Abstract
Introduction
Recent studies have revealed that microRNAs (miRNAs, miRs) are important for self-renewal, differentiation, and cellular reprogramming of somatic cells into induced pluripotent stem cells (iPSC); however, their functional roles and target genes that are regulated by human PSC–specific miRs including hsa-miR-302 clusters remain largely unknown. Analysis of their target gene will give us the opportunity to understand the functional roles of such miRs.
Methods
We analyzed the expression profiles of miRs in 4 somatic cell lines, 8 human iPSC lines derived from 4 different cell types, 3 human ESC lines, and embryoid bodies differentiated from the human ESCs to identify human PSC–specific miRs. We also analyzed the simultaneous expression profiles of miRs and mRNAs to identify candidate targets of human PSC–specific miRs. Then, we constructed a vector for overexpressing one of the target gene to dissect the functions of human PSC–specific miR in maintenance of self-renew and differentiation.
Results
We focused on hsa-miR-302 cluster as a human PSC–specific miR and identified 22 candidate targets of hsa-miR-302 cluster that were moderately expressed in undifferentiated human PSCs and up-regulated in differentiated cells. Deleted in azoospermia-associated protein 2 (DAZAP2), one such target, was directly repressed by hsa-miR-302a, -302b, -302c and -302d, but not by hsa-miR-367. Overexpression of DAZAP2 caused a decrease in cell proliferation of undifferentiated human iPSCs, although morphology and undifferentiated marker gene expression was not affected. In addition, neural differentiation was suppressed in DAZAP2-overexpressing human iPSCs.
Conclusion
Our study revealed that hsa-miR-302 cluster controls the cell proliferation of human PSCs and the neural differentiation of human PSCs by repression of DAZAP2, thereby highlighting an additional function of human PSC–specific miRs in maintaining pluripotency.
Keywords: MIR302 cluster (microRNA-302s, hsa-miR-302s); Human PSC; DAZAP2
1. Introduction
MicroRNAs (miRs) are small, non-coding RNAs, approximately 22 nucleotides in length, that negatively regulate the post-transcriptional expression of targeted mRNAs [1]. Recent studies have implicated miRs in the self-renewal and differentiation of embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) [[2], [3], [4], [5]]. Compared to differentiated cells, particular miR families are specifically upregulated in human pluripotent stem cells (PSCs) [[6], [7], [8], [9]], including the hsa-miR-302/367 [[10], [11], [12], [13]], hsa-miR-372 [12], hsa-miR-17-92 [14], and hsa-miR-200 families [15]. These miRs are also contained in extracellular vesicles derived from human PSCs [16]. The accurate identification of such miR targets aided in the understanding of the functional roles played by miRs and transcriptional regulatory networks in human PSCs. To date, only a few genes have been identified as target genes of human PSC-specific miRs. For instance, NR2F2 and MBD2 that are regulated by hsa-miR-302 family members inhibit POU5F1 [17,18] and NANOG [19] expression, respectively. The hsa-miR-302/367 cluster also inhibits DAZAP2, SLAIN1, and TOB2 expression and modulates bone morphogenetic protein signaling during differentiation [20]. The hsa-miR-302 also regulates BAF170 and BAF53a expression and regulates global chromatin structure that affects differentiation [21]. CDKN1C is a cell-cycle checkpoint regulator that is directly targeted by hsa-miR-92b, while CDKN1C mRNA downregulation by hsa-miR-92b likely contributes to the small fraction of human ESCs in G1 phase [22]. However, the functional roles of human PSC–specific miRs and the genes they regulate remain largely unknown.
Here, we analyzed miR expression profiles in 4 somatic cell lines and 8 human iPSC lines [23,24], derived from 4 different cell types, placental artery endothelium (PL), fetal lung fibroblast (MRC5), and amnion (AM936) [[25], [26], [27], [28]], 3 human ESC lines [29], and embryoid bodies (EBs) differentiated from the human ESCs. We also analyzed the simultaneous expression profiles of miRs and mRNAs and uncovered the human PSC–specific miR and 22 candidates of their target. Deleted in azoospermia-associated protein 2 (DAZAP2), one such target, was directly repressed by hsa-miR-302 members, but not by hsa-miR-367. We also showed that the DAZAP2 protein in human iPSCs caused a decrease in cell proliferation and suppressed neural differentiation by overexpressing DAZAP2 in human PSCs. Our study revealed that hsa-miR-302 cluster was important for self-renewal and differentiation by repressing DAZAP2, suggesting that overexpression of miR target genes partially recapitulated a functional deficit of miRs and may be a solution for dissecting miR functions.
2. Materials and methods
2.1. Cell culture
Human amnion (AM936), placental artery endothelium (PL), and menstrual blood (Edom22) cell lines were established in our laboratory under a protocol approved by the Institutional Review Board of the National Research Institute for Child Health and Development of Japan [25,26]. Human iPSCs were generated in our laboratory [[25], [26], [27], [28]] and maintained on irradiated mouse embryonic fibroblast (MEF) feeder layers in human iPSC medium, which contained KnockOut DMEM (Thermo Fisher Scientific, Waltham, MA, USA), 20% KnockOut Serum Replacement, GlutaMAX, sodium pyruvate, and non-essential amino acids (Thermo Fisher Scientific) supplemented with 10 ng/mL recombinant human basic fibroblast growth factor (bFGF; Wako Pure Chemical Industries, Ltd., Osaka, Japan) or on iMatrix-511 (Nippi. Inc., Tokyo, Japan) in StemFit medium (Reprocell Inc., Kanagawa, Japan). Human ESCs were generated in our laboratory [29] and maintained on irradiated MEF feeder layers in human iPSC medium or on iMatrix-511 in StemFit medium. The medium was changed every other day and cells were passaged approximately once per week by enzymatic (TrypLE Select Enzyme; Thermo Fisher Scientific) or mechanical methods.
2.2. Embryoid body (EB) formation
To form EBs, cells were grown in iPSC medium without bFGF. Human PSC colonies were detached with TrypLE Select Enzyme or StemPro Accutase (Thermo Fisher Scientific), washed in Dulbecco's phosphate-buffered saline (DPBS: Thermo Fisher Scientific), and seeded into Nunclon Sphera 96-well round (U) bottom plates (Thermo Fisher Scientific). The medium was changed every 2 days. The resulting EB cultures were maintained for 7 days and collected for microarray assay.
2.3. MiRNA extraction and genome-wide microRNA and mRNA profiling
Total RNA (including small RNA) was isolated using a miRNeasy Mini Kit (QIAGEN GmbH). Complementary DNA templates for quantitative reverse transcription polymerase chain reactions (qRT–PCR) analysis of miRNA expression was prepared using a TaqMan MicroRNA Reverse Transcription Kit and a Megaplex™ RT Primers, Human Pools Set v3.0 (Thermo Fisher Scientific), following the manufacturer's protocol. Global miRNA expression profiles were analyzed using a TaqMan Array Human MicroRNA A+B Cards Set v3.0 (Thermo Fisher Scientific) and TaqMan Universal Master Mix II, with UNG (Thermo Fisher Scientific), following the manufacturer's protocol. This set enables the accurate quantitation of 754 human miRNAs. Global miRNA expression profiles were also analyzed using an Agilent Human miRNA microarray v2 (Agilent, Santa Clara, CA, USA). Global mRNA expression profiles were analyzed using a Human Gene Expression 4 × 44K Microarray (Agilent). Hierarchical clustering analyses were performed using Ct values for gene expression data with MEV v4.8 statistical analysis software.
2.4. Transfection
To analyze the effects of the hsa-miR-302/367 cluster on DAZAP2 expression levels, Edom22 cells or human iPSCs were transfected with Lipofectamine 2000 and miRNA Mimics (Thermo Fisher Scientific) or anti-miR miRNA inhibitors (Thermo Fisher Scientific) at final concentrations of 50 nM added. Two or three days after transfection, DAZAP2 expression levels were compared with cells transfected with mirVana™ miRNA Inhibitor, and Negative Control #1 (Thermo Fisher Scientific) by qRT–PCR.
2.5. Quantitative RT-PCR analysis
Total RNA was isolated using the RNeasy Plus Mini Kit (QIAGEN GmbH). Single-stranded cDNA was synthesized from 0.1 to 2 μg of total RNA in 20 μL reactions containing random primers using a Superscript III First Strand cDNA Synthesis System (Thermo Fisher Scientific). For qRT–PCR, we used SYBR Green-based assays (Platinum SYBR Green qPCR SuperMix-UDG; Thermo Fisher Scientific). Transcript levels were determined using a QuantStudio 12K Flex Real-Time PCR System (Thermo Fisher Scientific). All qRT–PCR reactions using SYBR Green were conducted in triplicate or quadruplicate, and relative quantification was performed using GAPDH as a reference gene. In addition, we ran multiple gene expression assays with EBs derived from wild-type and DAZAP2-overexpressing Edom–iPSCs by qRT–PCR, using RT2 Profiler™ PCR Array Human Embryonic Stem Cells (QIAGEN GmbH). ACTB, and GAPDH were used as endogenous controls. Hierarchical clustering analyses were performed with MEV v4.8 statistical analysis software, using Ct values obtained from gene expression studies.
2.6. Lentivirus infection
To analyze DAZAP2 function in human iPSCs, we used lentiviral vectors to overexpress target genes. For the DAZAP2 overexpression assay, we cloned DAZAP2 isoform a (NM_014764) into the BamHI and SalI sites of the lentivirus expression vector pLV-EF1a-MCS-IRES-mCherry. For lentiviral vector preparation, Human embryonic kidney (HEK) 293T cells were cotransfected with a DAZAP2 isoform a construct and MISSION Lentiviral Packaging Mix (Sigma–Aldrich, MI, USA) using Lipofectamine 2000. Supernatants were collected 48 h post-transfection and filtered to exclude cellular debris. The resulting lentivirus was concentrated using a Lenti-X Concentrator (TaKaRa Bio Inc., Shiga, Japan).
2.7. Luciferase assays
The 3′-UTR of DAZAP2 was amplified from Edom22 cDNA and cloned into the SacI and NheI sites of the pmirGLO vector (Promega, WI, USA), downstream of the firefly luciferase gene. Humanized Renilla luciferase was used as a control reporter for normalization. HEK293 cells were seeded in 24-well plates and transfected with 50 nM miRNA mimics and 0.5 μg of pmirGLO encoding the DAZAP2 3′-UTR. Transfected cells were lysed 24–48 h post-transfection, and luciferase reporter activities were measured using the Dual-Luciferase Reporter Assay System (Promega) and GloMax-Multi Detection System (Promega).
2.8. Immunocytochemistry
Human iPSCs were grown on 35-mm glass coverslips (AGC TECHNO GLASS CO., LTD, Shizuoka, Japan), fixed in 4% paraformaldehyde, and permeabilized in PBS containing 0.5% Triton X-100. Subsequently, cells were blocked with PBS containing 5% normal serum appropriate for each antibody in preparation for immunohistochemistry studies. Cells were then incubated overnight at 4 °C with primary antibodies against POU5F1 (sc-5792; Santa Cruz Biotechnology, Inc., Dallas, TX, USA), NANOG (RCAB0003P; ReproCELL), beta III tubulin (G7121; Promega Corp.) or DAZAP2 (ab103317; Abcam, Cambridge, UK). Following overnight incubation, cells were washed with 5% normal serum in PBS. Next, cells were incubated in the dark for 1 h at room temperature in PBS containing 0.1% bovine serum albumin and appropriate secondary anti-mouse, anti-rabbit, or anti-goat antibodies coupled with Alexa 488 (BD Biosciences, Franklin Lakes, NJ, USA), as well as 4′, 6-diamidino-2-phenylindole (DAPI) for nuclei staining. Staining was observed under an LSM 510 Meta Laser Scanning Confocal Microscope (Carl Zeiss Microscopy GmbH, Jena, Germany).
2.9. Differentiation of iPSCs into neural stem cell and neural cell
The plates were coated with Geltrex matrix (Thermo Fisher Scientific) for neural stem cell induction and maintenance. The neural cells were induced from NSCs on poly-l-ornithine (Sigma) and laminin (Thermo Fisher Scientific) coated dishes. Human iPSCs were plated at 0.5–1 × 10ˆ5 cells/cm2 density on geltrex-coated dishes in StemFit medium with ROCK inhibitor. After 1 day, the medium containing ROCK inhibitor was changed to neural induction medium (Thermo Fisher Scientific). Fresh medium was changed every 2 days. One week after differentiation, generated NSCs were passed and maintained in neural expansion medium (Thermo Fisher Scientific). For neural cell induction, NSCs were passaged in neural differentiation medium containing DMEM/F12, Neurobasal Meidum, 2% B-27 supplement and 1% N-2 supplement (Thermo Fisher Scientific) onto the poly-l-ornithine and laminin-coated plates.
2.10. Primers
The following forward (F) and reverse (R) primers for qRT–PCR were used to amplify the indicated genes: OCT3/4, F: 5′- CTGGGTTGATCCTCGGACCT-3′ and R: 5′- CACAGAACTCATACGGCGGG-3′; NANOG, F: 5′- AAAGAATCTTCACCTATGCC-3′ and R: 5′- GAAGGAAGAGGAGAGACAGT-3′; SOX2, F: 5′- GCTTAGCCTCGTCGATGAAC-3′ and R: 5′- AACCCCAAGATGCACAACTC-3′; TRIM28, F: 5′-ACAAGGACCACCAGTACCAGTT-3′ and R: 5′-ATCTTGACATCCACTTGCACAC-3′; TDGF, F: 5′- TCCTTCTACGGACGGAACTG-3′ and R: 5′- AGAAATGCCTGAGGAAAGCA-3′; DNMT3B, F: 5′- ATAAGTCGAAGGTGCGTCGT-3′ and R: 5′- GGCAACATCTGAAGCCATTT-3′; STAT3, F: 5′- GGCCCCTCGTCATCAAGA-3′ and R: 5′- TTTGACCAGCAACCTGACTTTAGT-3′; DAZAP2, F: 5′-CGAACAGGAAGAGGACGAAA-3′ and R: 5′-CAGGGTAGGTTGGCTGTGTT-3′; hsa-miR-302/367 cluster pri-miRNA, F: 5′- TAACTTTATTGTATTGACCGCAGCTC-3′ and R: 5′- ACACAGTGTGGGCGTTAACG-3′; PAX6, F: 5′-GTCCATCTTTGCTTGGGAAA-3′ and R: 5′-TAGCCAGGTTGCGAAGAACT-3′; MAP2, F: 5′- TTCGTTGTGTCGTGTTCTCA-3′ and R: 5′- AACCGAGGAAGCATTGATTG-3′; NES, F: 5′- GAGGGAAGTCTTGGAGCCAC-3′ and R: 5′- AAGATGTCCCTCfAGCCTGG-3′; SOX1, F: 5′-CACAACTCGGAGATCAGCAA-3′ and R: 5′-GGTACTTGTAATCCGGGTGC-3′. To amplify the full-length DAZAP2 transcript, we used the following forward and reverse primers: F: 5′-AGAGTCGACATGAACAGCAAAGGTCAATAT-3′ and R: 5′-AGAGGATCCTCACCAGATGGTGTAGCCA-3′. Alternative forward (5′-TCCACCCTATACCGATGCTC-3′) and reverse (5′-CAGAGAGGCTCCAGGAAATG-3′) primers were used to detect DAZAP2 transcripts. The 3′-UTR of DAZAP2 was amplified from Edom22 cDNA using 5′-AGAGAGCTCTGGTTCAGATGGTGGCTACA-3′ as the forward primer and 5′-AGAGCTAGCAGGCTTGTACTCCTGCCTTT-3′ as the reverse primer.
3. Results
3.1. DAZAP2 is directly regulated by hsa-miR-302 members, but not hsa-miR-367
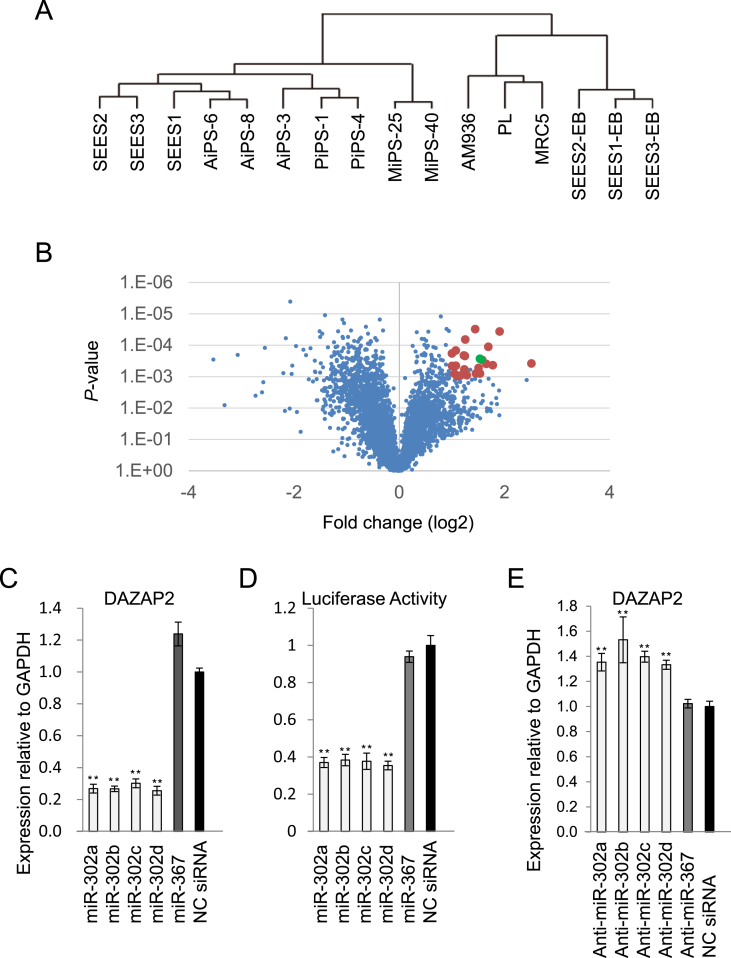
Recent analyses of miR expression profiles in human PSCs, such as ESCs and iPSCs, have shown that the hsa-miR-302 cluster was the most abundant and specific. First, we examined comprehensive miR expression profiles in human ESCs and iPSCs, their parental cells, and embryoid bodies (EBs) derived from human ESCs by microarray and/or TaqMan quantitative (q)PCR assay to confirm the importance of the hsa-miR-302 cluster in our human PSCs. Hierarchical clustering analysis of miR expression patterns revealed that human iPSCs clustered with human ESCs but were separate from the parental cells of human iPSC- and ESC-derived EBs (Fig. 1A, Supplemental Fig 1A), suggesting that human miRs are important for maintaining human PSC properties. Moreover, we could not distinguish human iPSCs and ESCs by miR profiling, suggesting miRs share a common role between such cells. Among the top 25 highly expressed miRs in human PSCs, only hsa-miR-302a, -302b, -302c, -302d and -367 were not expressed in parental cells and were clearly down-regulated after differentiation (Supplemental Fig. 1B). MiRs within in the hsa-miR-17-92 cluster, such as hsa-miR-17, -18a, -19a, -19b, -20a and -92a, were relatively highly expressed in human PSCs but further up-regulated by differentiation (Supplemental Fig. 1C). In this study, we focused on hsa-miR-302 cluster as human PSC-specific miRs and analyzed their functional role in the maintenance of human PSC properties.
Fig. 1.
DAZAP2 is a direct target of the hsa-miR-302 members. (A) Hierarchal clustering analysis was performed using 723 miR expression levels analyzed by Agilent Human miRNA microarray v2 in three somatic cell lines (placental artery endothelium [PL], fetal lung fibroblast [MRC5], and amnion [AM]), seven human induced pluripotent stem cell (iPSC) lines, three human embryonic stem cells (ESCs; SEES-1, SEES-2, and SEES-3) and three embryoid bodies (EBs) derived from human ESCs. AiPS-3, 6, and 8 are human iPSCs generated from the amnion. MiPS-25 and 40 are human iPSCs generated from fetal lung fibroblasts. PiPS-1 and 4 are human iPSCs generated from placental artery endothelium. Data clustering analysis were performed with MeV software, Ver. 4.8.1. (B) Among the 3723 probes that were expressed more than average in undifferentiated cells, 22 genes (red) were significantly upregulated, according to a t-test, and were more than two-fold upregulated in embryoid bodies (EBs). Deleted in azoospermia-associated protein 2 (DAZAP2) is indicated by a green dot. (C) Relative DAZAP2 expression in Edom22 cells following transfection with mature hsa-miR-302 members or -367, compared to negative control small interfering (si)RNA. (D) Relative luciferase activity in HEK293 cells cotransfected with an expression vector encoding firefly luciferase upstream of the DAZAP2 3′-untranslated region (UTR) with mature hsa-miR-302 members, -367, or a negative control siRNA. (E) Relative DAZAP2 expression in undifferentiated induced pluripotent stem cells (iPSCs; MiPS-25) transfected with anti–hsa-miR-302 members or -367 inhibitors, compared to undifferentiated iPSCs transfected with negative control siRNA. ∗∗P < 0.01.
Guo and colleagues reported that changes in mRNA levels closely reflect the impact of miRs on gene expression and indicate that destabilization of target mRNAs is the predominant mechanism for reducing protein expression [30]. Although negative correlations of miR and mRNA expression levels do not directly confirm functional targeting, it may predict a number of targets [31]. Thus, to identify candidate target genes of the hsa-miR-302 cluster, we examined global gene expression profiles in human PSCs and EBs by microarray. Expression patterns of target genes should be inversely proportional to those of hsa-miR-302 cluster. Also, we hypothesized that target genes of hsa-miR-302 cluster would be suppressed in undifferentiated human PSCs and up-regulated during differentiation by human PSC-specific miR down-regulation. Furthermore, because miRs negatively regulate their target mRNAs post-transcriptionally, actual target genes of hsa-miR-302 cluster would be transcriptionally active in undifferentiated human PSCs. Thus, we focused on mRNAs that were moderately expressed in an undifferentiated state and that were up-regulated during differentiation. Of 3724 probes that were detected above an average level (signal intensity: >2860) in undifferentiated cells, we found that 22 genes were significantly up-regulated (2-fold less, P-value: <eˆ-4) during differentiation. Consequently, 22 genes were identified as high-confidence candidate target genes of the hsa-miR-302 cluster (Fig. 1B, Table 1).
Table 1.
| Gene name | P-value | Fold change |
|---|---|---|
| H2AFY | 3.07E-05 | 2.717689 |
| FBLN1 | 3.66E-05 | 3.749952 |
| LSMD1 | 6.57E-05 | 2.386205 |
| ZNF395 | 1.12E-04 | 3.230262 |
| TPT1 | 1.48E-04 | 2.106364 |
| KLHL9 | 1.83E-04 | 2.002536 |
| HP1BP3 | 2.12E-04 | 2.343407 |
| HMBOX1 | 2.21E-04 | 2.372998 |
| DAZAP2 | 2.94E-04 | 2.9863 |
| SPARC | 3.83E-04 | 5.703576 |
| CAPN2 | 3.94E-04 | 3.146989 |
| VIM | 4.33E-04 | 3.425123 |
| GABARAP | 4.53E-04 | 2.106914 |
| RBM5 | 4.58E-04 | 2.004978 |
| GPSM1 | 5.30E-04 | 2.840752 |
| NDRG2 | 5.90E-04 | 2.357259 |
| EVL | 7.97E-04 | 2.745747 |
| NBPF15 | 8.00E-04 | 2.896611 |
| LEPROTL1 | 8.13E-04 | 2.071118 |
| LAMB2 | 9.03E-04 | 2.434277 |
| NBPF3 | 9.23E-04 | 2.24037 |
| SPEG | 9.68E-04 | 2.138279 |
Among the candidate genes mentioned above, we focused on DAZAP2, whose protein product was reported to be a top target of mmu-miR-291, which is one of the highly expressed miRs in mouse ESCs [32]. In addition, DAZAP2 is one of the targets of hsa-miR-302 cluster [20]. To experimentally validate our predictions, we employed assays based on mature miR overexpression, a luciferase, and mature miR inhibition. When Edom22 cells (primary epithelial cells from menstrual blood) were transfected with mature hsa-miR-302a, -302b, -302c, or -302d mimics, DAZAP2 expression levels significantly decreased by ≥ 70% compared to negative control (NC) small interfering (si)RNA transfectants (Fig. 1C). Unexpectedly, DAZAP2 expression did not change following transfection with a hsa-miR-367 mimic relative to NC siRNA transfectants. To determine whether DAZAP2 is directly targeted by human PSC-specific miRs, we cloned the 3′-untranslated region (UTR) of DAZAP2 isoform a (NM_014764) into a pmirGLO Dual–Luciferase Reporter Vector. The resultant vector was cotransfected with mature miR mimics or NC siRNA into HEK293 cells. Luciferase activities in cells transfected with the reporter vector containing the DAZAP2 3′-UTR were significantly decreased by > 60% following cotransfection with hsa-miR-302a, -302b, -302c, or -302d mimics, compared with luciferase activities in NC siRNA transfectants, but did not change following transfection with a hsa-miR-367 mimic (Fig. 1D). Furthermore, we transfected one of the human iPSC lines (MiPS-25) with anti-hsa-miR-302a, -302b, -302c, or -302d inhibitors or NC siRNA, and found that DAZAP2 expression levels in human iPSCs were significantly increased ~ 40%; however transfecting MiPS-25 cells with hsa-miR-367 inhibitor did not alter DAZAP2 expression (Fig. 1E). These data confirmed that DAZAP2 is a target of hsa-miR-302 members, but not a target of hsa-miR-367.
3.2. DAZAP2 overexpression affects human iPSC renewal
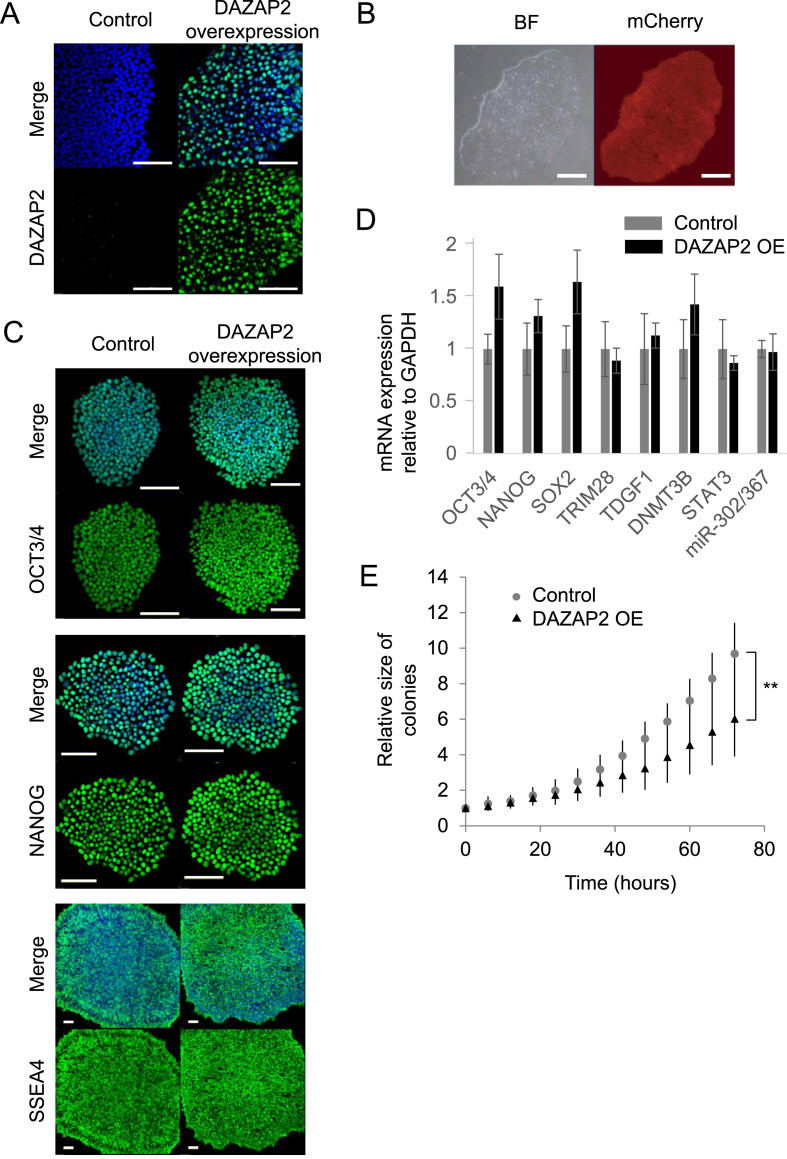
To investigate the functional roles of the hsa-miR-302 cluster, we first examined the sub-cellular localization of DAZAP2 in undifferentiated Edom-iPSC. Although we detected moderate expression of DAZAP2 at the RNA level by both microarray and qRT-PCR, DAZAP2 proteins were barely detected in undifferentiated human iPSCs (Fig. 2A, left panels), suggesting that an excess amount of hsa-miR-302 members strictly suppressed translation of DAZAP2 mRNA into protein. Next, we investigated the importance of DAZAP2 inhibition by hsa-miR-302 members in human PSCs using lentiviral vectors to direct the overexpression of transcripts of interest. We constructed a lentiviral vector containing DAZAP2 isoform a (NM_014764) with mCherry red fluorescent protein downstream of the elongation factor 1 alpha promoter. DAZAP2 proteins were predominantly located in the nucleus of Edom22-iPSC overexpressing DAZAP2 (Fig. 2A, right panels). DAZAP2-overexpressing Edom22-iPSCs had a normal morphology and were maintained for more than 10 passages (Fig. 2B). We also tested whether DAZAP2 expression levels exerted effects on pluripotent marker genes and found that the expression of pluripotent marker genes was not strongly affected at both mRNA and protein levels by DAZAP2 expression (Fig. 2C and D). Despite the importance of hsa-miR-302 members in the maintenance of human PSCs, overexpression of one target gene did not have significant effects on the gene expression of human PSC markers. Previous studies reported that hsa-miR-302 members regulated cell-cycle–related genes. To investigate the role of DAZAP2 inhibition in human PSC self-renewal, we performed time-lapse imaging of an area of undifferentiated colonies and compared the growth rate between control and DAZAP2-overexpressing human iPSCs. Expectedly, the growth rate significantly differed between the two lines, with DAZAP2 significantly decreasing cell proliferation in human iPSCs (Fig. 2E), suggesting that DAZAP2 inhibition by hsa-miR-302 members was required for normal cell proliferation.
Fig. 2.
DAZAP2 modified the cell proliferation rate in human iPSCs derived from Edom cells. (A) Immunofluorescence staining of DAZAP2 in undifferentiated induced pluripotent stem cells (iPSCs) derived from Edom22 cells. Green signals indicate DAZP2 protein. DAPI staining is shown in blue. Scale bar: 100 μm. (B) Colony of DAZAP2 overexpressing Edom–iPSCs with normal morphologies. Red signals indicated DAZAP2-overexpressing cells. Scale bar: 100 μm. (C) Undifferentiated markers, including OCT3/4, NANOG, and stage-specific embryonic antigen 4 (SSEA4), were normally observed in DAZAP2-overexpressing Edom–iPSCs. DAPI staining is shown in blue. Scale bar: 100 μm. (D) Undifferentiated marker gene expression, including OCT3/4, NANOG, SOX2, TRIM28, TDGF1, DNMT3B and STAT3, were comparable between control and DAZAP2-overexpressing (OE) Edom–iPSCs. Precursor of hsa-miR-302 cluster were also comparable between control and DAZAP2-overexpressing (OE) Edom–iPSCs. (E) The cell proliferation rate was significantly decreased in DAZAP2-overexpressing Edom–iPSCs compared to control Edom–iPSCs. ∗∗P < 0.01.
3.3. DAZAP2 overexpression modulates iPSC differentiation
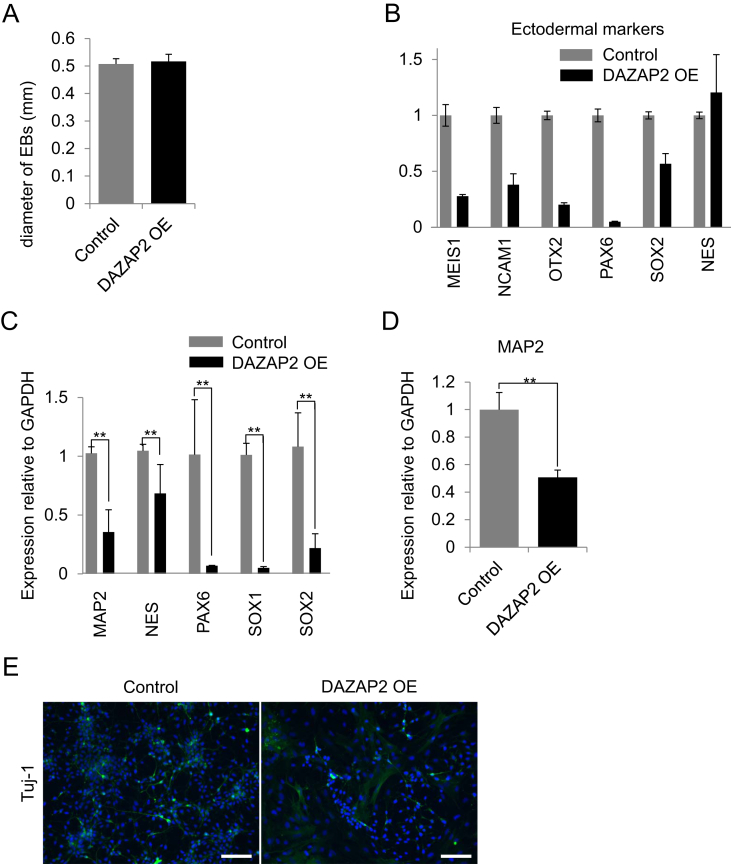
Next, to investigate the impact of DAZAP2 overexpression on differentiation, we compared differentiation properties between control and DAZAP2-overexpressing human iPSCs by lineage-specific marker gene expression. First, the spontaneous differentiation of human iPSCs via EB formation was used to observe differentiation properties by qPCR array analysis (QIAGEN, Hilden, Germany). The sizes of EBs were not significantly different despite a difference in cell proliferation when in the undifferentiated state (Fig. 3A). Ectodermal maker genes were down-regulated in DAZAP2-overexpressing iPSCs revealed by analyzing MEIS1, NCAM1, OTX2, PAX6, SOX2, and NESTIN expression (Fig. 3B). In addition, some mesodermal maker genes were up-regulated in DAZAP2-overexpressing iPSCs as revealed by DES and TAGLN expression (Supplemental Fig. 2A). In endodermal marker genes, only CD44 were up-regulated in DAZAP2-overexpressing iPSCs, however only slight changes were observed in ENG, THY1, SOX7 and TEK expression (Supplemental Fig. 2B). To observe ectodermal differentiation properties in more detail, we applied more lineage-specific differentiation methods such as differentiation into neural stem cells (NSCs), and NSCs to neurons. As expected, the expression of the NSC marker, PAX6, was significantly higher in the control compared to DAZAP2-overexpressing cells (Fig. 3C). We further differentiated NSCs into neurons and found that neural marker gene expression was significantly higher in the control, with reduced numbers of neurons observed in overexpressing-NSCs (Fig. 3D and E). Taken together, hsa-miR-302 members control the differentiation of human PSCs, especially to ectodermal lineage differentiation, by inhibiting of DAZAP2 protein expression.
Fig. 3.
DAZAP2-modified germ layer differentiation in Edom–iPSCs. (A) Diameters of embryoid bodies (EBs) of control and DAZAP2 overexpressing (OE) Edom–iPSCs did not differ. (B) Endodermal marker genes were decreased in DAZAP2-overexpressing Edom–iPSCs. (C) Neural stem cell (NSC) marker genes were decreased in NSCs derived from DAZAP2 overexpressing Edom-induced pluripotent stem cells (iPSCs). ∗∗P < 0.01 (D) MAP2 expression was significantly decreased in neurons derived from DAZAP2 overexpressing Edom–iPSCs. ∗∗P < 0.01 (E) Immunofluorescent staining of anti-beta tubulin 3 (Tuj-1) in neurons derived from control and DAZAP2-overexpressing Edom–iPSCs. DAPI staining is shown in blue. Scale bar: 100 μm.
4. Discussion
In this study, we employed a gene overexpression approach to dissect the role of such miRs in self-renewal and also in the differentiation of human PSCs. DAZAP2 overexpression affected both self-renewal and differentiation properties in human PSCs. Hereafter, we discuss the importance of hsa-miR-302 cluster for human PSCs.
Recent developments in genome engineering technologies enable us to delete specific regions of genomic DNA for the analysis of gene functions. Recent papers reported that Dicer (responsible for small RNA biogenesis, including that of miRs) knockout (KO) mouse ESCs were viable and morphologically normal, although they showed defects in cell proliferation and differentiation [2,33]. Dgcr8 (involved in miR-specific processing) KO mouse ESCs were also viable and morphologically normal, although they showed defects in cell proliferation and differentiation that were slightly different from those of Dicer mutant mouse ESCs [5]. In contrast to mouse ESCs, DICER1 deletion led to increased death receptor–mediated apoptosis and a failure of self-renewal in human ESCs [34]. The knockout of mmu-miR-290-295, the most abundant miR in mouse ESCs and early embryos, was partially lethal in embryos and compromised the fertility of females [35]. The knockout of mmu-miR-302 member, another miR cluster that shares a seed sequence with mmu-miR-290-295 and is enriched in mouse ESCs, was embryonically lethal due to an anterior neural tube closure defect [36,37]. In humans, the knockout of the miR-302 cluster in primary fibroblasts completely blocked iPSC generation [38]. Zhang and colleagues reported that the miR-302 cluster dually regulated the cell cycle and apoptosis in human ESCs by employing TALE-based transcriptional repressors of such miRs [39]. Since each miR inhibits a number of genes, it would be challenging to dissect miR functions by directly knocking out miRs. Further studies will be needed to investigate which miR target gene plays a role either in apoptosis or cell-cycle regulation.
Comparative genomics may be a valid approach for identifying miR target genes and the prediction of miR functions. Hsa-miR-302 members are evolutionarily related to other vertebrate miRNA genes, such as mir-427 in Xenopus [40] and mir-430 in zebrafish [41]. Xenopus mir-427 is required for correct endodermal and mesodermal specification in Xenopus embryos [40]. Zebrafish mir-430 is important for brain morphogenesis [41] and accelerates the clearance of several hundred maternal and maternal–zygotic mRNAs [42]. These findings suggest that the miR-430/427/302 family plays a crucial role in vertebrate embryogenesis by controlling germ layer specification. Also, the KO of either mmu-miR-290-295 or mmu-miR-302 cluster caused neurodevelopmental abnormalities. Taken together, previous works were consistent with our findings that hsa-miR-302 members are essential for human PSC maintenance and DAZAP2 overexpression had an impact in neural differentiation.
In summary, our study revealed that the hsa-miR-302 cluster controls the self-renewal and differentiation of human PSCs by repressing DAZAP2, suggesting that miR target gene overexpression may be useful in further investigating miR functions. However, further studies are needed to understand the molecular mechanisms that underlie neural differentiation with DAZAP2 overexpression.
Acknowledgement
We thank Masakazu Machida for providing technical assistance and Yoriko Takahashi for bioinformatics analysis. We also would like to thank Justin Ichida for reading this manuscript critically and for helpful discussion, as well as members of the laboratory for their suggestions. This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan to HA, AU; a grant from the Ministry of Health, Labour and Welfare (MHLW) to HA, AU; Grant-in-aids for Scientific Research (21390456), (26293364) and (25670710) to HA; grants from the National Center for Child Health and Development (26-1 and 24-6) to HA; a Grant-in-aid for Scientific Research (22770233) to TS; and a grant from JST-CREST given to HA. This research was supported by Japan Agency for Medical Research and Development (AMED) under Grant Number 19bm0704023h0002 to TS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Peer review under responsibility of the Japanese Society for Regenerative Medicine.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.reth.2020.03.011.
Authors Contribution
T.S.: Conception and design, Collection and/or assembly of data, Data analysis and interpretation, Manuscript writing, Final approval of manuscript; T.M.: Collection and/or assembly of data, Data analysis and interpretation; T.K.: Collection and/or assembly of data, Data analysis and interpretation; A.U.: Financial support, Administrative support, Provision of study material; H.A.: Conception and design, Financial support, Administrative support, Provision of study material, Data analysis and interpretation, Manuscript writing, Final approval of manuscript.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Kanellopoulou C., Muljo S.A., Kung A.L., Ganesan S., Drapkin R., Jenuwein T. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mallanna S.K., Rizzino A. Emerging roles of microRNAs in the control of embryonic stem cells and the generation of induced pluripotent stem cells. Dev Biol. 2010;344:16–25. doi: 10.1016/j.ydbio.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tiscornia G., Izpisúa Belmonte J.C. MicroRNAs in embryonic stem cell function and fate. Genes Dev. 2010;24:2732–2741. doi: 10.1101/gad.1982910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y., Medvid R., Melton C., Jaenisch R., Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bar M., Wyman S.K., Fritz B.R., Qi J., Garg K.S., Parkin R.K. MicroRNA discovery and profiling in human embryonic stem cells by deep sequencing of small RNA libraries. Stem Cell. 2008;26:2496–2505. doi: 10.1634/stemcells.2008-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lakshmipathy U., Love B., Goff L.A., Jörnsten R., Graichen R., Hart R.P. MicroRNA expression pattern of undifferentiated and differentiated human embryonic stem cells. Stem Cell Dev. 2007;16:1003–1016. doi: 10.1089/scd.2007.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suh M.R., Lee Y., Kim J.Y., Kim S.K., Moon S.H., Lee J.Y. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004;270:488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 9.Wilson K.D., Venkatasubrahmanyam S., Jia F., Sun N., Butte A.J., Wu J.C. MicroRNA profiling of human-induced pluripotent stem cells. Stem Cell Dev. 2009;18:749–758. doi: 10.1089/scd.2008.0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barroso-delJesus A., Romero-López C., Lucena-Aguilar G., Melen G.J., Sanchez L., Ligero G. Embryonic stem cell-specific miR-302-367 cluster: human gene structure and functional characterization of its core promoter. Mol Cell Biol. 2008;28:6609–6619. doi: 10.1128/MCB.00398-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marson A., Levine S.S., Cole M.F., Frampton G.M., Brambrink T., Johnstone S. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Subramanyam D., Lamouille S., Judson R.L., Liu J.Y., Bucay N., Derynck R. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat Biotechnol. 2011;29:443–448. doi: 10.1038/nbt.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parr C.J., Katayama S., Miki K., Kuang Y., Yoshida Y., Morizane A. MicroRNA-302 switch to identify and eliminate undifferentiated human pluripotent stem cells. Sci Rep. 2016;9:32532. doi: 10.1038/srep32532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He L., Thomson J.M., Hemann M.T., Hernando-Monge E., Mu D., Goodson S. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bracken C.P., Gregory P.A., Kolesnikoff N., Bert A.G., Wang J., Shannon M.F. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68:7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- 16.Kaur S., Abu-Shahba A.G., Paananen R.O., Hongisto H., Hiidenmaa H., Skottman H. Small non-coding RNA landscape of extracellular vesicles from human stem cells. Sci Rep. 2018;8:15503. doi: 10.1038/s41598-018-33899-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu S., Wilson K.D., Ghosh Z., Han L., Wang Y., Lan F. MicroRNA-302 increases reprogramming efficiency via repression of NR2F2. Stem Cell. 2013;31:259–268. doi: 10.1002/stem.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosa A., Brivanlou A.H. A regulatory circuitry comprised of miR-302 and the transcription factors OCT4 and NR2F2 regulates human embryonic stem cell differentiation. EMBO J. 2011;30:237–248. doi: 10.1038/emboj.2010.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee M.R., Prasain N., Chae H.D., Kim Y.J., Mantel C., Yoder M.C. Epigenetic regulation of NANOG by miR-302 cluster-MBD2 completes induced pluripotent stem cell reprogramming. Stem Cell. 2013;31:666–681. doi: 10.1002/stem.1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lipchina I., Elkabetz Y., Hafner M., Sheridan R., Mihailovic A., Tuschl T. Genome-wide identification of microRNA targets in human ES cells reveals a role for miR-302 in modulating BMP response. Genes Dev. 2011;25:2173–2186. doi: 10.1101/gad.17221311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wade S.L., Langer L.F., Ward J.M., Archer T.K. MiRNA-mediated regulation of the SWI/SNF chromatin remodeling complex controls pluripotency and endodermal differentiation in human ESCs. Stem Cell. 2015;33:2925–2935. doi: 10.1002/stem.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sengupta S., Nie J., Wagner R.J., Yang C., Stewart R., Thomson J.A. MicroRNA 92b controls the G1/S checkpoint gene p57 in human embryonic stem cells. Stem Cell. 2009;27:1524–1528. doi: 10.1002/stem.84. [DOI] [PubMed] [Google Scholar]
- 23.Nishino K., Toyoda M., Yamazaki-Inoue M., Fukawatase Y., Chikazawa E., Sakaguchi H. DNA methylation dynamics in human induced pluripotent stem cells over time. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishino K., Toyoda M., Yamazaki-Inoue M., Makino H., Fukawatase Y., Chikazawa E. Defining hypo-methylated regions of stem cell-specific promoters in human iPS cells derived from extra-embryonic amnions and lung fibroblasts. PloS One. 2010;5 doi: 10.1371/journal.pone.0013017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui C.H., Uyama T., Miyado K., Terai M., Kyo S., Kiyono T. Menstrual blood-derived cells confer human dystrophin expression in the murine model of Duchenne muscular dystrophy via cell fusion and myogenic transdifferentiation. Mol Biol Cell. 2007;18:1586–1594. doi: 10.1091/mbc.E06-09-0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Makino H., Toyoda M., Matsumoto K., Saito H., Nishino K., Fukawatase Y. Mesenchymal to embryonic incomplete transition of human cells by chimeric OCT4/3 (POU5F1) with physiological co-activator EWS. Exp Cell Res. 2009;315:2727–2740. doi: 10.1016/j.yexcr.2009.06.016. [DOI] [PubMed] [Google Scholar]
- 27.Nagata S., Toyoda M., Yamaguchi S., Hirano K., Makino H., Nishino K. Efficient reprogramming of human and mouse primary extra-embryonic cells to pluripotent stem cells. Gene Cell. 2009;14:1395–1404. doi: 10.1111/j.1365-2443.2009.01356.x. [DOI] [PubMed] [Google Scholar]
- 28.Toyoda M., Yamazaki-Inoue M., Itakura Y., Kuno A., Ogawa T., Yamada M. Lectin microarray analysis of pluripotent and multipotent stem cells. Gene Cell. 2011;16:1–11. doi: 10.1111/j.1365-2443.2010.01459.x. [DOI] [PubMed] [Google Scholar]
- 29.Akutsu H., Machida M., Kanzaki S., Sugawara T., Ohkura T., Nakamura N. A xenogenic free defined condition for derivation and expansion of human embryonic stem cells with mesenchymal stem cells. Regen Ther. 2014;1:18–29. doi: 10.1016/j.reth.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo H., Ingolia N.T., Weissman J.S., Bartel D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y., Hussein A.M., Somasundaram L., Sankar R., Detraux D., Mathieu J. MicroRNAs regulating human and mouse naïve pluripotency. Int J Mol Sci. 2019;20:E5864. doi: 10.3390/ijms20235864. pii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Melton C., Judson R.L., Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463:621–626. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murchison E.P., Partridge J.F., Tam O.H., Cheloufi S., Hannon G.J. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci USA. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teijeiro V., Yang D., Majumdar S., González F., Rickert R.W., Xu C. DICER1 is essential for self-renewal of human embryonic stem cells. Stem Cell Rep. 2018;11:616–625. doi: 10.1016/j.stemcr.2018.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Medeiros L.A., Dennis L.M., Gill M.E., Houbaviy H., Markoulaki S., Fu D. MiR-290-295 deficiency in mice results in partially penetrant embryonic lethality and germ cell defects. Proc Natl Acad Sci USA. 2011;108:14163–14168. doi: 10.1073/pnas.1111241108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parchem R.J., Moore N., Fish J.L., Parchem J.G., Braga T.T., Shenoy A. MiR-302 is required for timing of neural differentiation, neural tube closure, and embryonic viability. Cell Rep. 2015;12:760–773. doi: 10.1016/j.celrep.2015.06.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang S.L., Yang M., Herrlinger S., Liang C., Lai F., Chen J.F. MiR-302/367 regulate neural progenitor proliferation, differentiation timing, and survival in neurulation. Dev Biol. 2015;408:140–150. doi: 10.1016/j.ydbio.2015.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Z., Xiang D., Heriyanto F., Gao Y., Qian Z., Wu W.S. Dissecting the roles of miR-302/367 cluster in cellular reprogramming using TALE-based repressor and TALEN. Stem Cell Rep. 2013;1:218–225. doi: 10.1016/j.stemcr.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Z., Hong Y., Xiang D., Zhu P., Wu E., Li W. MicroRNA-302/367 cluster governs hESC self-renewal by dually regulating cell cycle and apoptosis pathways. Stem Cell Rep. 2015;4:645–657. doi: 10.1016/j.stemcr.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosa A., Spagnoli F.M., Brivanlou A.H. The miR-430/427/302 family controls mesendodermal fate specification via species-specific target selection. Dev Cell. 2009;16:517–527. doi: 10.1016/j.devcel.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 41.Giraldez A.J., Cinalli R.M., Glasner M.E., Enright A.J., Thomson J.M., Baskerville S. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 42.Giraldez A.J., Mishima Y., Rihel J., Grocock R.J., Van Dongen S., Inoue K. Zebrafish miR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.