Abstract
Objective
Laryngeal cancer is a common malignant tumor in the ENT, of which laryngeal squamous cell carcinoma (LSCC) accounts for more than 90% of laryngeal cancer. The purpose of this study is to investigate the regulatory mechanism of lncRNA SNHG16 in LSCC.
Materials and Methods
Real-time quantitative reverse transcription polymerase chain reaction (RT-qPCR) was used to measure mRNA expression. Cell Counting Kit (CCK-8), Transwell and luciferase reporter assays, flow cytometric analysis and Western blot analysis were used to investigate the function of lncRNA SNHG16 in LSCC.
Results
SNHG16 expression was increased in LSCC tissues and cells. The abnormal expression of SNHG16 was associated with clinical stage and lymph node metastasis in LSCC patients. In addition, knockdown of SNHG16 restrained cell proliferation, migration and invasion in LSCC. More importantly, SNHG16 acted as a competitive endogenous RNA in LSCC and regulated FOXP4 expression by making miR-877-5p sponge. Further, SNHG16 promoted LSCC progression by interacting with miR-877-5p and FOXP4.
Conclusion
LncRNA SNHG16 promotes the progression of LSCC by sponging miR-877-5p and upregulating FOXP4.
Keywords: SNHG16, laryngeal squamous cell carcinoma, miR-877-5p, FOXP4
Introduction
Laryngeal cancer is a malignant tumor that occurs in the larynx. It accounts for approximately 1% to 5% of the incidence of malignant tumors. The main pathological type is laryngeal squamous cell carcinoma (LSCC).1 Among the factors associated with the incidence of LSCC, smoking is the most relevant. In addition, air pollution and chemical pollution can also cause LSCC.2 Currently, the treatment of LSCC mainly includes surgery, radiation therapy, chemotherapy and biological therapy.3 The 5-year survival rate after early LSCC treatment is higher than 90%. And recurrence and metastasis are major factors affecting the prognosis of LSCC patients. LSCC patients with poor differentiation or metastatic lymph nodes have lower 5-year survival rate.4 Therefore, early diagnosis and treatment is very important to reduce the risk of laryngeal cancer.
As a new insight into the RNA world, long non-coding RNAs (lncRNAs) have been widely recognized by scholars. LncRNA is a class of RNA molecules with transcripts longer than 200 nt. They do not encode proteins, but can regulate gene expression levels through epigenetic regulation, transcriptional regulation, and post-transcriptional regulation. Many lncRNAs have been found to act as tumor promoters or inhibitors in human cancers, such as MIAT and ZFAS1.5,6 At the same time, different effects of lncRNAs were also found in LSCC. For example, lncRNA TMEM51-AS1 and RUSC1-AS1 functioned as ceRNAs to induce LSCC occurrence and predict prognosis.7 Functionally, lncRNA XIST enhanced the aggressiveness of LSCC by regulating miR-124-3p/EZH2.8 In addition, SNHG12 has been demonstrated to promote proliferation and invasion of LSCC cells via sponging miR-129-5p and potentiating WWP1 expression.9 Based on this study, we suspect that the small nucleolar RNA host gene 16 (SNHG16) may be involved in the regulation of LSCC. LncRNA SNHG16 has been extensively investigated in human cancers, For example, increased expression of lncRNA SNHG16 was associated with tumor progression and poor prognosis of non-small cell lung cancer.10 High expression of lncRNA SNHG16 contributed to the progression and metastasis of gastric cancer.11 However, there is no report on the role of lncRNA SNHG16 in LSCC. Therefore, this r study aims to solve the above problem.
Previous studies have shown that lncRNA SNHG16 is involved in tumorigenesis by regulating microRNAs (miRNAs). In this study, miR-877-5p was predicted to have a binding site with lncRNA SNHG16. The role of miR-877-5p has been investigated in human cancers, such as hepatocellular carcinoma and renal cell carcinoma.12,13 In particular, it was found that lncRNA TRG-AS1 promoted glioblastoma cell proliferation through competitive binding with miR-877-5p to regulate SUZ12 expression.14 However, the relationship between lncRNA SNHG16 and miR-877-5p has not been confirmed in previous studies and needs to be clarified. In addition, the interaction between lncRNA SNHG16 and Forkhead-box P4 (FOXP4) was investigated in LSCC. Upregulation of FOXP4 has been detected in malignant tumors such as prostate cancer and breast cancer.15,16 Functionally, FOXP4 promoted tumor growth of non-small cell lung cancer cells and was independently associated with miR-138.17 In our study, FOXP4 is predicted to be the target of miR-877-5p, but no reports have yet been reported.
Therefore, the regulatory mechanism of lncRNA SNHG16/miR-877-5p/FOXP4 was explored in LSCC. At the same time, this study initially defined the relationship between SNHG16, miR-877-5p and FOXP4. This study may exploit new targets for LSCC therapy.
Materials and Methods
Clinical Tissues
Tissue samples were obtained from 35 patients with LSCC in Qingdao Municipal Hospital. All patients did not receive radiotherapy or chemotherapy before surgery. Participants in this study provided written informed consents. This study was approved by the Institutional Ethics Committee of Qingdao Municipal Hospital. This study was conducted according to the Declaration of Helsinki.
Cell Culture and Transfection
Human normal bronchial epithelial cell line (16HBE) and LSCC cell line AMC-HN-8 were purchased from BNCC (Beijing, China). The cells were cultured in Dulbecco’s modified eagle medium (DMEM) (Thermo Fisher Scientific, Waltham, MA, USA) containing 10% fetal bovine serum (FBS) (Gibco, Rockville, MD, USA) at 37°C. Next, SNHG16 pcDNA3.1 plasmid, SNHG16 siRNA, miR-877-5p mimics, miR-877-5p inhibitor, and FOXP4 siRNA (GenePharma, Shanghai, China) were transfected into AMC-HN-8 cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), respectively.
RNA Isolation and Real-Time Quantitative Reverse Transcription Polymerase Chain Reaction (RT-qPCR)
Total RNA was isolated using TRIZOL reagent (Invitrogen, Carlsbad, CA, USA). The cDNA solution was synthesized using PrimeScript RT reagent kit (Takara, Dalian, China). RT-qPCR assay was performed using Real-time PCR Mixture assay (Takara, Dalian, China) according to the manufacturer’s instruction. The internal reference for SNHG16, FOXP4 or miR-877-5p is glyceraldehyde 3-phosphate dehydrogenase (GAPDH) or U6. The relative expression of RNA was calculated using the 2−ΔΔCq method. The primers used were: SNHG16 forward 5′-GCA GAA TGC CAT GGT TTC CC-3′and reverse 5′-GGA CAG CTG GCA AGA GAC TT-3′; miR-877-5p forward: 5′-TAG AGG AGA TGG CGC AG-3′ and reverse, 5′-GAA CAT GTC TGC GTA TCT C −3′; U6-forward: 5′-GCT TCG GCA GCA CAT ATA CTA AAA T-3′ and reverse, 5′-CGC TTC ACG AAT TTG CGT GTC AT-3′; FOXP4-forward: 5′-ATC GGC AGC TGA CGC TAA ATG AGA-3′ and reverse, 5′-AAA CAC TTG TGC AGG CTG AGG TT-3′; GAPDH forward: 5′-ACA ACT TTG GTA TCG TGG AAG G-3′, and reverse, 5′-GCC ATC ACG CCA CAG TTT C-3′.
Cell Counting Kit (CCK-8) Assay
Transfected AMC-HN-8 cells (5000 cells/well) were incubated at 37 °C in an atmosphere with 5% CO2 for 24, 48, 72 and 96 h, respectively. The cells were then incubated with CCK-8 (Dojindo, Kumamoto, Japan) for 4 h. The absorbance was observed at 450 nm using a microplate reader (Molecular Devices, Eugene, OR, USA).
Transwell Assay
Cell migration or invasion was measured in the upper chamber without or with Matrigel. Transfected AMC-HN-8 cells (density, 2×104) were added to the upper chamber. DMEM medium containing 10% FBS was added in the low chamber. After 24 h, the moving cells were stained with 0.1% crystal violet. The cells were counted and photographed under a light microscope.
Dual Luciferase Reporter Assay
The direct combination of miR-877-5p with FOXP4/SNHG16 was evaluated by dual luciferase reporter assay. To achieve this, the sequence of FOXP4/SNHG16, including binding sites of miR-877-5p (FOXP4-WT/SNHG16-WT), was cloned into the pmirGLO reporter plasmids (Promega, Madison, WI, USA), while FOXP4/SNHG16 mutated type (FOXP4-MUT/SNHG16-MUT) contained an unmatched binding sequence of miR-877-5p. Then, miR-877-5p mimics were transfected into AMC-HN-8 cells with FOXP4-WT/SNHG16-WT or FOXP4-MUT/SNHG16-MUT using Lipofectamine 3000. Luciferase activities standardized to Renilla were calculated using the Dual Luciferase Reporter Assay System (Promega, Madison, USA).
Western Blot Assay
Total proteins were extracted by lysing the cells with RIPA lysate (Beyotime, China). Then protein samples were subjected to SDS-PAGE and transferred to PVDF membranes (Millipore, USA). After blocking with 5% skim milk for 1 h at room temperature, the membranes were incubated with the primary antibody FOXP4 and GAPDH (Abcam, USA) at 4 °C for 12 h. Then the prepared membranes were incubated with secondary antibody for 2 h. Finally, the blots were detected by enhanced chemiluminescence kit (Pierce, Waltham, MA, USA).
Flow Cytometric Analysis
Flow cytometric analysis was performed to analyze the cell cycle. The transfected cells were cultured for 48 h and collected by centrifuging. After fixation with ice-cold 70% ethanol at 4°C for 12 h, the cell cycle was detected using Cell Cycle and Apoptosis Analysis Kit (Beyotime, Shanghai, China) according to the manufacturer’s instructions. All the samples were analyzed using the flow cytometry (Aceabio, San Diego, CA, USA).
Tumor Xenograft in Nude Mice
AMC-HN-8 cells transfected with sh-SNHG16 (2 × 106) were injected into the right flank of female nude mice (Guangdong Medical Laboratory Animal Center, China). Tumor growth was monitored using a vernier caliper every week. The tumors were excised on 5 weeks after the injection. The protocol was approved by Qingdao Municipal Hospital.
Statistical Analysis
Statistical analysis was performed using Statistical Product and Service Solutions (SPSS) 22. 0 software (IBM, Armonk, NY, USA). The data are expressed as mean ± SD (Standard Deviation). The relationship between RNA expression levels was analyzed using Pearson’s correlation test. Student’s t-test was used to analyze the differences between the two groups. Comparison between multiple groups was done using One-way ANOVA test followed by Post Hoc Test (Least Significant Difference). The statistical significance is considered as P < 0.05.
Results
SNHG16 Expression Is Increased in LSCC Tissues
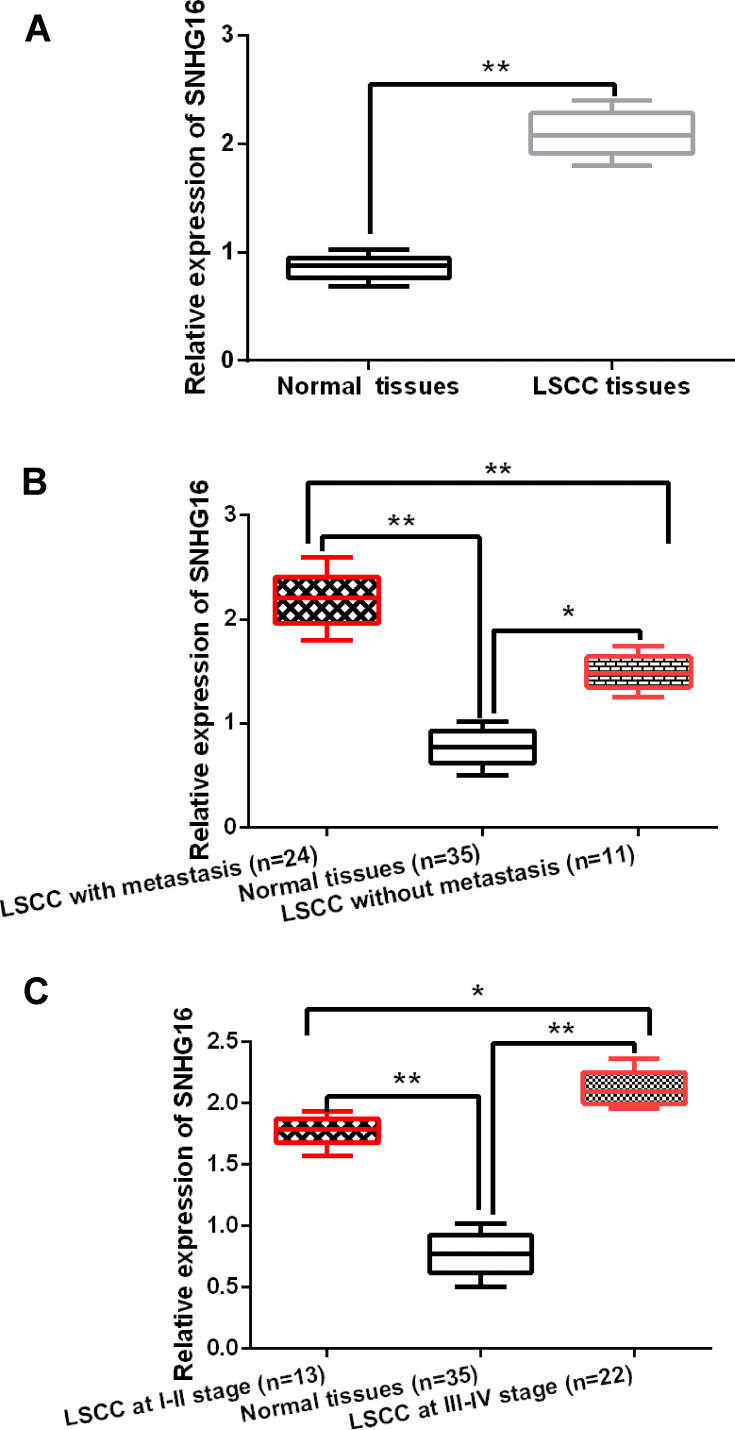
SNHG16 expression was measured in LSCC tissues. Compared with normal tissues, SNHG16 was found to be upregulated in LSCC tissues (P < 0.01, Figure 1A). In addition, we found that the expression of SNHG16 in LSCC patients with lymph node metastasis was higher than that in LSCC patients without lymph node metastasis (P < 0.01, Figure 1B). Meanwhile, higher expression of SNHG16 was identified in LSCC patients at advanced stage (III–IV, P < 0.01, Figure 1C). These results indicate that SNHG16 may be related to the clinical stage and lymph node metastasis of LSCC patients. Based on these results, we suspect that SNHG16 may be involved in the pathogenesis of LSCC.
Figure 1.
SNHG16 expression is increased in LSCC tissues. (A) SNHG16 expression was increased in LSCC tissues compared to normal tissues (n=35). (B) SNHG16 expression was higher in LSCC patients with lymph node metastasis (n=24). (C) SNHG16 was upregulated in LSCC patients at advanced stage (n=22). *P < 0.05, **P < 0.01.
SNHG16 Promotes LSCC Cell Proliferation, Migration and Invasion
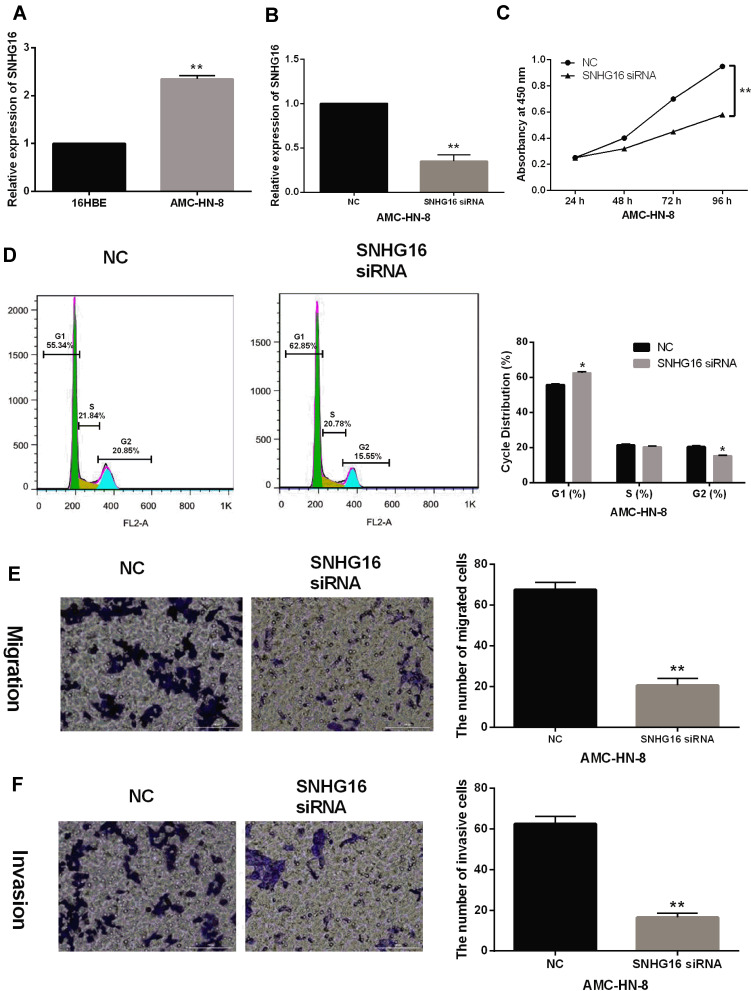
Next, upregulation of SNHG16 was detected in LSCC cells AMC-HN-8 compared to normal bronchial epithelial cells 16HBE (P < 0.01, Figure 2A). To explore the function of SNHG16 in the progression of LSCC, SNHG16 siRNA was transfected into AMC-HN-8 cells. RT-qPCR showed that the expression of SNHG16 was significantly reduced by its siRNA in AMC-HN-8 cells (P < 0.01, Figure 2B). CCK-8 assay showed that knockdown of SNHG16 restrained cell proliferation in AMC-HN-8 cells (P < 0.05, Figure 2C). In AMC-HN-8 cells with SNHG16 siRNA, the percentages of cells at the G2 and S phase cell cycle progression were decreased, while the cell percentage in the G1 phase was significantly increased (P < 0.01, Figure 2D). In addition, Transwell assay showed that downregulation of SNHG16 inhibited cell migration and invasion in AMC-HN-8 cells (P < 0.01, Figure 2E and F). These results suggest that knockdown of SNHG16 inhibits cell proliferation, migration and invasion in LSCC.
Figure 2.
SNHG16 promotes LSCC cell proliferation, migration and invasion. (A) SNHG16 expression was upregulated in AMC-HN-8 and 16HBE cells. (B) SNHG16 expression was reduced by its siRNA in AMC-HN-8 cells. (C–F) Knockdown of SNHG16 inhibited cell proliferation, migration and invasion in AMC-HN-8 cells. (D) Flow cytometric analysis for cell cycle progression of AMC-HN-8 was detected after transfection 48 h. *P < 0.05, **P < 0.01.
Reciprocal Suppression Between SNHG16 and MiR-877-5p
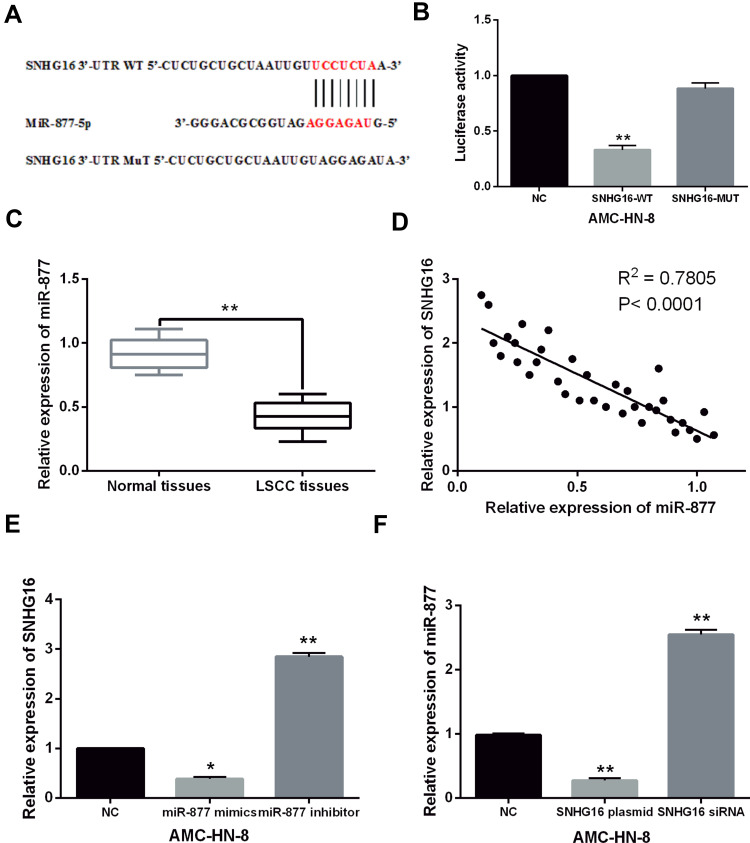
To further explain the regulatory mechanism of SNHG16 in LSCC, the starBase database (http://starbase.sysu.edu.cn/) was used to explore the downstream targets of SNHG16. It predicts that SNHG16 has a binding site to miR-877-5p (Figure 3A). Dual luciferase reporter assay showed that miR-877-5p mimics reduced the luciferase activity of WT-SNHG16, but had no effect on that of MuT-SNHG16 (P < 0.01, Figure 3B). This indicates that SNHG16 directly targets miR-877-5p. Then, downregulation of miR-877-5p was found in LSCC tissues compared to normal tissues (P < 0.01, Figure 3C). It was found that SNHG16 expression negatively regulated miR-877-5p expression in LSCC tissues (P < 0.01, Figure 3D). In addition, SNHG16 expression was found to be reduced by miR-877-5p overexpression and enhanced by miR-877-5p downregulation in AMC-HN-8 cells (P < 0.01, Figure 3E). At the same time, upregulation of SNHG16 suppressed the expression of miR-877-5p, while downregulation of SNHG16 promoted miR-877-5p expression in AMC-HN-8 cells (P < 0.01, Figure 3F). These results indicate that SNHG16 acts as a sponge for miR-877-5p in LSCC.
Figure 3.
Reciprocal suppression between SNHG16 and miR-877-5p was detected in LSCC. (A) The binding sites between SNHG16 with miR-877-5p. (B) MiR-877-5p mimics reduced the luciferase activity of WT-SNHG16, but had no effect on MuT-SNHG16. (C) MiR-877-5p expression was decreased in LSCC tissues compared to normal tissues. (D) SNHG16 expression negatively regulated miR-877-5p expression in LSCC tissues. (E) SNHG16 expression was reduced by miR-877-5p mimics and promoted by miR-877-5p inhibitor. (F) MiR-877-5p expression was downregulated by SNHG16 overexpression vector and upregulated by SNHG16 siRNA in AMC-HN-8 cells. *P <0.05, **P <0.01.
MiR-877-5p Directly Targets FOXP4
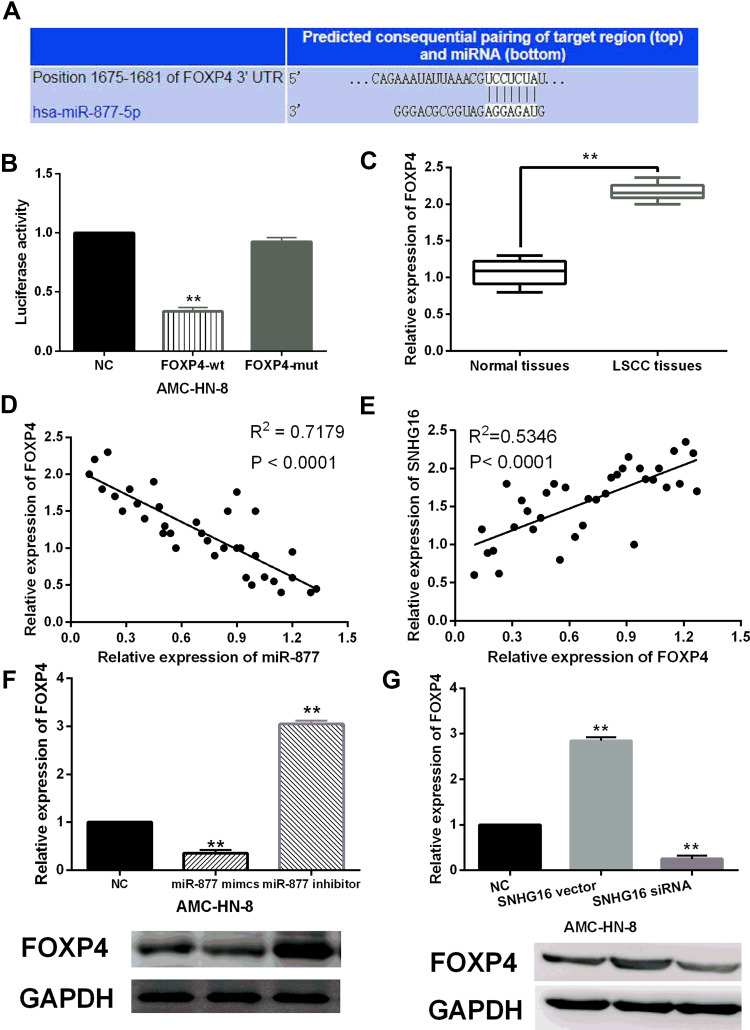
In addition, TargetScan (http://www.targetscan.org) predicts that FOXP4 is a potential target for miR-877-5p (Figure 4A). We found that miR-877-5p mimics reduced the luciferase activity of wt-FOXP4, but had no effect on the luciferase activity of mut-FOXP4 (P < 0.01, Figure 4B). It indicates that miR-877-5p can directly bind to FOXP4. Then, upregulation of FOXP4 was detected in LSCC tissues compared to normal tissues (P < 0.01, Figure 4C). A negative correlation between miR-877-5p and FOXP4 expression was found in LSCC tissues (P < 0.01, Figure 4D). Interestingly, a positive correlation between FOXP4 and SNHG16 expression was found in LSCC tissues (P < 0.01, Figure 4E). In AMC-HN-8 cells, the mRNA and protein expression of FOXP4 was reduced by miR-877-5p mimics and promoted by miR-877-5p inhibitor (P < 0.01, Figure 4F). However, the mRNA and protein expression of FOXP4 was promoted by the SNHG16 vector and inhibited by the SNHG16 siRNA in AMC-HN-8 cells (P < 0.01, Figure 4G). Based on these results, we speculate that FOXP4 may participate in the tumorigenesis of LSCC by regulating SNHG16 or miR-877-5p.
Figure 4.
MiR-877-5p directly targets FOXP4. (A) The binding sites between FOXP4 with miR-877-5p. (B) miR-877-5p mimics reduced the luciferase activity of wt-FOXP4, but had no effect on mut-FOXP4 luciferase activity. (C) FOXP4 expression was upregulated in LSCC tissues compared to normal tissues. (D) FOXP4 expression negatively regulated miR-877-5p expression in LSCC tissues. (E) FOXP4 expression positively regulated SNHG16 expression in LSCC tissues. (F) The mRNA and protein expression of FOXP4 was reduced by miR-877-5p mimics and increased by miR-877-5p inhibitor. (G) The mRNA and protein expression of FOXP4 was upregulated by SNHG16 overexpression vector and downregulated by SNHG16 siRNA in AMC-HN-8 cells. **P <0.01.
SNHG16 Is Involved in LSCC Tumorigenesis by Regulating MiR-877-5p/FOXP4 Axis
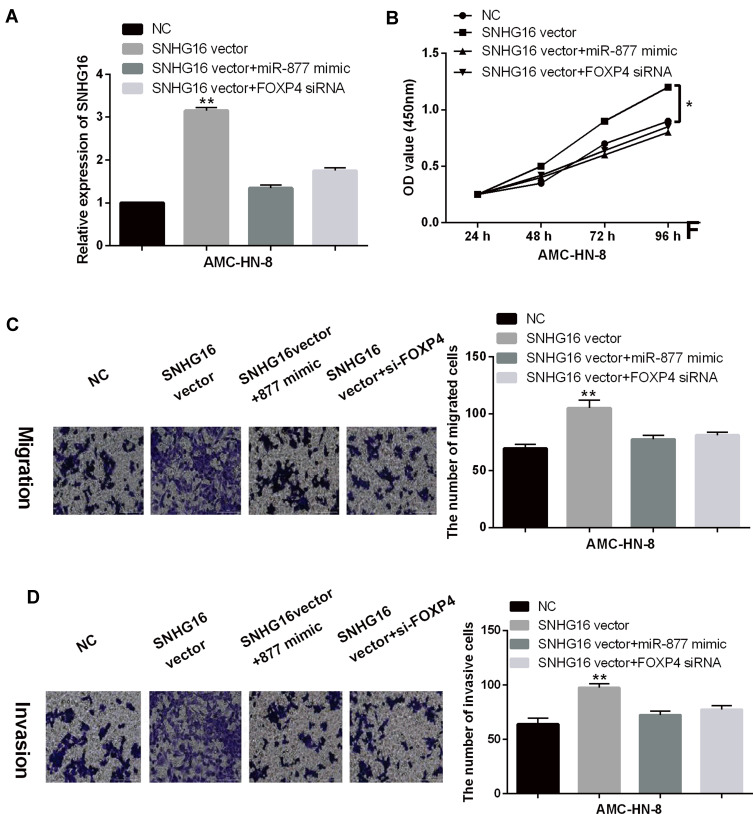
The above results indicate that SNHG16 may play an important role in LSCC tumorigenesis by affecting miR-877-5p or FOXP4. To verify their interaction, miR-877-5p mimics or FOXP4 siRNA were transfected into AMC-HN-8 cells with SNHG16 vector. RT-qPCR showed that the expression of SNHG16 was increased by its vector, but miR-877-5p mimics or FOXP4 siRNA reduced the increased expression of SNHG16 in AMC-HN-8 cells (Figure 5A). Functionally, upregulation of SNHG16 promoted the proliferation of AMC-HN-8 cells. In addition, FOXP4 downregulation or miR-877-5p overexpression attenuated the promoting effect of SNHG16 on cell proliferation in AMC-HN-8 cells (Figure 5B). Similarly, upregulation of SNHG16 also promoted cell migration and invasion in AMC-HN-8 cells. And the promotion of cell migration and invasion induced by SNHG16 vector was also reduced by miR-877-5p overexpression or FOXP4 downregulation in AMC-HN-8 cells (Figure 5C and D). Briefly, SNHG16 promotes LSCC progression by downregulating miR-877-5p and upregulating FOXP4.
Figure 5.
SNHG16 is involved in LSCC tumorigenesis by regulating miR-877-5p/FOXP4 axis. (A) MiR-877-5p mimics or FOXP4 siRNA reduced the increased expression of SNHG16 induced by its vector in AMC-HN-8 cells. (B–D) FOXP4 downregulation or miR-877-5p overexpression weakened the promoting effect of SNHG16 on cell proliferation, migration and invasion in AMC-HN-8 cells. *P < 0.05, **P < 0.01.
LncRNA SNHG16 Downregulation Reduces Tumor Growth in vivo
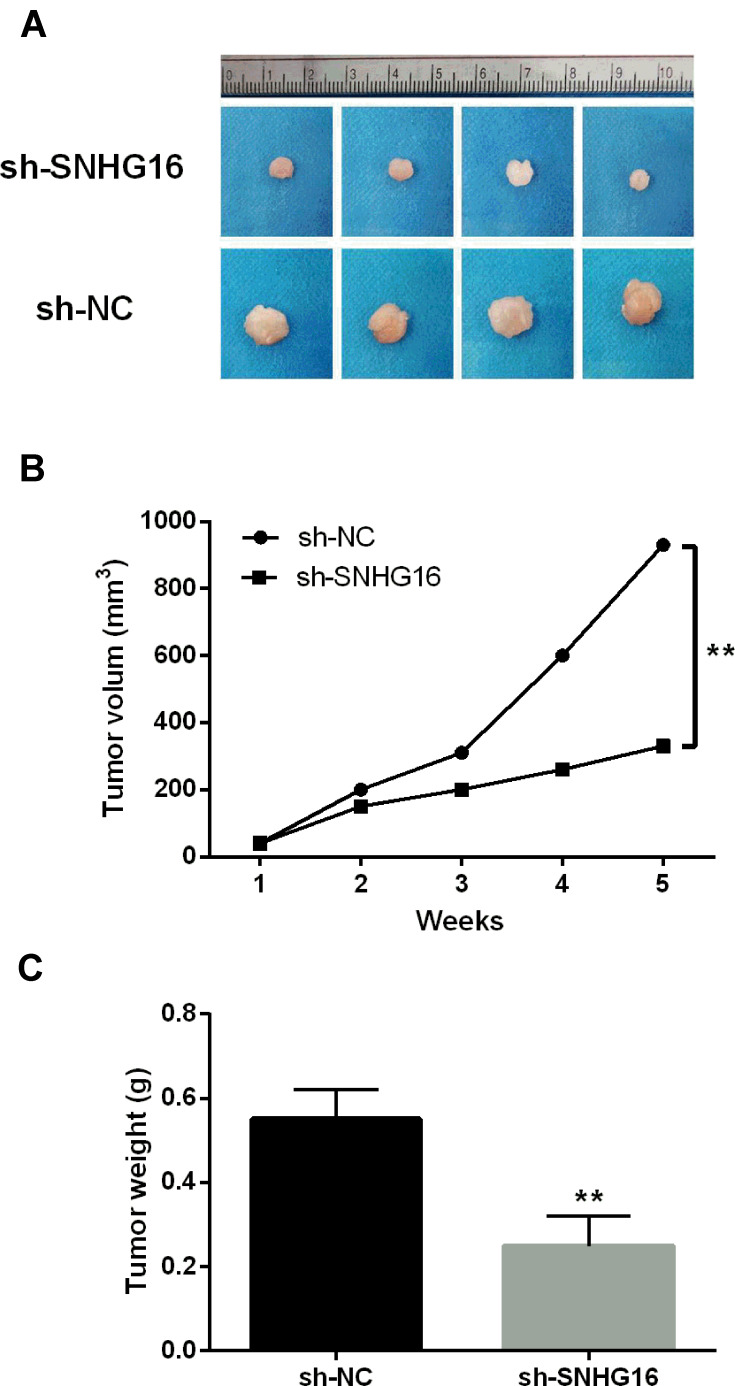
Finally, we explored the effect of lncRNA SNHG16 on tumor growth in vivo. The results showed that the tumor weight and volumes were obviously decreased in sh-SNHG16 group compared with the sh-NC group (P < 0.01, Figure 6A–C). These findings indicate that lncRNA SNHG16 promotes tumor growth in vivo.
Figure 6.
LncRNA SNHG16 depletion inhibited tumor growth in vivo. (A–C) Tumor growth rate was obviously reduced in sh-SNHG16 group. **P < 0.01.
Discussion
Early diagnosis and treatment are very important to reduce the harm of LSCC. On the one hand, it can improve the patient’s postoperative survival rate. And on the other hand, it can preserve the voice function of the larynx as much as possible, reducing postoperative complications. Therefore, it is necessary to find new molecular markers for early detection and treatment of LSCC. In recent years, the special functions of lncRNAs in LSCC have received more and more attention. Here, the regulatory mechanism of lncRNA SNHG16 was investigated in LSCC. We found that SNHG16 expression was increased in LSCC. The abnormal expression of SNHG16 was associated with the clinical stage and lymph node metastasis of LSCC patients. Functionally, it was found that knockdown of SNHG16 inhibited cell proliferation, migration and invasion in LSCC.
Upregulation of SNHG16 has been found in many human cancers, such as glioma and hepatocellular carcinoma,18,19 which is consistent with our results. In addition, it has been reported that lncRNA SNHG16 predicted the poor prognosis of esophageal squamous cell carcinoma patients and promoted cell proliferation and invasion.20 Cai et al demonstrated that SNHG16 promoted the migration of breast cancer cells through competitive binding miR-98.21 Here, the upregulation of SNHG16 was related to the poor clinical outcome of LSCC patients. The carcinogenesis of SNHG16 was also detected in LSCC tumorigenesis. In addition, we found that lncRNA SNHG16 promoted the progression of LSCC by regulating miR-877-5p.
Previous studies have shown that lncRNAs harbor miRNAs to act as a “sponge”. In this study, we also found that lncRNA SNHG16 is a “sponge” of miR-877-5p and negatively regulates miR-877-5p expression in LSCC. In addition, miR-877-5p overexpression attenuated the promoting effect of SNHG16 on the proliferation, migration and invasion of LSCC cells. It indicates that miR-877-5p can inhibit LSCC progression. Besides, downregulation of miR-877-5p was detected in LSCC tissues. Consistent with our results, the expression of miR-877-5p in non-small cell lung cancer and colorectal cancer was also reduced.22,23 Functionally, miR-877-5p has been proposed to suppress cell growth, migration and invasion in hepatocellular carcinoma.24 These findings indicate that miR-877-5p acts as a tumor suppressor in LSCC.
In addition, lncRNAs can regulate the expression and function of miRNA targets. Among the targets of miR-877-5p, FOXP4 has been found to be involved in human cancers, except for LSCC. In our study, upregulation of FOXP4 was observed in LSCC tissues. In addition, FOXP4 was confirmed to be a direct target of miR-877-5p and negatively regulated the expression of miR-877-5p in LSCC. Similar to our results, FOXP4 was upregulated in oral squamous cell carcinoma and osteosarcoma.25,26 The carcinogenic effect of FOXP4 has been found in hepatocellular carcinoma and Wilms’ tumor.27,28 Here, FOXP4 was positively correlated with miR-877-5p expression in LSCC. Knockdown of FOXP4 attenuated the carcinogenic role of SNHG16 in the progression of LSCC. It reflects that FOXP4 functions as an oncogene in LSCC. These findings are similar to previous studies and also reveal the novel mechanism of FOXP4.
Conclusions
In summary, SNHG16 expression is increased in LSCC, and promotes LSCC cell proliferation, migration and invasion. In addition, SNHG16 acts as a competitive endogenous RNA in LSCC and regulates FOXP4 expression by sponging miR-877-5p. Therefore, SNHG16 may have clinical potential for the treatment of LSCC.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Kreimer AR, Clifford GM, Boyle P, Franceschi S. Human papillomavirus types in head and neck squamous cell carcinomas worldwide: a systematic review. Cancer Epidemiol Biomarkers Prev. 2005;14(2):467–475. doi: 10.1158/1055-9965.EPI-04-0551 [DOI] [PubMed] [Google Scholar]
- 3.Marioni G, Marchese-Ragona R, Cartei G, Marchese F, Staffieri A. Current opinion in diagnosis and treatment of laryngeal carcinoma. Cancer Treat Rev. 2006;32(7):504–515. doi: 10.1016/j.ctrv.2006.07.002 [DOI] [PubMed] [Google Scholar]
- 4.Chu EA, Kim YJ. Laryngeal cancer: diagnosis and preoperative work-up. Otolaryngol Clin North Am. 2008;41(4):673–695. doi: 10.1016/j.otc.2008.01.016 [DOI] [PubMed] [Google Scholar]
- 5.Yang Y, Zhang Z, Wu Z, Lin W, Yu M. Downregulation of the expression of the lncRNA MIAT inhibits melanoma migration and invasion through the PI3K/AKT signaling pathway. Cancer Biomark. 2019;24(2):203–211. doi: 10.3233/CBM-181869 [DOI] [PubMed] [Google Scholar]
- 6.Wang M, Ji YQ, Song ZB, Ma XX, Zou YY, Li XS. Knockdown of lncRNA ZFAS1 inhibits progression of nasopharyngeal carcinoma by sponging miR-135a. Neoplasma. 2019;66(6):939–945. doi: 10.4149/neo_2018_181213N963 [DOI] [PubMed] [Google Scholar]
- 7.Hui L, Wang J, Zhang J, Long J. lncRNA TMEM51-AS1 and RUSC1-AS1 function as ceRNAs for induction of laryngeal squamous cell carcinoma and prediction of prognosis. PeerJ. 2019;7:e7456. doi: 10.7717/peerj.7456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao D, Cui X, Wang X. Long noncoding RNA XIST increases the aggressiveness of laryngeal squamous cell carcinoma by regulating miR-124-3p/EZH2. Exp Cell Res. 2019;381(2):172–178. doi: 10.1016/j.yexcr.2019.04.034 [DOI] [PubMed] [Google Scholar]
- 9.Li J, Sun S, Chen W, Yuan K. Small nucleolar RNA host gene 12 (SNHG12) promotes proliferation and invasion of laryngeal cancer cells via sponging miR-129-5p and potentiating WW domain-containing E3 ubiquitin protein ligase 1 (WWP1) expression. Med Sci Monit. 2019;25:5552–5560. doi: 10.12659/MSM.917088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han W, Du X, Liu M, Wang J, Sun L, Li Y. Increased expression of long non-coding RNA SNHG16 correlates with tumor progression and poor prognosis in non-small cell lung cancer. Int J Biol Macromol. 2019;121:270–278. doi: 10.1016/j.ijbiomac.2018.10.004 [DOI] [PubMed] [Google Scholar]
- 11.Lian D, Amin B, Du D, Yan W. Enhanced expression of the long non-coding RNA SNHG16 contributes to gastric cancer progression and metastasis. Cancer Biomark. 2017;21(1):151–160. doi: 10.3233/CBM-170462 [DOI] [PubMed] [Google Scholar]
- 12.Huang X, Qin J, Lu S. Up-regulation of miR-877 induced by paclitaxel inhibits hepatocellular carcinoma cell proliferation though targeting FOXM1. Int J Clin Exp Pathol. 2015;8(2):1515–1524. [PMC free article] [PubMed] [Google Scholar]
- 13.Shi Q, Xu X, Liu Q, Luo F, Shi J, He X. MicroRNA-877 acts as a tumor suppressor by directly targeting eEF2K in renal cell carcinoma. Oncol Lett. 2016;11(2):1474–1480. doi: 10.3892/ol.2015.4072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie H, Shi S, Chen Q, Chen Z. LncRNA TRG-AS1 promotes glioblastoma cell proliferation by competitively binding with miR-877-5p to regulate SUZ12 expression. Pathol Res Pract. 2019;215(8):152476. doi: 10.1016/j.prp.2019.152476 [DOI] [PubMed] [Google Scholar]
- 15.Huang C, Deng H, Wang Y, et al. Circular RNA circABCC4 as the ceRNA of miR-1182 facilitates prostate cancer progression by promoting FOXP4 expression. J Cell Mol Med. 2019;23(9):6112–6119. doi: 10.1111/jcmm.14477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang N, Gu Y, Li L, et al. Circular RNA circMYO9B facilitates breast cancer cell proliferation and invasiveness via upregulating FOXP4 expression by sponging miR-4316. Arch Biochem Biophys. 2018;653:63–70. doi: 10.1016/j.abb.2018.04.017 [DOI] [PubMed] [Google Scholar]
- 17.Yang T, Li H, Thakur A, et al. FOXP4 modulates tumor growth and independently associates with miR-138 in non-small cell lung cancer cells. Tumour Biol. 2015;36(10):8185–8191. doi: 10.1007/s13277-015-3498-8 [DOI] [PubMed] [Google Scholar]
- 18.Zhou XY, Liu H, Ding ZB, Xi HP, Wang GW. lncRNA SNHG16 promotes glioma tumorigenicity through miR-373/EGFR axis by activating PI3K/AKT pathway. Genomics. 2020;112(1):1021–1029. doi: 10.1016/j.ygeno.2019.06.017 [DOI] [PubMed] [Google Scholar]
- 19.Xie X, Xu X, Sun C, Yu Z. Long intergenic noncoding RNA SNHG16 interacts with miR-195 to promote proliferation, invasion and tumorigenesis in hepatocellular carcinoma. Exp Cell Res. 2019;383(1):111501. doi: 10.1016/j.yexcr.2019.111501 [DOI] [PubMed] [Google Scholar]
- 20.Han GH, Lu KJ, Wang P, Ye J, Ye YY, Huang JX. LncRNA SNHG16 predicts poor prognosis in ESCC and promotes cell proliferation and invasion by regulating Wnt/beta-catenin signaling pathway. Eur Rev Med Pharmacol Sci. 2018;22(12):3795–3803. doi: 10.26355/eurrev_201806_15262 [DOI] [PubMed] [Google Scholar]
- 21.Cai C, Huo Q, Wang X, Chen B, Yang Q. SNHG16 contributes to breast cancer cell migration by competitively binding miR-98 with E2F5. Biochem Biophys Res Commun. 2017;485(2):272–278. doi: 10.1016/j.bbrc.2017.02.094 [DOI] [PubMed] [Google Scholar]
- 22.Zhou G, Xie J, Gao Z, Yao W. MicroRNA-877 inhibits cell proliferation and invasion in non-small cell lung cancer by directly targeting IGF-1R. Exp Ther Med. 2019;18(2):1449–1457. doi: 10.3892/etm.2019.7676 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Zhang L, Li C, Cao L, et al. microRNA-877 inhibits malignant progression of colorectal cancer by directly targeting MTDH and regulating the PTEN/Akt pathway. Cancer Manag Res. 2019;11:2769–2781. doi: 10.2147/CMAR.S194073 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Yan TH, Qiu C, Sun J, Li WH. MiR-877-5p suppresses cell growth, migration and invasion by targeting cyclin dependent kinase 14 and predicts prognosis in hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2018;22(10):3038–3046. doi: 10.26355/eurrev_201805_15061 [DOI] [PubMed] [Google Scholar]
- 25.Xu Y, Liu Y, Xiao W, et al. MicroRNA-299-3p/FOXP4 axis regulates the proliferation and migration of oral squamous cell carcinoma. Technol Cancer Res Treat. 2019;18:1078142451. doi: 10.1177/1533033819874803 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Yin Z, Ding H, He E, Chen J, Li M. Up-regulation of microRNA-491-5p suppresses cell proliferation and promotes apoptosis by targeting FOXP4 in human osteosarcoma. Cell Prolif. 2017;50(1):e12308. doi: 10.1111/cpr.12308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang G, Sun Y, He Y, Ji C, Hu B, Sun Y. MicroRNA-338-3p inhibits cell proliferation in hepatocellular carcinoma by target forkhead box P4 (FOXP4). Int J Clin Exp Pathol. 2015;8(1):337–344. [PMC free article] [PubMed] [Google Scholar]
- 28.Ma L, Sun X, Kuai W, et al. Retracted: long noncoding RNA SOX2OT accelerates the carcinogenesis of wilms’ tumor through ceRNA through miR-363/FOXP4 axis. DNA Cell Biol. 2018. doi: 10.1089/dna.2018.4420 [DOI] [PubMed] [Google Scholar]