nudivirus-related sequences were obtained according to BlastX hits and ORF predictions and were used for the phylogenetic analysis. Sequences were aligned with closely related viral proteins using MUSCLE (Edgar 2004), and the conserved regions were selected using Gblocks (Castresana 2000). The appropriate substitution model for phylogenetic analysis was selected with ModelFinder (Kalyaanamoorthy et al. 2017) under the Bayesian information criterion. Maximum-likelihood (ML) phylogenies were estimated using MEGA5 (Tamura et al. 2011), with the substitution model LG + G and subtree-pruning-regrafting tree topologies. Support for node in the ML tree was evaluated using 1,000 nonparametric bootstrap replicates. The Bayesian inference (BI) tree was constructed with MrBayes 3.2 (Ronquist et al. 2012), using the LG + I + G4 model. MrBayes analyses were run across four Monte Carlo Markov chains for 1 million generations, sampling every 500 generations. The consensus tree was obtained after a burn-in of 500 generations, and the average standard deviation (SD) value of split frequencies was used as a proof of stationarity when this value was <0.01.
Results
Nudivirus-Related Sequences in the Genomes of Arthropods
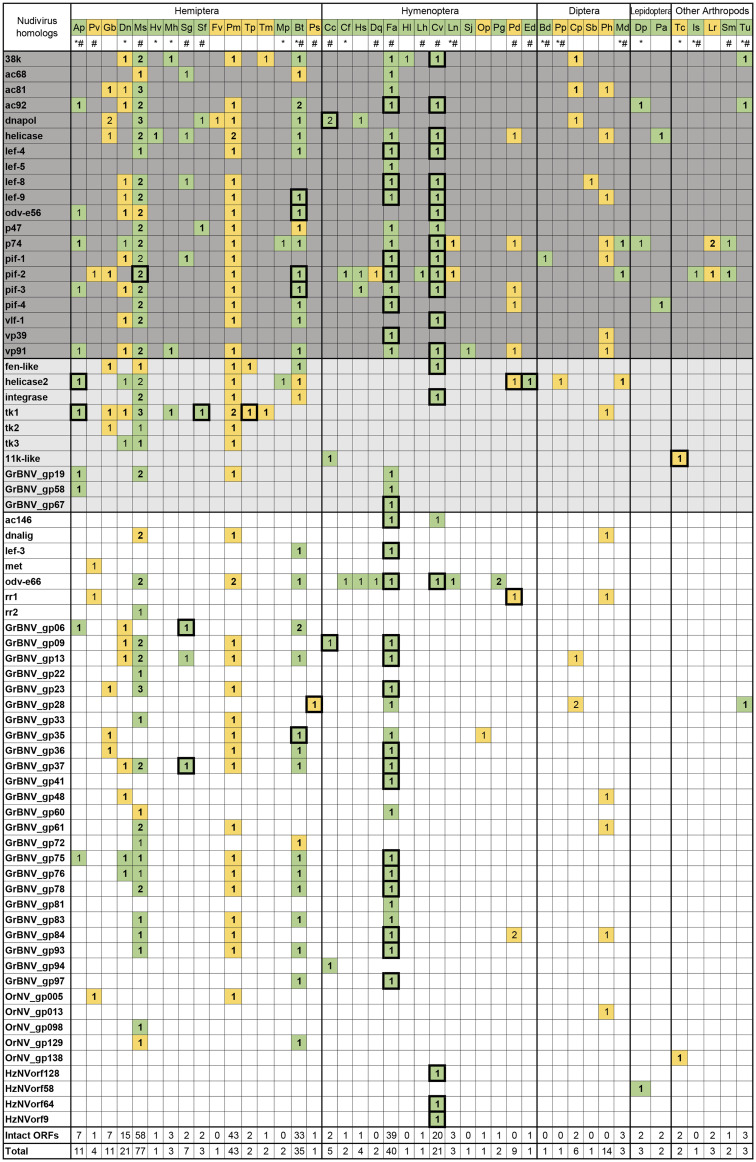
To screen for the presence of nudivirus-related EVEs in the arthropod genome, a library of nudiviral peptide sequences was constructed, and the TBlastN program was used to search genome assemblies in NCBI database. A total of 359 nudivirus-like sequences (best blast hits range from 21% to 89% amino acid identities) were identified from the genome assembly of 43 species (fig. 1). Most of these species were insects, including 16 hemipterans, 14 hymenopterans, 6 dipterans, and 2 lepidopterans. In addition, a few nudivirus-related sequences were identified in other arthropods, such as the tadpole shrimp (Triops cancriformis), the black-legged tick (Ixodes scapularis), the two-spotted spider mite (Tetranychus urticae), and two spider species (Loxosceles reclusa and Stegodyphus mimosarum).
Fig. 1.
—Nudivirus-like genes identified in arthropod genomes. Copy numbers for each nudivirus homolog are indicated. Core genes conserved between baculoviruses and nudiviruses are shown in dark gray, and nudivirus-specific core genes are shown in light gray. Sequences with or without flanking host sequences were colored in green and yellow, respectively. Ap, Acyrthosiphon pisum; Pv, Pachypsylla venusta; Gb, Gerris buenoi; Dn, Diuraphis noxia; Ms, Melanaphis sacchari; Hv, Homalodisca vitripennis; Mh, Maconellicoccus hirsutus; Sg, Schizaphis graminum; Sf, Sipha flava; Fv, Ferrisia virgate; Pm, Paracoccus marginatus; Tp, Trionymus perrisii; Tm, Trabutina mannipara; Mp, Myzus persicae; Bt, Bemisia tabaci; Ps, Philaenus spumarius; Cc, Cephus cinctus; Cf, Camponotus floridanus; Hs, Harpegnathos saltator; Dq, Dinoponera quadriceps; Fa, Fopius arisanus; Hl, Habropoda laboriosa; Lh, Linepithema humile; Cv, Cotesia vestalis; Ln, Lasius niger; Sj, Synergus japonicas; Op, Ormyrus pomaceus; Pg, Pseudomyrmex gracilis; Pd, Polistes dominula; Ed, Euglossa dilemma; Bd, Bactrocera dorsalis; Pp, Phlebotomus papatasi; Cp, Condylostylus patibulatus; Sb, Sphyracephala brevicornis; Ph, Phortica variegate; Md, Mayetiola destructor; Dp, Danaus plexippus; Pa, Papilio glaucus; Tc, Triops cancriformis; Is, Ixodes scapularis; Lr, Loxosceles reclusa; Sm, Stegodyphus mimosarum; and Tu, Tetranychus urticae. *EST data available; #TSA data available. Boxes surrounding the numbers indicate that gene expression was detected. Sequences with intact ORFs were indicated by boldface.
The numbers of candidate EVEs varied among different species. For example, the parasitoid wasp, Cotesia vestalis, which is a known host of bracovirus, contained 21 nudivirus-like genes. However, almost twice as many nudivirus-like sequences were identified in Fopius arisanus, another wasp species that parasitizes tephritid fruit flies, although no bracovirus had been reported in this species. A large amount of nudivirus-related sequences were discovered in hemipteran genomes. Among these, the sugarcane aphid, Melanaphis sacchari, had the highest number of nudivirus-like genes, many of which existed in two or more copies (fig. 1).
The 359 identified sequences corresponded to 70 nudivirus homologous genes. Twenty of these were core genes that are conserved between baculoviruses and nudiviruses, including all of the per os infectivity factors (pif-0 [p74], pif-1, pif-2, pif-3, pif-4 [19 kda], pif-5 [odv-e56], pif-6 [ac68], and pif-7 [vp91/95]), genes involved in DNA processing (dnapol and helicase), transcription (p47, lef-4, lef-8, lef-9, lef-5, and vlf-1), and viral packaging/assembly/morphogenesis (ac81, ac92 [p33], 38k, and vp39). In addition, 11 nudivirus-specific core genes were identified, which are conserved across all sequenced nudivirus genomes. These included helicase2, integrase, fen-1 (FLAP endonuclease), three thymidine kinase genes (tk1, tk2, and tk3), the 11K-like, and three other genes with unknown functions (GrBNV_gp19-like, GrBNV_gp58-like, and GrBNV_gp67-like).
Characteristics of Nudivirus-Related Sequences
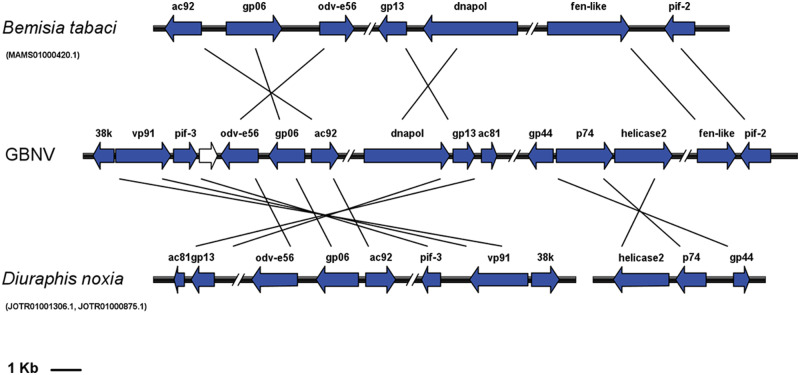
The scaffolds and contigs containing nudivirus-related sequences varied greatly in length, from 406 to 4,743,521 bp (supplementary table S2, Supplementary Material online). In some species, the virus-like sequences were located in the same scaffolds as host genomic sequences, whereas in some species, the sequences were identified in some short scaffolds or contigs that contained only one gene. Several scaffolds contained gene clusters that were arranged similarly to their orthologs in exogenous nudiviruses, such as the scaffold MAMS01000420.1 in Bemisia tabaci, and the scaffolds JOTR01001306.1 and JOTR01000875.1 of Diuraphis noxia (fig. 2 and supplementary table S2, Supplementary Material online). A core gene cluster containing helicase and pif-4, which are present in all sequenced nudiviruses and baculoviruses, was detected in the M. sacchari, Paracoccus marginatus, Polistes dominula, and Papilio glaucus genomes (supplementary table S2, Supplementary Material online). However, these two genes were separated from each other in the genomic sequences of B. tabaci and F. arisanus, whereas the pif-4 gene was absent in the C. vestalis genome.
Fig. 2.
—Conserved gene clusters identified in A. pisum and Di. noxia genomes. ORFs are indicated by boxed arrows, and conserved gene clusters are colored in blue; double slashes indicate the omitted genomic ranges. GbNV: Gryllus bimaculatus nudivirus.
Most of the sequences shared limited similarity (<50% amino acid identity) with known nudiviruses and were likely derived from previously undescribed virus species. A large proportion of scaffolds or contigs consisted of intact ORFs (263 out of the 359 putative EVEs, fig. 1), suggesting that they had the potential to generate functional proteins. However, some virus-like sequences (∼25%) appeared to be defective, containing numerous frameshifts, internal stop codons, and insertions or deletions (fig. 3 and supplementary table S2, Supplementary Material online). These sequences may represent degraded, nonfunctional pseudogenes. In addition, some ORFs were truncated by the ends of contigs or gaps in scaffolds and were annotated as “partial” (supplementary table S2, Supplementary Material online).
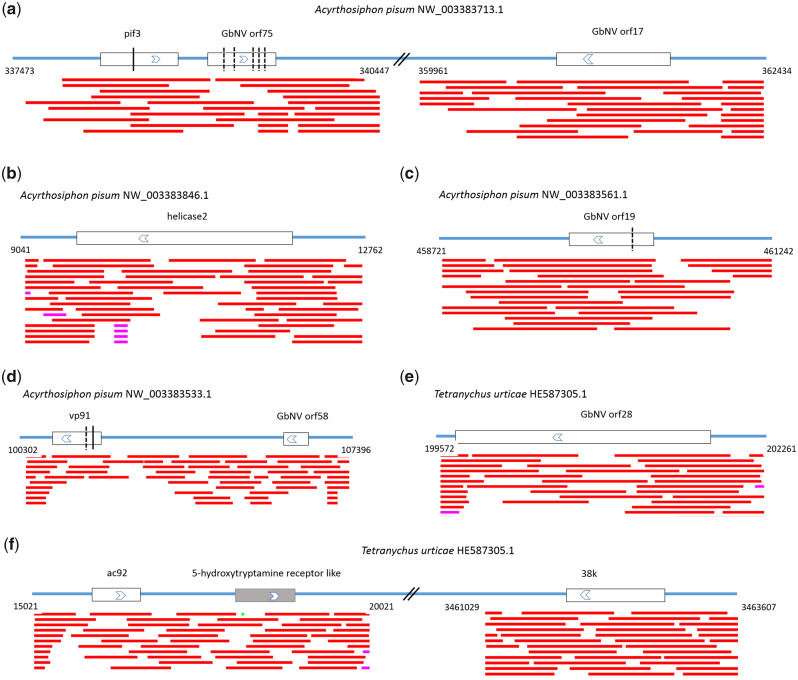
Fig. 3.
—Schematic representation of nudivirus-like genes with flanking regions. Predicted viral ORFs and their transcriptional directions are indicated as white boxes with arrows, and predicted host genes are shown as gray boxes with arrows. Double slashes indicate the omitted genomic ranges. The vertical lines indicate the predicted frameshift sites, and the dash lines indicate premature stop codons. The red lines below represent matched regions of a trace record.
To determine whether the virus-like sequences were integrated into host genomes, we carefully examined the raw sequence reads used for WGS assembly. The results showed that several trace records covered the junctions between viral sequences and adjacent cellular sequences (fig. 3). However, trace archives were not available for most genomes, and we could not rule out the possibility that the sequences were derived from exogenous viruses. In addition, some short scaffolds or contigs contained only viral sequences, such that determining whether they were part of the host genome was not possible (fig. 1 and supplementary table S2, Supplementary Material online). Actually, no convincing evidence of endogenization has yet been found in 15 species, suggesting that the sequenced hosts may have been suffering from a bona fide viral infection.
Expression of Viral Genes in Host Genomes
We searched the NCBI EST and TSA databases to identify corresponding mRNAs for the nudivirus-related sequences. A total of 77 transcripts were identified in 13 organisms (fig. 1 and supplementary table S3, Supplementary Material online), suggesting the mRNA level activity of these putative EVEs that may produce proteins. However, there were also some transcripts containing frameshifts or premature stop codons (supplementary table S3, Supplementary Material online). These sequences are unlikely to generate normal proteins but might function at the RNA level.
Transcripts for a large proportion of the identified nudivirus-like genes were not detected; however, the possibility of low level expressions or a stage/tissue-specific expression pattern cannot be excluded. On the other hand, the currently available EST and TSA data sets are still limited. For those host species without either EST or TSA sequence data (15 out of 43 species, fig. 1), it is not possible to determine whether the putative EVEs can be transcribed.
Genomic Context of Nudivirus-Derived EVEs
To determine the role of TEs in nudivirus endogenization, the 5-kb genomic regions flanking each side of the putative EVEs were extracted and scanned for adjacent TEs. Contigs without flanking host sequences were not included in the analysis. As a result, about 25% of the EVEs were flanked by TEs (supplementary table S4, Supplementary Material online), predominantly DNA transposons (supplementary table S5, Supplementary Material online). Then, we compared the percent TE occupancy between whole genome and EVE flanking regions to found out possible TE enrichment. However, in most species, no significant enrichment of TEs (P < 0.001) was observed using a cumulative binomial distribution (supplementary table S4, Supplementary Material online).
We also compared the EVE flanking regions between different species to detect orthologous copies of EVEs, but similarity was not found even in closely related species, suggesting that most (if not all) of the integration events occurred independently.
Phylogenetic Analysis of Nudivirus-Related Sequences
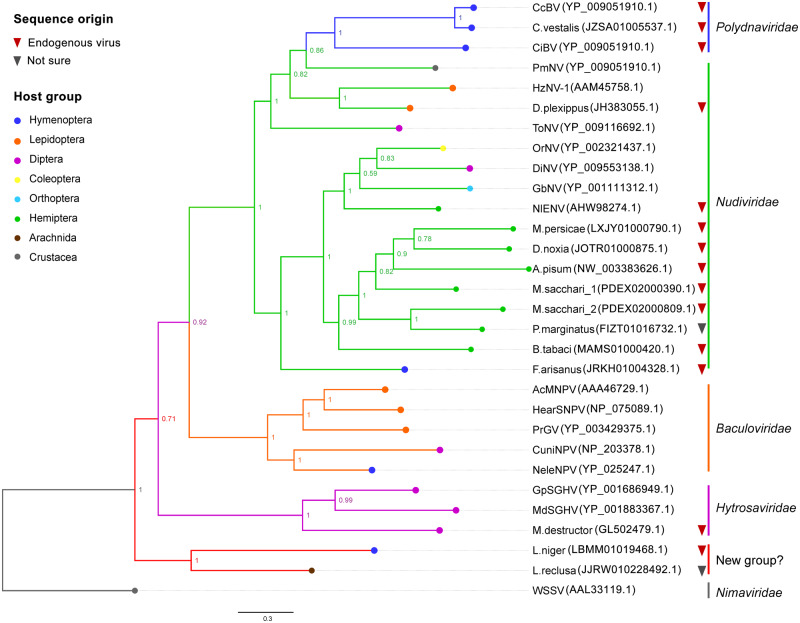
To understand the evolutionary relationships between the nudivirus homologs identified in this study and other exogenous viruses, genes encoding complete or near full-length sequences of P74 proteins were extracted and subjected to a phylogenetic analysis. Phylogenetic trees were calculated using ML and BI methods. Both methodologies resulted in similar tree topologies, and only the BI tree is presented here (fig. 4).
Fig. 4.
—Phylogenetic analysis of nudivirus-like genes in arthropod genomes. The tree is based on the amino acid sequences of P74 proteins and was constructed by MrBayes, using mixed models of amino acid substitutions. The BI posterior probabilities are presented at the nodes as percent values. Associated host groups are indicated by tip colors. The following viruses were included in this analysis, with abbreviated names: Autographa californica nucleopolyhedrovirus (AcMNPV), Helicoverpa armigera nucleopolyhedrovirus (HearSNPV), Pieris rapae granulosis virus (PrGV), Culex nigripalpus NPV (CuniNPV), Neodiprion lecontei NPV (NeleNPV), Cotesia congregata bracovirus (CcBV), Chelonus inanitus bracovirus (CiBV), Musca domestica salivary gland hypertrophy virus (MdSGHV), Glossina pallidipes salivary gland hypertrophy virus (GpSGHV), White spot syndrome virus (WSSV), Gryllus bimaculatus nudivirus (GbNV), Oryctes rhinoceros nudivirus (OrNV), Heliothis zea nudivirus 1 (HzNV-1), Drosophila innubila nudivirus (DiNV), Tipula oleracea nudivirus (ToNV), Penaeus monodon nudivirus (PmNV), and Nilaparvata lugens endogenous nudivirus (NlENV). WSSV was used to root the tree. GenBank accession number is given for each sequence.
As shown in figure 4, sequences from aphids (Acyrthosiphon pisum, Myzus persicae, M. sacchari, and Di. noxia), the papaya mealybug (Pa. marginatus), and the silverleaf whitefly (B. tabaci) were closely related and were placed within the subclades of genus Alphanudivirus. Surprisingly, two P74 sequences identified in M. sacchari did not cluster together, indicating that the coinfection of more than one virus might have occurred. The endogenous nudiviral sequence identified in N. lugens did not cluster with those of other hemipteran insects. Instead, it was more closely related to the exogenous alphanudiviruses. The F. arisanus P74 was also in the same cluster with Alphanudivirus, whereas the sequence identified from another braconid wasp, C. vestalis, was grouped together with bracoviruses, suggesting that the two sequences had different origins. PmNV, ToNV, and HzNV-1 were related to the bracovirus clade, and the sequence of Danaus plexippus formed a cluster with HzNV-1, the host of which was also a lepidopteran. In addition, P74 sequences from two distantly related species, Lasius niger and L. reclusa, formed a well-supported clade. It may be considered a new family of large dsDNA viruses, but the closely related exogenous viruses have not been reported yet.
In general, the putative EVEs clustered with different exogenous virus species, and the phylogenetic patterns of these EVEs were not consistent with the evolutionary relationships of their hosts, indicating that they were derived from multiple independent integration events, rather than a single insertion into a common ancestor.
Discussion
In animal genomes, the majority of EVEs are derived from retroviruses (Tristem 2000; Sperber et al. 2007); however, thorough screening of eukaryotic genomes has shown that any type of virus can become endogenous. EVEs derived from single-stranded DNA (ssDNA) virus families (Parvoviridae, Circoviridae, and Nanoviridae) have been identified in the genomes of mammals (Belyi et al. 2010; Katzourakis and Rj 2010), reptiles (Gilbert et al. 2014), amphibians (Liu et al. 2011), invertebrates (Thézé et al. 2014; Metegnier et al. 2015), as well as in plants, fungi, and protists (Liu et al. 2011). However, large dsDNA viruses are often not considered in paleovirology screenings.
Nudiviruses have dsDNA genomes that replicate and assemble in the nuclei of infected cells. HzNV-1 has been reported to be able to integrate its genome into host chromosomes during the infection process (Lin et al. 1999). Nudiviruses can also durably integrate into the genomes of their hosts, as described in the brown planthopper (Cheng et al. 2014). To determine whether the endogenization of nudiviruses is a common phenomenon, we searched the whole-genome assemblies of arthropods for virus-derived sequences. As expected, hundreds of nudivirus-like genes were discovered in 43 different species.
Nudiviruses are a diverse group of invertebrate large dsDNA viruses and represent an ancient sister group of the baculoviruses (Wang and Jehle 2009; Thézé et al. 2011). The sequenced nudiviruses have 32 genes in common, of which 21 are homologs of baculovirus core genes (Bézier et al. 2015). Most of these core genes were also identified in this study, with the exception of the DNA-binding protein P6.9. Because the sequences of P6.9 proteins are quite short and diverse, it was not characterized as a core gene of nudiviruses until recently. The screening strategy used here was based on homology searches, and less conserved sequences may have been overlooked. For those noncore genes that have lower identity and similarity, their homologous sequences might not be detected by this way.
Except for HzNV-1 and HzNV-2, the genome sizes and gene orders varied greatly among nudiviruses (Wang et al. 2012). Members of the genus Alphanudivirus (including NlENV) share a number of gene clusters composed of 2–7 collinearly arranged genes, whereas few gene clusters are shared between alphanudivirus and betanudiviruses. Only one organizationally conserved region was observed among the genomes of known nudiviruses, the core gene cluster that contains helicase and pif-4 (Cheng et al. 2014; Yang et al. 2014; Bézier et al. 2015). These two genes, together with 38K and lef-5, were found in synteny in all sequenced baculovirus genomes (Herniou et al. 2003). Homologs of 38K and lef-5 were also present in nudiviruses but were not grouped with helicase and pif-4. Two other conserved clusters have been reported for baculovirisues, p33-ac93-ac94 (Yuan et al. 2011) and ac142-ac143 (McCarthy and Theilmann 2008). Of these, only p33 was identified as a core gene of nudiviruses. In the genomes of B. tabaci and Di. noxia, the p33, GrBNV_gp06-like, and odv-e56 genes formed a syntenic cluster (supplementary table S2, Supplementary Material online), which corresponded with ORF5 to ORF7 in GbNV. However, this cluster was not observed in other nudiviruses.
The genomes of nudiviruses and other large dsDNA viruses often contain multiple gene families (Thézé et al. 2013). For example, the odv-e56 gene is likely to be duplicated in DiNV and Kallithea virus (Hill and Unckless 2018), whereas ToNV harbors three copies (Bézier et al. 2015). Many baculoviruses possess two or three homologs of ac66, and some have up to 16 bro genes (ac2) (Rohrmann 2008). Two variants of odv-e66 have been identified in several baculoviruses and in PmNV (Yang et al. 2014). This feature was also observed in some of the genomes that were searched in this study (fig. 1). In B. tabaci, two identical copies of ac92 and GrBNV_gp06-like homologs were detected; however, in most cases the paralogs within a genome showed different degrees of divergence. Two copies of the helicase, tk1, and odv-e66 genes were identified in the Pa. marginatus genome, and the identities between paralogs ranged from 36% to 60%. The genome of Pseudomyrmex gracilis also harbors two odv-e66 homologs, but one of these contains a frameshift. These two homologs share the same putative flanking regions (supplementary table S2, Supplementary Material online), indicating a postinsertional duplication. In M. sacchari, more than half of the nudivirus-like genes have two or three copies, which appears to be the result of multiple independent endogenization events because no similarity was observed among their flanking genomic sequences. The phylogenetic analysis of P74 sequences and the absence of homology in the EVE flanking regions between different species also suggested that more than one integration event may have occurred.
The integration of nonretroviral viral sequences can be mediated by nonhomologous recombination with chromosomal DNA (Arbuckle et al. 2010) or by interactions with TE-encoded enzymes (Geuking et al. 2009; Horie et al. 2010). Previous studies on mosquitoes have shown that nonretroviral integrated RNA virus in Aedes aegypti and A. albopictus genomes were predominantly associated with LTR elements (Umberto et al. 2017; Whitfield et al. 2017). In contrast, genomic screening of other arthropod species did not detect the association between EVEs and TEs (Thézé et el. 2014; Metegnier et al. 2015). In this study, inspections of regions flanking the putative EVEs have revealed several TEs including DNA transposons and LTR elements (supplementary tables S4 and S5, Supplementary Material online), but we have not observed significant enrichment of TEs in most host species. It is possible that some EVE-associated TEs were missed by RepeatMasker because of the lack of a fully representative library for the diverse genomes screened here, but de novo detection using LTR_retriever did not reveal additional LTRs in the EVE flanking regions, either. Besides, many EVEs identified were located on the same contigs, indicating the integration of large viral DNA fragments rather than individual RNA transcripts. All these results suggested that mechanisms other than LTR-mediated retrotransposition of viral mRNAs have been involved in nudivirus endogenization.
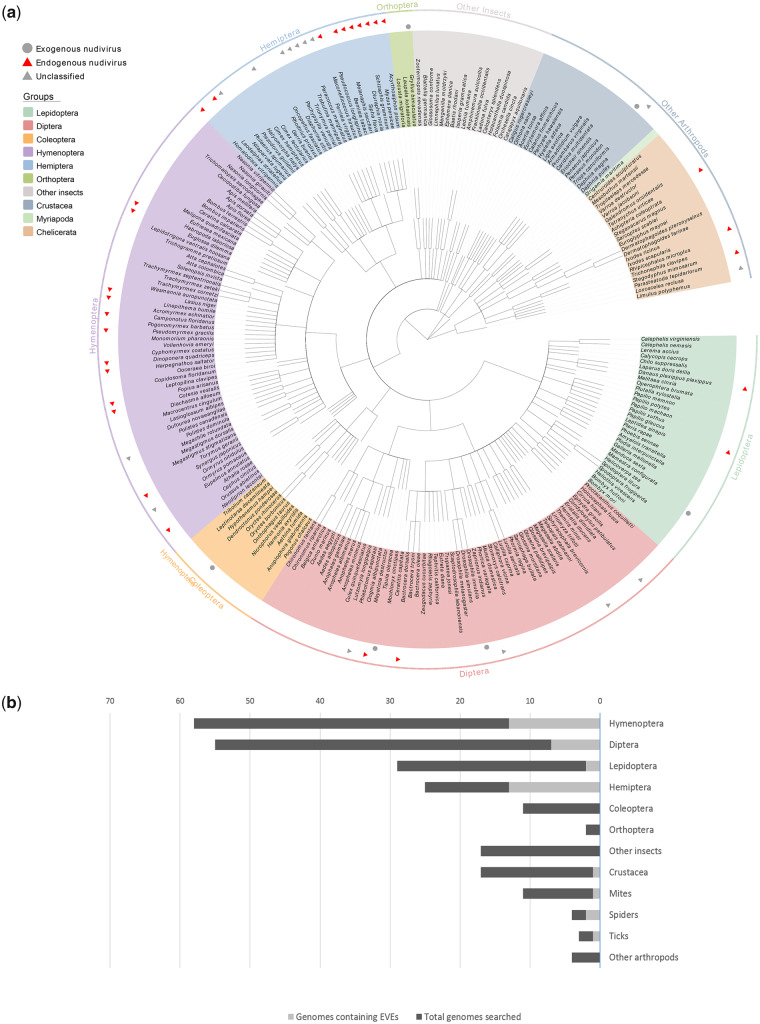
The known hosts of nudiviruses include insects in the orders Coleoptera, Diptera, Lepidoptera, and Orthoptera, and the crustacean order Decapoda. The identification of nudivirus-like genes in other arthropods has greatly expanded the host range of Nudiviridae. These sequences have provided evidence that nudiviruses are (or at least were at one time) capable of infecting insects from Hemiptera and Hymenoptera, as well as other arthropod species (fig. 5a). Specifically, nudivirus-related sequences were frequently found in hemipteran and hymenopteran genomes (fig. 5b), although no exogenous nudiviruses have been reported in these groups. The genome sequence data for four host species of known nudiviruses (H. zea, D. innubila, T. oleracea, and P. monodon) are available; however, no endogenous nudiviral sequences were identified among them. It should be noted that there is a clear subrepresentation of available genomic sequences for Coleoptera and Orthoptera, thus the data presented in figure 5 may not reflect the real distribution of nudivirus hosts.
Fig. 5.
—Phylogenetic diversity of nudivirus hosts. (a) Distribution of exogenous and endogenous nudiviruses in arthropods. A cladogram for the arthropods screened in this study was downloaded from the NCBI Taxonomy database. Red triangles indicate species in which nudivirus-like genes were identified, and gray circles indicate the known hosts of circulating nudiviruses. (b) Number of species from different arthropod groups screened in this study and number of genomes containing putative EVEs derived from nudiviruses.
According to the viral accommodation hypothesis, EVEs from both RNA and DNA viruses in arthropod genomes can randomly generate antisense RNA fragments which target the corresponding viral mRNAs, resulting in host tolerance to viral infection (Flegel 2009). A previous study of honeybees (Apis mellifera) showed that 30% of the tested populations carried EVEs derived from a dicistrovirus and became virus resistant (Maori et al. 2007). In addition, Utari et al. reported EVEs derived from the white spot syndrome virus (WSSV) in shrimp genomes and suggested that RNA expression from the putative EVEs might be associated with an RNAi defense mechanism (Utari et al. 2017). More recently, comprehensive analysis on mammals (Parrish et al. 2015), mosquitoes (Umberto et al. 2017; Whitfield et al. 2017), and other arthropods (ter Horst et al. 2019) has revealed that EVEs can give rise to PIWI-interacting RNAs which may contribute to an antiviral response. In this study, a potential correlation between the natural resistance to current nudiviruses and the presence of putative EVEs was observed (fig. 5a). However, whether these EVEs can generate PIWI-interacting RNAs and play a role in antiviral immunity remains unknown. Further studies incorporating the small RNA data sets of host species will provide a better insight into this issue.
In conclusion, the multitude of nudivirus-related sequences described in this report demonstrates current and past interactions between nudiviruses and a wide range of insects and other arthropods. Nudivirus infections may also have occurred in groups for which there is currently limited or no genome-wide sequencing data. The high degree of phylogenetic diversity among hosts suggests that nudiviruses have ancient origins and complex evolutionary histories. As more genomic data become available, the diversity, phylogeny, and origin of nudiviruses can be further explored.
Supplementary Material
Acknowledgments
This work was funded by the grant of Laboratory for Marine Biology and Biotechnology, Pilot National Laboratory for Marine Science and Technology (Qingdao) (No. OF2019NO05); Scientific Research Foundation of Third Institute of Oceanography, MNR (No. 2018022); and Natural Science Foundation of Fujian Province of China (No. 2019J05149).
Literature Cited
- Arbuckle JH, et al. 2010. The latent human herpesvirus-6A genome specifically integrates in telomeres of human chromosomes in vivo and in vitro. Proc Natl Acad Sci U S A. 107(12):5563–5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W, Kojima KK, Kohany O.. 2015. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob DNA 6:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyi VA, Levine AJ, Anna Marie S.. 2010. Sequences from ancestral single-stranded DNA viruses in vertebrate genomes: the Parvoviridae and Circoviridae are more than 40 to 50 million years old. J Virol. 84(23):12458–12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bézier A, et al. 2009. Polydnaviruses of braconid wasps derive from an ancestral nudivirus. Science 323(5916):926–930. [DOI] [PubMed] [Google Scholar]
- Bézier A, et al. 2015. The genome of the nucleopolyhedrosis-causing virus from Tipula oleracea sheds new light on the Nudiviridae family. J Virol. 89(6):3008–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burand JP, et al. 2012. Analysis of the genome of the sexually transmitted insect virus Helicoverpa zea nudivirus 2. Viruses 4(1):28–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17(4):540–552. [DOI] [PubMed] [Google Scholar]
- Cheng RL, et al. 2014. Brown planthopper nudivirus DNA integrated in its host genome. J Virol. 88(10):5310–5318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crochu S, et al. 2004. Sequences of flavivirus-related RNA viruses persist in DNA form integrated in the genome of Aedes spp. mosquitoes. J Gen Virol. 85(7):1971–1980. [DOI] [PubMed] [Google Scholar]
- Cui J, Holmes EC.. 2012. Endogenous RNA viruses of plants in insect genomes. Virology 427(2):77–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drezen JM, et al. 2017. Endogenous viruses of parasitic wasps: variations on a common theme. Curr Opin Virol. 25:41–48. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C, Gilbert C.. 2012. Endogenous viruses: insights into viral evolution and impact on host biology. Nat Rev Genet. 13(4):283–296. [DOI] [PubMed] [Google Scholar]
- Flegel TW. 2009. Hypothesis for heritable, anti-viral immunity in crustaceans and insects. Biol Direct 4:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuking MB, et al. 2009. Recombination of retrotransposon and exogenous RNA virus results in nonretroviral cDNA integration. Science 323(5912):393–396. [DOI] [PubMed] [Google Scholar]
- Gilbert C, et al. 2014. Endogenous hepadnaviruses, bornaviruses and circoviruses in snakes. Proc Biol Sci. 281(1791):20141122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herniou EA, Olszewski JA, Cory JS, O’Reilly DR.. 2003. The genome sequence and evolution of baculoviruses. Annu Rev Entomol. 48(1):211–234. [DOI] [PubMed] [Google Scholar]
- Hill T, Unckless RL.. 2018. The dynamic evolution of Drosophila innubila Nudivirus. Infect Genet Evol. 57:151–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie M, et al. 2010. Endogenous non-retroviral RNA virus elements in mammalian genomes. Nature 463(7277):84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huger AM, Krieg A.. 1991. Baculoviridae. Nonoccluded baculoviruses In: Atlas of invertebrate viruses. Boca Raton (FL: ): CRC Press; p. 287–319. [Google Scholar]
- Johnson WE. 2010. Endless forms most viral. PLoS Genet. 6(11):e1001210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS.. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzourakis A, Rj G.. 2010. Endogenous viral elements in animal genomes. PLoS Genet. 6(11):e1001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CL, et al. 1999. Persistent Hz-1 virus infection in insect cells: evidence for insertion of viral DNA into host chromosomes and viral infection in a latent status. J Virol. 73(1):128–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, et al. 2010. Widespread horizontal gene transfer from double-stranded RNA viruses to eukaryotic nuclear genomes. J Virol. 84(22):11876–11887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, et al. 2011. Widespread horizontal gene transfer from circular single-stranded DNA viruses to eukaryotic genomes. BMC Evol Biol. 11(1):276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maori E, Tanne E, Sela I.. 2007. Reciprocal sequence exchange between non-retro viruses and hosts leading to the appearance of new host phenotypes. Virology 362(2):342–349. [DOI] [PubMed] [Google Scholar]
- McCarthy CB, Theilmann DA.. 2008. AcMNPV ac143 (odv-e18) is essential for mediating budded virus production and is the 30th baculovirus core gene. Virology 375(1):277–291. [DOI] [PubMed] [Google Scholar]
- Metegnier G, et al. 2015. Comparative paleovirological analysis of crustaceans identifies multiple widespread viral groups. Mob DNA 6:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou SJ, Jiang N.. 2018. LTR_retriever: a highly accurate and sensitive program for identification of long terminal repeat retrotransposons. Plant Physiol. 176(2):1410–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish NF, et al. 2015. piRNAs derived from ancient viral processed pseudogenes as transgenerational sequence-specific immune memory in mammals. RNA 21(10):1691–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichon A, et al. 2015. Recurrent DNA virus domestication leading to different parasite virulence strategies. Sci Adv. 1(10):e1501150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinganum C, O’Loughlin GT, Hogan TW.. 1970. A nonoccluded virus of the field crickets Teleogryllus oceanicus and T. commodus (Orthoptera: Gryllidae). J Invertebr Pathol. 16(2):214–220. [Google Scholar]
- Rohrmann GF. 2008. Baculovirus molecular biology. Bethesda (MD: ): National Center for Biotechnology Information. [PubMed] [Google Scholar]
- Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotheram S. 1967. Immune surface of eggs of a parasitic insect. Nature 214(5089):700. [DOI] [PubMed] [Google Scholar]
- Sperber GO, Airola T, Jern P, Blomberg J.. 2007. Automated recognition of retroviral sequences in genomic data—RetroTector. Nucleic Acids Res. 35(15):4964–4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand MR, Burke GR.. 2012. Polydnaviruses as symbionts and gene delivery systems. PLoS Pathog. 8(7):e1002757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28(10):2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang KF, Lightner DV.. 2006. Infectious hypodermal and hematopoietic necrosis virus (IHHNV)-related sequences in the genome of the black tiger prawn Penaeus monodon from Africa and Australia. Virus Res. 118(1–2):185–191. [DOI] [PubMed] [Google Scholar]
- ter Horst AM, Nigg JC, Dekker FM, Falk BW.. 2019. Endogenous viral elements are widespread in arthropod genomes and commonly give rise to PIWI-Interacting RNAs. J Virol. 93(6):e02124–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thézé J, Bezier A, Periquet G, Drezen J-M, Herniou EA.. 2011. Paleozoic origin of insect large dsDNA viruses. Proc Natl Acad Sci U S A. 108(38):15931–15935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thézé J, Leclercq S, Moumen B, Cordaux R, Gilbert C.. 2014. Remarkable diversity of endogenous viruses in a crustacean genome. Genome Biol Evol. 6(8):2129–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thézé J, et al. 2013. New insights into the evolution of Entomopoxvirinae from the complete genome sequences of four entomopoxviruses infecting Adoxophyes honmai, Choristoneura biennis, Choristoneura rosaceana, and Mythimna separata. J Virol. 87(14):7992–8003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tristem M. 2000. Identification and characterization of novel human endogenous retrovirus families by phylogenetic screening of the human genome mapping project database. J Virol. 74(8):3715–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umberto P, et al. 2017. Comparative genomics shows that viral integrations are abundant and express piRNAs in the arboviral vectors Aedes aegypti and Aedes albopictus. BMC Genomics. 18(1):512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unckless RL. 2011. A DNA virus of Drosophila. PLoS One 6(10):e26564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utari HB, Soowannayan C, Flegel TW, Whityachumnarnkul B, Kruatrachue M.. 2017. Variable RNA expression from recently acquired, endogenous viral elements (EVE) of white spot syndrome virus (WSSV) in shrimp. Dev Comp Immunol. 76:370–379. [DOI] [PubMed] [Google Scholar]
- Wang Y, Bininda-Emonds ORP, Jehle JA.. 2012. Nudivirus genomics and phylogeny In: Garcia M, editor. Viral genomes-molecular structure, diversity, gene expression mechanisms and host–virus interactions. Rijeka, Croatia: InTech; p. 33–52. [Google Scholar]
- Wang Y, Bininda-Emonds ORP, van Oers MM, Vlak JM, Jehle JA.. 2011. The genome of Oryctes rhinoceros nudivirus provides novel insight into the evolution of nuclear arthropod-specific large circular double-stranded DNA viruses. Virus Genes 42(3):444–456. [DOI] [PubMed] [Google Scholar]
- Wang Y, Jehle JA.. 2009. Nudiviruses and other large, double-stranded circular DNA viruses of invertebrates: new insights on an old topic. J Invertebr Pathol. 101(3):187–193. [DOI] [PubMed] [Google Scholar]
- Wang Y-J, Burand JP, Jehle JA.. 2007. Nudivirus genomics: diversity and classification. Virol Sin. 22(2):128–136. [Google Scholar]
- Webster CL, et al. 2015. The discovery, distribution, and evolution of viruses associated with Drosophila melanogaster. PLoS Biol. 13(7):e1002210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler TJ, et al. 2013. Dfam: a database of repetitive DNA based on profile hidden Markov models. Nucleic Acids Res. 41(D1):D70–D82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield ZJ, et al. 2017. The diversity, structure, and function of heritable adaptive immunity sequences in the Aedes aegypti genome. Curr Biol. 27(22):3511–3519.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y-T, et al. 2014. The genome and occlusion bodies of marine Penaeus monodon nudivirus (PmNV, also known as MBV and PemoNPV) suggest that it should be assigned to a new nudivirus genus that is distinct from the terrestrial nudiviruses. BMC Genomics. 15(S11):1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, et al. 2011. Identification of Autographa californica nucleopolyhedrovirus ac93 as a core gene and its requirement for intranuclear microvesicle formation and nuclear egress of nucleocapsids. J Virol. 85(22):11664–11674. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.