Abstract
The immune mechanisms that cause tissue injury in lupus nephritis have been challenging to define. The advent of high-dimensional cellular analyses, such as single-cell RNA sequencing, has enabled detailed characterization of the cell populations present in small biopsy samples of affected kidney tissue. In parallel, the development of methods that cryopreserve kidney biopsy specimens in a manner that preserves intact, viable cells, has enabled the uniform analysis of tissue samples collected at multiple sites and across many geographic areas and demographic cohorts by high-dimensional platforms. The application of these methods to kidney biopsy samples from patients with lupus nephritis has begun to define the phenotypes of both infiltrating and resident immune cells, as well as parenchymal cells, present in nephritic kidneys. The detection of similar immune cell populations in urine suggests that it might be possible to non-invasively monitor immune activation in kidneys. Once applied to large patient cohorts, these high-dimensional studies might enable patient stratification according to patterns of immune cell activation in the kidney or identify disease features that can be used as surrogate measures of efficacy in clinical trials. Applied broadly across multiple inflammatory kidney diseases, these studies promise to enormously expand our understanding of renal inflammation in the next decade.
Introduction
Lupus nephritis is a common and serious manifestation of systemic lupus erythematosus (SLE). At least 50% of patients with SLE develop LN and, in 10% of these patients, LN progresses to end-stage renal disease (ESRD) within 5 years 1-8. Although mortality from LN has decreased over the past few decades owing to improvements in the treatment of comorbidities, more judicious use of immunosuppressive therapies and a greater willingness and ability to perform renal transplantation in patients with SLE, the morbidity and mortality associated with LN remain substantial. Advances in the treatment of LN have been hard to achieve and clinical trials in LN have frequently failed. Although many factors might explain these outcomes, three particular issues might be crucial.
First, our current classification of LN and, therefore, our identification of patients for inclusion or exclusion in clinical trials, is inconsistent with our knowledge of prognosis and progression in LN 9-12. The universally accepted classification system for LN from the International Society of Nephrology and Renal Pathology Society (ISN/RPS) is focused exclusively on glomerular pathology – the cellular composition and the presence of immune complexes in the glomeruli are evaluated by both light and electron microscopy 13. However, for several decades, data have suggested that the presence of infiltrating inflammatory cells in the interstitium correlates best with prognosis. Interstitial inflammation with associated tubular atrophy is the most important prognostic marker of disease progression to ESRD but is not scored in the current classification system 14-18. Of note, tubular atrophy secondary to glomerular disease and proteinuria may be present in the absence of interstitial inflammation, but the association of tubular atrophy with interstitial inflammation is what predicts poor prognosis in SLE 19. Thus, clinical trials currently include individuals with similar glomerular pathology but with potentially substantial differences in interstitial and tubular pathology. Expecting the same response to therapy from each of these patient subgroups might diminish the likelihood of positive outcomes in clinical trials. The development of standardized metrics for scoring interstitial inflammation would facilitate clinical studies aimed at defining the prognostic value of these histological features.
Second, our current clinical assessments do not always accurately reflect underlying changes in renal pathology 15, 20. In both clinical practice and clinical trials, we assess response to therapy based on reductions in proteinuria and the urine protein to creatinine ratio (UPCR), stabilization or improvement in serum creatinine levels, and successful tapering of systemic glucocorticoids. In two independent studies, investigators performed repeat renal biopsies in individuals with LN, 6 to 12 months after onset of standard immunosuppressive therapy 21, 22. Surprisingly, in approximately 50% of patients with a ‘complete’ clinical response (based on proteinuria and/or UPCR criteria), renal biopsy samples still had histological evidence of ongoing inflammation 20, 22. Moreover, approximately 50% of patients with persistent proteinuria had no residual inflammation 21. Thus, patients with continued renal inflammation might be clinical responders, and patients with markedly diminished inflammation might be clinical non-responders. Interestingly, although UCPR and proteinuria do not seem to accurately reflect renal histopathology findings, patients who achieve a clinical response according to these metrics are unlikely to progress to ESRD over 10 years 23, 24. Clarifying the mechanistic relationship between interstitial inflammation and glomerular injury requires further study. In addition, understanding whether kidney-infiltrating immune cells in clinical responders differ from those in non-responders will be of great importance.
Third, our choice of therapeutic targets in LN is based on notions of disease pathogenesis that are derived from mouse models and from analyses of blood rather than the kidney. For example, the identification of type 1 interferon (that is, IFNα and IFNβ) as a therapeutic target in SLE was based on a large amount of data demonstrating that patients with severe disease exhibited greater induction of interferon-stimulated genes (ISGs) in blood cells that those with less severe disease or healthy controls 25, 26. This observation alone cannot provide information on the presence of interferon in inflamed tissue or the contribution of interferon to tissue pathology such as lupus nephritis. The efficacy of targeting type 1 interferons to treat SLE remains under investigation in large clinical trials27. Furthermore, using mouse models to inform therapeutic strategies in SLE has had limited success 28. Although all mouse models of SLE are characterized by glomerular immunoglobulin deposition and proteinuria, the additional histopathologic characterization of kidney infiltrating cells and the mechanisms of disease vary among models29, 30. It is not known which models are most aligned with patient subsets.
To address these issues, a detailed analysis of the immune and parenchymal cells present within the kidneys of patients with LN might provide new insights into the cell types and pathways associated with tissue pathology, and might help characterize the heterogeneity of the disease. Such insights might inform therapeutic decisions and guide the development of novel drugs.
In this Review, we discuss methodological and clinical design considerations in implementing single-cell transcriptomic analyses of immune cells in lupus nephritis. We also discuss current findings from single-cell RNA sequencing (scRNA-seq) analyses of lupus nephritis kidneys and urine, including the immune cell populations identified and how such studies might help transform LN therapy. Given the ongoing efforts to use scRNA-seq to study multiple kidney diseases including diabetic nephropathy and allograft rejection, comparisons across diseases will be of major interest31, 32.
Design of scRNA-seq studies of LN
Bulk versus single-cell RNA-sequencing
One major limitation of the current histological assessments in LN is that the characterization of immune cells in the kidney is very limited, with little information gathered about the specific immune cell subsets present or their expression of functional molecules. For example, tissue histology can demonstrate the presence and localization of macrophages and lymphocytes in LN kidneys. However, both macrophages and lymphocytes have multiple differentiation states with starkly different functions that are mediated by expression of distinct effector molecules33, 34 – standard histological assessments cannot resolve this diversity of functions. Higher resolution analyses of the phenotypes and potential functions of immune cell types present in LN kidneys, and their variability across multiple patients, are likely to substantially improve the predictive value of kidney biopsy samples.
Global transcriptional profiling with RNA-seq allows genome-wide quantification of gene expression in tissue samples and can now be applied to individual cells within a tissue35. The application of RNA-seq to LN kidney biopsy samples thus offers the potential to quantify immune cell types and pathways at a resolution not previously possible. However, several issues need to be considered when conducting transcriptional analyses of infiltrating and kidney resident cells. The first consideration is whether to isolate cellular subsets from the kidney and perform bulk RNA-seq on small numbers of cells or, alternatively, to perform sc RNA-seq (TABLE 1).
Table 1:
Approaches to transcriptional analysis of LN kidneys.
| Input | Advantages | Drawbacks |
|---|---|---|
| Total kidney tissue | Global analysis without selective cell loss | Cannot discriminate the source of the differentially expressed genes |
| Can be applied to archived samples in tissue banks | ||
| Laser capture dissection of kidney tissue sections | Can select anatomic substructures or cell populations for analysis | Requires specialized microscopy, relatively low throughput |
| Can be applied to archived samples in tissue banks | Limited resolution complicates the ability to select individual cells within tissue | |
| Tissue compartments isolated by manual microdissection | Can distinguish signatures restricted to glomeruli versus interstitium | Requires microdissection of fresh tissue upon acquisition |
| Cannot discriminate the cellularsource of the signature within the structure | ||
| Isolated cell subsets | Analyzes signatures of specific cell populations of particular interest in detail | Only a limited number of cell populations can be analyzed and the remainder of the cells is lost. |
| Requires viable tissue for isolation of live cells | ||
| Requires flow cytometric cell sorting to isolate small populations with high purity | ||
| Cannot determine the localization of the cell populations | ||
| Individual cells | Potential to analyze almost all cells in kidney sample | Requires viable tissue for isolation of live cells |
| Difficult to detect low – abundance transcripts | ||
| Cannot determine the localization of the cells | ||
Transcriptomic analyses of bulk kidney tissue have provided some insights into mechanisms of kidney injury in LN. Transcriptional profiling of 32 archived LN biopsy samples demonstrated that the samples segregated into three groups using unsupervised clustering [G]36. One of the three clusters was characterized by the expression of adaptive immune cell genes, which is indicative of T cell and B cell infiltration; an increased frequency of B cells, CD4+ T cells and CD8+ T cells in these samples was confirmed by immunohistochemistry. Notably, patients with LN whose biopsy samples segregated into this group had a lower estimated glomerular filtration rate (eGFR) compared with the patients in the other two groups, consistent with the reported association between interstitial lymphocyte infiltrates and reduced kidney function in LN 37.
Another independent transcriptomic study of kidney biopsy samples from patients with diverse kidney disease, including LN, identified an association between reduced epidermal growth factor (EGF) expression and reduced eGFR 38, 39. This pattern of altered EGF expression across multiple chronic kidney diseases suggests that it might be a shared feature of kidney injury irrespective of the upstream mechanisms. Other studies quantified bulk gene expression in initial diagnostic kidney biopsies and in repeat biopsy samples in patients with LN, and also compared kidney biopsy expression profiles in patients who subsequently either had a complete response or did not respond to therapy40, 41. These reports suggested that gene expression patterns change following treatment of LN and identified differences in gene expression between complete responder and non-responder groups. Expression of neural cell adhesion molecule 1 (NCAM1), for example, was higher in the kidneys of patients with a complete response to treatment than in with patients who did not respond, whereas interleukin 1 receptor accessory protein (IL1RAP) and Fc fragment of IgA receptor (FCAR) were most highly expressed in the kidneys of patients with a poor response to treatment 40, 41.
The discrimination of pathways that affect distinct parts of the kidney is facilitated, for example, by laser capture microscopy [G], through which glomerular and interstitial tissue can be collected separately42 (TABLE 1). A focused transcriptional analysis of glomeruli from LN biopsy samples revealed the upregulation of multiple pathways compared to control kidney samples, including a prominent increase in myeloid lineage transcripts42. However, the precise identities of the myeloid cells present in these samples cannot be identified through this bulk RNA-seq approach.
Although transcriptomic studies of bulk tissue have begun to implicate critical immune pathways in the pathogenesis of LN, these analyses of intact tissue do not provide the resolution needed to identify which cell populations contribute the most to the observed altered gene expression pathways. Advances in computational deconvolution [G] methods, including some methods that rely on available scRNA-seq data, are improving the ability to predict cell composition in a sample based on bulk tissue transcriptomes43. However, such approaches are still limited, especially since LN kidneys might contain unique subsets of immune cells or cells with activation states that have not previously been described in blood, the typical source of reference samples used to inform deconvolution.
scRNA-seq is therefore an attractive alternative to bulk RNA-seq as it establishes a catalog of all the cells present in LN kidneys. This catalog can then inform analyses of studies that focus on intact glomeruli and interstitium. Single-cell transcriptomic analyses of murine and human kidneys have demonstrated the feasibility and promise of this approach. For example, a comprehensive analysis of adult mouse kidney cells by scRNA-seq identified 21 cell populations in the healthy mouse kidney and revealed a previously unrecognized cell population in the collecting duct that might represent a transitional cellular phenotype between principal cells and intercalated cells 44. scRNA-seq of over 72,000 cells from human kidneys, including samples of both fetal and adult tissue, as well as renal tumor cells, established the range of cell types in human kidney 45. Interestingly, this study suggested that both clear cell renal cell carcinoma and papillary renal cell carcinoma might derive from the same cell lineage as cells from both tumour types were most similar to one subpopulation of proximal convoluted tubule cells from healthy kidney.
Kidney tissue collection and processing
Studies attempting to establish a broad transcriptomic analysis of LN need to navigate multiple challenges related to tissue collection and processing (BOX 1). First, clinical research sites must obtain a segment of kidney biopsy tissue for research analysis. Because most scRNA-seq methods require intact, viable cells for analysis46, a fresh tissue specimen, distinct from the tissue allocated for histological analysis, is required. One approach is to collect an additional biopsy sample exclusively for research analysis. Investigators might be concerned about the potential morbidity associated with obtaining an additional renal core, yet available data indicate that, when collecting up to five cores, no incremental morbidity is associated with the collection of each additional core 47. Of note, collecting more than five cores is associated with an increase in the incidence of biopsy-related adverse events. The experience of the Accelerated Medicines Partnership (AMP) rheumatoid arthritis (RA)/SLE Network over the past 4 years corroborates the safety of obtaining a research core in LN. Minor bleeding events occurred in <10% of patients, yet all complications resolved, and no individuals who provided a research core required a blood transfusion after biopsy (B. Diamond, D. Wofsy, unpublished data). These safety data are important in enabling analogous studies in similar patient populations worldwide.
BOX 1: Major design considerations for scRNA-seq analysis of LN kidneys.
Key aspects of study design and implementation must be established for informative scRNA-seq analyses of kidney biopsy samples.
Patient selection and eligibility
The selection of patients with new-onset, proliferative LN who have not received immunosuppressive treatment might be considered ideal for studies of immune cells in lupus nephritis. However, since restricting patient selection to such a limited cohort substantially limits enrollment, alternative options should also be considered.
ISN classification: select only patients with proliferative GN or also include membranous GN
Immunosuppressant treatment: select only treatment-naïve patients or include cross-sectional cohort on varied medications
Disease course: select only patients with new-onset disease or include samples from re-biopsy of patients with known LN
Initial handling of kidney tissue
The ability to preserve and transport tissue samples for centralized processing greatly expands the number of samples analyzed and minimizes technical noise.
Analysis infrastructure: each collection site runs the final analysis (for example, RNA-seq) or collected samples are analyzed at a central site.
Tissue dissociation: each site dissociates tissue into single cells for preservation or intact tissue samples are immediately preserved with minimal handling.
Sample transport: samples are shipped on wet ice immediately after collection or cryopreserved for long-term storage and batched transport.
Tissue dissociation into single cells
Tissue dissociation protocols should be optimized to achieve maximum cell yield with minimal cell perturbation.
Enzymatic digestion: enzyme specificity and timing should be selected to maximize cell yields but limit loss of cell surface markers (if protein expression of these markers is to be assessed).
Temperature: use of proteolytic enzymes that are active at cold temperatures should be considered to reduce potential transcriptomic changes induced during tissue digestion104.
Culture conditions: the use of culture media and media supplementation should be optimized to maintain cell health and/or inhibit cell death.
Capture of single cells for transcriptomic analysis
Method selection depends on cell yield, cell populations of interest and the need to purify cells from tissue debris.
Cell capture: droplet-based, microfluidic or flow cytometric capture of single cells.
Cell selection: profile all cells in the sample or enrich for specific cell populations.
Removal of dead cells and debris: purify intact, live cells through magnetic column separation or flow sorting; alternatively cells can be directly analysed after tissue dissociation.
After collection, tissue samples need to be processed to obtain single cell suspensions for scRNA-seq analysis – rapid processing minimizes changes in the expression of transcriptionally active genes 48-50. Multiple technical variables, including handling time, incubation temperatures and dissociation conditions can alter the transcriptomes of isolated cells. Therefore, optimization of the processing of tissue samples is an important step
In multi-center studies, many enrollment sites do not have the capability to run scRNA-seq analyses. Even if it were feasible to perform RNA-seq at each individual site, this would likely introduce substantial technical noise due to inter-site variations in sample processing49. One approach to minimize technical variation is to centralize scRNA-seq analyses at a single site, which requires transporting dissociated cells, often across the long distances within the same country or even internationally. Following cryopreservation [G], samples can be transported either on wet ice or on dry ice (≤ −70ÐC). Transport of samples on wet ice imposes substantial logistical challenges, as the samples must be shipped immediately and the central lab site must be ready to receive them whenever they become available. This method of sample preservation for transport would itself introduce technical variation as each sample would constitute a separate batch. Alternatively, cryopreservation allows long-term storage of samples so that they can be shipped when convenient, and samples can be processed in batches in a controlled manner.
Biopsy tissue can also be frozen as intact tissue at the collection site using methods similar to those used to cryopreserve viable cells51, 52. Cryopreserved tissue can then be thawed, dissociated and analyzed at a later time. Freezing intact tissue samples offers several advantages: it minimizes the handling required at enrollment sites as tissue can be rapidly frozen after acquisition; it enables the accumulation of a biorepository of intact, viable tissue; and it allows the accumulation of samples at a central processing site, where they can be processed and analyzed in large batches to reduce batch-to-batch technical variation. The AMP RA/SLE Network, a multi-center effort to study tissue samples from patients with RA and SLE using single-cell technologies, developed and implemented this cryopreservation strategy for LN kidney biopsy samples collected across the network (FIG. 1)51 .
Figure 1: Implementation of single-cell RNA-seq to study lupus nephritis kidney biopsy samples.
Kidney tissue is routinely biopsied for histological evaluation of lupus nephritis (LN). Additional kidney tissue can be obtained at the time of biopsy for dedicated cellular analyses. The ability to store and transport these samples so that they can be analyzed in a uniform manner at a central processing site has enabled such analyses to be performed across multiple sites as a collaborative project. Intact kidney tissue, as well as cells from a pre-biopsy urine sample, can be cryopreserved immediately after acquisition and then transported to a central site. Kidney tissue can then be thawed and dissociated into a single cell suspension. For a focus on immune cells in LN, leukocytes from both kidney tissue or urine can be purified by flow cytometric cell sorting and then analyzed by single-cell RNA-seq. These molecular analyses, combined with clinical and histological features, might provide new biomarkers that predict progression of LN to end-stage renal disease or a good response to therapy. These data might also provide insight into potential novel therapeutic targets and the heterogeneity of LN pathogenesis.
In the AMP RA/SLE Network experience, cryopreservation of intact kidney tissue provided increased total cell yields compared with cryopreservation of dissociated kidney cells52. Whether tissue cryopreservation and subsequent dissociation is better than tissue dissociation followed by cryopreservation might vary depending on the tissue type although, in general, maintaining cells in their microenvironment is likely to induce less stress than removing cells from their native cell–cell contacts. Compared with freshly processed samples, in kidney samples subjected to cryopreservation and thawing, the temperature disturbance led to the upregulation of a specific set of genes. However, these transcriptomic changes are relatively small compared with the transcriptomic signatures that discriminate cell populations and also differ from the signatures that distinguish LN kidneys from control kidney samples. Notably, with the exception of neutrophils, leukocytes survived dissociation, freezing and thawing far better than both epithelial cells and endothelial cells and, as a result, their extracted gene expression data is of superior quality than that of other cell types51. Efforts to block caspase-mediated apoptosis with small peptide caspase inhibitors or to engage adhesion molecules with soluble ligands to mimic cell-cell contact failed to improve cell yields (B. Diamond, unpublished observations).
Following dissociation of tissue into single cells, it might be advantageous to purify intact, live cells from dead cells and debris, and this can be achieved with magnetic separation methods or through flow cytometric cell sorting53. One advantage of using cell sorting is that it enables the isolation of specific cell populations of interest; however, the additional handling time required for cell staining and sorting might impose additional stress on the cells. Single-nuclear RNA-seq (snRNA-seq) is an alternative RNA-seq approach in which nuclei are specifically isolated from cells for sequencing54. This method can be applied to intact frozen tissue samples, bypassing some of the challenges of tissue dissociation and the requirement for isolation of viable cells. snRNA-seq might reduce transcriptomic artefacts induced by cell stress and improve acquisition of data on renal resident cells 32, 55. While the number of transcripts detected with snRNA-seq is generally lower than that detected with scRNA-seq, data to date suggest that both methods detect similar numbers of genes in renal biopsy samples 32, 55, 56.
Quality control metrics
Determining the number of cells that provide high-quality single-cell transcriptomic data is not entirely straightforward because the number of cells considered suitable for analysis depends on the quality metrics and cutoffs used. Quality control metrics for scRNA-seq often include cutoffs for the number of genes detected per cell (a higher number indicates better quality) and the proportion of reads mapped to mitochondrial genes (a lower proportion indicates better quality)57. In one AMP RA/SLE Network analysis, only 10–15% of kidney epithelial cells met the cutoffs of more than 1,000 genes detected per cell with less than 25% of the transcripts derived from mitochondria 51. However, both the number of genes identified per cell and the proportion of mitochondrial reads might vary depending on the sequencing technology platform used and on the cell type studied. The 10X Genomics platform, for example, uses a droplet-based scRNA-seq method, which typically detects the expression of a smaller number of genes and therefore a lower threshold for this parameter is often applied58. In addition, RNA yields and quality may vary with cell type — leukocytes, for example, often demonstrate good viability whereas healthy parenchymal cells are more challenging to isolate and study51, 52. It is possible that this challenge is exacerbated in diseased kidneys, as they likely harbor stressed cells that might be especially vulnerable to the manipulations needed to preserve and dissociate tissue.
Acquisition of control kidney tissue
For studies focused on comparing different renal pathologies or stages of disease progression, the comparison between samples obtained from groups of patients with the relevant diseases might suffice. However, the inclusion of healthy kidney tissue addresses an additional set of questions about the differences between kidneys with and without pathology. Several options exist for obtaining healthy kidney tissue. For example, histologically normal kidney tissue can be dissected from surgical specimens obtained from tumor nephrectomies – such tissue is often available in relatively large quantities. However, the surgical procedure itself subjects nephrectomized kidney tissue to the stress of warm ischaemia and, typically, no information is available regarding the extent of cellular perturbations in tissue surrounding the tumor. An alternative is to use biopsy tissue from living donor kidneys as control tissue. The extensive evaluation of renal function of kidney donors prior to surgical removal of the kidney makes it very likely that the collected kidney tissue is healthy. However, kidneys obtained for transplant are perfused to remove blood cells, which potentially removes blood cells that accumulate within renal vessels and might introduce technical variation between these samples and those from biopsied native kidneys. Comparison of kidney biopsy samples taken before and after perfusion might reveal if particular blood cells accumulate in the vasculature of healthy glomeruli and enable their comparison with circulating blood cells; however, such datasets are not yet available.
Disease heterogeneity
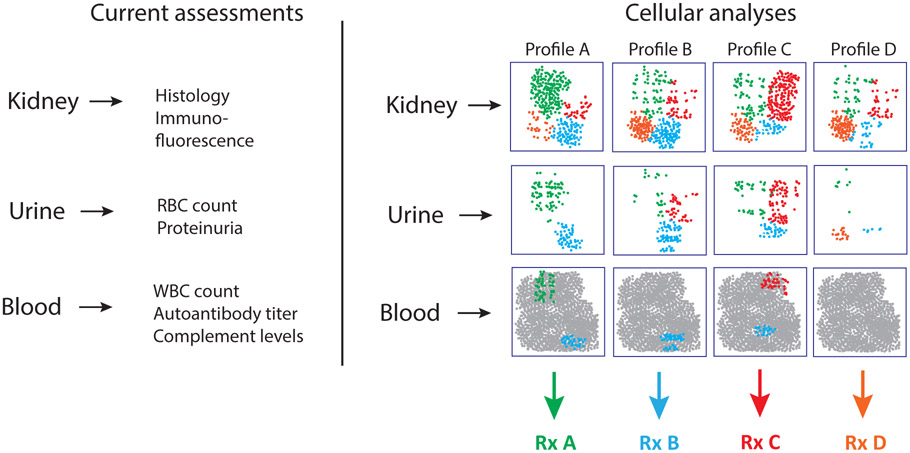
Multiple clinical features should be considered in the design of a study to interrogate cellular features of LN, including the histological class of glomerulonephritis, sex, ethnicity, drug treatment, and systemic disease activity. The ISN classification of LN includes 6 defined pathological states (classes 1–6) 13. Both the heterogeneity of disease mechanisms and whether patients with any class of disease can progress to a higher degree of pathology are poorly understood – assessing the minimum number of samples that must be analyzed to survey the spectrum of pathology in each class is therefore problematic. Class 6 disease, characterized by the presence of large numbers of sclerotic glomeruli, represents an advanced stage of pathology that is presumed to progress to ESRD and thought to be non-responsive to immune-targeted therapy due to irreversible tissue damage59. Patients with disease classes 3 or 4 (proliferative nephritis [G]), as well as those with class 5 disease (membranous nephritis [G]), are at risk of progression to ESRD but are thought to benefit the most from immunosuppressive therapy 1, 8. Proliferative forms of LN can be distinguished from membranous disease by light microscopy. The variability in disease progression and responses to treatment among patients, including in patients within the same ISN class of disease59, suggests that multiple immunopathological mechanisms exist and highlights the importance of additional predictive metrics. Detailed cellular analyses of LN offer the potential to yield insights into the molecular pathways and cell types involved in disease, and to identify predictive biomarkers (FIG. 2).
Figure 2. Towards a cellular classification of renal immunopathology.
Current clinical data evaluate metrics of immune activation and tissue injury in kidney, urine, and blood. However, these analyses do not assess the immune cell types that may mediate disease in lupus nephritis (LN). Detailed single-cell analyses of kidney tissue might enable more precise quantification of the major immune and parenchymal cell phenotypes present in LN kidneys – such analyses might distinguish subsets of patients with distinct patterns of immune activation in the kidney. Once these populations are defined in LN kidneys, it may be possible to detect disease-associated cell populations or cell states in urine as well, although the cell ratios and abundances are likely to differ. Selected populations enriched in LN kidneys might also be detectable in blood, where they would likely represent rare populations within the large pool of circulating mononuclear cells. Identification and detection of these cell populations or pathways might improve patient stratification and enable more precise selection of therapies that directly target the relevant cell types and better monitoring of response to therapy.
Some studies of blood cells have suggested that transcriptional differences exist between men and women and over the human lifespan 60, 61. Data also suggest that SLE might differ between sexes 62. Clinically, disease is often more severe in men but 90% of patients with SLE are female and an unselected population will thus have few males. It might therefore seem reasonable to study females first and determine an approximate number of patients who need to be studied to assess patient heterogeneity before initiating a study of male SLE patients.
Several studies have identified SLE risk alleles that pertain to particular racial or ethnic populations63. However, it is not known whether polymorphisms associated with race and/or ethnicity alter LN pathogenesis. Although African Americans who carry the APOL1 risk allele are more likely than individuals with non-risk alleles to develop LN 64, studies involving large numbers of genotyped individuals are needed to determine whether the LN of individuals with APOL1 risk alleles involves different cell types and pathogenic mechanisms. One reasonable approach is to include all racial and ethnic groups in exploratory studies and then evaluate if these variables associate with molecular or cellular features of LN.
The differences between new-onset LN and recurrent LN flares also need to be addressed. Data on whether the first occurrence of LN is the same or different from a renal flare are scarce. LN patients, especially those with an LN flare, are often receiving numerous medications that might affect the transcriptional profile of both infiltrating immune cells in the kidney and stromal cells. Corticosteroids diminish the interferon signature present in blood cells, as well as the expression of genes downstream of nuclear factor-κB (NF-κB)25, 26, and presumably in tissue infiltrating and tissue resident cells in LN,. The effects on gene expression of other medications used to treat patients with SLE are not well understood. However, studying only treatment-naive individuals is very challenging as therapy is often initiated even before biopsy.
The inclusion of a population that is potentially very heterogeneous with respect to demographic characteristics, disease manifestations and drug regimens means that some patient subsets will include very few individuals. For these patient subsets, it may not be possible to evaluate with confidence how single-cell transcriptional data from the kidney differ from that of other patient subsets; however, identification of such individuals will indicate the cohorts from whom more samples are needed.
Insights from studies of lupus nephritis
Leukocyte subsets in LN kidneys
Histological and bulk transcriptomic analyses of LN kidney biopsy samples suggest that the presence of lymphocyte infiltrates is associated with reduced renal function and poor prognosis 36, 37. To shed light on the composition and activation states of these immune infiltrates, as well as their effect on kidney epithelial cells, the AMP RA/SLE Network analyzed gene expression in single cells isolated from dissociated kidney samples, using two approaches. In one approach, total kidney cells were analysed without preselection, which mostly profiled kidney epithelial cells as these represent >90% of kidney cells. This analysis identified disease-associated features of kidney epithelial cells, including the expression of both ISGs and fibrosis-associated genes in patients with LN who had an inadequate response to treatment65 In parallel, immune cells were purified by flow cytometric single-cell sorting for an analysis that focused on the immune cell compartment in LN kidneys 51. A comprehensive assessment of the transcriptional profile of immune cells present in LN kidneys versus healthy kidney tissue begins to address multiple questions about active immune mechanisms in LN kidneys and their clinical implications, as detailed below (see also BOX 2).
BOX 2: Potential insights from scRNA-seq studies of immune cells in patients with lupus nephritis
Cellular identities in lupus nephritis kidneys
Number of identifiable distinct cell clusters (that is, populations)
Identity of cell types included in each cell cluster
Transitions between clusters indicative of multiple differentiation states
Cell types associated with disease
Identification and characterization of cell clusters that correspond to tissue resident cells in the healthy kidney
Identification of which cell clusters are over-represented in lupus nephritis kidneys compared with healthy tissue
Determination of cellular pathways that are activated by the cell clusters over-represented in lupus nephritis kidneys
Detection of migratory signals potentially linked to the recruitment of infiltrating immune cells
Immune correlates with clinical disease
Correlations between kidney cell types and ISN/RPS (International Society of Nephrology and /Renal Pathology Society) disease classes or histological features
Correlations between serological or blood metrics (for example, complement components, autoantibodies and interferon scores)
Identification of surrogate samples (for example blood or urine)
A total of 21 immune cell subsets were identified in LN kidneys51 – four macrophage clusters, including a tissue resident subset frequent in the living donor samples; ten clusters containing T cells or NK cells, including a tissue resident CD8+ T cell subset, a large populations of cytotoxic CD8 T cells and NK cells, regulatory T cells and a TFH-like population; two dendritic cell (DC) clusters (plasmacytoid and conventional); and four B cell clusters, including naïve, memory and antibody-secreting B cell populations. One cluster of proliferating cells included T cells, NK cells and myeloid cells. The infiltrating myeloid and lymphoid cells detected by scRNA-seq were consistent with prior immunohistochemical and immunofluorescent analyses37, 42, 66, 67. Although the total number of cells analyzed was relatively small (~2,700 leukocytes), a saturation analysis [G] showed that increasing the number of analyzed patients is unlikely to reveal new large cell clusters 51. These observations begin to define cell populations that contribute to immune injury in LN (FIG. 3).
Figure 3. Insights into composition of renal immune cell compartment obtained from single-cell RNA-seq analysis of lupus nephritis kidneys.
Single-cell RNA-seq analyses of leukocytes in lupus nephritis (LN) kidneys have revealed the presence of multiple immune cell subtypes with characteristic gene expression patterns. RNA expression patterns can suggest both potential effector functions and developmental trajectories. CD8+ T cells were segregated into two distinct effector populations, including a cytotoxic T cell (CTL) population with strong similarities to natural killer (NK) cells and a separate population with high expression of granzyme K. CD4+ T cell populations included an effector population, a population of T regulatory (Treg) cells and a T follicular helper (TFH)-like cell subset likely to help B cell responses. B cells in LN kidneys segregated into naive and activated populations, and trajectory analyses indicated that differentiation of naive B cells into activated cells and age-associated B cells (ABCs) likely occurs in situ. In contrast, trajectory analyses segregated plasmablasts and/or plasma cells from other B cells, suggesting that these cells develop elsewhere. The presence of B cells at different stages of differentiation, including memory B cells, ABCs, and plasma cells, as well as TFH-like cells, suggests local antibody production and antigen-specific T cell activation by B cells within the kidneys. Several monocyte and myeloid populations present in the kidney also seem to be developmentally related – trajectory analyses suggest that inflammatory monocytes might transition into phagocytic and alternatively activated phenotypes. Whether some cell populations are associated with the initiation of inflammation and others with the resolution of inflammation is unclear. Further analyses of the localization of these distinct cell populations within the kidney will be of major interest. CCR5, C-C motif chemokine receptor 5; CXCL13, C-X-C motif chemokine ligand 13; FCRL5, Fc receptor like 5; FOXP3, forkhead box P3; GNLY, granulysin; GZMB, granzyme B; GZMK, granzyme K; IGHD, immunoglobulin heavy constant delta; IGHM, immunoglobulin heavy constant mu; IKZF2, IKAROS family zinc finger 2; ITGAX, integrin subunit alpha X; MAF, MAF bZIP transcription factor; MERTK, MER proto-oncogene, tyrosine kinase; PDCD1, programmed cell death 1; PRF1, perforin 1; S100A8, S100 calcium binding protein A8; SCARB2, scavenger receptor class B member 2; STAB1, stabilin 1; TCL1A, T cell leukemia/lymphoma 1A; PTPRS, protein tyrosine phosphatase receptor type S; TCF7L2, transcription factor 7 like 2; TBX21, T-box transcription factor 21; TNF, tumour necrosis factor; VSIG4, V-set and immunoglobulin domain containing 4.
Normal kidney tissue from perfused living donors was dominated by two major immune cell populations: a myeloid population that is not highly similar to blood subsets and a population of effector memory CD4+ T cells 51. Essentially no B cells were present in healthy kidney, as assessed by both flow cytometry and scRNA-seq.
In situ activation of immune cells
A second important observation is that cells transition between activation states within the kidney 51, 68, 69. A clear continuum of cells exists among patrolling CD16+ macrophages, phagocytic macrophages and macrophages with a transcriptional profile of alternatively activated macrophages as well as high expression of genes associated with tissue repair. This represents a presumed transition from cells that remove debris in the vasculature to cells that remove debris in sites of inflammation to macrophages making soluble mediators that help suppress inflammation. Importantly, analysis of each cell at a single moment in time precludes a clear understanding of the directionality of the transition. However, because the macrophage population with the alternative activation–tissue repair gene signature is the least similar to blood monocytes, it seems likely that it corresponds to the last state in a differentiation pathway that occurs in the kidney, beginning with the patrolling, inflammatory CD16+ monocytes51, 70, Further studies will be required to test this hypothesis. It will also be interesting to ascertain if this macrophage differentiation pathway is present in other affected tissues in patients with SLE or if macrophage activation pathways are tissue-specific. Moreover, an analysis of repeat biopsies might elucidate whether a correlation exists between changes in macrophage subpopulations and patient responses to therapy.
A continuum of B cell phenotypes, which spanned naive B cells and activated B cells, also identified transitions among B cell subsets in the kidney 51, 71. Of note, the activated B cell subset included cells with a gene expression signature characteristic of age- and/or autoimmunity-associated B cells (ABCs)72, 73. Plasma cells were also present in the kidney but trajectory analyses of scRNA-seq data did not identify a continued differentiation of B cells into plasma cells within the kidney, suggesting that plasma cells might enter the kidney as plasmablasts [G] and might be clonally unrelated to the other B cells in the kidney. This finding is especially surprising as ABCs have been reported to be poised to become plasma cells 71, 74. The absence of transitions of B cells into plasma cells in LN kidneys might be due to a paucity of local availability of IL-21, a cytokine required for optimal plasma cell differentiation75, IL-21 was not abundantly detected in immune cells in the kidney in the AMP analysis; however, the limited sensitivity of scRNA-seq makes it difficult to detect expression low-abundance transcripts such as a cytokines. Thus it is possible that IL-21 is produced but not able to be detected. Alternatively, IL-21 might be produced by a small subset of T cells, which might not have been adequately sampled considering the number of analyzable cells. A third possibility is that the transcriptomic transition from ABC to plasma cell occurs abruptly with a few intervening steps, such that the intermediate states are not well captured by trajectory analysis. Surprisingly cell division was not observed among B cells in the scRNA-seq analysis, as clonal expansion of B cells has been reported in the kidney of patients with lupus nephritis66. These observations may be affected by the relatively small number of plasma cells and B cells profiled, or limitations in the ability to identify transcriptomic features that distinguish proliferating B cells from other B cells in scRNA-seq data. A more direct analysis of BCR specificities, which can be obtained at the single cell level with 5’ scRNA-seq [G] 76, is required to determine the clonal relation of the different subsets of B cells in LN kidneys and enable the analysis of the antigenic specificities of infiltrating B cells.
Kidney-infiltrating versus blood cells
Other unexpected findings included the absence of certain circulating cell subsets in the kidney, and of kidney cells previously identified in murine models of LN. For example, we identified a tissue resident macrophage population that does not have a blood counterpart. In addition, a second macrophage subset expressed an alternatively activated transcription profile distinct from all currently described blood subsets 77. These observations validate the importance of high-dimensional analysis of kidney immune cells to help understand renal pathology, as studies focused on circulating cells cannot reveal therapeutic targets unique to the kidney. It is now critical to develop protocols that can promote the differentiation of blood monocytes into the novel activation states found in the kidney, which would enable functional analyses of these cells.
Few, if any, TH17 cells were detected in LN kidneys, despite their presence in some, but not all, mouse models of the disease 78-82.In addition, a distinct CD8+ T cell population with an ‘exhausted’ signature was not detected in LN kidneys51. Exhausted T cells are a prominent feature of kidney T cells in murine SLE models83, and a global exhaustion signature in blood CD8+ T cells has been correlated with long-term disease quiescence and good responses to therapy in lupus patients84. Increased expression of exhaustion-related genes was detected in bulk CD8+ T cells from the blood of LN patients in the AMP cohort, compared to samples from healthy controls51. However, although expression of some ‘exhaustion’ related genes was detected in T cells from the LN kidneys, no single T cell cluster expressed high levels of these genes, which would be required to recognize a discrete population of exhausted T cells. Determining whether the ‘exhausted’ signature represents the gene expression profile of more than one T cell subset in kidneys of patients with LN has mechanistic and therapeutic implications and requires further analysis.
The AMP analysis also identified an upregulation of ISGs in all cell subsets in the LN kidneys, including both immune and non-immune cells 51, 65. The degree of upregulation of these genes was positively correlated between kidney and blood, suggesting that this activation pathway reflects a systemic process 51, 85. Induction of ISGs also occurs within the kidney, as tissue resident cells also upregulated genes induced by interferon. This upregulation might result from the production of type 1 interferons by kidney myeloid cells and plasmacytoid dendritic cells, perhaps stimulated by immune complexes containing nucleic acids and locally-secreted high mobility group protein B1 (HMGB1) that activate endosomal Toll-like receptors (TLRs), or through the activation of the STING (stimulator of interferon genes)-controlled innate immune pathway, which is induced by cytosolic DNA. IFN-γ production by infiltrating T cells or NK cells might also contribute to the local interferon signature.
scRNA-seq data and mechanistic insights
Almost every blood-borne cell population in the kidneys of patients with LN expressed CXC chemokine receptor type 4 (CXCR4), with >50% of the cells expressing CXCR4 in all but 2 of the leukocyte clusters 51. CXCL12, the ligand for CXCR486, 87 was expressed primarily by the alternatively activated macrophage population that appears to differentiate within the kidney, although it might also be produced by kidney epithelial cells51, 88. Although this macrophage population expresses some genes associated with tissue repair (e.g. CD169, C1Q), the upregulation of numerous chemokines in this cell subset might also sustain the inflammatory response in the kidney89. This observation suggests that disease mechanisms in LN are complex and that some cell subsets potentially contribute to both sustained inflammation and repair. Of note, CXCL12 has been shown to be an effective therapeutic target in murine lupus 90. In addition to CXCR4, the majority of myeloid cells (54–76%, depending on the specific cell subset), and more than 90% of NK cells and cytolytic T cells, expressed CX3C chemokine receptor 1 (CX3CR1)51; its ligand (CX3CL1), seems to be primarily produced by epithelial cells in LN kidneys data but is also known to be expressed by endothelial cells51, 91.
One important insight from the scRNA-seq data was that many of the same cell populations were found in multiple patient samples, although the relative proportions of the different leukocyte populations varied51. In this initial cohort of 24 patients, a distinct correlation between the composition of kidney immune cell compartment and LN classification was not identified. Perhaps it is not surprising that classes 3 and 4 could not be distinguished given the small number of samples from each group (n=7 class III samples, n=12 class IV samples) and their histological similarity. More surprisingly, pure membranous disease (class 5) could not be distinguished from proliferative disease (classes 3 and 4), even though class V biopsies lack an increased cellularity in glomeruli, which differs clearly from class III and class IV biopsies. The similar leukocyte yields obtained from biopsy samples corresponding to disease classes 3, 4 and 5 suggest that most of the leukocytes obtained from dissociated kidney biopsy samples, in particular lymphocytes, derive from the interstitium rather than the glomeruli. This is consistent with the histologic assessments of lymphocyte infiltrates, which show that most of the lymphocytes accumulate in the interstitial regions rather than in glomeruli67. It is also possible, even likely, that although SLE might be a heterogeneous disease with respect to mechanism of disease induction, the ensuing inflammatory response in the kidney may be more homogeneous. This might lead to a greater opportunity to discover therapeutic targets that would be broadly applicable across different subgroups of patients with SLE.
Limitations of scRNA-seq and next steps
The currently available RNA-seq datasets provide a framework to define the immune cell populations in LN kidneys but these analyses only provide a starting point for addressing multiple additional questions.
Relationships between cell types and clinical features over time.
Since data from biopsy tissue are obtained from a single moment in time, the directionality of transitions between cellular states observed in LN kidneys can be inferred but not demonstrated with certainty. Data from repeat biopsy samples may provide compelling information on the directionality of transitions as one subset may be absent in a sequential biopsy obtained during therapy. Analyses of repeat biopsies, which are now becoming standard in clinical practice, will determine which baseline features of the kidney infiltrating and resident cells are most predictive of response or non-response to therapy, or whether changes in cellular profiles are a better prognostic indicator. Finally, repeat biopsies might identify important therapeutic targets by identifying cell subsets that change in response to therapy.
Relationships between infiltrating immune cells and renal resident cells.
As leukocytes generally represent less than 10% of the cells obtained from a dissociated kidney biopsy, it may be necessary to isolate CD45+ cells in order to obtain the greatest possible insight into renal leukocytes. However, analysis should not focus exclusively on leukocytes as the examination of both immune and non-immune cells from the same tissue will provide important and perhaps surprising data. The identification of the leukocyte subsets present in the areas of renal tissue with the most tubular atrophy and stressed epithelial cells, for example, might provide insight into which populations contribute to renal tubular cell damage. Alternatively, it might be that the predictor of response to therapy lies more with how parenchymal cells respond to immune attack rather than with the infiltrating immune cells themselves.
Cellular features not captured by scRNA-seq
These scRNA-seq approaches do not provide direct measurements of microRNA, proteomic or metabolic data, although some insights can be inferred. Although such analyses have been performed on blood and urine of patients with SLE92-94, none have been performed on tissue. Technical advances that combine cell surface protein detection with scRNA-seq, such as REAP-seq and CITE-seq 95, 96, enable the correlation of protein expression with transcriptomic data and are of great interest in the analysis of tissue pathology. However, the proteolytic enzymes used to dissociate tissue for single cell analyses also can cleave membrane molecules and might lead to an artefactual loss of antibody binding97. The possibility of maintaining dissociated cells in culture for a short time to allow re-expression of cleaved surface proteins without altering transcriptional profile of the cells has not been explored.
Transcriptional analyses of LN have thus far not included an assessment of T and B cell antigen receptor repertoires so it has not yet been possible to detect clonal expansions in the kidney, which have been demonstrated for CD8 T cells and plasma cells through other methodologies 66, 67. It would be important to determine if B and T cell activation in the kidney is antigen-specific or is secondary to mitogenic signals.
Perhaps most importantly, information on the anatomic localization of the cells and their intercellular interactions is largely lost when analysing dissociated single cells37. Although these interactions can perhaps be deduced by analyses of receptor–ligand pairs for chemokines and cytokines, only the direct visualization of the spatial relationships among cells can really address this question. Because cellular localization cannot be determined with scRNA-seq, it is also not possible to discriminate leukocyte populations that infiltrate or reside in the kidney from those that may be circulating through the glomeruli or other vasculature of the kidney. Immunohistology, including the applications of highly multiparametric methods, as well as spatial transcriptomics, can assist in this analysis and might be enhanced by using markers identified through global transcriptomic data to localize individual cell populations 98, 99.
Pathological implications of immune cell phenotypes.
Deciphering whether a particular subset contributes to or limits pathology is complex issue, even when detailed descriptions of the transcriptomic features of specific cell populations, and perhaps even their anatomic localization, are available. For example, the macrophage population that is most dissimilar from blood populations and acquires its transcriptional profile within the kidney expresses molecules that are generally associated with tissue repair but is also a major source of the chemokines presumed to recruit blood cells into the kidney51. It is difficult, therefore, to assess whether this subset should be targeted therapeutically. Perhaps an analysis of the potential correlation between renal outcome and the presence of these cells in the kidney will be helpful, with the expectation that pathologic cell types will be highly represented in samples from patients with deteriorating renal function. A focus on infiltrating cell types that express likely pathological effector molecules such as granzymes might also be a reasonable approach to prioritize potential therapeutic targets. Using murine models that recapitulate the relevant cell phenotypes and effector functions may then allow further experimental manipulation.
Finally, it is also important not to over-extrapolate from the renal immune landscape. Although interstitial disease in the kidney correlates best with long term renal outcome 14-18, both human disease and murine models clearly indicate that glomerular immunoglobulin deposition is an initial trigger for LN100. Thus, the activation of extra-renal B cells that produce nephrogenic antibodies remains a rational therapeutic target whether or not such B cells can be found in the kidney. Targeted B cell therapies are an important focus of SLE therapeutics and include the approved biologic, belimumab, as well as agents that deplete B cells and plasma cells, agents that diminish B cell receptor signaling pathways, and agents that block B cell–T cell interactions101.
Neutrophil activation is also detectable in the blood of patients with LN but although histological analysis of LN kidneys indicates the presence of neutrophils in the tissue 102, these cells are unlikely to survive the tissue procurement and processing protocols required for analysis by RNA-seq. The transcriptional profile of neutrophils in LN kidneys is therefore not presently known. Neutrophils contribute to SLE pathogenesis, in part, through the extrusion of oxidized DNA, which is highly interferonogenic, and might thus be an appropriate therapeutic target in LN whether or not there are functional differences between circulating and infiltrating kidney neutrophils103. Advances in snRNA-seq technology might enable the analysis of neutrophil activation states in the kidney.
Conclusions
Global transcriptomic profiling of individual cells from LN kidneys has substantially improved the resolution with which we understand the immune cell infiltrates in LN, outlining many of the prominent myeloid, T cell, and B cell subsets that accumulate in LN kidneys and their characteristic gene expression programs. Although important limitations exist when interpreting scRNA-seq data, applying these approaches to informative cohorts of patients with LN, as well as other kidney diseases, may yield clinically useful metrics of prognosis and treatment response. Future studies should address whether the composition of the renal leukocyte infiltrate, or the transcriptomic signature of specific leukocyte populations in the kidney, correlates with kidney disease progression. Such observations might inform the development of predictive biomarkers based on simpler readouts of kidney biopsies such as immunohistochemistry (for example, staining for a specific cell population or cytokine), the identification of novel urine biomarkers (for example, detection of a cell population rather than measurement of UPCR) (BOX 3) and the development of therapeutic interventions that disrupt pathways enriched in LN kidneys. Perhaps one of the most important gains of a more widespread study of the transcriptional program of tissue resident and infiltrating cells in LN will be the creation of a new classification system that is based on the expression of prognostic markers that reflect molecular mechanisms of disease and tissue injury.
BOX 3: Urine as a surrogate marker of kidney pathology.
The development of a non-invasive method to detect and monitor kidney injury would be a major advance over the current need for a kidney biopsy. Numerous studies have attempted to study urine to understand renal pathology in lupus, generally with a focus on urine proteins, and have identified factors associated with lupus nephritis, including TWEAK and MCP1105-109. Immune cells accumulate in urine of patients with lupus nephritis51, 110-112, raising the possibility that these cells reflect renal pathology. Immune cells from urine can be collected by urine centrifugation and analyzed by flow cytometry, bulk RNA sequencing (RNA-seq) and single-cell RNA-seq (scRNA-seq)51, 52. The resulting cell profiles can be compared to those of immune cells isolated from kidney biopsy samples to identify similarities and differences, and to assess the potential utility of using urine to assess renal immune pathology. Data from the Accelerating Medicines Partnership (AMP) suggests that hematopoietic cells in the urine closely resemble those in kidney although their relative proportions differ in kidney and urine51. Additional studies are needed to determine whether urine will be a useful surrogate for kidney biopsy.
Advantages of analysis of cells from urine
Simple and non-invasive
Can be collected serially to monitor changes in cell composition over time
Potential limitations of analysis of cells from urine
Might select for only a subset of the immune infiltrate that can access urine from glomeruli or tubules
Might select for only a subset of cells that can survive in the unfavourable urine microenvironment
Urine may alter transcriptomes or function of immune cells and obscure their function in kidney
Cytometric analysis of lupus nephritis urine cells
Provides rapid, inexpensive quantification of urine immune cell composition
Careful fluorophore selection to avoid autofluorescence enables multi-parametric analysis
Immune cells in urine are enriched for myeloid cells and depleted of lymphocytes compared to LN kidneys51
The frequency of CXCR3+ CD4+ T cells in urine is associated with disease activity in lupus nephritis110
Transcriptomic analysis of lupus nephritis urine cells
Provides high-dimensional analysis of cell phenotypes and gene expression patterns.
Rapid collection and processing yields high-quality RNA for global transcriptomics
Initial scRNA-seq analyses show that the transcriptomes of immune cell populations from urine strongly resemble comparable populations in lupus nephritis kidneys
Key points:
Single-cell RNA-seq has begun to define the phenotypes of distinct immune cell populations that accumulate in the kidney in LN, which can be assessed across patients to identify molecularly distinct disease mechanisms and inform precision medicine strategies.
Design of cohort studies of LN using single cell RNA-seq must balance the desire for optimal samples with feasibility, and must consider aspects of clinical heterogeneity including disease duration, gender, race, and drug treatment in enrollment criteria.
Cryopreservation of tissue biopsies allows for accumulation of a biorepository of tissue containing viable cells, which can be analyzed uniformly in batches to reduce technical variation.
Comparison of immune cells in the kidney with cells from urine and blood might identify cellular markers that can be measured in urine or blood and reduce the need for invasive kidney biopsies.
Acknowledgements:
All authors are supported by the Accelerated Medicines Partnership (AMP) NIH UH2 AR06768. D.A.R. is supported by the Lupus Research Alliance, Burroughs Wellcome Fund Career Award for Medical Sciences and NIH NIAMS K08 AR-072791-01.
Glossary terms
- Unsupervised clustering
unbiased approach used to identify sets of samples that share similar gene expression signatures.
- Laser capture microscopy
a method that uses microscopy to identify and selectively collect specific regions of a tissue section for downstream analyses such as RNA expression measurement
- Computational deconvolution
in RNA-seq analysis, an effort to quantify or identify features of individual cell populations from bulk RNA-seq data using cell type-specific reference datasets.
- Cryopreservation
the process of freezing a sample (e.g. tissue, cells) for long-term storage, often using a controlled freezing method in the presence of a cryoprotectant such as dimethyl sulfoxide to preserve cell viability during freeze and thaw.
- Proliferative nephritis
a form of nephritis characterized histologically by endothelial and mesangial proliferation within glomeruli
- Membranous nephritis
a form of nephritis characterized by thickening of the glomerular capillary wall with subepithelial deposits but without cellular proliferative changes.
- Saturation analysis
a procedure to estimate the extent to which a given sample represents a studied population. Typically, this involves downsampling the original data, computing a statistic of interest (e.g. the number of discovered clusters) and estimating its rate of change across a sequence of sample sizes.
- Plasmablasts
a differentiated B cell population specialized for secretion of antibodies; often a precursor to plasma cells.
- 5’ scRNA-seq
single cell RNA-seq methods that sequence RNA transcripts from the 5’ end, allowing clearer analysis of the 5’ end of RNA transcripts, as opposed to the more common strategies of RNA-seq that analyze transcripts from the 3’ end.
Footnotes
Competing interests:
The authors declare no competing interests.
References
- 1.Almaani S, Meara A & Rovin BH Update on Lupus Nephritis. Clin J Am Soc Nephrol 12, 825–835 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Costenbader KH et al. Trends in the incidence, demographics, and outcomes of end-stage renal disease due to lupus nephritis in the US from 1995 to 2006. Arthritis Rheum 63, 1681–8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanly JG et al. The frequency and outcome of lupus nephritis: results from an international inception cohort study. Rheumatology (Oxford) 55, 252–62 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jorge A et al. All-Cause and Cause-Specific Mortality Trends of End-Stage Renal Disease Due to Lupus Nephritis From 1995 to 2014. Arthritis Rheumatol 71, 403–410 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Narain S & Furie R Update on clinical trials in systemic lupus erythematosus. Curr Opin Rheumatol 28, 477–87 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Vandepapeliere J et al. Prognosis of proliferative lupus nephritis subsets in the Louvain Lupus Nephritis inception Cohort. Lupus 23, 159–65 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Yap DY, Tang CS, Ma MK, Lam MF & Chan TM Survival analysis and causes of mortality in patients with lupus nephritis. Nephrol Dial Transplant 27, 3248–54 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Haridasan S, Sharma A Rathi M Treatment of membranous lupus nephritis. Clinical Quueries: Nephrology 3, 106–113 (2014). [Google Scholar]
- 9.Ayodele OE, Okpechi IG & Swanepoel CR Predictors of poor renal outcome in patients with biopsy-proven lupus nephritis. Nephrology (Carlton) 15, 482–90 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Broder A et al. Tubulointerstitial damage predicts end stage renal disease in lupus nephritis with preserved to moderately impaired renal function: A retrospective cohort study. Semin Arthritis Rheum 47, 545–551 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsieh C et al. Predicting outcomes of lupus nephritis with tubulointerstitial inflammation and scarring. Arthritis Care Res (Hoboken) 63, 865–74 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hill GS, Delahousse M, Nochy D, Mandet C & Bariety J Proteinuria and tubulointerstitial lesions in lupus nephritis. Kidney Int 60, 1893–903 (2001). [DOI] [PubMed] [Google Scholar]
- 13.Weening JJ et al. The classification of glomerulonephritis in systemic lupus erythematosus revisited. J Am Soc Nephrol 15, 241–50 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Kassianos AJ et al. Increased tubulointerstitial recruitment of human CD141(hi) CLEC9A(+) and CD1c(+) myeloid dendritic cell subsets in renal fibrosis and chronic kidney disease. Am J Physiol Renal Physiol 305, F1391–401 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Rovin BH, Parikh SV & Alvarado A The kidney biopsy in lupus nephritis: is it still relevant? Rheum Dis Clin North Am 40, 537–52, ix (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwartz MM et al. Irreproducibility of the activity and chronicity indices limits their utility in the management of lupus nephritis. Lupus Nephritis Collaborative Study Group. Am J Kidney Dis 21, 374–7 (1993). [DOI] [PubMed] [Google Scholar]
- 17.Teh CL et al. Causes and predictors of mortality in biopsy-proven lupus nephritis: the Sarawak experience. Clin Kidney J 11, 56–61 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen Y et al. Association of intrarenal B-cell infiltrates with clinical outcome in lupus nephritis: a study of 192 cases. Clin Dev Immunol 2012, 967584 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schelling JR Tubular atrophy in the pathogenesis of chronic kidney disease progression. Pediatr Nephrol 31, 693–706 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malvar A et al. Histologic versus clinical remission in proliferative lupus nephritis. Nephrol Dial Transplant 32, 1338–1344 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alvarado AS et al. The value of repeat kidney biopsy in quiescent Argentinian lupus nephritis patients. Lupus 23, 840–7 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Zickert A, Sundelin B, Svenungsson E & Gunnarsson I Role of early repeated renal biopsies in lupus nephritis. Lupus Sci Med 1, e000018 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dall’Era M et al. Predictors of long-term renal outcome in lupus nephritis trials: lessons learned from the Euro-Lupus Nephritis cohort. Arthritis Rheumatol 67, 1305–13 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Mackay M et al. Establishing Surrogate Kidney End Points for Lupus Nephritis Clinical Trials: Development and Validation of a Novel Approach to Predict Future Kidney Outcomes. Arthritis Rheumatol 71, 411–419 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Bennett L et al. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J Exp Med 197, 711–23 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baechler EC et al. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci U S A 100, 2610–5 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Felten R, Scher F, Sagez F, Chasset F & Arnaud L Spotlight on anifrolumab and its potential for the treatment of moderate-to-severe systemic lupus erythematosus: evidence to date. Drug Des Devel Ther 13, 1535–1543 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Group AT Treatment of lupus nephritis with abatacept: the Abatacept and Cyclophosphamide Combination Efficacy and Safety Study. Arthritis Rheumatol 66, 3096–104 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berthier CC et al. Cross-species transcriptional network analysis defines shared inflammatory responses in murine and human lupus nephritis. J Immunol 189, 988–1001 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bethunaickan R, Berthier CC, Zhang W, Kretzler M & Davidson A Comparative transcriptional profiling of 3 murine models of SLE nephritis reveals both unique and shared regulatory networks. PLoS One 8, e77489 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson PC et al. The single-cell transcriptomic landscape of early human diabetic nephropathy. Proc Natl Acad Sci U S A (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu H et al. Single-Cell Transcriptomics of a Human Kidney Allograft Biopsy Specimen Defines a Diverse Inflammatory Response. J Am Soc Nephrol 29, 2069–2080 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bassler K, Schulte-Schrepping J, Warnat-Herresthal S, Aschenbrenner AC & Schultze JL The Myeloid Cell Compartment-Cell by Cell. Annu Rev Immunol 37, 269–293 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Mueller SN, Gebhardt T, Carbone FR & Heath WR Memory T cell subsets, migration patterns, and tissue residence. Annu Rev Immunol 31, 137–61 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Birnbaum KD Power in Numbers: Single-Cell RNA-Seq Strategies to Dissect Complex Tissues. Annu Rev Genet 52, 203–221 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pamfil C et al. Intrarenal activation of adaptive immune effectors is associated with tubular damage and impaired renal function in lupus nephritis. Ann Rheum Dis 77, 1782–1789 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liarski VM et al. Cell distance mapping identifies functional T follicular helper cells in inflamed human renal tissue. Sci Transl Med 6, 230ra46 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ju W et al. Tissue transcriptome-driven identification of epidermal growth factor as a chronic kidney disease biomarker. Sci Transl Med 7, 316ra193 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berthier CC, Kretzler M & Davidson A A systems approach to renal inflammation in SLE. Clin Immunol 185, 109–118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parikh SV et al. Molecular imaging of the kidney in lupus nephritis to characterize response to treatment. Transl Res 182, 1–13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parikh SV et al. Characterising the immune profile of the kidney biopsy at lupus nephritis flare differentiates early treatment responders from non-responders. Lupus Sci Med 2, e000112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peterson KS et al. Characterization of heterogeneity in the molecular pathogenesis of lupus nephritis from transcriptional profiles of laser-captured glomeruli. J Clin Invest 113, 1722–33 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang X, Park J, Susztak K, Zhang NR & Li M Bulk tissue cell type deconvolution with multi-subject single-cell expression reference. Nat Commun 10, 380 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park J et al. Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science 360, 758–763 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Young MD et al. Single-cell transcriptomes from human kidneys reveal the cellular identity of renal tumors. Science 361, 594–599 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alles J et al. Cell fixation and preservation for droplet-based single-cell transcriptomics. BMC Biol 15, 44 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roccatello D et al. Outpatient percutaneous native renal biopsy: safety profile in a large monocentric cohort. BMJ Open 7, e015243 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leek JT et al. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev Genet 11, 733–9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nguyen QH, Pervolarakis N, Nee K & Kessenbrock K Experimental Considerations for Single-Cell RNA Sequencing Approaches. Front Cell Dev Biol 6, 108 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tung PY et al. Batch effects and the effective design of single-cell gene expression studies. Sci Rep 7, 39921 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arazi A et al. The immune cell landscape in kidneys of patients with lupus nephritis. Nat Immunol 20, 902–914 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rao DA et al. A protocol for single cell transcriptomics from cryopreserved renal tissue and urine for the Accelerating Medicines Partnership (AMP) RA/SLE network. bioRxiv 10.1101/275859 (2018). [DOI] [Google Scholar]
- 53.Beliakova-Bethell N et al. The effect of cell subset isolation method on gene expression in leukocytes. Cytometry A 85, 94–104 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krishnaswami SR et al. Using single nuclei for RNA-seq to capture the transcriptome of postmortem neurons. Nat Protoc 11, 499–524 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu H, Kirita Y, Donnelly EL & Humphreys BD Advantages of Single-Nucleus over Single-Cell RNA Sequencing of Adult Kidney: Rare Cell Types and Novel Cell States Revealed in Fibrosis. J Am Soc Nephrol 30, 23–32 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bakken TE et al. Single-nucleus and single-cell transcriptomes compared in matched cortical cell types. PLoS One 13, e0209648 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Luecken MD & Theis FJ Current best practices in single-cell RNA-seq analysis: a tutorial. Mol Syst Biol 15, e8746 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baran-Gale J, Chandra T & Kirschner K Experimental design for single-cell RNA sequencing. Brief Funct Genomics 17, 233–239 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hahn BH et al. American College of Rheumatology guidelines for screening, treatment, and management of lupus nephritis. Arthritis Care Res (Hoboken) 64, 797–808 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Debey S et al. A highly standardized, robust, and cost-effective method for genome-wide transcriptome analysis of peripheral blood applicable to large-scale clinical trials. Genomics 87, 653–64 (2006). [DOI] [PubMed] [Google Scholar]
- 61.Menon R et al. Gender-based blood transcriptomes and interactomes in multiple sclerosis: involvement of SP1 dependent gene transcription. J Autoimmun 38, J144–55 (2012). [DOI] [PubMed] [Google Scholar]
- 62.Schwartzman-Morris J & Putterman C Gender differences in the pathogenesis and outcome of lupus and of lupus nephritis. Clin Dev Immunol 2012, 604892 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lewis MJ & Jawad AS The effect of ethnicity and genetic ancestry on the epidemiology, clinical features and outcome of systemic lupus erythematosus. Rheumatology (Oxford) 56, i67–i77 (2017). [DOI] [PubMed] [Google Scholar]
- 64.Freedman BI et al. End-stage renal disease in African Americans with lupus nephritis is associated with APOL1. Arthritis Rheumatol 66, 390–6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Der E et al. Tubular cell and keratinocyte single-cell transcriptomics applied to lupus nephritis reveal type I IFN and fibrosis relevant pathways. Nat Immunol 20, 915–927 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chang A et al. In situ B cell-mediated immune responses and tubulointerstitial inflammation in human lupus nephritis. J Immunol 186, 1849–60 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Winchester R et al. Immunologic characteristics of intrarenal T cells: trafficking of expanded CD8+ T cell beta-chain clonotypes in progressive lupus nephritis. Arthritis Rheum 64, 1589–600 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Angerer P et al. destiny: diffusion maps for large-scale single-cell data in R. Bioinformatics 32, 1241–3 (2016). [DOI] [PubMed] [Google Scholar]
- 69.Qiu X et al. Reversed graph embedding resolves complex single-cell trajectories. Nat Methods 14, 979–982 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Meng XM, Mak TS & Lan HY Macrophages in Renal Fibrosis. Adv Exp Med Biol 1165, 285–303 (2019). [DOI] [PubMed] [Google Scholar]
- 71.Karnell JL et al. Role of CD11c(+) T-bet(+) B cells in human health and disease. Cell Immunol 321, 40–45 (2017). [DOI] [PubMed] [Google Scholar]
- 72.Jenks SA et al. Distinct Effector B Cells Induced by Unregulated Toll-like Receptor 7 Contribute to Pathogenic Responses in Systemic Lupus Erythematosus. Immunity 49, 725–739 e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Naradikian MS, Hao Y & Cancro MP Age-associated B cells: key mediators of both protective and autoreactive humoral responses. Immunol Rev 269, 118–29 (2016). [DOI] [PubMed] [Google Scholar]
- 74.Wang S et al. IL-21 drives expansion and plasma cell differentiation of autoreactive CD11c(hi)T-bet(+) B cells in SLE. Nat Commun 9, 1758 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zotos D et al. IL-21 regulates germinal center B cell differentiation and proliferation through a B cell-intrinsic mechanism. J Exp Med 207, 365–78 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cole C, Byrne A, Beaudin AE, Forsberg EC & Vollmers C Tn5Prime, a Tn5 based 5' capture method for single cell RNA-seq. Nucleic Acids Res 46, e62 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Villani AC et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 356 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang Z, Kyttaris VC & Tsokos GC The role of IL-23/IL-17 axis in lupus nephritis. J Immunol 183, 3160–9 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Amarilyo G, Lourenco EV, Shi FD & La Cava A IL-17 promotes murine lupus. J Immunol 193, 540–3 (2014). [DOI] [PubMed] [Google Scholar]
- 80.Jacob N et al. Accelerated pathological and clinical nephritis in systemic lupus erythematosus-prone New Zealand Mixed 2328 mice doubly deficient in TNF receptor 1 and TNF receptor 2 via a Th17-associated pathway. J Immunol 182, 2532–41 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee SY et al. Inhibition of IL-17 ameliorates systemic lupus erythematosus in Roquin(san/san) mice through regulating the balance of TFH cells, GC B cells, Treg and Breg. Sci Rep 9, 5227 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schmidt T et al. Function of the Th17/interleukin-17A immune response in murine lupus nephritis. Arthritis Rheumatol 67, 475–87 (2015). [DOI] [PubMed] [Google Scholar]
- 83.Tilstra JS et al. Kidney-infiltrating T cells in murine lupus nephritis are metabolically and functionally exhausted. J Clin Invest 128, 4884–4897 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McKinney EF, Lee JC, Jayne DR, Lyons PA & Smith KG T-cell exhaustion, co-stimulation and clinical outcome in autoimmunity and infection. Nature 523, 612–6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kok HM et al. Systemic and local granzyme B levels are associated with disease activity, kidney damage and interferon signature in systemic lupus erythematosus. Rheumatology (Oxford) 56, 2129–2134 (2017). [DOI] [PubMed] [Google Scholar]
- 86.Bleul CC et al. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature 382, 829–33 (1996). [DOI] [PubMed] [Google Scholar]
- 87.Oberlin E et al. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature 382, 833–5 (1996). [DOI] [PubMed] [Google Scholar]
- 88.Togel F, Isaac J, Hu Z, Weiss K & Westenfelder C Renal SDF-1 signals mobilization and homing of CXCR4-positive cells to the kidney after ischemic injury. Kidney Int 67, 1772–84 (2005). [DOI] [PubMed] [Google Scholar]
- 89.Griffith JW, Sokol CL & Luster AD Chemokines and chemokine receptors: positioning cells for host defense and immunity. Annu Rev Immunol 32, 659–702 (2014). [DOI] [PubMed] [Google Scholar]
- 90.Devarapu SK et al. Dual blockade of the pro-inflammatory chemokine CCL2 and the homeostatic chemokine CXCL12 is as effective as high dose cyclophosphamide in murine proliferative lupus nephritis. Clin Immunol 169, 139–147 (2016). [DOI] [PubMed] [Google Scholar]
- 91.Imai T & Yasuda N Therapeutic intervention of inflammatory/immune diseases by inhibition of the fractalkine (CX3CL1)-CX3CR1 pathway. Inflamm Regen 36, 9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Aljaberi N, Bennett M, Brunner HI & Devarajan P Proteomic profiling of urine: implications for lupus nephritis. Expert Rev Proteomics 16, 303–313 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Huang N & Perl A Metabolism as a Target for Modulation in Autoimmune Diseases. Trends Immunol 39, 562–576 (2018). [DOI] [PubMed] [Google Scholar]
- 94.Jafari Ghods F, Topal Sarikaya A, Arda N & Hamuryudan V MiRNA and mRNA Profiling in Systemic Lupus Reveals a Novel Set of Cytokine - Related miRNAs and their Target Genes in Cases With and Without Renal Involvement. Kidney Blood Press Res 42, 1322–1337 (2017). [DOI] [PubMed] [Google Scholar]
- 95.Peterson VM et al. Multiplexed quantification of proteins and transcripts in single cells. Nat Biotechnol 35, 936–939 (2017). [DOI] [PubMed] [Google Scholar]
- 96.Stoeckius M et al. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods 14, 865–868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gray DH, Chidgey AP & Boyd RL Analysis of thymic stromal cell populations using flow cytometry. J Immunol Methods 260, 15–28 (2002). [DOI] [PubMed] [Google Scholar]
- 98.Moor AE & Itzkovitz S Spatial transcriptomics: paving the way for tissue-level systems biology. Curr Opin Biotechnol 46, 126–133 (2017). [DOI] [PubMed] [Google Scholar]
- 99.Patel SS & Rodig SJ Overview of Tissue Imaging Methods. Methods Mol Biol 2055, 455–465 (2020). [DOI] [PubMed] [Google Scholar]
- 100.Nowling TK & Gilkeson GS Mechanisms of tissue injury in lupus nephritis. Arthritis Res Ther 13, 250 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Oleinika K, Mauri C & Salama AD Effector and regulatory B cells in immune-mediated kidney disease. Nat Rev Nephrol 15, 11–26 (2019). [DOI] [PubMed] [Google Scholar]
- 102.Lech M & Anders HJ The pathogenesis of lupus nephritis. J Am Soc Nephrol 24, 1357–66 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Daniel C et al. Extracellular DNA traps in inflammation, injury and healing. Nat Rev Nephrol 15, 559–575 (2019). [DOI] [PubMed] [Google Scholar]
- 104.Adam M, Potter AS & Potter SS Psychrophilic proteases dramatically reduce single-cell RNA-seq artifacts: a molecular atlas of kidney development. Development 144, 3625–3632 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Brunner HI et al. Urine biomarkers of chronic kidney damage and renal functional decline in childhood-onset systemic lupus erythematosus. Pediatr Nephrol 34, 117–128 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pejchinovski M et al. Urine peptidomic biomarkers for diagnosis of patients with systematic lupus erythematosus. Lupus 27, 6–16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rovin BH et al. Urine chemokines as biomarkers of human systemic lupus erythematosus activity. J Am Soc Nephrol 16, 467–73 (2005). [DOI] [PubMed] [Google Scholar]
- 108.Schwartz N et al. Urinary TWEAK as a biomarker of lupus nephritis: a multicenter cohort study. Arthritis Res Ther 11, R143 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Soliman S & Mohan C Lupus nephritis biomarkers. Clin Immunol 185, 10–20 (2017). [DOI] [PubMed] [Google Scholar]
- 110.Enghard P et al. CXCR3+CD4+ T cells are enriched in inflamed kidneys and urine and provide a new biomarker for acute nephritis flares in systemic lupus erythematosus patients. Arthritis Rheum 60, 199–206 (2009). [DOI] [PubMed] [Google Scholar]
- 111.Kopetschke K et al. The cellular signature of urinary immune cells in Lupus nephritis: new insights into potential biomarkers. Arthritis Res Ther 17, 94 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Scott E, Dooley MA, Vilen BJ & Clarke SH Immune cells and type 1 IFN in urine of SLE patients correlate with immunopathology in the kidney. Clin Immunol 168, 16–24 (2016). [DOI] [PubMed] [Google Scholar]