Abstract
The use of organochlorine pesticides, such as dichlorodiphenyltrichloroethane (DDT) and benzene hexachloride (BHC), have contributed substantially to the increase and stable supply of food production post-World War II. However, they have also become a major source of pollution on a global scale due to their persistence in the environment, high bioconcentration, toxicity, and their long-distance mobility. Although the use and production of these pesticides were banned over 45 years ago, they still present a risk to human health and ecosystems, and pose a threat to food safety. These pesticides were designated as persistent organic pollutants (POPs) by the Stockholm Convention in 2001, which urged the industry to reduce or eliminate them globally. The authors of this study have been involved in the research and development of bioaugmentation soil remediation technology to reduce the risk of environmental and crop contamination originating from POPs. In this paper, these studies are summarized, from basic studies (1, 2, 3) to an applied study (4), as follows: (1) use of the soil–charcoal perfusion method to explore POP-degrading bacteria, (2) bacteriological characteristics, metabolic pathways and dechlorination genes of the hexaclorobenzene (HCB)-mineralizing bacterial strain PD653, (3) characteristics and metabolic pathways of the dieldrin-degrading bacterial strain KSF27, and (4) application of these degrading bacteria for remediation of POPs-contaminated soil.
Keywords: persistent organic pollutants (POPs), soil–charcoal perfusion system, metabolic pathway, Nocardioides sp. PD653, novel dehalogenase genes, bioremediation
Introduction
Efforts to increase food production were escalating around the world during the second half of the 20th century, following the end of World War II. It is well known that the advent of chemical synthesis technologies led to the development of organochlorine pesticides, with typical examples including DDT and BHC (hexachlorocyclohexanes; HCHs), both of which have contributed substantially to the post-war enhancement of food production and the stability of the food supply. However, the manufacture and use of these pesticides were banned in many developed countries in the mid-1970s due to their persistence, bioconcentration, and toxicity, as highlighted in Rachel Carson’s book “Silent Spring.” Despite the regulatory efforts, these pesticides continue to cause pollution worldwide, even in the 21st century.1–3) Since they adversely affect both human health and the environment, the Stockholm Convention on Persistent Organic Pollutants (POPs) adopted in 2001 global mandates for the reduction and elimination of these pesticides. However, some of these POPs still remain in agricultural land in Japan, and contamination (detected POP values exceeding maximum residue levels) of crops such as Cucurbitaceae has been confirmed.4) Thus, there is a need for the development of soil remediation technologies to reduce the risk of contamination of crops by POPs. Among the technologies being developed for this purpose, bioaugmentation, in which POP-degrading bacteria are introduced from an external site into a contaminated site, is expected to provide a safe and inexpensive in situ strategy that does not adversely affect soil function. However, POP-degrading bacteria rarely exist in nature, and their enrichment and isolation have been extremely difficult. Accordingly, this paper introduces a method for the enrichment and isolation of POP-degrading bacteria using the soil–charcoal perfusion system developed by the authors, which has been used to enrich and isolate degrading bacteria for hexachlorobenzene (HCB), dieldrin, and HCHs. The characteristics, metabolic pathways, and degradation gene(s) of the bacterium are described. This paper also introduces our applied studies on the development of a novel material combining these degrading bacteria and carbonized woody material for the clean-up of soil contaminated with POPs.
1. Use of the soil–charcoal perfusion method to explore POPs-degrading bacteria
The soil–charcoal perfusion method uses a porous carbonized woody material as a microhabitat and is adsorbent of organic chemicals to quickly enrich persistent compound-degrading bacteria in soil and easily isolate the bacteria there.5,6) A key aspect of this method is the fact that the soil, to which the target compound had been repeatedly applied, is mixed with 0.5% (W/W) of the special charcoal A100 (pH, 7.8; Brunauer–Emmett–Teller (BET) specific surface area, 100 g/m2; the pore volume of fine pores with a diameter of 5–20 µm occupies ≥10% of the total pore volume). This is optimized as an adsorbent and a microhabitat for persistent organic compounds and degrading bacteria, respectively. By circulating a mineral medium (MM) containing only the target compound as a C or N source through the soil–charcoal layer, the compound absorbed on the charcoal, and then the degrading bacteria in the soil can be rapidly enriched in the carbonized woody material for assimilation. Further, if only the enriched charcoal is removed from the soil and inoculated into a new A100 specimen with the same circulating procedure, it is possible to further enrich the degrading bacteria, easily purify, and isolate them from the charcoal. Using this method, degrading bacteria have been isolated from materials such as triazine compounds,7–10) amide herbicides,11) organic arsenic,12) organochlorine fungicides (pentachloronitrobenzene (PCNB), pentachlorophenol (PCP)),6,13) and HCHs.14) However, in general, POPs (e.g., HCB and dieldrin) except HCHs have very low water solubility (<0.5 mg/L). Therefore, the enrichment of POPs-degrading bacteria is very difficult with this method due to the low bioavailability of the substrate. Thus, the authors used a structural analog with a common chlorine substituent moiety and water solubility of ≥0.5 mg/L as an enrichment substrate for POPs-degrading bacteria. For example, as an enrichment substrate for HCB (water solubility 0.005 mg/L)-degrading bacteria, PCNB (water solubility 0.5 mg/L),6) in which one Cl group was substituted by a NO2 group, was used. For dieldrin (water solubility 0.2 mg/L)-degrading bacteria, aldrin-trans-diol with a cleaved epoxy group (water solubility 5 mg/L)15) was used as the substrate. For poly chlorinated biphenyl (PCB, almost insoluble)-degrading bacteria, hydroxylated PCB (4OH-3PCB) was used.16) Thus, POPs-degrading bacteria can be enriched using the soil–charcoal perfusion method with a structural analog, which has greater bioavailability as an enrichment substrate.
2. Characteristics of the HCB-mineralizing bacterial strain PD6536)
2.1. Bacteriological characteristics and metabolic pathways of the HCB- and PCNB-degrading bacterial strain PD653
The strain PD653 was isolated from a PCNB-degrading bacteria consortium, PD3 (consisting of four bacterial species),13) that had been enriched and isolated from PCNB annually applied soil using the soil–charcoal perfusion method with PCNB as an enrichment substrate. This bacterium grows using HCB as a sole carbon source and eventually mineralizes HCB to CO2 and Cl−. This strain is a Gram-variable bacillus that forms pale yellow colonies. Based on phylogenetic analysis of 16S rRNA gene sequences, strain PD653 is a new species of the genus Nocardioides. HCB and PCNB are converted into PCP by the first oxidative dechlorination and denitration, followed by conversion into tetrachloro-p-hydroquinone (TeCH) by oxidative dechlorination, and then into 2,6-dichlorohydroquinone (DiCH) by reductive dechlorination, which is eventually mineralized (detoxified). Nocardioides sp. strain PD653 is the first known natural bacterium capable of aerobically mineralizing HCB via PCP. Also, this strain has the ability to dechlorinate β-HCH, which is the most recalcitrant isomer in the environment, among the four isomers of HCHs.17,18) Therefore, strain PD653 is a highly promising strain for utilization in the bioaugmentation of POPs-contaminated soil.
2.2. Novel catabolic genes involved in catalyzing the dechlorination of HCB and PCP
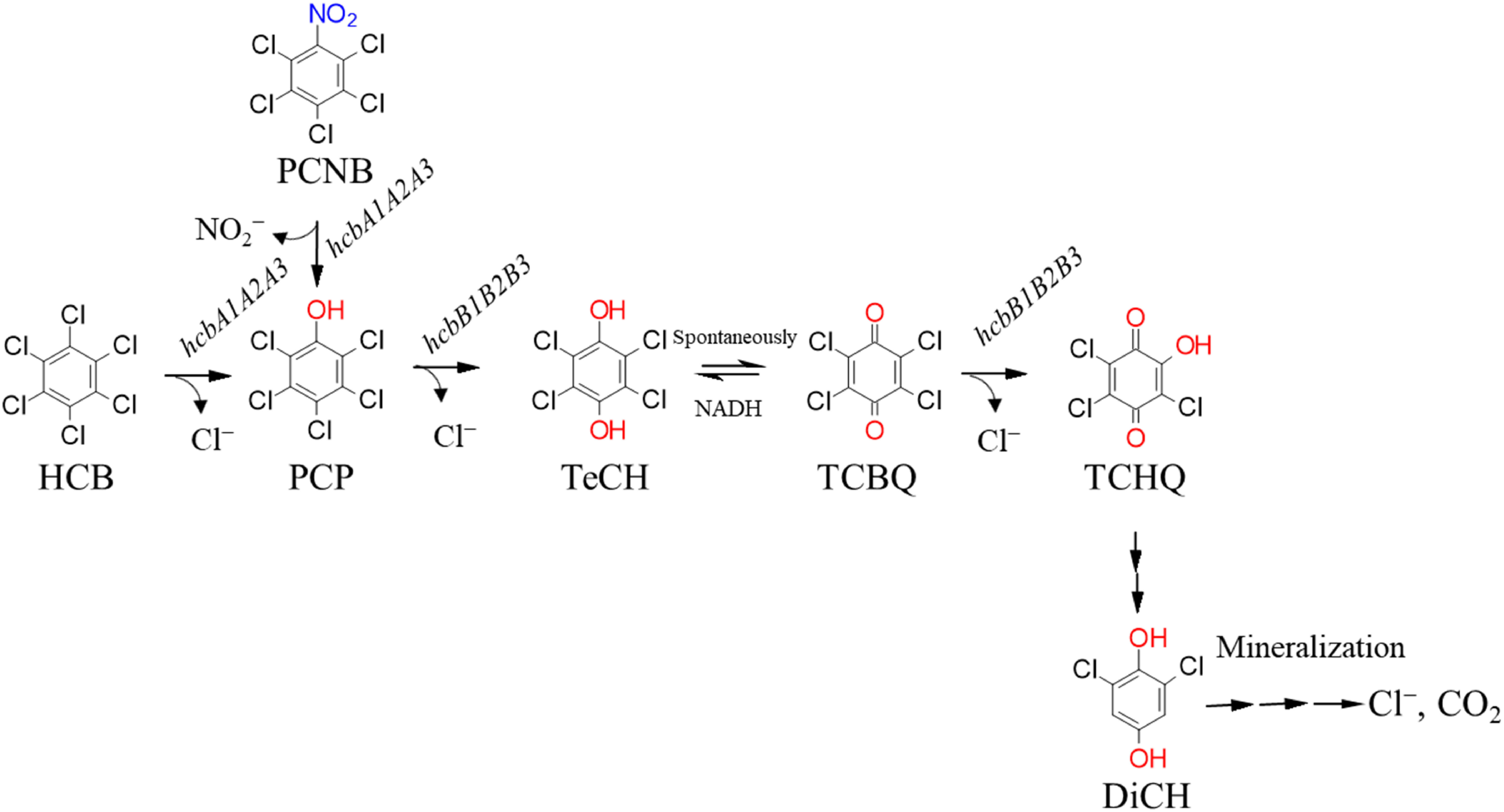
The authors conducted a comparative genomic analysis of the strain PD653 and the naturally occurring strain PD653-B2, which is deficient in the HCB dechlorination ability, and identified the hcbA1A2A3 encoding enzymes that catalyze the dechlorination and hydroxyl substitution reaction from HCB to PCP.19) In addition, RNA-seq analysis led to the identification of hcbB1B2B3, which encodes an enzyme that catalyzes a two-step hydrolytic dichlorination of PCP to 2,3,5-trichloro-6-hydroxy-p-benzoquinone (TCHQ).20) Based on the deduced amino acid sequence and the gene structure, it is strongly suggested that the protein involved in both the HCB and PCP dechlorination belongs to the two-component flavin diffusible monooxygenase family (TC-FDM),21) which reacts with flavin and O2 as substrates. To date, biocatalytic HCB dechlorination reactions under aerobic conditions have been studied using cytochrome P450,22) which is present in mammalian liver tissue, and the F87W/Y96F/L244A/V247L mutant gene of the Pseudomonas putida-derived CYP101 protein.23) Therefore, Cytochrome P450 monooxygenase is also believed to potentially play a role in the dechlorination reaction.24) However, our findings demonstrated that the TC-FDM possessed by the bacteria plays an important role in the dechlorination of halogenated aromatic compounds. In addition, genes derived from bacteria involved in PCP dechlorination have only been identified in Gram-negative bacteria25,26); Gram-positive bacteria, including the strain PD653, have a different dechlorination mechanism. However, this study led to the first discovery of novel PCP dechlorination genes (hcbB1B2B3) in Gram-positive bacteria.20) Figure 1 describes the dehalogenase genes associated with aerobic degradation pathways of PCNB and HCB by the strain PD653.
Fig. 1. Proposed pathway for the aerobic degradation of HCB and PCNB employed by the strain PD653. The genes involved at each catabolic step are indicated.
3. Characteristics and metabolic pathways of the dieldrin-degrading bacterial strain KSF2715)
The strain KSF27 was isolated from endosulfan (a structural analog of dieldrin) annually applied soil using the soil–charcoal perfusion method with aldrin-trans-diol as an enrichment substrate. This strain is a typical actinomycete (Gram-positive bacterium). The surface shape of colonies is flocculent, and aerial hyphae are orange–white on a yeast mold (YM) agar plate. Based on phylogenetic analysis of 16S rRNA gene sequences, strain KSF27 is a new species of the genus Pseudonocardia. When this strain was incubated at 30°C under shaking in MM containing 200 mg/L of sodium pyruvate as a carbon source for 10 days, it degraded dieldrin (5 mg/L) by 85%, although the degradation rate of known degrading bacteria was less than 50%. In addition, after examining the degradation capability of strain KSF27 for other POPs, it was revealed that heptachlor, heptachlor epoxide, and endosulfan sulfate remaining in the soil over a long period had been degraded by 84, 53 and 72%, respectively. Thus, this strain possesses a broad degradation spectrum of POPs. However, strain KSF27 is not a dechlorinating bacterium (assimilating bacterium), such as strain PD653, but a co-metabolizing bacterium that requires a carbon source, such as pyruvate. Figure 2 shows the putative metabolic pathway of dieldrin by the strain KSF27.15,27) Dieldrin was oxidized via aldrin-trans-diol (known metabolite) in which the epoxy group was hydrolyzed and converted into dihydrochlordene-1,3-dicarboxylic acid (a novel metabolite). The resulting dihydrochlordene-1,3-dicarboxylic acid was oxidatively decarboxylated, converted into 3-hydroxydihydrochlordene-1-carboxylic acid, and then into dehydrated chlordene-1-carboxylic acid. In addition, an additional pathway existed in which aldrin-trans-diol was hydroxylated and converted into 9-syn-hydroxyaldrin-trans-diol.
Fig. 2. Proposed pathways of dieldrin degradation employed by strain KSF27.
4. Bioremediation of HCHs-contaminated soil using charcoal, enriched with a constructed bacterial consortium
4.1. Characteristics of the HCH-degrading bacterial strain TSK-114)
The strain TSK-1 was isolated from γ-HCH+chlororhalonil (TPN) annually applied upland soil (collected from the Yayoi field at The University of Tokyo)28) using the soil–charcoal perfusion method with γ-HCH (water solubility 5 mg/L) as an enrichment substrate. This strain is a Gram-negative bacillus that forms yellow colonies. Based on phylogenetic tree analysis of 16S rRNA gene sequence, strain TSK-1 is a new species of the genus Sphingomonas, different from known HCHs-degrading bacteria. This strain possesses intact γ-HCH-degrading genes (linA, B, C, D, E), which can completely dechlorinate and assimilate γ-HCH, similar to other known HCHs-degrading bacteria.29) Strain TSK-1 could also completely dechlorinate α-, γ- and δ-HCH isomers. For the β-isomer, however, two chloride ions per molecule were released with 90% degradation. Accordingly, we constructed a two-bacterial consortium consisting of the β-HCH-assimilating bacterial strain PD653 and the strain TSK-1, in an attempt to clean-up HCHs-contaminated soil.
4.2. Development of enriched charcoal with a construction of a two-bacterial consortium and the performance evaluation using HCHs-contaminated soil
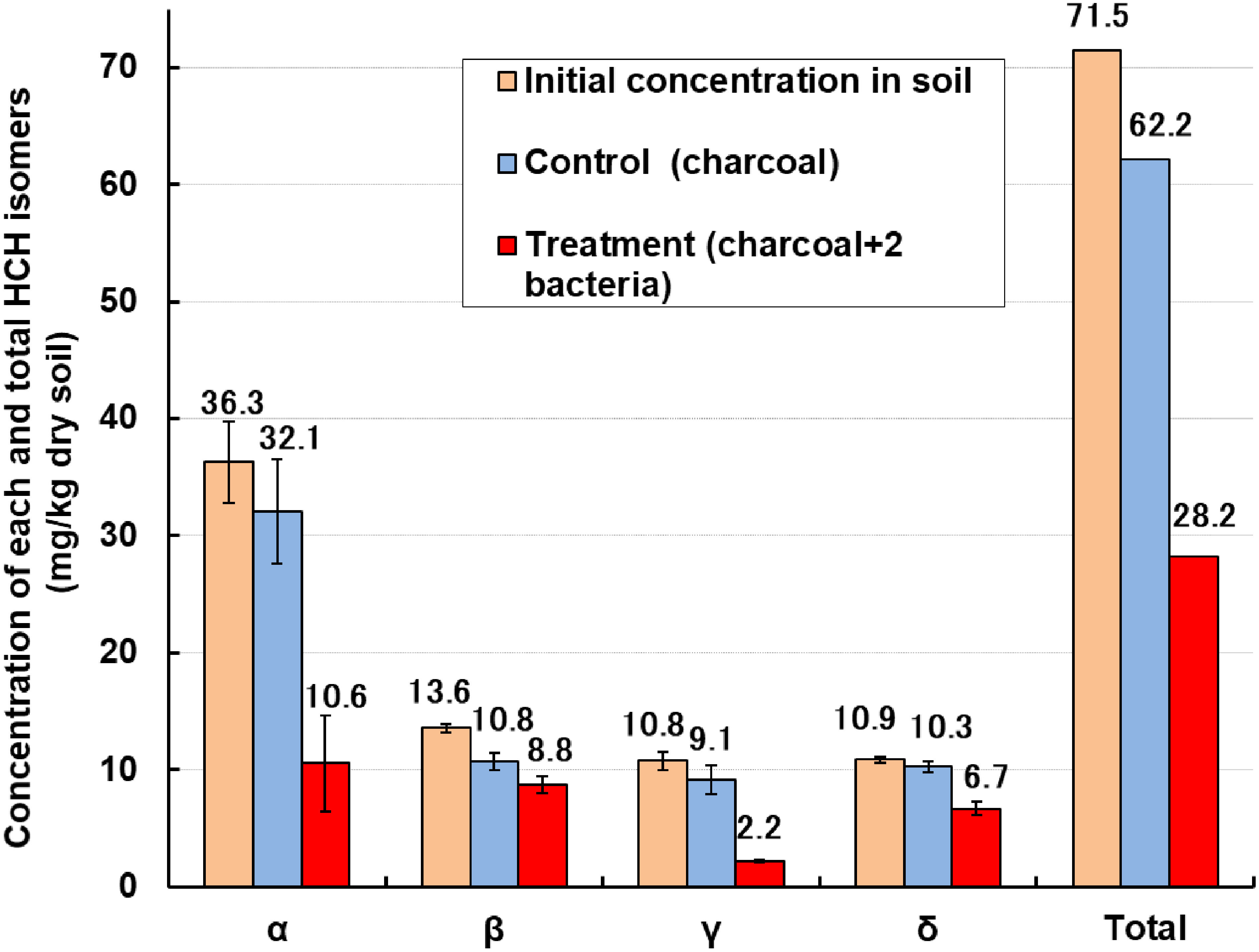
After mixing the R2A culture medium of strains TSK-1 (OD600=0.6) and PD653 (OD600=1.1) at a ratio of 6 : 1 (v/v), the mixture was immersed overnight in 20 g of coconut shell charcoal CC150 (pH, 7.8; specific surface area, 150 m2/g; particle size, 0.5–4 mm) to obtain an enriched type of composite carbonized material. Briefly, 30 g (equivalent to dry soil) of actually contaminated soil (pH, 5.8; total carbon (TC), 1.8%; residual concentrations of α-, β-, γ-, and δ-HCH isomers were 36.3, 13.6, 10.8, and 10.9 mg/kg dry soil, respectively) was mixed with 5% (w/w) of the enriched charcoal. The soil moisture was adjusted to 35% and the mixture was incubated at 25°C under dark conditions with a supply of water (1–2 mL/week) for 2 weeks. The degradation rates were 67.1, 18.5, 76.1, 34.6% for α-, β-, γ- and δ-HCH isomers, respectively, and in total, 55% of the HCHs were degraded compared to the control plot (with non-enriched charcoal) (Fig. 3).30) To the best of our knowledge, this is the first report to demonstrate that a charcoal enriched with a constructed bacterial consortium efficiently cleans up HCHs-contaminated soil. In addition, we conducted a field experiment by placing a 1-cm-thick layer of the charcoal A100 (pH, 7.8; BET specific surface area, 100 m2/g; particle size, 10 mm), enriched with the simazine-degrading bacterial consortium CD7 (consisting of three bacterial species),31,32) under the subsoil at 15 cm deep in a golf course for 2 years. This was to test the prevention of subsoils, rivers, and groundwater contamination with the herbicide simazine (water pollutant pesticide). Simazine is widely used in Japan and frequently detected in river water. Simazine, which was sprayed twice a year (4 times spraying in total) on the turf, leached down to the charcoal layer where it was then absorbed and degraded by 92, 70, 76 and 92% for each spraying in comparison with the control plot containing non-enriched charcoal.31,33) This result indicated that the carbonized woody material enriched with a degrading bacterial consortium was a novel practical material for remediating environments and soil contaminated with POPs and other recalcitrant organic chemicals.
Fig. 3. Simultaneous degradation of four HCH isomers in the contaminated sub-soil using coconut husk charcoal enriched with a constructed bacterial consortium (strainTSK1 + strain PD653). The mean±S.D. for triplicate samples is shown.

Concluding remarks
The study on microbial degradation of POPs aims to discover new knowledge by studying the past through scrutiny of the old. Research on organochlorine pesticides, whose production and use were banned over 40 years ago, has led to the discovery of novel aerobic POPs-degrading bacteria, novel metabolites, and dehalogenase genes. In general, the degradation of POPs by soil microorganisms produces more hydrophilic metabolites and consequently reduces their persistence in soil, their toxicity to mammals, including humans, as well as reduces environmental risks. In the future, we hope to accelerate our applied research concerning the formulation of these degrading bacteria or enzyme preparations to actively contribute to the safety and security of the food supply, and a healthy and sustainable environment. We also hope to dispel the negative image of pesticides through the remediation of POPs-contaminated soil, particularly arable soil.
Acknowledgments
All of the studies introduced in this review paper were conducted at my laboratory in the National Institute for Agro-Environmental Sciences (NIAES) under a system of linked graduate schools of the Tokyo University of Agriculture, laboratory of microbiology. At first, I would like thank Dr. Tai Uchimura and Dr. Sanae Okada for creating the system and inviting me as a professor. I also thank the former graduate students: Dr. Kenichi Yamazaki, Dr. Futa Sakakibara, Dr. Koji Ito, Mr. Kazuhito Sekiya, Mr. Fujimasa Kawashima, Mr. Yuichi Ikezawa, the former postdoctoral fellows: Dr. Ichiro Kamei, Dr. Ryota Kataoka, Dr. Takashi Hatakeyama, Dr. Youbin Si, Dr. Hirozumi Watanabe, Dr. Satomi Murata, research assistants and trainees of my laboratory. I would like to thank my collaborators: Dr. Akio Iwasaki, Dr. Hiromasa Kiyota, Dr. Naoki Harada, Mr. Yuichi Yoshioka, Dr. Kousuke Suyama, Dr. Koji Satuma, Dr. Fabrice Martin-Laurent, Dr. Ryuichiro Kondo, Dr. Arata Katayama and Dr. J. P. E. Anderson for their considerable contribution to my work and stimulating discussion. I express my sincere acknowledgement to the researchers of NIAES: Dr. Yasuhiro Yogo, Dr. Masako Ueji, Dr. Koji Hama, Dr. Yoichi Mayumi, Dr. Kiyotaka Miyashita, and Dr. Akira Hasebe for their support and kind guidance. Finally, I wish to acknowledge my wife and daughters for their assistance and warm encouragement.
This study was supported by a grant from the Ministry of Agriculture, Forestry and Fisheries of Japan (Research Project for Ensuring Food Safety from Farm to Table PO-2214), the JSPS Research Fellowship for Young Scientists (17J00825) and JSPS KAKENHI (Grant Number 18H02319).
References
- 1).J. L. Barber, A. J. Sweetman, D. van Wijk and K. C. Jones: Sci. Total Environ. 349, 1–442 (2005). [DOI] [PubMed] [Google Scholar]
- 2).E. Matsumoto, Y. Kawanaka, S.-J. Yun and H. Oyaizu: Appl. Microbiol. Biotechnol. 84, 205–216 (2009). [DOI] [PubMed] [Google Scholar]
- 3).J. Weber, C. J. Halsall, D. Muir, C. Teixeira, J. Small, K. Solomon, M. Hermanson, H. Hung and T. Bidleman: Sci. Total Environ. 408, 2966–2984 (2010). [DOI] [PubMed] [Google Scholar]
- 4).Y. Hashimoto: J. Pestic. Sci. 30, 397–402 (2005). [Google Scholar]
- 5).K. Takagi: “Nougyougijutsutaikei Dohihen Vol. 3 (11),” Noubunkyo, Tokyo, pp. 49–55, 2000. (in Japanese).
- 6).K. Takagi, A. Iwasaki, I. Kamei, K. Satsuma, Y. Yoshioka and N. Harada: Appl. Environ. Microbiol. 75, 4452–4458 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7).A. Iwasaki, K. Takagi, Y. Yoshioka, K. Fujii, Y. Kojima and N. Harada: Pest Manag. Sci. 63, 261–268 (2007). [DOI] [PubMed] [Google Scholar]
- 8).K. Yamazaki, K. Fujii, A. Iwasaki, K. Takagi, K. Satsuma, N. Harada and T. Uchimura: FEMS Microbiol. Lett. 286, 171–177 (2008). [DOI] [PubMed] [Google Scholar]
- 9).K. Takagi, K. Fujii, K. Yamazaki, N. Harada and A. Iwasaki: Appl. Microbiol. Biotechnol. 94, 1647–1656 (2012). [DOI] [PubMed] [Google Scholar]
- 10).T. Hatakeyama, K. Takagi, K. Yamazaki, F. Sakakibara, K. Ito, E. Takasu, T. Naokawa and K. Fujii: World J. Microbiol. Biotechnol. 31, 785–793 (2015). [DOI] [PubMed] [Google Scholar]
- 11).N. Harada, K. Takagi, A. Harazono, K. Fujii and A. Iwasaki: Soil Biol. Biochem. 38, 173–179 (2006). [Google Scholar]
- 12).N. Harada, K. Takagi, K. Baba, K. Fujii and A. Iwasaki: Biodegradation 21, 491–499 (2010). [DOI] [PubMed] [Google Scholar]
- 13).K. Takagi, Y. Yoshioka, A. Iwasaki, I. Kamei and N. Harada: Organohalogen Compd. 69, 254–260 (2007). [Google Scholar]
- 14).K. Takagi, W. Sasaki and R. Kataoka: Proceedings of the 4th International Conference on Soil Pollution and Remediation, Yantai, China, 186–189 (2012).
- 15).F. Sakakibara, K. Takagi, R. Kataoka, H. Kiyota, Y. Sato and S. Okada: Biochem. Biophys. Res. Commun. 411, 76–81 (2011). [DOI] [PubMed] [Google Scholar]
- 16).S. M. Murata, F. Sakakibara, K. Fujita, M. Fukuda, M. Kuramata and K. Takagi: Chemosphere 165, 173–182 (2016). [DOI] [PubMed] [Google Scholar]
- 17).K. Takagi, I. Kamei and A. Iwasaki: Proceedings of the 20th International Symposium on Environmental Biogeochemistry, Istanbul, Turkey, PS II-29 (2011).
- 18).K. Ito, K. Takagi, R. Kataoka, H. Kiyota and A. Iwasaki: J. Pestic. Sci. 44, 171–176 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19).K. Ito, K. Takagi, A. Iwasaki, N. Tanaka, Y. Kanesaki, F. Martin-Laurent and S. Igimi: Appl. Environ. Microbiol. 83, e00824-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20).K. Ito, K. Takagi, Y. Matsushima, A. Iwasaki, N. Tanaka, Y. Kanesaki, F. Martin-Laurent and S. Igimi: J. Pestic. Sci. 43, 1–8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21).L. Xun: J. Bacteriol. 178, 2645–2649 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22).H. M. Mehendal, M. Fields and H. B. Matthews: J. Agric. Food Chem. 23, 261–265 (1975). [DOI] [PubMed] [Google Scholar]
- 23).X. Chen, A. Christopher, L. P. Jones, S. G. Bell, Q. Guo, F. Xu, Z. Rao and L.-L. Wong: J. Biol. Chem. 277, 37519–37526 (2002). [DOI] [PubMed] [Google Scholar]
- 24).O. Uhlik, M. Strejcek, J. Vondracek, L. Musilova, J. Ridl, P. Lovecka and T. Macek: Chemoshere 113, 141–145 (2014). [DOI] [PubMed] [Google Scholar]
- 25).S. Chanama and R. L. Crawford: Appl. Environ. Microbiol. 63, 4833–4838 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26).M. Cai and L. Xun: J. Bacteriol. 184, 4672–4680 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27).F. Sakakibara: PhD Thesis, Tokyo University of Agriculture (2013) (in Japanese).
- 28).K. Takagi, H. Wada and S. Yamazaki: Soil Sci. Plant Nutr. 37, 583–590 (1991). [Google Scholar]
- 29).K. Miyauchi, H. S. Lee, M. Fukuda, M. Takagi and Y. Nagata: Appl. Environ. Microbiol. 68, 1803–1807 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30).K. Takagi, R. Kataoka and K. Ito: Abstract book of XIX International Plant Protection Congress (IPPC2019), Hyderabad, India, p. 288, Poster No. IPM2 P9 (2019).
- 31).K. Takagi, A. Iwasaki and Y. Yoshioka: Proceedings of the 2nd International Conference on Soil Pollution and Remediation, Nanjing, China, 228–230 (2004)
- 32).A. Iwasaki, K. Takagi, Y. Yoshioka, K. Fujii, Y. Kojima and N. Harada: Pest Manag. Sci. 63, 261–268 (2007). [DOI] [PubMed] [Google Scholar]
- 33).K. Takagi, R. Kataoka and K. Yamazaki: Jpn. Agric. Res. Q. 45, 129–136 (2011). [Google Scholar]


