Abstract
Allosteric behavior is central to the function of many proteins, enabling molecular machinery, metabolism, signaling and regulation. Recent years have shown that the intrinsic dynamics of allosteric proteins defined by their 3-dimensional architecture or by the topology of inter-residue contacts favors cooperative motions that bear close similarity to structural changes they undergo during their allosteric actions. These conformational motions are usually driven by energetically favorable or soft modes at the low frequency end of the mode spectrum, and they are evolutionarily conserved among orthologs. These observations brought into light evolutionary adaptation mechanisms that help maintain, optimize or regulate allosteric behavior as the evolution from bacterial to higher organisms introduces sequential heterogeneities and structural complexities.
Keywords: intrinsic dynamics, allosteric response, evolutionary adaptation, chaperonins, normal modes, elastic network models
Introduction
Allosteric regulation of protein function takes place in diverse biological processes, such as metabolism, signal transduction and molecular machinery. Two classical models of allostery have found broad utility in interpreting the behavior of allosteric proteins: the concerted (all-or-none) model and the sequential model, with respective underlying mechanisms of (i) selection from pre-existing states and (ii) induced fit, stabilized or elicited upon ligand binding [1]. However, recent experimental and computational data on the subject have exposed the need to go beyond simple phenomenological models and acquire a physics-based or mechanistic understanding of allostery as well as its evolutionary fine-tuning. As such, protein flexibility and structure-encoded collective dynamics have come forth in forming the link between folded structures and allosteric regulation. Furthermore, the need to understand the allosteric events in the light of the conformational space accessible to the protein under equilibrium conditions became clear [1–4]. While the thermodynamics-based concept of an ensemble of pre-existing states is plausible, the ease of undergoing allosteric switches/shifts triggered by local binding events calls attention to the role of conformational dynamics, or pre-existing paths that ought to be easily accessible on the conformational landscape, near the original equilibrium state [5].
The correlation between the allosteric mechanisms of conformational changes and the ‘soft’ modes of motions intrinsically accessible to folded structures, uniquely defined by the inter-residue contact topology, is now borne out by numerous studies [6,7]. Here ‘soft’ refers to minimal increase in energy for a given excursion away from the equilibrium state. Furthermore, given the functional significance of allosteric events, emerging topics of interest are the relationship of allosteric dynamics to evolution, the ability of proteins to evolve so as to favor/retain allosteric functions [7,8] (arrow 1 in Fig 1); or the possibility of designing allosteric proteins by learning from evolution [9,10].
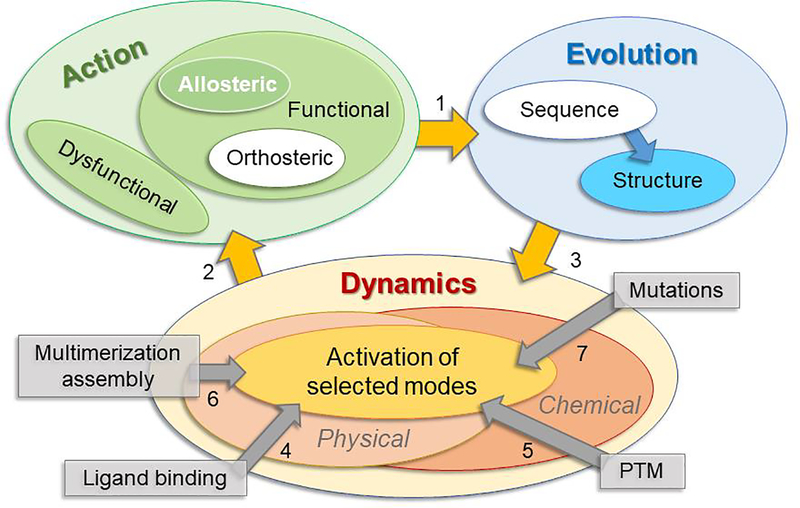
Figure 1.
Schematic representation of relationships between mechanisms of action, evolution and dynamics. Green round box represents the mechanisms of action. If an action is functional it may have allosteric and/or orthosteric effects. If it is dysfunctional, it will not be selected against by the evolutionary pressure (Blue round box). The protein sequences and structures filtered by evolution will then perform their functions defined by intrinsic dynamics, during which various perturbations act through either physical or chemical changes (orange round box). These changes may alter the favorability of selected modes through which the protein perform actions. Gray square boxes represent four main stimuli that trigger allosteric responses.
In this review, we focus on the relationship between biomolecular action, evolution and dynamics (Fig 1). Actions involve both chemical (e.g. catalytic, orthosteric) and physical (e.g., binding, allosteric) events. We focus here on the physical changes that underlie allosteric responses, how they are uniquely encoded by the structure, and how the structure (and sequences) that allow for these allosteric switches/shifts are uniquely selected by evolution.
Allosteric responses are triggered when stimuli exploit pre-existing dynamics
Conformational coupling between spatially distant functional sites, e.g. allosteric and orthosteric sites, is a key requirement for regulating allosteric function [11]. This way a first binding event (e.g. ATP binding) induces, or facilitates, a second (e.g. substrate binding) often taking place at a remote location on the structure. While classical examples of allostery have focused on ligand binding (e.g. oxygen binding to one of the subunits of hemoglobin that prompts cooperative binding to other subunits; or ATP-binding in ATP-regulated molecular machines), recent studies show that allosteric responses can be stimulated by a variety of mechanisms in addition to binding of an ion, small molecule, another protein, or DNA [12,13] (arrow 4 in Fig 1), such as chemical change or posttranslational modification (PTM)[14,15] (arrow 5), complex formation or assembly [16] (arrow 6), or mutations or amino acid substitutions [17–19] (arrow 7). In fact, all proteins, including monomers and complexes/assemblies, have been pointed out to be prone to allosteric modulation[20].
While the ability to exhibit allosteric responses has been observed for a broad range of proteins, including membrane proteins (transporters, receptors) in recent years, not all binding/complexation events result in allosteric responses. Allosteric responses are robustly dependent on the overall architecture [1], or topology of contacts often described by network models. The site, and even the direction, of application of the perturbation or alteration matters. The point is that the allosteric response would not be induced if the protein were not already predisposed to undergo that specific conformational change; and only those stimuli that exploit those pre-existing propensities do effectively induce cooperative, allosteric responses, hence the concept of selecting from an existing pool of collective modes (at the center of ‘dynamics’ box in Fig 1). Essentially, the first binding event acts on a site, and in a direction, that is highly sensitive to the specific perturbation, or should stimulate an energetically favorable movement in order to elicit an allosteric response [21,22]. In the energy landscape description of the conformational space, that direction is the first to be flooded/occupied upon a rise in energy. Perturbations due to energy input, such as that released by ATP hydrolysis, also fall in this category, as demonstrated in a study of GroEL [23], that the system is driven by the ATP hydrolysis-induced directional force to move along existing transition pathways on the free energy landscape.
Using these concepts, new methods have been developed for assessing the allosteric potential of individual residues or for predicting allosteric communication pathways [24–33]. A recent review [34] provides an overview of structure- or topology-based approaches for predicting allosteric mechanisms, including elastic network model (ENM)-based analyses, molecular dynamics (MD) simulations, and graph theoretical methods.
Allosteric changes in structure conform to evolutionarily conserved soft modes
The above described site- and direction-specific sensitivity to perturbations is an intrinsic property of the structure. Each structure encodes a spectrum of normal modes under equilibrium conditions. Among them the softest modes, also called global modes, are manifested in the largest changes in conformation (for a given energy increase), and they are often distinguished by their high collectivity, i.e. the motion is distributed over a large portion of the structure. Many studies, including those in recent years [35–38], demonstrated that these ‘cooperative’ modes are ‘used’ when the protein undergoes allosteric changes in its structure, i.e. the different types of perturbations (arrows 4–7 in Fig 1) elicit allosteric responses only if/when they selectively operate on those sites that stimulate the soft modes that lend themselves to allosteric behavior (arrow 2 in Fig 1). Likewise, alterations in these soft modes due to mutations or aberrant interactions may lead to dysfunction.
How can we determine these soft modes? A broadly used approach is to adopt an ENM for modeling the topology of inter-residue contacts. ENMs provide an analytical solution for the spectrum of normal modes uniquely accessible to each protein/complex, which is then decomposed to extract the soft modes. User-friendly interfaces have been developed to facilitate these computations; among them, DynOmics [39] also takes account of the environment such as the lipid bilayer for membrane proteins. More advanced methods combine ENM-based methods and MD simulations to determine the conformational subspace spanned by soft modes [40–42]. ClustENM [41] is such an unbiased hybrid method. Application to a broad range of proteins, from calmodulin to the ribosome, and to DNA-protein and protein-protein docking [43], as well as trigger-factor-ribosome complexation [44], provided evidence for the role of soft modes in sampling populated conformers, including those visited during allosteric machinery, binding and assembly.
A number of studies have shown that soft modes are conserved within protein families and “fine-tuned for specific function” [7] (arrow 3 in Fig 1). A recent systematic analysis of 116 CATH superfamilies showed that each superfamily is characterized by a signature dynamics, defined by 1–4 softest modes shared among all members [45]. Note that these modes include those that underlie, or facilitate, allosteric couplings. Detailed analysis for selected families further showed that the differentiation within families are mainly imparted by a second group of prominent modes, immediately following these highly conserved modes, termed low-to-intermediate frequency modes (LTIF) [45].
Interestingly, a few modes at the other end of the spectrum, the highest frequency modes, also exhibit a certain level of conservation among family members [45]. These usually refer to sites severely constrained, due to stability (e.g. folding nuclei) or chemical activity (orthosteric) requirements. A recent ENM analysis [11] highlighted the dynamic coupling between allosteric and orthosteric events. The coupling between these respective events is consistent with their concurrent evolutionary conservation.
Modularity and evolutionary adaptation mechanisms between orthologs facilitate allosteric responses
Many allosteric proteins contain conserved domains or subunits that serve as modular units for regulating their allosteric behavior. Examples of such domains in multidomain proteins are the ATPase domains, the Src module of cytoplasmic tyrosine kinases [46], the PDZ domains [47], the cAMP-binding domains of PKA [3], or the ATP-cones that regulate ribonuclease reductases [48]. Likewise, the individual subunits of allosteric complexes often possess intrinsic abilities to undergo conformational switches in accord with the allosteric transitions of the complex. The intrinsic abilities, or modular allostery, of these subunits are thus used, with suitable evolutionary adaptations originating from various effects (e.g. sequence changes, intermolecular interactions, PTMs), for enabling the more complex machinery of biological assemblies [49]. A recent example is the microseconds dynamics of an allosteric switch domain (M domain) in the AAA+ disaggregation machine, a hexameric chaperone that rescues aggregated proteins [50]. FRET experiments showed a broad population of conformers compatible with the active and inactive states of this domain, with interchanges much faster than the time scale of either ATP hydrolysis or disaggregation activity, thus prompting the AAA+ to its allosteric machinery.
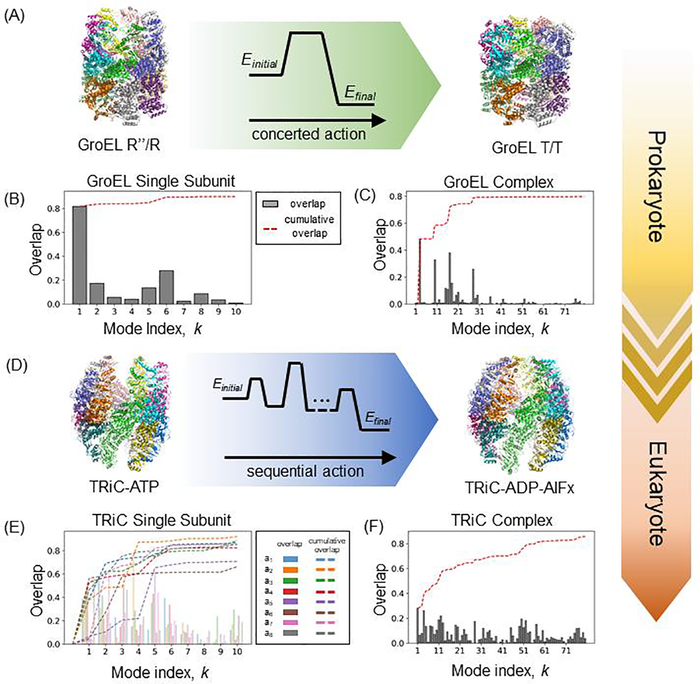
A closer look shows the role of these modular units in enabling allostery, and the level of conservation, or differentiation, by evolutionary adaptation mechanisms. Figure 2 compares the behavior of bacterial (GroEL) and mammalian (CCT) chaperonins, both functioning as allosteric machines. The individual subunits of the former readily favor the transition between R (ATP-bound) and T (-unbound) forms, as can be seen in panel B: a high overlap (correlation cosine > 0.80) is observed between the softest mode predicted by the ENM (k = 1) and the experimentally observed structural change between these two states [51]. This means that the individual GroEL subunits function as allosteric modules that predispose the heptameric ring to cooperatively undergo all-or-none transitions. In contrast, CCT exhibits a more complex behavior, consistent with the heterogeneity of its octameric rings, as shown in panel E. Certain subunits undergo slower transitions than others, presumably resulting in a sequential transition. In parallel, the overall transition of the CCT hetero-16-mer (panel F) is not as efficient as that of the GroEL 14-mer (panel C). Overall, this example demonstrates that (i) the individual subunits of GroEL operate as allosteric modules that enable an all-or-none transition of the rings in support of the GroEL allosteric behavior, and (ii) the heterogeneity of the CCT results in a distribution of soft modes resulting in a slower, sequential mechanism of allostery, consistent with the result from experiments [52–54] and an Arrhenius analysis [55].
Figure 2. Differential modes of action of prokaryotic and eukaryotic chaperonins.
(A and D) The transition of GroEL from cis-ring GroES-bound (R”) state (PDB id: 1gru) to apo (T) state (PDB id: 1gr5) follows a concerted mode of action, which surmounts a single energy barrier. Whereas the transition of TRiC/CCT from ATP-bound, open state (PDB id: 4a0v) to an ADP-P-bound, closed state (PDB id: 4a0w) follows a sequential mode of action, which involves several possibly smaller energy barriers. Co-chaperonin GroES is not included for the purpose of direct comparison between the two states of GroEL. (B) Overlaps between the ten softest modes predicted by ENM to be accessible to a single subunit of GroEL in the R form, and the experimentally observed deformation undergone by the subunit during its transition to the T form. The bars display the correlation cosines for each mode, and the red curve displays the cumulative overlap (summed over consecutive modes starting from mode k = 1. Panel (C) displays the behavior of softest 80 modes accessible to the entire complex. The cumulative contribution of ~ 30 soft modes is sufficient to reach a correlation cosine of ~ 0.80 with the experimentally observed transition between the two endpoints shown in panel (A). (D-F) Counterparts of the respective panels (A) -(C) shown for TRiC/CCT (mammalian chaperonin). CCT rings are each hetero-octameric, hence eight overlap curves and bar plots corresponding to each of the (sequentially different) subunits are displayed in (E). Overall the passage between the two end points necessitates a larger ensemble of modes of motions, compared to the bacterial chaperonin, suggesting a more controlled/restrained allosteric machinery evolutionarily endowed by sequence/structure divergence.
A dynamics-based regulation is therefore distinguished as the evolutionary adaptation mechanism that underlie the distinctive allosteric behavior of CCT compared to that of GroEL. A comparison of eukaryotic (Hsp70s, HspA1 and Hsc70) and bacterial (DnaK) chaperones also reveals the modulation of allosteric interactions by ‘evolutionary diversification’, so as to fine-tune Hsp70 function [56]. The significance of such dynamics-based fine-tuning of allosteric activity by evolution has been pointed by Hilser and coworkers in the context of the thermal adaptation of enzymes [57], and by Sjöberg and coworkers who noted the evolutionary flexibility of ATP-cones [48]. Likewise, Ozkan and coworkers invited attention to the evolving allosteric dynamics and resilience between ancient and extant thioredoxins [58], or to the evolution of β-lactamases to allosterically modulate resistance to antibiotics [59].
How does small molecule binding affect pre-existing states or dynamics?
The allosteric response triggered by small molecule binding can be viewed in the broadest sense as a change in the conformational energy landscape of the protein. The change in the energy landscape can be realized in multiple ways: the locations of existing minima do not change, but their population (or the corresponding depth or width/curvature) does; or their locations may also exhibit some shifts. Both effects were seen for example in the energy landscape generated for dopamine transporter in the presence and absence of its substrate, dopamine [60]. A change in the depth of the energy minimum could originate from an enthalpic effect such as electrostatic attractions with the ligand; whereas a change in the width or curvature of the energy minimum without altering the depth would be entropic. ENM modes are purely entropic. Their relevance to allosteric dynamics suggests that entropy is a major factor driving allosteric changes, or on the topology of inter-residue contacts. Yet, recent studies also invite attention to the importance of enthalpic effects that may also contribute to allostery [47,61].
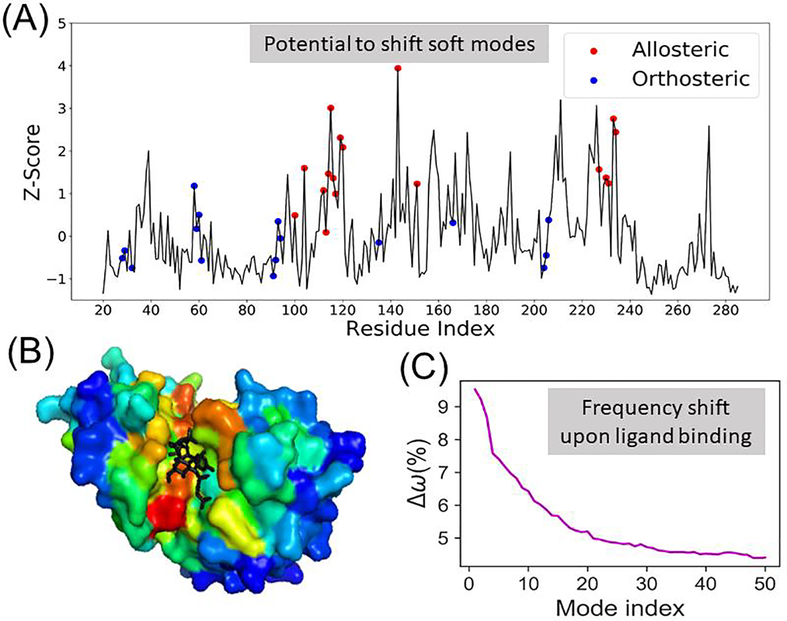
Figure 3 provides a view of the change induced in the frequency of the protein’s pre-existing soft modes upon binding an allosteric modulator. Shifts in the ENM mode frequency dispersion were computed by populating the local density in the vicinity of each residue, in analogy to an analysis on triosephosphate isomerase [62]. The frequency shift in the soft modes is illustrated for each residue of the holo-enzyme, glutamate racemase (Figure 3A). The residues interacting with the allosteric sites stand out in terms of their higher z-scores, as can be seen in the color-coded diagram in Fig 3B. Computations performed for a dataset of 315 proteins using RESPEC [63], a residue-specific ENM, showed that ligand-binding generally tends to shift the frequencies toward higher values, i.e., producing a constraining effect [64] on soft modes (Figure 3C).
Fig 3. Potential of residues to induce a shift (increase) in the frequency of soft modes if targeted by a ligand.
Panel (A) illustrates the results obtained by scanning all residues in glutamate racemase (PDB id: 2JFN). The ordinate, z-score, gives a measure of the extent of frequency shifts in the global modes (averaged over ten softest modes), with peaks referring to those sites that would induce the highest shift, if targeted. Experimentally known binding/coordination sites for allosteric ligands (residues within 4.5 Å from allosteric ligands) are indicated by red circles, and those for orthosteric ligands, by blue circles. (B) Glutamate racemase in complex with an allosteric activator (UMA, shown in black sticks) and the orthosteric ligand (L-Glu, not seen from this perspective). The enzyme is color-coded by z-scores, red and blue, representing highest and lowest values, respectively. The allosteric ligand binding cavity is distinguished by its high potential to induce a frequency shift. (C) Results for a dataset of 315 protein complexes resolved in the presence of orthosteric and/or allosteric ligands. The curve displays the percent shift in the frequency of each of the 50 softest modes, computed by RESPEC, between the complex and the holo forms of the protein, averaged over all members of the dataset. The frequencies shift toward higher values, indicating a stiffening in the soft modes.
Dynamics-based regulation of allostery as sequence and structure evolve
The above examples illustrate the significance of intrinsic dynamics in defining and evolutionarily conserving the allosteric function of proteins with suitable fine-tuning to mediate their sequence and structure diversification. Comparison of GroEL and CCT in Fig 2 showed how the distinctive allosteric dynamics of the bacterial and mammalian chaperonins can be traced to the intrinsic dynamics of their modular subunits, which in turn are encoded by their evolutionarily diverging sequences and structures. Fig 3 demonstrated how ligand binding regulates dynamics by changing the curvature of the energy landscape along the soft modes. On the other hand, mutations introduced through directed evolution, which increase the catalytic activity of tryptophan synthase, were reported to act allosterically through shifting the conformational ensemble [65]. Allosteric mutations located away from the subunit interface can modify the conformational dynamics, similar to allosteric ligands, and thereby induce changes in the oligomeric state of homologous proteins during evolution [66]. Thus, dynamics-based conservation or regulation of allosteric behavior emerges as an effective mechanism for maintaining or modulating function, as the protein sequence and structure evolve.
Acknowledgment
Research reported in this study was supported by the NIH awards P41GM103712 and P30DA035778 (to I.B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Thirumalai D, Hyeon C, Zhuravlev PI, Lorimer GH: Symmetry, Rigidity, and Allosteric Signaling: From Monomeric Proteins to Molecular Machines. Chem Rev 2019, 10.1021/acs.chemrev.8b00760. [DOI] [PubMed] [Google Scholar]
- 2.Guo J, Zhou HX: Protein Allostery and Conformational Dynamics. Chem Rev 2016, 116:6503–6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.England JP, Hao Y, Bai L, Glick V, Hodges HC, Taylor SS, Maillard RA: Switching of the folding-energy landscape governs the allosteric activation of protein kinase A. Proc Natl Acad Sci U S A 2018, 115:E7478–E7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wodak SJ, Paci E, Dokholyan NV, Berezovsky IN, Horovitz A, Li J, Hilser VJ, Bahar I,Karanicolas J, Stock G, et al. : Allostery in Its Many Disguises: From Theory to Applications. Structure 2019, 27:566–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meireles L, Gur M, Bakan A, Bahar I: Pre-existing soft modes of motion uniquely defined by native contact topology facilitate ligand binding to proteins. Protein Sci 2011, 20:1645–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tekpinar M, Yildirim A: Only a Subset of Normal Modes is Sufficient to Identify Linear Correlations in Proteins. J Chem Inf Model 2018, 58:1947–1961. [DOI] [PubMed] [Google Scholar]
- 7.Tiwari SP, Reuter N: Conservation of intrinsic dynamics in proteins-what have computational models taught us? Curr Opin Struct Biol 2018, 50:75–81.• This review describes a number of key studies comparing the dynamics of different proteins and draw conclusions about evolutionary conservation and allostery. They show that global modes are robustly conserved across proteins with similar structures and are largely insensitive to mutations.
- 8.Shah NH, Kuriyan J: Understanding molecular mechanisms in cell signaling through natural and artificial sequence variation. Nat Struct Mol Biol 2019, 26:25–34.•• This review illustrates the evolution of allostery using a few systems/proteins as examples. These examples provide a conclusive view how proteins evolve under selective pressure originating from the necessity to function under different environmental conditions.
- 9.Hocker B: Design of proteins from smaller fragments-learning from evolution. Curr Opin Struct Biol 2014, 27:56–62. [DOI] [PubMed] [Google Scholar]
- 10.Abrusan G, Marsh JA: Ligand-Binding-Site Structure Shapes Allosteric Signal Transduction and the Evolution of Allostery in Protein Complexes. Mol Biol Evol 2019, 36:1711–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma X, Meng H, Lai L: Motions of Allosteric and Orthosteric Ligand-Binding Sites in Proteins are Highly Correlated. J Chem Inf Model 2016, 56:1725–1733. [DOI] [PubMed] [Google Scholar]
- 12.Duro N, Varma S: Role of Structural Fluctuations in Allosteric Stimulation of Paramyxovirus Hemagglutinin-Neuraminidase. Structure 2019, 10.1016/j.str.2019.07.005. [DOI] [PubMed] [Google Scholar]
- 13.White AD, Fang F, Jean-Alphonse FG, Clark LJ, An HJ, Liu H, Zhao Y, Reynolds SL, Lee S,Xiao K, et al. : Ca(2+) allostery in PTH-receptor signaling. Proc Natl Acad Sci U S A 2019, 116:3294–3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang WYC, Ditlev JA, Chiang HK, Rosen MK, Groves JT: Allosteric Modulation of Grb2 Recruitment to the Intrinsically Disordered Scaffold Protein, LAT, by Remote Site Phosphorylation. J Am Chem Soc 2017, 139:18009–18015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Nam K: Dynamic, structural and thermodynamic basis of insulin-like growth factor 1 kinase allostery mediated by activation loop phosphorylation. Chem Sci 2017, 8:3453–3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stewart GD, Comps-Agrar L, Norskov-Lauritsen LB, Pin JP, Kniazeff J: Allosteric interactions between GABAB1 subunits control orthosteric binding sites occupancy within GABAB oligomers. Neuropharmacology 2018, 136:92–101. [DOI] [PubMed] [Google Scholar]
- 17.Li H, Sharma N, General IJ, Schreiber G, Bahar I: Dynamic Modulation of Binding Affinity as a Mechanism for Regulating Interferon Signaling. J Mol Biol 2017, 429:2571–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith IN, Thacker S, Seyfi M, Cheng F, Eng C: Conformational Dynamics and Allosteric Regulation Landscapes of Germline PTEN Mutations Associated with Autism Compared to Those Associated with Cancer. Am J Hum Genet 2019, 104:861–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naganathan AN: Modulation of allosteric coupling by mutations: from protein dynamics and packing to altered native ensembles and function. Curr Opin Struct Biol 2019, 54:1–9.• The author discusses a set of recent computational and experimental results and emphasizes that mutational perturbations effect not only on protein stability but also the native conformational ensemble, thereby impacting allosteric modulations.
- 20.Gunasekaran K, Ma B, Nussinov R: Is allostery an intrinsic property of all dynamic proteins? Proteins 2004, 57:433–443. [DOI] [PubMed] [Google Scholar]
- 21.Haliloglu T, Bahar I: Adaptability of protein structures to enable functional interactions and evolutionary implications. Curr Opin Struct Biol 2015, 35:17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krieger J, Lee JY, Greger IH, Bahar I: Activation and desensitization of ionotropic glutamate receptors by selectively triggering pre-existing motions. Neurosci Lett 2019, 700:22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J, Sankar K, Wang Y, Jia K, Jernigan RL: Directional Force Originating from ATP Hydrolysis Drives the GroEL Conformational Change. Biophys J 2017, 112:1561–1570.• In this study, a coarse-grained free energy landscape is constructed using 38 experimental structures of GroEL. A Monte Carlo method with random perturbing forces is applied to sample the closed-open transition, showing that the conformational change is driven by the ATP hydrolysis-induced directional force.
- 24.Kumar A, Glembo TJ, Ozkan SB: The Role of Conformational Dynamics and Allostery in the Disease Development of Human Ferritin. Biophys J 2015, 109:1273–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amor BR, Schaub MT, Yaliraki SN, Barahona M: Prediction of allosteric sites and mediating interactions through bond-to-bond propensities. Nat Commun 2016, 7:12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flechsig H: Design of Elastic Networks with Evolutionary Optimized Long-Range Communication as Mechanical Models of Allosteric Proteins. Biophys J 2017, 113:558–571.•• The principles of designing allosteric protein networks or toy models from scratch are studied to explore how allosteric proteins perform specified tasks. The author elaborates on the robustness of allosteric communications and optimization of evolution from the mechanical picture of allosteric systems based on random elastic network models with the aim of understanding the functional features of protein machinery.
- 27.Flechsig H, Togashi Y : Designed Elastic Networks: Models of Complex Protein Machinery. Int J Mol Sci 2018, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guarnera E, Berezovsky IN: On the perturbation nature of allostery: sites, mutations, and signal modulation. Curr Opin Struct Biol 2019, 56:18–27. [DOI] [PubMed] [Google Scholar]
- 29.Guzel P, Kurkcuoglu O: Identification of potential allosteric communication pathways between functional sites of the bacterial ribosome by graph and elastic network models. Biochim Biophys Acta Gen Subj 2017, 1861:3131–3141. [DOI] [PubMed] [Google Scholar]
- 30.Hacisuleyman A, Erman B: Entropy Transfer between Residue Pairs and Allostery in Proteins: Quantifying Allosteric Communication in Ubiquitin. PLoS Comput Biol 2017, 13:e1005319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hodges M, Barahona M, Yaliraki SN: Allostery and cooperativity in multimeric proteins: bond-to-bond propensities in ATCase. Sci Rep 2018, 8:11079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh S, Bowman GR: Quantifying Allosteric Communication via Both Concerted Structural Changes and Conformational Disorder with CARDS. J Chem Theory Comput 2017, 13:1509–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tee WV, Guarnera E, Berezovsky IN: Reversing allosteric communication: From detecting allosteric sites to inducing and tuning targeted allosteric response. PLoS Comp Biol 2018, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Greener JG, Sternberg MJE: Structure-based prediction of protein allostery. Curr Opin Struct Biol 2018, 50:1–8. [DOI] [PubMed] [Google Scholar]
- 35.Alfayate A, Caceres CR, Gomes Dos Santos H, Bastolla U: Predicted dynamical couplings of protein residues characterize catalysis, transport and allostery. Bioinformatics 2019, 10.1093/bioinformatics/btz301. [DOI] [PubMed] [Google Scholar]
- 36.Liang Z, Hu J, Yan W, Jiang H, Hu G, Luo C: Deciphering the role of dimer interface in intrinsic dynamics and allosteric pathways underlying the functional transformation of DNMT3A. Biochim Biophys Acta Gen Subj 2018, 1862:1667–1679. [DOI] [PubMed] [Google Scholar]
- 37.Ponzoni L, Zhang S, Cheng MH, Bahar I: Shared dynamics of LeuT superfamily members and allosteric differentiation by structural irregularities and multimerization. Philos Trans R Soc Lond B Biol Sci 2018, 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones PM, George AM: How Intrinsic Dynamics Mediates the Allosteric Mechanism in the ABC Transporter Nucleotide Binding Domain Dimer. J Chem Theory Comput 2017, 13:1712–1722. [DOI] [PubMed] [Google Scholar]
- 39.Li H, Chang YY, Lee JY, Bahar I, Yang LW: DynOmics: dynamics of structural proteome and beyond. Nucleic Acids Res 2017, 45:W374–W380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Costa MG, Batista PR, Bisch PM, Perahia D: Exploring free energy landscapes of large conformational changes: molecular dynamics with excited normal modes. J Chem Theory Comput 2015, 11:2755–2767. [DOI] [PubMed] [Google Scholar]
- 41.Kurkcuoglu Z, Bahar I, Doruker P: ClustENM: ENM-Based Sampling of Essential Conformational Space at Full Atomic Resolution. J Chem Theory Comput 2016, 12:4549–4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Togashi Y, Flechsig H: Coarse-Grained Protein Dynamics Studies Using Elastic Network Models. Int J Mol Sci 2018, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kurkcuoglu Z, Bonvin A: Pre- and post-docking sampling of conformational changes using ClustENM and HADDOCK for protein-protein and protein-DNA systems. Proteins 2019, 10.1002/prot.25802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Can MT, Kurkcuoglu Z, Ezeroglu G, Uyar A, Kurkcuoglu O, Doruker P: Conformational dynamics of bacterial trigger factor in apo and ribosome-bound states. PLoS One 2017, 12:e0176262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang S, Li H, Krieger JM, Bahar I: Shared Signature Dynamics Tempered by Local Fluctuations Enables Fold Adaptability and Specificity. Mol Biol Evol 2019, 36:2053–2068.•• This study systematically assesses how intrinsic dynamics are conserved across a large number of superfamilies and reveals a partitioning of the mode spectrum into different regions with different conservation profiles. In addition to showing that global modes at low frequency end of the spectrum are highly conserved, this study also shows that low-to-intermediate frequency modes are somewhat conserved and are important for subfamily specificity, and that many high frequency modes are also conserved.
- 46.Shah NH, Amacher JF, Nocka LM, Kuriyan J: The Src module: an ancient scaffold in the evolution of cytoplasmic tyrosine kinases. Crit Rev Biochem Mol Biol 2018, 53:535–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kumawat A, Chakrabarty S: Hidden electrostatic basis of dynamic allostery in a PDZ domain. Proc Natl Acad Sci U S A 2017, 114:E5825–E5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rozman Grinberg I, Lundin D, Hasan M, Crona M, Jonna VR, Loderer C, Sahlin M,Markova N, Borovok I, Berggren G, et al. : Novel ATP-cone-driven allosteric regulation of ribonucleotide reductase via the radical-generating subunit. Elife 2018, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berezovsky IN, Guarnera E, Zheng Z, Eisenhaber B, Eisenhaber F: Protein function machinery: from basic structural units to modulation of activity. Curr Opin Struct Biol 2017, 42:67–74. [DOI] [PubMed] [Google Scholar]
- 50.Mazal H, Iljina M, Barak Y, Elad N, Rosenzweig R, Goloubinoff P, Riven I, Haran G: Tunable microsecond dynamics of an allosteric switch regulate the activity of a AAA+ disaggregation machine. Nat Commun 2019, 10:1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang Z, Majek P, Bahar I: Allosteric transitions of supramolecular systems explored by network models: application to chaperonin GroEL. PLoS Comput Biol 2009, 5:e1000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamamoto YY, Uno Y, Sha E, Ikegami K, Ishii N, Dohmae N, Sekiguchi H, Sasaki YC, Yohda M: Asymmetry in the function and dynamics of the cytosolic group II chaperonin CCT/TRiC. PLoS One 2017, 12:e0176054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gestaut D, Limatola A, Joachimiak L, Frydman J: The ATP-powered gymnastics of TRiC/CCT: an asymmetric protein folding machine with a symmetric origin story. Curr Opin Struct Biol 2019, 55:50–58.• This review proposes that TRiC folds complex multi-domain proteins with subunit-specific substrate interactions and the segmental release into the chamber, driven by sequential ATP-binding and hydrolysis.
- 54.Jin M, Han W, Liu C, Zang Y, Li J, Wang F, Wang Y, Cong Y: An ensemble of cryo-EM structures of TRiC reveal its conformational landscape and subunit specificity. Proc Natl Acad Sci U S A 2019, 116:19513–19522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gruber R, Levitt M, Horovitz A: Sequential allosteric mechanism of ATP hydrolysis by the CCT/TRiC chaperone is revealed through Arrhenius analysis. Proc Natl Acad Sci U S A 2017, 114:5189–5194.•• The Arrhenius analysis used in this study revealed the sequential allosteric mechanism of TRiC triggered by differential ATP hydrolysis at the eight heterogeneous subunits within a ring. It outlines a free energy profile for the allosteric transition of a TRiC ring from the open to the closed form, with multiple but relatively small energy barriers, in contrast to a single barrier for a concerted action employed by GroEL.
- 56.Meng W, Clerico EM, McArthur N, Gierasch LM: Allosteric landscapes of eukaryotic cytoplasmic Hsp70s are shaped by evolutionary tuning of key interfaces. Proc Natl Acad Sci U S A 2018, 115:11970–11975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saavedra HG, Wrabl JO, Anderson JA, Li J, Hilser VJ: Dynamic allostery can drive cold adaptation in enzymes. Nature 2018, 558:324–328.• Dynamics-based regulation comes forth as an evolutionary adaptation mechanism, which could be used to fine-tune the biological function. Entropy-tuning changes/mutations at solvent-exposed sites modulate the activity of specific domains in adenylate kinase without altering the ground state structure.
- 58.Modi T, Huihui J, Ghosh K, Ozkan SB: Ancient thioredoxins evolved to modern-day stability-function requirement by altering native state ensemble. Philos Trans R Soc Lond B Biol Sci 2018, 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Modi T, Ozkan SB: Mutations Utilize Dynamic Allostery to Confer Resistance in TEM-1 beta-lactamase. Int J Mol Sci 2018, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cheng MH, Kaya C, Bahar I: Quantitative Assessment of the Energetics of Dopamine Translocation by Human Dopamine Transporter. J Phys Chem B 2018, 122:5336–5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu J, Nussinov R: Energetic redistribution in allostery to execute protein function. Proc Natl Acad Sci U S A 2017, 114:7480–7482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kurkcuoglu Z, Findik D, Akten ED, Doruker P: How an Inhibitor Bound to Subunit Interface Alters Triosephosphate Isomerase Dynamics. Biophys J 2015, 109:1169–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kaynak BT, Findik D, Doruker P: RESPEC Incorporates Residue Specificity and the Ligand Effect into the Elastic Network Model. J Phys Chem B 2018, 122:5347–5355. [DOI] [PubMed] [Google Scholar]
- 64.Kaynak BT, Doruker P: Protein–Ligand Complexes as Constrained Dynamical Systems. J Chem Inf Model 2019, 10.1021/acs.jcim.8b00946:acs.jcim.8b00946. [DOI] [PubMed] [Google Scholar]
- 65.Buller AR, van Roye P, Cahn JKB, Scheele RA, Herger M, Arnold FH: Directed Evolution Mimics Allosteric Activation by Stepwise Tuning of the Conformational Ensemble. J Am Chem Soc 2018, 140:7256–7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Perica T, Kondo Y, Tiwari SP, McLaughlin SH, Kemplen KR, Zhang X, Steward A, Reuter N,Clarke J, Teichmann SA: Evolution of oligomeric state through allosteric pathways that mimic ligand binding. Science 2014, 346:1254346. [DOI] [PMC free article] [PubMed] [Google Scholar]