Abstract
The SARS-CoV-2 pandemic that causes COVID-19 respiratory syndrome has caused global public health and economic crises, necessitating rapid development of vaccines and therapeutic countermeasures. The world-wide response to the COVID-19 pandemic has been unprecedented with government, academic, and private partnerships working together to rapidly develop vaccine and antibody countermeasures. Many of the technologies being used are derived from prior government-academic partnerships for response to other emerging infections.
Keywords: ▪▪▪
Collaboration between academic, government, and private institutions can enable the development and sustenance of a pandemic preparedness platform that can be readily deployed upon need, as is the case with the COVID-19 pandemic.
Main Text
Introduction
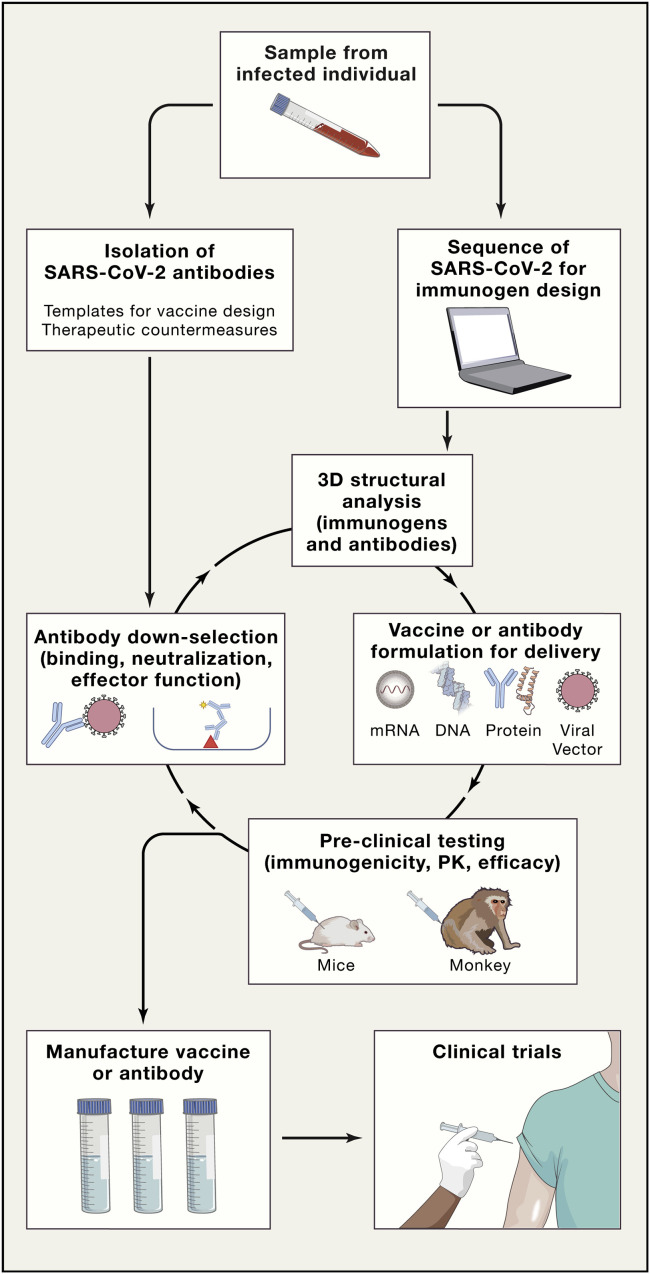
The novel coronavirus outbreak of SARS-CoV-2, as of May 14, 2020, has resulted in more than 4,300,000 individuals infected and over 290,000 deaths worldwide (Dong et al., 2020). The current pandemic, as well as the potential of future pandemics based on estimates of undiscovered zoonotic infections (Carroll et al., 2018), has brought to the forefront urgency and necessity for rapid development of pandemic countermeasures. Two countermeasures with promise for controlling the current SARS-CoV-2 pandemic are recombinant neutralizing antibodies (Ju et al., 2020, Walker and Burton, 2018) and vaccines (Graham, 2020, Graham et al., 2018, Graham and Sullivan, 2018) directed against the virus that causes COVID-19, SARS-CoV-2. In particular, over the past 15 years, the NIAID Center for HIV/AIDS Vaccine Immunology (CHAVI) program (Burton et al., 2012, Haynes et al., 2016), the NIH Vaccine Research Center (Kwong and Mascola, 2012) as well as others, and, for the past three years, the DARPA Pandemic Prevention Program (P3) program (Cable et al., 2020, DARPA, 2017, Kose et al., 2019) have worked to define the platforms and enable technology for HIV vaccine development and rapid response to viral pandemics. Although an HIV vaccine has not yet been developed, much of the technology the HIV vaccine field has developed is now being used to fight the COVID-19 pandemic. From the HIV field and the DARPA preparedness programs have come teams and technologies that are now responding to the COVID-19 epidemic to both isolate SARS-CoV-2 neutralizing antibodies and develop SARS-CoV-2 vaccine candidates. Here we comment on some of the strategies that are being used to develop antibody and vaccine countermeasures for SARS CoV-2 (Figure 1 ).
Figure 1.
Schema of Iterative and Synergistic Approaches Being Used to Simultaneously Develop Both Vaccines and Antibody Countermeasures for SARS-CoV-2/COVID-19
Neutralizing antibodies. Antibodies isolated from a single B cell are called monoclonal antibodies (mAbs) and have become an effective new biologic class in our pharmacopeia with a wide-range of FDA-approved mAbs for indications such as arthritis and other inflammatory diseases, heart disease, hypercholesterolemia, osteoporosis, cancer, and infectious diseases (Shepard et al., 2017). Recombinant human or humanized monoclonal antibodies are proving to be safe, effective, and highly specific in their ability to target a pathway, process, or invading pathogen. More than 70 recombinant monoclonal antibodies have now been approved by the FDA for use in the treatment of infectious, autoimmune and inflammatory, malignant, or cardiovascular diseases (Carter and Lazar, 2018, Shepard et al., 2017). Specifically, recombinant neutralizing antibodies for infectious diseases, such as for protection from anthrax toxin and for the prevention of respiratory syncytial virus infection (Empey et al., 2010, Shepard et al., 2017), have been approved by the FDA. Neutralizing antibodies are currently in development for prevention and/or treatment of HIV (Caskey et al., 2019, Gaudinski et al., 2019) and pending approval for Ebola (Saphire et al., 2018).
Thus, recombinant neutralizing antibodies isolated from those infected with SARS-CoV-2 are the most rapid and readily manufacturable immune intervention for passive administration that might be developed to either prevent or treat COVID-19 disease (Andreano et al., 2020, Brouwer et al., 2020, Ju et al., 2020, Rogers et al., 2020, Seydoux et al., 2020). SARS-CoV-2 antibody countermeasures will benefit from the last 20 years of antibody optimization research that has discovered point mutations in the Fc portion of antibodies that finetune antibody function and circulation half-life (Saunders, 2019). Such mutations have been described for the Fc region of IgG that have prolonged antibody half-life for up to 6–7 weeks (Gaudinski et al., 2019, Robbie et al., 2013, Yu et al., 2016). Additionally, mutations are known that can increase antibody-dependent infected cell killing and antibody-dependent complement activation (Idusogie et al., 2001, Richards et al., 2008). Given the ability of certain antibodies to facilitate SARS-CoV-1 virus entry via engagement of Fc receptors on host cells (Jaume et al., 2011), the introduction of mutations that inhibit Fc binding to Fc receptors could also be important for successful development of SARS-CoV-2 neutralizing antibody treatments.
Neutralizing antibodies to the spike protein receptor binding domain (RBD) protect mice from MERS, SARS-CoV-1, and SARS-CoV-2 infection (Quinlan et al., 2020, Wang et al., 2018, Zhou et al., 2018). Thus, neutralizing antibodies are under development as proteins or gene-delivered formulations to prevent or treat SARS-CoV-2 infection. One example of technology now brought to bear on SARS-CoV-2 countermeasure work is the strategy developed to isolate and screen for HIV neutralizing antibodies without antibody gene cloning (Liao et al., 2009), and this technology was recently used in China to isolate the first neutralizing antibodies to SARS-CoV-2 (Ju et al., 2020). As seen with SARS-CoV-1 and HIV, viruses resistant to a particular antibody can emerge among the circulating virus variants (Caskey et al., 2017, ter Meulen et al., 2006). In these cases, combinations of two of more neutralizing antibodies can broaden the efficacy of neutralizing antibody prophylaxis or treatment (Mendoza et al., 2018, ter Meulen et al., 2006). Thus, large numbers of effective neutralizing antibodies are needed so that antibody cocktails can be formulated to provide optimal coverage of circulating SARS-CoV-2 isolates.
The Defense Advanced Research Projects Agency (DARPA) Pandemic Prevention Program (P3) within the Department of Defense has played a key role in funding and driving public/private development of end-to-end platforms to rapidly develop antibody countermeasures. The DARPA P3 program aims to identify and propagate viral pathogens, isolate human neutralizing antibody sequences, develop effective approaches to deliver neutralizing antibodies as nucleic acids, and develop good manufacturing practices for a final drug product to submit to the Agency for Phase I approval within 60 days of receiving an outbreak blood specimen (Cable et al., 2020, DARPA, 2017).
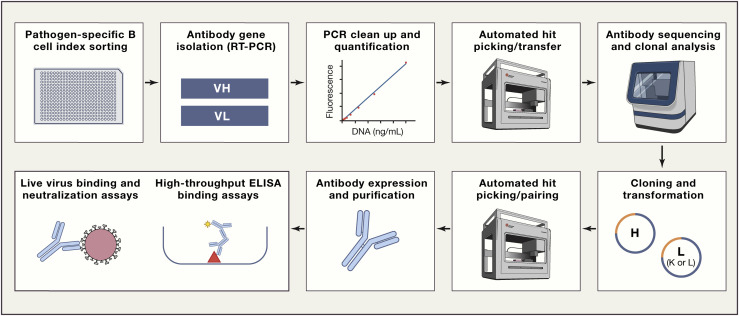
To meet this challenge, the Duke DARPA P3 program developed a “thaw and infect” permissive cell line array to rapidly grow Risk Group 2 and 3 viruses, such as SARS-CoV-2. This team also developed platform technologies for real-time virus growth monitoring, fluorescently labeling viruses, antibody focus/plaque-reduction neutralization assays, and whole virus binding assays. By using innovative and time-tested approaches from the HIV field (Klein et al., 2013, Liao et al., 2009), the Duke P3 group developed a rapid, virus-independent, high-throughput, semi-automated Ab sequence isolation and screening pipeline in high-containment (Figure 2 ). This pipeline has recently isolated a panel of H3N2 influenza neutralizing antibodies that provide protection in mouse challenge models and is now fully engaged in isolating a cocktail of neutralizing SARS-CoV-2 human monoclonal antibodies. The human angiotensin-converting enzyme 2 (ACE2) transgenic mouse (Bao et al., 2020) will be incorporated into the pipeline as well as rhesus and cynomolgus macaque models (Lu et al., 2020, Rockx et al., 2020) as SARS-CoV-2 acquisition models to rapidly evaluate the ability of antibodies to protect from SARS-CoV-2 challenge and the possibility of antibody-induced enhancement of disease (Peeples, 2020). Rapid movement of potent viral neutralizing antibodies to testing in the clinic requires an alternative to typical recombinant antibody protein production. DARPA P3 performer teams and others are therefore developing gene-delivered approaches (Figure 1) for rapid cGMP-scalable manufacturing. One approach being used is modified RNA encapsulated in lipid nanoparticles (LNPs) due to its proven utility and potential for rapid manufacturing (Kose et al., 2019, Pardi et al., 2018). In pre-clinical models, protective concentrations of recombinant neutralizing antibodies have been successfully expressed from intravenously and intramuscularly delivered LNP-encapsulated mRNA, supporting this approach for use in response to a rapidly spreading pandemic (Kose et al., 2019, Pardi et al., 2017b). Moreover, Crowe at al. (Vanderbilt P3 Performer Site), in collaboration with Moderna, has successfully isolated and delivered a Chikungunya neutralizing antibody by using mRNA in LNPs in a Phase 1 human study (Kose et al., 2019). In addition to mRNA-LNP, DNA or viral vector approaches are also being rapidly developed for pandemic prevention antibody delivery (Balazs et al., 2011, Muthumani et al., 2013).
Figure 2.
Accelerated Platform Technology for Rapid B Cell Screening and Isolation of Pathogen Neutralizing Antibodies Being Used to Isolate SARS-CoV-2 Antibodies
SARS-CoV-2 vaccine development. Vaccines are the time-honored method for establishing long-lived immune memory for controlling infectious diseases, and technologies have been developed such that vaccines can now be developed faster than in previous times (Figure 1) (Graham, 2020, Graham et al., 2018, Graham and Sullivan, 2018). Over 100 companies or academic institutions are working on COVID-19 vaccines with strategies that include recombinant vectors, mRNA in lipid nanoparticles, DNA, inactivated virus, live attenuated virus, virus-like particles, and protein subunits (Thanh Le et al., 2020, WHO, 2020b). Three vaccine candidates have already advanced to Phase II testing that include an mRNA vaccine encoding the viral spike protein from Moderna, an Adeno-type 5 vector vaccine expressing the S protein from CanSino Biologicals, and a chimpanzee adenovirus encoding the spike protein from the Jenner Institute in Oxford, UK. Five other vaccine candidates are also now in phase I trials including other mRNA/LNP or DNA vaccines as well as three forms of whole inactivated vaccines (WHO, 2020b).
As seen from this rapid movement of SARS-CoV-2 vaccine candidates into human trials, the time it takes to develop vaccines for emerging pathogens is decreasing from that in the past for traditional childhood vaccines. Recently, a DNA vaccine for the original SARS (SARS-CoV-1) was developed in 20 months, a vaccine for H5 influenza A/Indonesia/2006 in 11 months, a vaccine for H1 influenza A/California/2009 in 4 months, and a Zika virus vaccine in 3.5 months (Graham et al., 2018). These successes have been brought about by innovative technology and approaches that have allowed for rapid identification and sequencing of new viral pathogens and new technologies for vaccine delivery (Graham and Sullivan, 2018). For the past 15 years, the HIV vaccine field has pioneered development and use of recombinant antibody technology, advanced computational methods, novel animal models, and new vaccine delivery approaches to accelerate HIV vaccine immunogen design and development (Burton et al., 2012, Caskey et al., 2019, Haynes et al., 2019, Haynes et al., 2016, Klein et al., 2013, Kwong and Mascola, 2018, Liao et al., 2009). This work has deciphered the roadblocks for this most difficult-to-develop HIV vaccine (Haynes et al., 2019). It is expected that the timeline for a SARS-CoV-2 vaccine will be much faster and much easier than for HIV-1. Investigators have worked to integrate these iterative approaches for vaccine and antibody countermeasure development and applied them to a rapid response to the COVID-19 disease epidemic caused by SARS-CoV-2 (Figure 1).
The COVID-19 pandemic caused by SARS-CoV-2 was first widely recognized in December 2019, and the first virus sequence published online in January 2020. By March 16, 2020, the first mRNA/LNP vaccine trial developed by the VRC in collaboration with Moderna had begun (NIH, 2020). A rapid SARS-CoV2 vaccine development approach involves the integration of computational and structural-based immunogen design strategies; production of immunogens as inactivated virus; DNA, mRNA, vectored or protein subunits; and immunogenic profiling in animal models prior to vaccine manufacturing and testing in clinical trials (Figure 1). Computational biology techniques have facilitated the rapid analysis of antibody and virus sequences for influenza, HIV, and now SARS-CoV-2 to enable vaccine development (GISAID, 2020, Los Alamos National Laboratory, 2020a, Saunders et al., 2019, Wiehe et al., 2018). Monitoring of HIV evolution by using the Los Alamos HIV Sequence Database (Los Alamos National Laboratory, 2020a) has been critical for HIV vaccine immunogen designs. The SARS-CoV-2 virus is evolving, albeit at a slower rate than HIV, and virus evolution is a concern for successful COVID-19 vaccine development. The SARS-CoV-2 spike protein RBD is the prime target for vaccine-induced neutralizing antibodies although other spike protein neutralizing epitopes are of interest. However, comparison of the SARS-CoV2 RBD with that of SARS-COV-1 reveals only partial homology although both retain the ability to bind to the ACE2 as a receptor (Wrapp et al., 2020). The GISAID database is proving to be helpful information for monitoring the viral evolution of SARS-CoV-2 (GISAID, 2020), and the Los Alamos National Laboratory is developing a website with tools for analysis of global SARS-CoV-2 spike protein sequences (Korber et al., 2020, Los Alamos National Laboratory, 2020b).
Structural determination of the primary targets of neutralizing antibodies, for example, hemagglutinin in influenza, Env in HIV, and now the spike protein in SARS-CoV-2, has provided valuable atomic-level insight for vaccine design strategies. In particular, cryo-electron microscopy has enabled the rapid solution of the structure of the SARS-CoV-2 spike protein (Walls et al., 2020, Wrapp et al., 2020). Structural biology analysis of pathogens combines structural, computational, biophysical, and biochemical methods to understand interactions of pathogens with the immune system (Henderson et al., 2020, LaBranche et al., 2019, Murin et al., 2019, Saunders et al., 2019). Early this year, structural biologists pivoted to apply technology developed for HIV-1 envelope or respiratory syncytial virus (RSV) structural biology to fast-track structure-based vaccine design for COVID-19 (Lan et al., 2020, Walls et al., 2020, Wrapp et al., 2020, Yuan et al., 2020). Currently, established pipelines for high-resolution cryo-EM structural determination of the SARS-CoV-2 spike are integrated with the computational teams, thus providing atomic-level feedback to COVID-19 vaccine designs.
The past eight years of HIV-1 antibody discovery has provided templates for HIV-1 vaccine design aiming to elicit broadly reactive neutralizing antibodies (Kwong and Mascola, 2018, Sok and Burton, 2018). From the study of the ontogeny of HIV neutralizing antibodies it has become clear that an effective vaccine will likely require multiple immunogens administered in a specific order to facilitate proper antibody development to multiple neutralizing targets on HIV (Haynes et al., 2019). Hopefully, the development of SARS-CoV-2 neutralizing antibodies will require a much simpler vaccination regimen like the Zika vaccine, where one immunization with one immunogen was sufficient to elicit protective neutralizing antibodies (Pardi et al., 2017a). Such a vaccine would be amenable to rapid development, large-scale manufacturing, and global administration.
What will follow rapidly now for prevention of COVID-19 will be a number of mRNA/LNP (e.g., from Moderna/NIAID, BioNTech/Fosum, Pharma/Pfizer) or DNA (e.g., Inovio) vaccines as well as attenuated viruses, proteins, nanoparticles, and viral vectors containing SARS-CoV-2 viral genes as vaccine candidates moving through safety and immunogenicity trials, and a smaller subset of vaccine candidates will be tested in Phase III or efficacy trials to continue to determine if they are safe, as well to determine their efficacy. In parallel now with Phase I and II trials, it is important to develop capacity for large-scale vaccine production, in the event of a successful efficacy trial (Corey et al., 2020, Graham, 2020). It is possible that genetic immunization strategies such as DNA or mRNA in LNPs can be manufactured more rapidly than proteins or viral vectors and can be more cost effective.
In addition to efficacy being the primary goal of SARS-CoV-2 vaccine development, safety is also a major concern (Peeples, 2020). Immunization with a SARS-CoV-1 vaccine has induced vaccine-associated immunopathology in the lung (Bolles et al., 2011, Liu et al., 2019, Tseng et al., 2012). Both CD4 and CD8 T cell responses have been suggested to also be protective for SARS-CoV-1 (Zhao et al., 2016). Thus, careful animal preclinical studies as well as intense monitoring of human clinical trials will be of critical importance to developing safe and effective anti-COVID-19 antibody and vaccine countermeasures.
Finally, collaboration and coordination will be essential to ending the pandemic. Globally, one example of private support for COVID-19 research is the Coalition for Epidemic Preparedness Innovations (CEPI). CEPI is raising funds for COVID-19 vaccine development and as well is funding vaccine development projects (Gouglas et al., 2019). The Bill & Melinda Gates Foundation has made a $250 million commitment to fight COVID-19 and has established a Coronavirus Immunotherapy Consortium, or CoVIC, to foster sharing and comparison of SARS-CoV-2 antibodies to speed therapeutic antibody development (Bill & Melinda Gates Foundation, 2020). The recent formation of an NIH-organized public-private partnership, termed Accelerating COVID-19 Therapeutic Interventions and Vaccines (ACTIV) (Corey et al., 2020, Kaiser, 2020), is necessary and will facilitate a coordinated COVID-19 pandemic response. Globally, the World Health Organization is playing a critical multinational coordination and informational role (WHO, 2020a).
Summary
Government-funded and private initiatives have synergized to provide countermeasure platforms to rapidly respond to the SARS-CoV-2 pandemic. Continued cooperation among public and private institutions coupled with speed of development of antibody countermeasures and vaccines, with rapid evaluation of their safety and efficacy, and early planning for scale-up and manufacture will be critical for expeditious control of the global COVID-19 pandemic.
Acknowledgments
Funded by NIH grants AI142596 (B.F.H.), AI145687 and AI150415 (P.A.), AI058607 (G.D.S.); Department of Defense HR0011-17-2-0069 (G.D.S.); and the Translating Duke Health Initiative (P.A.). All authors wrote and edited the manuscript. We thank Megan Averill for editorial assistance and David Easterhoff and QiFeng Han for their contributions to the DARPA P3 program.
Declaration of Interests
B.F.H. and G.D.S. have a patent submitted for the emerging infections preparedness platform and influenza antibodies (No. 62_902705).
References
- Andreano E., Nicastri E., Paciello I., Pileri P., Manganaro N., Piccini G., Manenti A., Pantano E., Kabanova A., Troisi M. Identification of neutralizing human monoclonal antibodies from Italian Covid-19 convalescent patients. bioRxiv. 2020 doi: 10.1101/2020.05.05.078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazs A.B., Chen J., Hong C.M., Rao D.S., Yang L., Baltimore D. Antibody-based protection against HIV infection by vectored immunoprophylaxis. Nature. 2011;481:81–84. doi: 10.1038/nature10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao L., Deng W., Huang B., Gao H., Liu J., Ren L., Wei Q., Yu P., Xu Y., Qi F. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020 doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- Bill & Melinda Gates Foundation . 2020. Press Release: COVID-19 Therapeutics Accelerator Awards $20 Million in Initial Grants to Fund Clinical Trials. [Google Scholar]
- Bolles M., Deming D., Long K., Agnihothram S., Whitmore A., Ferris M., Funkhouser W., Gralinski L., Totura A., Heise M., Baric R.S. A double-inactivated severe acute respiratory syndrome coronavirus vaccine provides incomplete protection in mice and induces increased eosinophilic proinflammatory pulmonary response upon challenge. J. Virol. 2011;85:12201–12215. doi: 10.1128/JVI.06048-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer P., Caniels T., van Straten K., Snitselaar J., Aldon Y., Bangaru S., Torres J., Okba N., Claireaux M., Kerster G. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. bioRxiv. 2020 doi: 10.1101/2020.05.12.088716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton D.R., Ahmed R., Barouch D.H., Butera S.T., Crotty S., Godzik A., Kaufmann D.E., McElrath M.J., Nussenzweig M.C., Pulendran B. A Blueprint for HIV Vaccine Discovery. Cell Host Microbe. 2012;12:396–407. doi: 10.1016/j.chom.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cable J., Srikantiah P., Crowe J.E., Jr., Pulendran B., Hill A., Ginsberg A., Koff W., Mathew A., Ng T., Jansen K. Vaccine innovations for emerging infectious diseases-a symposium report. Ann. N Y Acad. Sci. 2020;1462:14–26. doi: 10.1111/nyas.14235. [DOI] [PubMed] [Google Scholar]
- Carroll D., Daszak P., Wolfe N.D., Gao G.F., Morel C.M., Morzaria S., Pablos-Méndez A., Tomori O., Mazet J.A.K. The Global Virome Project. Science. 2018;359:872–874. doi: 10.1126/science.aap7463. [DOI] [PubMed] [Google Scholar]
- Carter P.J., Lazar G.A. Next generation antibody drugs: pursuit of the ‘high-hanging fruit’. Nat. Rev. Drug Discov. 2018;17:197–223. doi: 10.1038/nrd.2017.227. [DOI] [PubMed] [Google Scholar]
- Caskey M., Schoofs T., Gruell H., Settler A., Karagounis T., Kreider E.F., Murrell B., Pfeifer N., Nogueira L., Oliveira T.Y. Antibody 10-1074 suppresses viremia in HIV-1-infected individuals. Nat. Med. 2017;23:185–191. doi: 10.1038/nm.4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caskey M., Klein F., Nussenzweig M.C. Broadly neutralizing anti-HIV-1 monoclonal antibodies in the clinic. Nat. Med. 2019;25:547–553. doi: 10.1038/s41591-019-0412-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corey B.L., Mascola J.R., Fauci A.S., Collins F.S. A strategic approach to COVID-19 vaccine R&D. Science. 2020:eabc5312. doi: 10.1126/science.abc5312. [DOI] [PubMed] [Google Scholar]
- DARPA . 2017. Pandemic Prevention Platform (P3)https://www.darpa.mil/program/pandemic-prevention-platform [Google Scholar]
- Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Empey K.M., Peebles R.S., Jr., Kolls J.K. Pharmacologic advances in the treatment and prevention of respiratory syncytial virus. Clin. Infect. Dis. 2010;50:1258–1267. doi: 10.1086/651603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudinski M.R., Houser K.V., Doria-Rose N.A., Chen G.L., Rothwell R.S.S., Berkowitz N., Costner P., Holman L.A., Gordon I.J., Hendel C.S., VRC 605 study team Safety and pharmacokinetics of broadly neutralising human monoclonal antibody VRC07-523LS in healthy adults: a phase 1 dose-escalation clinical trial. Lancet HIV. 2019;6:e667–e679. doi: 10.1016/S2352-3018(19)30181-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GISAID . Munich; Germany: 2020. Next hCoV-19 App.https://www.gisaid.org/epiflu-applications/next-hcov-19-app/ [Google Scholar]
- Gouglas D., Christodoulou M., Plotkin S.A., Hatchett R. CEPI: Driving Progress Toward Epidemic Preparedness and Response. Epidemiol. Rev. 2019;41:28–33. doi: 10.1093/epirev/mxz012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham B.S. Rapid COVID-19 vaccine development. Science. 2020:eabb8923. doi: 10.1126/science.abb8923. [DOI] [PubMed] [Google Scholar]
- Graham B.S., Sullivan N.J. Emerging viral diseases from a vaccinology perspective: preparing for the next pandemic. Nat. Immunol. 2018;19:20–28. doi: 10.1038/s41590-017-0007-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham B.S., Mascola J.R., Fauci A.S. Novel Vaccine Technologies: Essential Components of an Adequate Response to Emerging Viral Diseases. JAMA. 2018;319:1431–1432. doi: 10.1001/jama.2018.0345. [DOI] [PubMed] [Google Scholar]
- Haynes B.F., Shaw G.M., Korber B., Kelsoe G., Sodroski J., Hahn B.H., Borrow P., McMichael A.J. HIV-Host Interactions: Implications for Vaccine Design. Cell Host Microbe. 2016;19:292–303. doi: 10.1016/j.chom.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes B.F., Burton D.R., Mascola J.R. Multiple roles for HIV broadly neutralizing antibodies. Sci. Transl. Med. 2019;11:eaaz2686. doi: 10.1126/scitranslmed.aaz2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson R., Lu M., Zhou Y., Mu Z., Parks R., Han Q., Hsu A.L., Carter E., Blanchard S.C., Edwards R.J. Disruption of the HIV-1 Envelope allosteric network blocks CD4-induced rearrangements. Nat. Commun. 2020;11:520. doi: 10.1038/s41467-019-14196-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idusogie E.E., Wong P.Y., Presta L.G., Gazzano-Santoro H., Totpal K., Ultsch M., Mulkerrin M.G. Engineered antibodies with increased activity to recruit complement. J. Immunol. 2001;166:2571–2575. doi: 10.4049/jimmunol.166.4.2571. [DOI] [PubMed] [Google Scholar]
- Jaume M., Yip M.S., Cheung C.Y., Leung H.L., Li P.H., Kien F., Dutry I., Callendret B., Escriou N., Altmeyer R. Anti-severe acute respiratory syndrome coronavirus spike antibodies trigger infection of human immune cells via a pH- and cysteine protease-independent FcγR pathway. J. Virol. 2011;85:10582–10597. doi: 10.1128/JVI.00671-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju B., Zhang Q., Ge X., Wang R., Yu J., Shan S., Zhou B., Song S., Tang X., Yu J. Potent human neutralizing antibodies elicited by SARS-CoV-2 infection. bioRxiv. 2020 doi: 10.1101/2020.03.21.990770. [DOI] [PubMed] [Google Scholar]
- Kaiser J. NIH organizes hunt for drugs. Science. 2020;368:351. doi: 10.1126/science.368.6489.351. 351. [DOI] [PubMed] [Google Scholar]
- Klein F., Mouquet H., Dosenovic P., Scheid J.F., Scharf L., Nussenzweig M.C. Antibodies in HIV-1 vaccine development and therapy. Science. 2013;341:1199–1204. doi: 10.1126/science.1241144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Foley B., Giorgi E., Bhattacharya T., Parker M. Spike mutation pipeline reveals the emergence of a more transmissible form of SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.04.29.069054. [DOI] [Google Scholar]
- Kose N., Fox J.M., Sapparapu G., Bombardi R., Tennekoon R.N., de Silva A.D., Elbashir S.M., Theisen M.A., Humphris-Narayanan E., Ciaramella G. A lipid-encapsulated mRNA encoding a potently neutralizing human monoclonal antibody protects against chikungunya infection. Sci. Immunol. 2019;4:eaaw6647. doi: 10.1126/sciimmunol.aaw6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong P.D., Mascola J.R. Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity. 2012;37:412–425. doi: 10.1016/j.immuni.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong P.D., Mascola J.R. HIV-1 Vaccines Based on Antibody Identification, B Cell Ontogeny, and Epitope Structure. Immunity. 2018;48:855–871. doi: 10.1016/j.immuni.2018.04.029. [DOI] [PubMed] [Google Scholar]
- LaBranche C.C., Henderson R., Hsu A., Behrens S., Chen X., Zhou T., Wiehe K., Saunders K.O., Alam S.M., Bonsignori M. Neutralization-guided design of HIV-1 envelope trimers with high affinity for the unmutated common ancestor of CH235 lineage CD4bs broadly neutralizing antibodies. PLoS Pathog. 2019;15:e1008026. doi: 10.1371/journal.ppat.1008026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Liao H.X., Levesque M.C., Nagel A., Dixon A., Zhang R., Walter E., Parks R., Whitesides J., Marshall D.J., Hwang K.K. High-throughput isolation of immunoglobulin genes from single human B cells and expression as monoclonal antibodies. J. Virol. Methods. 2009;158:171–179. doi: 10.1016/j.jviromet.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wei Q., Lin Q., Fang J., Wang H., Kwok H., Tang H., Nishiura K., Peng J., Tan Z. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight. 2019;4:e123158. doi: 10.1172/jci.insight.123158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Los Alamos National Laboratory . Los Alamos, NM; US: 2020. Los Alamos HIV Sequence Database.http://www.hiv.lanl.gov/ [Google Scholar]
- Los Alamos National Laboratory . 2020. SARS-CoV-2 Sequence Analysis Pipeline.https://cov.lanl.gov/ [Google Scholar]
- Lu S., Zhao Y., Yu W., Yang Y., Gao J., Wang J., Kuang D., Yang M., Yang J., Ma C. Comparison of SARS-CoV-2 infections among 3 species of non-human primates. bioRxiv. 2020 doi: 10.1101/2020.04.08.031807. [DOI] [Google Scholar]
- Mendoza P., Gruell H., Nogueira L., Pai J.A., Butler A.L., Millard K., Lehmann C., Suárez I., Oliveira T.Y., Lorenzi J.C.C. Combination therapy with anti-HIV-1 antibodies maintains viral suppression. Nature. 2018;561:479–484. doi: 10.1038/s41586-018-0531-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murin C.D., Wilson I.A., Ward A.B. Antibody responses to viral infections: a structural perspective across three different enveloped viruses. Nat. Microbiol. 2019;4:734–747. doi: 10.1038/s41564-019-0392-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthumani K., Flingai S., Wise M., Tingey C., Ugen K.E., Weiner D.B. Optimized and enhanced DNA plasmid vector based in vivo construction of a neutralizing anti-HIV-1 envelope glycoprotein Fab. Hum. Vaccin. Immunother. 2013;9:2253–2262. doi: 10.4161/hv.26498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIH . 2020. Press Release: NIH clinical trial of investigational vaccine for COVID-19 begins. [Google Scholar]
- Pardi N., Hogan M.J., Pelc R.S., Muramatsu H., Andersen H., DeMaso C.R., Dowd K.A., Sutherland L.L., Scearce R.M., Parks R. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature. 2017;543:248–251. doi: 10.1038/nature21428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N., Secreto A.J., Shan X., Debonera F., Glover J., Yi Y., Muramatsu H., Ni H., Mui B.L., Tam Y.K. Administration of nucleoside-modified mRNA encoding broadly neutralizing antibody protects humanized mice from HIV-1 challenge. Nat. Commun. 2017;8:14630. doi: 10.1038/ncomms14630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines - a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeples L. News Feature: Avoiding pitfalls in the pursuit of a COVID-19 vaccine. Proc. Natl. Acad. Sci. USA. 2020;117:8218–8221. doi: 10.1073/pnas.2005456117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan B.D., Mou H., Zhang L., Guo Y., He W., Ojha A., Parcells M.S., Luo G., Li W., Zhong G. The SARS-CoV-2 receptor-binding domain elicits a potent neutralizing response without antibody-dependent enhancement. bioRxiv. 2020 doi: 10.1101/2020.04.10.036418. [DOI] [Google Scholar]
- Richards J.O., Karki S., Lazar G.A., Chen H., Dang W., Desjarlais J.R. Optimization of antibody binding to FcgammaRIIa enhances macrophage phagocytosis of tumor cells. Mol. Cancer Ther. 2008;7:2517–2527. doi: 10.1158/1535-7163.MCT-08-0201. [DOI] [PubMed] [Google Scholar]
- Robbie G.J., Criste R., Dall’acqua W.F., Jensen K., Patel N.K., Losonsky G.A., Griffin M.P. A novel investigational Fc-modified humanized monoclonal antibody, motavizumab-YTE, has an extended half-life in healthy adults. Antimicrob. Agents Chemother. 2013;57:6147–6153. doi: 10.1128/AAC.01285-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockx B., Kuiken T., Herfst S., Bestebroer T., Lamers M.M., Oude Munnink B.B., de Meulder D., van Amerongen G., van den Brand J., Okba N.M.A. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science. 2020:eabb7314. doi: 10.1126/science.abb7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers T.F., Zhao F., Huang D., Beutler N., Abbott R.K., Callaghan S., Garcia E., He W.-t., Hurtado J., Limbo O. Rapid isolation of potent SARS-CoV-2 neutralizing antibodies and protection in a small animal model. bioRxiv. 2020 doi: 10.1101/2020.05.11.088674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saphire E.O., Schendel S.L., Fusco M.L., Gangavarapu K., Gunn B.M., Wec A.Z., Halfmann P.J., Brannan J.M., Herbert A.S., Qiu X., Viral Hemorrhagic Fever Immunotherapeutic Consortium Systematic Analysis of Monoclonal Antibodies against Ebola Virus GP Defines Features that Contribute to Protection. Cell. 2018;174:938–952.e13, e913. doi: 10.1016/j.cell.2018.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders K.O. Conceptual Approaches to Modulating Antibody Effector Functions and Circulation Half-Life. Front. Immunol. 2019;10:1296. doi: 10.3389/fimmu.2019.01296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders K.O., Wiehe K., Tian M., Acharya P., Bradley T., Alam S.M., Go E.P., Scearce R., Sutherland L., Henderson R. Targeted selection of HIV-specific antibody mutations by engineering B cell maturation. Science. 2019;366:eaay7199. doi: 10.1126/science.aay7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux E., Homad L.J., MacCamy A.J., Parks K.R., Hurlburt N.K., Jennewein M.F., Akins N.R., Stuart A.B., Wan Y.-H., Feng J. Characterization of neutralizing antibodies from a SARS-CoV-2 infected individual. bioRxiv. 2020 doi: 10.1101/2020.05.12.091298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard H.M., Phillips G.L., D Thanos C., Feldmann M. Developments in therapy with monoclonal antibodies and related proteins. Clin. Med. (Lond.) 2017;17:220–232. doi: 10.7861/clinmedicine.17-3-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sok D., Burton D.R. Recent progress in broadly neutralizing antibodies to HIV. Nat. Immunol. 2018;19:1179–1188. doi: 10.1038/s41590-018-0235-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Meulen J., van den Brink E.N., Poon L.L., Marissen W.E., Leung C.S., Cox F., Cheung C.Y., Bakker A.Q., Bogaards J.A., van Deventer E. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med. 2006;3:e237. doi: 10.1371/journal.pmed.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanh Le T., Andreadakis Z., Kumar A., Gómez Román R., Tollefsen S., Saville M., Mayhew S. The COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020;19:305–306. doi: 10.1038/d41573-020-00073-5. [DOI] [PubMed] [Google Scholar]
- Tseng C.T., Sbrana E., Iwata-Yoshikawa N., Newman P.C., Garron T., Atmar R.L., Peters C.J., Couch R.B. Immunization with SARS coronavirus vaccines leads to pulmonary immunopathology on challenge with the SARS virus. PLoS ONE. 2012;7:e35421. doi: 10.1371/journal.pone.0035421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker L.M., Burton D.R. Passive immunotherapy of viral infections: ‘super-antibodies’ enter the fray. Nat. Rev. Immunol. 2018;18:297–308. doi: 10.1038/nri.2017.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292.e6, e286. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Shi W., Chappell J.D., Joyce M.G., Zhang Y., Kanekiyo M., Becker M.M., van Doremalen N., Fischer R., Wang N. Importance of Neutralizing Monoclonal Antibodies Targeting Multiple Antigenic Sites on the Middle East Respiratory Syndrome Coronavirus Spike Glycoprotein To Avoid Neutralization Escape. J. Virol. 2018;92:e02002–e02017. doi: 10.1128/JVI.02002-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2020. Coronavirus disease (COVID-2019) technical guidance.https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance World Health Organization. [Google Scholar]
- WHO . 2020. Draft landscape of COVID-19 candidate vaccines – 20 March 2020.https://www.who.int/blueprint/priority-diseases/key-action/novel-coronavirus-landscape-ncov.pdf?ua=1 [Google Scholar]
- Wiehe K., Bradley T., Meyerhoff R.R., Hart C., Williams W.B., Easterhoff D., Faison W.J., Kepler T.B., Saunders K.O., Alam S.M. Functional Relevance of Improbable Antibody Mutations for HIV Broadly Neutralizing Antibody Development. Cell Host Microbe. 2018;23:759–765.e6, e756. doi: 10.1016/j.chom.2018.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X.Q., Robbie G.J., Wu Y., Esser M.T., Jensen K., Schwartz H.I., Bellamy T., Hernandez-Illas M., Jafri H.S. Safety, Tolerability, and Pharmacokinetics of MEDI4893, an Investigational, Extended-Half-Life, Anti-Staphylococcus aureus Alpha-Toxin Human Monoclonal Antibody, in Healthy Adults. Antimicrob. Agents Chemother. 2016;61:61. doi: 10.1128/AAC.01020-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Zhao J., Mangalam A.K., Channappanavar R., Fett C., Meyerholz D.K., Agnihothram S., Baric R.S., David C.S., Perlman S. Airway Memory CD4(+) T Cells Mediate Protective Immunity against Emerging Respiratory Coronaviruses. Immunity. 2016;44:1379–1391. doi: 10.1016/j.immuni.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Jiang S., Du L. Prospects for a MERS-CoV spike vaccine. Expert Rev. Vaccines. 2018;17:677–686. doi: 10.1080/14760584.2018.1506702. [DOI] [PMC free article] [PubMed] [Google Scholar]