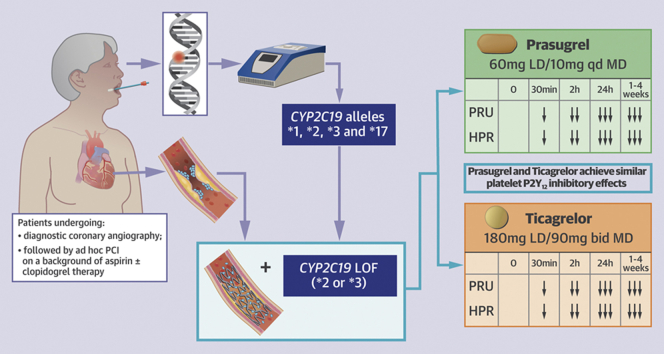
Visual Abstract
Key Words: genotyping, percutaneous coronary intervention, pharmacodynamics, prasugrel, ticagrelor
Abbreviations and Acronyms: ACS, acute coronary syndrome; CI, confidence interval; CYP, cytochrome P450; DAPT, dual antiplatelet therapy; HPR, high on-treatment platelet reactivity; LOF, loss of function; PD, pharmacodynamic; PCI, percutaneous coronary intervention; PRU, P2Y12 reaction unit
Highlights
-
•
Our study supports the feasibility of using rapid CYP2C19 genotyping among both patients with stable and acute coronary syndrome undergoing diagnostic coronary angiography, with intent to undergo ad hoc PCI, in real-world clinical practice.
-
•
Rapid bedside genetic testing assay allows for very rapid turnaround times of results, with patients approached the same day of their procedure and availability of CYP2C19 genotypes within 1 h of sampling and before patients undergoing PCI.
-
•
Among carriers of CYP2C19 loss-of-function alleles undergoing PCI there were no differences in levels of platelet inhibition between prasugrel and ticagrelor (loading and maintenance dosing).
Summary
The feasibility of rapid genetic testing in patients undergoing percutaneous coronary intervention (PCI) and the comparison of the pharmacodynamic effects of prasugrel versus ticagrelor among carriers of cytochrome P450 2C19 loss-of-function alleles treated with PCI has been poorly explored. Rapid genetic testing using the Spartan assay was shown to be feasible and provides results in a timely fashion in a real-world setting of patients undergoing coronary angiography (n = 781). Among patients (n = 223, 28.5%), carriers of at least 1 loss-of-function allele treated with PCI (n = 65), prasugrel, and ticagrelor achieve similar levels of platelet inhibition. (A Pharmacodynamic Study Comparing Prasugrel Versus Ticagrelor in Patients Undergoing PCI With CYP2C19 Loss-of-function [NCT02065479])
Dual antiplatelet therapy (DAPT) with aspirin and a P2Y12 inhibitor is the cornerstone of treatment for the prevention of thrombotic events in patients undergoing percutaneous coronary intervention (PCI) (1). Although clopidogrel is the most commonly used P2Y12 inhibitor, its pharmacodynamic (PD) effects are nonuniform, and patients with high on-treatment platelet reactivity (HPR) are at increased thrombotic risk (2, 3, 4, 5, 6). Such PD variability is in part attributed to genetic polymorphisms of the cytochrome P450 (CYP) 2C19 enzyme, a key modulator of clopidogrel metabolism (7, 8, 9, 10). In particular, carriers of loss-of-function (LOF) alleles of the CYP2C19 gene are associated with reduced generation of clopidogrel’s active metabolite, diminished platelet inhibition, and increased rates of thrombotic events (7, 8, 9, 10, 11, 12, 13). Consequently, drug-regulating authorities have issued a boxed warning on the reduced efficacy of clopidogrel among CYP2C19 LOF carriers and suggest that alternative P2Y12-inhibiting therapies (i.e., prasugrel or ticagrelor) be used in these individuals (14,15). However, in clinical practice, implementing a strategy of genotype-guided selection of oral P2Y12 inhibitor has been limited by turnaround times of test results (16, 17, 18).
Prasugrel and ticagrelor are characterized by more potent platelet inhibitory effects and greater efficacy in reducing thrombotic complications, albeit at the expense of increased bleeding, compared with clopidogrel (19, 20, 21). Post hoc analyses of large-scale investigations have not shown any interaction between CYP2C19 LOF polymorphisms and the clinical effects of prasugrel and ticagrelor (22, 23, 24, 25). However, prasugrel and ticagrelor differ considerably regarding their pharmacokinetic properties, and studies assessing the comparative PD effects between prasugrel and ticagrelor have yielded conflicting findings, with earlier investigations suggesting ticagrelor to have enhanced and less variable P2Y12 inhibitory effects with lower rates of HPR compared with prasugrel (19,26). Of note, although ticagrelor is a direct-acting P2Y12 inhibitor, prasugrel is a prodrug that needs to be metabolized by CYP enzymes to generate an active metabolite to exert its effects (19). Hence, it had been suggested that genetic polymorphisms regulating CYP enzyme activity could have contributed to these findings (26). Although subsequent studies have failed to demonstrate greater P2Y12 inhibitory effects of ticagrelor over prasugrel, or for these effects to be potentially modulated by CYP2C19 genetic status (19,22,23,27), to date there are no studies that have prospectively compared these agents specifically among CYP2C19 LOF carriers.
The aim of this investigation was to assess the feasibility of implementing a rapid bedside CYP2C19 genetic testing assay in real-world clinical practice of patients undergoing coronary angiography and to compare the PD effects of prasugrel and ticagrelor selectively among those identified as having a CYP2C19 LOF allele and undergoing PCI.
Methods
Study design and participants
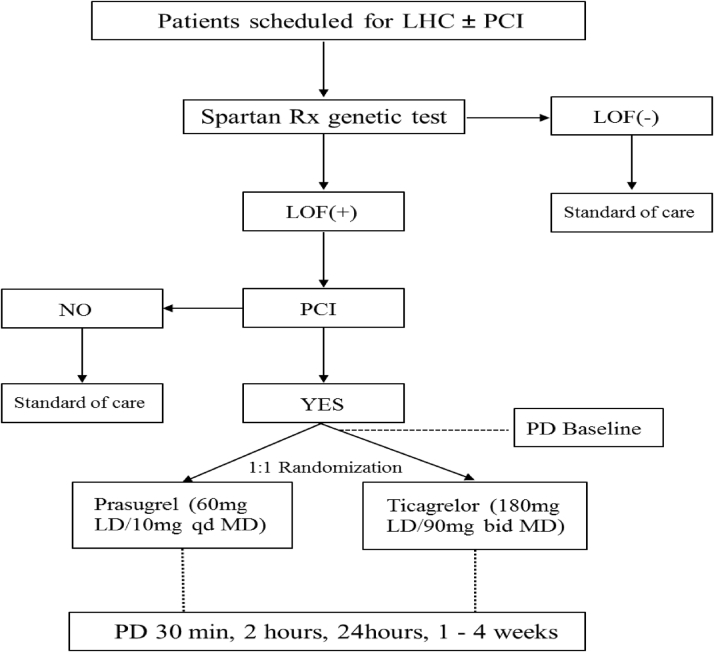
This was a prospective, randomized, parallel design, open-label investigation conducted in patients scheduled to undergo diagnostic coronary angiography with intent to undergo ad hoc PCI (Clinicaltrials.gov identifier: NCT02065479). The study was performed at the University of Florida Health–Jacksonville (Jacksonville, Florida). Patients were screened the same day of the scheduled procedure and before going to the interventional suite. Specific study inclusion and exclusion criteria are provided in the Supplemental Appendix. In brief, patients age 18 to 75 years scheduled for diagnostic coronary angiography with intent to undergo ad hoc PCI and who did not have any contraindications to treatment with prasugrel or ticagrelor were considered for CYP2C19 genetic testing. Patients could have been on aspirin monotherapy (81 mg every day) or on DAPT with aspirin (81 mg every day) and clopidogrel (75 mg every day); patients who were not on any antiplatelet medication were treated with aspirin 325 mg the morning of the procedure. Patients with stable ischemic heart disease and patients with non–ST-segment elevation acute coronary syndrome (ACS) were eligible. Only patients undergoing urgent/emergent coronary angiography that would not allow for genetic testing results to be available at the time of PCI were excluded (e.g., patients undergoing primary PCI, cardiogenic shock). Patients meeting study entry criteria underwent rapid genetic testing using the Spartan RX assay (Spartan Bioscience, Ottawa, Ontario, Canada). The Spartan RX assay identifies the following CYP2C19 alleles: ∗1, ∗2, ∗3, and ∗17. The most common LOF alleles are ∗2 and ∗3. Therefore, carriers of ∗2 or ∗3 LOF carrier status (homozygotes [∗2/∗2, ∗3/∗3, or ∗2/∗3] or heterozygotes [∗1/∗2, ∗1/∗3, ∗2/∗17, ∗3/∗17]) were considered eligible for randomization if they proceeded with PCI. Patients who were noncarriers of LOF alleles (∗1/∗1, ∗1/∗17, or ∗17/∗17) were not eligible for randomization and considered as screen failures and treated per standard of care; similarly, patients with CYP2C19 LOF alleles who did not undergo PCI were considered as screen failures and treated per standard of care.
Patients identified to be CYP2C19 LOF allele carriers and undergoing PCI were randomly assigned 1:1 using a computer-based randomization system to either prasugrel (60 mg loading dose to 10 mg daily maintenance dose) or ticagrelor (180 mg loading dose to 90 mg twice a day maintenance dose). Randomization was stratified according to baseline antiplatelet therapy (aspirin alone vs. DAPT with aspirin and clopidogrel). Loading dose administration was given immediately after PCI as per local standard of care. Randomized patients underwent PD testing at 5 time points: 1) baseline (before initiating the PCI procedure and loading dose administration of antiplatelet therapy); 2) 30 mi after loading dose administration; 3) 2 h after loading dose administration; 4) 24 h after loading dose administration or at hospital discharge (whichever came first); 5) during routine clinical follow-up 1 to 4 weeks after PCI while on maintenance dose antiplatelet therapy. At 24 h and at follow-up, blood was collected before the morning dose of prasugrel or ticagrelor to measure trough levels of platelet inhibition. Laboratory personnel were blinded to treatment assignments. Compliance to randomized treatment was assessed by pill count and patient interview. After completing the study, the choice and length of DAPT were left at the discretion of the treating cardiologist. The study complied with the Declaration of Helsinki, was approved by the University of Florida Institutional Review Board, and all patients gave their written informed consent. A flow diagram of the study design is illustrated in Figure 1.
Figure 1.
Study Design
bid = twice a day; LD = loading dose; LHC = left heart catheterization; LOF = loss of function; MD = maintenance dose; PCI = percutaneous coronary intervention; PD = pharmacodynamics; qd = every day.
Genetic and PD testing
Spartan RX rapid genotyping
Spartan RX (Spartan Bioscience Inc.) defines CYP2C19 (∗1, ∗2, ∗3, ∗17) allele status within 1 h. This test consists of 4 separate steps intended to be done in <8 min: acquisition of a buccal swab; insertion of the swab into the cartridge; insertion of the reaction solution into the device; and analysis of CYP2C19 genotype triggered by a button on the device. In this study, patients with the ∗2 or ∗3 LOF carrier status (homozygotes [∗2/∗2, ∗3/∗3, or ∗2/∗3] or heterozygotes [∗1/∗2, ∗1/∗3, ∗2/∗17, ∗3/∗17]) were considered eligible for randomization, whereas noncarriers of LOF alleles (∗1/∗1, ∗1/∗17, or ∗17/∗17) were not (28, 29, 30).
VerifyNow point-of-care testing
The VerifyNow System is a turbidimetric-based optical detection system that measures platelet-induced aggregation as an increase in light transmittance (Accriva, San Diego, California), and was used according to the manufacturer’s instructions (29). The assay is based on microbead agglutination and uses specific reagents for the pathways of interest. The instrument measures this change in optical signal and reports results in P2Y12 reaction units (PRUs). HPR was defined by PRU >208 (31).
Study endpoints and determination of sample size
The primary endpoint of the study was the noninferiority in platelet reactivity, measured as PRU, at 24 h or hospital discharge (whichever came first) of prasugrel versus ticagrelor among LOF allele carriers. Under the assumption of 0 difference in mean PRU between treatments and a common standard deviation of 50 PRU, a sample size of 60 patients with valid data would allow for the 95% confidence interval (CI) to stay within 40 PRU with an 85% power and alpha = 0.025 (27,32). Considering up to a 25% rate of invalid results due to hemolysis or drop-out, we planned to randomize up to a total of 80 patients to ensure complete data for analysis. Noninferiority was assessed using a 95% CI of the difference in mean PRU between the 2 groups. The 40 PRU noninferiority margin was defined according to previously published studies (33). Exploratory endpoints included assessment of PD differences between prasugrel and ticagrelor at 30 min, 2 h, and 1 to 4 weeks, and rates of HPR at all study time points.
Statistical analysis
Continuous variables are expressed as a mean ± SD or median (interquartile range) as appropriate. Categorical variables are expressed as frequencies and percentages. Comparisons between continuous variables was performed using Student’s t-test or Wilcoxon rank-sum test. Comparisons between categorical variables were performed using chi-square test or Fisher exact test. Missing data were not imputed. An analysis of variance method with a general linear model, with treatment as the main effect, was used to evaluate the primary noninferiority endpoint as well as all superiority between-group comparisons at each time point. Least squares mean differences in PRU between groups and the corresponding 2-sided 95% CI for the difference was obtained based on the analysis of variance model and used to assess noninferiority. The p values are used to report superiority testing, and a 2-tailed p value of <0.05 is considered to indicate a statistically significant difference for all the analyses performed. The PD population included all patients with PD data and without a major protocol deviation thought to significantly affect the effects of ticagrelor or prasugrel. The PD population was used for analysis of all primary and exploratory PD variables. Statistical analysis was performed by our group using SPSS v24.0 software (SPSS Inc., Chicago, Illinois).
Results
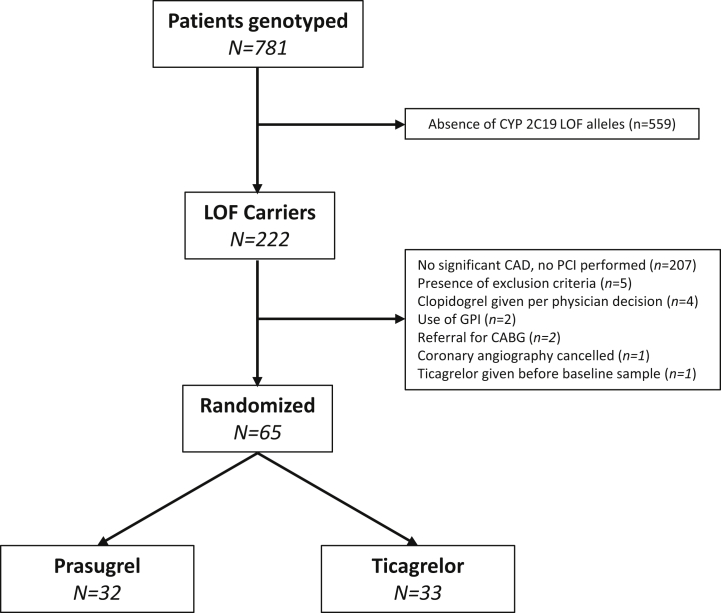
Between March 2014 and September 2018, a total of 781 consecutive patients scheduled for left heart catheterization with the intent to undergo PCI were genotyped. Of these, 222 (28.5%) patients were carriers of at least 1 LOF: 9% were homozygotes (∗2/∗2, n = 20) and 91% were heterozygotes (∗1/∗2, n = 189; ∗1/∗3, n = 1; ∗2/∗17, n = 32). Of the cohort of CYP2C19 LOF carriers, 157 patients did not meet criteria to be randomized. Thus, a total 65 patients underwent PCI and were randomized to either prasugrel (n = 32) or ticagrelor (n = 33). These patients represented the PD population of the study (Figure 2). Baseline characteristics of the PD population are summarized in Table 1. CYP2C19 LOF carriers who were not randomized were more likely to be female and less likely to have ACS compared with those who were randomized (Supplemental Table 1).
Figure 2.
Trial Profile
CABG = coronary artery bypass graft; CAD = coronary artery disease; CYP = cytochrome P450; GPI = glycoprotein IIb/IIIa inhibitor; LOF = loss of function; PCI = percutaneous coronary intervention.
Table 1.
Baseline Characteristics of the PD Population
| Prasugrel (n = 32) | Ticagrelor (n = 33) | p Value | |
|---|---|---|---|
| Age, yrs | 60 ± 9 | 58 ± 8 | 0.593 |
| Gender, male | 25 (78) | 26 (79) | 1.000 |
| BMI, kg/m2 | 31 ± 5 | 31 ± 5 | 0.986 |
| Race | 0.690 | ||
| White | 21 (66) | 23 (70) | |
| Black | 9 (28) | 8 (24) | |
| Other | 2 (6) | 2 (6) | |
| Genotype | 0.398 | ||
| ∗1/∗2 | 24 (75) | 27 (82) | |
| ∗1/∗3 | 0 (0) | 1 (3) | |
| ∗2∗/17 | 4 (12.5) | 1 (3.0) | |
| ∗2/∗2 | 4 (12.5) | 4 (12.0) | |
| ACS | 17 (53) | 18 (54) | 1.000 |
| SIHD | 15 (47) | 15 (46) | |
| Cohort | 1.000 | ||
| Aspirin | 25 (78) | 26 (79) | |
| DAPT | 7 (22) | 7 (21) | |
| Hypertension | 28 (87) | 29 (88) | 1.000 |
| Dyslipidemia | 20 (62) | 24 (73) | 0.434 |
| Active smoking | 11 (34) | 9 (27) | 0.599 |
| Diabetes mellitus | 14 (44) | 10 (30) | 0.310 |
| Prior MI | 10 (31) | 5 (15) | 0.150 |
| Prior PCI | 11 (34) | 15 (45) | 0.450 |
| Prior CABG | 7 (22) | 8 (24) | 1.000 |
| Creatinine, mg/dl | 1.0 ± 0.3 | 0.9 ± 0.2 | 0.373 |
| Platelet count, × 103/μl | 232 ± 70 | 213 ± 45 | 0.206 |
| Hematocrit, % | 41 ± 5 | 42 ± 3 | 0.182 |
| Medications∗ | |||
| Insulin therapy | 6 (19) | 5 (15) | 0.751 |
| OAD | 8 (25) | 9 (27) | 0.358 |
| Beta-blockers | 14 (44) | 20 (61) | 0.218 |
| ACE-I/ARB | 16 (50) | 19 (58) | 0.622 |
| Statins | 20 (62.5) | 25 (76) | 0.290 |
Values are mean ± SD and or n (%).
ACE-I = angiotensin converting enzyme inhibitor; ACS = acute coronary syndrome; ARB = angiotensin receptor blocker; BMI = body mass index; CABG = coronary artery bypass graft; DAPT = dual antiplatelet therapy; MI = myocardial infarction; OAD = oral antidiabetic drugs; PD = pharmacodynamics; PCI = percutaneous coronary intervention; SIHD = stable ischemic heart disease.
Medications at the time of coronary angiography.
In the PD population, 8 patients (12%) were homozygotes for ∗2/∗2, and 57 (88%) were heterozygotes (∗1/∗2: 78.5%; ∗1/∗3: 1.5%; ∗2/∗17: 8%). Fifty-one (78.5%) and 14 (21.5%) patients were on aspirin or DAPT before randomization, respectively. No ischemic or Bleeding Academic Research Consortium type 2 to 5 bleeding events were observed; 3 patients (9%) receiving ticagrelor experienced dyspnea, which led to drug discontinuation in 1 patient, versus none of those receiving prasugrel; 1 patient receiving prasugrel had a stroke; 2 patients had chest pain during follow-up that did not require any intervention (prasugrel, n = 1; ticagrelor, n = 1).
Pharmacodynamic results
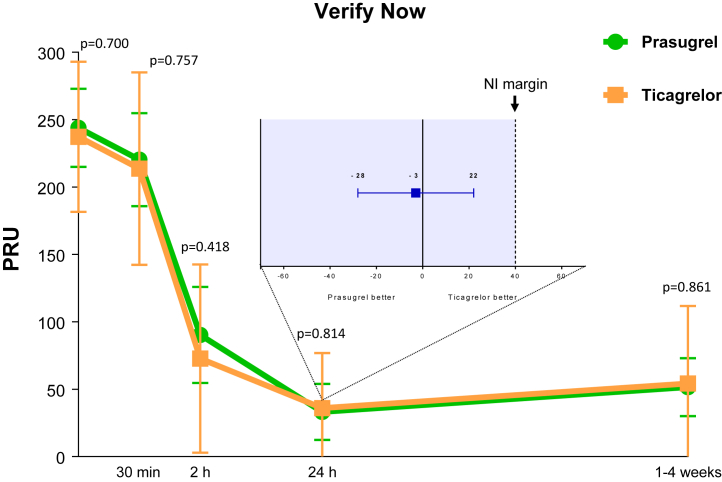
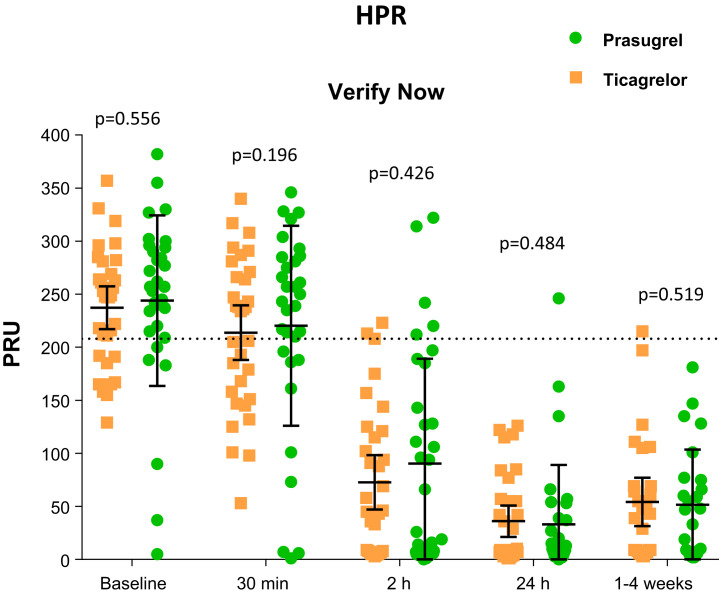
At baseline, PRU levels were similar between groups. A significant reduction in PRU was observed as early as 30 min following loading dose for both prasugrel (p = 0.018) and ticagrelor (p = 0.029). PRU levels continued to markedly reduce to a similar extent with no differences between groups at 2 h and 24 h (or hospital discharge) after loading dose administrations and remained low at follow-up while on maintenance therapy (Figure 3). Median time between study drug loading dose and primary end point sample was 20 h (interquartile range: 7 to 22 h). Mean PRU levels at this time point were 33 ± 56 for prasugrel versus 36 ± 41 for ticagrelor (mean difference = −3; 95% CI: −28 to 22; p = 0.814 for superiority) meeting the primary endpoint of noninferiority. HPR rates also significantly reduced over time, with no differences between groups at any time point (Figure 4). Results were consistent irrespective of baseline antiplatelet treatment regimen (aspirin monotherapy or DAPT; Supplemental Figure 1), with no cohort by treatment group interaction (p for interaction >0.05 for all time points)
Figure 3.
Pharmacodynamic Assessment Measured by VerifyNow P2Y12 After Administration of Prasugrel Versus Ticagrelor
The main figure represents P2Y12 reaction units (PRUs) measured by the VerifyNow P2Y12 assay. Values are expressed as means. Error bars indicate SDs; p values indicate the comparisons for superiority between groups at each time point. The box represents the primary endpoint: 24-h PRU absolute difference and 2-sided 95% confidence interval between prasugrel and ticagrelor. Tinted area indicates zone of noninferiority (NI). The dotted line represents the prespecified limit of noninferiority of +40.
Figure 4.
Individual Values of Platelet Reactivity Measured by VerifyNow P2Y12 After Administration of Prasugrel Versus Ticagrelor
Comparisons of P2Y12 reaction units (PRUs) measured by the VerifyNow P2Y12 assay. Data are presented as individual values. The dashed line indicates threshold for high on-treatment platelet reactivity. Solid lineswith error bars indicate mean ± SD. HPR = high on-treatment platelet reactivity.
Discussion
The implementation of genotype-guided selection of P2Y12-inhibiting therapy in patients undergoing PCI has been limited in real-world clinical practice by the availability of assays able to provide results of CYP2C19 genotypes in a timely fashion (16, 17, 18). Patients most commonly undergo ad hoc PCI immediately following diagnostic angiography, which further emphasizes the importance of having readily available genotyping results. Earlier small-scale investigations have suggested the clinical utility of genotype-guided selection of oral P2Y12 inhibitors in patients undergoing PCI (34, 35, 36, 37). However, some of these were conducted using genotyping approaches that would require several days, making results unavailable until after hospital discharge (34,35). This has important implications. First, switching antiplatelet treatment after hospital discharge is of limited practicality; second, the early post-PCI period is when the risk for thrombotic complication is highest, underscoring the importance of optimized antiplatelet therapy during this time frame; third, adherence is improved when medications are prescribed before hospital discharge (38,39). The results of our investigation overcome these limitations and support the feasibility of using a rapid CYP2C19 genotyping assay among both patients with stable coronary syndrome and patients with ACS undergoing diagnostic coronary angiography, with intent to undergo ad hoc PCI, in real-world clinical practice. In particular, such rapid bedside genetic testing assay allowed for very rapid turnaround times of results, with patients approached the same day of their procedure and availability of CYP2C19 genotypes within 1 h and before patients undergo PCI. This in contrast to other studies assessing the feasibility of rapid bedside genetic testing, in which results were not available before the start of the PCI procedure (29,30,40). Therefore, similar to other standard laboratory tests (e.g., complete blood count, creatinine, liver function, coagulation panel) performed in these patients, in our study, genotyping results were readily available when the decision on choice of oral P2Y12-inhibiting therapy most commonly occurs (i.e., at the time of PCI).
The use of platelet function or genetic testing to tailor the selection of P2Y12-inhibiting therapy as a strategy to improve outcomes has been a subject of investigation for more than a decade (31). Although earlier studies of platelet function testing, mostly evaluating a strategy of escalation of P2Y12-inhibiting therapy, have failed to show any clinical benefit, the TROPICAL ACS (Testing Responsiveness to Platelet Inhibition on Chronic Antiplatelet Treatment for Acute Coronary Syndromes) trial demonstrated that a strategy of platelet function guided de-escalation was noninferior to maintaining conventional nonguided treatment at 12 months in patients with high-risk ACS undergoing PCI (41). The results of this study led an update in revascularization guidelines that now indicate that platelet function testing can be considered to help guide the decision on choice of antiplatelet therapy in patients with ACS who cannot take the more potent P2Y12 inhibitors (42). However, there are some limitations associated with platelet function guided de-escalation. First, following de-escalation, clopidogrel maintenance therapy needs to be maintained for a certain period for it to achieve its full antiplatelet effects before performing platelet function testing to assess responsiveness, and if patients have inadequate platelet inhibition they would need to be switched back to a more potent P2Y12 inhibitor; second, patients with inadequate response to clopidogrel during this time frame are exposed to an increased risk of thrombotic complications (43). The presence of inadequate clopidogrel response can be further exacerbated by a drug-drug interaction that may occur with de-escalation from ticagrelor to clopidogrel (33). Indeed, using a genotype-guided approach overcomes these shortcomings, as also endorsed by expert consensus recommendations (31). Importantly, the clinical impact of a genotype-guide de-escalation approach was recently demonstrated in the POPular Genetics (Patient Outcome after primary PCI Genetics) study (40). In particular, this study was the first adequately powered randomized study of a CYP2C19 genotype-guided strategy for selection of oral P2Y12 inhibitor (LOF carriers received ticagrelor or prasugrel and noncarriers received clopidogrel) and showed this strategy to be noninferior to standard treatment with ticagrelor or prasugrel at 12 months with respect to thrombotic events and resulted in a lower incidence of major or minor bleeding events among patients with ST elevation myocardial infarction undergoing primary PCI (40). Indeed, it may be argued that this and other studies support the feasibility of performing rapid bedside genotyping; however, none of these executed the test routinely before undergoing diagnostic coronary angiography enabling in-laboratory guidance of P2Y12-inhibiting therapy in patients requiring PCI (29,30,40).
Although prior investigations have assessed the comparative PD effects of prasugrel and ticagrelor, yielding conflicting findings (26,27), this is the first study to assess this specifically among CYP2C19 LOF carriers. We observed no differences in platelet reactivity between prasugrel and ticagrelor among CYP2C19 LOF carriers undergoing PCI during the entire study time-course assessing the effects of both the loading and maintenance doses of these agents. Although CYP enzymes are involved in prasugrel metabolism, which could potentially affect its PD effects, the contribution of CYP2C19 is minimal, as also supported by the very high levels of platelet inhibition achieved with prasugrel in our study (44). Moreover, our investigation confirms results of prior studies on the prompt and enhanced platelet inhibitory effects associated with escalation from clopidogrel to prasugrel or ticagrelor, leading to a reduction in HPR rates when a loading dose is used (27,38).
In addition to the POPular Genetics study, in which ticagrelor was the most commonly used potent P2Y12 inhibitor among patients who were CYP2C19 LOF carriers, our study observations also have important clinical implications in light of the recently reported ISAR REACT 5 (Intracoronary Stenting and Antithrombotic Regimen: Rapid Early Action for Coronary Treatment 5) study (45). ISAR REACT 5 had hypothesized that ticagrelor would reduce ischemic events to a greater extent than prasugrel in patients with ACS undergoing invasive management in part based on the assumption of the enhanced antiplatelet effects of ticagrelor (45). However, ISAR REACT 5 failed to demonstrate its study hypothesis and actually showed significantly reduced ischemic events with prasugrel over ticagrelor at 12 months without differences in bleeding. A number of reasons have been provided to explain such study findings, including compliance issues (once-daily administration of prasugrel vs. twice-daily with ticagrelor), increased rate of nonbleeding side effects with ticagrelor (i.e., dyspnea) leading to drug discontinuation, and differential pharmacologic profile (short half-life, reversibility of action, and drug interactions with ticagrelor). Our current investigation, showing no differences in PD effects between prasugrel and ticagrelor among CYP2C19 LOF carriers, is in line with prior investigations from our group assessing the comparative PD effects between these agents not stratified according to CYP2C19 genotype (27). Overall, these observations may have practical and cost implications, given that prasugrel is now available in a generic formulation in many countries, making it an attractive treatment option, including among physicians who elect an alternative agent to clopidogrel based on results of genetic testing.
Study limitations
This study was not designed to assess the clinical outcomes of a genotype-guided strategy in patients undergoing PCI, which is the objective of a number of ongoing investigations (23, 24, 25). Moreover, it was not designed to compare the safety and efficacy of prasugrel versus ticagrelor. Our study used only the VerifyNow assay for PD assessments. However, the choice to use only this rapid bedside assay was in line with the overall scope of this investigation to consider tests, both PD and genetic, of simple and practical utility, as well as fast turnaround. Ultimately, there are more genetic polymorphisms of CYP2C19 than those assessed by the Spartan RX assay. However, their prevalence is extremely rare and of unclear clinical significance (31).
Conclusions
Rapid CYP2C19 genotyping using the Spartan assay is feasible in providing results in a timely fashion in a real-world clinical practice of patients undergoing coronary angiography. Among patients with CYP2C19 LOF carrier status undergoing ad hoc PCI in this setting, prasugrel and ticagrelor markedly reduce levels of platelet reactivity to a similar extent with no differences between groups. Ongoing large-scale clinical investigations will help define the safety and efficacy of using genetic testing to individualize the choice of oral P2Y12-inhibiting therapy in patients undergoing PCI.
Perspectives.
COMPETENCY IN MEDICAL KNOWLEDGE: The implementation of genotype-guided selection of P2Y12-inhibiting therapy in patients undergoing PCI has been limited in real-world clinical practice by the availability of assays able to provide results of CYP2C19 genotypes in a timely fashion. The results of our investigation overcome these limitations and support the feasibility of using a rapid genotyping assay among both patients with stable coronary syndrome and patients with ACS undergoing diagnostic coronary angiography, with intent to undergo ad hoc PCI, in real-world clinical practice. In our study, genotyping results were readily available when the decision on choice of oral P2Y12-inhibiting therapy most commonly occurs (i.e., at the time of PCI). The potential clinical benefits of a genotype-guided approach have been recently demonstrated. However, none of these performed the test routinely before undergoing diagnostic coronary angiography, enabling in-laboratory guidance of P2Y12-inhibiting therapy in patients requiring PCI. Although CYP enzymes are involved in prasugrel metabolism, which could potentially affect its PD effects, our study showed no differences in platelet reactivity between prasugrel and ticagrelor among CYP2C19 LOF carriers undergoing PCI during the entire study time-course, assessing the effects of both the loading and maintenance doses of these agents. These observations may have practical and cost implications, given that prasugrel is now available in a generic formulation in many countries, making it an attractive treatment option, including among physicians who elect an alternative agent to clopidogrel based on results of genetic testing.
TRANSLATIONAL OUTLOOK: Our current investigation demonstrating no differences in PD effects between prasugrel and ticagrelor in this subset of patients, which is in line with prior investigations from our group assessing the comparative PD effects between these agents not stratified according to CYP2C19 genotype, support that recent trial findings showing significantly reduced ischemic events with prasugrel over ticagrelor cannot be attributed to differential levels of P2Y12 inhibition. However, our study was not designed to assess the clinical outcomes of a rapid genotype-guided strategy in patients undergoing PCI. Moreover, it was not designed to compare the safety and efficacy of prasugrel versus ticagrelor in CYP2C19 LOF carriers, which would also warrant further investigation in large-scale clinical trials.
Footnotes
The present study was funded by an investigator-initiated grant from the Scott R. MacKenzie Foundation for genome research. Spartan Bioscience provided the Spartan RX system and the reagents used free of charge. These entities had no role in study design conception, conduct of the study or decision to publish these results. Dr. Franchi has received payment as an individual for a consulting fee or honorarium from AstraZeneca and Sanofi. Dr. Rollini has received payment as an individual for a consulting fee or honorarium from Chiesi. Dr. Pineda has received payment as an individual for consulting from Edwards Lifesciences; and educational institutional grants from Abbott, Boston Scientific, Medtronic, Edwards Lifesciences, and Gore Medical. Dr. Angiolillo has received consulting fees or honorarium from Abbott, Amgen, Aralez, AstraZeneca, Bayer, Biosensors, Boehringer Ingelheim, Bristol-Myers Squibb, Chiesi, Daiichi-Sankyo, Eli Lilly, Haemonetics, Janssen, Merck, PhaseBio, PLx Pharma, Pfizer, Sanofi, and The Medicines Company; participation in review activities from CeloNova and St. Jude Medical; and has received institutional payments for grants from Amgen, AstraZeneca, Bayer, Biosensors, CeloNova, CSL Behring, Daiichi-Sankyo, Eisai, Eli Lilly, Gilead, Idorsia, Janssen, Matsutani Chemical Industry Co., Merck, Novartis, Osprey Medical, and Renal Guard Solutions. All other authors have reported that they have no relationships relevant to the contents of this paper to disclose.
The authors attest they are in compliance with human studies committees and animal welfare regulations of the authors’ institutions and Food and Drug Administration guidelines, including patient consent where appropriate. For more information, visit the JACC: Basic to Translational Scienceauthor instructions page.
Appendix
For a supplemental table and figure, and other material, please see the online version of this paper.
Appendix
References
- 1.Capodanno D., Alfonso F., Levine G.N., Valgimigli M., Angiolillo D.J. ACC/AHA versus ESC guidelines on dual antiplatelet therapy: JACC guideline comparison. J Am Coll Cardiol. 2018;72:2915–2931. doi: 10.1016/j.jacc.2018.09.057. [DOI] [PubMed] [Google Scholar]
- 2.Angiolillo D.J., Fernandez-Ortiz A., Bernardo E. Variability in individual responsiveness to clopidogrel. J Am Coll Cardiol. 2007;49:1505–1516. doi: 10.1016/j.jacc.2006.11.044. [DOI] [PubMed] [Google Scholar]
- 3.Price M.J., Angiolillo D.J., Teirstein P.S. Platelet reactivity and cardiovascular outcomes after percutaneous coronary intervention: a time-dependent analysis of the Gauging Responsiveness with a VerifyNow P2Y12 assay: Impact on Thrombosis and Safety (GRAVITAS) trial. Circulation. 2011;124:1132–1137. doi: 10.1161/CIRCULATIONAHA.111.029165. [DOI] [PubMed] [Google Scholar]
- 4.Stone G.W., Witzenbichler B., Weisz G. Platelet reactivity and clinical outcomes after coronary artery implantation of drug-eluting stents (ADAPT-DES): a prospective multicentre registry study. Lancet. 2013;382:614–623. doi: 10.1016/S0140-6736(13)61170-8. [DOI] [PubMed] [Google Scholar]
- 5.Aradi D., Kirtane A., Bonello L. Bleeding and stent thrombosis on P2Y12-inhibitors: collaborative analysis on the role of platelet reactivity for risk stratification after percutaneous coronary intervention. Eur Heart J. 2015;36:1762–1771. doi: 10.1093/eurheartj/ehv104. [DOI] [PubMed] [Google Scholar]
- 6.Aradi D., Gross L., Trenk D. Platelet reactivity and clinical outcomes in acute coronary syndrome patients treated with prasugrel and clopidogrel:a pre-specified exploratory analysis from the TROPICAL-ACS trial. Eur Heart J. 2019;40:1942–1951. doi: 10.1093/eurheartj/ehz202. [DOI] [PubMed] [Google Scholar]
- 7.Marín F., González-Conejero R., Capranzano P., Bass T.A., Roldán V., Angiolillo D.J. Pharmacogenetics in cardiovascular antithrombotic therapy. J Am Coll Cardiol. 2009;54:1041–1057. doi: 10.1016/j.jacc.2009.04.084. [DOI] [PubMed] [Google Scholar]
- 8.Simon T., Bhatt D.L., Bergougnan L. Genetic polymorphisms and the impact of a higher clopidogrel dose regimen on active metabolite exposure and antiplatelet response in healthy subjects. Clin Pharmacol Ther. 2011;90:287–295. doi: 10.1038/clpt.2011.127. [DOI] [PubMed] [Google Scholar]
- 9.Hochholzer W., Trenk D., Fromm M.F. Impact of cytochrome P450 2C19 loss-of-function polymorphism and of major demographic characteristics on residual platelet function after loading and maintenance treatment with clopidogrel in patients undergoing elective coronary stent placement. J Am Coll Cardiol. 2010;55:2427–2434. doi: 10.1016/j.jacc.2010.02.031. [DOI] [PubMed] [Google Scholar]
- 10.Mega J.L., Close S.L., Wiviott S.D. Cytochrome p-450 polymorphisms and response to clopidogrel. N Engl J Med. 2009;360:354–362. doi: 10.1056/NEJMoa0809171. [DOI] [PubMed] [Google Scholar]
- 11.Simon T., Verstuyft C., Mary-Krause M. Genetic determinants of response to clopidogrel and cardiovascular events. N Engl J Med. 2009;360:363–375. doi: 10.1056/NEJMoa0808227. [DOI] [PubMed] [Google Scholar]
- 12.Shuldiner A.R., O’Connell J.R., Bliden K.P. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. JAMA. 2009;302:849–857. doi: 10.1001/jama.2009.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mega J.L., Simon T., Collet J.-P. Reduced-function CYP2C19 genotype and risk of adverse clinical outcomes among patients treated with clopidogrel predominantly for PCI: a meta-analysis. JAMA. 2010;304:1821–1830. doi: 10.1001/jama.2010.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.FDA Drug Safety Communication: reduced effectiveness of Plavix (clopidogrel) in patients who are poor metabolizers of the drug. http://www.fda.gov/Drugs/DrugSafety/PostmarketDrugSafetyInformationforPatientsandProviders/ucm203888.htm Available at:
- 15.European Medicines Agency Summary of product characteristics (Plavix) http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Product_Information/human/000174/WC500042189.pdf Available at.
- 16.Moon J.Y., Franchi F., Rollini F. Role of genetic testing in patients undergoing percutaneous coronary intervention. Expert Rev Clin Pharmacol. 2018;11:151–164. doi: 10.1080/17512433.2017.1353909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Empey P.E., Stevenson J.M., Tuteja S. Multisite investigation of strategies for the implementation of CYP2C19 genotype-guided antiplatelet therapy. Clin Pharmacol Ther. 2018;104:664–674. doi: 10.1002/cpt.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pereira N.L., Rihal C.S., So D.Y.F. Clopidogrel pharmacogenetics. Circ Cardiovasc Interv. 2019;12 doi: 10.1161/CIRCINTERVENTIONS.119.007811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franchi F., Angiolillo D.J. Novel antiplatelet agents in acute coronary syndrome. Nat Rev Cardiol. 2015;12:30–47. doi: 10.1038/nrcardio.2014.156. [DOI] [PubMed] [Google Scholar]
- 20.Wiviott S.D., Braunwald E., McCabe C.H. Prasugrel versus clopidogrel in patients with acute coronary syndromes. N Engl J Med. 2007;357:2001–2015. doi: 10.1056/NEJMoa0706482. [DOI] [PubMed] [Google Scholar]
- 21.Wallentin L., Becker R.C., Budaj A. Ticagrelor versus clopidogrel in patients with acute coronary syndromes. N Engl J Med. 2009;361:1045–1057. doi: 10.1056/NEJMoa0904327. [DOI] [PubMed] [Google Scholar]
- 22.Brandt J.T., Close S.L., Iturria S.J. Common polymorphisms of CYP2C19 and CYP2C9 affect the pharmacokinetic and pharmacodynamic response to clopidogrel but not prasugrel. J Thromb Haemost. 2007;5:2429–2436. doi: 10.1111/j.1538-7836.2007.02775.x. [DOI] [PubMed] [Google Scholar]
- 23.Tantry U.S., Bliden K.P., Wei C. First analysis of the relation between CYP2C19 genotype and pharmacodynamics in patients treated with ticagrelor versus clopidogrel: the ONSET/OFFSET and RESPOND genotype studies. Circ Cardiovasc Genet. 2010;3:556–566. doi: 10.1161/CIRCGENETICS.110.958561. [DOI] [PubMed] [Google Scholar]
- 24.Mega J.L., Close S.L., Wiviott S.D. Genetic variants in ABCB1 and CYP2C19 and cardiovascular outcomes after treatment with clopidogrel and prasugrel in the TRITON-TIMI 38 trial: a pharmacogenetic analysis. Lancet. 2010;376:1312–1319. doi: 10.1016/S0140-6736(10)61273-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wallentin L., James S., Storey R.F. Effect of CYP2C19 and ABCB1 single nucleotide polymorphisms on outcomes of treatment with ticagrelor versus clopidogrel for acute coronary syndromes: a genetic substudy of the PLATO trial. Lancet. 2010;376:1320–1328. doi: 10.1016/S0140-6736(10)61274-3. [DOI] [PubMed] [Google Scholar]
- 26.Lemesle G., Schurtz G., Bauters C., Hamon M. High on-treatment platelet reactivity with ticagrelor versus prasugrel: a systematic review and meta-analysis. J Thromb Haemost. 2015;13:931–942. doi: 10.1111/jth.12907. [DOI] [PubMed] [Google Scholar]
- 27.Rollini F., Franchi F., Cho J.R. A head-to-head pharmacodynamic comparison of prasugrel vs. ticagrelor after switching from clopidogrel in patients with coronary artery disease: results of a prospective randomized study. Eur Heart J. 2016;37:2722–2730. doi: 10.1093/eurheartj/ehv744. [DOI] [PubMed] [Google Scholar]
- 28.Cavallari L.H., Franchi F., Rollini F. Clinical implementation of rapid CYP2C19 genotyping to guide antiplatelet therapy after percutaneous coronary intervention. J Transl Med. 2018;16:92. doi: 10.1186/s12967-018-1469-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bergmeijer T.O., Vos G.J., Claassens D.M. Feasibility and implementation of CYP2C19 genotyping in patients using antiplatelet therapy. Pharmacogenomics. 2018;19:621–628. doi: 10.2217/pgs-2018-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roberts J.D., Wells G.A., Le May M.R. Point-of-care genetic testing for personalisation of antiplatelet treatment (RAPID GENE):a prospective, randomised, proof-of-concept trial. Lancet. 2012;379:1705–1711. doi: 10.1016/S0140-6736(12)60161-5. [DOI] [PubMed] [Google Scholar]
- 31.Sibbing D., Aradi D., Alexopoulos D. Updated expert consensus statement on platelet function and genetic testing for guiding P2Y12 receptor inhibitor treatment in percutaneous coronary intervention. J Am Coll Cardiol Interv. 2019;12:1521–1537. doi: 10.1016/j.jcin.2019.03.034. [DOI] [PubMed] [Google Scholar]
- 32.Franchi F., Rollini F., Aggarwal N. Pharmacodynamic comparison of prasugrel versus ticagrelor in patients with type 2 diabetes mellitus and coronary artery disease: the OPTIMUS (Optimizing Antiplatelet Therapy in Diabetes Mellitus)-4 study. Circulation. 2016;134:780–792. doi: 10.1161/CIRCULATIONAHA.116.023402. [DOI] [PubMed] [Google Scholar]
- 33.Franchi F., Rollini F., Rivas Rios J. Pharmacodynamic effects of switching from ticagrelor to clopidogrel in patients with coronary artery disease: results of the SWAP-4 study. Circulation. 2018;137:2450–2462. doi: 10.1161/CIRCULATIONAHA.118.033983. [DOI] [PubMed] [Google Scholar]
- 34.Cavallari L.H., Lee C.R., Beitelshees A.L. Multisite investigation of outcomes with implementation of CYP2C19 genotype-guided antiplatelet therapy after percutaneous coronary intervention. J Am Coll Cardiol Interv. 2018;11:181–191. doi: 10.1016/j.jcin.2017.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sánchez-Ramos J., Dávila-Fajardo C.L., Toledo Frías P. Results of genotype-guided antiplatelet therapy in patients who undergone percutaneous coronary intervention with stent. Int J Cardiol. 2016;225:289–295. doi: 10.1016/j.ijcard.2016.09.088. [DOI] [PubMed] [Google Scholar]
- 36.Notarangelo F.M., Maglietta G., Bevilacqua P. Pharmacogenomic approach to selecting antiplatelet therapy in patients with acute coronary syndromes: the PHARMCLO trial. J Am Coll Cardiol. 2018;71:1869–1877. doi: 10.1016/j.jacc.2018.02.029. [DOI] [PubMed] [Google Scholar]
- 37.Janssen P.W.A., Bergmeijer T.O., Vos G.A. Tailored P2Y12 inhibitor treatment in patients undergoing non-urgent PCI-the POPular Risk Score study. Eur J Clin Pharmacol. 2019;75:1201–1210. doi: 10.1007/s00228-019-02696-z. [DOI] [PubMed] [Google Scholar]
- 38.Angiolillo D.J., Rollini F., Storey R.F. International expert consensus on switching platelet P2Y12 receptor-inhibiting therapies. Circulation. 2017;136:1955–1975. doi: 10.1161/CIRCULATIONAHA.117.031164. [DOI] [PubMed] [Google Scholar]
- 39.Mehta R.H., Chen A.Y., Alexander K.P., Ohman E.M., Roe M.T., Peterson E.D. Doing the right things and doing them the right way: association between hospital guideline adherence, dosing safety, and outcomes among patients with acute coronary syndrome. Circulation. 2015;131:980–987. doi: 10.1161/CIRCULATIONAHA.114.013451. [DOI] [PubMed] [Google Scholar]
- 40.Claassens D.M.F., Vos G.J.A., Bergmeijer T.O. A genotype-guided strategy for oral P2Y12 inhibitors in primary PCI. N Engl J Med. 2019;381:1621–1631. doi: 10.1056/NEJMoa1907096. [DOI] [PubMed] [Google Scholar]
- 41.Sibbing D., Aradi D., Jacobshagen C. Guided de-escalation of antiplatelet treatment in patients with acute coronary syndrome undergoing percutaneous coronary intervention (TROPICAL-ACS): a randomised, open-label, multicentre trial. Lancet. 2017;390:1747–1757. doi: 10.1016/S0140-6736(17)32155-4. [DOI] [PubMed] [Google Scholar]
- 42.Neumann F.-J., Sousa-Uva M., Ahlsson A. 2018 ESC/EACTS guidelines on myocardial revascularization. Eur Heart J. 2019;40:87–165. doi: 10.1093/eurheartj/ehy855. [DOI] [PubMed] [Google Scholar]
- 43.Angiolillo D.J. Dual antiplatelet therapy guided by platelet function testing. Lancet. 2017;390:1718–1720. doi: 10.1016/S0140-6736(17)32279-1. [DOI] [PubMed] [Google Scholar]
- 44.Farid N.A., Kurihara A., Wrighton S.A. Metabolism and disposition of the thienopyridine antiplatelet drugs ticlopidine, clopidogrel, and prasugrel in humans. J Clin Pharmacol. 2010;50:126–142. doi: 10.1177/0091270009343005. [DOI] [PubMed] [Google Scholar]
- 45.Schüpke S., Neumann F.J., Menichelli M. Ticagrelor or prasugrel in patients with acute coronary syndromes. N Engl J Med. 2019;381:1524–1534. doi: 10.1056/NEJMoa1908973. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.