Abstract
The increasing availability of scholarly metadata in the form of Knowledge Graphs (KG) offers opportunities for studying the structure of scholarly communication and evolution of science. Such KGs build the foundation for knowledge-driven tasks e.g., link discovery, prediction and entity classification which allow to provide recommendation services. Knowledge graph embedding (KGE) models have been investigated for such knowledge-driven tasks in different application domains. One of the applications of KGE models is to provide link predictions, which can also be viewed as a foundation for recommendation service, e.g. high confidence “co-author” links in a scholarly knowledge graph can be seen as suggested collaborations. In this paper, KGEs are reconciled with a specific loss function (Soft Margin) and examined with respect to their performance for co-authorship link prediction task on scholarly KGs. The results show a significant improvement in the accuracy of the experimented KGE models on the considered scholarly KGs using this specific loss. TransE with Soft Margin (TransE-SM) obtains a score of 79.5% Hits@10 for co-authorship link prediction task while the original TransE obtains 77.2%, on the same task. In terms of accuracy and Hits@10, TransE-SM also outperforms other state-of-the-art embedding models such as ComplEx, ConvE and RotatE in this setting. The predicted co-authorship links have been validated by evaluating profile of scholars.
Keywords: Scholarly knowledge graph, Author recommendation, Knowledge graph embedding, Scholarly communication, Science graph, Metaresearch queries, Link prediction, Research of research
Introduction
With the rapid growth of digital publishing, researchers are increasingly exposed to an incredible amount of scholarly artifacts and their metadata. The complexity of science in its nature is reflected in such heterogeneously interconnected information. Knowledge Graphs (KGs), viewed as a form of information representation in a semantic graph, have proven to be extremely useful in modeling and representing such complex domains [8]. KG technologies provide the backbone for many AI-driven applications which are employed in a number of use cases, e.g. in the scholarly communication domain. Therefore, to facilitate acquisition, integration and utilization of such metadata, Scholarly Knowledge Graphs (SKGs) have gained attention [3, 25] in recent years. Formally, a SKG is a collection of scholarly facts represented in triples including entities and a relation between them, e.g. (Albert Einstein, co-author, Boris Podolsky). Such representation of data has influenced the quality of services which have already been provided across disciplines such as Google Scholar1, Semantic Scholar [10], OpenAIRE [1], AMiner [17], ResearchGate [26]. The ultimate objective of such attempts ranges from service development to measuring research impact and accelerating science. Recommendation services, e.g. finding potential collaboration partners, relevant venues, relevant papers to read or cite are among the most desirable services in research of research enquiries [9, 25]. So far, most of the approaches addressing such services for scholarly domains use semantic similarity and graph clustering techniques [2, 6, 27].
The heterogeneous nature of such metadata and variety of sources plugging metadata to scholarly KGs [14, 18, 22] keeps complex metaresearch enquiries (research of research) challenging to analyse. This influences the quality of the services relying only on the explicitly represented information. Link prediction in KGs, i.e. the task of finding (not explicitly represented) connections between entities, draws on the detection of existing patterns in the KG. A wide range of methods has been introduced for link prediction [13]. The most recent successful methods try to capture the semantic and structural properties of a KG by encoding information as multi-dimensional vectors (embeddings). Such methods are known as knowledge graph embedding (KGE) models in the literature [23]. However, despite the importance of link prediction for the scholarly domains, it has rarely been studied with KGEs [12, 24] for the scholarly domain.
In a preliminary version of this work [11], we tested a set of embedding models (in their original version) on top of a SKG in order to analyse suitability of KGEs for the use case of scholarly domain. The primary insights derived from results have proved the effectiveness of applying KGE models on scholarly knowledge graphs. However, further exploration of the results proved that the many-to-many characteristic of the focused relation, co-authorship, causes restrictions in negative sampling which is a mandatory step in the learning process of KGE models. Negative sampling is used to balance discrimination from the positive samples in KGs. A negative sample is generated by a replacement of either subject or object with a random entity in the KG e.g., (Albert Einstein, co-author, Trump) is a negative sample for (Albert Einstein, co-author, Boris Podolsky). To illustrate the negative sampling problem, consider the following case: Assuming that 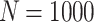 is the number of all authors in a SKG, the probability of generating false negatives for an author with 100 true or sensible but unknown collaborations becomes
is the number of all authors in a SKG, the probability of generating false negatives for an author with 100 true or sensible but unknown collaborations becomes 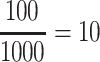 %. This problem is particularly relevant when the in/out-degree of entities in a KG is very high. This is not limited to, but particularly relevant, in scholarly KGs with its network of authors, venues and papers. To tackle this problem, we propose a modified version of the Margin Ranking Loss (MRL) to train the KGE models such as TransE and RotatE. The model is dubbed SM (Soft Margins), which considers margins as soft boundaries in its optimization. Soft margin loss allows false negative samples to move slightly inside the margin, mitigating the adverse effects of false negative samples. Our main contributions are:
%. This problem is particularly relevant when the in/out-degree of entities in a KG is very high. This is not limited to, but particularly relevant, in scholarly KGs with its network of authors, venues and papers. To tackle this problem, we propose a modified version of the Margin Ranking Loss (MRL) to train the KGE models such as TransE and RotatE. The model is dubbed SM (Soft Margins), which considers margins as soft boundaries in its optimization. Soft margin loss allows false negative samples to move slightly inside the margin, mitigating the adverse effects of false negative samples. Our main contributions are:
proposing a novel loss function explicitly designed for KGs with many-to-many relations (present in co-authorship relation of scholarly KGs),
showcasing the effect of the proposed loss function for KGE models,
providing co-authorship recommendations on scholarly KGs,
evaluating the effectiveness of the approach and the recommended links on scholarly KGs with favorable results,
validating the predicted co-authorship links by a profile check of scholars.
The remaining part of this paper proceeds as follows. Section 2 represents details of the scholarly knowledge graph that is created for the purpose of applying link discovery tools. Section 3 provides a summary of preliminaries required about the embedding models and presents some of the focused embedding models of this paper, TransE and RotatE. Moreover, other related works in the domain of knowledge graph embeddings are reviewed in Sect. 3.2. Section 4 contains the given approach and description of the changes to the MRL. An evaluation of the proposed model on the represented scholarly knowledge graph is shown in Sect. 5. In Sect. 6, we lay out the insights and provide a conjunction of this research work.
A Scholarly Knowledge Graph
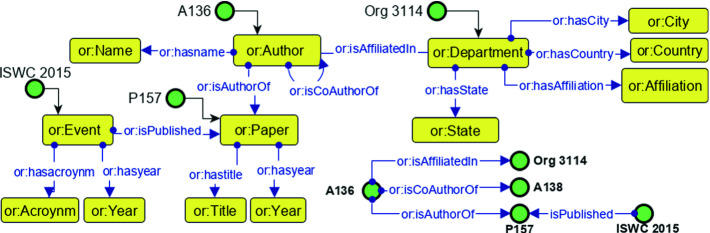
A specific scholarly knowledge graphs has been constructed in order to provide effective recommendations for the selected use case (co-authorship). This knowledge graph is created after a systematic analysis of the scholarly metadata resources on the Web (mostly RDF data). The list of resources includes DBLP2, Springer Nature SciGraph Explorer3, Semantic Scholar4 and the Global Research Identifier Database (GRID)5 with metadata about institutes. A preliminary version of this KG has been used for experiments of the previous work [11] where suitability of embedding models have been tested of such use cases. Through this research work we will point to this KG as SKGOLD. Towards this objective, a domain conceptualization has been done to define the classes and relations of focus. Figure 1 shows the ontology that is used for the creation of these knowledge graphs. In order to define the terms, the OpenResearch [20] ontology is reused.
Fig. 1.
Ontology of a scholarly knowledge graph for experimenting embedding models in co-authorship link prediction.
Each instance in the scholarly knowledge graph is equipped with a unique ID to enable the identification and association of the KG elements. The knowledge graphs consist of the following core entities of Papers, Events, Authors, and Departments.
In the creation of the our KG6 which will be denoted as SKGNEW a set of 7 conference series have been selected (namely ISWC, ESWC, AAAI, NeurIPS, CIKM, ACI, KCAP and HCAI have been considered in the initial step of retrieving raw metadata from the source). In addition, the metadata flitted for the temporal interval of 2013–2018. The second version of the same KG has been generated directly from Semantic Scholar.
The datasets, used for model training, which in total comprise 70,682 triples where 29,469 triples are coming from the SKGOLD and 41,213 triples are generated in SKGNEW. In each set of experiments, both datasets are split into triples of training/validation/test sets. Table 1 includes the detailed statistics about the datasets only considering three relationships between entities namely hasAuthor (paper - author), hasCoauthor (author - author), hasVenue (author/paper - venue). Due to the low volume of data, isAffiliated (author - organization) relationship is eliminated due in SKGNEW.
Table 1.
Dataset statistics. The number of triples that are used in different datasets are shown per each entity and relationship.
| Dataset | Entities | Relations | ||||
|---|---|---|---|---|---|---|
| Author | Publication | Venue | hasAuthor | hasCoauthor | hasVenue | |
| SKGOLD | 4,660 | 2,870 | 7 | 9,934 | 12,921 | 6,614 |
| SKGNEW | 12,472 | 5,001 | 42 | 14,933 | 21,279 | 5,001 |
Preliminaries and Related Work
In this section we focus on providing required preliminaries for this work as well as the related work. The definitions required to understand our approach are:
Knowledge Graph. Let
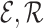 be the sets of entities and relations respectively. A Kg is roughly represented as a set
be the sets of entities and relations respectively. A Kg is roughly represented as a set 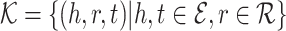 in which h, t, r refer to the subject and object and relation respectively.
in which h, t, r refer to the subject and object and relation respectively.Embedding Vectors. The vector representation of symbolic entities and relations in a KG are considered as embeddings. The vectors of a triple h, r, t are depicted as
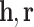 ,
, 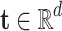 , where d refers to the dimension of the embedding space.
, where d refers to the dimension of the embedding space.Score Function. Each KGE model defines an score function
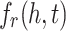 . The score function gets the embedding vectors of a triple (h, r, t) and returns a value determining if the triple is a fact or not. A lower value for the score function indicates that the triple is more plausible comparing to those triples with higher values.
. The score function gets the embedding vectors of a triple (h, r, t) and returns a value determining if the triple is a fact or not. A lower value for the score function indicates that the triple is more plausible comparing to those triples with higher values.Loss Function. Each KGE model utilizes a loss function to adjust embedding. In the beginning of the learning process, the model initializes the embedding vectors randomly. Then it updates the vectors by optimizing a loss function
 . Since typically many variables should be adjusted in the learning process, Stochastic Gradient Descent (SGD) method is commonly used for the optimization of the loss function.
. Since typically many variables should be adjusted in the learning process, Stochastic Gradient Descent (SGD) method is commonly used for the optimization of the loss function.Negative Sampling. KGs contain only positive samples. Most of KGE models generate artificial negative samples to have a better discrimination from positive ones. Uniform negative sampling (unif) is the most widely used negative sampling technique in which a negative sample is generated for a triple (h, r, t) by replacement of either h or t with a random entity (
 or
or 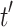 ) existing in
) existing in 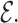
Review of TransE and RotatE Models
The proposed loss is trained on a classical translation-based embedding models named TransE and a model for complex space as RotatE. Therefore, we mainly provide a description of TransE and RotatE and further focus on other state-of-the-art models.
TransE. It is reported that TransE [4], as one of the simplest translation based models, outperformed more complicated KGEs in [11].
The initial idea of TransE model is to enforce embedding of entities and relation in a positive triple (h, r, t) to satisfy the following equality:
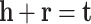 |
1 |
where h, r and t are embedding vectors of head, relation and tail respectively. TransE model defines the following scoring function:
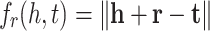 |
2 |
RotatE. Here, we address RotatE [16] which is a model designed to rotate the head to the tail entity by using relation. This model embeds entities and relations in Complex space. By inclusion of constraints on the norm of entity vectors, the model would be degenerated to TransE. The scoring function of RotatE is
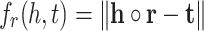 |
in which  is the element-wise product.
is the element-wise product.
Loss Function. Margin ranking loss (MRL) is one of the most used loss functions which optimizes the embedding vectors of entities and relations. MRL computes embedding of entities and relations in a way that a positive triple gets lower score value than its corresponding negative triple. The least difference value between the score of positive and negative samples is margin (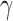 ). The MRL is defined as follows:
). The MRL is defined as follows:
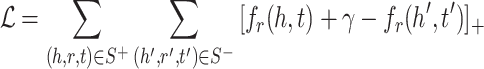 |
3 |
where 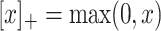 and
and 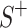 and
and 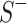 are respectively the set of positive and negative samples.
are respectively the set of positive and negative samples.
MRL has two disadvantages: 1) the margin can slide, 2) embeddings are adversely affected by false negative samples. More precisely, the issue of margin sliding is described with an example. Assume that 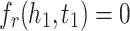 and
and 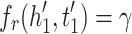 , or
, or 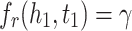 and
and 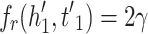 are two possible scores for a triple and its negative sample. Both of these scores get minimum value for the optimization causing the model to become vulnerable to a undesirable solution. To tackle this problem, Limited-based score [28] revises the MRL by adding a term to limit maximum value of positive score:
are two possible scores for a triple and its negative sample. Both of these scores get minimum value for the optimization causing the model to become vulnerable to a undesirable solution. To tackle this problem, Limited-based score [28] revises the MRL by adding a term to limit maximum value of positive score:
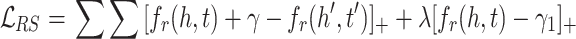 |
4 |
It shows 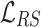 significantly improves the performance of TransE. Authors in [28] denote TransE which is trained by
significantly improves the performance of TransE. Authors in [28] denote TransE which is trained by 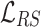 as TransE-RS. Regarding the second disadvantage, MRL enforces a hard margin in the side of negative samples. However, using relations with many-to-many characteristic (e.g., co-author), the rate of false negative samples is high. Therefore, using a hard boundary for discrimination adversely affects the performance of a KGE model.
as TransE-RS. Regarding the second disadvantage, MRL enforces a hard margin in the side of negative samples. However, using relations with many-to-many characteristic (e.g., co-author), the rate of false negative samples is high. Therefore, using a hard boundary for discrimination adversely affects the performance of a KGE model.
Review of Other State-of-the-Art Models
With a systematic evaluation (performance under reasonable set up) of suitable embedding models to be considered in our evaluations, we have selected two other models that are described here.
ComplEx. One of the embedding models focusing on semantic matching model is ComplEx [19]. In semantic matching models, the plausibility of facts are measured by matching the similarity of their latent representation, in other words it is assumed that similar entities have common characteristics i.e. are connected through similar relationships [13, 23]. In ComplEx the entities are embedded in the complex space. The score function of ComplEx is given as follows:
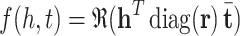 |
in which 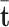 is the conjugate of the vector
is the conjugate of the vector 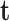 .
.
ConvE. Here we present a multi-layer convolutional network model for link prediction named as ConvE. The score function of the ConvE is defined as below:
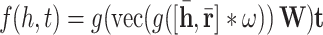 |
in which g denotes a non-linear function, 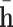 and
and 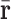 are 2D reshape of head and relation vectors respectively,
are 2D reshape of head and relation vectors respectively, 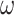 is a filter and
is a filter and 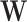 is a linear transformation matrix. The core idea behind the ConvE model is to use 2D convolutions over embeddings to predict links. ConvE consists of a single convolution layer, a projection layer to the embedding dimension as well as an inner product layer.
is a linear transformation matrix. The core idea behind the ConvE model is to use 2D convolutions over embeddings to predict links. ConvE consists of a single convolution layer, a projection layer to the embedding dimension as well as an inner product layer.
Soft Marginal Loss
This section proposes a new model independent optimization framework for training KGE models. The framework fixes the second problem of MRL and its extension mentioned in the previous section. The optimization utilizes slack variables to mitigate the negative effect of the generated false negative samples. In contrast to margin ranking loss, our optimization uses soft margin. Therefore, uncertain negative samples are allowed to slide inside of margin.
Figure 2 visualizes the separation of positive and negative samples using margin ranking loss and our optimization problem. It shows that the proposed optimization problem allows false negative samples to slide inside the margin by using slack variables (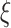 ). In contrast, margin ranking loss doesn’t allow false negative samples to slide inside of the margin. Therefore, embedding vectors of entities and relations are adversely affected by false negative samples. The mathematical formulation of our optimization problem is as follows:
). In contrast, margin ranking loss doesn’t allow false negative samples to slide inside of the margin. Therefore, embedding vectors of entities and relations are adversely affected by false negative samples. The mathematical formulation of our optimization problem is as follows:
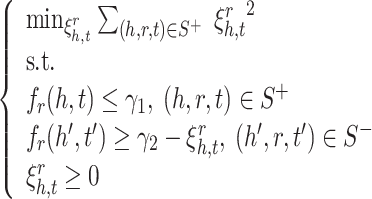 |
5 |
where 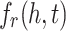 is the score function of a KGE model (e.g., TransE or RotatE),
is the score function of a KGE model (e.g., TransE or RotatE), 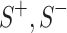 are positive and negative samples sets.
are positive and negative samples sets. 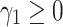 is the upper bound of score of positive samples and
is the upper bound of score of positive samples and 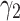 is the lower bound of negative samples.
is the lower bound of negative samples.  is margin (
is margin (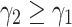 ).
). 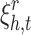 is slack variable for a negative sample that allows it to slide in the margin.
is slack variable for a negative sample that allows it to slide in the margin. 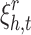 helps the optimization to better handle uncertainty resulted from negative sampling.
helps the optimization to better handle uncertainty resulted from negative sampling.
Fig. 2.
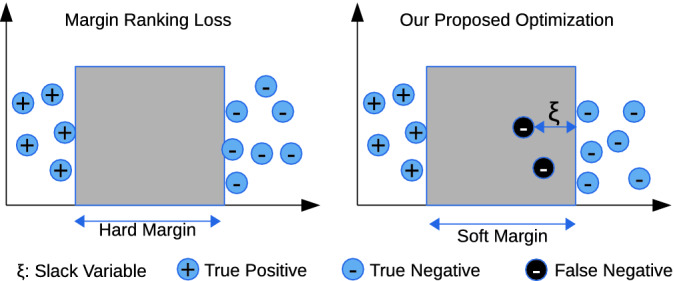
Optimization of margin ranking loss.
The term (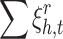 ) represented in the problem 5 is quadratic. Therefore, it is convex which results in a unique and optimal solution. Moreover, all three constraints can be represented as convex sets. The constrained optimization problem (5) is convex. As a conclusion, it has a unique optimal solution. The optimal solution can be obtained by using different standard methods e.g. penalty method [5]. The goal of the problem (5) is to adjust embedding vectors of entities and relations. A lot of variables participate in optimization. In this condition, using batch learning with stochastic gradient descent (SGD) is preferred. In order to use SGD, constrained optimization problem (5) should be converted to unconstrained optimization problem. The following unconstrained optimization problem is proposed instead of (5).
) represented in the problem 5 is quadratic. Therefore, it is convex which results in a unique and optimal solution. Moreover, all three constraints can be represented as convex sets. The constrained optimization problem (5) is convex. As a conclusion, it has a unique optimal solution. The optimal solution can be obtained by using different standard methods e.g. penalty method [5]. The goal of the problem (5) is to adjust embedding vectors of entities and relations. A lot of variables participate in optimization. In this condition, using batch learning with stochastic gradient descent (SGD) is preferred. In order to use SGD, constrained optimization problem (5) should be converted to unconstrained optimization problem. The following unconstrained optimization problem is proposed instead of (5).
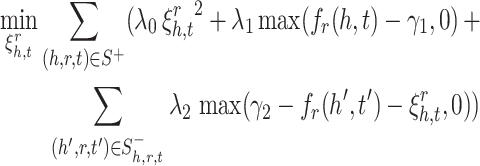 |
6 |
The problem (5) and (6) may not have the same solution. However, we experimentally see that if 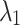 and
and 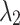 are properly selected, the results would be improved comparing to margin ranking loss.
are properly selected, the results would be improved comparing to margin ranking loss.
Evaluation
This section presents the evaluations of TransE-SM and RotatE-SM (TransE and RotatE trained by SM loss), over a scholarly knowledge graph. The evaluations are motivated for a link prediction task in the domain of scholarly communication in order to explore the ability of embedding models in support of metaresearch enquiries. In addition, we provide a comparison of our model with other state-of-the-art embedding models (selected by performance under a reasonable set up) on two standard benchmarks (FreeBase and WordNet). Four different evaluation methods have been performed in order to approve: 1) better performance and effect of the proposed loss, 2) quality and soundness of the results, 3) validity of the discovered co-authorship links and 4) sensitivity of the proposed model to the selected hyperparameters. More details about each of these analyses are discussed in the remaining part of this section.
Performance Analysis
The proposed loss is model independent, however, we prove its functionality and effectiveness by applying it on different embedding models. In the first evaluation method, we run experiments and assess performance of TransE-SM model as well as RotatE-SM in comparison to the other models and the original loss functions. In order to discuss this evaluation further, let (h, r, t) be a triple fact with an assumption that either head or tail entity is missing (e.g., (?, r, t) or (h, r, ?)). The task is to aim at completing either of these triples (h, r, ?) or (?, r, t) by predicting head (h) or tail (t) entity. Mean Rank (MR), Mean Reciprocal Rank (MRR) [23] and Hits@10 have been extensively used as standard metrics for evaluation of KGE models on link prediction.
In computation of Mean Rank, a set of pre-processing steps have been done such as:
head and tail of each test triple are replaced by all entities in the dataset,
scores of the generated triples are computed and sorted,
the average rank of correct test triples is reported as MR.
Let rank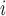 refers to the rank of the
refers to the rank of the 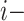 th triple in the test set obtained by a KGE model. The MRR is obtained as follows:
th triple in the test set obtained by a KGE model. The MRR is obtained as follows:
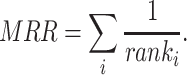 |
The computation of Hits@10 is obtained by replacing all entities in the dataset in terms of head and tail of each test triples. The result is a sorted list of triples based on their scores. The average number of triples that are ranked at most 10 is reported as Hits@10 as represented in Table 2.
Table 2.
Link prediction results. Results of TransE (reported from [11]), TransRS, and our proposed model (TransE-SM) are obtained. The others are obtained from original code. Dashes: results could not be obtained. The underlined values show the best competitor model and the bold results refer to the cases where our model outperforms other competitors.
| SKGOLD – Filtered | SKGNEW – Filtered | |||||
|---|---|---|---|---|---|---|
| FMR | FHits@10 | FMRR | FMR | FHits@10 | FMRR | |
| TransE [4] | 647 | 50.7 | – | 1150 | 77.2 | – |
| ComplEx [19] | – | 56.2 | 0.326 | – | 73.9 | 0.499 |
| ConvE [7] | 1215 | 49.3 | 0.282 | 1893 | 71.3 | 0.442 |
| RotatE [15] | 993 | 60.6 | 0.346 | 1780 | 69.5 | 0.486 |
| TransE-RS [28] | – | – | – | 762 | 75.8 | 0.443 |
| TransE-SM (our work) | 910 | 61.4 | 0.347 | 550 | 79.5 | 0.430 |
| RotatE-SM (our work) | 990 | 60.9 | 0.347 | 1713 | 76.7 | 0.522 |
Experimental Setup. A Python-based computing package called PyTorch7 has been used for the implementation of TransE-SM and RotatE-SM8. Adagrad was selected as an optimizer. The whole training set is reshuffled in each epoch. Then 100 mini-batches are generated on the reshuffled samples. Batches are taken sequentially and the parameters of the model are optimized on the selected batches in each iteration. The parameters 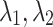 are set to one for simplicity of our experiments. Sub-optimal embedding dimension (d) is selected among the values in
are set to one for simplicity of our experiments. Sub-optimal embedding dimension (d) is selected among the values in 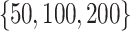 . Upper bound of positive samples (
. Upper bound of positive samples (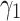 ) and lower bound of negative samples (
) and lower bound of negative samples (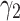 ) are selected from the sets
) are selected from the sets 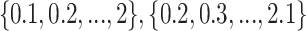 respectively. It should be noted that
respectively. It should be noted that 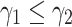 . The regularization term (
. The regularization term (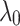 ) is adjusted among the set
) is adjusted among the set 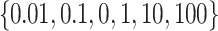 . For each positive sample in a batch, we generate a set of
. For each positive sample in a batch, we generate a set of 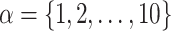 negative samples.
negative samples.
Both for TransE-SM and RotatE-SM, the optimal configurations are 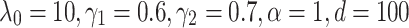 for SKGOLD and
for SKGOLD and 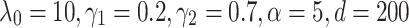 for SKGNEW. The results of TransE and TransE-RS are obtained by our implementation. The results corresponding to ConvE, ComplEx are obtained by running their codes.
for SKGNEW. The results of TransE and TransE-RS are obtained by our implementation. The results corresponding to ConvE, ComplEx are obtained by running their codes.
The results mentioned in the Table 2 validate that TransE-SM and RotatE-SM significantly outperformed other embedding models in all metrics.
In addition, evaluation of the state-of-the-art models have been performed over the two benchmark datasets namely FB15K and WN18. While our focus has been resolving problem of KGEs in presence of many-to-many relationships, the evaluations of the proposed loss function (SM) on other datasets show the effectiveness of SM in addressing other types of relationships.
Table 3 shows the results of experiments for TransE, ComplEx, ConvE, RotatE, TransE-RS, TransE-SM and RotatE-SM. The proposed model significantly outperforms the other models with an accuracy of 87.2% on FB15K. The evaluations on WN18 shows that RotatE-SM outperforms other evaluated models. The optimal settings for our proposed model corresponding to this part of the evaluation are 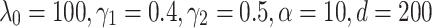 for FB15K and
for FB15K and 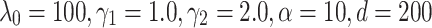 for WN18.
for WN18.
Table 3.
Experimental results for FB15K and WN18. Results of TransE-RS and TransE-SM are based on our code. For RotatE we ran the code of authors. Results of other models are taken from the original papers.
| FB15k | WN18 | |||||
|---|---|---|---|---|---|---|
| FMR | FMRR | FHits@10 | FMR | FMRR | FHits@10 | |
| TransE [4] | 125 | – | 47.1 | 251 | – | 89.2 |
| ComplEx [19] | 106 | 67.5 | 82.6 | 543 | 94.1 | 94.7 |
| ConvE [7] | 51 | 68.9 | 85.1 | 504 | 94.2 | 95.5 |
| RotatE [15] | 49 | 68.8 | 85.9 | 388 | 94.6 | 95.5 |
| TransE-RS [28] | 38 | 57.2 | 82.8 | 189 | 47.9 | 95.1 |
| TransE-SM | 46 | 64.8 | 87.2 | 201 | 47.8 | 95.2 |
| RotatE-SM | 40 | 70.4 | 87.2 | 213 | 94.7 | 96.1 |
Quality and Soundness Analysis
With the second evaluation method, we aim at approving quality and soundness of the results. In order to do so, we additionally investigate the quality of the recommendation of our model. A sample set of 9 researchers associated with the Linked Data and Information Retrieval communities [21] are selected as the foundation for the experiments of the predicted recommendations. Table 4 shows the number of recommendations and their ranks among the top 50 predictions for all of the 9 selected researchers. These top 50 predictions are filtered for a closer look. The results are validated by checking the research profile of the recommended researchers and the track history of co-authorship. In the profile check, we only kept the triples which are indicating:
close match in research domain interests of scholars by checking profiles,
none-existing scholarly relation (e.g., supervisor, student),
none-existing affiliation in the same organization,
none-existing co-authorship.
Table 4.
Co-authorship recommendations. The rank links of discovered potential co-authorship for 9 sample researchers.
| Author | #Recom. | Rank of Recom. |
|---|---|---|
| A136 | 10 | 23, 26, 31, 32, 34, 35, 37, 38, 47, 49 |
| A88 | 4 | 2, 19, 30, 50 |
| A816 | 10 | 3, 7, 8, 9, 12, 13, 15, 44, 48 |
| A1437 | 1 | 21 |
| A138 | 6 | 5, 27, 28, 29, 36, 40 |
| A128 | 1 | 24 |
| A295 | 7 | 1, 11, 14, 18, 22, 39, 41 |
| A940 | 3 | 1, 16, 17 |
| A976 | 8 | 6, 20, 25, 33, 42, 43, 45, 46 |
For example, out of all the recommendations that our approach has provided for researcher with id A136, 10 of them have been identified sound and new collaboration target. The rank of each recommended connection is shown in the third column.
Validity Analysis
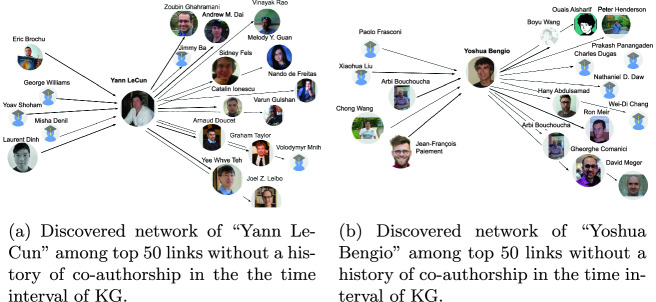
Furthermore, the discovered links for co-authorship recommendations have been examined with a closer look to the online scientific profile of two top machine learning researchers, Yoshua Bengio9, A860 and Yann LeCun10, A2261. The recommended triples have been created in two patterns of (A860, r, ?) and (?, r, A860) and deduplicated for the same answer. The triples are ranked based on scores obtained from TransE-SM and RotatE-SM. For evaluations, a list of top 50 recommendations has been selected per considered researcher, Bengio and LeCun. In order to validate the profile similarity in research and approval of not existing earlier co-authorship, we analyzed the profile of each recommended author to “Yoshua Bengio” and “Yann LeCun” as well as their own profiles. We analyzed the scientific profiles of the selected researchers provided by the most used scholarly search engine, Google Citation11. Due to author name-ambiguity problem, this validation task required human involvement. First, the research areas indicated in the profiles of researchers have been validated to be similar by finding matches. In the next step, some of the highlighted publications with high citations and their recency have been controlled to make sure that the profiles of the selected researchers match in the machine learning community close to the interest of “Yoshua Bengio” – to make sure the researchers can be considered in the same community. As mentioned before, the knowledge graphs that are used for evaluations consist of metadata from 2013 till 2018. In checking the suggested recommendations, a co-authorship relation which has happened before or after this temporal interval is considered valid for the recommendation. Therefore, the other highly ranked links with none-existed co-authorship are counted as valid recommendations for collaboration. Figure 4b shows a visualization of such links found by analyzing top 50 recommendations to and from “Yoshua Bengio” and Fig. 4a shows the same for “Yann LeCun”.
Fig. 4.
Example of co-authorship recommendations.
Out of the 50 discovered triples for “Yoshua Bengio” being head, 12 of them have been approved to be a valid recommendation (relevant but never happened before) and 8 triples have been showing an already existing co-authorship. Profiles of 5 other researchers have not been made available by Google Citation. Among the triples with “Yoshua Bengio” considered in the tail, 8 of triples have been already discovered by the previous pattern. Profile of 5 researchers were not available and 7 researchers have been in contact and co-authorship with “Yoshua Bengio”. Finally, 5 new profiles have been added as recommendations.
Out of 50 triples (YannLeCun, r, ?), 14 recommendations have been discovered as new collaboration cases for “Yann LeCun”. In analyzing the triples with a pattern of the fixed tail (?, r, YannLeCun), there have been cases either without profiles on Google Citations or have had an already existing co-authorship. By excluding these examples as well as the already discovered ones from the other triple pattern, 5 new researchers have remained as valid recommendations.
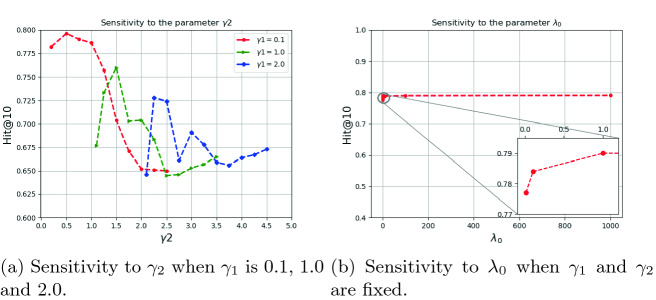
Sensitivity Analysis
In this part we investigate the sensitivity of our model to the hyperparameters (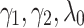 ). To analyze sensitivity of the model to the parameters
). To analyze sensitivity of the model to the parameters 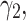 we fix
we fix 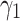 to 0.1, 1 and 2. Moreover,
to 0.1, 1 and 2. Moreover, 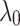 is also fixed to one. Then different values for
is also fixed to one. Then different values for 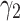 are tested and visualized. Regarding the red dotted line in Fig. 3a, the parameter
are tested and visualized. Regarding the red dotted line in Fig. 3a, the parameter 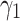 is set to 0.1 and
is set to 0.1 and 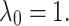 It is shown that by changing
It is shown that by changing 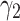 from 0.2 to 3, the performance increases to reach the peak and then decreases by around 15%. Therefore, the model is sensitive to
from 0.2 to 3, the performance increases to reach the peak and then decreases by around 15%. Therefore, the model is sensitive to 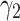 . The significant waving of results can be seen when
. The significant waving of results can be seen when 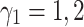 as well (see Fig. 3a). Therefore, proper selection of
as well (see Fig. 3a). Therefore, proper selection of 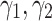 is important in our model.
is important in our model.
Fig. 3.
Sensitivity analysis of TransE-EM to the parameter 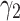 (with fixed values of
(with fixed values of 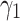 ) and
) and 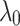 .
.
We also analyze the sensitivity of the performance of our model on the parameter 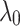 . To do so, we take the optimal configuration of our model corresponding to the fixed
. To do so, we take the optimal configuration of our model corresponding to the fixed 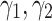 . Then the performance of our model is investigated in different setting where the
. Then the performance of our model is investigated in different setting where the 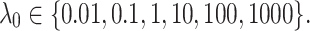 According to Fig. 3b, the model is less sensitive to the parameter
According to Fig. 3b, the model is less sensitive to the parameter 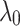 . Therefore, to obtain hyper parameters of the model, it is recommended that first (
. Therefore, to obtain hyper parameters of the model, it is recommended that first (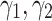 ) are adjusted by validation when
) are adjusted by validation when 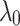 is fixed to a value (e.g., 1). Then the parameter
is fixed to a value (e.g., 1). Then the parameter 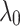 is adjusted while (
is adjusted while (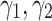 ) are fixed.
) are fixed.
Conclusion and Future Work
The aim of the present research was to develop a novel loss function for embedding models used on KGs with a lot of many-to-many relationships. Our use case is scholarly knowledge graphs with the objective of providing predicted links as recommendations. We train the proposed loss on embedding model and examine it for graph completion of a real-world knowledge graph in the example of scholarly domain. This study has identified a successful application of a model free loss function namely SM. The results show the robustness of our model using SM loss function to deal with uncertainty in negative samples. This reduces the negative effects of false negative samples on the computation of embeddings. We could show that the performance of the embedding model on the knowledge graph completion task for scholarly domain could be significantly improved when applied on a scholarly knowledge graph. The focus has been to discover (possible but never happened) co-author links between researchers indicating a potential for close scientific collaboration. The identified links have been proposed as collaboration recommendations and validated by looking into the profile of a list of selected researchers from the semantic web and machine learning communities. As future work, we plan to apply the model on a broader scholarly knowledge graph and consider other different types of links for recommendations e.g, recommend events for researchers, recommend publications to be read or cited.
Acknowledgement
This work is supported by the EPSRC grant EP/M025268/1, the WWTF grant VRG18-013, the EC Horizon 2020 grant LAMBDA (GA no. 809965), the CLEOPATRA project (GA no. 812997), and the German national funded BmBF project MLwin.
Footnotes
The datasets created for SKGs are available here: https://github.com/SmartDataAnalytics/OpenResearch.
The code for Soft margin loss is available here: https://github.com/mojtabanayyeri/Soft-Margin-Loss.
Contributor Information
Andreas Harth, Email: andreas.harth@fau.de.
Sabrina Kirrane, Email: sabrina.kirrane@wu.ac.at.
Axel-Cyrille Ngonga Ngomo, Email: axel.ngonga@upb.de.
Heiko Paulheim, Email: heiko@informatik.uni-mannheim.de.
Anisa Rula, Email: anisa.rula@unimib.it.
Anna Lisa Gentile, Email: annalisa.gentile@ibm.com.
Peter Haase, Email: ph@metaphacts.com.
Michael Cochez, Email: m.cochez@vu.nl.
Mojtaba Nayyeri, Email: nayyeri@cs.uni-bonn.de.
Sahar Vahdati, Email: sahar.vahdati@cs.ox.ac.uk.
Xiaotian Zhou, Email: 6xizhou@uni-bonn.de.
Hamed Shariat Yazdi, Email: shariatyazdi@gmail.com.
Jens Lehmann, Email: jens.lehmann@cs.uni-bonn.de, Email: jens.lehmann@iais.fraunhofer.de.
References
- 1.Alexiou G, Vahdati S, Lange C, Papastefanatos G, Lohmann S. OpenAIRE LOD services: scholarly communication data as linked data. In: González-Beltrán A, Osborne F, Peroni S, editors. Semantics, Analytics, Visualization. Enhancing Scholarly Data; Cham: Springer; 2016. pp. 45–50. [Google Scholar]
- 2.Ammar, W., et al.: Construction of the literature graph in semantic scholar. arXiv preprint arXiv:1805.02262 (2018)
- 3.Auer, S., Kovtun, V., Prinz, M., Kasprzik, A., Stocker, M., Vidal, M.E.: Towards a knowledge graph for science. In: Proceedings of the 8th International Conference on Web Intelligence, Mining and Semantics, p. 1. ACM (2018)
- 4.Bordes, A., Usunier, N., Garcia-Duran, A., Weston, J., Yakhnenko, O.: Translating embeddings for modeling multi-relational data. In: Advances in NIPS (2013)
- 5.Boyd S, Vandenberghe L. Convex Optimization. Cambridge: Cambridge University Press; 2004. [Google Scholar]
- 6.Cai X, Han J, Li W, Zhang R, Pan S, Yang L. A three-layered mutually reinforced model for personalized citation recommendation. IEEE Trans. Neural Netw. Learn. Syst. 2018;99:1–12. doi: 10.1109/TNNLS.2018.2817245. [DOI] [PubMed] [Google Scholar]
- 7.Dettmers, T., Minervini, P., Stenetorp, P., Riedel, S.: Convolutional 2D knowledge graph embeddings. In: AAAI (2018)
- 8.Färber M, Ell B, Menne C, Rettinger A. A comparative survey of DBpedia, Freebase, OpenCyc, Wikidata, and YAGO. Semant. Web J. 2015;1(1):1–5. [Google Scholar]
- 9.Fortunato S, et al. Science of science. Science. 2018;359(6379):eaao0185. doi: 10.1126/science.aao0185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fricke S. Semantic scholar. J. Med. Libr. Assoc. JMLA. 2018;106(1):145. [Google Scholar]
- 11.Henk, V., Vahdati, S., Nayyeri, M., Ali, M., Yazdi, H.S., Lehmann, J.: Metaresearch recommendations using knowledge graph embeddings. In: RecNLP Workshop of AAAI Conference (2019)
- 12.Mai, G., Janowicz, K., Yan, B.: Combining text embedding and knowledge graph embedding techniques for academic search engines. In: Semdeep/NLIWoD@ ISWC, pp. 77–88 (2018)
- 13.Nickel M, Murphy K, Tresp V, Gabrilovich E. A review of relational machine learning for knowledge graphs. Proc. IEEE. 2016;104(1):11–33. [Google Scholar]
- 14.Schirrwagen J, Manghi P, Manola N, Bolikowski L, Rettberg N, Schmidt B. Data curation in the openaire scholarly communication infrastructure. Inf. Stand. Q. 2013;25(3):13–19. [Google Scholar]
- 15.Sun, Z., Deng, Z.H., Nie, J.Y., Tang, J.: Factorizing YAGO: scalable machine learning for linked data. In: ICLR, pp. 271–280 (2019)
- 16.Sun, Z., Deng, Z.H., Nie, J.Y., Tang, J.: Rotate: knowledge graph embedding by relational rotation in complex space. In: International Conference on Learning Representations (2019). https://openreview.net/forum?id=HkgEQnRqYQ
- 17.Tang, J., Zhang, J., Yao, L., Li, J., Zhang, L., Su, Z.: ArnetMiner: extraction and mining of academic social networks. In: Proceedings of the 14th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, pp. 990–998. ACM (2008)
- 18.Tharani K. Linked data in libraries: a case study of harvesting and sharing bibliographic metadata with BIBFRAME. Inf. Technol. Libr. 2015;34(1):5–19. [Google Scholar]
- 19.Trouillon, T., Welbl, J., Riedel, S., Gaussier, É., Bouchard, G.: Complex embeddings for simple link prediction. In: International Conference on Machine Learning, pp. 2071–2080 (2016)
- 20.Vahdati S, Arndt N, Auer S, Lange C. OpenResearch: collaborative management of scholarly communication metadata. In: Blomqvist E, Ciancarini P, Poggi F, Vitali F, editors. Knowledge Engineering and Knowledge Management; Cham: Springer; 2016. pp. 778–793. [Google Scholar]
- 21.Vahdati S, Palma G, Nath RJ, Lange C, Auer S, Vidal M-E. Unveiling scholarly communities over knowledge graphs. In: Méndez E, Crestani F, Ribeiro C, David G, Lopes JC, editors. Digital Libraries for Open Knowledge; Cham: Springer; 2018. pp. 103–115. [Google Scholar]
- 22.Wan H, Zhang Y, Zhang J, Tang J. AMiner: search and mining of academic social networks. Data Intell. 2019;1(1):58–76. [Google Scholar]
- 23.Wang Q, Mao Z, Wang B, Guo L. Knowledge graph embedding: a survey of approaches and applications. IEEE TKDE. 2017;29(12):2724–2743. [Google Scholar]
- 24.Wang, R., et al.: AceKG: a large-scale knowledge graph for academic data mining. ACM (2018)
- 25.Xia F, Wang W, Bekele TM, Liu H. Big scholarly data: a survey. IEEE Trans. Big Data. 2017;3(1):18–35. [Google Scholar]
- 26.Yu MC, Wu YCJ, Alhalabi W, Kao HY, Wu WH. ResearchGate: an effective altmetric indicator for active researchers? Comput. Hum. Behav. 2016;55:1001–1006. [Google Scholar]
- 27.Yu S, et al. PAVE: personalized academic venue recommendation exploiting co-publication networks. J. Netw. Comput. Appl. 2018;104:38–47. [Google Scholar]
- 28.Zhou, X., Zhu, Q., Liu, P., Guo, L.: Learning knowledge embeddings by combining limit-based scoring loss. In: Proceedings of the 2017 ACM on Conference on Information and Knowledge Management, pp. 1009–1018. ACM (2017)