Abstract
In most organisms, cells typically maintain genome integrity, as radical genome reorganization leads to dramatic consequences. However, certain organisms, ranging from unicellular ciliates to vertebrates, are able to selectively eliminate specific parts of their genome during certain stages of development. Moreover, partial or complete elimination of one of the parental genomes occurs in interspecies hybrids reproducing asexually. Although several examples of this phenomenon are known, the molecular and cellular processes involved in selective elimination of genetic material remain largely undescribed for the majority of such organisms. Here, we elucidate the process of selective genome elimination in water frog hybrids from the Pelophylax esculentus complex reproducing through hybridogenesis. Specifically, in the gonads of diploid and triploid hybrids, but not those of the parental species, we revealed micronuclei in the cytoplasm of germ cells. In each micronucleus, only one centromere was detected with antibodies against kinetochore proteins, suggesting that each micronucleus comprises a single chromosome. Using 3D-FISH with species-specific centromeric probe, we determined the role of micronuclei in selective genome elimination. We found that in triploid LLR hybrids, micronuclei preferentially contain P. ridibundus chromosomes, while in diploid hybrids, micronuclei preferentially contain P. lessonae chromosomes. The number of centromere signals in the nuclei suggested that germ cells were aneuploid until they eliminate the whole chromosomal set of one of the parental species. Furthermore, in diploid hybrids, misaligned P. lessonae chromosomes were observed during the metaphase stage of germ cells division, suggesting their possible elimination due to the inability to attach to the spindle and segregate properly. Additionally, we described gonocytes with an increased number of P. ridibundus centromeres, indicating duplication of the genetic material. We conclude that selective genome elimination from germ cells of diploid and triploid hybrids occurs via the gradual elimination of individual chromosomes of one of the parental genomes, which are enclosed within micronuclei.
Subject terms: Chromosomes, Nuclear organization, Germline development, Cytogenetics, Polyploidy, Cell division, Genetic hybridization
Introduction
The maintenance of genome integrity is thought to be a crucial characteristic of eukaryotic organisms. However, some organisms have developed sophisticated mechanisms allowing them to selectively eliminate certain parts of their genome (sometimes up to 90%)1–3. The selective elimination of genetic material has been observed in a diverse range of animal groups, which includes chromatin diminution1, programmed DNA rearrangements2,4–7, the elimination of accessory or sex chromosomes8,9, and paternal genome elimination10. Additionally, selective elimination of genetic material in some interspecies hybrids seems to share similarities with the abovementioned processes, as genetic material destined for elimination is generally recognized, marked and selectively eliminated during specific stages of ontogenesis3,11,12. Interspecies hybrids exploit selective genome elimination to reproduce and are known to occur among animals that reproduce without recombination, collectively termed asexuals12–14. In such animals, gametogenesis is modified to produce gametes with an unreduced genome composition, and in the vast majority of these cases the genome of one parental species (usually paternal) is selectively excluded after fertilization (androgenesis, kleptogenesis, gynogenesis) or during gonocyte multiplication (hybridogenesis and triploid hybridogenesis)11,13,15,16. Nevertheless, the mechanisms underlying selective genome elimination are still a mystery.
Genome elimination that accompanies hybridogenetic reproduction is observed in interspecies hybrids of the European water frog (Pelophylax esculentus) complex17,18. Crossings of two parental species – the marsh frog P. ridibundus (RR) and the pool frog P. lessonae (LL) – produce the hybrid edible frog P. esculentus, in which adult individuals are either diploid (2n, RL) or triploid (3n, RRL and LLR)19–22. During gametogenesis, the genomes of diploid hybrid males and females undergo massive reorganization18,23–25. In particular, one parental genome (usually that of P. lessonae) is eliminated, while the remaining genome is duplicated to bypass normal meiosis, which leads to the production of haploid gametes18,23,26–28. Moreover, genome elimination has been observed in triploid hybrid frogs in a reproductive mode known as triploid hybridogenesis13,29. During triploid hybridogenesis, a single-copy genome is eliminated, and the two remaining genomes presumably undergo meiosis without duplication, leading to the formation of haploid gametes (i.e., in LLR hybrids, the R genome is usually eliminated; in RRL hybrids, the L genome is usually eliminated)13,27,30.
Reproduction via hybridogenesis (observed in Poeciliopsis31, Hypseleotris32, Hexagrammos33, Bacillus34, Pelophylax17,26) and triploid hybridogenesis (observed in Misgurnus35, Squalius29,36, Cobitis37, Bufotes38, Pelophylax26,30) has been found in hybrid organisms from various taxonomic groups. Nevertheless, the cytological mechanisms of parental genome elimination vary in each of these organisms, as elimination of the whole chromosomal set occurs either during gonocyte (germ cell) mitotic divisions24,39,40 or meiosis41,42. However, in studies of European water frogs, different groups have reported an absence of unipolar premeiotic genome elimination and observed aneuploid metaphase and occasional chromosomal lagging in dividing germ cells23–25,43,44. Moreover, in diploid and triploid hybrid frogs from various localities, the cytoplasm of the germ cells harbors micronuclei (previously known as nucleus-like bodies24) that contain DNA, probably from the eliminated genome24,25,44,45. A recent study showed that micronuclei are not associated with cell death but accumulate heterochromatin markers and degrade inside autophagosomes25. Moreover, Chmielewska et al.25 suggested the role of micronuclei in programmed DNA elimination through budding off the interphase nucleus. However, neither presence of chromosomes inside micronuclei nor their genomic identity (P. ridibundus or P. lessonae) has been proven so far for diploid and triploid hybrids. Taken together, mechanisms of selective genome elimination in water frog hybrids remain disputed. Here, we aimed to verify whether micronuclei act as vectors of selective genome elimination in diploid and triploid hybrids.
In current work we focused on diploid and triploid hybrid tadpoles obtained from artificial crosses of hybrid frogs with one another or the parental species. Using FISH with a species-specific centromeric probe, we identified the genomes in the micronuclei and gonocyte nuclei in gonads of diploid and triploid hybrid tadpoles. To confirm our FISH results, we detected kinetochores inside micronuclei in the cytoplasm of germ cells. Additionally, to describe possible mechanisms of genome elimination we analyzed gonocytes during metaphase. The obtained results allowed us to estimate the number and origin of the eliminated chromosomes in each micronucleus in the gonads of hybrid tadpoles.
Results
We analysed the gonads of 106 diploid and triploid hybrid tadpoles obtained from 22 artificial crosses of diploid and triploid hybrids with each other or the parental species (Table S1, Table S2). The genome composition of the tadpoles was identified by FISH with centromeric or telomeric probes performed on metaphase plates from somatic tissues. Triploid tadpoles originated from crosses between diploid hybrid females and either triploid hybrid males with the LLR genotype or P. lessonae males (2 crosses). Diploid tadpoles were produced from various crossings of diploid and triploid hybrids as well as P. lessonae and P. ridibundus individuals, involving both females and males of all taxa: (1) both parents were hybrids, one 2n and one 3n (5 crosses); (2) one parent was a 2n (10 crosses) or 3n (2 crosses) hybrid, while the second parent came from a parental species; or (3) both parents came from the parental species (one crossing).
Genome elimination in germ cells of diploid and triploid hybrid tadpoles occurs gradually
In the gonads of hybrid tadpoles and P. ridibundus individuals, gonocytes were identified with antibodies against the Vasa protein (Fig. S1a,b) and appeared as large cells with multiple nucleoli and less intensive chromatin staining compared to somatic cells. In all analysed hybrid tadpoles, we observed micronuclei (DAPI-positive chromatin bodies surrounded by the membrane) in the cytoplasm of germ cells. The frequency of germ cells with micronuclei ranged from 10 to 30% (Table S1). The number of micronuclei varied from 1 to 4 per individual germ cell.
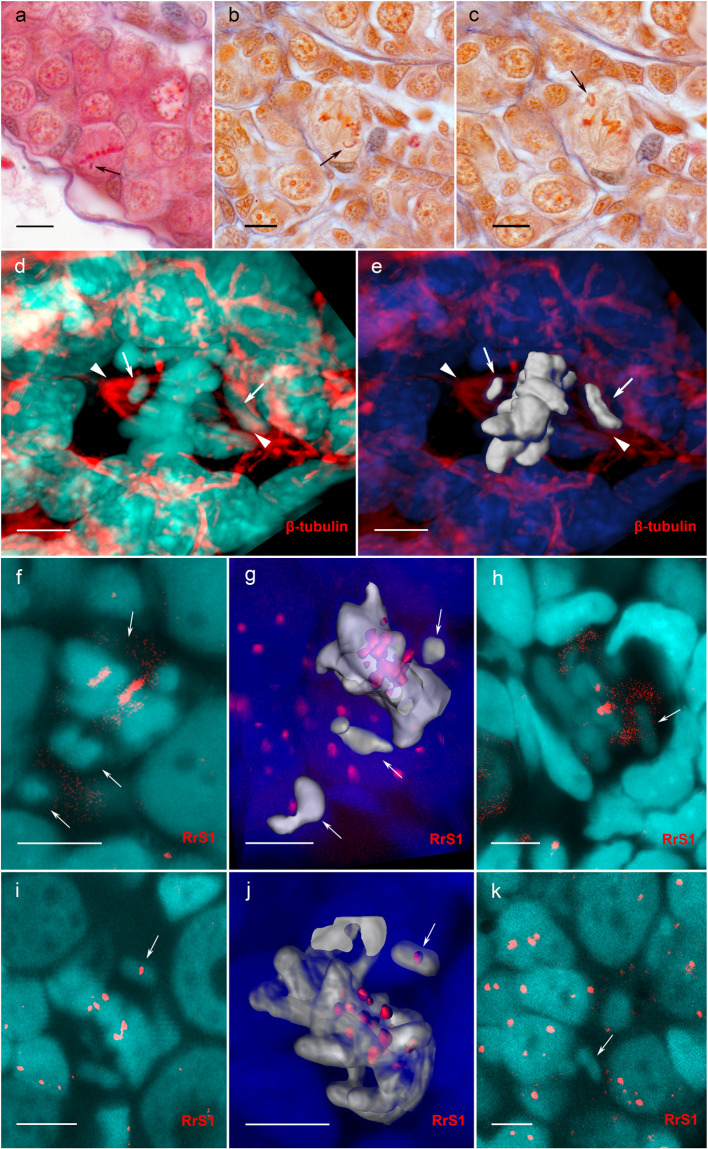
Whole-mount immunostaining with anticentromere CREST serum against kinetochore proteins revealed centromere regions in the vast majority of micronuclei in all hybrid tadpoles studied (11 diploid and 2 triploid tadpoles) (Fig. 1a,b; Table S1). Furthermore, we usually observed one centromere in each micronucleus, and only 4% of micronuclei either lacked a centromere or exhibited two or three centromeres (Fig. 1a,b; Table S1). Thus, we concluded that each micronucleus usually comprises only one chromosome. This result indicates a gradual process of genome elimination, since the whole chromosome set of one of the parental species (n = 13 chromosomes) should be eliminated.
Figure 1.
Micronuclei in germ cells (gonocytes) of tadpoles contain individual chromosomes selectively eliminated in diploid (a,c,e,g) and triploid (b,d,f,h) hybrids. (a,b) Whole-mount immunofluorescent staining with CREST serum in tadpole gonads revealed one kinetochore in each micronucleus. (d,f) 3D-FISH with a probe specific to the P. ridibundus RrS1 repeat revealed P. ridibundus centromeres in the cell nuclei in the gonads of diploid and triploid hybrids as well as in the micronuclei in the gonads of triploid hybrids. (c,e) P. ridibundus chromosomes were usually absent from the micronuclei in the gonads of diploid hybrids. Images (a–f) are single confocal sections of 0.8 µm in thickness. (g,h) 3D surface reconstruction of the germ cells and micronuclei depicted in (e,f). (g) 3D surface reconstruction clearly demonstrates the absence of signals in the micronuclei and 13 P. ridibundus centromeres (red) in the interphase nuclei (gray) of diploid hybrids. (h) 3D surface reconstruction shows the presence of 1 P. ridibundus centromere (red) in each micronucleus and 9 P. ridibundus centromeres (red) in the interphase nucleus (gray) of triploid LLR hybrids. Arrowheads indicate micronuclei. Scale bars for images (a–d) = 20 µm. Scale bars for images (e–h) = 10 µm.
In the germ cells of diploid hybrid tadpoles, micronuclei preferentially incorporate P. lessonae chromosomes
Here, a probe to the RrS1 centromeric repeat served as a species-specific marker allowing identification of the centromeres of P. ridibundus but not P. lessonae chromosomes by FISH. In previous studies, a probe to the RrS1 centromeric repeat marked 5 large chromosomes and 1 small chromosome43,46,47. According to our FISH protocol (see methods), we detected strong signals on 6–7 chromosomes, while the others exhibited weaker but detectable signals, suggesting differences in the accumulation of centromeric repeats between various P. ridibundus chromosomes (Fig. S2). The differences between our results and previous studies can be explained by either methodological modifications or a high level of heterochromatin polymorphism between different P. ridibundus populations.
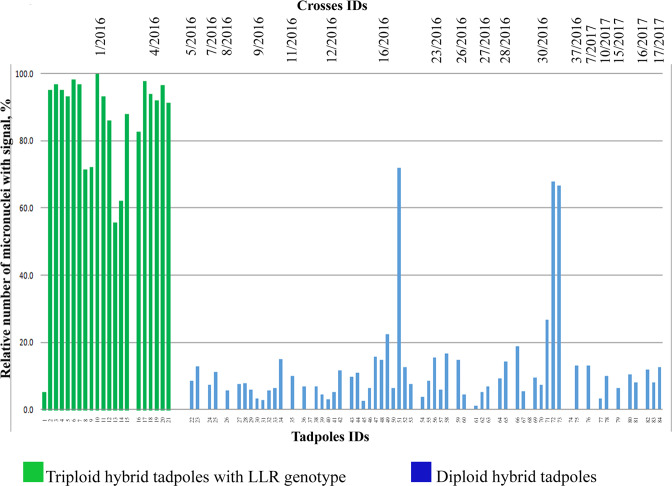
FISH with an RrS1 centromeric probe specific to P. ridibundus centromeres was performed on the gonads of 63 diploid hybrids (Gosner48 stages 28–38) obtained from 18 different crosses (Table S1, Table S2). According to previous results and the current findings, centromere heterochromatin in the interphase nuclei of water frogs usually does not fuse (Fig. 1a,c,e,g)46,47,49. FISH with a centromeric probe or immunostaining with antibodies against kinetochore components revealed individual centromeres in interphase nuclei (Fig. 1a,c,e,g). Accordingly, in the majority of the interphase nuclei of gonocytes, we observed approximately 13 signals corresponding to P. ridibundus chromosomes (n = 13) (Fig. 1g). We examined 38,521 gonocytes and found 8,540 micronuclei in the gonads from 63 diploid tadpoles (Table S1). Among the micronuclei of 60 studied diploid hybrid tadpoles, we observed P. ridibundus centromeres in 694 micronuclei (approximately 9%), but did not reveal any signals in the remaining 7,449 micronuclei (91%) (Fig. 1c,e,g; Fig. 2; Table S1). Taking into account that each micronucleus usually comprises one centromere (and very likely one chromosome), we suggest that in diploid hybrid tadpoles P. lessonae chromosomes are preferentially eliminated via micronuclei. However, on average, 9% of the micronuclei from these tadpoles contained P. ridibundus chromosomes, indicating nonselective genome elimination in some populations of germ cells (Fig. 2). Accordingly, a small portion of the germ cell nuclei harboured fewer than 13 P. ridibundus centromeres. In the gonads of three tadpoles, we observed P. ridibundus chromosomes in approximately 70% of all analysed micronuclei, suggesting either an absence of selective genome elimination or the elimination of different genomes in various germ cell populations (Fig. 2; Table S1).
Figure 2.
The relative number of micronuclei containing P. ridibundus chromosomes in diploid (marked blue) and triploid (marked green) hybrids obtained from various crosses.
Notably, in at least 10 diploid female hybrid tadpoles in developmental stages 33–38 (according to Gosner48) and gonadal development stages 4–5 according to Ogielska and Kotusz50, we detected individual cells (approximately 4–8 per individual) with 26 P. ridibundus centromeres (Fig. 3a,b). We suggest that such gonocytes with a doubled number of P. ridibundus chromosomes appeared as a result of genome duplication.
Figure 3.
Germ cell interphase nucleus (a) and metaphase chromosome plate (b) with a doubled number of P. ridibundus chromosomes detected by FISH with a probe to centromeric RrS1 repeat. The 3D surface reconstruction indicates approximately 26 P. ridibundus centromeres in the gonocytes of diploid hybrid tadpoles (indicated by arrows). Scale bars = 10 µm.
In the germ cells of triploid hybrid tadpoles, micronuclei preferentially incorporate P. ridibundus chromosomes
FISH with an RrS1 centromeric probe specific to P. ridibundus centromeres was performed on the gonads of 21 triploid hybrid tadpoles with LLR genotype obtained from 2 different crosses (Table S1, Table S2). We found that the number of micronuclei containing P. ridibundus chromosomes was significantly higher in triploid hybrids as compared to diploid hybrid tadpoles (p < 0.01). We investigated 9,546 germ cells from the gonads of all triploid hybrid tadpoles with an LLR genotype and detected 2,375 micronuclei (Table S1). Detailed analysis showed that the gonads of 18 triploid hybrid tadpoles with LLR genotype contained 1,898 micronuclei (92%) with P. ridibundus chromosomes (Fig. 1d,f,h; Fig. 2; Table S1). Approximately 8% of micronuclei did not contain P. ridibundus chromosomes (Fig. 1d,f,h; Fig. 2; Table S1). Moreover, in germ cell nuclei, the number of P. ridibundus chromosomes was almost always lower than 13 and usually ranged from 4 to 11 (Fig. 1h). In two tadpoles, we detected P. ridibundus chromosomes in approximately 59% of all observed micronuclei, suggesting either an absence of selective genome elimination or the elimination of different genomes in different germ cells (Fig. 2; Table S1). Only one triploid tadpole with LLR genotype contained small number of P. ridibundus chromosomes, which were found in 5% of all observed micronuclei, suggesting the preferential elimination of P. lessonae chromosomes (Fig. 2; Table S1).
Misaligned chromosomes can be selectively eliminated during metaphase
Although numerous micronuclei were present within the gonads of hybrid tadpoles and were observed in approximately 30% of germ cells, we found 19 cases of misaligned chromosomes during the mitotic division of germ cells in diploid hybrids (Fig. 4a–j; Table S1). Misaligned chromosomes exhibited an abnormal or no connection to the spindle (Fig. 4a–e). Notably, the misaligned chromosomes corresponded to P. lessonae chromosomes in 13 cases, whereas they corresponded to P. ridibundus chromosomes in only two cases (Fig. 4f–j). Moreover, in germ cells of two tadpoles, we observed individual P. lessonae chromosomes located in the cytoplasm and separated from the interphase nucleus (Fig. 4k).
Figure 4.
Metaphases with misaligned chromosomes in the gonads of diploid hybrids. (a–c) Histological sections of gonads show metaphase plates with misaligned chromosomes. (d,e) 3D surface reconstruction of the metaphase plate after immunofluorescent staining with antibodies against b-tubulin performed in frozen tissue sections from the gonads of hybrid females. Misaligned chromosomes (indicated by arrows) unable to attach to the spindle (indicated by arrowheads). (f–j) Misaligned P. lessonae chromosomes (f,g,h) and P. ridibundus chromosomes (i,j) during the metaphase stage identified by FISH with a probe to centromeric RrS1 repeat. Images (f,h,i,k) are single confocal sections of 0.8 µm in thickness. (g,j) 3D surface reconstructions of metaphase chromosomes depicted in panels (f,i), which specifically show misaligned P. lessonae (g) and P. ridibundus (j) chromosomes. (k) Individual P. lessonae chromosomes located in the cytoplasm of the cells during interphase. Scale bars for images A-C = 20 µm. Scale bars for images D-K = 10 µm.
Discussion
Here, we clearly demonstrated that in hybrid European water frogs chromosomes are gradually eliminated from the gonocyte nuclei via micronucleus formation. Each micronucleus contains only one chromosome, while the majority of gonocytes are aneuploid until they eliminate all chromosomes of one of the parental genomes. We showed that in the majority of cases, the elimination of chromosomes is selective; i.e., the chromosomes of one of the parental species are eliminated. Several cases of misaligned chromosomes during gonocyte mitosis were observed. Genome identity of misaligned chromosomes coincided with the chromosome composition of micronuclei. In addition, genome duplication in the germ cells of hybrid tadpoles was revealed.
Genome duplication in germ cells of diploid hybrids
In addition to genome elimination, we observed germ cells with an increased number of P. ridibundus chromosomes in diploid hybrids (Fig. 3a), suggesting the doubling of P. ridibundus genome, similar to observations made by Heppich and Tunner18. Such cells were detected in tadpoles at stages 33–38 according to Gosner48, which corresponds to stages 4–5 of gonadal development according to Ogielska and Kotusz50. In accordance with suggestions that in diploid hybrids premeiotic genome doubling occurs after the elimination of the P. lessonae chromosomes23,26, we detected 26 P. ridibundus but no P. lessonae chromosomes in certain gonocyte metaphases (Fig. 3b). However, proper cytogenetic markers are still unavailable for the P. lessonae genome; thus, we cannot check whether some P. lessonae chromosomes are still present in the gonocyte nuclei during interphase.
According to the classical scheme of hybridogenesis in water frogs, genome endoreplication is usually absent in triploids27. In triploid hybrid tadpoles, the number of P. ridibundus chromosomes usually ranged from 4 to 11 suggesting the absence of cells with duplicated genomes. However, oocytes with a duplicated genome were occasionally found among triploid hybrids from R-E populations51. Thus, the frequency of genome duplication in the gonocytes of triploid hybrids requires further investigation.
Premeiotic genome doubling has been suggested to be an almost universal mechanism of unreduced egg formation in the asexual hybrids of vertebrates, invertebrates and some plants11,12,52. Indeed, in the majority of cases, genome replication has been inferred after the identification of the genome composition in gametes11–14. In contrast, an alternative mechanism has been described for Carassius hybrids: germ cells have been shown to fuse with each other during gametogenesis, leading to the formation of gametes with doubled chromosome set53. However, in diploid and triploid P. esculentus tadpoles, neither we nor other researchers24,25,44,45 have observed the fusion of germ cells. Thus, we propose that the most likely mechanism of the formation of germ cells with a doubled number of P. ridibundus chromosomes is either genome endoreplication (cell cycle without mitosis) or endomitosis (mitosis without chromosome segregation).
Nucleus budding and chromosome lagging as mechanisms of genome elimination in P. esculentus
In the present study, we demonstrated for the first time the correspondence between the genome origin of misaligned chromosomes and the genome composition of micronuclei in the same gonocytes of diploid hybrids. Together with our recent results showing that micronuclei can form via budding from interphase nuclei25, our current finding that misaligned P. lessonae chromosomes appear during the mitosis of gonocytes of diploid hybrids confirms their loss during anaphase (Fig. 4). Moreover, in studied hybrids, misaligned chromosomes show inability to attach to the spindle and are located outside the metaphase plate during mitosis (Fig. 4). Such chromosomes often tend to lag during anaphase and are not incorporated into the nuclei after mitotic division, causing aneuploidy in different cell cultures, moreover, they are observed at a high frequency in the presence of stress factors54,55. Furthermore, misaligned chromosomes in plant hybrids are often incorporated in micronuclei during selective genome elimination56–59. Thus, in accordance with other studies23–25, here we suggest that in water frog hybrids, the genome can be eliminated not only via the budding off the interphase nucleus but also through the inability of individual chromosomes to attach to the spindle, causing them to lag during the mitotic divisions of gonocytes.
The earlier suggestion that the chromosomes of P. lessonae can lag during the mitotic division of gonocytes was rooted to the fact that chromosomes of parental species exhibit different number of copies of the centromeric repeat44. Furthermore, the divergence of species-specific centromere-binding proteins frequently leads to the inability of chromosomes of one parental species to attach to the spindle, causing them to lag and eliminate in interspecies hybrids57–60. Selective chromosomal loss was also observed in artificial and natural plant hybrids between species with different variants of centromeric histone CENP-A57,61,62. Moreover, the elimination of such chromosomes can be gradual via aberrant loading of centromeric proteins from one parental species to the centromeres of the another61. Failure to attach to the spindle caused by the aberrant uploading of centromeric histones has also been documented for the fragments of holocentric chromosomes that are lost during chromatin diminution in somatic cells of Parascaris univalens63. Another aberration affecting centromere stability and function is known to be related to centromere-associated noncoding RNAs60,64,65. In addition to misaligned chromosomes, we also detected individual chromosomes in the cytoplasm of certain germ cells while the main nucleus was in interphase, which could indicate an inability of such chromosomes to incorporate to the main nucleus after mitosis. Although the underlying mechanism of misaligned chromosome formation in water frog hybrids requires further investigation, it seems that there are many complex avenues causing selective genome elimination.
Micronuclei include single chromosomes that are gradually eliminated from the nucleus
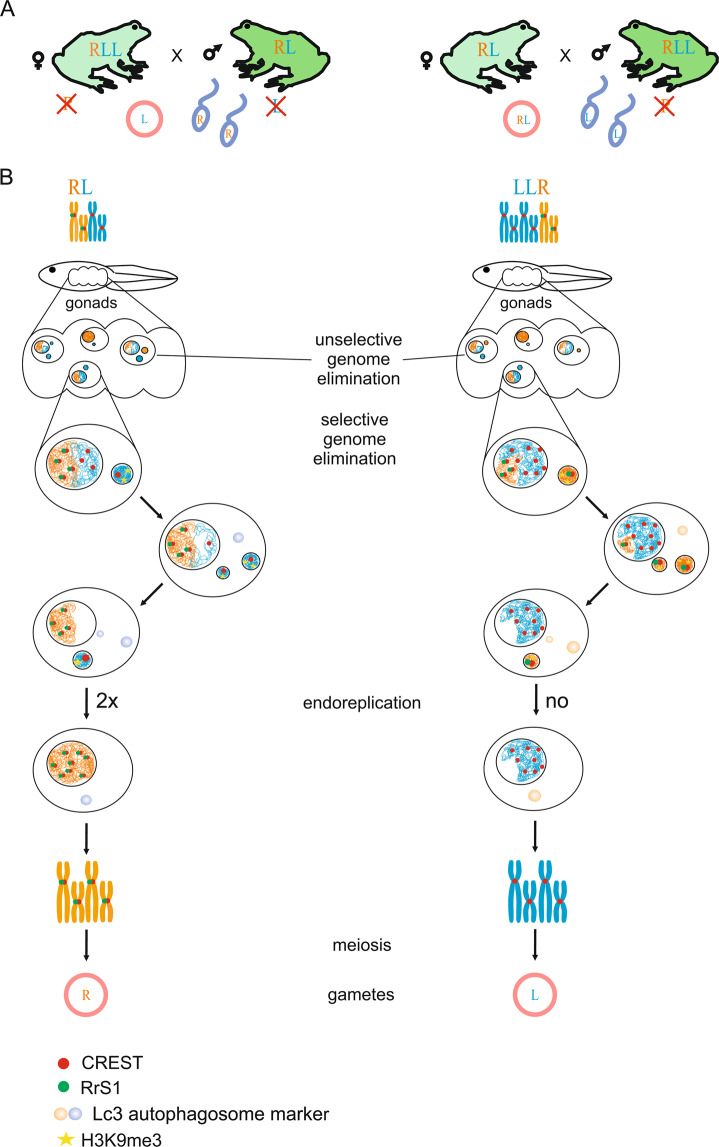
By the identification of the chromosomes contained within micronuclei and interphase nuclei of the same gonocytes, we confirmed the role of micronuclei in selective genome elimination during early gametogenesis in hybridogenetic diploid and triploid water frog hybrids for the first time (Fig. 5). Using FISH with the species-specific probe to the centromeric region and immunofluorescent detection of kinetochore proteins we showed that micronuclei usually contained only one chromosome. Regardless of the cross type, all diploid hybrid tadpoles preferentially eliminated P. lessonae chromosomes, while triploid LLR hybrids preferentially eliminated P. ridibundus chromosomes (Fig. 5; Table S1, Table S2). In diploid hybrids, we also observed misalignment of P. lessonae chromosomes that corresponds to the genome composition of the majority of micronuclei. Moreover, the genome composition of interphase nuclei was in agreement with the genome composition of micronuclei. In triploid LLR hybrids, we observed germ cells with a decreased number of P. ridibundus chromosomes usually ranged from 4 to 13 chromosomes, while in diploid hybrids, we observed germ cells with all P. ridibundus chromosomes. Our results suggest that during genome elimination gonocytes are aneuploid until one of the parental genomes will be completely eliminated from the nuclei. As we usually observe from 1 to 3 micronuclei (this study and24,25,44,45) and/or misaligned chromosomes as well as decreased number of P. ridibundus chromosomes in gonocytes of triploid LLR hybrids, we assume that several (up to 13) gonocyte divisions are required to eliminate one of the parental genomes. However, chromosome elimination through budding of the interphase nucleus24,25 can speed up this process. Continuous chromosome elimination through several mitotic divisions was detected in somatic cells of plant hybrids56–59, during elimination of supernumerary chromosomes66 and through programmed DNA rearrangement1,2,7,67.
Figure 5.
Schematic overview of selective genome elimination in diploid (left) and triploid (right) hybrid tadpoles. (A) Classical scheme of the reproduction of diploid and triploid hybrid frogs. (B) Suggested simplified scheme of gradual chromosome elimination via micronucleus formation in diploid and triploid hybrids. Individual chromosomes were detected via the visualization of centromeric regions with CREST antibodies (red circle) and FISH with a probe specific to the P. ridibundus RrS1 centromeric repeat (green circle). Each micronucleus usually comprises only one chromosome. In the micronuclei, chromatin accumulates heterochromatin markers, indicated by the yellow asterisk (according to Chmielewska et al.25). Subsequently, micronuclei accumulate autophagosome marker LC3 (transparent blue micronuclei) and undergo degradation (according to Chmielewska et al.25). In diploid hybrid frogs, after the elimination of the P. lessonae genome, P. ridibundus chromosomes undergo endoreplication, leading to a doubled number of chromosomes. Cells in which both genomes are eliminated probably die.
Both chromosomal budding and lagging have been frequently observed in somatic cells of plant hybrids during early development58,59. In contrast to hybrid plants, genome elimination in water frog hybrids occurs only in germ cells during the specific stage of their gonadal development while genomes of somatic cells remain stable. In hybrid plants, Gernand et al.58 observed lagging chromosomes and chromosomal fragments without centromeres, while centric chromosomal fragments were usually extruded via budding from the interphase nuclei. Such chromosomes can be rearranged within micronuclei to form new chromosomes which can be inherited68. However, our findings suggest that in water frog hybrids, the vast majority of micronuclei contain centromeres, indicating the encapsulation of the entire chromosomes but not their fragments in micronuclei. This observation, together with chromatin condensation in the micronuclei, could explain the observed variation in the size of micronuclei (this study25,44,45).
In contrast to plant hybrids, gradual chromosome elimination occurs rarely among hybrid vertebrates and is well described only for water frog hybrids to date. In the majority of known asexual hybrids and cases of paternal genome elimination, whole sets of parental chromosomes are eliminated via a single mitotic division immediately after insemination (ambystomes69, molluscs from the Corbicula genus70, hybrid Carassius species Carassius gibelio71), during the mitotic division of gonial cells (hybridogenetic Poeciliopsis39) or during meiosis (in the loach Misgurnus anguillicaudatus42 and probably Squalius alburnoides41). In the other asexuals (Bufotes38 and stick insects from the Bacillus genus40), the genome elimination occurs before meiosis but its cellular mechanisms remain undiscovered.
The results for the majority of diploid and triploid hybrid tadpoles were highly concordant with the genome predicted to be eliminated according to the accepted model of hybridogenesis26,27 in adult hybrid frogs. Nevertheless, in both diploid and triploid LLR hybrids, we detected a small portion (around 9%) of micronuclei including different chromosomes than expected from the classical scheme of hybridogenesis. Presence of such micronuclei indicates irregularities in genome elimination in some gonocytes and may lead to the previously detected increased level of gonial cell death in the gonads of hybrids72. On the other hand, the potential ability of such gonocytes to proceed beyond meiosis can lead to a simultaneous production of gametes with various genome composition by individual hybrid frogs frequently detected in various populations20,26,30,43,44,73,74.
We stress that the following findings imply complicated mechanisms involved in genome elimination in water frog hybrids (Fig. 5): (1) chromatin heterochromatinization occurs within micronuclei25; (2) kinetochores are still present within micronuclei; and (3) P. ridibundus chromosomes are preferentially eliminated in triploid LLR hybrids, while P. lessonae chromosomes are usually eliminated in diploid RL and triploid RRL hybrids20,26,44,45,51,74.
Recognition of genetic material is crucial for its selective elimination
The molecular mechanisms of selective genome elimination remain mostly unknown. Similar to the majority of the known cases of the selective elimination of genetic material, we suggest that in water frog hybrids one of the parental genomes should be recognized, marked, eliminated from the nucleus, and finally degraded. According to recent studies and the current work, selective genome elimination in P. esculentus hybrids involves chromosomal budding from the interphase nucleus and lagging during gonocyte divisions, leading to the formation of micronuclei, which are likely degraded via autophagy24,25,44,45. However, key components of genome elimination are not known for hybrid animals as well as almost all other cases in which the selective elimination of genetic material has been observed. The crucial role of noncoding RNA in recognizing DNA sequences designed for removal (in Oligohymenophorea4) or retention (in Spirotrichea5) has been previously demonstrated for only two classes of ciliate protozoa. To explain the mechanism of selective genome elimination, we previously hypothesized that in hybrid germ cells, retrotransposons can become activated in the genome, which is not protected by maternally inherited short noncoding RNAs from the oocyte51. However, in the current study, we have shown that regardless of the parent-of-origin effect, diploid hybrid tadpoles usually eliminate the L genome, while triploid LLR hybrid tadpoles usually eliminate the R genome. Based on studies of selective paternal genome elimination in Coccilla insects9,75, we propose the possibility of competition between the P. ridibundus and P. lessonae genomes that induces elimination and the development of anti-elimination mechanisms, but only in germ line cells. We can hypothesize that noncoding RNA from one genome can distinguish DNA sequences from the other genome as alien and prevent them from functioning, leading to their removal. Alternatively, the RNA of one genome can protect its genome from elimination and lead to the elimination of the unprotected genome.
Materials and Methods
Samples studied
European water frogs were collected from drainage areas in northwestern and southwestern Poland. P. ridibundus (N = 4) and P. lessonae (N = 10) individuals were captured from R-E (Ruda Milicka) and L-E systems (Sanie, surroundings of Wroclaw and Urwitalt in Mazury), respectively. Diploid hybrids (N = 13) and triploid hybrids with an LLR genome composition (N = 7) were collected from two separate E-E systems (Wysoka Kamieńska and Uciechów). The parental species and all hybrids except for triploid frogs with the LLR genotype were represented by both sexes. All manipulations with animals were carried out in accordance with national and international guidelines.
Crossing experiments
Artificial crosses were carried out according to the protocol described in Berger et al.76 after hormonal stimulation. Females were injected intraperitoneally with salmon luteinizing hormone-releasing hormone (LHRH, H-7525.0001, Bachem) in amphibian PBS (APBS, pH 7.4, 11.2 mM NaCl, 0.22 mM KCl, 0.8 mM Na2HPO4, 0.14 mM KH2PO4) at a dose of 6.25 mg/kg of body weight. After the females had spawned, their eggs were fertilized with a homogenate of dissected testes from male frogs. Males and females were euthanized after being anaesthetized in a 0.5% solution of ethyl 3-aminobenzoate methane sulfonate (MS-222, Sigma Chemical Co.) in APBS, followed by tissue collection for chromosomal analysis. From stage 28 according to Gosner48 (hind limb bud development) to stage 42 (forelimb emergence), 5–20 tadpoles were randomly collected from each clutch each week. The tadpoles were placed in an anaesthetic solution (0.15% MS 222) preceding the dissection of their gonads, gills and intestines. Gonadal tissues were fixed in 2% paraformaldehyde in 1× PBS for 90 min at room temperature (RT) and then kept in 1× PBS with 0.02% NaN3 until use.
We analysed 106 diploid and triploid hybrid tadpoles obtained from 22 artificial crosses of diploid and triploid hybrids with each other and parental species (Supplementary Table 1). The tadpole genome composition was identified by FISH with centromeric or telomeric probes performed on metaphase plates from somatic tissues according to44,49.
Probe labelling
The labelling of the RrS1 probe specific to the pericentromere tandem repeat of P. ridibundus46, not P. lessonae, was carried out by PCR (annealing temperature 62 °C) with the genomic DNA of P. ridibundus and the following primers according to a previously published protocol49:
Forward: 5′-AAGCCGATTTTAGACAAGATTGC-3′
Reverse: 5′-GGCCTTTGGTTACCAAATGC-3′
The probes were labelled with biotin-16-dUTP (Roche). In addition, a Cy3-labelled oligonucleotide probe (5′- AAGCCGATTTTAGACAAGATTGC-3′) specific to the P. ridibundus RrS1 pericentromeric sequence was used. A single-stranded oligonucleotide telomeric probe (TTAGGG)5 conjugated with biotin or fluorochrome Cy3 was also used to identify species.
Fluorescent in situ hybridization
Fluorescent in situ hybridization was performed according to previously published protocols44,45,49. For hybridization, we used either a labelled probe generated from genomic DNA by PCR or a directly labelled oligonucleotide probe for the RrS1 repeat; however, the best results were provided by a PCR-labelled probe. The hybridization mixture contained 50% formamide, 10% dextran sulphate, 2× ЅЅС, 5 ng/μl labelled probe and a 10–50-fold excess of tRNA. In the case of the oligonucleotide probe, the hybridization mixture contained 40% formamide, 10% dextran sulphate, 2× ЅЅС, 5 ng/μl labelled probe and a 10–50-fold excess of tRNA. Joint denaturation of the probe and chromosomal DNA on the slides was performed at 75 °C for five minutes. The slides were incubated for 12–24 hours at room temperature (RT). After hybridization, the slides were washed in 0.2× SSC at 50 °C. Oligonucleotide-probed slides were washed in 2× ЅЅС at 42 °C. Biotin was detected with avidin conjugated with the Cy3 fluorochrome (Jackson ImmunoResearch Laboratories). Thereafter, the slides were dehydrated in a graded ethanol solutions (50°, 70° and 96°), dried, and mounted in DABCO (Merck) antifade solution containing 1 μg/ml DAPI.
Histology
After dissection, the gonads were fixed in Bouin’s solution (Sigma), dehydrated in graded ethanol solutions (70–100%), immersed in xylene and mounted in paraffin. Tissue sections of 7 µm in thickness were deparaffinized in xylene, rehydrated and stained via a routine protocol according to the Mallory method77. Imaging was performed with an Axioskop microscope (Zeiss) using AxioVision Rel 4.8 software (Zeiss).
Whole-mount fluorescence in situ hybridization
Prior to whole mount FISH, tissues were incubated in a 0.5% solution of Triton X100 in 1× PBS for 4–5 hours at RT, washed in 1× PBS for 15 min followed by impregnation with 50% formamide, 10% dextran sulphate, and 2× SSC (saline-sodium citrate buffer; 20 × SSC – 3 M NaCl 300 mМ Na3C6H5O7) for 3–4 hours at 37 °C. The hybridization mixture for the PCR-labelled probe contained 50% formamide, 2× SSC, 10% dextran sulphate, 20 ng/µl probe and a 10- to 50-fold excess salmon sperm DNA. For the oligonucleotide probe, the hybridization mixture contained 40% formamide, 2× SSC and 10% dextran sulphate, 20 ng/µl probe and a 10 to 50-fold excess of tRNA. Gonadal tissues were denatured at 82 °C for 15 min and incubated for 24 hours at RT. In the case of the directly labelled oligonucleotide probe, the tissues were washed in three changes of 2× SSC at 42 °C for 15 min each, followed by staining with DAPI (1 µg/µl) (Sigma) prepared in 1× PBS. In the case of the PCR-labelled probe, the tissues were washed in three changes of 0.2× SSC at 42 °C for 15 minutes each and blocked in 4× SSC containing 1% blocking reagent (Roche) in 4× SSC for 1 hour at RT. Biotin was detected by incubation with avidin conjugated with Cy3 or Alexa 488 (Jackson ImmunoResearch Laboratories) for 12 hours. The tissues were stained with DAPI (1 µg/µl) (Sigma) prepared in 1× PBS at RT overnight.
Whole-mount immunofluorescence staining
Kinetochore proteins were detected by human CREST serum (Antibodies Incorporated), and Vasa was detected with the rabbit polyclonal DDX4 antibody [C1C3] (GeneTex) via 3D immunofluorescence staining. Prior to the addition of the antibodies, the tissues were incubated in a 0.5% solution of Triton X100 in 1× PBS for 4–5 hours at RT, washed in 1× PBS at RT and incubated for 1–2 hours in a 1% blocking solution (Roche) in 1× PBS. Incubation with the primary antibodies (concentration 1:40) was carried out at RT overnight, followed by washing in 1× PBS with 0.01% Tween (ICN Biomedical Inc). Secondary antibodies conjugated with the Alexa-488 fluorochrome were applied for 12 hours at RT. The tissues were then washed in 1× PBS with 0.01% Tween (ICN Biomedical Inc) and stained with DAPI (1 µg/µl) (Sigma) in 1× PBS at RT overnight.
Immunofluorescence in tissue sections
Beta tubulin staining was carried out using rabbit polyclonal antibodies (ab6046, Abcam) following the protocol described in Chmielewska et al.25. Briefly, 20 µm-thick cryostat sections were mounted on Superfrost Plus microscope slides (Thermo Scientific), thawed and permeabilized for 1 hr and 15 min at RT in 0.5% Triton X-100 (Sigma) in 1× PBS after washing in PBST (pH 7.4, 14 mM NaCl, 0.27 mM KCl, 1 mM Na2HPO4, 0.18 mM KH2PO4, 0.05% Tween 20). Next, the tissues were blocked in 6% bovine serum albumin (BSA) diluted in PBS for 30 min at RT. Then, primary antibodies diluted 1:200 in 3% BSA in PBST were applied for 2 days at 4 °C, followed by washing in PBST with gentle shaking. Donkey anti-Rabbit Rhodamine Red-X secondary antibodies (1:100, Jackson ImmunoResearch) diluted in 3% BSA in PBST containing 0.5 µg/ml DAPI were applied overnight at RT, followed by washes in PBST. Sections were mounted in Vectashield antifade mounting medium (Vector Laboratories, Thermo Fisher Scientific).
Confocal laser scanning microscopy
Tissues were placed in a drop of DABCO antifade solution containing 1 mg/ml DAPI, and confocal laser scanning microscopy was carried out using a Leica TCS SP5 microscope based on the Leica DMI 6000 CS inverted microscope (Leica Microsystems, Germany). Specimens were analysed using an HC PL APO 40× objective. Diode, argon and helium-neon lasers were used to excite the DAPI fluorescent dye and the Alexa488 and Cy3 fluorochromes, respectively. Images were captured using LAS AF software (Leica Microsystems, Germany). Confocal imaging of the tissue sections was performed using an Olympus FV1000 confocal microscope equipped with a UPLSAPO 60× oil lens. The 3D-volume rendering and surface reconstruction of confocal image stacks were performed in Imaris 7.7.1 (Bitplane). The image stacks used for reconstruction were cropped to the region of interest (ROI), preserving the image voxel dimensions set during image acquisition. Isosurfaces of multichannel images were created for each channel separately, applying automated thresholding parameters for channel intensity cutoffs. ROI isosurfaces were split into separate surface objects corresponding to individual nuclei (DAPI channel) or FISH signals (Cy3 channel). In some cases when two adjacent nuclei were inseparable as individual surface objects, isosurface reconstruction was performed via a “manual creation” tab. For the highlighted visualization of germ cells, only surface objects belonging to individual germ cells were retained in the reconstruction.
Ethic Statement
Collected species are not listed by the IUCN Red list or CITES. Collection of adult frogs was approved by the Polish General and Regional Directorates for Environmental Protection (DOP-oz.6401.02.2.2013.JRO, DOP-oz.6401.02.2.2013.2014.JRO.as, WPN.6205.28.2014.IW.2, DZP-WG.6401.02.5.2015.JRO, WPN.6401.177.2016.IL). Techniques used in the capture, breeding, tissue sampling and euthanasia sought to minimize animal suffering and were in accordance with recommendations of the Herpetological Animal Care and Use Committee (HACC) of the American Society of Ichthyologists and Herpetologists. Each individual was anaesthetized by submersion in a 0,5% solution of 3-aminobenzoic acid ethyl ester (MS 222). All experimental procedures were accepted by the Local Commission for Ethics in Experiments on Animals in Wrocław, Poland (7/2013, 27/2016).
Supplementary information
Acknowledgements
Authors would like to thank Antonina Maslova (Saint-Petersburg State University) for the help with 3D imaging. The authors acknowledge resource centers “Environmental Safety Observatory” and “Molecular and Cell Technologies” (Saint-Petersburg State University) for the access to experimental equipment. The work of DD and SR was supported by grants from the Russian Science Foundation № 18–74–00115 (including expenses connected with whole-mount in situ hybridization and whole mount immunofluorescence staining) and Russian Foundation of Basic Research № 18–34–00514 (including expenses connected with cytogenetic experiments). The work of MO, MC, and BR-K was supported by a grant from the Polish National Science Centre no. 2012/07/B/NZ3/02563. The authors declare that the funding body had no role in the design of the study or the collection, analysis, and interpretation of the data or in writing the manuscript.
Author contributions
A.K. and D.D. conceived the study and designed the experiments. M.O., M.C., B.R.-K., M.K., K.K., D.D. performed crosses experiments and preparation of tissue collection. D.D., S.R., M.K. and K.K. made species identification. D.D. and S.R. carried out cytogenetic experiments, whole-mount in situ hybridization and whole mount immunofluorescence staining. M.C., B.R.-K. and A.D. implemented histology and immunocytochemistry on tissue sections. D.D., S.R., M.C. and A.D. obtained and analyzed confocal images. D.D. and S.R. performed quantitative analysis of germ cells and micronuclei. D.D. and A.K. have written the manuscript with the help of M.O., M.C. and B.R.-K. and improved by all other authors.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-64977-3.
References
- 1.Tobler H. Results and Problems in Cell Differentiation. Berlin, Heidelberg: Springer Berlin Heidelberg; 1986. The Differentiation of Germ and Somatic Cell Lines in Nematodes; pp. 1–69. [DOI] [PubMed] [Google Scholar]
- 2.Wang J, Davis RE. Programmed DNA elimination in multicellular organisms. Curr.Opin. Genet. Devel. 2014;27:26–34. doi: 10.1016/j.gde.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kloc M, Zagrodzinska B. Chromatin elimination – an oddity or a common mechanism in differentiation and development? Differentiation. 2001;68:84–91. doi: 10.1046/j.1432-0436.2001.680202.x. [DOI] [PubMed] [Google Scholar]
- 4.Mochizuki K. DNA rearrangements directed by non-coding RNAs in ciliates. WIR: RNA. 2010;1:376–387. doi: 10.1002/wrna.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fang W, Wang X, Bracht JR, Nowacki M, Landweber LF. Piwi-interacting RNAs protect DNA against loss during Oxytricha genome rearrangement. Cell. 2012;151:1243–1255. doi: 10.1016/j.cell.2012.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith JJ, Antonacci F, Eichler EE, Amemiya CT. Programmed loss of millions of base pairs from a vertebrate genome. PNAS. 2009;106:11212–11217. doi: 10.1073/pnas.0902358106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kojima NF, et al. Whole chromosome elimination and chromosome terminus elimination both contribute to somatic differentiation in Taiwanese hagfish Paramyxine sheni. Chromosome Res. 2010;18:383–400. doi: 10.1007/s10577-010-9122-2. [DOI] [PubMed] [Google Scholar]
- 8.Gerbi S. A. Results and Problems in Cell Differentiation. Berlin, Heidelberg: Springer Berlin Heidelberg; 1986. Unusual Chromosome Movements in Sciarid Flies; pp. 71–104. [DOI] [PubMed] [Google Scholar]
- 9.Herrick G, Seger J. Imprinting and paternal genome elimination in insects. Results Probl Cell Differ. 1999;25:41–71. doi: 10.1007/978-3-540-69111-2_3. [DOI] [PubMed] [Google Scholar]
- 10.Burt, A. & Trivers, R. Genes in Conflict. The biology of selfish genetic elements. (2006).
- 11.Schön, K M & P van Dijk. Lost Sex: The Evolutionary Biology of Parthenogenesis.132, (Springer Netherlands, 2009).
- 12.Stenberg P, Saura A. Meiosis and its deviations in polyploid animals. Cytogenet. Genome Res. 2013;140:185–203. doi: 10.1159/000351731. [DOI] [PubMed] [Google Scholar]
- 13.Dawley, R. M. & Bogart, J. P. Evolution and ecology of unisexual vertebrates. (University of the State of New York, State Education Department, New York State Museum, 1989).
- 14.Neaves WB, Baumann P. Unisexual reproduction among vertebrates. Trends Genet. 2011;27:81–88. doi: 10.1016/j.tig.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 15.Schwander, T. & Oldroyd, B. P. Androgenesis: where males hijack eggs to clone themselves. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 371, (2016). [DOI] [PMC free article] [PubMed]
- 16.Bogart JP, Bi K, Fu J, Noble DWA, Niedzwiecki J. Unisexual salamanders (genus Ambystoma) present a new reproductive mode for eukaryotes. Genome. 2007;50:119–136. doi: 10.1139/g06-152. [DOI] [PubMed] [Google Scholar]
- 17.Tunner HG. Die klonale Struktur einer Wasserfroschpopulation1. Journal of Zoological Systematics and Evolutionary Research. 1974;12:309–314. [Google Scholar]
- 18.Tunner HG, Heppich S. Premeiotic genome exclusion during oogenesis in the common edible frog, Rana esculenta. Naturwissenschaften. 1981;68:207–208. doi: 10.1007/BF01047207. [DOI] [PubMed] [Google Scholar]
- 19.Berger, L. Morphology of the F1 generation of various crosses within Rana esculenta-complex. (Zakład Zoologii Systematycznej Polskiej Akademii Nauk, 1968).
- 20.Berger L. Viability, sex and morphology of F2 generation within forms of Rana esculenta complex. Zool. Pol. 1971;21:345–393. [Google Scholar]
- 21.Berger L, Roguski H. Ploidy of progeny from different egg size classes of Rana esculenta L. Folia Biol. (Krakow) 1978;26:231–248. [PubMed] [Google Scholar]
- 22.Arioli, M. Reproductive patterns and population genetics in pure hybridogenetic water frog populations of Rana esculenta. PhD thesis, University of Zurich, Zurich, Switzerland, http://www.dissertationen.uzh.ch (2007).
- 23.Tunner HG, Heppich-Tunner S. Genome exclusion and two strategies of chromosome duplication in oogenesis of a hybrid frog. Naturwissenschaften. 1991;78:32–34. [Google Scholar]
- 24.Ogielska M. Nucleus-like bodies in gonial cells of Rana esculenta [Amphibia, Anura] tadpoles—a putative way of chromosome elimination. ZoolPol. 1994;39:461–474. [Google Scholar]
- 25.Chmielewska M, et al. The programmed DNA elimination and formation of micronuclei in germ line cells of the natural hybridogenetic water frog Pelophylax esculentus. Sci Rep. 2018;8:7870. doi: 10.1038/s41598-018-26168-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Graf, J.-D. & Polls Pelaz, M. Evolutionary genetics of the Rana esculenta complex. in Evolution and ecology of unisexual vertebrates 289–302.
- 27.Plötner, J. Die westpaläarktischen Wasserfrösche: von Märtyrern der Wissenschaft zur biologischen Sensation; mit 9 Tabellen. (Laurenti-Verlag, 2005).
- 28.Graf J-D, Müller WP. Experimental gynogenesis provides evidence of hybridogenetic reproduction in the Rana esculenta complex. Experientia. 1979;35:1574–1576. doi: 10.1007/BF01953200. [DOI] [PubMed] [Google Scholar]
- 29.Alves MJ, Coelho MM, Collares-Pereira MJ. Diversity in the reproductive modes of females of the Rutilus alburnoides complex (Teleostei, Cyprinidae): a way to avoid the genetic constraints of uniparentalism. Mol Biol Evol. 1998;15:1233–1233. [Google Scholar]
- 30.Vinogradov AE, Borkin LJ, Günther R, Rosanov JM. Genome elimination in diploid and triploid Rana esculenta males: cytological evidence from DNA flow cytometry. Genome. 1990;33:619–627. doi: 10.1139/g90-092. [DOI] [PubMed] [Google Scholar]
- 31.Schultz RJ. Hybridization, unisexuality, and polyploidy in the Teleost Poeciliopsis (Poeciliidae) and other vertebrates. The American Naturalist. 1969;103:605–619. [Google Scholar]
- 32.Schmidt DJ, Bond NR, Adams M, Hughes JM. Cytonuclear evidence for hybridogenetic reproduction in natural populations of the Australian carp gudgeon (Hypseleotris: Eleotridae) Mol. Ecol. 2011;20:3367–3380. doi: 10.1111/j.1365-294X.2011.05206.x. [DOI] [PubMed] [Google Scholar]
- 33.Munehara H, Horita M, Kimura‐Kawaguchi MR, Yamazaki A. Origins of two hemiclonal hybrids among three Hexagrammos species (Teleostei: Hexagrammidae): genetic diversification through host switching. Ecol. Evol. 2016;6:7126–7140. doi: 10.1002/ece3.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mantovani B, Scali V. Hybridogenesis and androgenesis in the stick-insects Bacillus rossius-grandii benazzii (Insecta, Phasmotodea) Evolution. 1992;46:783–796. doi: 10.1111/j.1558-5646.1992.tb02084.x. [DOI] [PubMed] [Google Scholar]
- 35.Morishima K, et al. Cryptic clonal lineages and genetic diversity in the loach Misgurnus anguillicaudatus (Teleostei: Cobitidae) inferred from nuclear and mitochondrial DNA analyses. Genetica. 2008;132:159–171. doi: 10.1007/s10709-007-9158-1. [DOI] [PubMed] [Google Scholar]
- 36.Alves MJ, Coelho MM, Collares-Pereira MJ. Evolution in action through hybridisation and polyploidy in an Iberian freshwater fish: a genetic review. Genetica. 2001;111:375–385. doi: 10.1023/a:1013783029921. [DOI] [PubMed] [Google Scholar]
- 37.Saitoh K, Kim I-S, Lee E-H. Mitochondrial gene introgression between spined loaches via hybridogenesis. Zool. Sci. 2004;21:795–798. doi: 10.2108/zsj.21.795. [DOI] [PubMed] [Google Scholar]
- 38.Stöck M, et al. Simultaneous Mendelian and clonal genome transmission in a sexually reproducing, all-triploid vertebrate. Proc. Biol.Sci. 2012;279:1293–1299. doi: 10.1098/rspb.2011.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cimino MC. Egg-Production, Polyploidization and evolution in a diploid all-female fish of the genus Poeciliopsis. Evolution. 1972;26:294–306. doi: 10.1111/j.1558-5646.1972.tb00195.x. [DOI] [PubMed] [Google Scholar]
- 40.Scali V, Passamonti M, Marescalchi O, Mantovani B. Linkage between sexual and asexual lineages: genome evolution in Bacillus stick insects. Biol. J. Linn. Soc. 2003;79:137–150. [Google Scholar]
- 41.Nabais C, Pereira C, Cuñado N, Collares-Pereira MJ. Synaptonemal complexes in the hybridogenetic Squalius alburnoides fish complex: new insights on the gametogenesis of allopolyploids. CGR. 2012;138:31–35. doi: 10.1159/000339522. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Q, Arai K, Yamashita M. Cytogenetic mechanisms for triploid and haploid egg formation in the triploid loach Misgurnus anguillicaudatus. Journal of Experimental Zoology. 1998;281:608–619. [Google Scholar]
- 43.Ragghianti M, et al. Gametogenesis of intergroup hybrids of hemiclonal frogs. Genet. Res. 2007;89:39–45. doi: 10.1017/S0016672307008610. [DOI] [PubMed] [Google Scholar]
- 44.Dedukh, D., Litvinchuk, S., Rosanov, J., Shabanov, D. & Krasikova, A. Mutual maintenance of di- and triploid Pelophylax esculentus hybrids in R-E systems: results from artificial crossings experiments. BMC Evol. Biol. 17, (2017). [DOI] [PMC free article] [PubMed]
- 45.Dedukh D, et al. Variation in hybridogenetic hybrid emergence between populations of water frogs from the Pelophylax esculentus complex. PLoS ONE. 2019;14(11):e0224759. doi: 10.1371/journal.pone.0224759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ragghianti M, et al. Molecular characterization of a centromeric satellite DNA in the hemiclonal hybrid frog Rana esculenta and its parental species. Chromosome Res. 1995;3:497–506. doi: 10.1007/BF00713965. [DOI] [PubMed] [Google Scholar]
- 47.Marracci S, et al. RrS1-like sequences of water frogs from Central Europe and around the Aegean Sea: chromosomal organization, evolution, possible function. J. Mol. Evol. 2011;72:368–382. doi: 10.1007/s00239-011-9436-5. [DOI] [PubMed] [Google Scholar]
- 48.Gosner KL. A simplified table for staging anuran embryos and larvae with notes on identification. Herpetologica. 1960;16:183–190. [Google Scholar]
- 49.Dedukh D, et al. Cytological maps of lampbrush chromosomes of European water frogs (Pelophylax esculentus complex) from the Eastern Ukraine. BMC Genet. 2013;14:26. doi: 10.1186/1471-2156-14-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ogielska M, Kotusz A. Pattern and rate of ovary differentiation with reference to somatic development in anuran amphibians. J. Morphol. 2004;259:41–54. doi: 10.1002/jmor.10162. [DOI] [PubMed] [Google Scholar]
- 51.Dedukh D, et al. Optional endoreplication and selective elimination of parental genomes during oogenesis in diploid and triploid hybrid European water frogs. PLoS ONE. 2015;10:e0123304. doi: 10.1371/journal.pone.0123304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Storme N, Geelen D. Sexual polyploidization in plants - cytological mechanisms and molecular regulation. New Phytologist. 2013;198:670–684. doi: 10.1111/nph.12184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang J, et al. Cell fusion as the formation mechanism of unreduced gametes in the gynogenetic diploid hybrid fish. Sci. Rep. 2016;6:31658. doi: 10.1038/srep31658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Potapova, T. & Gorbsky, G. J. The consequences of chromosome segregation errors in mitosis and meiosis. Biology (Basel) 6, (2017). [DOI] [PMC free article] [PubMed]
- 55.Schvartzman J-M, Sotillo R, Benezra R. Mitotic chromosomal instability and cancer: mouse modelling of the human disease. Nat. Rev. Cancer. 2010;10:102–115. doi: 10.1038/nrc2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Subrahmanyam NC, Kasha KJ. Selective chromosomal elimination during haploid formation in barley following interspecific hybridization. Chromosoma. 1973;42:111–125. [Google Scholar]
- 57.Ishii T, Karimi-Ashtiyani R, Houben A. Haploidization via chromosome elimination: means and mechanisms. Ann. Rev. Plant Biol. 2016;67:1–18. doi: 10.1146/annurev-arplant-043014-114714. [DOI] [PubMed] [Google Scholar]
- 58.Gernand D, et al. Uniparental chromosome elimination at mitosis and interphase in wheat and pearl millet crosses involves micronucleus formation, progressive heterochromatinization, and DNA fragmentation. Plant Cell. 2005;17:2431–2438. doi: 10.1105/tpc.105.034249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gernand D, Rutten T, Pickering R, Houben A. Elimination of chromosomes in Hordeum vulgare×H. bulbosum crosses at mitosis and interphase involves micronucleus formation and progressive heterochromatinization. Cytogenet. Genome Res. 2006;114:169–174. doi: 10.1159/000093334. [DOI] [PubMed] [Google Scholar]
- 60.Brown JD, O’Neill RJ. Chromosomes, conflict, and epigenetics: chromosomal speciation revisited. Annu. Rev. Genom. Hum. G. 2010;11:291–316. doi: 10.1146/annurev-genom-082509-141554. [DOI] [PubMed] [Google Scholar]
- 61.Sanei M, Pickering R, Kumke K, Nasuda S, Houben A. Loss of centromeric histone H3 (CENH3) from centromeres precedes uniparental chromosome elimination in interspecific barley hybrids. PNAS. 2011;108:13373–13374. doi: 10.1073/pnas.1103190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ravi M, Chan S. Haploid plants produced by centromere-mediated genome elimination. Nature. 2010;464:615–618. doi: 10.1038/nature08842. [DOI] [PubMed] [Google Scholar]
- 63.Kang Y, et al. Differential chromosomal localization of centromeric histone CENP-A contributes to nematode programmed DNA elimination. Cell Rep. 2016;16:2308–2316. doi: 10.1016/j.celrep.2016.07.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Grenfell AW, Heald R, Strzelecka M. Mitotic noncoding RNA processing promotes kinetochore and spindle assembly in Xenopus. J. Cell Biol. 2016;214:133–141. doi: 10.1083/jcb.201604029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rošić S, Köhler F, Erhardt S. Repetitive centromeric satellite RNA is essential for kinetochore formation and cell division. J. Cell Biol. 2014;207:335–349. doi: 10.1083/jcb.201404097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Staiber W. Chromosome elimination in germ line-soma differentiation of Acricotopus lucidus (Diptera, Chironomidae) Genome. 2006;49(3):269–274. doi: 10.1139/g05-103. [DOI] [PubMed] [Google Scholar]
- 67.Timoshevskiy VA, Herdy JR, Keinath MC, Smith JJ. Cellular and molecular features of developmentally programmed genome rearrangement in a vertebrate (Sea lamprey: Petromyzon marinus) PLoS Genet. 2016;12(6):e1006103. doi: 10.1371/journal.pgen.1006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tan EH, et al. Catastrophic chromosomal restructuring during genome elimination in plants. eLife. 2015;4:e06516. doi: 10.7554/eLife.06516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Elinson RP, Bogart JP, Licht LE, Lowcock LA. Gynogenetic mechanisms in polyploid hybrid salamanders. J. Exp. Zool. 1992;264:93–99. [Google Scholar]
- 70.Komaru A, Ookubo K, Kiyomoto M. All meiotic chromosomes and both centrosomes at spindle pole in the zygotes discarded as two polar bodies in clam Corbicula leana: unusual polar body formation observed by antitubulin immunofluorescence. Dev. Genes Evol. 2000;210:263–269. doi: 10.1007/s004270050313. [DOI] [PubMed] [Google Scholar]
- 71.Zhang J, et al. Meiosis completion and various sperm responses lead to unisexual and sexual reproduction modes in one clone of polyploid Carassius gibelio. Sci.Rep.s. 2015;5:10898. doi: 10.1038/srep10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Szydłowski P, Chmielewska M, Rozenblut-Kościsty B, Ogielska M. The frequency of degenerating germ cells in the ovaries of water frogs (Pelophylax esculentus complex) Zoomorphology. 2017;136:75–83. [Google Scholar]
- 73.Biriuk OV, et al. Gamete production patterns and mating systems in water frogs of the hybridogenetic Pelophylax esculentus complex in north-eastern Ukraine. J. Zool. Syst. Evol. Res. 2016;54:215–225. [Google Scholar]
- 74.Christiansen DG. Gamete types, sex determination and stable equilibria of all-hybrid populations of diploid and triploid edible frogs (Pelophylax esculentus) BMC Evol. Biol. 2009;9:135. doi: 10.1186/1471-2148-9-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brown SW. Automatic frequency response in the evolution of male haploidy and other coccid chromosome systems. Genetics. 1964;49:797–817. doi: 10.1093/genetics/49.5.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Berger, L., Uzzell, T. & Hotz, H. Postzygotic reproductive isolation between mendelian species of European water frogs. Zool.Pol. 39, (1994).
- 77.Kiernan, J. A. Histological and histochemical methods. Theory and practice. 3rd ed. Bloxham 502p., (UK: Scion; 1999).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.