Abstract
Accumulating studies have confirmed that mammary serine protease inhibitor (MASPIN) plays an essential role in non-small cell lung cancer (NSCLC). However, results are still controversial or inconsistent. In the present study, we attempted to identify the clinical significance of MASPIN and its potential molecular roles in NSCLC. The correlation of MASPIN with prognosis and clinicopathological characteristics was assessed by meta-analysis. Additionally, the potential molecular mechanisms of MASPIN in NSCLC was also investigated through several online databases. A total of 2220 NSCLC patients from 12 high quality studies were included and the results indicated that up-regulated MASPIN nucleus and cytoplasm expression was associated with poor overall survival (OS) (hazard ratio (HR) = 1.43, 95% confidence interval (CI) = 1.01–2.04, P<0.05), elevated MASPIN cytoplasm expression was associated with poor OS (HR = 1.45, 95% CI = 1.01–2.07, P<0.05), disease-free survival (DFS) (HR = 1.95, 95% CI = 1.31–2.88, P=0.001), and disease-specific survival (DSS) (HR = 2.17, 95% CI = 1.18–3.99, P=0.013). MASPIN both nucleus and cytoplasm location were associated with clinicopathological characteristics. Bioinformatics analysis validated the above results and suggested that human serpin family B member 5 (SERPINB5) hypomethylated levels were negatively correlated with its mRNA expression. Bioinformatics analysis also revealed the 85 most frequently altered neighboring genes of SERPINB5, and gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis revealed 20 GO terms and 3 KEGG pathways with statistical significance. MASPIN had a statistically negative correlation with NSCLC prognosis, functioning as an oncoprotein by hypomethylation and influencing specific pathways involving the 85 genes identified herein. MASPIN might be a promising prognostic signature in NSCLC.
Keywords: MASPIN, non-small cell lung cancer, prognosis, SERPINB5
Introduction
The mammary serine protease inhibitor (MASPIN), encoded by the human serpin family B member 5 (SERPINB5) gene, was first reported in 1994 as a tumor suppressor [1]. MASPIN is highly expressed in normal mammary epithelial cells but is often down-regulated or not expressed in the progression of breast malignant tumors [1,2]. Advanced research found that MASPIN inhibits cell invasion, promotes apoptosis, and inhibits angiogenesis, so it is considered to have class II tumor-suppressive properties [3]. MASPIN localizes in not only primarily cytoplasm (in the mammary epithelial cells), but also nucleus (in the myoepithelial cells), secretory vesicles, and cell surface [4]. MASPIN subcellular localization appears linked to its biological functions [5], and its nuclear localization indicates a more favorable prognosis in human malignancies, such as breast cancer [6], pancreatic cancer [4], and ovarian carcinoma [7]. This phenomenon also appears in non-small cell lung cancer (NSCLC) [8], which is one of the most familiar malignant tumors [9,10] and can be mainly divided into lung squamous cell carcinoma (LUSC) and lung adenocarcinoma (LUAD), and lung large cell carcinoma, pathologically [11]. One study shows emaciated cytoplasmic MASPIN expression could consider as an independent positive predictor in primary NSCLC [12], whereas another study shows that dominant nuclear expression was related to positive prognosis in resectable NSCLC [13]. This paradoxical localization and expression of MASPIN might have cell-specific characteristics [14] and play different roles in NSCLC. Moreover, MASPIN may have an ability to limit malignancy cell phenotypic plasticity and affect the cancer cell response to drugs [15], so it might be a valuable marker and potential therapeutic agent [5]. Thus, we conducted present study to clarify the roles of MASPIN expression and subcellular localization in NSCLC.
Materials and methods
Studies search and selection
We performed a systematic search on Medline, Embase, and Web of Science (WOS) for relevant published studies from inception to 31 January 2019, with language restricted to English. The following keywords were used in different combinations: ‘Carcinoma’, ‘Non-Small-Cell Lung’, ‘MASPIN’, ‘SERPIN-B5’, ‘Prognosis’. Search strategies in Medline were available in Supplementary Table S1.
Inclusion criteria for studies: (1) participants were pathologically diagnosed with NSCLC; (2) the expression and subcellular localization were detected by immunohistochemistry; (3) patients were divided into high and low expression groups according to MASPIN expression and the relationship between MASPIN and NSCLC prognosis (such as overall survival (OS), disease-free survival (DFS), progression-free survival (PFS), and disease-specific survival (DSS)) and clinicopathological characteristics were described; (iv) relevant data were sufficient to obtain hazard ratio (HR) and corresponding 95% confidence interval (CI); (v) cohort studies. Exclusion criteria included reviews, case reports, comments, expert opinions, meta-analysis, and conference summaries. When studies or data duplication were found, only the latest or most complete studies were included.
Data extraction and management
Citations were screened at the title and abstract level by two independent reviewers, and the full text was retrieved for those potentially eligible. For studies that did not provide HRs and 95% CIs directly, we estimated them through indirect methods [16]. If only Kaplan–Meier curves were available, HRs were extracted from graphics by Engauge Digitizer [17]. One reviewer extracted data of interest into a predesigned excel spreadsheet. Then, another two reviewers checked received data and contradictory cases were resolved by the final author.
Quality assessment
Two reviewers critically and independently assessed included studies using the Newcastle–Ottawa Scale (NOS) [18,19]. The assessments include selection, comparability, and outcome. The studies were divided into three grades: low, medium, and high, with NOS scores ranging from 0 to 3, 4 to 6, and 7 to 9, respectively. All data were finally shown in the predesigned excel spreadsheet and discrepancies, contradictories were resolved by the third author.
Data synthesis and analysis
The STATA, statistical software for data science, was adopted to synthesize and analyze data. Similar to the method described previously [20], heterogeneity was assessed by I2 tests. If the data were substantially homogeneous, a synthesis was directly conducted with the fixed-effect model. Otherwise, a synthesis was directly conducted with the random-effect model, successively subgroup analysis was performed. Sensitivity analysis was additionally conducted to investigate the robustness of our results. At last, the funnel plot and Egger’s tests were applied to assess for publication bias. Later, the different subcellular location was also analyzed.
Bioinformatics analysis
The SERPINB5 mRNA and protein expression between NSCLC and normal tissue were studied using UALCAN [21] and Human Pathology Atlas [22], respectively. The association between SERPINB5 and OS or PFS and the clinicopathological significances of SERPINB5 in NSCLC were all conducted in the Kaplan–Meier plotter [23]. The patients were grouped by median SERPINB5 expression and HR with 95% CI were calculated. In addition, MethHC [24] was utilized to compare the SERPINB5 promoter region methylation between NSCLC and normal tissue as well as the relationship between mRNA expression and methylation in NSCLC patients. One-way ANOVA was used to analyze differential expression. Moreover, the genes co-expressed with SERPINB in TCGA-LUAD were identified using the cBioPortal [25]. Only those genes with Spearman’s correlation coefficients of more than 0.4 were identified as SERPINB-related genes. Finally, SERPINB-related genes were loaded into DAVID [26,27] for gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis, and the illustration of the biological significance was performed using the clusterProfiler package in R [28].
Results
Characteristics of eligible studies
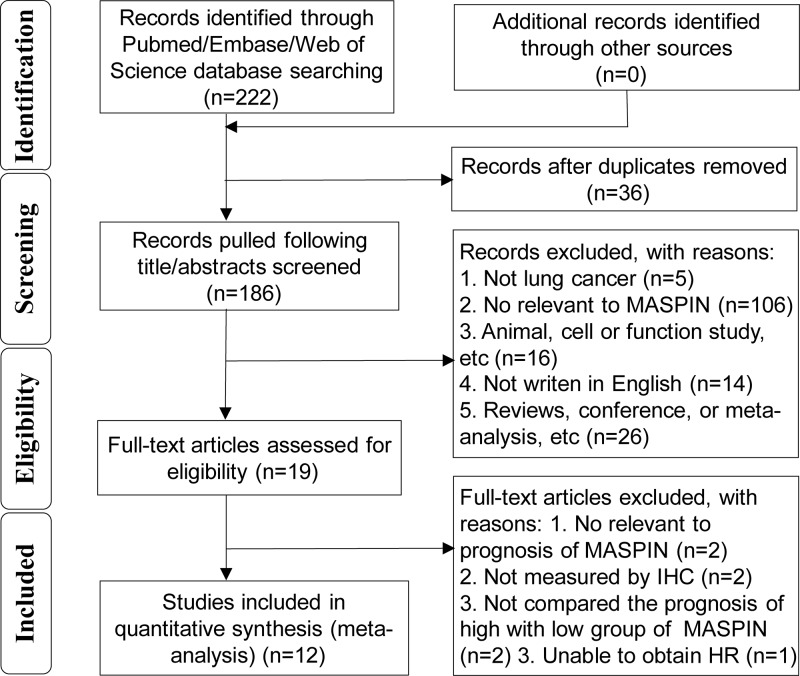
The primary search of all sources yielded 222 articles eligible for identification. A total of 186 title/abstracts were screened for eligibility after removing 36 duplicates. Of these, 19 full-text articles were assessed for eligibility. Finally, a total of 12 studies [12,29–39] were included in the present study (Figure 1).
Figure 1. Flow diagram of the selection process in this meta-analysis.
Twelve studies comprising 2220 patients contained information regarding OS, PFS, DFS, and DSS. All included studies were published in 2005 and later, and more than half of the studies were conducted in Japan [12,31–35,39], only three studies involved LUSC or LUAD [31,33,34]. Only one study did not report gender composition [37]. Two studies did not report the median age [31,37], and the median age of the included individuals was approximately 65 years old or so. Only two studies had no staging information for NSCLC [36,37]. As for subcellular localization, three studies [12,30,34] only involved cytoplasm, while others included both cytoplasm and nucleus. Some HRs were extracted directly from corresponding studies [29–35], and some were extracted from corresponding curve [12,32,35–39], and some were not reported [29,32,38]. The included studies also had different definitions of the cut-off value. NOS scores of all eligible studies were greater than 7. The main characteristics of the included studies are shown in Table 1.
Table 1. Main characteristics of eligible studies.
| Author, year | Country | Type | No. (M/F) | Median age | Stage | Location | Cut-off | Positive (%) | Median follow-up | Outcome | HR estimation | NOS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hirai, 2005 | Japan | A94/S38 | 132 (90/42) | 65.5 | I-II78/III54 | C | ≥40% | 73 (55.3%) | NR (1–42 m) | Negative (NSCLC+C) | OS curve | 8 |
| Nakagawa, 2006 | Japan | A116/S76/O18 | 210 (151/59) | 62.6 | I-II142/III68 | T | intensity+-++ | 73 (34.8%) | NR (1–60 m) | NO (NSCLC+T)/Positive (S+T) | OS NR/OS R | 8 |
| Woenckhaus, 2007 | Germany | A74/S209/O69 | 487 (NR) | NR | NR | T (C+N) | ≥10% | 352 (72.3%) | 27 m (0–200 m) | NO (NSCLC+C)/NO (NSCLC+N) | OS curve/OS curve | 7 |
| Zheng, 2008 | China+Japan | A86/S37/O32 | 155 (105/50) | 69.5 | I-II115/III-IV37 | T (C+N) | >5% | 91 (58.7%)/21 (13.5%) | 20.4 m (1–144 m) | NO (NSCLC+C)/Positive (A+C) | OS curve/OS curve | 7 |
| Takanami, 2008 | Japan | A105/S70/O6 | 181 (130/51) | 65.5 | I-II126/III55 | T | ≥5% | 74 (40.9%) | 75.2 m (60–108 m) | NO (NSCLC+T)/Positive (S+T) | PFS, OS curve/PFS, OS R | 8 |
| Bircan, 2010 | Turkey | A18/S28 | 46 (42/4) | 61.4 | II3/III-IV37 | C | >5% | 39 (84.8%) | 261.8 d (10–800 d) | NO (NSCLC+C) | OS R | 7 |
| Berardi, 2012 | Italy | A192/S172/O75 | 439 (374/65) | 68 | I-II333/III106 | T (C+N) | >5% | 284 (67%)/79 (18.6%) | 42.5 m (10.6–87.2 m) | Negative (NSCLC+C)/Positive (NSCLC+N) | OS NR/OS R | 7 |
| Wang, 2014 | China | A71/S23/O4 | 98 (60/38) | 63 | NR | T | intensity++ | 26 (26.5%) | 42.8 m (6.87–69 m) | NO (NSCLC+T) | DFS curve | 8 |
| Takagi, 2015 | Japan | A110 | 110 (55/55) | 67.9 | I-II101/III9 | C | >10% | 27 (24.5%) | 71 m (6–88 m) | Negative (A+C) | DFS, OS R | 7 |
| Yaman, 2015 | Turkey | A45/S23/O12 | 80 (71/9) | 59 | I-II50/III-IV30 | T | >5% | 26 (32.1%) | 41.4 m (1–91 m) | NO (NSCLC+T)/Negative (A+T) | OS NR/OS Curve | 8 |
| Matsuoka, 2016 | Japan | S101 | 101 (92/9) | NR | I-II90/III11 | T (C+N) | >10% | 25 (24.6%) | 50 m (2–119 m) | Negative (S+C) | DFS, DSS R | 9 |
| Ohno, 2018 | Japan | A181 | 181 (87/94) | 69.7 | I181 | T (C+N) | >10% | 45 (24.9%) | 74 m (12–151 m) | Negative (A+C) | DFS, DSS R | 9 |
Abbreviations: A, adenocarcinoma; C, cytoplasm; F, female; m, month; M, male; No., number; N, nucleus; NR, not reported; O, other type non-small lung cancer; R, reported in text; S, squamous cell carcinoma; T, total, T = C+N.
Association between MASPIN and NSCLC prognosis
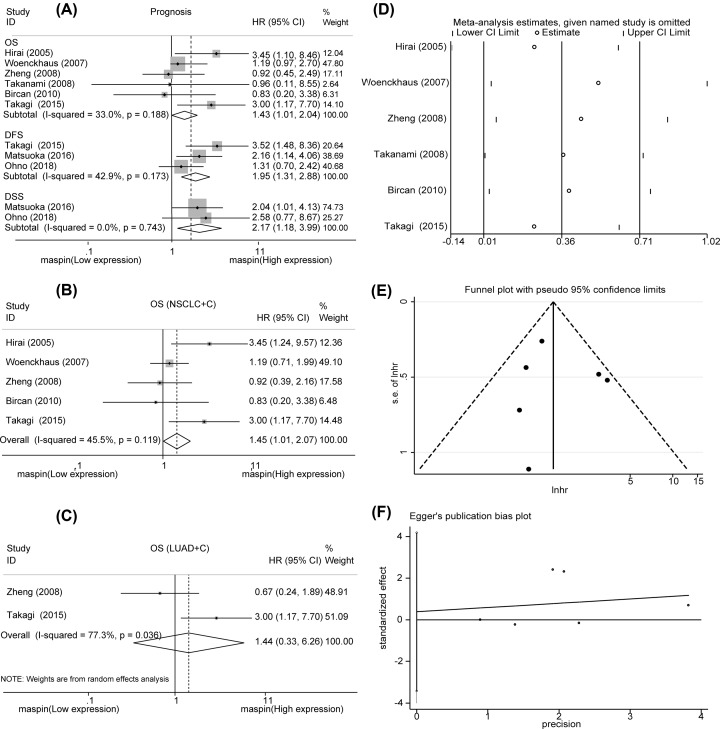
The differences in MASPIN expression and subcellular localization were analyzed in six studies. As a result, we found that up-regulated MASPIN nucleus and cytoplasm expression was associated with poor OS in NSCLC (HR = 1.43, 95% CI = 1.01–2.04, P<0.05) without obvious heterogeneity (I2 = 33.00%, Ph<0.05, Figure 2A). The results also showed that elevated MASPIN cytoplasm expression was associated with poor DFS in NSCLC (HR = 1.95, 95% CI = 1.31–2.88, P=0.001) without obvious heterogeneity (I2 = 42.90%, Ph=0.001, Figure 2A). Additionally, similar results were observed with DSS in NSCLC (HR = 2.17, 95% CI = 1.18–3.99, P=0.013) without obvious heterogeneity (I2 = 0.00%, Ph=0.013, Figure 2A). In order to uncover the effect of MASPIN cytoplasm localization on OS, we conducted a subgroup analysis, and the results showed that MASPIN overexpression was associated with poor OS in NSCLC (HR = 1.45, 95% CI = 1.01–2.07, P<0.05, Figure 2B) and LUAD (HR = 1.44, 95% CI = 0.33–6.26, P=0.626, Figure 2C). In summary, our results showed a statistically negative interaction between MASPIN and NSCLC prognosis, and MASPIN might be a promising prognostic signature in NSCLC.
Figure 2. Forest plot of the HR for the correlation between survival rate and MASPIN expression.
(A) The association of MASPIN expression with OS, PFS, DFS, and DSS. (B,C) The association of MASPIN expression with OS by subgroup analysis. (D) Sensitivity analysis of OS. (E) Publication bias analysis of OS by funnel plot. (F) Publication bias analysis of OS by Egger’s test. Abbreviations: C, cytoplasm; CI, confidence interval; N, nucleus; T, total, T = C+N.
Association between MASPIN and clinicopathological characteristics of NSCLC
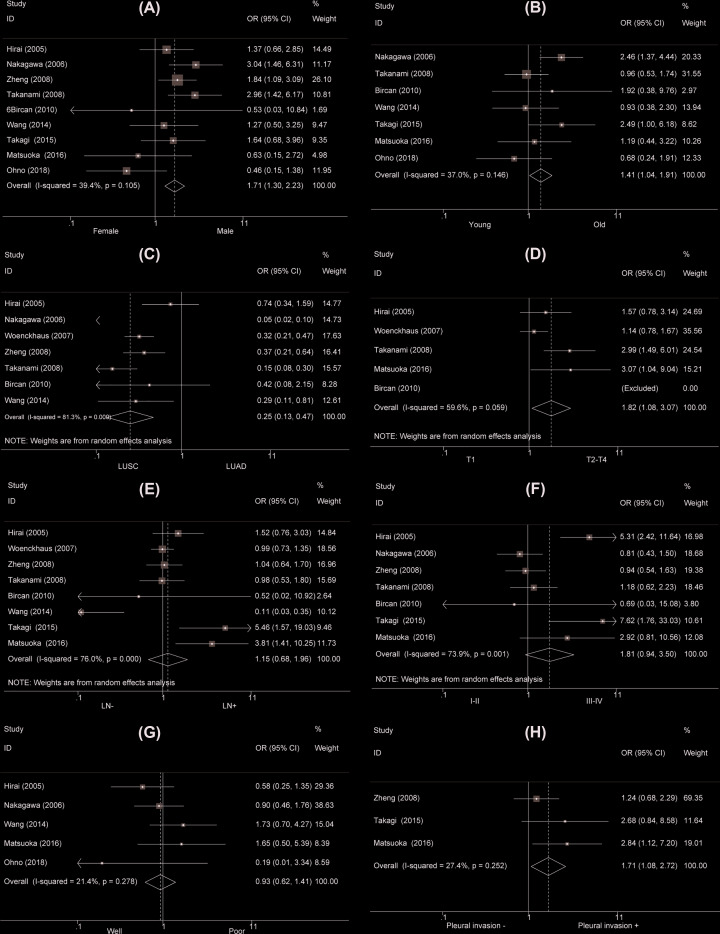
Not all the included studies discussed the interaction between MASPIN and clinicopathological characteristics of NSCLC. Nine studies showed a higher MASPIN nucleus and cytoplasm expression was detected in male NSCLC patients than female NSCLC patients (OR = 1.71, 95% CI = 1.31–2.23, P<0.001, Supplementary Table S2, Figure 3A). The old NSCLC patients had an elevated MASPIN nucleus and cytoplasm expression than young patients (OR = 1.41, 95% CI = 1.04–1.91, P=0.027, Supplementary Table S2, Figure 3B). LUSC patients had a lower MASPIN nucleus and cytoplasm expression than LUAD patients (OR = 0.25, 95% CI = 0.13–0.47, P<0.001, Supplementary Table S2, Figure 3C). The patients with stage T1 exposed MASPIN nucleus and cytoplasm underexpression compared with those with stages T2–T4 (OR = 1.82, 95% CI = 1.08–3.07, P=0.024, Supplementary Table S2, Figure 3D). MASPIN nucleus and cytoplasm expression was not related to clinical stage, differentiation, and lymph node metastasis of NSCLC (OR = 1.81, 95% CI = 0.94–3.50, P=0.078; OR = 0.93, 95% CI = 0.62–1.41, P=0.74 and OR = 1.15, 95% CI = 0.68–1.96, P=0.604, respectively, Supplementary Table S2, Figure 3E–G). Moreover, NSCLC patients with pleural invasion displayed MASPIN nucleus and cytoplasm overexpression than those without with pleural invasion (OR = 1.71, 95% CI = 1.08–2.72, P=0.023, Supplementary Table S2, Figure 3H). As for MASPIN cytoplasm expression, the subgroup analysis results were generally consistent with the MASPIN nucleus and cytoplasm expression (Supplementary Table S2, Figure 4).
Figure 3. Forest plot for the relationship between total (cytoplasmic + nuclear) MASPIN expression and clinicopathological parameters of NSCLC.
(A) Gender (male versus female); (B) age (old versus young); (C) type (LUAD versus LUSC); (D) T stage (T2–4 versus T1); (E) TNM stage (III-IV vs I-II); (F) lymph node metastasis (LN+ vs LN−); (G) differentiation (poor versus well); (H) pleural invasion (yes versus no).
Figure 4. Forest plot for the relationship between cytoplasmic MASPIN expression and clinicopathological parameters of NSCLC.
(A) Gender (male versus female); (B) age (old versus young); (C) type (LUAD versus LUSC); (D) T stage (T2–4 versus T1); (E) TNM staging (III-IV vs I-II); (F) lymph node metastasis (LN+ vs LN−); (G) differentiation (poor versus well); (H) pleural invasion (yes versus no).
Sensitivity analysis and publication bias
A sensitivity analysis of OS was performed to verify the robustness of consequences. The results disclosed there had no significant heterogeneity (Figure 2D). The sensitivity analysis and assessment of publication bias with PFS, DFS, and DSS were not conducted because there were few numbers of included studies. The funnel plot indicated no publication bias analysis of OS (Figure 2E). Moreover, Egger’s test revealed no obvious publication bias (P=0.795, Figure 2F).
The prognostic significances of SERPINB5 in NSCLC
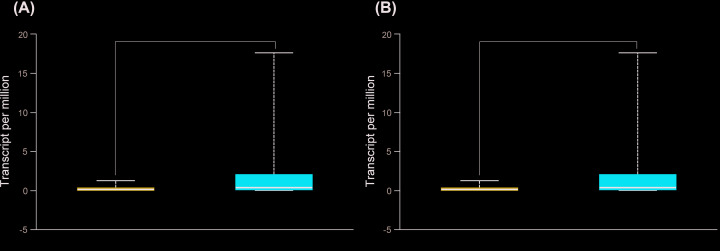
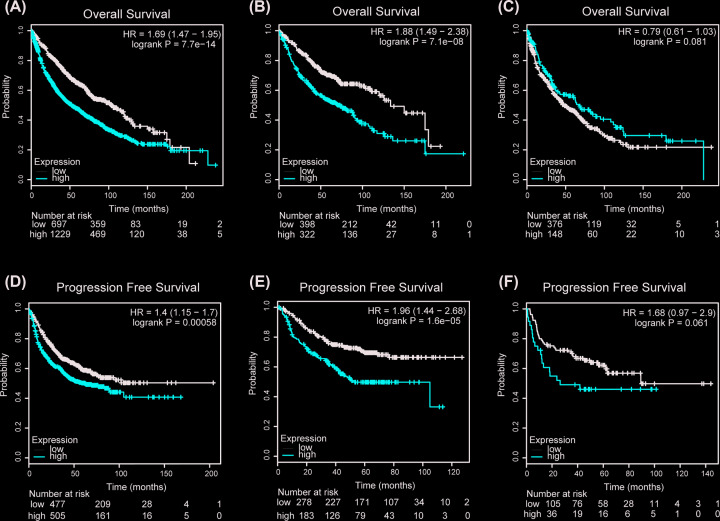
The bioinformatics analysis results, perform by UALCAN, showed that SERPINB5 mRNA expression was higher in both LUAD and LUSC patients than the normal (P<0.001, Figure 5). The Human Pathology Atlas illustrated that SERPINB5 protein expression was significantly higher in both LUAD and LUSC than normal lung (Figure 6). Next, OS was analyzed in NSCLC, LUAD, and LUSC related to SERPINB5 mRNA expression, and we found that NSCLC and LUAD patients with high expression of SERPINB5 mRNA had poorer prognosis (HR = 1.69, 95% CI = 1.47–1.95, P<0.001; HR = 1.88, 95% CI = 1.49–2.38, P<0.001; respectively, Figure 7A,B), and there was no statistically difference in LUSC patients (HR = 0.79, 95% CI = 0.61–1.03, P=0.081, Figure 7C). Additionally, the same trend was observed during the PFS analysis (Figure 7D–F).
Figure 5. Transcriptional expression of SERPINB5 in LUAD and LUSC (UALCAN).
(A) LUAD; (B) LUSC. ***P<0.001.
Figure 6. Representative proteins expressions of IHC images of distinct MASPIN were detected in LUAD, LUSC, and normal tissues (Human Protein Atlas).
(A) MASPIN proteins were found not or low expressed in normal lung tissues. (B,C) Significantly high staining expressions were observed in LUAD and LUSC.
Figure 7. SERPINB5 expression is correlated with the survival rate of non-small lung cancer patients using the Kaplan–Meier plotter website.
(A–C) The OS rate was related to SERPINB5 mRNA expression in NSCLC, LUAD, and LUSC. (D–F) PFS was related to SERPINB5 mRNA expression in NSCLC, LUAD, and LUSC.
The clinicopathological significances of SERPINB5 in NSCLC
According to Kaplan–Meier plotter, higher SERPINB5 mRNA expression was negatively correlated with OS of LUSC, T3 and T4 and stage III (P<0.05, Table 2), and positively correlated with gender (male and female), smoking and non-smoking, LUAD, T1 and T2, N and stages I and II, (non-) chemotherapy, (non-) radiotherapy and surgical margins negative (P<0.05, Table 2). There appeared a positive correlation between SERPINB5 mRNA overexpression and PFS in gender (male and female), smoking and non-smoking, histology (LUAD and LUSC), T1, N0 and N2, stage I, (non-) chemotherapy, (non-) radiotherapy and surgical margins negative (P<0.05, Table 2), whereas negative relationship in T2 and T3, N1 and stage II (P<0.05, Table 2).
Table 2. The prognostic significance of SERPINB5 mRNA in non-small lung cancer by Kaplan–Meier plotter.
| Clinicopathological features | OS | PFS | ||
|---|---|---|---|---|
| HR | P | HR | P | |
| Sex | ||||
| Male | 1.69 (1.41–2.03) | 9.40E-09 | 1.59 (1.21–2.09) | 8.20E-04 |
| Female | 1.55 (1.23–1.96) | 1.90E-04 | 1.32 (0.94–1.87) | 1.10E-01 |
| Smoking | ||||
| Yes | 1.7 (1.38–2.09) | 3.90E-07 | 1.49 (1.12–1.97) | 5.60E-03 |
| No | 2.23 (1.27–3.89) | 3.90E-03 | 1.83 (1.13–2.96) | 1.30E-02 |
| Histology | ||||
| Adenocarcinoma | 1.88 (1.49–2.38) | 7.10E-08 | 1.96 (1.44–2.68) | 1.60E-05 |
| Squamous | 0.79 (0.617–1.03) | 8.10E-02 | 1.68 (0.97–2.9) | 6.10E-02 |
| T | ||||
| 1 | 1.71 (1.28–2.3) | 2.80E-04 | 1.49 (0.81–2.75) | 2.00E-01 |
| 2 | 1.53 (1.17–1.99) | 1.70E-03 | 0.82 (0.6–1.11) | 1.90E-01 |
| 3 | 0.66 (0.37–1.19) | 1.67E-01 | 0.62 (0.22–1.76) | 3.60E-01 |
| 4 | 0.64 (0.34–1.23) | 1.80E-01 | ||
| N | ||||
| 0 | 1.53 (1.24–1.89) | 7.50E-05 | 1.31 (0.91–1.87) | 1.40E-01 |
| 1 | 1.37 (1–1.87) | 5.00E-02 | 0.74 (0.47–1.17) | 1.90E-01 |
| 2 | 1.48 (0.91–2.41) | 1.10E-01 | 2.73 (1.22–6.09) | 1.10E-02 |
| TNM stage | ||||
| I | 2.08 (1.59–2.72) | 4.80E-08 | 2.05 (1.31–3.2) | 1.30E-03 |
| II | 1.23 (0.86–1.78) | 2.60E-01 | 0.4 (0.22–0.73) | 1.70E-03 |
| III | 0.76 (0.43–1.34) | 3.40E-01 | ||
| Chemotherapy | ||||
| Yes | 2.41 (1.58–3.68) | 2.60E-05 | 1.66 (1.08–2.57) | 2.00E-02 |
| No | 2.26 (1.62–3.17) | 1.00E-06 | 1.57 (1.03–2.37) | 3.30E-02 |
| Radiotherapy | ||||
| Yes | 3.94 (2.2–7.05) | 9.30E-07 | 2.16 (1.24–3.78) | 5.70E-03 |
| No | 1.76 (1.22–2.53) | 2.00E-03 | 1.37 (0.89–2.12) | 1.50E-01 |
| Surgery success | ||||
| Surgical margins negative | 2.25 (1.79–2.84) | 1.40E-12 | 1.85 (1.39–2.46) | 2.20E-05 |
DNA methylation of SERPINB5 and its correlation with mRNA expression in NSCLC patients
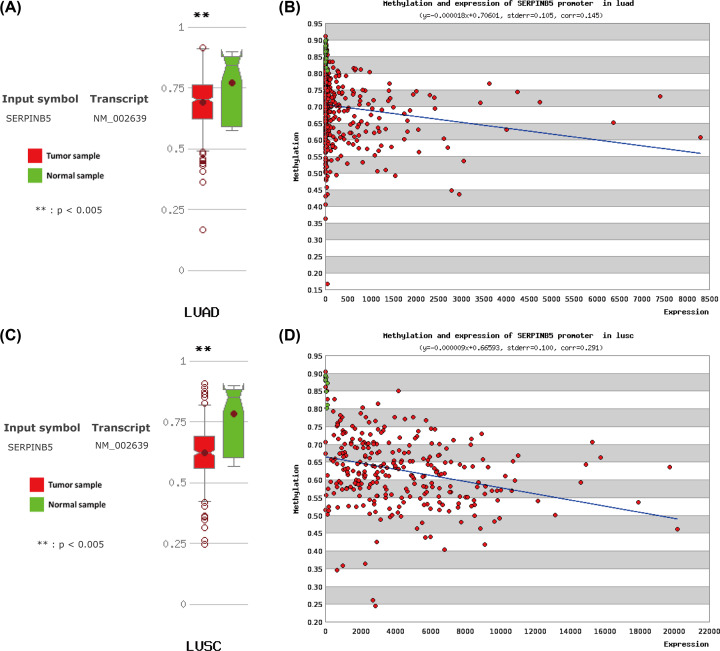
To recognize the role of methylation in regulating SERPINB5 expression in NSCLC patients, MethHC was applied to explore the level of DNA methylation in the SERPINB5 gene promoter region and its correlation with mRNA expression. The difference of the SERPINB5 methylation level between LUAD and normal samples was statistically significant (P<0.005, Figure 8A). Additionally, SERPINB5 methylation was negatively correlated with its mRNA expression in LUAD (r = −0.128, P<0.001, Figure 8B). The statistically significant results were also indicated in LUSC patients (Figure 8C,D).
Figure 8. DNA methylation of the SERPINB5 gene promoter region in non-small lung cancer (MethHC).
(A) Comparison of DNA methylation levels of the SERPINB5 gene in LUAD with normal tissues. (B) The correlation between DNA methylation and mRNA expression in the SERPINB5 gene of LUAD. (C) Comparison of DNA methylation levels of the SERPINB5 gene in LUSC with normal tissues. (D) The correlation between DNA methylation and mRNA expression in the SERPINB5 gene of LUSC. Green indicates normal lung tissues, red represents non-small lung cancer tissues.
Function and pathway enrichment analysis of SERPINB5-related genes in LUAD
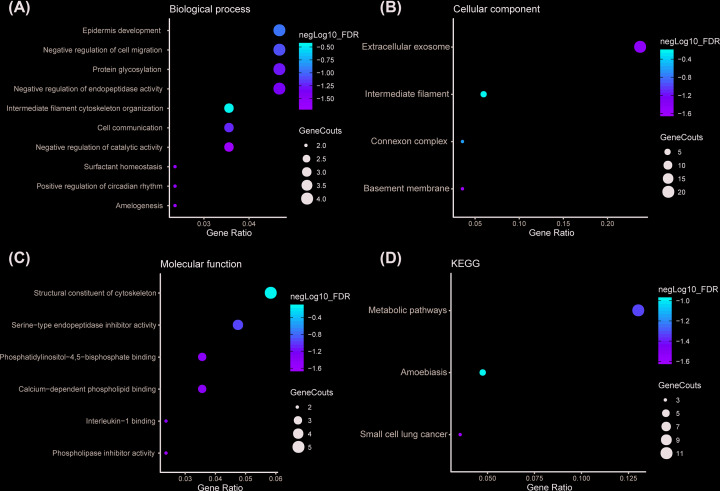
To comprehend the functions and potential mechanisms of SERPINB5 in NSCLC, cBioPortal [25] was used to identify SERPINB5-related genes in LUAD. There were 85 genes with Spearman’s correlation coefficients more than 0.4 and were identified as SERPINB-related genes in TCGA-LUAD. Besides, 85 genes were put into the DAVID [26,27] for GO and KEGG pathway enrichment analysis, revealing 20 GO terms (Supplementary Table S3) and 3 KEGG pathways (Supplementary Table S4) with statistical significance. These corresponding GO terms were considered as the most specific and useful for describing the concrete function of SERPINB5 in LUAD, the interaction between different GO terms were visualized as three GO maps (Figure 9A–C). KEGG pathway enrichment analysis results subjected that the genes were enriched in amoebiasis, metabolic pathways, and small cell lung cancer (Figure 9D).
Figure 9. Dot plot of GO enrichment and KEGG pathways analyses from the predicted SERPINB5-related genes.
(A) Biological process; (B) Cellular component; (C) Molecular function; (D) KEGG.
Discussion
Up to date, more and more studies are emerging to investigate MASPIN expression and its roles in malignant tumors, and carry out in-depth researches on different mechanisms of MASPIN about inhibiting tumor cell plasticity and affecting the context-dependent response to drugs [15]. Nevertheless, only a minority of MASPIN expression and subcellular localization were evaluated in NSCLC and no unanimous agreement was reached. One study announced that MASPIN cytoplasm expression alone could be a useful negative prognostic effect in p-stage IA LUAD [33]. Other studies indicated strong MASPIN nuclear expression was an independent favorable prognostic factor in NSCLC [29,40], and it improved differentiated epithelial phenotypes, decreased tumor angiogenesis, and increased tumor sensitivity to drug-induced apoptosis [41]. In contrast, Bircan et al. indicated MASPIN had no prognostic significance although it was higher in both LUSC and LUAD [42]. There is no doubt that larger studies are urgently needed to reveal the exact role of MASPIN in the occurrence, development, and prognosis of NSCLC. Hence, we summarized 12 studies with 2220 patients, which met preordained inclusion criteria and had high quality according to NOS scores, to obtain more accurate results and further explored the mechanism of MASPIN in NSCLC through different online databases.
Clinical data conflict on the prognostic utility of MASPIN [29,36,43], and we found that MASPIN expression was up-regulated in NSCLC and was closely related to prognosis. Importantly, we not only analyzed the relationship between MASPIN and NSCLC prognosis but also applied different subcellular localization. Our results suggested that MASPIN was statistically correlated with poor prognosis (OS, DFS, and DSS) regarding different subcellular localization (nucleus and cytoplasm or cytoplasm alone), which indicated MASPIN might be a promising prognostic signature in NSCLC, although the nuclear-cytoplasmic expression pattern might have stronger influences on metastasis and proliferation than nuclear expression alone [36]. Our subgroup analysis showed that rising MASPIN protein was associated with poor OS in LUAD patients with no statistical difference (P=0.626) while the results from the Kaplan–Meier plotter uncovered that negative correlation between SERPINB5 mRNA and OS and PFS was statistically significant in LUAD patients (P<0.001). In addition, the results of LUAD and LUSC were not identical. The discrepancies might be largely attributable to background heterogeneity, which suggests that larger sample size of clinically relevant studies may be needed to verify this phenomenon. But although LUSC and LUAD account for a large proportion of NSCLC [44], they still do not fully reflect all NSCLC.
In addition, we analyzed the relationship between MASPIN and clinicopathological characteristics from the perspective of meta-analysis and Kaplan–Meier plotter. The results were complementary, as one focused on subcellular localization and the other on prognosis; one is the protein condition and the other is the RNA condition. However, it is found that the expression of both SERPINB5 protein and mRNA is significantly correlated with various clinicopathological characteristics and survival outcomes. Many studies have revealed the relevant mechanism between MASPIN and the occurrence, development, treatment, and prognosis of lung cancer. In LUAD cells, MASPIN can not only down-regulate the expression of matrix metalloproteinase-2 and integrin β1 to significantly inhibit cell migration and invasion but also affect cell growth mainly by targeting AKT signaling molecules. The different functions of MASPIN may be due to its different molecular characteristics [45]. One study suggested that cytoplasmic MASPIN may play an important role in LUAD by regulating apoptosis and, thus, is a favorable prognostic marker for LUAD patients, whereas the nuclear location may be linked to the promotion of angiogenesis [46]. Others hold MASPIN inhibits cell motility by suppressing Rac1 and PAK1 activity [47], promotes cell adhesion via the PI3K/ERK pathway [47,48], and suppresses survival of lung cancer cells through modulation of AKT pathway [49,50]. It was suggested that the loss-expression of MASPIN may participate in the invasion and metastasis of NSCLC and it has a positive relationship to vasculogenic mimicry in NSCLC [51]. About the treatment, resistance to osimertinib partially arises through MASPIN [52] and a novel statistical methodology for detecting splicing changes in exon array data presented that SERPINB5 showed alternative splicing in NSCLC patients treated with bevacizumab/erlotinib [53].
To uncover additional mechanisms, we performed methylation analysis. Online databases from MethHC showed that the underlying mechanism of MASPIN affecting NSCLC survival outcomes may be related to hypomethylation and this was following a previous study [54], and this difference may be critical for epigenetic regulation of radiosensitivity [55]. And then, in this study, we demonstrated that SERPINB5 in both LUAD and LUSC were lowly methylated in the promoter region and that there was an inverse correlation between DNA methylation and mRNA expression. Hypomethylated status means activation of gene expression, therefore, it is not difficult to infer that SERPINB5 methylation partially contributes to its increased expression and plays an important role in the occurrence and development in NSCLC. Given the statistical difference between SERPINB5 mRNA and prognosis in LUAD and not in LUAC, TCGA-LUAD was selected for the subsequent analysis of co-expressed genes and enrichment to confirmed that SERPINB5 influenced related pathways involving the neighboring genes, as is mentioned in a previous study [53,56], and future investigations and validation are needed.
Sensitivity analysis demonstrated that the correlation between MASPIN and NSCLC prognosis was stable and unchanged after removing any one study. Funnel plot and Egger’s test were used to detect publication bias and events indicated no obvious publication bias.
However, the present study was not without limitations. Foremost, all the data analyzed in our study were obtained from other published studies or different online databases, which might cause background heterogeneity, as mentioned earlier. Additionally, eight studies were conducted in Asia (China or Japan) which may result in an excursion in race. Later multinational and multiracial studies for this specific population are needed. Third, the included studies had different definitions of MASPIN cut-off value. Therefore, before carrying out relevant researches, it may be necessary to identify the cut-off value in a unified way. Fourth, according to Kaplan–Meier plotter, higher SERPINB5 mRNA expression was positively correlated with OS and PFS of surgery success (surgical margins negative) (all P<0.0001). Due to the limitations of the data obtained, we could only divide the surgery success into one group, which was bound to cause the loss of information, and the later data with richer dimensions is warranted. Finally, a series of silico-bioinformatic analysis was performed to uncover the promising molecular mechanisms of MASPIN in NSCLC, and more elaborate studies are required to be designed and conducted to validate these results in the future.
In conclusion, our study demonstrated that MASPIN had a statistically negative correlation with NSCLC prognosis, functioning as an oncoprotein by hypomethylation and influencing specific pathways, which lays a foundation for MASPIN to develop into an independent prognostic factor for NSCLC patients.
Supplementary Material
Acknowledgements
We thank relevant published studies and databases for their support.
Abbreviations
- CI
confidence interval
- DFS
disease-free survival
- DSS
disease-specific survival
- GO
gene ontology
- HR
hazard ratio
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LUAD
lung adenocarcinoma
- LUSC
lung squamous cell carcinoma
- MASPIN
mammary serine protease inhibitor
- NOS
Newcastle–Ottawa Scale
- NSCLC
non-small cell lung cancer
- OS
overall survival
- PFS
progression-free survival
- SERPINB5
human serpin family B member 5
- WOS
web of science
Contributor Information
Ning Gu, Email: 20193122@njucm.edu.cn.
Ye-Han Zhu, Email: zhuyehansz@sina.com.
Competing Interests
X.-F.W. and B.L. were members of Morning Star Academic Cooperation, Helixlife (Shanghai, China) when they did the work described in the article.
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China [grant number 81774229]; the Jiangsu Leading Talents Project of Traditional Chinese Medicine [grant number Jiangsu TCM 2018 No. 4]; the Major Projects of Nanjing Medical Science and Technology Development During the 13th Five-Year Plan [grant number ZDX16013]; the Clinical Key Speciality Project of China; the Clinical Medicine Center of Suzhou [grant number SZZX201502]; the Gusu Youth Medical Talent [grant number 3101030342000318]; and the Science and Education of Public Health Project for Young Medical Talents of Jiangsu Province [grant number QNRC2016747].
Author Contribution
X.-F.W., B.L., Y.-H.Z., and N.G. conceived, designed, or planned the study. X.-F.W., B.L., D.-X.Z., W.L., C.C., and Y.-B.C. acquired data. X.-F.W., B.L., Y.-H.Z., N.G., and J.-A.H. analyzed the data. Y.-H.Z. and N.G. helped interpret the results. X.-F.W. and B.L. drafted the manuscript. All authors revised and reviewed this work, and all authors gave their final approval of the submitted manuscript.
References
- 1.Zou Z., Anisowicz A., Hendrix M.J.C. et al. (1994) Maspin, a serpin with tumor-suppressing activity in human mammary epithelial cells. Science 263, 526–529 10.1126/science.8290962 [DOI] [PubMed] [Google Scholar]
- 2.Berardi R., Morgese F., Onofri A. et al. (2013) Role of maspin in cancer. Clin. Transl. Med. 2, 8 10.1186/2001-1326-2-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ning J., Yonghong M., Suliang Z. et al. (2002) Maspin sensitizes breast carcinoma cells to induced apoptosis. Oncogene 21, 4089–4098 [DOI] [PubMed] [Google Scholar]
- 4.Maass N., Ziebart M., Nagasaki K. et al. (2002) Maspin locates to the nucleus in certain cell types. J. Pathol. 197, 274–275 10.1002/path.1103 [DOI] [Google Scholar]
- 5.Bailey C.M., Khalkhali-Ellis Z., Seftor E.A. et al. (2006) Biological functions of maspin. J. Cell. Physiol. 209, 617–624 10.1002/jcp.20782 [DOI] [PubMed] [Google Scholar]
- 6.Mohsin S.K., Zhang M., Clark G.M. et al. (2003) Maspin expression in invasive breast cancer: association with other prognostic factors. J. Pathol. 199, 432–435 10.1002/path.1319 [DOI] [PubMed] [Google Scholar]
- 7.Sood A.K., Fletcher M.S., Gruman L.M. et al. (2002) The paradoxical expression of maspin in ovarian carcinoma. Clin. Cancer Res. 8, 2924–2932 [PubMed] [Google Scholar]
- 8.Lonardo F., Li X., Siddiq F. et al. (2006) Maspin nuclear localization is linked to favorable morphological features in pulmonary adenocarcinoma. Lung Cancer 51, 31–39 10.1016/j.lungcan.2005.07.011 [DOI] [PubMed] [Google Scholar]
- 9.Bray F., Ferlay J., Soerjomataram I. et al. (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424 [DOI] [PubMed] [Google Scholar]
- 10.Antonia S.J., Borghaei H., Ramalingam S.S. et al. (2019) Four-year survival with nivolumab in patients with previously treated advanced non-small-cell lung cancer: a pooled analysis. Lancet Oncol. 20, 1395–1408 10.1016/S1470-2045(19)30407-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ettinger D.S., Wood D.E., Aggarwal C. et al. (2019) Non-small cell lung cancer, version 4.2019. J. Natl. Compr. Cancer Netw. 12, 1738–1761 10.6004/jnccn.2014.0176 [DOI] [PubMed] [Google Scholar]
- 12.Hirai K., Koizumi K., Haraguchi S. et al. (2005) Prognostic significance of the tumor suppressor gene maspin in non-small cell lung cancer. Ann. Thorac. Surg. 79, 248–253 10.1016/j.athoracsur.2004.06.118 [DOI] [PubMed] [Google Scholar]
- 13.Smith S.L., Ratschiller S.G., Watson D., Gugger M. et al. (2003) Maspin - the most commonly-expressed gene of the 18q21.3 serpin cluster in lung cancer - is strongly expressed in preneoplastic bronchial lesions. Oncogene 22, 8677–8687 10.1038/sj.onc.1207127 [DOI] [PubMed] [Google Scholar]
- 14.Zheng H.-C. and Gong B.-C. (2017) The roles of maspin expression in gastric cancer: a meta- and bioinformatics analysis. Oncotarget 8, 66476–66490 10.18632/oncotarget.20192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sheng S., Bernardo M.M., Dzinic S.H. et al. (2018) Tackling tumor heterogeneity and phenotypic plasticity in cancer precision medicine: our experience and a literature review. Cancer Metastasis Rev. 37, 655–663 10.1007/s10555-018-9767-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tierney J.F., Stewart L.A., Ghersi D. et al. (2007) Practical methods for incorporating summary time-to-event data into meta-analysis. Trials 8, 16 10.1186/1745-6215-8-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parmar M.K., Torri V. and Stewart L. (1998) Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat. Med. 17, 2815–2834 [DOI] [PubMed] [Google Scholar]
- 18.Stang A. (2010) Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur. J. Epidemiol. 25, 603–605 10.1007/s10654-010-9491-z [DOI] [PubMed] [Google Scholar]
- 19.Chuling F., Hui H. and Zuojun X. (2016) The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomized studies [Google Scholar]
- 20.Liang B., Zhao L.-Z., Liao H.-L. et al. (2019) Rivaroxaban for cancer-associated venous thromboembolism: a systematic review and meta-analysis protocol. Medicine (Baltimore) 98, e18087 10.1097/MD.0000000000018087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chandrashekar D.S., Bashel B., Balasubramanya S.A.H. et al. (2017) UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 19, 649–658 10.1016/j.neo.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Asplund A., Edqvist P.-H.D., Schwenk J.M. et al. (2012) Antibodies for profiling the human proteome-The Human Protein Atlas as a resource for cancer research. Proteomics 12, 2067–2077 10.1002/pmic.201100504 [DOI] [PubMed] [Google Scholar]
- 23.Nagy Á., Lánczky A., Menyhárt O. et al. (2018) Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci. Rep. 8, 9227 10.1038/s41598-018-27521-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang W.-Y., Hsu S.-D., Huang H.-Y. et al. (2015) MethHC: a database of DNA methylation and gene expression in human cancer. Nucleic Acids Res. 43, D856–D861 10.1093/nar/gku1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cerami E., Gao J., Dogrusoz U. et al. (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang D.W., Sherman B.T. and Lempicki R.A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 27.Huang D.W., Sherman B.T. and Lempicki R.A. (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu G.-C., Wang L.-G., Han Y. et al. (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berardi R., Santinelli A., Onofri A. et al. (2012) Maspin expression is a favorable prognostic factor in non-small cell lung cancer. Anal. Quant. Cytol. Histol. 34, 72–78 [PubMed] [Google Scholar]
- 30.Bircan A., Bircan S., Kapucuoglu N. et al. (2010) Maspin, VEGF and p53 expression in small biopsies of primary advanced lung cancer and relationship with clinicopathologic parameters. Pathol. Oncol. Res. 16, 553–561 10.1007/s12253-010-9259-5 [DOI] [PubMed] [Google Scholar]
- 31.Matsuoka Y., Takagi Y., Nosaka K. et al. (2016) Cytoplasmic expression of maspin predicts unfavourable prognosis in patients with squamous cell carcinoma of the lung. Histopathology 69, 114–120 10.1111/his.12921 [DOI] [PubMed] [Google Scholar]
- 32.Nakagawa M., Katakura H., Adachi M. et al. (2006) Maspin expression and its clinical significance in non-small cell lung cancer. Ann. Surg. Oncol. 13, 1517–1523 10.1245/s10434-006-9030-z [DOI] [PubMed] [Google Scholar]
- 33.Ohno T., Kubouchi Y., Wakahara M. et al. (2018) Clinical significance of subcellular localization of maspin in patients with pathological stage IA lung adenocarcinoma. Anticancer Res. 38, 3001–3007 [DOI] [PubMed] [Google Scholar]
- 34.Takagi Y., Matsuoka Y., Shiomi T. et al. (2015) Cytoplasmic maspin expression is a predictor of poor prognosis in patients with lung adenocarcinoma measuring <3 cm. Histopathology 66, 732–739 [DOI] [PubMed] [Google Scholar]
- 35.Takanami I., Abiko T. and Koizumi S. (2008) Expression of maspin in non-small-cell lung cancer: correlation with clinical features. Clin. Lung Cancer 9, 361–366 10.3816/CLC.2008.n.052 [DOI] [PubMed] [Google Scholar]
- 36.Wang X., Wang Y., Li S. et al. (2014) Decreased maspin combined with elevated vascular endothelial growth factor C is associated with poor prognosis in non-small cell lung cancer. Thorac. Cancer 5, 383–390 10.1111/1759-7714.12104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Woenckhaus M., Bubendorf L., Dalquen P. et al. (2007) Nuclear and cytoplasmic Maspin expression in primary non-small cell lung cancer. J. Clin. Pathol. 60, 483–486 10.1136/jcp.2005.033407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yaman B., Nart D., Ekren P.K. et al. (2015) Expression of p63, TTF-1 and Maspin in non-small cell lung carcinoma and their effect on the prognosis and differential diagnosis. Turk. Patoloji Derg. 31, 163–174 [DOI] [PubMed] [Google Scholar]
- 39.Zheng H.-C., Saito H., Masuda S. et al. (2008) Cytoplasmic and nuclear maspin expression in lung carcinomas: an immunohistochemical study using tissue microarrays. Appl. Immunohistochem. Mol. Morphol. 16, 459–465 [DOI] [PubMed] [Google Scholar]
- 40.Takanami I., Abiko T. and Koizumi S. (2008) Expression of Maspin in non–small-cell lung cancer: correlation with clinical features. Clin. Lung Cancer 9, 361–366 10.3816/CLC.2008.n.052 [DOI] [PubMed] [Google Scholar]
- 41.Dzinic S.H., Bernardo M.M., Li X. et al. (2017) An essential role of Maspin in embryogenesis and tumor suppression. Cancer Res. 77, 886–896 10.1158/0008-5472.CAN-16-2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bircan A., Bircan S., Kapucuoglu N. et al. (2010) Maspin, VEGF and p53 expression in small biopsies of primary advanced lung cancer and relationship with clinicopathologic parameters. Pathol. Oncol. Res. 16, 553–561 [DOI] [PubMed] [Google Scholar]
- 43.Teoh S.S., Vieusseux J., Prakash M. et al. (2014) Maspin is not required for embryonic development or tumour suppression. Nat. Commun. 5, 3164 10.1038/ncomms4164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skoulidis F. and Heymach J.V. (2019) Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat. Rev. Cancer 19, 495–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou J., Hualong Q., Zhou P. et al. (2015) Different maspin functions in the lung adenocarcinoma A549 and SPC-A1 cell lines. Int. J. Mol. Med. 36, 1440–1448 10.3892/ijmm.2015.2336 [DOI] [PubMed] [Google Scholar]
- 46.Zheng H.-C., Saito H., Masuda S. et al. (2008) Cytoplasmic and nuclear maspin expression in lung carcinomas: an immunohistochemical study using tissue microarrays. Appl. Immunohistochem. Mol. Morphol. 16, 459–465 [DOI] [PubMed] [Google Scholar]
- 47.Odero-Marah V.A., Khalkhali-Ellis Z., Chunthapong J. et al. (2003) Maspin regulates different signaling pathways for motility and adhesion in aggressive breast cancer cells. Cancer Biol. Ther. 2, 398–403 [DOI] [PubMed] [Google Scholar]
- 48.Jenkinson S.E., Brown L.J., Ombor J. et al. (2017) Identification of novel peptide motifs in the serpin maspin that affect vascular smooth muscle cell function. Biochim. Biophys. Acta Mol. Cell Res. 1864, 336–344 10.1016/j.bbamcr.2016.11.019 [DOI] [PubMed] [Google Scholar]
- 49.Nam E. and Park C. (2010) Maspin suppresses survival of lung cancer cells through modulation of Akt pathway. Cancer Res. Treat. 42, 42–47 10.4143/crt.2010.42.1.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou J., Hualong Q., Zhou P. et al. (2015) Different maspin functions in the lung adenocarcinoma A549 and SPC-A1 cell lines. Int. J. Mol. Med. 36, 1440–1448 10.3892/ijmm.2015.2336 [DOI] [PubMed] [Google Scholar]
- 51.Qiao L., Liang N., Zhang J. et al. (2015) Advanced research on vasculogenic mimicry in cancer. J. Cell. Mol. Med. 19, 315–326 10.1111/jcmm.12496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ku B.M., Choi M.K., Sun J.-M. et al. (2018) Acquired resistance to AZD9291 as an upfront treatment is dependent on ERK signaling in a preclinical model. PLoS ONE 13, e0194730 10.1371/journal.pone.0194730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baty F., Klingbiel D., Zappa F. et al. (2015) High-throughput alternative splicing detection using dually constrained correspondence analysis (DCCA). J. Biomed. Inform. 58, 175–185 10.1016/j.jbi.2015.10.002 [DOI] [PubMed] [Google Scholar]
- 54.Kwon Y.-J., Lee S.J., Koh J.S. et al. (2012) Genome-wide analysis of DNA methylation and the gene expression change in lung cancer. J. Thorac. Oncol. 7, 20–33 10.1097/JTO.0b013e3182307f62 [DOI] [PubMed] [Google Scholar]
- 55.Kim E.H., Park A.K., Dong S.M. et al. (2010) Global analysis of CpG methylation reveals epigenetic control of the radiosensitivity in lung cancer cell lines. Oncogene 29, 4725–4731 10.1038/onc.2010.223 [DOI] [PubMed] [Google Scholar]
- 56.Yuan K., Xie K., Fox J. et al. (2013) Decreased levels of miR-224 and the passenger strand of miR-221 increase MBD2, suppressing maspin and promoting colorectal tumor growth and metastasis in mice. Gastroenterology 145, 853–864.e859 10.1053/j.gastro.2013.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.