Abstract
Extensive use of gallium arsenide (GaAs) has led to increased exposure to humans working in the semiconductor industry. This study employed physicochemical characterization of GaAs obtained from a workplace, cytotoxicity analysis of damage induced by GaAs in 16HBE cells, RNA-seq and related bioinformatic analysis, qRT-PCR verification and survival analysis to comprehensively understand the potential mechanism leading to lung toxicity induced by GaAs. We found that GaAs-induced abnormal gene expression was mainly related to the cellular response to chemical stimuli, the regulation of signalling, cell differentiation and the cell cycle, which are involved in transcriptional misregulation in cancer, the MAPK signalling pathway, the TGF-β signalling pathway and pulmonary disease-related pathways. Ten upregulated genes (FOS, JUN, HSP90AA1, CDKN1A, ESR1, MYC, RAC1, CTNNB1, MAPK8 and FOXO1) and 7 downregulated genes (TP53, AKT1, NFKB1, SMAD3, CDK1, E2F1 and PLK1) related to GaAs-induced pulmonary toxicity were identified. High expression of HSP90AA1, RAC1 and CDKN1A was significantly associated with a lower rate of overall survival in lung cancers. The results of this study indicate that GaAs-associated toxicities affected the misregulation of oncogenes and tumour suppressing genes, activation of the TGF-β/MAPK pathway, and regulation of cell differentiation and the cell cycle. These results help to elucidate the molecular mechanism underlying GaAs-induced pulmonary injury.
Subject terms: Respiratory tract diseases, Risk factors
Introduction
Gallium arsenide (GaAs) is a semiconductor material widely used in electronic devices, which are in particularly high demand as electronic components for communication equipment1–3. As one of the largest production and processing companies of GaAs, the increasing utilization of GaAs in electronic devices has raised concerns regarding its potential risks to worker health in China. Occupational exposure to GaAs occurs predominantly in the microelectronics industry when workers come into contact with the production of GaAs crystals, ingots and wafers, grinding and sawing operations, device fabrication, and sandblasting and clean-up activities4–6.
GaAs is a crystalline intermetallic solid composed of arsenic (As) and gallium (Ga)7. Biomonitoring of exposure to GaAs, primarily by measuring As in human tissues or body fluids, has several limitations because occupational exposure limits for arsenic have been established in many countries, and the analytical methods available for measuring GaAs are more sensitive than those for gallium6. GaAs is partly dissociated in vivo into inorganic As and Ga, but GaAs are found to have lower solubilities than dissolved arsenic6. There have been sporadic reports of health effects among workers exposed to this highly toxic arsenic compound8. Studies have shown that GaAs are toxic and carcinogenic9–12. Because of the ability of GaAs to cause extensive pulmonary damage in the rat, a TLV-TWA of 0.3 μg/m3 recommended with an A3-Confirmed Animal Carcinogen with Unknown Relevance to Humans, designation13. GaAs particles were observed in the alveolar spaces and in macrophages with significant elevations of lung lipids and proteins with intratracheal instillation of GaAs particulates14. Despite the increasing utilization of GaAs in electronic devices, the potential mechanism governing the effect of GaAs on workers through respiratory exposure under workplace conditions has not been elucidated.
An in vitro model is an important means to study the adverse effects of particulate pollutants on the respiratory tract and the means of action. To assess the effects of particle pollution on the respiratory tract, it is important to develop systems and methods that allow cultured cells to be repeatedly exposed to particles in the mixture. Human bronchial epithelioid cells (16HBE cells) have been successfully used in lung toxicity tests to evaluate the cytotoxicity and metabolism of PM2.5 and gaseous pollutants15.
The objective of this study was to evaluate the cytotoxic effects of GaAs particles on 16HBE cells, to obtain differentially expressed genes induced by GaAs particles, and to reveal potential mechanisms of pulmonary injury after exposure. To better understand the potential molecular mechanisms underlying the pulmonary toxicity of the GaAs particles, 16HBE cells were chosen as the in vitro exposure model, and then RNA-seq, related bioinformatic analysis, qRT-PCR verified analysis and survival curve analysis using The Cancer Genome Atlas (TCGA) data were conducted.
Results
Cytotoxicity induced by GaAs
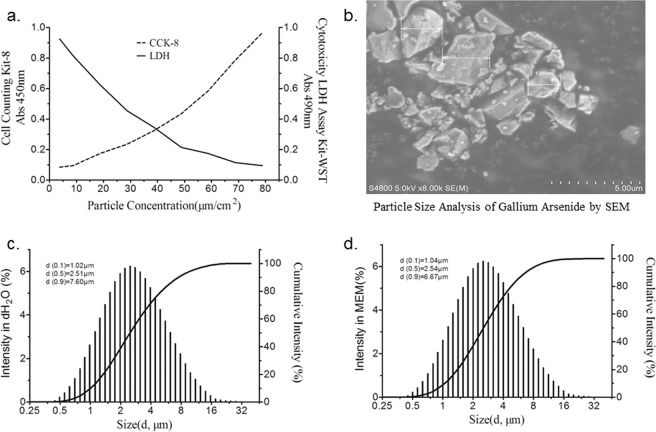
To evaluate the possible cytotoxicity of GaAs particles on 16HBE cells, cell viability was measured after exposing the cells to 5.0, 10.0, 20.0, 30.0, 40.0, 50.0, 60.0, 70.0, and 80.0 μg/cm2 of GaAs particles for 24 h in 96-well plates. As shown in Fig. 1a, cell viability decreased and cell mortality increased gradually in a dose-dependent manner as the concentration increased. As the dose increased to 20.0 μg/cm2, the cell viability lost nearly 15%, as measured with LDH, and cell mortality increased nearly 20%, as measured with CCK-8, which was significant compared with that in the control group. Therefore, 20.0 μg/cm2 was chosen as the experimental dose for the following analysis.
Figure 1.
Assessment of cell viability and characterization of GaAs particles. (a) Cell viability measured with LDH and CCK-8. (b) Scanning electron microscope (SEM) images of the GaAs particles. Scale bar, 0.5 μm. (c) The hydrodynamic sizes of GaAs particles in dH2O. (d) The hydrodynamic sizes of GaAs particles in MEM culture medium.
Physicochemical characterization of GaAs
The characteristics of GaAs particles are given in Fig. 1. GaAs particles appeared as agglomerates of different sizes, which were mainly composed of regular or irregularly shaped ultrafine particles and fine particles. The morphology of the particles obtained from SEM is mostly irregular, as shown in Fig. 1b. The hydrodynamic diameter of GaAs particles measured in dH2O and in MEM culture medium are shown in Fig. 1c,d. The hydrodynamic size (median) of GaAs particles is 2.51 μm or 2.54 μm when GaAs particles are suspended in dH2O or MEM, respectively.
The average concentrations of As and Ga in MEM culture medium after exposure for 24 h were 1.0 and 0.8 μg/mL. In the negative control, the concentrations were less than 0.02 μg/mL. Other heavy metals, including Be, Al, Ti, V, Cr, Mn, Co, Ni, Cu, Se, Sr, Zr, Mo, Pd, Ag, Cd, Cs, and Pb, were detected in MEM culture medium after exposure for 24 h. Only Mo and Pd were significantly different from the NC group, which was significantly higher than the control group. The average concentrations of Mo and Pd were 18.5 and 8.0 μg/L, respectively.
Differential gene expression induced by GaAs
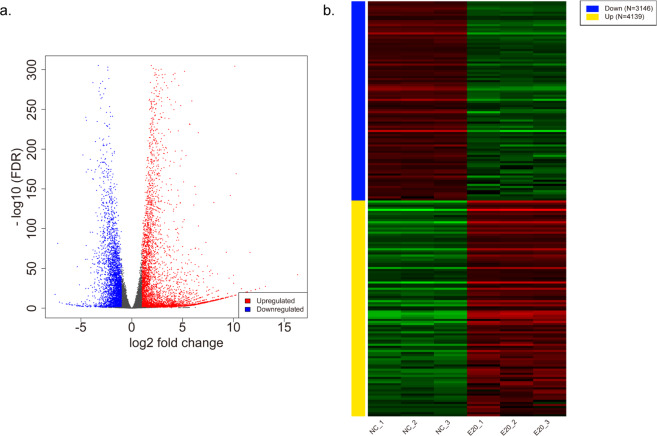
We performed RNA-seq to investigate the possible gene expression change in 16HBE cells induced by GaAs. After quality control of raw sequencing data, the clean reads were mapped to the human reference genome (hg19), and the median total mapping rates were 94.63% in the NC group (93.9%–94.79%) and 87.06% in the E20 group (86.85%–87.72%). The mRNA expression levels and transcripts were estimated with FPKMs. A total of 7285 mRNA transcripts, including 4139 upregulated and 3146 downregulated transcripts (p < 0.05), were differentially expressed in the EC20 group relative to the NC group (Fig. 2a). These differentially expressed mRNAs were used for subsequent analysis. Cluster analysis of differentially expressed mRNAs was conducted with heat maps (Fig. 2b). The top 20 differentially expressed upregulated and downregulated genes between the NC group and GaAs-treated group are listed in Table 1.
Figure 2.
Differentially expressed genes (DEGs) induced by GaAs in 16HBE cells. (a) Volcano plot showing that 3146 genes were downregulated and 4139 genes were upregulated between the GaAs and NC groups. DEGs with log2FoldChange (log2FC)>1 are labelled in red; DEMs with log2FoldChange (log2FC) <−1 are labelled in blue (P < 0.05). (b) The DEG expression profiles of mRNAs are presented as a heatmap.
Table 1.
The top 20 differentially expressed genes between GaAs and NC groups in 16HBE.
| Gene Symbol | Mean of readcount (GaAs) | Mean of readcount (NC) | log2FoldChange | padj | |
|---|---|---|---|---|---|
| Up regulated genes | CRYAB | 13354.47715 | 0 | 16.71152296 | 1.34E-44 |
| HSPA6 | 570082.8219 | 15.59458786 | 15.1533678 | 0 | |
| RFPL4A | 2874.493451 | 0.190242569 | 13.53369186 | 1.39E-29 | |
| CT45A1 | 992.143666 | 0 | 12.96069412 | 4.14E-27 | |
| SERPINA7 | 758.5311161 | 0 | 12.57396271 | 1.72E-25 | |
| CCL26 | 1455.726687 | 0.228898208 | 12.55200595 | 1.52E-25 | |
| NEFM | 554.9236214 | 0 | 12.1230176 | 1.04E-23 | |
| RFPL4AL1 | 433.9097466 | 0 | 11.7672136 | 2.42E-22 | |
| KRTAP2-3 | 428.0256979 | 0 | 11.74782879 | 2.78E-22 | |
| PRR9 | 781.3895624 | 0.190242569 | 11.65379393 | 5.04E-22 | |
| FMR1NB | 392.3592048 | 0 | 11.62220187 | 8.46E-22 | |
| CD300LB | 358.4648122 | 0 | 11.49310827 | 3.14E-21 | |
| FGF19 | 336.4592526 | 0 | 11.40062334 | 5.94E-21 | |
| ARC | 4688.385657 | 1.676563105 | 11.35752989 | 4.56E-106 | |
| MLC1 | 324.872728 | 0 | 11.35041552 | 8.96E-21 | |
| ZNF556 | 314.8386264 | 0 | 11.30504573 | 1.32E-20 | |
| TEX19 | 1042.109777 | 0.453565577 | 11.18744001 | 8.36E-27 | |
| LBH | 480.1538676 | 0.263323009 | 10.95168561 | 1.55E-19 | |
| PRSS55 | 198.501012 | 0 | 10.63995065 | 3.96E-18 | |
| ITK | 190.6265953 | 0 | 10.5810697 | 5.95E-18 | |
| Down regulated genes | PLEKHS1 | 0.914321697 | 209.7064732 | −7.761268855 | 4.09E-13 |
| PCSK9 | 5.228998183 | 932.7962655 | −7.454393368 | 4.88E-61 | |
| GBP4 | 0.457160848 | 55.93539395 | −6.748976858 | 1.85E-07 | |
| SLC16A7 | 0 | 24.04444067 | −6.491580583 | 3.12E-06 | |
| PRR15L | 0 | 18.22963028 | −6.092787428 | 2.85E-05 | |
| KCNIP2 | 0 | 17.03020545 | −5.990095124 | 5.25E-05 | |
| VASH1 | 0.517292735 | 30.63114054 | −5.878827459 | 1.56E-05 | |
| UGT1A6 | 0.457160848 | 29.38882924 | −5.816345507 | 2.16E-05 | |
| CHST4 | 0 | 14.2640851 | −5.738229644 | 0.000142003 | |
| TNFSF10 | 0.937671191 | 51.45654047 | −5.735989035 | 5.74E-07 | |
| JMJD7 | 0 | 14.0599548 | −5.714919551 | 0.000168574 | |
| LCN12 | 0 | 12.61771595 | −5.559550172 | 0.000360681 | |
| FGD3 | 7.852244248 | 353.8214632 | −5.512552741 | 9.16E-42 | |
| DEFB131B | 0 | 11.8362086 | −5.468792097 | 0.000480387 | |
| ALDH3B2 | 4.508505043 | 186.9671612 | −5.420111415 | 1.07E-23 | |
| GOLGA8O | 0 | 11.38144781 | −5.409920859 | 0.000662611 | |
| FAM198B | 0.517292735 | 20.6261176 | −5.308205572 | 0.000235478 | |
| EDDM13 | 0.457160848 | 19.45743674 | −5.225385183 | 0.00028589 | |
| MXRA5 | 0.517292735 | 17.57855567 | −5.080106293 | 0.000506725 | |
| TMX2-CTNND1 | 0 | 8.823688589 | −5.052477188 | 0.003958624 |
GO analysis of differential gene expression induced by GaAs
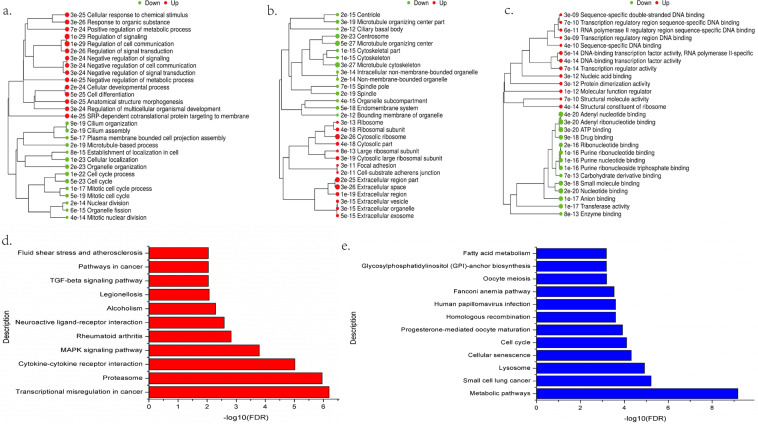
First, GO and KEGG analyses were performed on 7285 significantly dysregulated mRNAs in GaAs vs NC. We derived 85 highly enriched GO terms (adj. P val <0.05) and 27 significantly enriched pathways (adj. P val <0.05). The top 10 significant GO terms during biological process (BP), cellular component (CC) and molecular function (MF) induced by GaAs are displayed in Fig. 3a–c. Several GO terms, such as cellular response to chemical stimulus, regulation of signalling, cell differentiation, cell cycle, focal adhesion, transcription regulator activity and small molecule binding, were closely related to GaAs exposure. As shown in Fig. 3d, the significantly upregulated pathways induced by GaAs included transcriptional misregulation in cancer, cytokine-cytokine receptor interaction, MAPK signalling pathway, TGF-β signalling pathway and pathways in cancer. Significantly downregulated pathways, such as metabolic pathways, small cell lung cancer, cell cycle, and fatty acid metabolism, were detected (Fig. 3e). The summaries of genes involved in the significant up- and downregulation pathways are listed in Supplementary Table S2.
Figure 3.
Significantly changed up- and downregulated GO terms and pathways of differentially expressed genes (DEGs) induced by GaAs in 16HBE cells. (a) Significant up- and downregulation GO terms during BP; (b) Significant up- and downregulation GO terms during CC; (c) Significant up- and downregulation GO terms during MF; (d) Significantly changed pathways of differentially expressed upregulated genes based on the KEGG database. −log10(FDR), negative logarithm of the adjusted P value. FDR < 0.05 was identified as a significantly changed pathway. (e) Significantly changed pathways of differentially expressed downregulated genes based on the KEGG database.
DO analysis of differential gene expression induced by GaAs
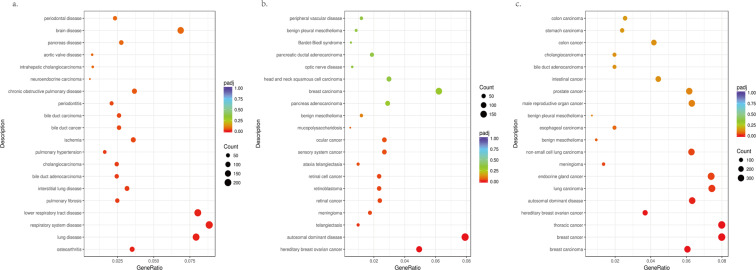
DO (Disease Ontology) is a database describing human gene function and disease that can be used to consider the interactions and functions of differentially expressed genes with disease. The top 20 enrichment terms related to human disease were recognized based on the DO database. The P < 0.05 gene list was also analysed for DO annotation, enabling us to study gene-disease relationships. Significantly upregulated genes induced by GaAs in 16HBE using DO analysis may be related to lung disease, respiratory system disease, lower respiratory tract disease, pulmonary fibrosis, interstitial lung disease and chronic obstructive pulmonary disease (Fig. 4a). For example, hereditary breast ovarian cancer, retinal cancer and ocular cancer were relevant to the downregulation of DO-related diseases (Fig. 4b). Taken together, these results indicate that dysregulated DO of differentially expressed genes induced by GaAs in 16HBE may be related to lung carcinoma, non-small-cell lung carcinoma and thoracic cancer (Fig. 4c).
Figure 4.
Disease ontology (DO) enrichment analysis of differentially expressed genes (DEGs) induced by GaAs in 16HBE cells. The x-axis indicates the number of enriched genes in the given DO category mapping to the size of the dots. The colour-coding indicates the adjusted p value. (a) Significantly upregulated Dos; (b) Significantly downregulated Dos; (c) Significantly dysregulated regulatory Dos.
Protein-protein interaction (PPI) analysis revealed the key genes triggered by GaAs
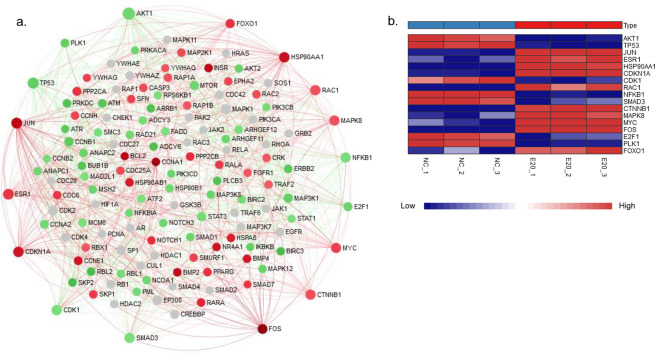
PPI analysis of the STRING interactome was used to screen the key genes involved in GaAs-induced toxicity in 16HBE cells. According to the significant genes from the significant pathway and DO analysis, we next focused on the DEGs in the cell cycle pathway, pathways in cancer, transcriptional misregulation in cancer, small-cell lung cancer, the MAPK signalling pathway, the TGF-β signalling pathway and non-small-cell lung cancer, and the corresponding significant genes were mapped to the corresponding molecular interaction database (Fig. 5a). Seventeen key genes with the top degree (>20) involved in the above 6 pathway interaction network were screened (Table 2), which may play an important role in GaAs-induced pulmonary toxicity in 16HBE cells. These 17 key genes consisted of 10 upregulated genes (FOS, JUN, HSP90AA1, CDKN1A, ESR1, MYC, RAC1, CTNNB1, MAPK8 and FOXO1) and 7 downregulated genes (TP53, AKT1, NFKB1, SMAD3, CDK1, E2F1 and PLK1) (Fig. 5b).
Figure 5.
Protein-protein interaction (PPI) network of differentially expressed genes (DEGs) induced by GaAs in 16HBE cells. The PPI network was drawn using the NetworkAnalyst platform based on the STRING interactome. (a) The red circles represent upregulated genes, the green circles represent downregulated genes, and the grey circles indicate no DEGs. The top degree (>20) involved in the interaction network was screened; (b) The heatmap shows the expression level of the 17 key genes in the RNA-seq data.
Table 2.
The top genes ranked by degree over 20 in PPI analysis.
| Gene symbol | Description | KEGG Pathways | gene_chr | Degree | Betweenness | log2FoldChange (GaAs vs.NC) |
|---|---|---|---|---|---|---|
| AKT1 | v-akt murine thymoma viral oncogene homolog 1, protein_coding | Small cell lung cancer | 14 | 57 | 972.3 | −1.961368001 |
| TP53 | tumor protein p53, protein_coding | Small cell lung cancer, Cell cycle | 17 | 41 | 470.66 | −2.592326592 |
| JUN | jun proto-oncogene, protein_coding | Pathways in cancer, MAPK signaling pathway | 1 | 39 | 282.42 | 4.837850274 |
| ESR1 | estrogen receptor 1, protein_coding | Pathways in cancer | 6 | 37 | 337.46 | 1.787520803 |
| HSP90AA1 | heat shock protein 90 kDa alpha (cytosolic), class A member 1, protein_coding | Pathways in cancer | 14 | 34 | 417.35 | 3.926057435 |
| CDKN1A | cyclin-dependent kinase inhibitor 1A (p21, Cip1), protein_coding | Pathways in cancer, Transcriptional misregulation in cancer | 6 | 34 | 301.04 | 3.632760311 |
| CDK1 | cyclin-dependent kinase 1, protein_coding | Cell cycle | 10 | 33 | 346.53 | −1.423420996 |
| RAC1 | ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1), protein_coding | Pathways in cancer, MAPK signaling pathway | 7 | 33 | 297.85 | 1.045045893 |
| NFKB1 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1, protein_coding | Small cell lung cancer | 4 | 32 | 372.86 | −1.465472937 |
| SMAD3 | SMAD family member 3, protein_coding | Cell cycle | 15 | 32 | 242.91 | −1.428252719 |
| CTNNB1 | catenin (cadherin-associated protein), beta 1, 88 kDa, protein_coding | Pathways in cancer | 3 | 32 | 264.91 | 1.011091149 |
| MAPK8 | mitogen-activated protein kinase 8, protein_coding | Pathways in cancer, MAPK signaling pathway | 10 | 32 | 260.71 | 1.011586254 |
| MYC | v-myc myelocytomatosis viral oncogene homolog (avian), protein_coding | Pathways in cancer, MAPK signaling pathway, Transcriptional misregulation in cancer, TGF-beta signaling pathway | 8 | 31 | 190.63 | 1.439621447 |
| FOS | FBJ murine osteosarcoma viral oncogene homolog, protein_coding | Pathways in cancer, MAPK signaling pathway | 14 | 28 | 98.25 | 7.328134023 |
| E2F1 | E2F transcription factor 1, protein_coding | Small cell lung cancer, Cell cycle | 20 | 21 | 72.8 | −2.011988593 |
| PLK1 | polo-like kinase 1, protein_coding | Cell cycle | 16 | 20 | 322.92 | −2.341394476 |
| FOXO1 | forkhead box O1, protein_coding | Pathways in cancer, Transcriptional misregulation in cancer | 13 | 23 | 93.93 | 1.906896342 |
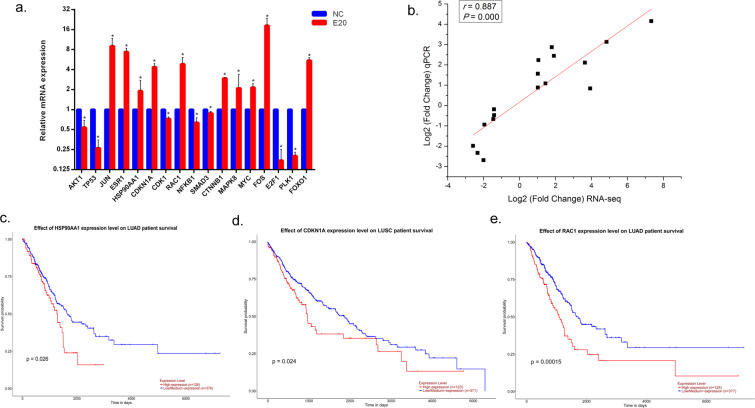
qRT-PCR verification and survival curve analysis
To verify the RNA-seq analysis results, the expression of the above 17 significantly dysregulated genes selected from PPIs was verified using qRT-PCR (Fig. 6a). These selected genes were related to various pathways (Table 2), such as small-cell lung cancer (AKT1, TP53, NFKB1 and E2F1),
Figure 6.
qRT-PCR verification and survival curve analysis. (a) Gene expression by qRT-PCR analysis. The fold changes were calculated by using the 2−ΔΔCt method comparing the GaAs-treated group to the NC group. Data are expressed as the means ± S.D. from 3 biological repeats and 3 technical repeats. *P < 0.05 compared to control groups by Mann-Whitney test. (b) The potential correlation between the expression of dysregulated genes from RNA-seq and qRT-PCR verification was analysed using Spearman’s rank test. A P value less than 0.05 was considered statistically significant. (c) TCGA data from UALCAN demonstrated that high expression of HSP90AA1 predicted a significantly poor prognosis of lung adenocarcinoma (LUAD) patients (P < 0.05); (d) TCGA data from UALCAN demonstrated that high expression of RAC1 predicted a significantly poor prognosis of lung squamous cell carcinoma (LUSC) patients (P < 0.05); (e) TCGA data from UALCAN demonstrated that high expression of CDKN1A predicted a significantly poor prognosis of LUAD patients (P < 0.05).
MAPK signalling pathway (JUN, RAC1, MAPK8, MYC and FOS) and transcriptional misregulation in cancer (CDKN1A, MYC and FOXO1). The potential correlation between the expression of the above 17 significantly dysregulated genes from RNA-seq and qRT-PCR verification was analysed with the Spearman rank order correlation test. We found that the gene expression results of qPCR were positively correlated with the corresponding results of RNA-seq (Fig. 6b; r = 0.887, P = 0.000). The results suggest that good consistency was obtained between RNA-seq analysis and qRT-PCR verification.
Then, we performed 10 upregulated genes for survival curve analysis using TCGA data by UALCAN. Further survival analyses on these key genes were employed to evaluate their effects on the survival of lung cancer. Figure 6c–e shows that HSP90AA1, RAC1 and CDKN1A expression levels were clearly related to the prognosis of lung adenocarcinoma (LUAD) or lung squamous cell carcinoma (LUSC) patients, which indicated that high expression of the three genes was significantly associated with a lower rate of overall survival.
Discussion
Workers in the semiconductor manufacturing industry are potentially exposed to inhalable GaAs dusts. Because GaAs has a lower solubility than any other arsenic compound and is associated with lung toxicity, even linked to lung neoplasm risk, exploring the toxicogenomic effects of GaAs on human bronchial epithelial cells is critical to understanding the mechanisms of these adverse health effects7,16. In this study, we first examined the physicochemical characterization of GaAs from a workplace and cytotoxicity induced by GaAs and then conducted RNA-seq, related bioinformatic analysis, qRT-PCR verification and survival analysis to comprehensively understand potential genes and pathways that lead to lung toxicity induced by GaAs. Our aim was to elucidate the role of GaAs in 16HBE cells and to provide a mechanistic explanation for GaAs exposure increasing pulmonary injury risk in humans. This research was the first attempt to perform transcriptome sequencing analysis to fully understand GaAs-induced toxicity in 16HBE cells.
GaAs could release gallium and arsenic moieties in vitro though a relatively lower dissolution rate17. In our study, the dissolution rate of GaAs particles was approximately 2.3% after 24 h of exposure to 16HBE cells in the cell supernatant. In addition to dissolution of the above two metals, the toxic effect depends on particle size, exposure duration and exposure route18. We collected the GaAs from a workplace that reflects the real environmental exposure, in which the hydrodynamic median sizes of GaAs particles are 2.51 μm or 2.54 μm when suspended in dH2O or MEM, respectively. It was demonstrated that micron-sized GaAs particles can result in hazardous pulmonary effects in animal studies, which indicates that a smaller fraction of GaAs is a relatively more severe pneumotoxicant19,20. It was reported that a strong inflammatory response induced by solid GaAs in the lungs disappeared when the gallium and arsenic oxides dissolved, as shown in animal models, in which the unchanged GaAs particles were cleared from the lung20. Then, the toxic effects of micron-sized GaAs particles exposed to 16HBE were evaluated.
The lung is one of the target organs for the toxic effects of GaAs, as this organ has higher absorption after intratracheal administration than is observed after oral dosing5. In inhalation studies, lung retention of inhaled GaAs dust has been shown to be influenced by toxic effects from GaAs itself19. GaAs is a compound of gallium and arsenic that is responsible for the toxicity properties of these two metals. Gallium is considered mildly toxic because most of this metal is excreted through urine or faeces, which does not contribute significantly to the lung toxicity of GaAs21. Arsenic exposure has been shown to induce human tumourigenesis, and the lung is one of the main targets22. Toxicities of arsenic-induced carcinogenicity, including generation of oxidative stress, altered cell proliferation, changes in DNA methylation and co-carcinogenesis, though the exact molecular mechanism governing this toxicity is not well understood18,23,24. The toxicity of GaAs in pulmonary tissue includes inflammation, lung weight increase, fibrosis, seropurulent pneumonia and pneumocyte hyperplasia18,20,25.
Our results indicated that GaAs induced abnormal gene expression mainly related to cellular response to chemical stimulus, regulation of signalling, cell differentiation, the cell cycle and regulation of signalling. GaAs exposure in 16HBE cells induced the upregulation of cell differentiation-related genes and the downregulation of cell cycle-related genes, which indicated that GaAs is an inducer of cell differentiation. In 16HBE cells after 24 h of GaAs exposure, we also observed morphological changes, such as irregular shapes, smaller volumes and abnormal nuclei. GaAs-related dysregulated genes are involved in signal transduction pathways, such as transcriptional misregulation in cancer, cytokine-cytokine receptor interactions, the MAPK signalling pathway and the TGF-β signalling pathway. It has been summarized that arsenic-induced lung tumours may, through disruption of the PI3K/AKT signalling pathway, activate the EGFR signalling pathway and affect the NRF2 signalling pathway to play a role in carcinogenesis22. The TGF-β and MAPK pathways play critical roles in cell development and cell cycle regulation, even in tumour formation and metastasis. It has been shown that activation of MAPK and TGF-β could be induced by ROS accumulation, and the MAPK pathway and TGF-β pathways were closely related to lung fibrosis26,27. TGF-β can activate MAPK signalling, and inhibition of the TGF-β/MAPK pathway could protect against lung fibrosis28,29, which may explain the GaAs toxicity in pulmonary tissue. Then, a view of the global gene-disease relationships of GaAs exposure in 16HBE was obtained by DO analysis. Lung disease and respiratory system disease were found in significantly upregulated genes induced by GaAs in 16HBE. All dysregulated genes in the significant DO term were clustered into tumour-related diseases, including lung carcinoma and non-small-cell lung carcinoma. These results indicated that pulmonary disease-related pathways were affected when GaAs exposure occurred in 16HBE.
Then, we screened the 17 key genes with the top degree (>20) involved in significantly related pathways of GaAs-induced pulmonary toxicity in 16HBE cells, whose gene expression levels were verified by qPCR. Among these key genes, several oncogenes (FOS, JUN, MYC) and tumour suppressor genes (TP53) were associated with lung cancer development. ESR1/2 might directly or indirectly regulate oncogenic pathways in non-small-cell lung cancer, and FOXO1 expression is a favourable prognostic factor in non-small-cell lung cancer30,31. CTNNB1 has been found to be genetically mutated in various human cancers, including lung adenocarcinoma, whose gene expression was increased in lung tissue with pulmonary fibrosis32. MAPK8 was found to be potentially related to lung cancer33. Finally, the survival curve analysis of 10 upregulated genes using TCGA data indicated that high expression of HSP90AA1, RAC1 and CDKN1A induced by GaAs was clearly related to the lower survival probability in LUAD or LUSC patients. HSP90AA1 was found to be directly associated with lung cancer34. The overexpression of Rac1 was linked to aggressive growth and other malignant characteristics of tumours, and a high level of Rac1 could predict a poor prognosis in different types of cancer35. CDKN1A expression is upregulated in long-term oxidative stress-induced experimental bronchopulmonary dysplasia and in a hyperoxia-induced lung injury rat model36,37. The results of this study indicate that GaAs-associated toxicities affect the misregulation of oncogenes and tumour suppressing genes, activation of the TGF-β/MAPK pathway, and regulation of cell differentiation and the cell cycle.
Methods
Particle sample collection
GaAs particles, obtained from a manufacturer of gallium arsenide crystals in Beijing, were collected using an IOM personal sampler (SKC USA) equipped with a cylindrical body, 37 mm cassette, and PVC filter with enough time. Then, the samples were rinsed with distilled water, and the weight change was recorded to calculate the collection quality of the particles. We chose the wafer manufacturing process as the sampling site in which workers are only exposed to GaAs particles.
Cell culture
The human bronchial epithelial cell line (16HBE14O-, abbreviated as 16HBE) was a gift from Dr D.C. Gruenert (University of California, San Francisco, USA). The cells were maintained in MEM culture medium (Gibco, USA) supplemented with 10% foetal bovine serum (Gibco, USA), 100 U/mL penicillin and 100 μg/mL streptomycin and cultured at 37 °C in a 5% CO2 humidified environment.
Assessment of cell viability
To evaluate the cytotoxicity and cell viability induced by GaAs particles, the Cell Counting Kit-8 (CCK-8) assay and Cytotoxicity LDH Assay Kit-WST (LDH)38 assay were employed to assess mitochondrial dehydrogenase activity and loss of cell integrity, respectively. Three biological replicates were performed. Cytotoxicity EC20 (effective concentration resulting in 20%) of 16HBE cells exposed to GaAs particles was determined. The cells were exposed to GaAs particles at 5.0, 10.0, 20.0, 30.0, 40.0, 50.0, 60.0, 70.0, and 80.0 μg/cm2 for 24 h in a 96-well orifice plate. The cells were incubated for an additional 1 h at 37 °C. The detailed procedures of the CCK-8 assay and LDH assay are available as the product operation instructions. Medium-only treated cells were used as negative control exposures. Optical density at 450 nm and 490 nm was detected by microplate reader (Themo Multiskan MK3, USA), respectively. These data were used to determine appropriate GaAs particle exposure concentrations for subsequent assays.
Physicochemical characterization of GaAs particles
Scanning electron microscopy (SEM HITACHI S4800) was used to examine the particle morphology. Characterization of GaAs particles was performed at a concentration identical to the cytotoxicity EC20. The hydrodynamic diameter of GaAs particles was determined by dynamic light scattering (DLS) using a MASTERSIZER 2000 (Malvern Instruments, Malvern, UK). GaAs particles were suspended in dH2O and in MEM at the concentration of cytotoxicity EC20 and sonicated prior to measurement.
Dissolution of GaAs particles was assessed in full MEM. GaAs particles were incubated at the concentration of Cytotoxicity EC20 for 24 h at 37 °C. Three biological replicates were performed. After incubation, the samples were centrifuged at 25,000 × g for 30 min, and the supernatant was used for Ga and As determination by inductively coupled plasma mass spectrometry (ICP-MS, Thermo).
GaAs particle exposure
For experiments, the cells were seeded in culture plates at a density of 1 × 105 cells/mL, allowed to attach for 24 h, and treated with GaAs particles suspended in MEM culture medium of certain concentrations for another 24 h. A suspension of GaAs particles was dispersed by a sonicator (Bioruptor UDC-200, Belgium), diluted to EC20 concentrations, and immediately added to 16HBE cells. In this study, 6-well culture plates were used, the bottom area of which was 9.6 cm2. To ensure that each unit area of cells was exposed to the same amount of GaAs particles, the exposure volume of the GaAs particle suspension was calculated according to the bottom area of the cell culture plate. Cells maintained in MEM culture medium without GaAs particles were used as the control group. Three biological replicates were performed.
Total RNA extraction and RNA-seq
Total RNA was extracted from the 16HBE cells exposed to EC20 of GaAs particles and negative control (NC) using TRIzol reagent (Invitrogen, San Diego, CA, USA) according to the manufacturer’s instructions. Both groups were conducted in triplicate. A total amount of 3 μg RNA per sample was used as input material for the RNA sample preparations. Sequencing libraries were generated using the NEBNext® UltraTM RNA Library Prep Kit for Illumina® (NEB, USA), and index codes were added to attribute sequences to each sample. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumina). After cluster generation, the library preparations were sequenced on an Illumina HiSeq platform to generate 150 bp paired-end reads (Novogene, Beijing).
Bioinformatic analysis
The raw sequence files generated from 6 files (bam) in this study has been deposited to NCBI’s Sequence Read Archive (SRA) database with the accession number PRJNA623863. Clean data (clean reads) were obtained by removing reads containing adapters, reads containing poly-N and low-quality reads from raw data using in-house Perl scripts (Novogene, Beijing). FPKM (fragments per kilobase of exon per million fragments) of each gene was calculated based on the length of the gene and read count mapped to this gene. Differential expression analysis of two groups (three biological replicates per group) was performed using the DESeq. 2 R package (1.16.1). The resulting P-values were adjusted using Benjamini and Hochberg’s approach for controlling the false discovery rate. Genes with an adjusted P-value <0.05 found by DESeq. 2 were assigned as differentially expressed.
Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of differentially expressed genes was implemented using the clusterProfiler R package and iDEP program39, in which gene length bias was corrected. Identified GO terms and KEGG pathways with a corrected p < 0.05 were considered significantly enriched40. Disease ontology (DO) annotates human gene-disease relationships, which is important annotation in translating molecular findings from high-throughput data to clinical relevance41. DO disease terms and semantic associations were obtained through the DOSE R package, which provided DO terms with a corrected p < 0.05 were considered significantly enriched. Protein-protein interaction (PPI) analysis of differentially expressed genes was based on the STRING database, and the images were examined using the NetworkAnalyst platform42.
Quantitative real-time PCR (qRT-PCR) analysis
The RNeasy Mini Kit (Qiagen, Hilden, Germany) was used to isolate total RNA from 16HBE cells exposed to EC20 of GaAs particles and NC, as described above for the GaAs particle exposure method. Reverse transcription to synthesize first-strand cDNA was conducted using the SuperScript II First-stand Synthesis System for RT-PCR (Invitrogen, USA). The gene expression levels were assessed with the use of TaqMan qRT-PCR assays to validate the GaAs particle-related key genes acquired from RNA-seq. All TaqMan qRT-PCR reactions were carried out using TaqMan® Fast Advanced Master Mix (Applied Biosystems, USA) on a ViiA 7 Real-Time PCR system (Applied Biosystems, USA) in 384-well plates. The relative mRNA expression levels of the target genes were normalized to GAPDH (housekeeping gene), and the fold change was calculated by using the 2−ΔΔCt method. The primers and reaction conditions are listed in Supplementary Table S1.
Survival analysis
The survival analysis of key GaAs particle exposure-related genes was performed on UALCAN (http://ualcan.path.uab.edu/) to conduct the Kaplan–Meier estimator43. The survival curves of samples with high gene expression and low/medium gene expression were compared by the log rank test. P values <0.05 were considered to be significant.
Statistical analysis
Statistical analysis was performed using SPSS 17.0 software (SPSS, Chicago, IL, USA). The Mann-Whitney test was used to compare the qPCR results between groups. The potential correlation between the expression of dysregulated genes from RNA-seq and qRT-PCR verification was analysed using Spearman’s rank test. P values less than 0.05 were considered to be significant.
Supplementary information
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81601796), the Beijing Talents Fund Supported Young Backbone project (2015000021469G183) and Capital Health Research and Development Special grants (2018-1-1151).
Author contributions
Y.O. performed and analyzed the RNA-seq results, drafted and wrote the manuscript. X.L., H.L. and S.C. determined the hydrodynamic diameter of GaAs particles using a MASTERSIZER 2000, and assessed the dissolution of GaAs particles in full MEM after exposed using ICP-MS. H.Y. contributed to the conception and design of the study and analysis of the data. X.P. contributed to the conception and design of the study, assisted in writing the manuscript, and gave final approval of the manuscript submitted for publication. All authors read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Huifang Yan, Email: yan_huifang@hotmail.com.
Xingfu Pan, Email: pan_xingfu@hotmail.com.
Supplementary information
is available for this paper at 10.1038/s41598-020-65518-8.
References
- 1.Chakrabarti NB. Gaas Integrated Circuits. Iete J. Res. 1992;38:16. doi: 10.1080/03772063.1992.11437044. [DOI] [Google Scholar]
- 2.Gray, F., Kramer, D. A. & Bliss, J. D. Gallium and Gallium Compounds, 2000).
- 3.Adriana, R. R., A, F. J. & Wenjie, S. Gallium Arsenide (Gaas) Leaching Behavior and Surface Chemistry Changes in Response to Ph and O-2. Waste Manage. 1–9 (2018). [DOI] [PubMed]
- 4.Harrison RJ. Gallium Arsenide. Occup Med. 1986;1:49. [PubMed] [Google Scholar]
- 5.Webb DR, Sipes IG, Carter DE. In Vitro Solubility and in Vivo Toxicity of Gallium Arsenide. Toxicology & Applied Pharmacology. 1984;76:96–104. doi: 10.1016/0041-008X(84)90032-2. [DOI] [PubMed] [Google Scholar]
- 6.Zhou, Q. & Xi, S. A Review On Arsenic Carcinogenesis: Epidemiology, Metabolism, Genotoxicity and Epigenetic Changes. Regul. Toxicol. Pharm. 78–88 (2018). [DOI] [PubMed]
- 7.Carter DE, Aposhian HV, Gandolfi AJ. The Metabolism of Inorganic Arsenic Oxides, Gallium Arsenide, and Arsine: A Toxicochemical Review. Toxicology & Applied Pharmacology. 2003;193:309–334. doi: 10.1016/j.taap.2003.07.009. [DOI] [PubMed] [Google Scholar]
- 8.Burns LA, Sikorski EE, Saady JJ, Munson AE. Evidence for Arsenic as the Immunosuppressive Component of Gallium Arsenide. Toxicology & Applied Pharmacology. 1991;110:157–169. doi: 10.1016/0041-008X(91)90298-S. [DOI] [PubMed] [Google Scholar]
- 9.Hartmann CB, Mccoy KL. Gallium Arsenide Augments Antigen Processing by Peritoneal Macrophages for Cd4 + Helper T Cell Stimulation. Toxicology & Applied Pharmacology. 1996;141:365–372. doi: 10.1006/taap.1996.0301. [DOI] [PubMed] [Google Scholar]
- 10.Omura M, et al. Testicular Toxicity Evaluation of Arsenic-Containing Binary Compound Semiconductors, Gallium Arsenide and Indium Arsenide, in Hamsters. Toxicol. Lett. 1996;89:123–129. doi: 10.1016/S0378-4274(96)03796-4. [DOI] [PubMed] [Google Scholar]
- 11.Omura M, et al. Testicular Toxicity of Gallium Arsenide, Indium Arsenide, and Arsenic Oxide in Rats by Repetitive Intratracheal Instillation. Fundamental & Applied Toxicology. 1996;32:72–78. doi: 10.1006/faat.1996.0108. [DOI] [PubMed] [Google Scholar]
- 12.Burns LA, Munson AE. Gallium Arsenide Selectively Inhibits T Cell Proliferation and Alters Expression of Cd25 (Il-2R/P55) Journal of Pharmacology & Experimental Therapeutics. 1993;265:178–186. [PubMed] [Google Scholar]
- 13.ACGIH. Documentation of the Tlvs and Beis with Other Worldwide Occupational Exposure Values, 2015).
- 14.Yamauchi, H. Arsenic Contamination in Asia:Biological Effects and Preventive Measures. 1st edition. edn, 2018).
- 15.Jin X, Xue B, Zhou Q, Su R, Li Z. Mitochondrial Damage Mediated by Ros Incurs Bronchial Epithelial Cell Apoptosis upon Ambient Pm2.5 Exposure. J. Toxicol. Sci. 2018;43:101–111. doi: 10.2131/jts.43.101. [DOI] [PubMed] [Google Scholar]
- 16.Hong Wen C. Exposure and Health Risk of Gallium, Indium, and Arsenic From Semiconductor Manufacturing Industry Workers. Bulletin of Environmental Contamination & Toxicology. 2007;78:5–9. doi: 10.1007/s00128-007-9037-6. [DOI] [PubMed] [Google Scholar]
- 17.Pierson B, Wagenen SV, Nebesny KW, Fernando Q, Carter NSDE. Dissolution of Crystalline Gallium Arsenide in Aqueous Solutions Containing Complexing Agents. American Industrial Hygiene Association Journal. 1989;50:455–459. doi: 10.1080/15298668991374985. [DOI] [PubMed] [Google Scholar]
- 18.Flora, S. J. S. & Dwivedi, N. A Toxicochemical Review of Gallium Arsenide (Review Paper). Defence Sci. J. 62, (2012).
- 19.National Toxicology Program. Ntp Toxicology and Carcinogenesis Studies of Gallium Arsenide (Cas No. 1303-00-0) in F344/N Rats and B6C3F1 Mice (Inhalation Studies). National Toxicology Program Technical Report. 492, (2000). [PubMed]
- 20.Webb DR, Wilson SE, Carter DE. Pulmonary Clearance and Toxicity of Respirable Gallium Arsenide Particulates Intratracheally Instilled Into Rats. Am Ind Hyg Assoc J. 1987;48:660–667. doi: 10.1080/15298668791385372. [DOI] [PubMed] [Google Scholar]
- 21.Chitambar CR. Medical Applications and Toxicities of Gallium Compounds. Int J Environ Res Public Health. 2010;7:2337–2361. doi: 10.3390/ijerph7052337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hubaux R, et al. Molecular Features in Arsenic-Induced Lung Tumors. Mol. Cancer. 2013;12:20. doi: 10.1186/1476-4598-12-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abdul KS, Jayasinghe SS, Chandana EP, Jayasumana C, De Silva PM. Arsenic and Human Health Effects: A Review. Environ Toxicol Pharmacol. 2015;40:828–846. doi: 10.1016/j.etap.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 24.Khairul I, Wang QQ, Jiang YH, Wang C, Naranmandura H. Metabolism, Toxicity and Anticancer Activities of Arsenic Compounds. Oncotarget. 2017;8:23905–23926. doi: 10.18632/oncotarget.14733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang W, et al. Pro-Inflammatory and Pro-Fibrogenic Effects of Ionic and Particulate Arsenide and Indium-Containing Semiconductor Materials in the Murine Lung. Acs Nano. 2017;11:1869–1883. doi: 10.1021/acsnano.6b07895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chanda D, et al. Developmental Pathways in the Pathogenesis of Lung Fibrosis. Mol. Aspects Med. 2019;65:56–69. doi: 10.1016/j.mam.2018.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chapnick DA, Warner L, Bernet J, Rao T, Liu X. Partners in Crime: The Tgfbeta and Mapk Pathways in Cancer Progression. Cell Biosci. 2011;1:42. doi: 10.1186/2045-3701-1-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Derynck R, Zhang YE. Smad-Dependent and Smad-Independent Pathways in Tgf-Beta Family Signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 29.Wang D, et al. Protective Effect of Peptide Dr8 On Bleomycin-Induced Pulmonary Fibrosis by Regulating the Tgf-Beta/Mapk Signaling Pathway and Oxidative Stress. Toxicol Appl Pharmacol. 2019;382:114703. doi: 10.1016/j.taap.2019.114703. [DOI] [PubMed] [Google Scholar]
- 30.Gao X, et al. Estrogen Receptors Promote Nsclc Progression by Modulating the Membrane Receptor Signaling Network: A Systems Biology Perspective. J. Transl. Med. 2019;17:308. doi: 10.1186/s12967-019-2056-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maekawa T, et al. Expression and Localization of Foxo1 in Non-Small Cell Lung Cancer. Oncol. Rep. 2009;22:57–64. [PubMed] [Google Scholar]
- 32.Baarsma HA, Konigshoff M. ‘Wnt-Er is Coming’: Wnt Signalling in Chronic Lung Diseases. Thorax. 2017;72:746–759. doi: 10.1136/thoraxjnl-2016-209753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.El-Aarag SA, et al. In Silico Identification of Potential Key Regulatory Factors in Smoking-Induced Lung Cancer. Bmc Med. Genomics. 2017;10:40. doi: 10.1186/s12920-017-0284-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hasan AN, Ahmad MW, Madar IH, Grace BL, Hasan TN. An in Silico Analytical Study of Lung Cancer and Smokers Datasets From Gene Expression Omnibus (Geo) for Prediction of Differentially Expressed Genes. Bioinformation. 2015;11:229–235. doi: 10.6026/97320630011229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma J. Prognostic and Clinicopathological Value of Rac1 in Cancer Survival: Evidence From a Meta-Analysis. J. Cancer. 2018;9:2571–2579. doi: 10.7150/jca.24824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wagenaar GT, et al. Gene Expression Profile and Histopathology of Experimental Bronchopulmonary Dysplasia Induced by Prolonged Oxidative Stress. Free Radic. Biol. Med. 2004;36:782–801. doi: 10.1016/j.freeradbiomed.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 37.Pan YQ, Hou AN. Hyperoxia-Induced Lung Injury Increases Cdkn1a Levels in a Newborn Rat Model of Bronchopulmonary Dysplasia. Exp. Lung Res. 2018;44:424–432. doi: 10.1080/01902148.2018.1479898. [DOI] [PubMed] [Google Scholar]
- 38.Dekkers Susan, Williams Tim D., Zhang Jinkang, Zhou Jiarui (Albert), Vandebriel Rob J., De La Fonteyne Liset J. J., Gremmer Eric R., He Shan, Guggenheim Emily J., Lynch Iseult, Cassee Flemming R., De Jong Wim H., Viant Mark R. Multi-omics approaches confirm metal ions mediate the main toxicological pathways of metal-bearing nanoparticles in lung epithelial A549 cells. Environmental Science: Nano. 2018;5(6):1506–1517. [Google Scholar]
- 39.Ge SX, Son EW, Yao R. Idep: An Integrated Web Application for Differential Expression and Pathway Analysis of Rna-Seq Data. Bmc Bioinformatics. 2018;19:534. doi: 10.1186/s12859-018-2486-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mao X, Tao CJGO, Wei L. Automated Genome Annotation and Pathway Identification Using the Kegg Orthology (Ko) as a Controlled Vocabulary. Bioinformatics. 2005;21:3787–3793. doi: 10.1093/bioinformatics/bti430. [DOI] [PubMed] [Google Scholar]
- 41.Schriml LM, et al. Human Disease Ontology 2018 Update: Classification, Content and Workflow Expansion. Nucleic Acids Res. 2019;47:D955–D962. doi: 10.1093/nar/gky1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jianguo X, Benner MJ, Hancock REW. Networkanalyst–Integrative Approaches for Protein-Protein Interaction Network Analysis and Visual Exploration. Nucleic Acids Res. 2014;42:W167. doi: 10.1093/nar/gku443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chandrashekar DS, et al. Ualcan: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.